Abstract
The cellular protein AMF-1 (Gps2) positively modulates gene expression by the papillomavirus E2 protein (D. E. Breiding et al., Mol. Cell. Biol. 17:7208–7219, 1997). We show here that AMF-1 also binds the transcriptional coactivator p300 in vitro and in vivo. E2 interacted weakly with p300. These observations led to a model in which AMF-1 recruits p300 into a complex with E2. Cotransfection of AMF-1 or p300 stimulated levels of E2-dependent transcription, while cotransfection of both AMF-1 and p300 showed an additive effect. The functional significance of p300 recruitment for E2 transactivation was evidenced by repression of E2-activated transcription by adenovirus E1A, which inhibits both coactivator and acetylase activities of p300. Antibodies to AMF-1 or E2 immunoprecipitated histone acetylase activity from cell lysates. Western blotting using antibody against acetyl-lysine failed to detect acetylation of AMF-1 or E2 in complex with p300. These results suggest that AMF-1 facilitates the recruitment of p300 and its histone acetylase activity into complexes with E2 and represents a novel mechanism of transcriptional activation.
The bovine papillomavirus type 1 (BPV-1) E2 protein serves multiple functions for the virus. Binding of its carboxy-terminal sequence-specific DNA binding domain (DBD) to recognition elements in the papillomavirus genome regulates viral gene expression and replication of the viral genome (1, 5, 8, 16, 19, 57). The amino-terminal activation domain (AD) of E2 mediates protein-protein interactions with the cellular transcription machinery. This region also binds the papillomavirus E1 protein, a DNA binding helicase essential for viral DNA replication (17, 27, 54). E2 targets E1 to the origin of replication. It is also believed that E2 plays an additional role in DNA replication, as several E2 mutants retained E1 and DNA binding were unable to stimulate replication (16). Because E2 is required for replication of viral nucleosomal DNA but not naked plasmid DNA in vitro (33, 53), it has been proposed that E2 may relieve chromatin suppression and allowing entry of cellular replication and transcription factors to the origin and promoter, respectively.
The cooperation of at least two functional units within the large 220-amino-acid E2 AD is essential for transcriptional activation. The crystal structure of the human papillomavirus (HPV) E2 AD reveals multiple surfaces available for protein-protein interactions (2, 25). A transcription factor IIB (TFIIB) interaction domain has been mapped to residues 74 to 134 (56). Amino acids 134 to 216 mediate interaction with the activation domain modulation factor AMF-1 (9). AMF-1 is identical to Gps2, which was initially reported as a human cDNA that repressed lethality of a G-protein mutation in Saccharomyces cerevisiae (46). Gps2 also interacts with the human T-cell leukemia virus type 1 (HTLV-1) Tax protein (30). Gps2/AMF-1 has been shown to influence the transcriptional activities of E2, Tax, and c-Jun (9, 30, 46). E2 mutants phenotypically selected for inability to bind to AMF-1, while able to bind the E1 protein, exhibited defects in both transcriptional activation and DNA replication (9). This suggested that AMF-1 mediates a function required for both DNA replication and transcriptional activation.
Given that the function of E2 in transcriptional activation and the initiation of DNA replication involves formation of E2-dependent complexes on viral nucleosomal DNA, efficient assembly of these complexes in vivo might involve modification of nucleosome structure. The coactivators p300/CREB binding protein (CBP) and p300/CBP-associated cofactors (e.g., P/CAF, P/CIP-ACTR, and SRC-1) contain an intrinsic histone acetyltransferase (HAT) activity (4, 11, 39, 47, 50, 55). Both p300 and CBP were demonstrated to modify chromatin structures by acetylation of histones (12). It is postulated that recruitment of coactivators bearing HAT activity by promoter-bound transcription factors results in histone acetylation of nearby nucleosomes, thus enhancing access of the transcriptional or replication machinery to DNA (21, 36, 48). A class II transactivator was shown to mediate interaction between a DNA-bound factor (RFX5) and p300 (32, 43), suggesting that additional pathways leading to recruitment of p300 by transcriptional activators remain to be identified.
Recruitment of p300/CBP can also affect other aspects of transcription carried out by RNA polymerase II (6, 12, 29, 44). p300 and CBP function as coactivators by mediating protein-protein interactions with the transcriptional apparatus (12, 44). Many cellular transcription factors have been identified to interact with p300/CBP, in a signal-dependent and sometimes mutually exclusive fashion (18). p300/CBP can also directly acetylate TFIIE and TFIIF (29). The activities of p53, GATA-1, and human immunodeficiency virus type 1 Tat proteins have been shown to be regulated by p300/CBP acetylation (7, 22, 29, 31, 35, 40, 42). On the other hand, viral regulatory proteins target p300 and CBP. The adenovirus E1A oncoprotein binds to p300 and inhibits both its transcriptional coactivator and HAT activity (10, 24, 38). Tax protein of HTLV-1 inhibits p300/CBP-mediated transcription by interfering with recruitment of p300/CBP onto DNA (49, 51). Recently, HPV E6 proteins were shown to bind CBP at the same site as E1A (58). While both low- and high-risk E6 proteins were shown to associate with p300, HPV type 16 (HPV-16) E6 interacted at three sites, whereas HPV-6 E6 interacted only with the CH1 domain (41).
Based on the genetic requirement for the AMF-1 interaction for E2 activity in transcription and DNA replication, a possible role for AMF-1 in recruitment of p300 by the E2 AD was examined. We found that AMF-1 enhances the interaction of p300 with E2 and that the complexes were highly active for histone acetylation.
MATERIALS AND METHODS
Protein expression and purification.
For the production of p53, BPV-1 E2, BPV-1 E1, AMF-1, and Flag-tagged p300, Sf9 cells were grown to 80% confluence in 150-mm-diameter tissue culture dishes, infected with recombinant baculoviruses expressing each of the proteins, and incubated for 40 to 48 h. Cells were harvested, washed with phosphate-buffered saline, and frozen at −80°C. Cells were extracted with lysis buffer (50 mM Tris [pH 8.0], 200 mM NaCl [320 mM NaCl in the case of Flag-p300], 0.2% NP-40, 5% glycerol, 1 mM EDTA, 1 mM dithiothreitol [DTT], phenylmethylsulfonyl fluoride [PMSF], leupeptin, pepstatin), and sonicated for 10 s followed by 30 min on ice to resuspend and lyse the cells. Cell lysates were clarified by centrifugation at 40,000 rpm for 20 min. Recombinant baculovirus expressing AMF-1 was constructed by cotransfection of insect cells with the nonviable viral DNA (Pharmingen) and a complementing transfer vector containing AU1-tagged AMF-1 (pVL1392-AU1-AMF-1). The baculovirus strain expressing Flag-p300 is a gift from Lou Schiltz and Yoshihiro Nakatani (39).
The glutathione S-transferase (GST)–p300 fusion proteins and histidine-tagged p53 were synthesized in Escherichia coli and purified as previously described (9). GST-p300 fusion expression vectors were kindly provided by Steven R. Grossman and David M. Livingston. Induction of His6-AMF-1 expression in S. cerevisiae was done as described previously (9). Yeast cell pellets were resuspended in a mixture of 20 mM Tris (pH 8.0), 400 mM NaCl, 1% Tween 20, PMSF, leupeptin, and pepstatin and lysed by the glass bead method. The yeast expression vector for His6-AMF-1 (YEplac112G6His:AMF-1) was constructed by cloning the His-AMF-1 open reading frame into YEplac112G (9), placing the His6-AMF-1 fusion under control of the GAL promoter.
Transcriptional activation.
Transactivation assays with E2 were done essentially as described elsewhere (9). All transfections included pSVβ-gal, which was used to standardize E2 activation values for transfection efficiency. C33A cells were transfected with 100 ng of E2 expression construct pCGE2B, pcDNALexA-E2(1-216) (9), pCG16E2 (13), or pcDNALexA-16E2(1-208) along with 0, 250, or 1,000 ng of pCMVE1A12S (52). These transfections included 500 ng of the E2-dependent reporter pE2-4SVluc or LexA-dependent reporter pDBL8, where appropriate (9). VP16 AD fusions to the LexA DBD (pcDNALexAVP16; 100 ng) and the minimal E2 DBD (pCGVP16E2 125; 200 ng) were used to demonstrate the specificity of E1A inhibition for E2. For AMF-1 and p300 stimulation of E2-dependent transcription (Fig. 1), 500 ng of the E2-dependent reporter pE2-4SVluc was cotransfected into C33A cells with or without 100 ng of E2 expression vector pCGE2B, 3 μg of AMF-1 expression vector (9), and 0, 100, or 200 ng of p300 expression vector pCMVp300NHA (14). Backbone pCG vector was added to each sample to bring the total amount of cytomegalovirus promoter-containing DNA to 4 μg. At 48 h after transfection, the luciferase activities in cell lysates were measured with the luciferase assay system (Promega) and presented as the increase in activation over reporter alone.
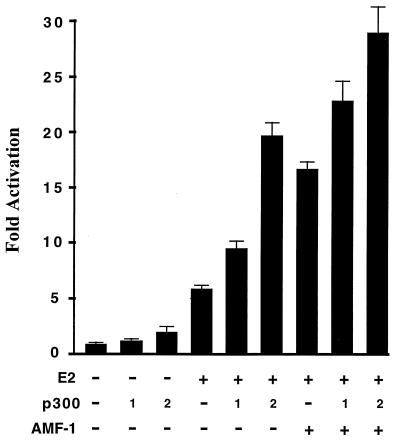
FIG. 1.
p300 and AMF-1 additively stimulate BPV-1 E2 transcriptional activation. An E2-dependent luciferase reporter was cotransfected into C33A cells with or without vectors expressing BPV-1 E2, AMF-1, and p300 as indicated; 0, 100, or 200 ng of p300 expression vector was used when the effect of p300 was evaluated. Two days after transfection, luciferase activities were measured and are presented as the increase in activation over reporter alone. Each sample was analyzed in triplicate, and standard deviations are shown. The experiments were repeated several times with similar results.
Protein interaction assays.
Human C33A cells stably expressing His6-AMF-1 or His6-β-galactosidase (β-Gal) were established by transfection of C33A cells with pcDNA3.1/6H:AMF-1 or pcDNA3.1/6H:LacZ DNA. Transfected cells were maintained in medium containing G418 (300 μg/ml; GIBCO/BRL) for 2 weeks, and positive colonies were selected for expression of His6-AMF-1 or His6-β-Gal by Western blotting. Transient expression of hemagglutinin epitope-tagged p300 (HA-p300) in C33A cells expressing His6-AMF-1 or His6-β-Gal was accomplished by transfecting HA-p300-expressing plasmid pCMVp300NHA and harvesting cells 48 h after transfection. For coprecipitation of HA-p300 with His6-AMF-1 or His6-β-Gal, the cell extract was adjusted to 40 mM imidazole. Nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen) were added in the presence or absence of 10 mM EDTA. Binding was allowed to proceed for 2 h at 4°C. Bound proteins were eluted with extraction buffer plus 250 mM imidazole. Concentrated eluates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 6% gel and subjected to Western blotting analysis using anti-p300 monoclonal antibody MAb RW128 (14, 15). For coimmunoprecipitation of AMF-1 with p300, cell extract expressing His6-AMF-1 was incubated with anti-HA MAb 12CA5 (Boehringer Mannheim) or anti-HPV-16 E6 ascites fluid. The immunocomplexes were pulled down with protein A-Sepharose beads (PAS) (Pharmacia) and washed with lysis buffer. Proteins were released from PAS beads by boiling in SDS-PAGE sample buffer and Western blotted with anti-AMF-1 serum.
For precipitation of proteins from Sf9 insect cell lysates, the cell lysates were diluted 1:1 in a buffer containing 50 mM Tris (pH 8.0), 100 mM KCl, 0.1 mM EDTA, 2 mM DTT, 0.2% NP-40, 0.1% nonfat milk, 2.5% glycerol, PMSF (100 μg/ml), leupeptin (0.5 μg/ml), and pepstatin A (1 μg/ml) before addition of antibody and PAS beads. The reactions were incubated at 4°C for 3 h. Beads were collected afterwards and washed three times in 1 ml of LSAB buffer (100 mM Tris [pH 8], 100 mM NaCl, 1% NP-40, 2 mM DTT, 100 μg of PMSF per ml). Proteins remained on beads were then analyzed by Western blotting using the Super Signal Ultra chemiluminescent reagent (Pierce). GST pull-down experiments were done as described elsewhere (9).
Acetylation analysis.
For histone acetylation assays, immunopurified E2-p300 or AMF-1–p300 complexes were mixed with 500 ng of histones (Sigma) in a 30-μl reaction buffer containing 50 mM Tris (pH 8.0), 10% glycerol, 0.1 mM EDTA, 1 mM DTT, PMSF (100 μg/ml), leupeptin (0.5 μg/ml), pepstatin A (1 μg/ml), 10 mM sodium butyrate, and 0.3 μl of [14C]acetyl coenzyme A (100 μCi/ml, 1.54 nmol/μl) and incubated for 30 min at 30°C followed by an additional 10 min on ice. Histone proteins were resolved by SDS-PAGE on a 15% gel and quantitated with a Bio-Rad GS-250 molecular imager.
Acetylation of p53, AMF-1, E2, and E1 by p300 was analyzed both in vivo and in vitro. In vivo analysis was carried out by coinfection of Flag-p300-expressing baculovirus with one or more of the baculoviruses expressing p53, AMF-1, E2, or E1 to Sf9 cells. At 24 h after infection, 5 mM sodium butyrate and 5 μM trichostatin A were added into culture medium. Cells were harvested at 40 h and lysed as described for protein production in insect cells. Target proteins were immunoprecipitated and Western blotted first with anti-acetyl-lysine antibody (Upstate Biotechnology) and then with antibody against the corresponding protein. Between the two blottings, the polyvinylidene difluoride (PVDF) membranes were incubated at 55°C for 30 min in Tris-buffered saline plus 0.1% Tween 20 (TBST) with 1% SDS and 50 mM β-mercaptoethanol, washed in TBST, and reblocked with 5% nonfat milk. In vitro acetylation was carried out in a similar way except that p300 and other target proteins were made separately in Sf9 cells, and cell lysates were combined in vitro followed by a 30-min incubation at 30°C in the presence of 0.05 mM acetyl coenzyme A (Sigma) and 10 mM sodium butyrate, before immunoprecipitation.
RESULTS
p300 and AMF-1 stimulate transcriptional activation by BPV-1 E2.
Previously we demonstrated that a novel cellular protein, AMF-1, interacted with an 82-amino-acid subdomain of the BPV-1 E2 AD (9). Mutations in this region that abolished E2 interaction with AMF-1 were found to be defective for transcriptional activation, suggesting that AMF-1 is a coactivator of E2-dependent transcription. It has been well established that p300 is a transcriptional coactivator (29, 34). To determine whether p300 is involved in the E2 transcription activation pathway, an E2-dependent luciferase reporter was transfected into human C33A cells with or without E2 and p300 expression vectors. As shown in Fig. 1, exogenous expression of p300 stimulated transactivation by E2 ∼ 2-fold. In other studies, p300 was found to enhance p53 transactivation by a similar magnitude (3, 34). Notably, coexpression of AMF-1 with p300 stimulated E2 transactivation approximately fourfold (Fig. 1). Previously we showed that AMF-1 alone did not stimulate the basal expression of this E2-dependent reporter construct (9). The observations that AMF-1 and p300 stimulate E2 transactivation in an additive but not synergistic fashion indicated that AMF-1 and p300 affect the same activation pathway. Overexpression of p300 did not restore activity of two transcriptionally inactive E2 mutants (9, 56), E2:W99C and E2:W145R, which are unable to bind TFIIB and AMF-1, respectively (data not shown).
Adenovirus E1A inhibits E2-activated transcription.
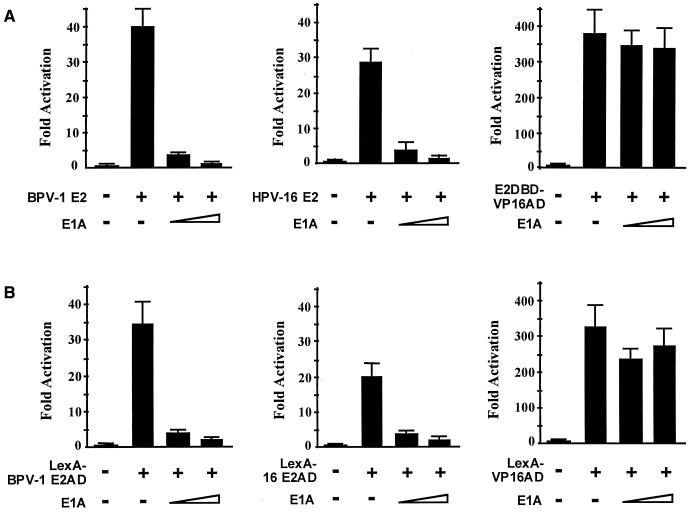
The functional significance of p300 recruitment for E2 transactivation was evaluated by determining whether activation by E2 is inhibited by the adenovirus E1A 12S protein (10, 23, 45). The E1A protein binds p300 and inhibits both its acetylase activity and coactivator functions through recruitment of RNA polymerase II (see references 10, 24, and 38 and references therein). In transient expression assays, E1A substantially inhibited activation of an E2-dependent reporter by the BPV-1 E2 and HPV-16 E2 proteins (Fig. 2A) but did not significantly affect activation by E2DBD-VP16AD, a fusion of the E2 DBD domain to the VP16 AD (amino acids 410 to 490). We previously found that AMF-1 does not interact with this VP16 AD fragment, and the E2-VP16 fusion lacked the region of E2 that interacts with AMF-1 (9). It has been reported that the VP16 does not interact directly with p300 (37). E1A mutations that are defective for Rb binding showed partial inhibition of E2, as did an N-terminal CR1 mutation that alters p300 association (data not shown). It has been reported that another region of E1A also interacts with p300 (10), and thus partial repression may be due to residual p300 association. These results suggested that p300 participates in E2 transactivation.
FIG. 2.
Adenovirus E1A inhibits E2-activated transcription. (A) The E2-dependent luciferase reporter was cotransfected into C33A cells in the presence or absence of wild-type (BPV-1 or HPV-16) E2, or E2DBD-VP16AD fusion, and E1A expression vectors. Luciferase activities were measured 2 days after transfection and are presented as the increase in activation over reporter alone; 250 and 1,000 ng of E1A expression vector were used. (B) The E2 AD confers sensitivity to E1A inhibition. Conditions were as for panel A except that the luciferase reporter is LexA dependent, and transcriptional activators are fusions of the LexA DBD to the BPV-1 E2 AD (left), HPV-16 E2 AD (middle), and VP16 AD (right).
We also performed E1A inhibition experiments using fusions of the BPV-1 and HPV-16 E2 ADs to the LexA DBD to confirm that the inhibition by E1A did not require the E2 DBD. E1A inhibited transactivation of a LexA operator reporter by the fusions LexA–BPV-1–E2AD(1-216) and LexA-16E2AD(1-208) but had little effect on activation by a LexA-VP16AD fusion protein (Fig. 2B). The specificity of E1A inhibition for the E2 AD implies that p300 recruitment functions independently of the E2 DBD. Since the E2 AD functionally interacts with both AMF-1 and p300, we then examined whether AMF-1 and p300 interact each other in vivo.
Coprecipitation of p300 with AMF-1 from C33A cells.
Human C33A cervical carcinoma cells were modified to stably express His6-β-Gal or His6-AMF-1 (Fig. 3A). As shown by quantitative Western blotting, only the His6-AMF-1 species was detected in cells transfected with His6-AMF-1, with the endogenous AMF-1 protein below the detection level of the assay (Fig. 3A, lane 3). Endogenous AMF-1 could be easily detected in the same amount of extract from the parental C33A cell line (lane 4) and in His6-β-Gal-expressing C33A cells (data not shown). The level of His6-AMF-1 in the transfected cells was similar to the level of endogenous AMF-1 in the parental C33A cells, implying that the AMF-1 level is finely modulated in vivo. Furthermore, BPV-1 E2 activated transcription of an E2-dependent reporter efficiently in cells expressing His6-AMF-1 (data not shown), indicating that the His6-AMF-1 protein is able to carry out the function of endogenous AMF-1. For binding experiments, the His6-β-Gal and His6-AMF-1 proteins were quantitatively exhausted from cell extracts by chelating to a Ni-NTA resin. After extensive washing, both His6-β-Gal and His6-AMF-1 proteins could be eluted from the resin by imidazole (data not shown).
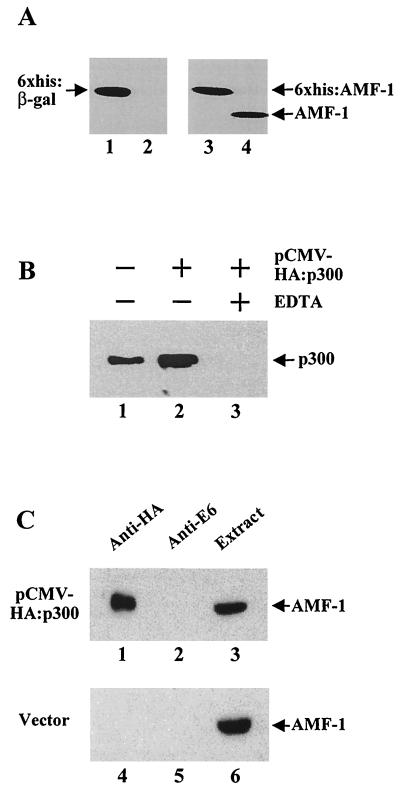
FIG. 3.
Coprecipitation of p300 with AMF-1. (A) Stable expression of His6-β-Gal or His6-AMF-1 in human C33A cells. Equal amounts of total cell protein extracts from parental C33A (lanes 2 and 4), His6-β-Gal-expressing C33A (lane 1), and His6-AMF-1-expressing C33A (lane 3) cells were subjected to Western blotting with either MAb against β-Gal (Boehringer Mannheim) (lanes 1 and 2) or polyclonal rabbit sera against AMF-1 (lanes 3 and 4). (B) Human C33A cells expressing His6-AMF-1 were transfected with pCMV-HA:p300 or vector alone; 48 h later, extracts were prepared and incubated with Ni-NTA resin in the presence (lane 3) or absence (lanes 1 and 2) of 10 mM EDTA. After extensive washing, His6-AMF-1 was eluted and concentrated. Copurification of p300 with His6-AMF-1 was probed by Western blotting with MAb RW128 (14, 15). Both endogenous (lane 1) and HA-tagged (lane 2) p300 copurified with His6-AMF-1. (C) The same cell extracts as in panel B were incubated with anti-HA (lanes 1 and 4) or anti-BPV-1 E6 (lanes 2 and 5, as negative controls) antiserum. Immunoprecipitates were resolved by SDS-PAGE and Western blotted with anti-AMF-1 serum. Equal aliquots of each extract were run in the gel as an indication of AMF-1 expression (lanes 3 and 6).
The stable expression cell lines were then transfected with a p300 expression vector. The His6-AMF-1 in the cell extract was bound to Ni-NTA resin, washed extensively, and eluted with imidazole as shown in Fig. 3B. Both endogenous (lane 1) and HA-tagged p300 (lane 2) were present in the eluate, indicating that p300 copurified with the His6-AMF-1 protein. No p300 was detected when binding was performed in the presence of 10 mM EDTA (lane 3), which blocks binding of the His6 tag protein to the Ni-NTA resin (26). Nonspecific binding of p300 to the Ni-NTA resin in the absence of EDTA was ruled out by the use of p300-transfected cell extract expressing His6-β-Gal. His6-β-Gal was quantitatively recovered using the Ni-NTA resin, but p300 was not detected in the eluate (data not shown).
The association of AMF-1 and p300 was confirmed in a reciprocal assay. HA-tagged p300 was transfected into the C33A cell line that expressed His6-AMF-1. His6-AMF-1 was coimmunoprecipitated with HA-p300 using the anti-HA MAb 12CA5 (Fig. 3C, lane 1, top gel). His6-AMF-1 was not precipitated from the same extract using a control antibody (Fig. 3C, lane 2). His6-AMF-1 was also not detected in control immunoprecipitation reactions using the HA antibody and control extracts prepared from His6-AMF-1 cells transfected with vector alone (Fig. 3C, lane 4, bottom gel). These experiments demonstrated that AMF-1 and p300 exist in a complex in vivo.
In vitro complex formation of AMF-1 and E2 with p300.
To further explore the relationships between E2, AMF-1, and p300, we produced these proteins in Sf9 cells using recombinant baculoviruses. In these experiments, p300 had an amino-terminal Flag tag that allowed its isolation from insect cell extracts using the anti-Flag MAb M2 conjugated to Sepharose beads. Cell extracts containing AMF-1, E2, or E1 were combined with extract containing Flag-p300 (Fig. 4, lane 3). Immunoprecipitations with M2 antibody-coated beads consistently pulled down AMF-1 and E2, but not BPV-1 E1, in complex with Flag-p300. Neither AMF-1 nor E2 was present in M2 complexes from uninfected cell extract that did not contain Flag-p300 (lane 2), indicating that the AMF-1–p300 and E2-p300 interactions are specific. Immunoprecipitation of p300 was more efficient with AMF-1 than with E2 (Fig. 4). Typically, approximately 30% of input AMF-1 could be coimmunoprecipitated with p300, while only 5% of input E2 was pulled down with the same amount of Flag-p300 (Fig. 4). In experiments using bacterially produced GST-p300 fusion and E2 proteins, association of these proteins was marginally above background levels (data not shown).
FIG. 4.
In vitro complex formation of AMF-1 and E2 with p300. Sf9 cells were infected by recombinant baculoviruses expressing each of the proteins, harvested 48 h postinfection, and lysed as described in Materials and Methods. Cell extract with (lane 3) or without (lane 2, from uninfected Sf9 cells) Flag-p300 was incubated with extract containing AMF-1, E2, or E1. Flag-p300 was immunoprecipitated with anti-Flag MAb M2 conjugated to Sepharose beads (Sigma). After washing, the immunoprecipitates were analyzed by Western blotting with rabbit polyclonal antibody against AMF-1 or E1 or MAb against E2. The heavy chain of M2 (M2 HC) was detected by anti-mouse secondary antibody. Lane 1 shows 10% of input cell extracts containing AMF-1, E2, or E1.
Histone acetylation by E2-p300 and AMF-1–p300 complexes.
E2 or AMF-1 complexes were collected from Sf9 cell lysates, using an antibody against each protein. The activity of p300 in the complexes was tested by a histone acetylation assay (Fig. 5). Both E2 and AMF-1 immunoprecipitations showed acetylase activity only when combined with extracts containing p300 (lanes 3 and 7), not in the absence of p300 (lanes 1, 2, 5, and 6). Immunoprecipitations using antibodies against AMF-1 resulted in more acetylase activity than immunoprecipitations with antibody against E2 when incubated with equal amounts of p300-containing cell extract (compare lanes 3 and 7). The presence of AMF-1 in the immunoprecipitations with E2 and p300 increased the acetylase activity about twofold (compare lanes 3 and 4), indicating that AMF-1 enhanced the interaction between E2 and p300.
FIG. 5.
Histone acetylation by AMF-1–p300 and E2-p300 complexes. Immunoprecipitation reactions were set up with or without Sf9 cell extracts containing p300, E2, or AMF-1, as indicated. A minus sign represents equal amount of extract from uninfected Sf9 cells. Protein complexes were immunoprecipitated with either MAb against E2 or polyclonal antibody against AMF-1. Precipitates were washed and added to histone acetylation reactions as described in Materials and Methods. Histone proteins were resolved on an SDS–15% polyacrylamide gel and analyzed with a Bio-Rad molecular imager.
AMF-1 interacts with the N-terminal portion of p300.
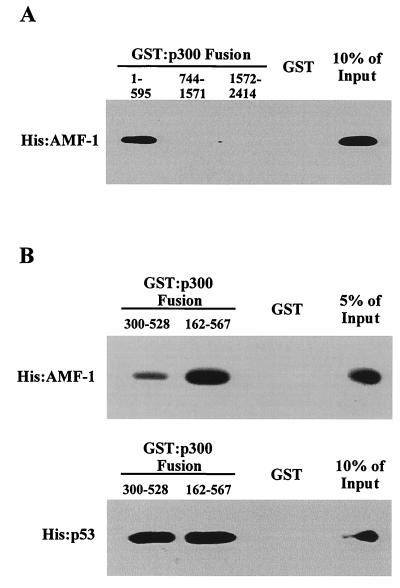
In vitro binding assays using GST-p300 fusions expressed in E. coli and recombinant His6-AMF-1 purified from S. cerevisiae were performed to determine if p300 and AMF-1 interacted directly and to identify the domain on p300 that binds AMF-1. In a GST pull-down assay, about 10% of the input His6-AMF-1 was bound to GST-p300(1-595), while none was retained by GST alone or GST-p300(744-1571), GST-p300(1572-2414) (Fig. 6A). Amino acids 1 to 595 of p300 include the CH1 domain that binds p53 (20). To determine whether AMF-1 and p53 bind to the same region of p300, interaction between His-AMF-1 and the CH1 domain of p300 [GST-p300(300-528)] was examined. The results showed that AMF-1 binding to the CH1 domain was weak, with only 1.9% of input bound (Fig. 6B). A larger fusion [GST-p300(162-567)] bound AMF-1 more efficiently, retaining 8.4% of input (Fig. 6B). As expected, the two fusion proteins bound to p53 with similar efficiencies [GST-p300(300-528), 31% input; GST-p300(162-567), 38% input] (Fig. 6B). These results indicated that the requirements for AMF-1 and p53 binding to the N terminus of p300 are not identical. A similar situation was observed when mdm2 and p53 binding to this region of p300 was examined. While mdm2 bound essentially to the CH1 domain, p53 binding to CH1 required additional residues (20).
FIG. 6.
Direct interaction of AMF-1 with p300. GST or GST-p300 fusions were incubated with His-AMF-1 or His-p53 and washed extensively. Bound proteins were resolved by SDS-PAGE followed by Western blotting. (A) AMF-1 binds to the amino-terminal 595 amino acids of p300 but not GST alone or GST-p300 fusions containing amino acid residues 744 to 1571 and 1572 to 2414. (B) Comparison of p300 CH1 domain (amino acids 300 to 528) binding by AMF-1 and p53.
Acetylation analysis of AMF-1, E2, and E1 by p300.
The preceding results suggested that histones are a potential target for acetylation by the p300 complexes with AMF-1 and E2. To determine whether AMF-1 or E2 can also be acetylated by p300, Sf9 cell lysates containing each of p53, AMF-1, E2, and E1 were mixed with cell lysates containing p300 in the presence of acetyl coenzyme A and deacetylase inhibitors. After 30 min of incubation at 30°C, target proteins (p53, AMF-1, E2, and E1) were immunoprecipitated and analyzed for the presence of acetyl-lysine by Western blotting. Neither AMF-1, E2, nor E1 reacted with acetyl-lysine antibodies, while p53 showed strong acetyl-lysine reactivity after incubation with p300 (Fig. 7). To examine the effect of E2 on acetylation of AMF-1, cell lysate containing E2 was added into the reaction when acetylation of AMF-1 was tested; vice versa, AMF-1 was added into E2 acetylation reaction to test its effect on E2 acetylation. The combination of E2 with AMF-1 and p300 did not lead to their acetylation on lysine (Fig. 7). Similarly, E2 had no effect on acetylation of E1 (Fig. 7). The acetylation was also performed in vivo by coinfection of Sf9 cells with recombinant baculovirus expressing p300 and one or more baculoviruses expressing p53, AMF-1, E2, and E1. Target proteins (p53, AMF-1, E2, and E1) were immunoprecipitated directly from cell lysates and tested for acetyl-lysine by Western blotting. The results were similar to those of in vitro assays (data not shown). These data indicated that AMF-1, E2, and E1 are probably not targets of p300 lysine acetylase activity.
FIG. 7.
Acetylation analysis of p53, AMF-1, E2, and E1 by p300. In vitro acetylation of p53, AMF-1, E2, and E1 was tested using Sf9 cell extracts containing each of these proteins and extract containing p300. Cell extract containing p300 was mixed with lysate containing each of p53, AMF-1, E2, and E1 at 30°C for 30 min, in the presence of 0.05 mM acetyl coenzyme A and 10 mM sodium butyrate. p53, AMF-1, E2, and E1 were then immunoprecipitated (IP) by incubation with antibodies (Ab) against each protein. Immunoprecipitates were washed, resolved on an SDS–10% polyacrylamide gel, and transferred to PVDF membranes. After Western blotting with antibody against acetyl-lysine, the PVDF membranes were stripped and Western blotted with antibody against p53, AMF-1, E2, or E1. In each panel, results of anti-acetyl-lysine blotting are shown at the top. Acetylation reactions were also performed by mixing AMF-1, E2, and E1 cell extracts. In vivo acetylation assays were carried out by coinfecting Sf9 cells with baculovirus expressing p300 and one or more baculoviruses expressing p53, AMF-1, E2, or E1, as indicated; 24 h after infection, 5 mM sodium butyrate and 5 μM trichostatin A were added to the cell culture medium. Cell extracts were prepared 48 h postinfection. p53, AMF-1, E2, and E1 proteins were immunoprecipitated from 200 μl of cell extracts, followed by the same Western blotting procedure as for in vitro assays.
DISCUSSION
We previously identified AMF-1/Gps2 as an E2-interacting factor by a yeast two-hybrid screen. AMF-1 binds the E2 AD and increases its transcriptional activity. E2 mutants that were unable to bind AMF-1 were severely crippled for activation of transcription and replication. We questioned whether AMF-1 affected a putative common step such as chromatin remodeling, and by analogy with other transcriptional activators, we suspected that E2 might interact with p300. While we observed only weak interactions between E2 and p300, direct complex formation between the AMF-1 and p300 proteins was more efficient (Fig. 4). We attempted to show complex formation between the endogenous AMF-1 and p300 in different cell lines, but those experiments were not successful (data not shown). It is possible that the endogenous AMF-1 bound to p300 is below detectable amounts; AMF-1–p300 complex formation may also be transient or result in a complex inaccessible to antibodies.
AMF-1 enhanced the interaction of E2 with p300, and overexpression of both AMF-1 and p300 showed additive stimulation of E2-dependent transcription (Fig. 1). Overexpression of p300 did not increase the transcriptional activity of E2: W145R, a mutant that is defective for AMF-1 binding (data not shown). The sensitivity of BPV-1 E2 transactivation to inhibition by adenovirus E1A and stimulation of E2 activation by expression of exogenous p300 imply that p300 recruitment plays a role in transcriptional activation by E2. The recruitment of p300 in this fashion is likely a general property of papillomavirus E2 proteins, because residues of the BPV-1 E2 AD critical for AMF-1 interaction are highly conserved in other E2 proteins (9). We also demonstrated direct interaction of AMF-1 with HPV E2 proteins using GST pull-down and mammalian two-hybrid assays (unpublished data).
The AMF-1 interaction has been mapped to residues 162 to 567 of p300 (Fig. 6). This region of p300 also participates in both direct and mdm2-mediated interactions with p53 (20) and plays an important role in regulating turnover of p53 (20). A GST-p300 fusion protein [GST:p300(300-528)] containing the CH1 region bound p53 with high avidity but interacted weakly with AMF-1, indicating that AMF-1 and p53 interaction domains on p300 are not identical (Fig. 6). While the CH1 domain may play a role in AMF-1–p300 interaction, additional residues appear to be required for efficient binding. These results suggest that AMF-1 may regulate p53 through p300. We have observed that AMF-1 significantly stimulates the activity of p53 (unpublished data).
The dual nature of the p53-p300 interaction prompted us to search for direct interaction of BPV-1 E2 with p300. Our initial attempts to detect E2-p300 complexes in extracts prepared from p300- and E2-transfected C33A cells failed, suggesting that direct E2-p300 interaction is not sufficient for p300 recruitment (data not shown). However, weak binding of E2 to p300 could be detected using recombinant E2 and p300 proteins produced in insect cells (Fig. 4). We cannot exclude the possibility that E2 protein association with Flag-p300 is indirect. Histone acetylation experiments also showed that antibody against E2 could immunoprecipitate acetylase activity from cell extract containing both E2 and p300 proteins but not from cell extract containing E2 or p300 alone (Fig. 5). We hypothesize that AMF-1 binds p300 and enhances its normally weak interaction with E2. Currently we are screening our E2 mutations for the ability to bind recombinant p300 and testing whether these overlap with the AMF-1 binding domain.
Genetic evidence indicates that recruitment of RNA polymerase II by the interaction of AMF-1 with p300 would be necessary but not sufficient for transcriptional activation by E2. For example, a TFIIB binding domain has been mapped to a region of the E2 AD distinct from the AMF-1 binding domain, and TFIIB binding-defective mutants within this domain were identified (56). These TFIIB binding-defective mutants were unable to activate transcription but retained the ability to associate with AMF-1, suggesting that TFIIB binding, as well as AMF-1 binding, is necessary for transcriptional activation (unpublished data). Nakajima et al. also found a dual requirement for CREB: activation of transcription requires both recruitment of the RNA polymerase II through interactions of the CREB KIX domain with CBP and recruitment of TFIID by the glutamine-rich Q2 domain of CREB by interaction with human TAFII 130 (38). These authors also found that CREB activation and recruitment of RNA polymerase II by CBP was inhibited by E1A. The inhibition of RNA polymerase II recruitment by p300/CBP might also explain the sensitivity of E2 transactivation to E1A.
E1A has been recently found to inhibit the acetyltransferase activity of p300/CBP as well as recruitment of RNA polymerase II (10, 24). Recruitment of histone acetylase activity (4, 39) by AMF-1 could result in modification of the chromatin structure and promote formation of transcription and replication initiation complexes. Although we could not find any report showing that the HAT activity of p300/CBP is required for DNA replication, it was recently demonstrated that histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human DNA replication initiator protein (28). E2 targets the papillomavirus E1 DNA helicase to the origin of replication, which contains E2 binding sites. The phenotype of the AMF-1 binding-defective mutants (9) is consistent with a defect in the recruitment of histone acetylase activity that results in an inability to modulate chromatin remodeling. It will be of interest to see if p300 and CBP are directly involved in papillomavirus DNA replication.
Our results showed that fusions of the BPV-1 and HPV-16 E2 ADs to the LexA DBD retain sensitivity to inhibition by E1A (Fig. 2), suggesting that the mechanism by which p300 influences E2 transcriptional activation is independent of the E2 DBD. Acetylation of p53 and GATA-1 by p300/CBP has been found to modulate the function of these transcription factors at the level of DNA binding (22, 35, 42). The DNA binding activity of p53 is regulated in vivo via acetylation by p300/CBP and P/CAF in response to DNA damage (42). Acetylation of human immunodeficiency virus type 1 Tat by p300 or P/CAF at two lysine residues (Lys50 and Lys28) regulates its activities in transcription, binding to an RNA polymerase II C-terminal domain (CTD) kinase and release from trans-activation response region (TAR) RNA (31). It is unlikely that p300 regulates the E2 DNA binding domain in an analogous manner, as we showed that p300 stimulated the chimera of the E2 AD to the LexA DBD. Furthermore, acetylation experiments with antibody against acetyl-lysine failed to show acetylation of AMF-1, E2, and E1 by p300 (Fig. 7). However, we cannot exclude the possibility that these proteins are acetylated, perhaps on another amino acid, in vivo. Further experiments are necessary to find out what properties of E2 may be affected upon binding to p300 and AMF-1.
ACKNOWLEDGMENTS
Y.-C. Peng and D. E. Breiding contributed equally to the work.
We are grateful to L. Banks, M. Botchan, S. Grossman, D. Livingston, Y. Nakatani, and L. Schiltz for providing reagents. We thank the members of the lab for many useful discussions.
This work is supported by NIH grants R01 CA58376 and U01 AI38001 to E.J.A.
REFERENCES
- 1.Abroi A, Kurg R, Ustav M. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J Virol. 1996;70:6169–6179. doi: 10.1128/jvi.70.9.6169-6179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antson A A, Burns J E, Moroz O V, Scott D J, Sanders C M, Bronstein I B, Dodson G G, Wilson K S, Maitland N J. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature. 2000;403:805–809. doi: 10.1038/35001638. [DOI] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Berg M, Stenlund A. Functional interactions between papillomavirus E1 and E2 proteins. J Virol. 1997;71:3853–3863. doi: 10.1128/jvi.71.5.3853-3863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bevilacqua M A, Faniello M C, Russo T, Cimino F, Costanzo F. P/CAF/p300 complex binds the promoter for the heavy subunit of ferritin and contributes to its tissue-specific expression. Biochem J. 1998;335:521–525. doi: 10.1042/bj3350521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 8.Breiding D E, Grossel M J, Androphy E J. Genetic analysis of the bovine papillomavirus E2 transcriptional activation domain. Virology. 1996;221:34–43. doi: 10.1006/viro.1996.0350. [DOI] [PubMed] [Google Scholar]
- 9.Breiding D E, Sverdrup F, Grossel M J, Moscufo N, Boonchai W, Androphy E J. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol Cell Biol. 1997;17:7208–7219. doi: 10.1128/mcb.17.12.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 12.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cripe T P, Haugen T H, Turk J P, Tabatabai F, Schmid P G d, Durst M, Gissmann L, Roman A, Turek L P. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 1987;6:3745–3753. doi: 10.1002/j.1460-2075.1987.tb02709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 15.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson M K, Botchan M R. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J Virol. 1996;70:4193–4199. doi: 10.1128/jvi.70.7.4193-4199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouts E T, Yu X, Egelman E H, Botchan M R. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J Biol Chem. 1999;274:4447–4458. doi: 10.1074/jbc.274.7.4447. [DOI] [PubMed] [Google Scholar]
- 18.Giordano A, Avantaggiati M L. p300 and CBP: partners for life and death. J Cell Physiol. 1999;181:218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 19.Grossel M J, Sverdrup F, Breiding D E, Androphy E J. Transcriptional activation function is not required for stimulation of DNA replication by bovine papillomavirus type 1 E2. J Virol. 1996;70:7264–7269. doi: 10.1128/jvi.70.10.7264-7269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 21.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 22.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 24.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 25.Harris S F, Botchan M R. Crystal structure of the human papillomavirus type 18 E2 activation domain. Science. 1999;284:1673–1677. doi: 10.1126/science.284.5420.1673. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann A, Roeder R G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991;19:6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes F J, Romanos M A. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res. 1993;21:5817–5823. doi: 10.1093/nar/21.25.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- 29.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 30.Jin D Y, Teramoto H, Giam C Z, Chun R F, Gutkind J S, Jeang K T. A human suppressor of c-Jun N-terminal kinase 1 activation by tumor necrosis factor alpha. J Biol Chem. 1997;272:25816–25823. doi: 10.1074/jbc.272.41.25816. [DOI] [PubMed] [Google Scholar]
- 31.Kiernan R E, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang K T, Benkirane M, Van Lint C. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li R, Botchan M R. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc Natl Acad Sci USA. 1994;91:7051–7055. doi: 10.1073/pnas.91.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizzen C A, Allis C D. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 38.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 39.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 40.Ott M, Schnolzer M, Garnica J, Fischle W, Emiliani S, Rackwitz H R, Verdin E. Acetylation of the HIV-1 tat protein by p300 is important for its transcriptional activity. Curr Biol. 1999;9:1489–1492. doi: 10.1016/s0960-9822(00)80120-7. [DOI] [PubMed] [Google Scholar]
- 41.Patel D, Huang S M, Baglia L A, McCance D J. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scholl T, Mahanta S K, Strominger J L. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci USA. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverman E S, Du J, Williams A J, Wadgaonkar R, Drazen J M, Collins T. cAMP-response-element-binding-protein-binding protein (CBP) and p300 are transcriptional co-activators of early growth response factor-1 (Egr-1) Biochem J. 1998;336:183–189. doi: 10.1042/bj3360183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Somasundaram K, El-Deiry W S. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 46.Spain B H, Bowdish K S, Pacal A R, Staub S F, Koo D, Chang C Y, Xie W, Colicelli J. Two human cDNAs, including a homolog of Arabidopsis FUS6 (COP11), suppress G-protein- and mitogen-activated protein kinase-mediated signal transduction in yeast and mammalian cells. Mol Cell Biol. 1996;16:6698–6706. doi: 10.1128/mcb.16.12.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 48.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki T, Uchida-Toita M, Yoshida M. Tax protein of HTLV-1 inhibits CBP/p300-mediated transcription by interfering with recruitment of CBP/p300 onto DNA element of E-box or p53 binding site. Oncogene. 1999;18:4137–4143. doi: 10.1038/sj.onc.1202766. [DOI] [PubMed] [Google Scholar]
- 50.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 51.Van Orden K, Yan J P, Ulloa A, Nyborg J K. Binding of the human T-cell leukemia virus Tax protein to the coactivator CBP interferes with CBP-mediated transcriptional control. Oncogene. 1999;18:3766–3772. doi: 10.1038/sj.onc.1202703. [DOI] [PubMed] [Google Scholar]
- 52.White E, Cipriani R, Sabbatini P, Denton A. Adenovirus E1B 19-kilodalton protein overcomes the cytotoxicity of E1A proteins. J Virol. 1991;65:2968–2978. doi: 10.1128/jvi.65.6.2968-2978.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 54.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 56.Yao J M, Breiding D E, Androphy E J. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J Virol. 1998;72:1013–1019. doi: 10.1128/jvi.72.2.1013-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasugi T, Benson J D, Sakai H, Vidal M, Howley P M. Mapping and characterization of the interaction domains of human papillomavirus type 16 E1 and E2 proteins. J Virol. 1997;71:891–899. doi: 10.1128/jvi.71.2.891-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmermann H, Degenkolbe R, Bernard H U, O'Connor M J. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73:6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]