Abstract
The M2 gene of respiratory syncytial (RS) virus has two open reading frames (ORFs). ORF1 encodes a 22-kDa protein termed M2-1. The M2-1 protein contains a Cys3-His1 motif (C-X7-C-X5-C-X3-H) near the amino terminus. This motif is conserved in all human, bovine, and ovine strains of RS virus. A similar motif found in the mammalian transcription factor Nup475 has been shown to bind zinc. The M2-1 protein of human RS virus functions as a transcription factor which increases polymerase processivity, and it enhances readthrough of intergenic junctions during RS virus transcription, thereby acting as a transcription antiterminator. The M2-1 protein also interacts with the nucleocapsid protein. We examined the effects of mutations of cysteine and histidine residues predicted to coordinate zinc in the Cys3-His1 motif on transcription antitermination and N protein binding. We found that mutating the predicted zinc-coordinating residues, the cysteine residues at amino acid positions 7 and 15 and the histidine residue at position 25, prevented M2-1 from enhancing transcriptional readthrough. In contrast, mutations of amino acids within this motif not predicted to coordinate zinc had no effect. Mutations of the predicted zinc-coordinating residues in the Cys3-His1 motif also prevented M2-1 from interacting with the nucleocapsid protein. One mutation of a noncoordinating residue in the motif which did not affect readthrough during transcription, E10G, prevented interaction with the nucleocapsid protein. This suggests that M2-1 does not require interaction with the nucleocapsid protein in order to function during transcription. Analysis of the M2-1 protein in reducing sodium dodecyl sulfate-polyacrylamide gels revealed two major forms distinguished by their mobilities. The slower migrating form was shown to be phosphorylated, whereas the faster migrating form was not. Mutations in the Cys3-His1 motif caused a change in distribution of the M2-1 protein from the slower to the faster migrating form. The data presented here show that the Cys3-His1 motif of M2-1 is essential for maintaining the functional integrity of the protein.
Human respiratory syncytial virus (RS virus) is a member of the Pneumovirus genus of the Paramyxoviridae family. It is the leading viral cause of pediatric lower respiratory tract disease and is a significant cause of morbidity and mortality worldwide. The genome of RS virus is a single strand of negative-sense RNA 15,222 nucleotides in length, having 10 genes encoding 11 proteins (4, 14, 24, 32). Each mRNA encodes one of the viral proteins observed in infected cells. As with all nonsegmented, negative-sense RNA viruses, RNA synthesis requires a genomic RNA encapsidated with nucleocapsid (N) protein and the virus-encoded components of the RNA-dependent RNA polymerase, the phosphoprotein (P) and the large polymerase protein (L) (7, 37). The N, P, and L proteins are sufficient for replication of the genomic RNA (10, 37). However, RS virus, unlike other nonsegmented negative-sense RNA viruses, encodes an additional protein, M2-1, which functions during transcription of the viral mRNAs (3). This protein has been shown to increase the processivity of the viral polymerase, thus preventing premature termination during transcription. Additionally, we have shown that the M2-1 protein enhances readthrough of transcription termination signals and thus functions as a transcription antiterminator (8, 12). Further, the RS virus gene end sequences vary, and we have shown that the M2-1 protein acts differentially at the different gene ends (11).
The M2-1 protein is found only in pneumoviruses. It is encoded by the next-to-last gene of the RS virus genome and is 194 amino acids in length (calculated molecular mass, 22,150 Da) (5). M2-1 is a hydrophilic protein with a predicted pI of 9.6. Examination of the predicted amino acid sequence led to the identification of a Cys3-His1 motif (C-X7-C-X5-C-X3-H) located near the amino terminus of the protein, from residues 7 to 25. This motif is found in the M2-1 protein of all the pneumoviruses examined to date (19, 36, 38; R. W. Hardy and G. W. Wertz, unpublished data). A similar motif is found in VP30 of the filoviruses (28). A number of cysteine-rich motifs have been characterized and grouped according to the arrangement and number of cysteine and histidine residues involved in coordinating a zinc ion. Many proteins bind zinc, and in a number of enzymes, zinc has been shown to play a role in catalysis (15). However, in other cases, zinc plays a purely structural role (23). The Cys3-His1 motif has been characterized for only one protein, Nup475, a mammalian transcription factor in which it was demonstrated to bind zinc, and a structure for this motif has been proposed using nuclear magnetic resonance and photometric analyses (35).
Six species of the RS virus M2-1 protein have been observed by two-dimensional electrophoresis (26). When the M2-1 protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions, the majority of the protein was found in two forms distinguished by their electrophoretic mobilities. The cause of the differences in migration of these species or whether the different species have different functions is unknown. The M2-1 protein has been reported to be phosphorylated, but the relationship between phosphorylation and the different forms of the protein has not been investigated (17).
The M2-1 protein has also been shown to interact with the N protein in RS virus-infected cells or when the two proteins are coexpressed in cells from plasmid vectors (9, 27). The significance of this interaction and the role it may play are currently unknown.
In this report, we examine the role of the predicted zinc-coordinating residues of the Cys3-His1 motif in the gene end antitermination activity of M2-1, in its ability to interact with N, and in its phosphorylation state. We found that mutations of the residues predicted to coordinate zinc prevented the M2-1 protein from enhancing transcriptional readthrough and interacting with the nucleocapsid protein. We also found that the two major species of the M2-1 protein distinguished by their mobilities in reducing SDS-PAGE differed according to whether they were phosphorylated. This work demonstrates the requirement for conservation of the Cys3-His1 motif, a potential zinc binding domain, to maintain the functional integrity of the M2-1 protein.
MATERIALS AND METHODS
cDNA constructs.
We generated vectors for expressing the M2-1 protein in eukaryotic cells, using a vaccinia virus-T7 expression system. Generation of cDNAs expressing N, P, L, and wild-type M2-1 proteins (pN, pP, pL, and pORF1, respectively) has been described previously (12, 37). Mutations in the Cys3-His1 motif of the M2-1 protein were generated by PCR mutagenesis with primers containing coding changes and cloned into the BamHI-HindIII sites of pGEM3 behind the T7 promoter. Nucleotide sequences of cDNA constructs were determined by dideoxy nucleotide chain termination DNA sequencing. The generation of pM/SH (encoding an RS virus dicistronic subgenomic replicon) was described previously (12).
Virus and cells.
HEp-2 cells were grown in minimum essential medium (MEM) (GIBCO Laboratories) supplemented with 5% heat-inactivated fetal bovine serum (FBS) in 60-mm-diameter dishes.
The A-2 strain of human RS virus was propagated in HEp-2 cells. RS virus was added to the cells at a multiplicity of infection of 1 PFU/cell. Virus was allowed to adsorb for 2 h at 37°C. Fresh medium (2 ml of MEM supplemented with 5% FBS) was added, and cells were incubated at 37°C for 18 h. The medium was removed, and medium deficient in methionine and cysteine or phosphate, as required, was added for 30 min. Proteins were labeled for 2 h, using 66 μCi of [35S]methionine and [35S]cysteine (Tran35S-label; ICN) per ml or 100 μCi of [33P]inorganic phosphate (ICN) per ml. Cells were harvested, and cytoplasmic extracts were prepared as previously described (34). RS virus-specific proteins were detected by immunoprecipitation followed by SDS-PAGE.
cDNA transfections.
RS virus RNA synthesis, programmed by subgenomic replicons, was assayed by using a recombinant vaccinia virus-T7 expression system. HEp-2 cells infected with recombinant MVA vaccinia virus expressing T7 RNA polymerase were transfected with 6 μg of pM/SH, 5 μg of pN, 2 μg of pP, 2 μg of pL, and 0.3 μg pORF1 (encoding wild-type M2-1 protein) or pGEM-based plasmid bearing the gene encoding mutant M2 protein. RS virus-specific RNAs were labeled with [3H]uridine (33 μCi/ml; Moravek) in the presence of actinomycin D (10 μg/ml; Sigma) and cytosine arabinoside (50 μg/ml; Sigma) at 16 h posttransfection. After a 5-h labeling period, cells were harvested and cytoplasmic extracts were prepared as previously described (25). RNAs were purified by phenol extraction followed by ethanol precipitation. RNAs were analyzed by electrophoresis in 1.75% agarose–urea gels and detected by fluorography (18, 33).
Expression of M2-1 mutant proteins was analyzed by transfecting vTF7-3-infected HEp-2 cells with 2 μg of pGEM-based plasmids bearing the gene encoding the wild-type M2-1 protein or mutant M2-1 proteins. The interaction of the M2-1 protein with the nucleocapsid protein was analyzed for cells cotransfected with the M2-1-bearing plasmids with wild-type or mutant M2-1 proteins and 5 μg of pN. Sixteen hours posttransfection, cells were incubated in methionine- and cysteine-free medium or phosphate-free medium (ICN) for 30 min and then exposed to [35S]methionine and [35S]cysteine (66 μCi/ml, Tran35S-label; ICN) or [33P]inorganic phosphate (100 μCi/ml; ICN). Following a 2-h labeling period, cells were harvested and cytoplasmic extracts were prepared as previously described.
Pulse-chase analysis of M2 protein.
M2-1 protein maturation and stability were analyzed by metabolic labeling with a short exposure to [35S]methionine and [35S]cysteine (pulse) followed by various incubation times postlabeling (chase). HEp-2 cells infected with vTF7-3 were transfected with a plasmid bearing the gene encoding the wild-type M2-1 protein as described above. Sixteen-hour-posttransfection cells were incubated in methionine- and cysteine-free medium for 30 min and then exposed to [35S]methionine and [35S]cysteine (100 μCi/ml, Tran35S-label; ICN) for 15 min. Following the labeling period, either cells were harvested and cytoplasmic extracts were prepared or the label was removed, cells were washed, and fresh medium containing excess unlabeled methionine and cysteine (10 mM) was added for 15, 30, or 60 min prior to harvest.
Immunoprecipitations of labeled proteins.
Immunoprecipitation of RS virus-specific proteins from cytoplasmic extracts was performed using an M2-1 protein-specific monoclonal antibody (MAb), 5H5 or 1C13 (kind gift from G. Toms), or a polyclonal anti-RS virus serum (Chemicon International) and protein G-Sepharose (Pharmacia Biotech). Immunoprecipitated proteins were analyzed by SDS-PAGE in 11% polyacrylamide gels under reducing conditions and detected by fluorography (2, 16).
RESULTS
Effect of mutations in the Cys3-His1 motif on RS virus transcription.
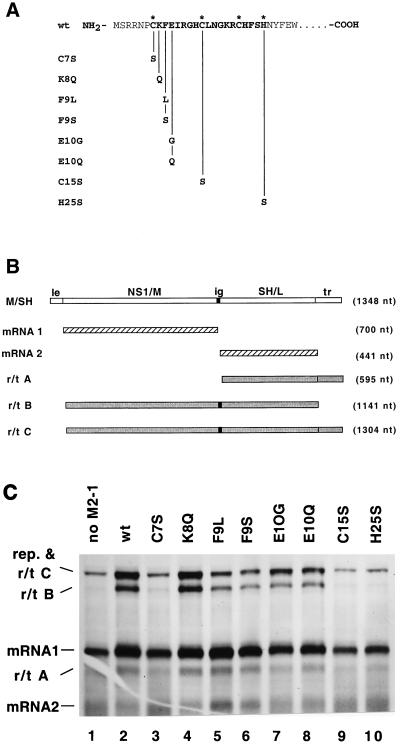
The M2-1 protein causes a decrease in the efficiency of transcription termination at RS virus gene junctions, resulting in an increased production of readthrough transcripts and thus functioning as a transcription antiterminator (8, 12). Sequence analysis revealed that the M2-1 protein contains a Cys3-His1 amino acid sequence motif (C-X7-C-X5-C-X3-H). A similar motif has been shown to bind zinc in Nup475 protein, a mammalian transcription factor (35). To determine whether this motif was important for the M2-1 protein to function as an antiterminator during viral transcription, point mutations were generated in the cDNA encoding the M2-1 protein (Fig. 1A). Three of the residues predicted to coordinate a zinc ion, the cysteine residues at positions 7 and 15 and the histidine at position 25, were each changed to serines. The mutants were named the C7S, C15S, and H25S mutants, respectively. In addition, three other residues within the motif which would not be predicted to be involved in coordinating a zinc ion were mutated as controls: K8Q, F9L, F9S, E10G, and E10Q (Fig. 1A). The mutant M2-1 ORFs were cloned behind the T7 promoter in pGEM3.
FIG. 1.
Analysis of the effects of mutations in the Cys3-His1 motif of the M2-1 protein on RS virus transcription. (A) Diagram of the mutations made in the Cys3-His1 motif. NH2 signifies the amino terminus of the M2 protein. The predicted zinc-coordinating residues are identified by an asterisk. (B) Diagram of the RS virus dicistronic subgenomic replicon containing the M/SH intergenic junction used in the transcription assay and the potential products of transcription. le, leader; ig, intergenic junction; tr, trailer; nt, nucleotide. (C) Products of RNA synthesis from the M/SH subgenomic replicon in the presence of wild-type (wt) or mutated M2-1 proteins. Cells infected with recombinant MVA vaccinia virus expressing T7 RNA polymerase were transfected with pM/SH, pN, pP, pL, and plasmids bearing genes expressing wild-type protein or mutant M2-1 proteins as indicated. Cells were exposed to [3H]uridine in the presence of actinomycin D and cytosine arabinoside. Total RNA was phenol extracted, ethanol precipitated, and analyzed by agarose-urea gel electrophoresis followed by fluorography. rep, replication products.
The effects of mutations in the Cys3-His1 motif of the M2-1 protein on RS virus transcription were assayed by using an RS virus subgenomic replicon supported by the recombinant vaccinia virus-T7 expression system (12, 37). An RS virus subgenomic replicon containing two genes separated by the M/SH gene junction (Fig. 1B) was expressed from cDNA in cells also expressing the N, P, L, and M2-1 proteins from T7 expression plasmids. The construction of this subgenomic replicon has been described previously (12). The effect of the wild-type and mutant M2-1 proteins on RS virus transcription was determined by direct metabolic labeling of RNA. The synthesis of discrete monocistronic mRNA1 and mRNA2 (Fig. 1B) was analyzed in comparison to the synthesis of the dicistronic mRNA, termed readthrough product B [r/t B], generated by the failure of the polymerase to terminate transcription at the end of mRNA1. The wild-type M2-1 protein decreased transcriptional termination and increased readthrough transcription as previously reported (11, 12). The increase in readthrough transcription can be seen by comparing lanes 1 and 2 of Fig. 1C. In the absence of M2-1, primarily the products of replication and mRNA1, mRNA2, and a small amount of readthrough from mRNA2 into the trailer (r/t A) were synthesized. In the presence of M2-1, a significant increase in the products of readthrough transcription (polycistronic mRNAs) occurred, shown most strikingly by the presence of r/t B (a dicistronic mRNA consisting of the sequences of mRNA1 and mRNA2). Detailed identification of the RNAs in Fig. 1C has been presented in previous work (12). The data in Fig. 1C show that the M2 proteins in which residues which are not predicted to coordinate the binding of a zinc ion were changed (K8Q, F9L, F9S, E10G, and E10Q) functioned to cause an increase in readthrough transcription (Fig. 1C, lanes 2 and 4 to 8). However, mutations of the residues of M2-1 which are predicted to coordinate the binding of a zinc ion inhibited the ability of the protein from increasing readthrough transcription (Fig. 1C, lanes 3, 9, and 10). Quantitation of readthrough transcripts and mRNA1 revealed that not all mutations in noncoordinating residues increased readthrough to the same extent. Mutations of the phenylalanine at residue 9 caused less readthrough transcription than that caused by wild-type M2-1. Thus, the integrity of the Cys3-His1 motif was important for maintaining the function of the M2-1 protein as an antiterminator during RS virus transcription.
Effects of mutations in the Cys3-His1 motif on M2-1 protein mobility in SDS-PAGE.
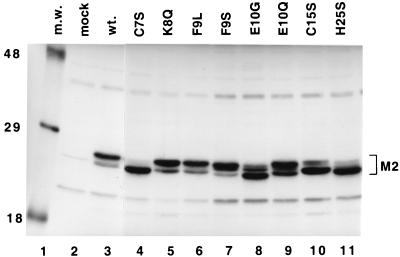
Expression of each of the mutant M2-1 proteins was analyzed by immunoprecipitation followed by reducing SDS-PAGE to test whether each of the mutant proteins was expressed and stable. We also examined the effects of the mutations on electrophoretic mobility, as previous reports showed that the M2 protein migrated as a doublet (26). The wild-type and mutant M2-1 proteins were expressed in HEp-2 cells from cDNAs using the recombinant vaccinia virus-T7 expression system. Proteins were labeled with [35S]methionine and [35S]cysteine and immunoprecipitated from cell lysates using an M2-specific MAb, 5H5. Immunoprecipitated proteins were analyzed by SDS-PAGE under reducing conditions. In Fig. 2, it can be seen that specific mutations affected the mobility of the M2 protein. The wild-type protein migrated as a doublet, with the majority of the protein being present as the slower migrating form (Fig. 2, lane 3). This was also true of K8Q, F9L, F9S, and E10Q (Fig. 2, lanes 5 to 7 and 9). The mutation E10G caused a shift in the distribution of protein from the slower to the faster migrating form and a slight increase in mobility (Fig. 2, lane 8). Mutations of the residues predicted to coordinate the binding of zinc (C7S, C15S, and H25S) caused a significant shift from the slower to the faster migrating form of the protein (Fig. 2, lanes 4, 10, and 11). Thus, mutations in this region had a significant effect on the electrophoretic mobility of the M2-1 protein, specifically altering the distribution of protein between a slower and a faster migrating form.
FIG. 2.
Analysis of expression of M2-1 proteins with mutations in the Cys3-His1 motif. HEp-2 cells were infected with vTF7-3 and then transfected with plasmids bearing genes encoding the wild-type protein or mutant M2-1 proteins. Cells were labeled with [35S]methionine and [35S]cysteine for 2 h at 16 h posttransfection. Cytoplasmic extracts were prepared, and proteins were immunoprecipitated with the M2-specific MAb 5H5. Labeled proteins from mock-transfected cells (lane 2) and cells transfected with plasmids bearing genes encoding the M2-1 proteins (lanes 3 to 11) were analyzed by SDS-PAGE in 11% polyacrylamide gels, followed by fluorography. Lane 1, [14C]-labeled molecular weight markers (m.w.) (molecular sizes in thousands are shown on the left of the gel). The position of the M2-1 protein(s) is indicated. wt., wild type.
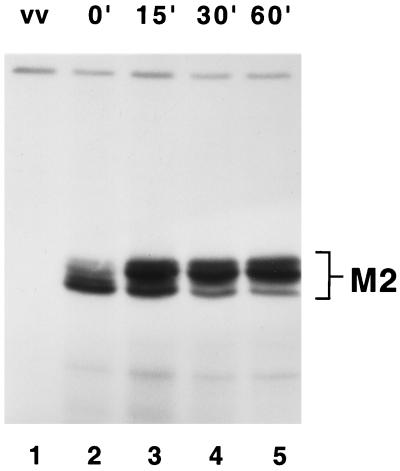
The nature of the two forms of the M2 protein, seen as a doublet in reducing SDS-PAGE, was investigated by metabolic pulse-labeling with [35S]methionine and [35S]cysteine, followed by a chase with excess unlabeled amino acids. The faster-migrating form of M2-1 was labeled initially (Fig. 3, lane 2) and then over time chased into the slower migrating form (Fig. 3, lanes 3 to 5). These results suggest that the slower migrating form was a posttranslationally modified form of the M2-1 protein (Fig. 3).
FIG. 3.
Pulse-chase analysis of wild-type M2-1 protein expression. HEp-2 cells infected with vTF7-3 were transfected with a plasmid bearing the gene expressing the wild-type M2-1 protein. Cells were exposed to [35S]methionine and [35S]cysteine for 15 min at 16 h posttransfection. Following the 15-min labeling period, either cells were harvested (lanes 1 and 2) or the medium was removed and medium containing unlabeled methionine and cysteine was added for 15 (lane 3), 30 (lane 4), or 60 (lane 5) min prior to harvest. Labeled proteins were immunoprecipitated from cytoplasmic extracts using M2-1-specific MAb 1C13. Immunoprecipitated proteins were analyzed by SDS-PAGE under reducing conditions in 11% polyacrylamide gels followed by fluorography. vv, mock transfection.
Phosphorylation of M2-1.
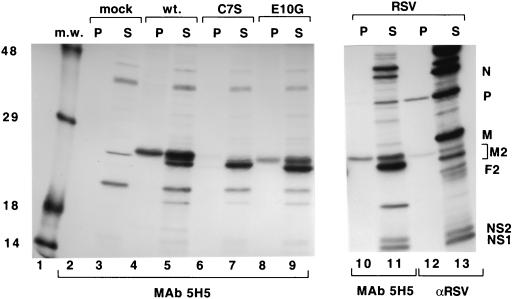
The M2-1 protein has previously been reported to be phosphorylated in RS virus-infected cells (17). Results of the above-described pulse-chase analysis (Fig. 3) suggested that the cause of the difference in the mobilities of the two forms of M2-1 was due to a posttranslational modification. Therefore, we investigated whether the mobility difference of the M2-1 protein in SDS-PAGE was related to its phosphorylation state. Wild-type M2-1 and C7S and E10G mutant M2-1 proteins were expressed in vTF7-3-infected cells and labeled with [35S]methionine and [35S]cysteine or with [33P]inorganic phosphate. Proteins synthesized in RS virus-infected cells were labeled under the same conditions. Labeled proteins were immunoprecipitated with MAb 5H5 or polyclonal antiserum to RS virus and analyzed by SDS-PAGE under reducing conditions.
Analysis of the 35S-labeled M2-1 protein produced in RS virus-infected cells showed it was present as a doublet, with the majority of the protein (approximately 75%) found in the faster migrating form (Fig. 4, lanes 11 and 13). When labeled with [33P]inorganic phosphate, only a single labeled band which corresponded to the slower migrating form of M2-1 was observed (Fig. 4, lanes 10 and 12). Expression of the wild-type M2-1 protein alone from a plasmid in cells also showed that it was the slower form of M2-1 that was phosphorylated and that the faster form was not (Fig. 4, lanes 4 and 5). This demonstrated that the M2-1 protein could be phosphorylated in the absence of other RS virus proteins. In contrast, the majority of the mutant C7S M2-1 protein migrated as the faster form, which was not detectably phosphorylated (Fig. 4, lanes 6 and 7). Analysis of the E10G M2-1 mutant protein showed a shift in the distribution toward the faster migrating form which corresponded with an approximately 50% decrease in phosphorylation compared to that of wild-type M2-1 protein (Fig. 4, lanes 8 and 9).
FIG. 4.
Analysis of the effects of mutations in the Cys3-His1 motif of M2-1 on phosphorylation. HEp-2 cells infected with vTF7-3 were mock transfected (mock lanes 2 and 3) or transfected with a plasmid bearing a gene encoding the wild-type or a mutant M2-1 protein as indicated (lanes 4 to 9), or HEp-2 cells were infected with RS virus (RSV lanes 10 to 13). Cells were exposed to [35S]methionine and [35S]cysteine (S lanes 3, 5, 7, 9, 11, and 13) or [33P]inorganic phosphate (P lanes 2, 4, 6, 8, 10, and 12) for 2 h at 16 h posttransfection or 20 h postinfection. Labeled proteins were immunoprecipitated using M2-1-specific MAb 5H5 (lanes 2 to 11) or anti-RS virus polyclonal serum (αRSV lanes 12 and 13) and analyzed by SDS-PAGE in 11% polyacrylamide gels followed by fluorography. Positions of RS virus proteins are shown (lane 13). The exposure time of lanes 10 to 13 was two times that of lanes 1 to 9. m.w., molecular weight markers (sizes in thousands are noted at the left); wt., wild type.
These results showed that the faster and slower migrating forms of M2-1 protein observed by SDS-PAGE under reducing conditions were differentiated by their phosphorylation state; the slower migrating form was phosphorylated, and the faster migrating form was not. During 24 h of an RS virus infection, the relative ratios of these two forms remained constant (data not shown). In addition, the mutations of the predicted zinc-coordinating residues in the Cys3-His1 motif prevented efficient phosphorylation of the M2-1 protein. Treatment of the wild-type M2-1 protein with calf alkaline phosphatase confirmed these results, as digestion with calf alkaline phosphatase resulted in a shift of the slower migrating form to the faster form (T. Cartee and G. W. Wertz, unpublished data).
Effects of mutations in the Cys3-His1 motif of M2-1 on interaction with N protein.
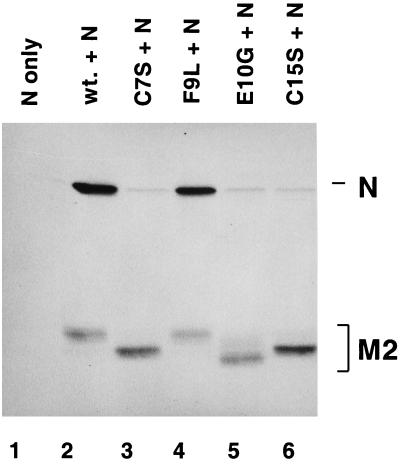
The M2-1 and N proteins have been demonstrated to interact in RS virus-infected cells and when the two proteins are coexpressed (9). The effects of mutations in the Cys3-His1 motif of the M2-1 protein on its interaction with the N protein were assayed by coexpression of wild-type and mutant M2-1 proteins with the N protein in HEp-2 cells. Proteins labeled with [35S]methionine and [35S]cysteine were immunoprecipitated from cell lysates by using the M2-1-specific MAb 5H5 and analyzed by SDS-PAGE under reducing conditions. The N protein was coprecipitated with the wild-type and F9L M2-1 proteins (Fig. 5, lanes 2 and 4). Mutation of the cysteine residues at positions 7 and 15 (Fig. 5, lanes 3 and 6) and the histidine at position 25 (data not shown) of the M2-1 protein severely decreased the efficiency with which N protein was coprecipitated, demonstrating the importance of the predicted zinc-coordinating residues for maintenance of the M2-1 interaction with N. Surprisingly, the E10G mutation, which did not prevent M2-1 protein-mediated antitermination during RS virus transcription, prevented the M2-1 protein from interacting with the N protein. The reason for this is not clear but may be related to the presence of a glycine residue rather than the loss of the glutamate, as the E10Q mutation in the M2-1 protein did interact with the N protein (data not shown).
FIG. 5.
Analysis of the effects of mutations in the Cys3-His1 motif of the M2-1 protein on interaction with the N protein. HEp-2 cells infected with vTF7-3 were transfected with pN alone (lane 1) or pN and a plasmid bearing a gene encoding the wild-type or a mutated M2-1 protein as indicated (lanes 2 to 6). Cells were exposed to [35S]methionine and [35S]cysteine for 2 h at 16 h posttransfection. Labeled proteins were immunoprecipitated from cytoplasmic extracts by using M2-1-specific MAb 5H5. Immunoprecipitated proteins were analyzed by SDS-PAGE in 11% polyacrylamide gels followed by fluorography. Positions of the M2-1 and N proteins are indicated. wt., wild type.
These results demonstrate the requirement for maintaining the Cys3-His1 motif in order for M2-1 protein to interact with N protein. In addition, the phenotype of the E10G mutation suggested that the interaction is specific in that it can be disrupted by a single mutation in a noncoordinating residue of the predicted zinc binding domain. This mutation separates the function of M2-1 protein in transcription from its ability to interact with N protein, implying that this interaction is not required for M2 to function during transcription. However, the conditions under which the immunoprecipitations were performed were stringent and may disrupt weak interactions which may be functionally relevant in cells. Additionally, it should be noted that these results do not rule out the possibility that this interaction is mediated via another molecule with which both N and M2-1 proteins interact.
DISCUSSION
Sequence analysis of the RS virus M2-1 protein revealed a Cys3-His1 motif, which had been shown to bind zinc in another protein (5, 35). Mutational analysis of the Cys3-His1 motif demonstrated that maintaining the cysteine and histidine residues predicted to coordinate zinc was essential for the functional integrity of the M2-1 protein. Alteration of the predicted zinc-coordinating residues resulted in an M2-1 protein which was unable to enhance transcriptional readthrough at RS virus gene ends. In addition, mutations of the predicted zinc-coordinating residues resulted in an alteration in the migration pattern of the M2-1 protein in SDS-PAGE from a relatively slow to a relatively fast migrating form. The two forms of M2-1 protein differed in phosphorylation, the slower form being phosphorylated and the faster form not being phosphorylated.
Mutation of the predicted zinc-coordinating residues also inhibited the interaction of the M2-1 protein with the N protein. Mutating any one of the residues of the M2-1 protein predicted to coordinate zinc resulted in the same phenotype, whereas mutating other noncoordinating residues in the Cys3-His1 motif had little if any effect on the antitermination function of the M2-1 protein. Thus, maintaining the potential zinc-coordinating residues of the Cys3-His1 motif was essential for M2-1 function. These results are consistent with the idea that the Cys3-His1 motif coordinates the binding of an ion of zinc. Studies are under way to directly demonstrate that M2-1 binds zinc by various techniques.
The data presented here show that the two forms of M2-1 protein separated by SDS-PAGE under reducing conditions are discriminated by whether they are phosphorylated. The slower migrating form of M2-1 was phosphorylated, whereas the faster form was not. Multiple forms of the M2-1 protein can be seen in Fig. 2 and 3 in two major bands and at least two other minor species. These minor species may correspond to those observed by Routledge et al. in infected cells (26). Additionally, the extent to which the M2-1 protein is phosphorylated has not yet been determined and the less prevalent species may represent differentially phosphorylated forms of M2-1.
The integrity of the Cys3-His1 motif was important for phosphorylation. A protein-folding study has shown that zinc binding and protein folding are tightly coupled with zinc binding conferring structural stability (6). One could hypothesize that M2-1, which has zinc bound, is folded and can be recognized by the appropriate cellular kinase but that mutants which prevent the binding of zinc are not properly folded and are not recognized by the kinase. Such a model would mean that zinc binding is essential for phosphorylation, which, in turn, may play a role in M2-1 function. In such a scenario, zinc binding may play a crucial role in the regulation of RS virus transcription.
The role of the M2-1–N protein-protein interaction in the RS virus replication cycle is currently unknown. The E10G M2-1 protein is active in transcription, but our results indicate that it does not interact with the N protein, suggesting that this interaction is not required for M2-1 to enhance transcriptional readthrough. This interaction may be important at another point during virus replication. The M2-1 protein was initially characterized as a matrix-like protein, as it dissociated from nucleocapsids under conditions similar to those under which the matrix protein, M, dissociated (13). If M2-1 can function as a matrix-like protein, in addition to its activity during transcription, it is possible that the interaction between M2-1 and N may be involved in virus assembly.
Zinc-binding motifs have been found in a number of proteins and, in many cases, mediate protein-protein or protein-nucleic acid interactions (1, 20, 21, 22, 29, 30, 31). Often the binding of zinc plays a purely structural role by holding the protein in a conformation which is functional (23). We hypothesize that the Cys3-His1 motif of M2-1 binds zinc in order for the protein to be in a conformation which allows it to function in RS virus transcription, to interact with the N protein, and to be efficiently phosphorylated. In conclusion, we have demonstrated that the predicted zinc-binding motif of M2-1 is essential for maintaining the functional integrity of the protein and that there are at least two forms of M2-1 produced in infected cells which can be distinguished by their phosphorylation state.
ACKNOWLEDGMENTS
We thank Tara Cartee for making available data prior to publication and Shawn Harmon for aid with data analysis. We also thank the members of our laboratory and the laboratories of L. A. Ball, W. Sullender, and A. C. R. Samson for advice and constructive criticism.
This work was supported by Public Health Service grants AI12464 and AI20181 from the NIH and NIAID to G.W.W.
REFERENCES
- 1.Bowles N E. Effect of rearrangements and duplications of the Cys-His motifs of Rous sarcoma virus nucleocapsid protein. J Virol. 1993;67:623–631. doi: 10.1128/jvi.67.2.623-631.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chamberlain J P. Fluorographic detection of radioactivity in polyacrylamide gels with water soluble fluor, sodium salicylate. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 3.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a non-segmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins P L, Huang Y T, Wertz G W. Identification of a tenth mRNA of respiratory syncytial virus and assignment of polypeptides to the 10 viral genes. J Virol. 1984;49:572–578. doi: 10.1128/jvi.49.2.572-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins P L, Wertz G W. The envelope-associated 22K protein of human respiratory syncytial virus: nucleotide sequence of the mRNA and a related polytranscript. J Virol. 1985;54:65–71. doi: 10.1128/jvi.54.1.65-71.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eis P S, Lakowicz J R. Time-resolved energy transfer measurements of donor-acceptor distance distributions and intramolecular flexibility of a CCHH zinc-finger peptide. Biochemistry. 1993;32:7981–7993. doi: 10.1021/bi00082a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerson S U, Wagner R R. Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J Virol. 1972;10:297–309. doi: 10.1128/jvi.10.2.297-309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fearns R, Collins P L. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J Virol. 1999;73:5852–5864. doi: 10.1128/jvi.73.7.5852-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia J, Garcia-Barreno B, Vivo A, Melero J. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that co-express the nucleoprotein, the phosphoprotein, and the 22K protein. Virology. 1993;195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- 10.Grosfeld H, Hill M, Collins P L. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy R W, Harmon S B, Wertz G W. The diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J Virol. 1999;73:170–176. doi: 10.1128/jvi.73.1.170-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy R W, Wertz G W. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J Virol. 1998;72:520–526. doi: 10.1128/jvi.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y T, Collins P L, Wertz G W. Characterization of the 10 proteins of human respiratory syncytial virus: identification of a fourth envelope associated protein. Virus Res. 1985;2:157–173. doi: 10.1016/0168-1702(85)90246-1. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y T, Wertz G W. The genome of respiratory syncytial virus is a negative-stranded RNA that codes for at least seven mRNA species. J Virol. 1982;43:150–157. doi: 10.1128/jvi.43.1.150-157.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keilin D, Mann T. Carbonic anhydrase. Purification and nature of the enzyme. Biochem J. 1940;34:1163–1176. doi: 10.1042/bj0341163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lambert D M, Hambor J, Diebold M, Galinski B. Kinetics of synthesis and phosphorylation of respiratory syncytial virus polypeptides. J Gen Virol. 1988;69:313–323. doi: 10.1099/0022-1317-69-2-313. [DOI] [PubMed] [Google Scholar]
- 18.Laskey R. The use of intensifying screens or organic scintillators for visualizing radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65:363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- 19.Ling R, Easton A J, Pringle C R. Sequence analysis of the 22K, SH and G genes of the turkey rhinotracheitis virus and their intergenic regions reveals a gene order different from that of other pneumoviruses. J Gen Virol. 1992;73:1709–1715. doi: 10.1099/0022-1317-73-7-1709. [DOI] [PubMed] [Google Scholar]
- 20.Mabrouk T, Lemay G. Mutations in a CCHC zinc binding motif of the reovirus ς3 protein decrease its intracellular stability. J Virol. 1994;68:5287–5290. doi: 10.1128/jvi.68.8.5287-5290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Méric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meric C, Gouilloud E, Sphar P-F. Mutations in Rous sarcoma virus nucleocapsid protein p12(NC): deletions of Cys-His boxes. J Virol. 1988;62:3328–3333. doi: 10.1128/jvi.62.9.3328-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J, McLachlan A D, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mink M A, Stec D S, Collins P L. Nucleotide sequences of the 3′ leader and 5′ trailer regions of human respiratory syncytial virus genomic RNA. Virology. 1991;185:615–624. doi: 10.1016/0042-6822(91)90532-g. [DOI] [PubMed] [Google Scholar]
- 25.Pattnaik A K, Wertz G W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990;64:2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Routledge E G, Willcocks M M, Morgan L, Samson A C R, Scott R, Toms G L. Heterogeneity of the respiratory syncytial virus 22K protein revealed by Western blotting with monoclonal antibodies. J Gen Virol. 1987;68:1209–1215. doi: 10.1099/0022-1317-68-4-1209. [DOI] [PubMed] [Google Scholar]
- 27.Samal S K, Pastey M K, McPhillips T H, Mohanty S B. Bovine respiratory syncytial virus nucleocapsid protein expressed in insect cells specifically interacts with the phosphoprotein and the M2 protein. Virology. 1993;193:470–473. doi: 10.1006/viro.1993.1148. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez A, Kiley M P, Holloway B P, Auperin D D. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 29.Sands M S, Bogenhagen D F. Two zinc-finger proteins from Xenopus laevis bind the same region of 5S RNA but with different nuclease protection patterns. Nucleic Acids Res. 1991;19:1797–1803. doi: 10.1093/nar/19.8.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shepard D A, Ehnstrom J G, Skinner P J, Schiff L A. Mutations in the zinc-binding motif of the reovirus capsid protein ς3 eliminates its ability to associate with capsid protein μ1. J Virol. 1996;70:2065–2068. doi: 10.1128/jvi.70.3.2065-2068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somers W, Ultsch M, De Vos A, Kossiakoff A A. The X-ray structure of a growth hormone-prolactin complex. Nature. 1994;372:478–481. doi: 10.1038/372478a0. [DOI] [PubMed] [Google Scholar]
- 32.Stec D S, Hill M G, Collins P L. Sequence analysis of the polymerase L gene of human respiratory syncytial virus and predicted phylogeny of nonsegmented negative strand viruses. Virology. 1991;183:273–287. doi: 10.1016/0042-6822(91)90140-7. [DOI] [PubMed] [Google Scholar]
- 33.Wertz G W, Davis N L. Characterization and mapping of RNaseIII cleavage sites in vesicular stomatitis virus genome RNA. Nucleic Acids Res. 1981;9:6487–6503. doi: 10.1093/nar/9.23.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wertz G W, Kreiger M, Ball L A. Structure and cell surface maturation of the attachment glycoprotein of human respiratory syncytial virus in a cell line deficient in O glycosylation. J Virol. 1989;63:4767–4776. doi: 10.1128/jvi.63.11.4767-4776.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worthington M T, Amann B T, Nathans D, Berg J M. Metal binding properties and secondary structure of the zinc binding domain of Nup475. Proc Natl Acad Sci USA. 1996;93:13754–13759. doi: 10.1073/pnas.93.24.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Q, Davis P J, Brown T D K, Cavanagh D. Sequence and in vitro expression of the M2 gene of turkey rhinotracheitis pneumovirus. J Gen Virol. 1992;73:1355–1363. doi: 10.1099/0022-1317-73-6-1355. [DOI] [PubMed] [Google Scholar]
- 37.Yu Q, Hardy R W, Wertz G W. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define the minimal trans-acting requirements for RNA replication. J Virol. 1995;69:2412–2419. doi: 10.1128/jvi.69.4.2412-2419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamora M, Samal S K. Sequence analysis of M2 mRNA of bovine respiratory syncytial virus obtained from an F-M2 dicistronic mRNA suggests structural homology with that of human respiratory syncytial virus. J Gen Virol. 1992;73:737–741. doi: 10.1099/0022-1317-73-3-737. [DOI] [PubMed] [Google Scholar]