Abstract
There is now convincing evidence that the human Tap protein plays a critical role in mediating the nuclear export of mRNAs that contain the Mason-Pfizer monkey virus constitutive transport element (CTE) and significant evidence that Tap also participates in global poly(A)+ RNA export. Previously, we had mapped carboxy-terminal sequences in Tap that serve as an essential nucleocytoplasmic shuttling domain, while others had defined an overlapping Tap sequence that can bind to the FG repeat domains of certain nucleoporins. Here, we demonstrate that these two biological activities are functionally correlated. Specifically, mutations in Tap that block nucleoporin binding also block both nucleocytoplasmic shuttling and the Tap-dependent nuclear export of CTE-containing RNAs. In contrast, mutations that do not inhibit nucleoporin binding also fail to affect Tap shuttling. Together, these data indicate that Tap belongs to a novel class of RNA export factors that can target bound RNA molecules directly to the nuclear pore without the assistance of an importin β-like cofactor. In addition to nucleoporins, Tap has also been proposed to interact with a cellular cofactor termed p15. Although we were able to confirm that Tap can indeed bind p15 specifically both in vivo and in vitro, a mutation in Tap that blocked p15 binding only modestly inhibited CTE-dependent nuclear RNA export. However, p15 did significantly enhance the affinity of Tap for the CTE in vitro and readily formed a ternary complex with Tap on the CTE. This result suggests that p15 may play a significant role in the recruitment of the Tap nuclear export factor to target RNA molecules in vivo.
While there has been considerable recent progress in understanding the mechanisms underlying the nucleocytoplasmic transport of proteins and noncoding RNAs, the pathway(s) used for nuclear export of cellular mRNAs remains undefined (reviewed in references 13 and 30). However, recent data strongly implicate the Tap protein expressed in vertebrate cells and its yeast ortholog Mex67p as critical participants in the process of mRNA export in both higher and lower eukaryotes (6, 14, 18–20, 25, 26).
The identification of the human Tap protein as a possible mRNA export factor initially resulted from efforts to identify cellular proteins that specifically bind to functional forms of the Mason-Pfizer monkey virus (MPMV) constitutive transport element (CTE), an RNA element that can activate the nuclear export of incompletely spliced mRNAs when present in cis (7, 14). Human Tap has been reported to significantly enhance CTE function upon microinjection into Xenopus oocytes and can rescue MPMV CTE function when expressed in the otherwise nonpermissive quail cell line QCl-3 (6, 14, 19). Tap has also been shown to shuttle between the nucleus and cytoplasm, a general characteristic of nuclear transport factors, and to partially colocalize with nuclear pore complexes, the portals used for all nucleocytoplasmic transport (2, 19, 20).
While the evidence that Tap is a critical cofactor for CTE function is therefore compelling, the evidence obtained from vertebrate cells for a role for Tap in global mRNA export is more circumstantial. The possibility that a cofactor required for CTE-dependent nuclear RNA export might also be critical for cellular mRNA export initially arose from the observation that microinjection of a CTE competitor into Xenopus oocyte nuclei inhibited export of not only CTE-containing RNAs but also of mRNAs (23, 24). In contrast, the nuclear export of tRNAs, as well as the export of RNAs via the Crm1 pathway, remained unaffected. Importantly, coinjection of human Tap rescued both CTE-dependent and CTE-independent mRNA transport (14), thus strongly suggesting that Tap is a critical participant in both processes. Subsequently, Tap was shown to be associated with global poly(A)+ RNA in human cells and to bind RNA nonspecifically in vitro (20). However, the affinity of Tap for nonspecific RNAs is much lower than its affinity for CTE RNA (14, 18, 19), and the mechanism of recruitment of Tap to global poly(A)+ RNA in vivo is therefore unclear.
A more compelling case for a role in global mRNA export has been made for Mex67p, the Saccharomyces cerevisiae ortholog of Tap. Like human Tap, Mex67p is associated with both total poly(A)+ RNA and with nuclear pore complexes in vivo (26). More importantly, however, Mex67p is essential for viability in yeast, and a shift to a nonpermissive temperature in cells bearing a temperature-sensitive allele of mex67 results in the rapid nuclear accumulation of poly(A)+ RNA (25, 26). Both the association of Mex67p with nuclear pores and the nuclear export of yeast poly(A)+ RNA are dependent on a second protein, termed Mtr2p, which forms a heterodimer with Mex67p (17, 25). However, Mtr2p can localize to nuclear pores independently of Mex67p and directly binds to at least one yeast nucleoporin. Therefore, it has been suggested that the Mex67p-Mtr2p heterodimer might represent a novel mRNA export complex that can bind to both poly(A)+ RNA, via the Mex67p subunit, and to nuclear pores, via the Mtr2p subunit (25).
Efforts to define the functional domain organization of the 619-amino-acid human Tap protein have led to the mapping of a CTE binding domain (approximately residues 80 to 372) and a transportin-dependent nuclear localization signal (NLS; residues 61 to 102) (1, 2, 6, 19, 20, 32). A third domain, extending from approximately residue 540 to the Tap carboxy terminus, has been shown to directly interact with nucleoporins, including CAN/Nup214, and to promote the nuclear pore association of Tap (2, 20). Independently, this third Tap domain was shown to act as both an NLS and as a nuclear export signal (NES) (19). Although mutational inactivation of the nucleocytoplasmic shuttling activity of this carboxy-terminal domain blocked the ability of human Tap to rescue MPMV CTE function in quail cells, this domain could be functionally replaced by the leucine-rich NES found in the human immunodeficiency virus type 1 (HIV-1) Rev protein (19). Based on these data, it appeared possible that the carboxy-terminal domain of Tap may target ribonucleoprotein complexes to the nuclear pore, and hence to the cytoplasm, by directly binding to nucleoporins. However, because the nucleoporin binding activity and nucleocytoplasmic shuttling activity of this Tap domain were demonstrated independently, it has remained unclear whether these two activities are indeed functionally related.
A fourth domain in Tap, extending from approximately residue 352 to 550, was recently shown to bind to a cellular factor, termed p15, that displays homology to the Ran-specific nuclear import factor NTF2 but not obviously to yeast Mtr2p (20). Surprisingly, while Tap expression cannot restore the viability of Mex67p-deficient yeast, the combination of both Tap and p15 rescued not only Mex67p-deficient yeast but also Mex67p- and Mtr2p-deficient yeast (20). These data strongly suggest (i) that human Tap-p15 can at least partially substitute for Mex67p and Mtr2p in mediating mRNA export in yeast cells, (ii) that the Tap/Mex67p mRNA export pathway has been evolutionarily conserved, and (iii) that p15 is required for non-sequence-specific mRNA export by Tap.
In this paper, we have further analyzed the interaction of Tap with both CAN/Nup214 and p15 and have, in particular, examined whether these interactions are important for the Tap-induced export of mRNAs containing the MPMV CTE. While nucleoporin binding was found to be absolutely critical for Tap function, p15 binding only moderately enhanced Tap-dependent CTE export. However, p15 did detectably increase the in vitro binding affinity of Tap for CTE, and we were able to readily demonstrate the formation of a specific ternary complex containing CTE RNA, Tap, and the p15 protein. These data demonstrate that nuclear export of RNA by Tap is critically dependent on carboxy-terminal sequences that act both as a nucleoporin binding motif and as a nucleocytoplasmic shuttle domain and raise the possibility that the primary biological role of p15 may lie in mediating the non-sequence-specific recruitment of Tap to cellular mRNA species.
MATERIALS AND METHODS
Plasmid construction.
The following plasmids have been previously described: the pBC12/CMV (8)-based metazoan expression plasmids pcTat, pBC12/CMV/β-gal (31), pcHA-Tap, pcHA-TapΔ-Rev NES, and pcTat-Tap (19); the indicator constructs pDM128/CTE (4), pTAR/CAT (31), and pCTE/CAT (19); the yeast expression plasmids pVP16-HA-Tap (19), pIII/MS2/CTE (18), and pGAL4/CAS (16); and the bacterial expression plasmid pGST-Tap(61–619) (19).
A DNA sequence encoding amino acids 61 to 619 of Tap [Tap(61–619)] was inserted into the EcoRI and SalI sites of the yeast two-hybrid plasmid pGBT9 (Clontech) to generate pGBT9-Tap(61–619), which expresses the GAL4 DNA binding domain fused to Tap(61–619). Wild-type Tap was expressed as a nonfused protein in yeast cells using the pPGK expression plasmid (5). pGBT9-Crm1 was produced by inserting the Crm1 coding sequence (11) into the BamHI and SalI sites of pGBT9. Yeast expression plasmids pAD-CAN(1864–2090), pAD-CAN(1805–2090), and pAD-CAN(1600–2090), expressing N-terminally deleted forms of the nucleoporin CAN/Nup214 fused to the GAL4 activation domain (AD), were derived from pACTII (Clontech) by inserting the corresponding CAN coding sequence into the NcoI and XhoI sites of pACTII (20).
A DNA sequence encoding the human p15 protein was amplified by PCR from a Clontech human T-cell cDNA library and cloned between the EcoRI and XhoI sites of pGEX4T-1 to generate pGST-p15. This same p15 sequence was also cloned into the EcoRI/SalI sites of pGBT9 or the EcoRI/XhoI sites of pVP16-HA (18) to produce the yeast expression plasmids pGBT9-p15 and pVP16-HA-p15, respectively.
Mutated metazoan, yeast, and bacterial Tap expression plasmids were generated by mutating targeted sequences in pcHA-Tap, pcHA-TapΔ-RevNES, pcTat-Tap, pGST-Tap, pPGK-Tap, and pGBT9-Tap(61–619) to 5′-GCGGCCGCC-3′, which encodes triple alanine and forms a NotI site, using a Quickchange kit (Stratagene).
A two-nucleotide mutation (GA to CC) in nucleotides 5 and 6 downstream of the transcription start site was introduced into the in vitro transcription plasmid pGEM-3fZ(+) (Promega), thereby introducing a unique Bsp120I site. A synthetic DNA sequence encoding the upper half of the CTE (nucleotides 8064 to 8120) (19) was then cloned into the Bsp120I and AvaI sites. The 67-nucleotide RNA produced by in vitro transcription of this plasmid, after digestion with SmaI, contains an extended double helical stem that may stabilize the secondary structure of the half-CTE RNA.
Cell culture and transfection.
Quail QCl-3 cells and human 293T cells were maintained as previously described (4, 8) and were transfected using DEAE-dextran (8) or Lipofectamine (Life Technologies), respectively. All transfections were performed on cell cultures in 35-mm-diameter plates, with pBC12/CMV/β-gal (12) included as an internal control. The parental eukaryotic expression plasmid pBC12/CMV (8) was used as a fill-in plasmid or a negative control. In all transfection experiments, chloramphenicol acetyltransferase (CAT) enzyme levels were determined ∼48 h after transfection, as previously described, and were normalized to the level of β-galactosidase (β-Gal) activity present in the same cell lysate (4).
Yeast two-hybrid analysis.
Plasmids encoding the appropriate GAL4(1–147) DNA binding domain and VP16 or GAL4(768–881) AD fusion proteins were transformed into the yeast indicator strain Y190 (15) by standard techniques. After 3 days of yeast growth at 30°C on selective culture plates, double transformants were transferred to selective medium for overnight culture. The following day, cultures were lysed and assayed for β-Gal activity as previously described (18).
Western blot analyses.
Western blot analyses of protein expression levels in transfected 293T cells or transformed yeast cells were performed as previously described (3, 12) using a mouse monoclonal antibody specific for the hemagglutinin (HA) tag (Roche Molecular Biochemicals) or the GAL4 DNA binding domain (Santa Cruz Biotechnology).
HeLa cell microinjection.
HeLa cells were maintained and microinjected as previously described (16, 19, 32). Briefly, 2 days before microinjection, HeLa cells were seeded onto CELLocate microgrid coverslips (Eppendorf Scientific) at a density of 2 × 105 per 35-mm-diameter dish. The test proteins (final concentration in phosphate-buffered saline [PBS], ∼2 μg/μl) were coinjected with a previously described (32) tetramethylrhodamine isothiocyanate-conjugated maltose binding protein-simian virus 40 T-antigen (T-NLS) fusion protein as a tracer (final concentration, 1.5 μg/μl) to verify the site of injection and as a negative control. After injection, cells were incubated at 37°C for 40 min and then fixed with 3% paraformaldehyde in PBS. The glutathione S-transferase (GST) fusion proteins were visualized by indirect immunofluorescence using a polyclonal affinity-purified rabbit anti-GST antibody and a fluorescein isothiocyanate-conjugated donkey anti-rabbit antiserum. The subcellular localization of the injected proteins was visualized using a Leica DMRB fluorescence microscope at ×100 magnification.
Protein purification and gel shift analysis.
GST fusion proteins, encoding wild-type or mutant forms of Tap residues 61 to 619, were expressed and purified on glutathione affinity resin as previously described (19). To further purify the Tap fusion protein, the eluate from the glutathione beads was then loaded onto a Bio-Rex 70 resin column and washed with equilibration buffer (20 mM HEPES [pH 7.0], 1 mM EDTA, 0.1 mM dithiothreitol [DTT], 10% glycerol). Full-length protein bound to the column was eluted with 400 mM sodium chloride in equilibration buffer and concentrated using a Centricon 10 concentrator (Amicon, Beverly, Mass.). The GST-p15 fusion protein was affinity purified using the same conditions as those previously described (19), and nonfused p15 protein was then produced by cleavage with thrombin (Amersham Pharmacia Biotech).
The half-CTE RNA probe was labeled with [α-32P]CTP using the Riboprobe in vitro transcription system (Promega), and the total isotope incorporation was determined by scintillation counting after column purification. The binding reaction was carried out with ∼104 cpm (∼0.1 ng) of the probe and ∼50 ng of GST-Tap and/or p15 protein in 20 μl of binding buffer (150 mM KCl, 10 mM HEPES [pH 7.6], 0.5 mM EGTA, 2 mM MgCl2, 1 mM DTT, 10% glycerol) containing 4 μg of bacterial rRNA and 1 μg of yeast tRNA. Binding was allowed to proceed for 20 min at 4°C, and the reaction products were then resolved on a 5% (40:1) native polyacrylamide gel and visualized by autoradiography.
RESULTS
Tap function requires nucleoporin binding.
Previously, we have reported that residues 540 to 619 of Tap form a nucleocytoplasmic shuttling domain that is essential for Tap-dependent nuclear export of CTE-containing RNAs (19). Independently, Katahira et al. (20) reported that residues 507 to 619 of Tap are able to directly bind to the FG repeat domain of CAN/Nup214 both in vivo and in vitro, as well as to a second FG repeat nucleoporin termed hCG1. These observations raised the possibility that this domain might target Tap, and hence any bound mRNA, to the cytoplasm by directly binding to nucleoporins rather than to an intermediate export receptor comparable to Crm1 or CAS.
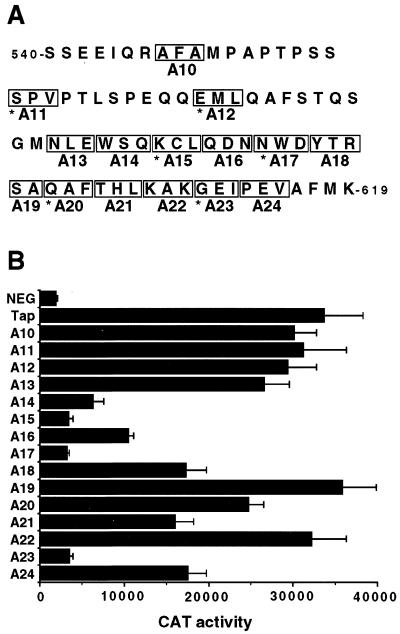
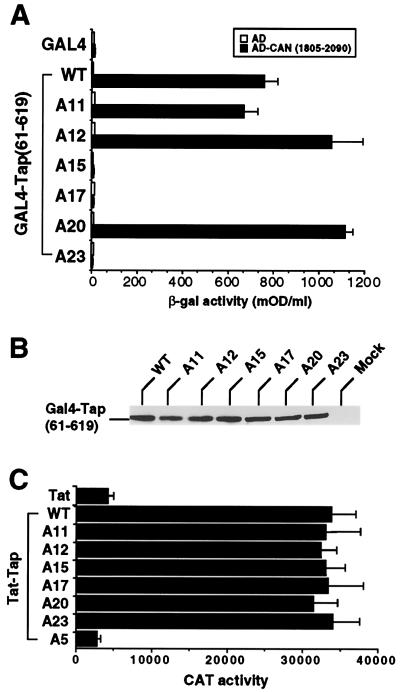
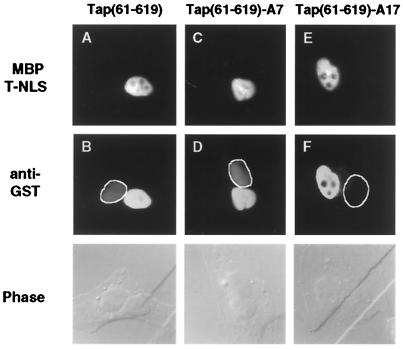
To test this hypothesis, we performed alanine-scanning mutagenesis of the Tap sequence between residues 540 and 619 in the context of full-length Tap, as shown in Fig. 1A. The resultant Tap mutants were then assayed for their abilities to rescue CTE-dependent RNA export in QCl-3 cells, as previously described (18, 19), using the cat-based pDM128/CTE indicator construct. As shown in Fig. 1B, three mutants (A15, A17, and A23) proved inactive while the remainder were partially active (A14, A16, A18, A21, and A24) or essentially fully active (A10 through A13, A19, A20, and A22). We next selected the three inactive Tap mutants and three fully active mutants and compared their abilities to bind to a segment of CAN/Nup214, i.e., residues 1805 to 2090, that was previously reported to interact with Tap in the yeast two-hybrid assay (9, 20). As shown in Fig. 2A, the three active Tap mutants A11, A12, and A20 were equivalent to wild-type Tap in their ability to bind to CAN/Nup214 while the three inactive Tap mutants A15, A17, and A23 proved unable to detectably bind to this nucleoporin. This lack of activity does not reflect destabilization of Tap by introduction of these mutations, as Western analysis showed equivalent levels of expression of all seven GAL4-Tap fusion proteins in yeast cells (Fig. 2B). Further, these proteins are not simply misfolded, as all seven Tap mutants proved fully able to bind to the CTE RNA target when expressed as HIV-1 Tat fusion proteins (Fig. 2C). The assay used relies on the ability of the Tat protein to activate transcription from the HIV-1 long terminal repeat (LTR) when tethered to an introduced, heterologous RNA target (27, 31). The previously described pCTE/CAT indicator construct (19), which contains the CTE inserted in place of the TAR RNA target found in the wild-type HIV-1 LTR, is activated by the wild-type Tat-Tap fusion protein when coexpressed in human 293T cells, but not by unfused Tat or Tap or by a Tat-Tap fusion containing the A5 mutation previously shown (19) to block CTE binding (Fig. 2C). In contrast, all mutations introduced into the carboxy-terminal domain of Tap resulted in mutants that bound to the CTE RNA indistinguishably from wild-type Tap. From these data we conclude that the C-terminal nucleocytoplasmic shuttling domain of Tap can indeed bind FG repeat nucleoporins, as first reported by Katahira et al. (20), and that this interaction is critical for the ability of Tap to mediate nuclear export of target mRNA species. Analysis of nucleocytoplasmic shuttling by these Tap mutants, by microinjection of recombinant GST fusion proteins into one nucleus in a binuclear HeLa cell, showed that wild-type Tap and Tap mutants A11 and A12 all shuttled while mutants A17 and A23 lacked any detectable NES activity (Fig. 3 and data not shown). Therefore, nucleocytoplasmic shuttling and nucleoporin binding by Tap are indeed functionally correlated.
FIG. 1.
Mutational analysis of the carboxy-terminal domain of human Tap. (A) The 15 indicated alanine-scanning mutations were introduced into pcHA-Tap, which expresses full-length human Tap bearing an amino-terminal HA epitope tag. Each mutation results from introduction of the sequence 5′-GCGGCCGCC-3′, which encodes three alanine residues. In cases where the mutated sequence already encoded one or more alanines, e.g., A22, only one or two encoded amino acid residues were changed, even though the underlying nucleotide sequence was different. Asterisks denote Tap mutants used for subsequent analysis. (B) The biological activity of the indicated Tap mutants was analyzed by cotransfection into quail QCl-3 cells along with the indicator construct pDM128/CTE and the internal control plasmid pBC12/CMV/β-gal, as previously described (19). The parental pBC12/CMV plasmid served as a negative control (NEG). Cultures were harvested at ∼48 h posttransfection, and the induced CAT and β-Gal activities were determined (4). The indicated data represent the averages of three independent transfections, with standard deviations indicated, after correction for minor variations in the internal control. CAT activities are given as counts per minute.
FIG. 2.
Nucleoporin binding by Tap. (A) The ability of wild-type and mutant Tap proteins to bind to the FG repeat domain of CAN/Nup214 was assayed by yeast two-hybrid analysis (9, 20). Tap proteins (residues 61 to 619) were expressed fused to the GAL4 DNA binding domain, while residues 1805 to 2090 of CAN/Nup214 were expressed fused to the GAL4 AD. After selection for transformants, induced β-Gal activities were measured as previously described. Results are the averages of three experiments. mOD, milli-optical density units. (B) The yeast lysates generated (A) were examined for GAL4-Tap fusion protein expression levels by Western analysis using an antibody directed against the GAL4 DNA binding domain. Mock, extract from nontransformed yeast cells. (C) Abilities of the indicated Tap mutants to bind to the MPMV CTE were determined in transfected 293T cells using a previously described HIV-1 Tat-based RNA binding assay (4, 19, 31). Briefly, the indicated Tap proteins were expressed as HIV-1 Tat-Tap fusions. The pCTE/CAT indicator plasmid contains the HIV-1 LTR linked to the CTE target sequence. Recruitment of the Tat activation domain to the LTR-proximal CTE by the fused Tap protein resulted in transcriptional activation and, hence, enhanced CAT expression in cotransfected cells. Unfused Tat and the mutant Tat-TapA5 fusion protein, both of which fail to bind the CTE, served as negative controls. The pBC12/CMV/β-gal plasmid served as an internal control. Data are averages of three experiments. CAT activity is expressed in counts per minute. WT, wild type.
FIG. 3.
Nucleocytoplasmic shuttling by Tap fusion proteins. Recombinant wild-type or mutant GST-Tap(61–619) fusion proteins were expressed in bacteria and purified by affinity and ion-exchange chromatography. Each GST-Tap(61–619) fusion protein was then mixed with a rhodamine-labeled fusion protein consisting of the maltose binding protein (MBP) fused to the simian virus 40 T-antigen NLS (T-NLS), which served as an internal control. After microinjection into one nucleus in a binuclear HeLa cell, the culture was incubated at 37°C for 40 min and fixed and the subcellular locations of the two injected proteins were determined by fluorescence microscopy, as previously described (16, 19, 32). The second, noninjected nucleus in each binuclear cell is highlighted in the middle row and is also visible in the parallel phase-contrast images shown in the bottom row. These data are representative of several microinjected binuclear cells.
Comparison of the abilities of nuclear export factors to bind to CAN/Nup214.
Nuclear export of mRNAs containing the cis-acting HIV-1 Rev response element (RRE) RNA target requires both recruitment of HIV-1 Rev to the RRE and recruitment of the Crm1 nuclear export factor to the leucine-rich Rev NES (4, 10, 22, 29). The Crm1 protein in turn directly binds to the FG repeat domains of certain nucleoporins, including CAN/Nup214, during the subsequent mRNA export process (22). The minimal sequence in CAN/Nup214 able to bind to Crm1 has been mapped to residues 1864 to 2090, and overexpression of this fragment of CAN/Nup214, which has been termed ΔCAN, causes a relocalization of Crm1 from the nuclear periphery to the nucleoplasm and strongly inhibits the function of HIV-1 Rev and of other viral RNA export factors that contain a leucine-rich NES (4, 11, 19, 33). This inhibition is specific, as ΔCAN inhibits Rev function but not either CTE-dependent mRNA export or cellular mRNA export. However, if Tap, the cellular cofactor for CTE-dependent mRNA export, also binds to the FG repeat domain of CAN/Nup214, then why is ΔCAN a specific inhibitor of Crm1 function?
While ΔCAN, the CAN/Nup214 fragment that blocks Crm1 function, extends from residue 1864 to 2090, the carboxy-terminal fragments of CAN/Nup214 that were identified during the original yeast two-hybrid screen for Tap binding proteins extended minimally from residue 1805 to 2090 (20). Therefore, it seemed possible that the specific inhibition of Crm1 function by ΔCAN(1864–2090) might reflect a difference between the ability of this nucleoporin fragment to bind to Crm1 and its ability to bind to Tap. As shown in Table 1, the fragment comprising residues 1864 to 2090 of CAN/Nup214 indeed bound only very weakly to the GAL4-Tap(61–619) fusion protein, even though this same ΔCAN fragment gave strong binding to a GAL4-Crm1 fusion protein. While extension of the tested CAN/Nup214 sequence to residue 1805 increased the interaction with GAL4-Tap(61–619) by ∼28-fold, extension to residue 1600 had no further enhancing effect. As a negative control, a fusion protein consisting of the GAL4 DNA binding domain linked to the CAS nuclear export factor did not bind any tested CAN/Nup214 fragment, although binding to importin α2, a known export substrate for CAS, was readily detected (Table 1) (16, 21). Using an in vitro binding assay, Bachi et al. (1) have very recently also demonstrated that Crm1 can bind to fragments of CAN/Nup214 extending from either residue 1690 to 2090 or from residue 1983 to 2090, while Tap is only able to bind to the longer segment of CAN comprising residues 1690 to 2090. We therefore conclude that although both Crm1 and Tap can bind to the FG repeat domain of CAN/Nup214, effective binding by Crm1 clearly requires a smaller number of FG repeat elements. This difference presumably explains the ability of ΔCAN to act as a selective inhibitor of Crm1 function in vivo. It is also important to note that it is not presently clear whether CAN/Nup214 is indeed a relevant target for Tap binding in vivo or whether it is instead simply serving as a surrogate, in the two-hybrid assay, for another FG repeat nucleoporin(s) that is the physiologically relevant target. Several other nucleoporins have in fact also been shown to specifically bind the Tap C-terminal domain (1, 20). In contrast, Crm1 and CAN/Nup214 can be coimmunoprecipitated from expressing cells (11), and this interaction is therefore clearly relevant.
TABLE 1.
Interaction of nuclear export receptors with nucleoporin CAN in yeast cellsa
| DNA binding domain fusion | Induced β-Gal activityb withc:
|
|||
|---|---|---|---|---|
| AD-CAN (1864–2090) | AD-CAN (1805–2090) | AD-CAN (1600–2090) | AD-importin α2 | |
| GAL4 | <10 | <10 | <10 | <10 |
| GAL4-Crm1 | 9,936 | 28,064 | 9,880 | <10 |
| GAL4-Tap(61–619) | 32 | 902 | 232 | <10 |
| GAL4-CAS | <10 | <10 | <10 | 8,234 |
The ability of the indicated GAL4 DNA binding domain fusions and GAL4-AD fusions to interact in vivo was measured using the yeast two-hybrid assay as described for Fig. 2A.
In milli-optical density units per milliliter of lysate.
The residues from each protein used in the relevant fusions are indicated. If no residue numbers are shown, then the full-length protein was analyzed.
Binding to p15 facilitates, but is not essential for, Tap-dependent CTE export.
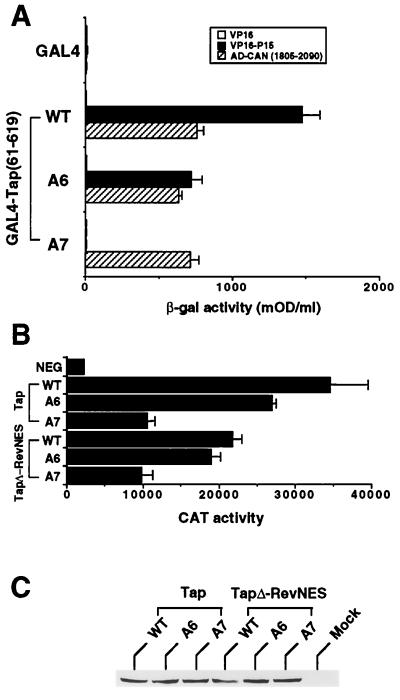
To determine whether p15 binding plays a role in mediating the nuclear export of CTE-containing RNAs, we next introduced a number of alanine-scanning mutations into the region of Tap shown by Katahira et al. (20) to bind to p15, i.e., residues 352 to 550. Two of these mutations, termed A6 (381-ENL-383→AAA) and A7 (415-CCS-417→AAA), reduced the binding of Tap to p15, as assessed by a two-hybrid assay, but did not affect binding to CAN/Nup214. Specifically, mutant A6 reduced binding by approximately twofold, while A7 blocked p15 binding entirely (Fig. 4A).
FIG. 4.
The p15 protein is not required for Tap-dependent CTE RNA export. (A) The abilities of wild-type (WT) and mutant forms of Tap to bind to the human p15 protein were determined by a yeast two-hybrid assay, as described for Fig. 2A. AD-CAN (1805–2090), positive control. The A6 mutant contains three alanines in place of Tap residues 381 to 383, while A7 contains alanines in place of residues 415 to 417. mOD, milli-optical density units. (B) The A6 and A7 mutations were introduced into an amino-terminally HA epitope-tagged form of wild-type Tap or into a previously described chimeric protein consisting of HA epitope-tagged Tap residues 1 to 581 linked carboxy terminally to the HIV-1 Rev NES (TapΔ-RevNES) (19). The abilities of these Tap derivatives to induce CTE-dependent mRNA export from the nuclei of quail cells were then assessed by cotransfection into QCl-3 cells, as described for Fig. 1B. (C) The expression levels of the indicated Tap derivatives were quantified in transfected 293T cells by Western analysis using an anti-HA epitope tag antibody. Mock, mock-transfected 293T cells.
An analysis of the ability of these mutations to affect the rescue by human Tap of CTE-containing RNA export in quail cells revealed only a modest effect for the A6 mutation and an approximately threefold inhibition for the A7 mutation, which produces a mutant that entirely fails to bind p15 (Fig. 4B). Cotransfection of a human p15 expression plasmid into the QCl-3 cells did not exert any detectable phenotypic effect in this assay (data not shown). A possible explanation for this modest inhibition, suggested particularly by the known role of Mtr2p in targeting Mex67p to the yeast nuclear pore (25), is that loss of p15 binding might affect the ability of Tap to exit the nucleus. To test this hypothesis, we introduced the A6 and A7 mutations into the previously described TapΔ-RevNES fusion protein, in which the essential carboxy-terminal Tap shuttling domain has been replaced with the Rev NES (19). We reasoned that, if p15 is indeed acting to specifically enhance nucleocytoplasmic shuttling by this Tap domain, then replacement with the heterologous Rev NES should rescue the modest but significant inhibition exerted by the A7 mutation. In fact, however, the inhibitory effects exerted by the A7 mutation in the context of Tap and of the TapΔ-RevNES chimera were comparable (Fig. 4B). This modest inhibitory effect did not reflect reduced protein expression, as all six Tap proteins were expressed at comparable levels in transfected cells as assessed by Western blot assay (Fig. 4C).
As a direct test of the effect of p15 on Tap nucleocytoplasmic shuttling, we also expressed the A7 mutant in bacteria in the context of the GST-Tap(61–619) fusion protein and then examined shuttling by microinjection into one nucleus in a binuclear HeLa cell. As shown in Fig. 3, the A7 mutation, which blocks p15 binding, failed to detectably inhibit nucleocytoplasmic shuttling by Tap.
The p15 protein enhances CTE binding by Tap.
We next considered the possibility that the inhibition of Tap function exerted by, particularly, the A7 mutation might reflect reduced binding to the CTE. This would be an unexpected result, as we and others have previously mapped the Tap CTE binding domain to between residues 80 and 372 using in vivo and in vitro assays (6, 19). To test whether p15 can form a ternary complex with the CTE and wild-type Tap, we analyzed this RNA binding event in vitro using exclusively recombinant proteins, including fusion proteins consisting of GST linked to the wild-type or A6 or A7 mutant forms of Tap residues 61 to 619. This amino-terminally truncated form of Tap, whose first residue is encoded by the second methionine codon in the Tap gene and which was originally thought to be full-length (6, 14, 19), has been shown to effectively rescue CTE RNA export in quail cells (19) and is more easily expressed in intact form in bacteria (data not shown). In addition, the p15 protein was also expressed in bacteria and prepared as a purified unfused protein. These reagents were then used to examine CTE binding by Tap in vitro by electrophoretic mobility shift assay using a 32P-labeled half-CTE RNA probe.
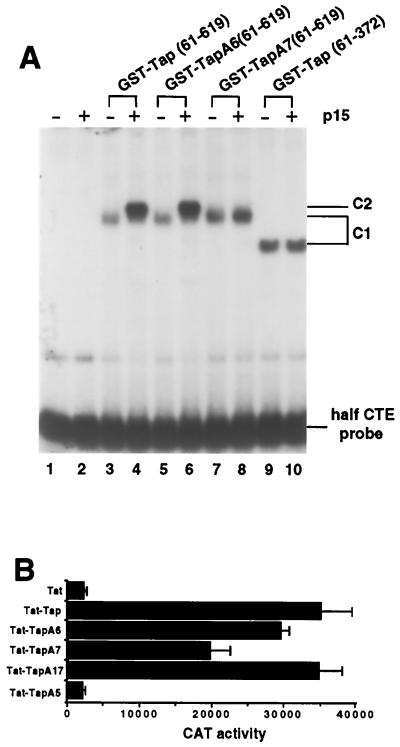
As shown in Fig. 5A, the wild-type and mutant GST-Tap(61–619) proteins all bound the half-CTE equivalently in the absence of p15 (lanes 3, 5, and 7) and the A6 and A7 mutations, both of which lie outside of the mutationally defined minimal Tap RNA binding domain, therefore do not directly affect CTE binding by Tap. While no RNA binding by p15 alone was detected (lane 2), the addition of wild-type GST-Tap(61–619) or of the GST-TapA6(61–619) mutant together with p15 resulted in a supershifted complex (Fig. 5A, C2) and, more importantly, increased the level of CTE binding by 5- to 10-fold (compare lanes 3 and 5 with lanes 4 and 6). No supershift or increase in CTE binding was observed with the GST–TapA7(61–619) protein, which is not predicted to bind to p15 (Fig. 5A, lanes 7 and 8). Similarly, a fusion protein consisting of GST linked to the Tap RNA binding domain (residues 61 to 372), which lacks Tap sequences required for p15 binding, also was unaffected by the addition of p15 to the binding assay mixture (Fig. 5A, lanes 9 and 10). We therefore conclude that Tap and p15 can form a ternary complex with the CTE RNA and that a Tap-p15 heterodimer or higher-order multimer can bind to the CTE with a higher affinity than Tap alone. The ability of Tap to induce the specific recruitment of the p15 protein to the MPMV CTE was also confirmed in vivo using the yeast three-hybrid assay (18, 28) (data not shown).
FIG. 5.
The p15 protein enhances CTE binding by Tap. (A) The effect of purified nonfused p15 protein on the abilities of wild-type and mutant forms of the GST-Tap(61–619) fusion protein to bind the CTE in vitro was assessed using an electrophoretic mobility shift assay. The probe used was a 32P-labeled half-CTE probe. Incubations were performed in the presence of 5 μg of nonspecific RNA competitor and included 50 ng of GST-Tap(61–619) and/or p15 per reaction, as indicated. The GST-Tap(61–372) fusion protein, which contained the minimal Tap CTE binding domain but which lacked Tap sequences required for p15 binding, was used in lanes 9 and 10. C1, complexes formed by Tap proteins and the CTE; C2, proposed ternary complex containing CTE, GST-Tap, and p15. Due to the small size of p15, this complex migrates only slightly more slowly than the C1 complex. (B) Inactivation of p15 binding only modestly reduces CTE binding by Tap in vivo, as assessed using the Tat-based RNA binding assay described in Fig. 2C and previously (19). The A5 mutant of Tap, which has lost the ability to bind the CTE, served as a negative control. This experiment was performed in triplicate in transfected human 293T cells and relies on endogenous human p15 protein. CAT activity is expressed in counts per minute.
Because of the observed homology of p15 to NTF2, which is known to interact with the GDP-bound form of Ran (13, 20), we also tested whether addition of either the GTP- or the GDP-bound form of Ran would affect complex formation on the CTE RNA probe. However, neither Ran-GDP nor Ran-GTP exerted any obvious effect on the observed mobility or efficiency of formation of the CTE-Tap-p15 ternary complex (data not shown).
To examine whether p15 can also enhance CTE binding by Tap in vivo, we used the Tat-based assay for analyzing RNA-protein interactions in the mammalian cell nucleus (Fig. 2C) to ask whether Tap mutants that have reduced (A6) or no (A7) ability to interact with p15 would bind the CTE as effectively as wild-type Tap in vivo. As shown in Fig. 5B, the A7 mutation indeed reduced CTE binding by approximately twofold. While modest, this effect is nevertheless comparable to the effect exerted by the A7 mutation on the ability of Tap to support MPMV CTE function in vivo (Fig. 4B).
DISCUSSION
It is increasingly apparent that the vertebrate Tap protein and its yeast ortholog Mex67p are likely to play a critical role in mediating the nuclear export of cellular mRNAs. There is now convincing evidence that Tap mediates the sequence-specific nuclear export of CTE-containing mRNAs (1, 3, 6, 16, 18, 19) and equally persuasive data that Mex67p is critical for global poly(A)+ RNA export in yeast cells (20, 24, 25). The finding that Tap, together with its cofactor p15, can at least in part functionally substitute for Mex67p and its cofactor Mtr2p in mediating poly(A)+ RNA export in yeast cells strongly suggests that Tap and Mex67p are likely to be critical participants in an mRNA export pathway that has been conserved through much of eukaryotic evolution (20). A more complete understanding of the role of these proteins in this export pathway is therefore of obvious importance.
Previously, we had identified several functional domains in Tap that play a role in the sequence-specific nuclear export of MPMV CTE-containing mRNA species, a goal that became feasible with our finding that the quail cell line QCl-3 is normally essentially nonpermissive for MPMV CTE function but can be rendered fully permissive by expression of human Tap (19). In particular, these earlier experiments allowed us to mutationally define an essential nucleocytoplasmic shuttle sequence located carboxy-terminal to Tap residue 540 (Fig. 6A). This finding raised the possibility that the carboxy terminus of Tap might serve as the binding domain for a nuclear export factor belonging to the importin β family of nuclear transport factors. If this were the case, then the role of Tap would simply be to serve as an adapter molecule linking the CTE RNA to a β-like export factor, just as HIV-1 Rev serves as an adapter between the RRE RNA and Crm1 (10, 13, 22, 29, 30). The observation that the essential Tap nucleocytoplasmic shuttle domain can be, at least in part, functionally replaced by the HIV-1 Rev NES (19) could be viewed as evidence in favor of this hypothesis.
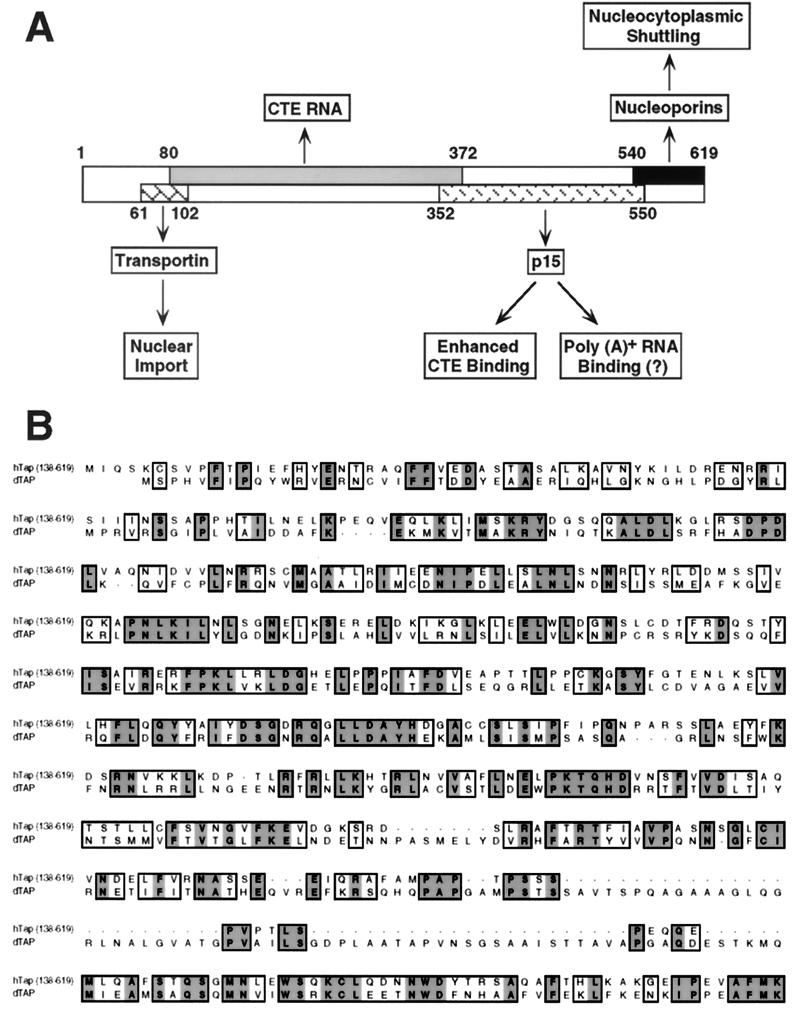
FIG. 6.
Functional domain organization of the human Tap nuclear RNA export factor. (A) Schematic overview of the known cellular targets and correlated functions of different domains in human Tap. The proposed role of p15 in poly(A)+ RNA binding is hypothetical. (B) Sequence alignment of residues 138 to 619 of human Tap with the sequence of what may be a partial clone of the Drosophila Tap protein (EMBL accession no., AJ251947).
While our research identified the carboxy-terminal domain of Tap as a nucleocytoplasmic shuttling domain, Katahira et al. (20) independently showed that these same sequences could also directly bind to the FG repeat domain of the nucleoporins CAN/Nup214 and hCG1. One possible interpretation of the latter result, by analogy to β-like nuclear transport factors (13), is that Tap shuttles between the nucleus and cytoplasm due to its ability to directly bind to components of the nuclear pore complex. To test this hypothesis, we performed an alanine-scanning mutagenesis of the carboxy-terminal region of human Tap and identified mutants that were either inactive or fully active in mediating CTE-dependent RNA export in transfected QC1-3 cells (Fig. 1). As shown in Fig. 2A, all Tap mutants inactive for CTE RNA export had also lost the ability to bind to the nucleoporin CAN/Nup214, while all mutants that retained activity also retained nucleoporin binding. All these Tap mutants were stable (Fig. 2B) and remained fully able to bind to the CTE RNA target (Fig. 2C). Yet, the mutants that had lost the ability to bind to CAN/Nup214 also lacked the ability to undergo nucleocytoplasmic shuttling (Fig. 3 and data not shown). Based on these results, we therefore propose that the RNA export activity of Tap is dependent on a functionally autonomous carboxy-terminal nucleocytoplasmic shuttling domain that is able to directly target ribonucleoprotein complexes containing Tap to the nuclear pore complex (Fig. 6A). If this Tap domain is indeed critical for Tap function, then one would predict that it would have been conserved during evolution. A comparison of the sequence of the human Tap protein with the sequence of what appears to be a partial cDNA clone of the Drosophila melanogaster Tap protein, recently deposited in the database (G. S. Wilkie, Drosophila melanogaster mRNA for tip-associated protein, EMBL accession no. AJ251947, 1999), fully supports the functional importance of this Tap domain. Specifically, the last ∼50 residues of human Tap are highly conserved in the Drosophila homolog while flanking, more amino-terminal sequences are quite divergent (Fig. 6B). Of interest, several of the residues that are conserved from Drosophila to humans are coincident with residues whose mutagenesis blocks both nucleoporin binding and Tap-dependent CTE RNA export (Fig. 1 and 2).
Recently, Bear et al. (2) also reported a mutational analysis of human Tap function. These workers identified an NLS coincident with the transportin-dependent NLS reported by ourselves and others (Fig. 6) (19, 20) and also reported that the Tap carboxy-terminal domain could target Tap to nuclear pores and could also act as an NES in at least some experimental contexts. However, these workers also reported a second Tap NES, which they mapped to between residues 83 and 110. We were not able to observe this NES in our previously published work (19) and again failed to detect this NES in our present work (e.g., Fig. 3). If Tap indeed contains an NES between residues 83 and 110, then it is unclear why the essential Tap nucleocytoplasmic shuttle domain can be functionally replaced by the Rev NES (Fig. 4B). While it seems possible that this Tap sequence could represent a cryptic NES whose activity is only uncovered in certain deletion mutants of Tap, this issue clearly needs to be more fully addressed.
As noted above, Katahira et al. (20) reported not only an interaction between Tap and nucleoporins but also an interaction with a protein termed p15. The functional relevance of this interaction, at least for non-sequence-specific nuclear mRNA export, is strongly supported by the finding that p15 is required for the ability of Tap to rescue the viability of mex67− or mex67− mtr2− yeast cells. However, the Tap domain identified by Katahira et al. (20) as the binding site for p15, which extends approximately from residues 352 to 550 in Tap, did not coincide with any Tap sequence shown to be required for CTE-dependent mRNA export (Fig. 6A). Specifically, the Tap nucleocytoplasmic shuttle domain (residues 540 to 619), the RNA binding domain (residues 80 to 372), and the Tap NLS (residues 61 to 102) all were found to be functional in the absence of sequences critical for p15 binding (2, 6, 19).
To test whether p15 might nevertheless play a role in the Tap-dependent nuclear export of CTE-containing RNAs, we constructed two Tap mutants that were attenuated (A6) or inactivated (A7) for p15 binding (Fig. 4A). While both proteins, and particularly A7, were indeed less effective than wild-type Tap in mediating CTE RNA export (Fig. 4B), the observed inhibition was quite modest, and it is therefore apparent that p15 is not an essential cofactor for Tap-dependent CTE RNA export. Efforts to define the step in Tap function affected by p15 binding suggested that p15 did not play a role in mediating Tap nuclear export (Fig. 3). Rather, it appeared that p15 instead enhanced the binding of Tap to the CTE (Fig. 5). Specifically, recombinant p15 protein not only proved able to supershift the CTE-Tap complex, thus indicating the in vitro formation of a ternary complex on the CTE containing both Tap and p15, but also significantly enhanced the amount of the CTE probe that was bound (Fig. 5A). Therefore, it appears that p15 enhances the binding of Tap to the CTE either by causing a conformational change in Tap or by directly contacting the CTE RNA itself. However, we did not observe any evidence for CTE binding by p15 in the absence of Tap (Fig. 5A).
Immediately prior to submission of this paper, Bachi et al. (1) published a series of experiments that examined nucleoporin and p15 binding by Tap in vitro. Consistent with our experimental observations and previously published results (19, 20), they showed that the carboxy-terminal domain of Tap directly interacts with several FG repeat nucleoporins and is critical for MPMV CTE-dependent nuclear RNA export. While Bachi et al. (1) also observed that p15 binding is not critical for Tap-dependent CTE RNA export, their results differ from ours in that Bachi et al. (1) reported that the binding of Tap to the MPMV CTE blocked the interaction with p15. In contrast, we were able to readily detect formation of a ternary complex containing Tap, p15, and the MPMV CTE both in vitro and in vivo (Fig. 5A and data not shown). Of note, Bachi et al. (1) used a GST-p15 fusion protein in their in vitro binding assays rather than nonfused p15, and it is therefore possible that the GST moiety, which is significantly larger than p15, might have sterically hindered the formation of a ternary complex on the CTE. Consistent with this interpretation, we have found that GST-p15 differs from nonfused p15 in being unable to bind to both the CTE and Tap simultaneously in vitro (data not shown). More importantly, however, our data demonstrate that p15 enhances both CTE binding by Tap (Fig. 5A) and Tap-dependent CTE RNA export (Fig. 4B) and therefore raise the possibility that p15 may have a similar, but more critical, role in mediating the non-sequence-specific recruitment of Tap to cellular mRNAs. This hypothesis is consistent with the finding that human Tap is unable to functionally substitute for Mex67p in mediating mRNA export in yeast cells unless the human p15 protein is also coexpressed (20). Clearly, it is likely that recruitment of the Tap-p15 complex to mRNA is a tightly regulated process that probably also requires specific protein-protein interactions. How mRNA is specifically targeted for nuclear export by the Tap-p15 heterodimer, while pre-mRNAs and introns are retained in the nucleus, is a question of considerable future interest.
ACKNOWLEDGMENTS
We thank Ed Hurt for the pAD-CAN(1805–2090) expression plasmid, Gerard Grosveld for the CAN/Nup214 cDNA clone, and Jae Jung for the Tap cDNA clone.
This research was supported by the Howard Hughes Medical Institute.
REFERENCES
- 1.Bachi A, Braun I C, Rodrigues J P, Panté N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Görlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bear J, Tan W, Zolotukhin A S, Tabernero C, Hudson E A, Felber B K. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair W S, Cullen B R. A yeast TATA-binding protein mutant that selectively enhances gene expression from weak RNA polymerase II promoters. Mol Cell Biol. 1997;17:2888–2896. doi: 10.1128/mcb.17.5.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd H P, Echarri A, Ross T M, Cullen B R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd H P, Wiegand H L, Bieniasz P D, Cullen B R. Functional differences between the human and the bovine immunodeficiency virus Tat transcription factors. J Virol. 2000;74:4666–4671. doi: 10.1128/jvi.74.10.4666-4671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun I C, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjöld M-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 9.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 10.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 11.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridell R A, Bogerd H P, Cullen B R. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 14.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 15.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 16.Herold A, Truant R, Wiegand H, Cullen B R. Determination of the functional domain organization of the importin α nuclear import factor. J Cell Biol. 1998;143:309–318. doi: 10.1083/jcb.143.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadowaki T, Hitomi M, Chen S, Tartakoff A M. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol Biol Cell. 1994;5:1253–1263. doi: 10.1091/mbc.5.11.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y, Bogerd H P, Yang J, Cullen B R. Analysis of the RNA binding specificity of the human Tap protein, a constitutive transport element-specific nuclear RNA export factor. Virology. 1999;262:200–209. doi: 10.1006/viro.1999.9906. [DOI] [PubMed] [Google Scholar]
- 19.Kang Y, Cullen B R. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katahira J, Sträßer K, Podtelejnikov A, Mann M, Jung J U, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutay U, Bischoff F R, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 22.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 23.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjöld M-L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, utilises factors required for the export of cellular mRNAs. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 25.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Lührmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selby M J, Peterlin B M. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell. 1990;62:769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- 28.Sengupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 [Crm1p] is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 30.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 31.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 32.Truant R, Kang Y, Cullen B R. The human Tap nuclear RNA export factor contains a novel transportin-dependent NLS that lacks NES function. J Biol Chem. 1999;274:32167–32171. doi: 10.1074/jbc.274.45.32167. [DOI] [PubMed] [Google Scholar]
- 33.Zolotukhin A S, Felber B K. Nucleoporins Nup98 and Nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]