Abstract
EBNA-3A, -3B, and -3C are three latent infection nuclear proteins important for Epstein-Barr virus (EBV)-induced B-cell immortalization and the immune response to EBV infection. All three are hypothesized to function as transcriptional transactivators, but little is known about their precise mechanism of action or their role in EBV pathogenesis. We have cloned and studied the three EBNA-3 homologues from a closely related lymphocryptovirus (LCV) which naturally infects rhesus monkeys. The rhesus LCV EBNA-3A, -3B, and -3C homologues have 37, 40, and 36% amino acid identity with the EBV genes, respectively. Function, as measured by in vitro assays, also appears to be conserved with the EBV genes, since the rhesus LCV EBNA-3s can interact with the transcription factor RBP-Jκ and the rhesus LCV EBNA-3C encodes a Q/P-rich domain with transcriptional activation properties. In order to better understand the relationship between these EBV and rhesus LCV latent infection genes, we asked if the rhesus LCV EBNA-3 locus could be recombined into the EBV genome and if it could substitute for the EBV EBNA-3s when assayed for human B-cell immortalization. Recombination between the EBV genome and rhesus LCV DNA was reasonably efficient. However, these studies suggest that the rhesus LCV EBNA-3 locus was not completely interchangeable with the EBV EBNA-3 locus for B-cell immortalization and that at least one determinant of the species restriction for LCV-induced B-cell immortalization maps to the EBNA-3 locus. The overall conservation of EBNA-3 structure and function between EBV and rhesus LCV indicates that rhesus LCV infection of rhesus monkeys can provide an important animal model for studying the role of the EBNA-3 genes in LCV pathogenesis.
Epstein-Barr virus (EBV) is a gammaherpesvirus in the lymphocryptovirus (LCV) subgroup, which infects and persists in nearly all humans. EBV infection is also associated with several malignant diseases, including Burkitt's lymphoma, posttransplant lymphoproliferative disease, and nasopharyngeal carcinoma (16). EBV's malignant potential can be demonstrated in vitro by immortalization of B-cell growth in tissue culture. The importance of the immune response for control of acute and persistent EBV infection is highlighted by the development of B-cell lymphomas in immunosuppressed AIDS and transplant patients and their response to adoptive T-cell immunotherapy (23, 33). Three related nuclear proteins expressed during latent EBV infection, EBNA-3A, -3B, and -3C, are important for EBV-induced B-cell immortalization in vitro (38, 39) and are immunodominant targets for the immune response to EBV infection in vivo (15, 36). In order to better understand the relationship between these EBV and rhesus LCV latent-infection genes and to develop tools necessary for the study of the EBNA-3 genes in vivo, we have examined and compared the EBNA-3 locus encoded by the LCV which naturally infects rhesus monkeys.
Old World primates are naturally infected with LCV, which have significant genetic and biologic similarity to EBV (10). The rhesus and baboon LCV have been the most thoroughly studied at a molecular level, and these simian viruses appear to have the same repertoire of lytic and latent infection genes as EBV. The lytic infection genes generally show a high degree of nucleotide and amino acid homology (∼60 to 90% amino acid identity [21, 42; P. Rao, H. Jiang, and F. Wang, submitted for publication]). Lytic gene function is also well conserved functionally, as the rhesus LCV BZLF1 homologue can efficiently induce viral replication in EBV-infected human B cells (21, 22). In contrast, the latent infection genes identified to date have shown much greater sequence divergence than the lytic infection genes, from 50% amino acid identity in the baboon and rhesus LCV EBNA-1 homologues to less than 30% amino acid identity for the baboon and rhesus LCV LMP1 homologues (2, 5, 8, 9, 19, 24, 29, 42). In most instances, simian LCV latent infection gene function has also been functionally conserved when measured using in vitro assays designed for the EBV latent infection genes. The only difference discovered to date is the failure of the rhesus and baboon LCV EBNA-1 glycine-alanine repeats to protect cytotoxic T-cell epitopes from antigen presentation as described for the EBV EBNA-1 Gly-Ala repeats (2). In this case, differences between EBV and simian LCV suggest that protection from antigen presentation by EBNA-1 may not be an essential mechanism for persistence by all LCV.
The strong functional conservation between simian and EBV latent infection genes has provided important insights. For example, the baboon and rhesus LCV LMP1 homologues can also induce NF-κB and bind to tumor necrosis factor receptor-associated factors in human cells. The marked sequence divergence among the LMP1s provided for rapid identification of the conserved PXQXT/S tumor necrosis factor receptor-associated factor binding motif in the proximal transformation effector site (TES-1) and highlighted the remarkably conserved terminal 13 amino acids in the distal transformation effector site (TES-2) that interacts with the tumor necrosis factor receptor 1-associated death domain protein (9).
Despite this apparently strong conservation of LCV latent infection gene function, the rhesus and baboon LCV are incapable of efficiently immortalizing human B lymphocytes. Previous studies indicate that this species restriction for B-cell immortalization occurs beyond the step of virus binding and penetration (21). In these studies, simian LCV can infect, persist, and replicate in human B cells but are incapable of immortalizing human B cells without EBV coinfection. We hypothesize that one or more of the latent infection genes may interact with cell proteins in a species-specific manner. This species restriction may identify mechanisms important for cell growth transformation and may have potential clinical relevance, since xenotransplantation of baboon bone marrow carries a high risk of introducing baboon LCV into humans.
Genetic experiments show that the EBNA-3A and -3C genes are essential for EBV immortalization, whereas EBNA-3B is dispensable in vitro (38, 39). These three latent infection nuclear proteins have a common exon-intron structure, are encoded in tandem fashion in the middle of the EBV genome, and share distant homology (12, 13, 25). All three proteins are hypothesized to function as transcriptional transactivators (20, 27, 30). EBNA-3C can regulate cell CD21 expression and viral LMP1 expression, and all three EBNA-3s can act together to induce pleckstrin expression (1, 17, 40). All three latent nuclear proteins interact with the transcription factor RBP-Jκ, and EBNA-3C has been reported to have an SP1-like transcriptional transactivation domain (20, 30). Since EBNA-3B is not necessary for EBV-induced B-cell immortalization in vitro, one hypothesis is that EBNA-3B is important for successful EBV infection in vivo, perhaps by regulating expression of cell genes important for acute or persistent EBV infection. However, the precise mechanism of these viral latent genes and how they contribute to B-cell growth transformation or successful EBV infection in vivo remain to be elucidated. In order to better understand these important latent infection nuclear proteins and the potential utility of the rhesus animal model for studying the role of these genes in vivo, we have cloned the rhesus LCV EBNA-3A, -3B, and -3C homologues and compared them to the EBV genes. We have also applied a genetic approach to test whether the EBNA-3 locus contributes to the species restriction for LCV-induced B-cell growth transformation.
MATERIALS AND METHODS
Cells and cell lines.
LCL8664 is a rhesus LCV-infected (cercopithicine herpesvirus 15) cell line derived from a B-cell tumor in a rhesus monkey (28). 278LCL is a cell line derived by in vitro infection of rhesus monkey peripheral blood mononuclear cells (PBMC) with rhesus LCV. P3HR-1 is a human B-cell line containing a type 2 EBV with a spontaneous deletion of EBNA-2 and part of the EBNA-LP gene. Louckes is an EBV-negative Burkitt's lymphoma cell line. Human PBMC were isolated on Ficoll-Hypaque gradients from blood donated by healthy individuals. All cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum.
Cosmid library and plasmids.
Cosmid clones of LCL8664 genomic DNA were constructed and screened as previously described (29). A cosmid clone, RcosL12, encoding the rhesus LCV EBNA-3 locus was identified by cohybridization with rhesus LCV probes for the gp350 and EBNA-1 homologues (2, 21), since the EBV EBNA-3 genes are encoded between the gp350 and EBNA-1 genes and the simian LCV genomes are colinear with EBV. RcosL12 BamHI fragments were subcloned into pCR2.1ΔRI, a plasmid derived from pCR2.1 (Invitrogen), by EcoRI digestion and self-religation. The nucleotide sequences for the rhesus LCV EBNA-3A, -3B, and -3C homologues (GenBank accession no. AF159308, AF159309, and AF159310, respectively) were assembled, and sequence analyses were performed using DNAstar software. pSVNaeI-Z is an expression vector for EBV BZLF1 used to induce EBV and rhesus LCV replication, and the EcoRI-A cosmid is an EBV DNA clone used to complement the P3HR-1 deletion as previously described (7).
pGal4-RhE3C and pGal4-RhE3CR contain a SmaI DNA fragment containing rhesus LCV EBNA-3C amino acid residues 745 to 922 inserted either in frame or in opposite orientation into the SmaI site of pGal4(1–147) (34). pM3-VP16 is a plasmid with the VP16 transcriptional activation domain cloned into pGal4(1–147). pFR-Luc (Stratagene) is a luciferase reporter plasmid with five GAL4 DNA-binding sites.
RBP-Jκ binding assay with GST fusion proteins.
The portions of the rhesus LCV genome encoding EBNA-3A amino acids 143 to 588 (NcoI fragment), amino acids 129 to 303 (BamHI-StuI fragment), and amino acids 304 to 691 (StuI-EcoRI fragment); EBNA-3B amino acids 156 to 390 (BamHI-BsrBI fragment); and EBNA-3C amino acids 132 to 326 (HindIII-EcoRI fragment) were cloned in frame into pGEX-2TK or pGEX-3X (Pharmacia Biotech). The portion of the rhesus LCV genome encoding EBNA-3A amino acids 506 to 601 were PCR amplified using primers 3A2720 (5′-CGG GAT CCC CCA CTG GGC ATT TTG TTA G-3′) and 3A2824 (5′-GGA ATT CCT AGG CAA TGG AGC AGG TCT T-3′) and cloned in frame into pGEX-2TK. GST-EBNA-2(243–336) and RBP-Jκ expression plasmids have been described previously (11). RBP-Jκ was in vitro translated (TNT in vitro translation system; Promega) with [35S]methionine. Binding assays for glutathione-S-transferase (GST) fusion proteins with in vitro-translated RBP-Jκ were performed as previously described at 4°C for 1 h in 300 μl of binding buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 10% glycerol, 0.5% NP-40, 0.5 mM dithiothreitol, 1 μg of aprotinin per ml, 0.5 μg of leupeptin per ml, 0.7 μg of pepstatin per ml, 1 mM phenylmethylsulfonyl fluoride) (9). Equal amounts of GST fusion proteins were used for the binding assays, and this was confirmed by Coomassie stain after polyacrylamide gel electrophoresis (PAGE).
Luciferase reporter assay.
A total of 107 Louckes cells were transfected by electroporation (Bio-Rad Gene Pulser) in 0.4 ml of RPMI with 10 μg of expression constructs and 10 μg of reporter plasmid. As an internal control, 2 μg of pCMV-βGal was cotransfected with each test sample. Cells were harvested 48 h later, and luciferase activity was assayed by using a luciferase assay system (Promega). β-Galactosidase activity was assayed using the Galacto-Light kit (Tropix). Luciferase activity was normalized to β-galactosidase activity, and all assays were repeated with at least three independent transfections.
Generation of EBV recombinants.
A total of 107 P3HR-1 cells were transfected with 10 μg of EcoRI-A cosmid and 25 μg of pSVNaeI-Z, with or without 50 μg of RcosL12 cosmid in 0.4 ml of RPMI by electroporation at 0.2 kV (Bio-Rad Gene Pulser). Viral supernatant was collected 72 h after transfection by filtration through a 0.45-μm filter. Human PBMC (2 × 107) were infected with 1 ml of viral supernatant at 37°C for 2 h. Infected cells were then resuspended in RPMI with 10% fetal calf serum, 0.5 μg of cyclosporin A per ml, and 10 μg of gentamicin per ml at 106 cells/ml in 96-well microtiter plates. Viral passage assays were carried out by transfecting B cells with 25 μg of pSVNaeI-Z to induce lytic replication. Viral supernatants were collected and used to infect human PBMC as described above.
PCR assays.
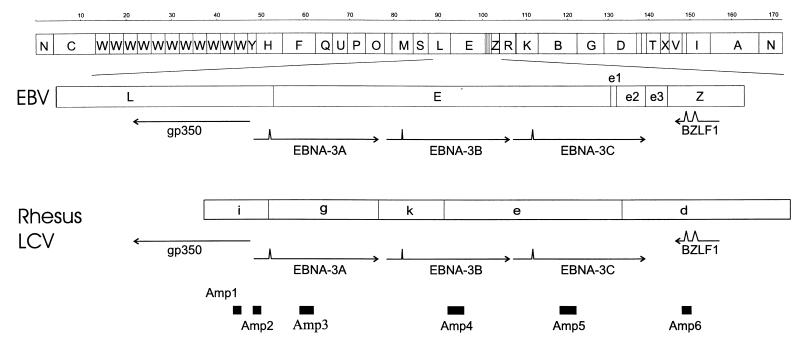
Species-specific PCR primers (designated EBV for EBV specific and Rh for rhesus LCV specific) were made to amplify regions of gp350, EBNA-3A, -3B, and -3C, and BZLF1 (Fig. 1). The amplicon 1 primer set (Amp1) amplifies the gp350 region with Rhamp1F (5′-GGC ATG TCC TGA ATA GTG G-3′) and Rhamp1R (5′-AAA TGA CAA GCG AGG G-3′). The amplicon 2 primer set (Amp2) amplifies a 123-bp segment containing the rhesus LCV EBNA-3A initiation codon with Rhamp2F (5′-AGC CGC TTC CAT TGT TTC AGT GC-3′) and Rhamp2R (5′-GCC CGG CCT TTC TTC CTC CTA-3′). The amplicon 3 primer sets (Amp3) amplify EBNA-3A regions with EBVamp3F (5′-GAA ACC AAG ACC AGA GGT CC-3′) and EBVamp3R (5′-CCC AGG GCC GGA CAA TAG G-3′) and Rhamp3F (5′-CCC ACC GAG GCT CCG TTG TCT-3′) and Rhamp3R (5′-GGC CTT CGT GTC TCC CAT TCG TTA-3′). The amplicon 4 primer sets (Amp4) amplify EBNA-3B regions with EBVamp4F (5′-CCC TTG CGG ATG CAG CCA AT-3′) and EBVamp4R (5′-GGC TGA TAT GGA ATG TGC CC-3′) and Rhamp4F (5′-ACC AAC CTC CAG CGC AAG TCA GTG-3′) and Rhamp4R (5′-AAA CGC AGG GAG CAG CTA TTG TGG-3′). The amplicon 5 primer sets (Amp5) amplify EBNA-3C regions with EBVamp5F (5′-AGA AGG GGA GCG TGT GTT GT-3′) and EBVamp5R (5′-GGC TCG TTT TTG ACG TCG GC-3′) and Rhamp5F (5′-GGG CAA GTC GTG ATG GTA GT-3′) and Rhamp5R (5′-AGT TGT ATC CTG GGG CTC CT-3′). The amplicon 6 primer sets (Amp6) amplify BZLF1 regions with EBVamp6F (5′-CCG CTG CCG CCC CTC CAT-3′) and EBVamp6R (5′-CCC GGC ACG ACG CAC ACG-3′) and Rhamp6F (5′-GCG CCG TGG CAG GTG GTT G-3′) and Rhamp6R (5′-GTG CTT TGG CCG GTG CTG TCG-3′). All PCR assays were carried out at an annealing temperature of 65°C for 35 cycles with 1.5 mM MgCl2 using Ampli-Taq Gold (Perkin-Elmer).
FIG. 1.
BamHI restriction map of the EBV genome, the EBV EBNA-3 locus, and the corresponding rhesus LCV EBNA-3 locus. Coding regions are represented by arrows, and the relative positions of the species-specific PCR amplicons are shown as solid boxes.
EBV- and rhesus LCV-specific PCR products were hybridized on Southern blots with the following species-specific internal probes: rhesus LCV EBNA-3A internal probe, 5′-GAA TTC CTG TAA TGA GGC CG-3′, and EBV EBNA-3A internal probe, 5′-GTT GAG GGC TAG TAT GGG CC-3′. Hybridization was carried out at 55°C, and the blots were washed with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate (SDS) at room temperature and then at 55°C.
RESULTS
Cloning and sequencing of rhesus LCV EBNA-3A, EBNA-3B, and EBNA-3C homologues.
Rhesus LCV BamHI DNA fragments, labeled arbitrarily a to k based on decreasing size, were subcloned from a cosmid clone, RCosL12, encoding the rhesus LCV EBNA-3 locus. Rhesus LCV BamHI DNA fragments cross-hybridizing at low stringency with EBV BamHI-E DNA containing most of the EBNA-3A, -3B, and -3C coding regions were identified (RcosL12 g, k, and e). RcosL12 i and d were identified as potential flanking fragments by cross-hybridization with the rhesus LCV gp350 and EBV BZLF1 DNA probes (Fig. 1). RcosL12 i, g, k, e, and d were sequenced, and colinearity was confirmed by PCR amplification across each BamHI site from viral DNA.
Rhesus LCV genes with 57, 61, and 59% nucleotide homology to EBNA-3A, EBNA-3B, and EBNA-3C, respectively, were identified. Each of the rhesus LCV EBNA-3 homologues was composed of a short first exon, a short intron, and a long second exon, similar to each of the EBV genes. Splice sites were confirmed by nucleotide sequencing the rhesus LCV EBNA-3A, -3B, and -3C cDNA clones obtained by reverse transcription-PCR amplification. The rhesus LCV EBNA-3A, -3B, and -3C genes encode 955, 938, and 1,167 amino acids, respectively, with overall amino acid sequence identities of 37, 40, and 36% with the respective EBV genes. The translated amino acid sequences of rhesus LCV EBNA-3A, -3B, and -3C and the alignments with the type 1 EBV homologues are presented in Fig. 2, 3, and 4.
FIG. 2.
Amino acid sequence alignment of rhesus LCV EBNA-3A (RhE3A, bottom line) with that of type 1 EBV EBNA-3A (E3A, top line). Identical (:) and similar (.) amino acids are shown. The RBP-Jκ binding region in EBV EBNA-3A is identified by the arrows. The repeated elements in rhesus LCV EBNA-3A are indicated by alternating single and double underlines.
FIG. 3.
Amino acid sequence alignment of rhesus LCV EBNA-3B (RhE3B, bottom line) with that of type 1 EBV EBNA-3B (E3B, top line). Identical (:) and similar (.) amino acids are shown. The RBP-Jκ binding region in EBV EBNA-3B is identified by the arrows. The repeated elements in rhesus LCV EBNA-3B are indicated by alternating single and double underlines. An additional repeat element is differentiated in italics.
FIG. 4.
Amino acid sequence alignment of rhesus LCV EBNA-3C (RhE3C, bottom line) with that of type 1 EBV EBNA-3C (E3C, top line). Identical (:) and similar (.) amino acids are shown. The RBP-Jκ binding region in EBV EBNA-3C is identified by the arrows. The repeated elements in rhesus LCV EBNA-3C are indicated by alternating single and double underlines. The conserved leucine residues of a leucine zipper domain are boxed. The Q/P-rich transcription transactivating region in EBV EBNA-3C is shown in bold.
In all three rhesus LCV EBNA-3 genes, the N-terminal region is better conserved, with 47, 48, and 49% amino acid identity to the respective EBV genes. A previously identified leucine repeat which forms a basic leucine zipper domain (bZIP) in the amino terminus of EBV EBNA-3C (1) is conserved in the rhesus LCV EBNA-3C (Fig. 4, boxed). RBP-Jκ interaction domains have been grossly defined in the amino terminus of each EBV EBNA-3 protein, but the alignments of the EBV and rhesus LCV EBNA-3 homologues do not provide any obvious evidence of well-conserved RBP-Jκ binding motifs in these proteins.
There is more sequence divergence in the carboxy-terminal two-thirds of each rhesus LCV EBNA-3, but the formation of direct repeat structures is a common theme conserved in each protein. In rhesus LCV EBNA-3A, there are eight direct repeats of an 8-amino-acid motif (Fig. 2, alternating single and double underlines). The sequence of this 8-amino-acid motif is not well conserved with the three direct repeat elements identified in the carboxy terminus of EBV EBNA-3A, which are well conserved between type 1 and type 2 EBV (35). Similarly, the rhesus LCV EBNA-3B encodes a 10-amino-acid motif that is directly repeated 9.5 times (Fig. 3, alternating single and double underlines). The position of this direct repeat element is similar to, but the sequence is divergent from, a 20-amino-acid element repeated three times in EBV EBNA-3B which is not highly conserved between type 1 and type 2 EBV (35). In rhesus LCV EBNA-3C, there is a 16- to 18-amino-acid element which is repeated five times (Fig. 4, alternating single and double underlines). Again, the sequence of the rhesus LCV repeats is different from the 10× 5-amino-acid and 3× 13-amino-acid repeats in EBV EBNA-3C, which are not well conserved between type 1 and type 2 EBV (35). Thus, as in the EBV EBNA-3 proteins, directly repeated elements can be identified in each rhesus LCV EBNA-3 protein, but the amino acid sequence of the direct repeats is not well conserved between an EBV EBNA-3 and its rhesus LCV homologue. The significance of these repeated elements remains to be determined, but an important functional role, e.g., protein conformation or interaction with other cell proteins, is suggested by the conserved evolution of direct repeat structures in the EBNA-3 homologues.
Interactions with RBP-Jκ are conserved in rhesus LCV EBNA-3A, -3B, and -3C.
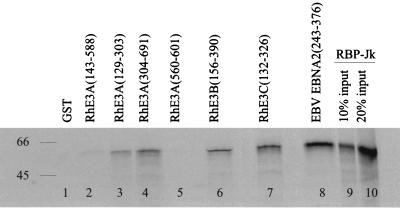
EBV EBNA-3 binding to the transcription factor RBP-Jκ is hypothesized to be one mechanism by which these proteins regulate cell and viral gene transcription during latent EBV infection (26, 31, 43). To test whether interaction with RBP-Jκ is conserved in the rhesus LCV homologues, GST fusion proteins containing various N-terminal regions of rhesus LCV EBNA-3A, -3B, and -3C were made to test their interactions with in vitro-translated human RBP-Jκ (Fig. 5). Amino-terminal fusions of all three rhesus LCV EBNA-3 proteins with GST were able to bind to RBP-Jκ in vitro, suggesting an important functional role for the conserved RBP-Jκ interaction. The interaction was at comparable levels to the EBV homologues (data not shown) and less efficient than EBV EBNA-2. The presence of two nonoverlapping RBP-Jκ binding domains in the rhesus LCV EBNA-3A (RhE3A129–303 and RhE3A304–691) is similar to the finding with EBV EBNA-3A (6).
FIG. 5.
Rhesus LCV EBNA-3A (RhE3A), -3B (RhE3B), and -3C (RhE3C) bind to RBP-Jκ in vitro. GST proteins fused to various portions of the rhesus LCV EBNA-3s (specific amino acid residues shown in parentheses) were incubated with in vitro-translated [35S]RBP-Jκ and analyzed on SDS–15% PAGE gels. Equal amounts of GST fusion proteins were loaded and confirmed by Coomassie staining (data not shown). Representations of the in vitro-translated [35S]RBP-Jκ input are shown in lanes 9 and 10. Sizes are shown at the left (in kilodaltons).
Conserved transcriptional transactivating activity in the rhesus LCV EBNA-3C carboxy terminus.
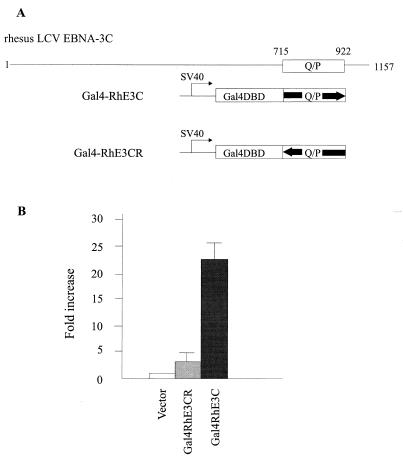
The 16- to 18-amino-acid repeat in the rhesus LCV EBNA-3C carboxy terminus is rich in glutamine and proline residues and reminiscent of an EBV EBNA-3C Q/P-rich domain with transcriptional transactivating properties (20). To test whether a transcriptional transactivating domain was functionally conserved in the rhesus LCV EBNA-3C carboxy terminus, we fused amino acid residues 716 to 923 in the sense and antisense directions with the Gal4 DNA-binding domain under the control of the simian virus 40 early promoter (Fig. 6A). These expression constructs were transiently transfected with a Gal4-dependent reporter construct into Louckes cells. Transfection of the Gal4 construct with the rhesus LCV EBNA-3C Q/P domain in the sense orientation showed an average of greater than 20-fold activation, whereas the Gal4 construct with the rhesus LCV EBNA-3C Q/P domain in the antisense orientation gave an average of only 3-fold activation over the Gal4 construct alone (Fig. 6). Thus, a Q/P-rich transcriptional transactivating domain has also been conserved in the rhesus LCV EBNA-3C.
FIG. 6.
The rhesus LCV EBNA-3C (RhE3C) Q/P-rich region has transcriptional transactivation activity. (A) Schematic diagram of the rhesus LCV EBNA-3C and its Q/P-rich region (box). The Q/P-rich region was fused to the Gal4 DNA-binding domain (Gal4DBD) in the sense orientation (Gal4-RhE3C) and antisense orientation (Gal4-RhE3CR). (B) Gal4-responsive luciferase reporter activity in Louckes cells transfected with the rhesus LCV EBNA-3C Gal4 fusion constructs. Mean values and standard deviations of fold increase versus vector control from three independent experiments are shown.
Construction of chimeric EBV with rhesus LCV EBNA-3 genes.
In order to test whether the functional similarities between the rhesus LCV and EBV EBNA-3 genes were sufficient to support human B-cell immortalization with either EBNA-3 locus, we initiated a series of experiments to generate recombinant EBV in which the EBNA-3 locus had been replaced with the rhesus LCV EBNA-3 locus. To generate the chimeric EBV, we modified the second-site recombination strategy originally used by Tomkinson et al. to introduce mutations into the EBV EBNA-3 genes (37). P3HR-1 cells were cotransfected with the EcoRI-A cosmid to restore the EBNA-LP/EBNA-2 deletion and the RcosL12 cosmid containing the rhesus LCV EBNA-3 genes to potentially replace the EBV EBNA-3 genes by homologous recombination. Tomkinson et al. showed that a second EBV cosmid clone containing the EBNA-3 locus was recombined in approximately 30% of immortalizing viruses in which the EBNA-LP and EBNA-2 genes had been restored by recombination with the EcoRI-A cosmid (37). We hypothesized that a similar second recombination event could occur with the rhesus LCV RcosL12 cosmid, albeit perhaps at a lower frequency due to the sequence divergence.
Tomkinson et al. also transfected a cosmid (type 1 EBNA-3 genes) which was different from the P3HR-1 EBV EBNA-3 genes (type 2 EBV) to generate the homologous recombination (37). The EBV type 1 and type 2 EBNA-3 genes A, B, and C have 90, 88, and 81% nucleotide homology, respectively, and the regions immediately flanking the EBNA-3 locus are also highly conserved (96%) (35). The rhesus LCV gp350 homologue immediately 5′ and the BZLF1 homologue immediately 3′ to the rhesus LCV EBNA-3 locus have 66 and 76% nucleotide homology to the respective late and immediate-early EBV lytic genes. The ends of the rhesus LCV RcosL12 cosmid have also been sequenced (∼200 bp from each end) and align with 77% nucleotide homology to the EBV genome at coordinate 66124 and with 89% homology to coordinate 110586. Thus, there are approximately 26 kb 5′ to the EBNA-3 locus and approximately 9 kb 3′ to the EBNA-3 locus in the rhesus LCV RcosL12 cosmid where homologous recombination with P3HR-1 might occur.
Screening of recombinant viruses for second-site recombination of the rhesus LCV EBNA-3 locus.
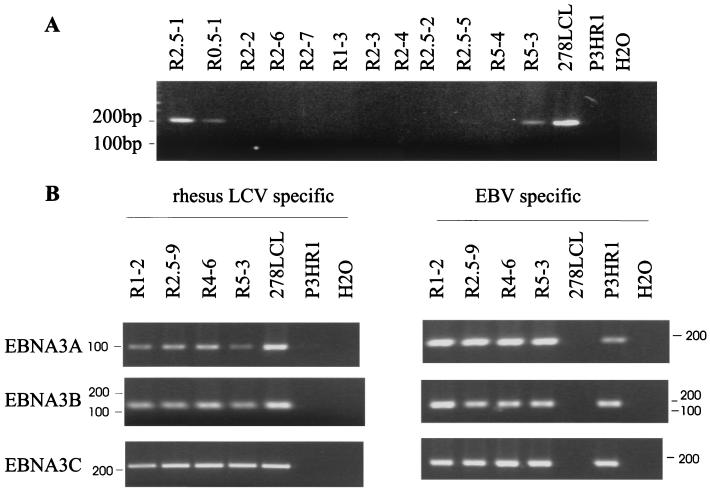
Filtered supernatants were harvested from P3HR-1 cells transfected with EcoRI-A alone or EcoRI-A plus rhesus LCV RcosL12. The viral supernatants were used to infect human peripheral blood B cells, and similar frequencies of immortalized B cells were recovered with viral supernatants from EcoRI-A-transfected P3HR-1 cells with and without RcosL12 cosmid cotransfection (18.6% versus 16%) (Table 1). Forty-five cell clones immortalized with virus from P3HR-1 cells cotransfected with EcoRI-A and RcosL12 were screened by PCR for rhesus LCV DNA in order to identify cells infected with chimeric viruses. Seven of 45 cell clones (15.6%) were positive by PCR with primers specific for rhesus LCV EBNA-3C DNA (Amp5, Fig. 1). Three representative clones positive for rhesus LCV EBNA-3C DNA by PCR are shown in Fig. 7A. Specificity of the PCR primers could be demonstrated by positive amplification with DNA from rhesus LCV-infected cells (278LCL) and negative amplification with cell DNA from P3HR-1 cells (Fig. 7A). These preliminary screening tests do not address whether the recombination has been homologous. However, these results do indicate that rhesus LCV DNA can be recombined into EBV with reasonable efficiency.
TABLE 1.
Frequency of transforming viruses from P3HR-1 transfected with EBV and rhesus LCV cosmids
| Cosmid DNA | No. of wells with immortalized cells | Total no. of wells plated | % of wells with immortalized cells |
|---|---|---|---|
| None | 0 | 96 | 0 |
| EcoRI-A | 46 | 288 | 16.0 |
| EcoRI-A + RcosL12 | 139 | 744 | 18.7 |
FIG. 7.
Species-specific PCR analysis of chimeric P3HR-1 viruses. (A) Representative rhesus LCV EBNA-3C-specific screening assay of 12 immortalized human B-cell clones established by infection with recombinant P3HR-1 viral supernatants after transfection with the EBV EcoRI-A and rhesus LCV RcosL12 cosmids. The specificity of the rhesus LCV-specific primers is demonstrated by using a rhesus LCV-infected B-cell line (278LCL) and EBV-infected B-cell line (P3HR-1). Three clones infected with chimeric rhesus LCV-P3HR-1 viruses are shown (R2.5-1, R0.5-1, and R5-3). H2O indicates control reactions with no DNA template. (B) Rhesus LCV- and EBV-specific PCR analysis for the EBNA-3A, EBNA-3B, and EBNA-3C locus in four clones infected with chimeric viruses (R1-2, R2.5-9, R4-6, and R5-3). PCR with rhesus LCV-specific primers is shown in the left panel, and PCR with EBV-specific primers is shown in the right panel.
Mapping the extent of rhesus LCV DNA recombined in chimeric viruses.
To characterize the amount of rhesus LCV DNA recombined in each clone, all seven clones positive for rhesus LCV EBNA-3C DNA were screened with additional PCR primers specific for rhesus LCV gp350, EBNA-3A, EBNA-3B, and BZLF1 (Amp1, -2, -3, -4, and -6) (Fig. 1). The results of these experiments are summarized in Table 2. One clone failed to score positive with any other primers and only scored positive with the original rhesus LCV EBNA-3C PCR primers (clone R2.5-1). One clone (clone R0.5-1) scored positive with rhesus LCV EBNA-3B PCR primers (Amp4), and the remaining five clones scored positive with the rhesus LCV EBNA-3A PCR primers (Amp3). In order to test whether the entire rhesus LCV EBNA-3A coding region (i.e., 5′ end) had been recombined, four of the rhesus LCV EBNA-3A-positive clones were first tested with rhesus LCV gp350 PCR primers (Amp1). However, all clones were negative by rhesus LCV gp350 PCR, suggesting that the recombination event took place between the Amp1 and Amp3 primers in gp350 and EBNA-3A. In order to test whether the complete rhesus LCV EBNA-3A had been recombined, PCR primers were designed which spanned the rhesus LCV EBNA-3A initiation codon (Amp2). Among the five EBNA-3A-positive clones, four clones were positive with the rhesus LCV EBNA-3A initiation codon PCR primers, suggesting that these clones had a complete coding sequence for rhesus LCV EBNA-3A (R1-2, R2.5-9, R5-3, and R1.5-9). In all four of these clones (R1-2, R2.5-9, R5-3, and R1.5-9), a complete rhesus LCV EBNA-3B coding region has probably been recombined, since PCR results with rhesus LCV EBNA-3A, EBNA-3B, and EBNA-3C PCR primers were all positive.
TABLE 2.
Rhesus LCV-specific PCR profile of chimeric P3HR-1 virusesa
| Clone | PCR result with amplicon (rhesus LCV region):
|
|||||
|---|---|---|---|---|---|---|
| Amp1 (gp350) | Amp2 (EBNA-3A ATG site) | Amp3 (EBNA-3A) | Amp4 (EBNA-3B) | Amp5 (EBNA-3C) | Amp6 (BZLF1) | |
| R2.5-1 | Neg | ND | Neg | Neg | Pos | Neg |
| R0.5-1 | Neg | ND | Neg | Pos | Pos | Pos |
| R4-6 | Neg | Neg | Pos | Pos | Pos | Pos |
| R1-2 | Neg | Pos | Pos | Pos | Pos | Neg |
| R2.5-9 | Neg | Pos | Pos | Pos | Pos | Pos |
| R5-3 | Neg | Pos | Pos | Pos | Pos | Pos |
| R1.5-9 | ND | Pos | Pos | Pos | Pos | Pos |
Neg, negative; Pos, positive; ND, not done.
In order to determine which clones contained an intact rhesus LCV EBNA-3C coding region, PCR primers (Amp6) specific for the rhesus BZLF1 homologue, located immediately 3′ to the EBNA-3C locus, were used. Five of seven clones were positive by rhesus LCV BZLF1 PCR, suggesting a complete rhesus LCV EBNA-3C gene and a recombination event 3′ to the Amp6 BZLF1 primers in these clones.
Thus, overall there were three clones (R2.5-9, R5-3, and R1.5-9) in which the PCR analysis suggested that complete rhesus LCV EBNA-3A, -3B, and -3C genes were recombined and transferred to immortalized human B cells by viral infection. Two additional clones were PCR positive for rhesus LCV EBNA-3A, -3B, and -3C. Since one clone (R4-6) failed to amplify with the rhesus LCV EBNA-3A initiation codon primers, we cannot rule out a frameshift or illegitimate recombination event without additional studies (e.g., direct sequencing of the recombinant junction). In the other clone (R1-2), there is evidence for an intact rhesus LCV EBNA-3A coding region, but the screening experiments cannot rule out an adverse recombination event in the EBNA-3C coding region, since amplifications with the rhesus LCV BZLF1 primers were negative. Of the two remaining clones, one (R2.5-1) was positive only with rhesus LCV EBNA-3C primers, suggesting a small recombination event which was unlikely to be useful. The other clone (R0.5-1) was PCR positive for EBNA-3B, EBNA-3C, and BZLF1. In this case, a complete rhesus LCV EBNA-3C may have been recombined, but there is potential for disrupting either EBNA-3B or BZLF1.
Studies for coinfection with wild-type P3HR-1 genomes and persistence of chimeric viruses.
Clones were tested with EBV-specific primers to determine if there was evidence for coinfection with wild-type P3HR-1 viruses. The species specificity of the EBV EBNA-3A, -3B, and -3C PCR primers could be demonstrated by positive amplification with DNA from P3HR-1 cells and negative amplification with DNA from rhesus LCV-infected cells, 278LCL (Fig. 7B). All clones positive for rhesus LCV DNA were also PCR positive for EBV EBNA-3A, -3B, and -3C. The most straightforward interpretation was coinfection with two viruses in one cell, (i) a transforming virus in which two recombination events have occurred with the EcoRI-A and RcosL12 cosmids and (ii) coinfection with wild-type P3HR-1, which is produced in vast excess relative to recombinant viruses. However, alternative combinations were possible, since the two recombination events do not necessarily have to occur in the same virus and recombination may not necessarily be homologous. First, clones may have been infected with two recombinant viruses, one virus with an EcoRI-A recombination and a second virus with an RcosL12 recombination. In this case, the rhesus LCV EBNA-3 locus would be recombined into an EBNA-2-negative, nontransforming P3HR-1 virus. Another possibility was that the RcosL12 DNA may have recombined illegitimately into a site different from the EBNA-3 locus. This could occur either as a second-site recombination in an EBNA-2-positive transforming virus or as a single recombination event into P3HR-1 if there is coinfection with an EBNA-2-positive transforming virus.
To begin addressing these possibilities, we evaluated the effect of long-term culture on the stability of the rhesus LCV DNA in the immortalized cell lines. We hypothesized that if rhesus LCV DNA were recombined into nontransforming, EBNA-2-negative P3HR-1 viruses, rhesus LCV DNA might be lost over time since there would be little selective pressure for preserving rhesus LCV DNA. PCR assays were repeated after 18 weeks in culture, and two clones (R2.5-1 and R0.5-1) became PCR negative for rhesus LCV DNA, suggesting that the rhesus LCV DNA had recombined into a nontransforming, EBNA-2-negative P3HR-1 virus. It was also possible, though perhaps less likely, that the rhesus LCV DNA recombined into a transforming, EBNA-2-positive virus and was lost over time because the rhesus LCV EBNA-3C-positive virus grows less well than a coinfecting EBNA-2-positive P3HR-1 virus. Unfortunately, without a positive selection marker to maintain and isolate the recombinant virus, it was impossible to resolve these possibilities.
Passage and purification of recombinant chimeric viruses.
In the remaining five clones, rhesus LCV DNA persisted after prolonged tissue culture. Three of these clones had evidence of recombination of complete rhesus LCV EBNA-3A, -3B, and -3C genes by PCR screening. In order to determine whether the rhesus LCV DNA recombination was homologous and whether the rhesus LCV EBNA-3 locus could substitute for the EBV EBNA-3 locus, the recombinant viruses must be passaged and purified of coinfecting P3HR-1 viruses. Two clones which had evidence of recombination of the complete EBNA-3 locus (R5-3 and R1-2) were selected, and cells were transfected with a BZLF1 expression vector to induce viral replication. gp350 expression on the cell surface could be detected in 1 to 5% of the cells (data not shown). Cell-free virus supernatants were used to infect human PBMC, and the infected PBMC were plated in 96-well plates.
Only one immortalized cell clone was obtained from passage of clone R1-2 virus. This clone was negative for rhesus LCV EBNA-3 DNA and positive for the P3HR-1 EBV EBNA-3 locus (data not shown). The most likely explanation for the failure to recover immortalizing viruses with the rhesus LCV EBNA-3 locus was that the chimeric P3HR-1 with rhesus LCV DNA was nontransforming, i.e., the rhesus LCV EBNA-3 is unable to completely replace the EBV EBNA-3 locus for human B-cell immortalization. The single transforming event recovered may have been due to recombination between an EBNA-2-positive chimeric virus and the coinfecting wild-type P3HR-1, thereby restoring the EBV EBNA-3C locus and efficient human B-cell immortalization. However, it is difficult to make definitive conclusions from the negative results with this clone.
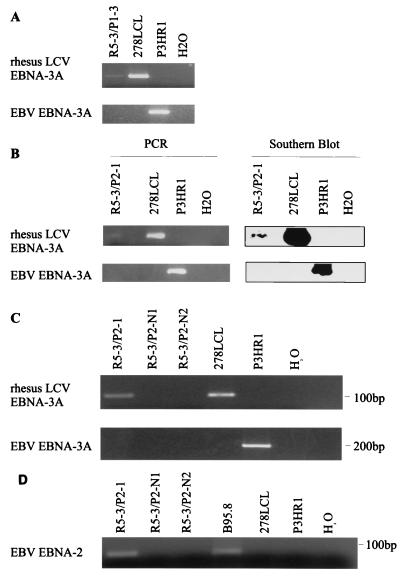
Passage of clone R5-3 virus supernatants resulted in 7 of 384 wells with macroscopically visible cell growth sufficient for PCR testing after 8 weeks of incubation. Cell DNA was prepared from these wells, and all seven clones were PCR positive for rhesus LCV EBNA-3A DNA, suggesting that the chimeric recombinant virus containing the rhesus LCV EBNA-3 locus had been successfully passaged (Fig. 8). The same DNA samples were PCR negative for EBV EBNA-3A DNA, suggesting that the chimeric virus had been purified of a wild-type P3HR-1 virus and that the rhesus LCV DNA had homologously recombined and replaced the EBNA-3 locus. All seven clones maintained a stationary phase for weeks 8 to 12 but failed to expand any further or sustain cell growth beyond 12 weeks. These results suggest that the rhesus LCV EBNA-3 locus was not capable of fully replacing the EBV EBNA-3 locus for B-cell immortalization.
FIG. 8.
Separation of chimeric viruses by passage of viral supernatant into human PBMC. (A) PCR of cell DNA from a representative clone (R5-3/P1-3) resulting from infection and passage of viral supernatants from the R5-3 clone coinfected with chimeric and wild-type P3HR-1 viruses. PCR with primers specific for rhesus LCV EBNA-3A (top panel) and primers specific for EBV EBNA-3A (bottom panel) are shown. H2O indicates control reactions with no DNA template. (B) Rhesus LCV EBNA-3A-specific (top panel) and EBV EBNA-3A-specific (bottom panel) PCR of viral DNA from a representative human B-cell clone (R5-3/P2-1) derived from a second experiment to passage virus from the R5-3 clone. The specificity of the PCR products was confirmed by Southern blot hybridization with rhesus LCV EBNA-3A and EBV EBNA-3A internal oligonucleotide probes. (C) PCR signals are specific for wells with evidence of cell growth, since no PCR signal was obtained from random wells with no cell growth at 8 weeks (R5-3/P2-N1 and R5-3/P2-N2). (D) EBNA-2 amplifications with EBV EBNA-2-specific PCR primers from viral DNA of clone R5-3/P2-1. B95-8 and P3HR-1 are B-cell lines infected with EBNA-2-positive and -negative EBV strains, respectively.
To confirm these findings, the experiment was repeated with fresh induction of virus from clone R2-5. In this instance, two wells showed cell growth after 8 weeks. In order to see if cell growth would continue when cells were maintained at a higher cell density, viral DNA for PCR testing was collected from culture supernatants in order to keep as many cells in culture as possible. Rhesus LCV EBNA-3A DNA was again detected by PCR in wells with visible cell growth, while EBV EBNA-3A DNA was no longer detected from cells after R5-3 virus passage, confirming the results in the previous experiment (Fig. 8B). In addition, EBV EBNA-2 DNA was detected in the same R5-3 passage DNA samples, consistent with recombination of the EBV EBNA-2 and rhesus LCV EBNA-3 in the same virus (Fig. 8C). As an additional control, samples from wells with no visible cell growth on the same microtiter plate were prepared in parallel and were PCR negative for rhesus LCV EBNA-3A DNA (Fig. 8C). Thus, these experiments provided evidence for homologous recombination of the rhesus LCV EBNA-3 locus into an EBNA-2-positive P3HR-1 virus, since the EBV EBNA-3 locus was no longer detected after the virus was passaged. However, cells infected with these chimeric viruses also failed to sustain cell growth even when maintained at high cell densities.
DISCUSSION
This is the first description of the EBNA-3 homologues in a nonhuman oncogenic LCV. The initial observation that all three latent infection genes are conserved in the rhesus LCV is not unexpected but does highlight the importance of these three nuclear proteins. The importance of EBNA-3A and -3C is obvious from their essential role in EBV-induced B-cell immortalization in tissue culture (39), but the significance of EBNA-3B is less obvious. One presumes that EBNA-3B must be essential for successful LCV infection in the natural host. Despite strong immune recognition of EBNA-3B, there appears to be strong selective pressure for conservation of an EBNA-3B gene in the nonhuman LCV. One hypothesis is that EBNA-3B induces cell genes which may downmodulate the immune response to assist virus infection or enhance the immune response to limit uncontrolled and otherwise lethal virus infection. The identification of a conserved rhesus LCV EBNA-3B gene in the current study and development of recombinant genetic techniques for manipulating rhesus LCV are important steps for formal testing of this hypothesis. The rhesus animal model provides a system in which the effect of EBNA-3B on acute and persistent LCV infection can be studied by infection of natural hosts with wild-type or EBNA-3B-deleted viruses. It will also be important to identify cell genes regulated by EBNA-3B and to correlate how EBNA-3B regulation of specific cell genes in vitro might contribute to the pathogenesis of LCV infection in vivo.
Despite the common interactions with RBP-Jκ, the sequence comparisons do not readily identify a consensus RBP-Jκ binding motif between a given EBV EBNA-3 and its rhesus LCV homologue, other than the fact that two separable RBP-Jκ domains can be identified in the EBV and rhesus LCV EBNA-3As. The RBP-Jκ interaction sites in each of the EBV EBNA-3s have not been precisely delineated but appear to be different from each other (6, 26, 31, 43). These regions share little similarity with the RBP-Jκ binding site in EBV EBNA-2, which has been reasonably conserved in the baboon and rhesus LCV EBNA-2-surrounding tryptophan residues (19). Similarly, there does not appear to be a consensus RBP-Jκ binding site common among the rhesus LCV EBNA-3s.
These studies also provide the first genetic experiments with rhesus LCV. We used a rhesus LCV cosmid (homologous to EBV coordinates 66124 to 110586) which was similar in size to the SalE/C cosmid (62201 to 105296) used by Tomkinson et al. for the first second-site EBV recombination studies (37). We screened for recombination in the EBNA-3C locus, and the frequency of second-site recombination for rhesus LCV EBNA-3C DNA into the EBV genome was 15.6%, versus 30% for a similar strategy using an EBV cosmid. These experiments formally demonstrate that simian LCV and EBV can recombine relatively efficiently. It is almost certain that recombination in other, more homologous regions of the cosmid also occurred and could be documented if one were to screen specifically for those regions. Thus, in a clinical setting in which simian LCV may be introduced into humans, e.g., xenogenic bone marrow transplantation, there is now laboratory evidence that simian LCV can infect human B cells (21) and that homologous recombination between simian LCV and EBV can occur. While coinfection of simian LCV and EBV might be a rare event in vivo, there is also evidence that coinfection and homologous recombination between EBV-1 and EBV-2 do occur in immunosuppressed AIDS patients (3, 41).
These first recombination studies establish that recombination between EBV and rhesus LCV can occur. The results suggest that the EBNA-3 locus is not totally interchangeable between EBV and rhesus LCV and that the species restriction for LCV-induced B-cell immortalization maps to at least one or more of the rhesus LCV EBNA-3s. One would expect that if the EBNA-3 locus were interchangeable, these transforming chimeric viruses would become apparent during the selection process for B-cell immortalization. It is recognized that studying mutants which lack transforming ability is difficult in the P3HR-1 system and that repeated failure to obtain transforming mutants in these studies does not absolutely exclude the possibility that the rhesus LCV EBNA-3 locus can be interchanged. However, these first-generation studies establish that rhesus LCV sequences can be recombined and will help guide the design of second-level genetic studies aimed at isolating chimeric viruses which may not have full transforming capacity.
It remains to be determined which EBNA-3 genes contribute to the species restriction for B-cell immortalization. One would predict that swapping EBNA-3Bs would be irrelevant, since EBNA-3B is not required for EBV-induced B-cell immortalization. EBNA-3A and -3C are more likely to be important for the species-specific effects, since they are essential for EBV-induced B-cell immortalization (39). We suspect that EBNA-3C may contribute significantly to the species restriction, since we are unable to generate immortalizing recombinant viruses from Raji cells coinfected with rhesus LCV (P. Rao and F. Wang, unpublished results). In these experiments, Raji cells are coinfected with rhesus LCV, and the full lytic infection cycle, i.e., gp350 expression, can be induced, presumably because the rhesus LCV can complement the block to lytic infection associated with the Raji BALF2 deletion (32). Rhesus LCV is replicated in these cells, since transforming rhesus LCV can be readily recovered in rhesus monkey PBMC. The current experiments predict that in some instances, homologous recombination between rhesus LCV and Raji genomes will restore the Raji EBNA-3C deletion with the rhesus LCV EBNA-3C locus. However, we have been unable to recover transforming virus in human B cells, consistent with the hypothesis that rhesus LCV EBNA-3C is unable to replace EBV EBNA-3C for human B-cell immortalization.
We did not ask whether the EBV-rhesus LCV EBNA-3 chimeras generated in the current study were able to immortalize rhesus monkey B cells; i.e., is an EBNA-3 locus from the same species sufficient for B-cell immortalization? However, in other experiments using P3HR-1 cells coinfected with rhesus LCV, we have also been unable to recover transforming virus in human B cells (Rao and Wang, unpublished). The rhesus LCV regions corresponding to both sides of the P3HR-1 deletion have a high degree of nucleotide homology (80 to 90%), and the experience from the current studies would again suggest that recombination between the rhesus LCV and P3HR-1 genomes should occur (24). The failure to recover viruses capable of transforming human B cells suggests that the rhesus LCV EBNA-LP and EBNA-2 may also contribute to the species restriction for LCV-induced B-cell immortalization. Thus, multiple latent infection genes may be important for determining which B-cell species can be immortalized by a given LCV. This may make it more difficult to recreate an appropriate EBV-rhesus LCV chimeric virus for animal studies similar to the simian-human immunodeficiency virus created previously (18).
Why the latent infection genes are not easily interchangeable for B-cell immortalization is unclear. It is particularly surprising given that virtually all of the rhesus and baboon LCV latent infection genes have been functionally similar to the EBV homologues using in vitro assays in human cells. Cells infected with chimeric virus appeared to be capable of initial cell growth but were unable to expand and sustain cell growth. These results are reminiscent of the phenotype recently described for EBV with LMP1 mutations, in which the amino-terminal 231 amino acids are sufficient for initial growth transformation but the carboxyl-terminal 155 amino acids are necessary for efficient long-term outgrowth (14).
The rhesus LCV EBNA-3 genes might be considered natural mutants defective for a transformation-essential pathway in human B cells. Thus, it may be interesting to find human B-cell proteins which interact well with an EBV EBNA-3 but perhaps not as well as with the rhesus LCV EBNA-3 and vice versa. In this light, it is curious to note how repetitive elements have been conserved in the rhesus LCV EBNA-3s, although with very different sequences. Alternatively, the mechanism for the species-specific differences may be subtle qualitative differences in existing interactions. For example, the rhesus LCV EBNA-3C has a Q/P-rich region and direct transactivating activity in an in vitro assay, but the activity we observed was significantly higher than that reported for EBV EBNA-3C (20). Perhaps this represents excessive transactivation activity in human cells versus rhesus cells, which may be detrimental to efficient human B-cell immortalization, but no dominant negative effect was observed in these studies.
The cloning of the rhesus LCV EBNA-3s, in combination with all of the other latent infection homologues, now allows us to address the question of whether the EBNA-3s are an immunodominant target for the host response to LCV infection in a different species. In humans, the EBNA-3s are immunodominant latent infection targets for the cytotoxic T lymphocyte (CTL) response during acute EBV infection, i.e., infectious mononucleosis, as well as persistent, asymptomatic infection (15, 36). However, it remains to be determined which CTL responses are important for control of EBV infection or pathogenesis of acute infection. It will be interesting to test whether the CTL repertoire in rhesus LCV-infected hosts is also skewed toward the EBNA-3s. The rhesus LCV animal model also provides a method for formal testing of potential EBNA-3-specific CTL vaccines. Study of naturally infected rhesus monkeys may be slightly complicated by the recent discovery of two types of rhesus LCV, similar to EBV-1 and EBV-2 (5). It will be interesting to see whether the type-specific sequence heterogeneity in rhesus LCV extends to the EBNA-3 genes, as it does in humans. Similarly, the cloning of the rhesus LCV BZLF1 homologue in these studies sets the stage to ask if there is as robust a CTL response to this lytic infection target as recently described in humans with infectious mononucleosis and to what degree immune responses to lytic infection targets such as BZLF1 might be important for protection or control of LCV infection (4).
ACKNOWLEDGMENTS
This work was funded by grants from the American Heart Association and the U.S. Public Health Service (CA68051 and CA65319).
REFERENCES
- 1.Allday M J, Crawford D H, Thomas J A. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J Gen Virol. 1993;74:361–369. doi: 10.1099/0022-1317-74-3-361. [DOI] [PubMed] [Google Scholar]
- 2.Blake N W, Moghaddam A, Rao P, Kaur A, Glickman R, Cho Y G, Marchini A, Haigh T, Johnson R P, Rickinson A B, Wang F. Inhibition of antigen presentation by the glycine/alanine repeat domain is not conserved in simian homologues of Epstein-Barr virus nuclear antigen 1. J Virol. 1999;73:7381–7389. doi: 10.1128/jvi.73.9.7381-7389.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrows J M, Khanna R, Sculley T B, Alpers M P, Moss D J, Burrows S R. Identification of a naturally occurring recombinant Epstein-Barr virus isolate from New Guinea that encodes both type 1 and type 2 nuclear antigen sequences. J Virol. 1996;70:4829–4833. doi: 10.1128/jvi.70.7.4829-4833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callan M F, Tan L, Annels N, Ogg G S, Wilson J D, O'Callaghan C A, Steven N, McMichael A J, Rickinson A B. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus in vivo. J Exp Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho Y G, Gordadze A V, Ling P D, Wang F. Evolution of two types of rhesus lymphocryptovirus similar to type 1 and type 2 Epstein-Barr virus. J Virol. 1999;73:9206–9212. doi: 10.1128/jvi.73.11.9206-9212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cludts I, Farrell P J. Multiple functions within the Epstein-Barr virus EBNA-3A protein. J Virol. 1998;72:1862–1869. doi: 10.1128/jvi.72.3.1862-1869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franken M, Annis B, Ali A N, Wang F. 5′ coding and regulatory region sequence divergence with conserved function of the Epstein-Barr virus LMP2A homolog in herpesvirus papio. J Virol. 1995;69:8011–8019. doi: 10.1128/jvi.69.12.8011-8019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J Virol. 1996;70:7819–7826. doi: 10.1128/jvi.70.11.7819-7826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerber P, Kalter S S, Schidlovsky G, Peterson W J, Daniel M D. Biologic and antigenic characteristics of Epstein-Barr virus-related herpesviruses of chimpanzees and baboons. Int J Cancer. 1977;20:448–459. doi: 10.1002/ijc.2910200318. [DOI] [PubMed] [Google Scholar]
- 11.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennessy K, Fennewald S, Kieff E. A third viral nuclear protein in lymphoblasts immortalized by Epstein-Barr virus. Proc Natl Acad Sci USA. 1985;82:5944–5948. doi: 10.1073/pnas.82.17.5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennessy K, Wang F, Bushman E W, Kieff E. Definitive identification of a member of the Epstein-Barr virus nuclear protein 3 family. Proc Natl Acad Sci USA. 1986;83:5693–5697. doi: 10.1073/pnas.83.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaye K M, Izumi K M, Li H, Johannsen E, Davidson D, Longnecker R, Kieff E. An Epstein-Barr virus that expresses only the first 231 LMP1 amino acids efficiently initiates primary B-lymphocyte growth transformation. J Virol. 1999;73:10525–10530. doi: 10.1128/jvi.73.12.10525-10530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khanna R, Burrows S R, Kurilla M G, Jacob C A, Misko I S, Sculley T B, Kieff E, Moss D J. Localization of Epstein-Barr virus cytotoxic T cell epitopes using recombinant vaccinia: implications for vaccine development. J Exp Med. 1992;176:169–176. doi: 10.1084/jem.176.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieff E. Epstein-Barr virus and its replication. In: Fields B, Knipe D, Howley P, editors. Fields virology. 3rd ed. Philadelphia, Pa: Raven Press; 1996. pp. 2343–2396. [Google Scholar]
- 17.Kienzle N, Young D, Silins S L, Sculley T B. Induction of pleckstrin by the Epstein-Barr virus nuclear antigen 3 family. Virology. 1996;224:167–174. doi: 10.1006/viro.1996.0518. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Lord C I, Haseltine W, Letvin N L, Sodroski J. Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus that expresses the HIV-1 envelope glycoproteins. J Acquir Immune Defic Syndr. 1992;5:639–646. [PubMed] [Google Scholar]
- 19.Ling P D, Ryon J J, Hayward S D. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall D, Sample C. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moghaddam A, Koch J, Annis B, Wang F. Infection of human B lymphocytes with lymphocryptoviruses related to Epstein-Barr virus. J Virol. 1998;72:3205–3212. doi: 10.1128/jvi.72.4.3205-3212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moghaddam A, Rosenzweig M, Lee-Parritz D, Annis B, Johnson R P, Wang F. An animal model for acute and persistent Epstein-Barr virus infection. Science. 1997;276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 23.Papadopoulos E B, Ladanyi M, Emanuel D, Mackinnon S, Boulad F, Carabasi M H, Castro-Malaspina H, Childs B H, Gillio A P, Small T N, et al. Infusions of donor leukocytes to treat Epstein-Barr virus-associated lymphoproliferative disorders after allogeneic bone marrow transplantation. N Engl J Med. 1994;330:1185–1191. doi: 10.1056/NEJM199404283301703. [DOI] [PubMed] [Google Scholar]
- 24.Peng R, Gordadze A V, Fuentes Panana E M, Wang F, Zong J, Hayward G S, Tan J, Ling P D. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J Virol. 2000;74:379–389. doi: 10.1128/jvi.74.1.379-389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petti L, Sample J, Wang F, Kieff E. A fifth Epstein-Barr virus nuclear protein (EBNA3C) is expressed in latently infected growth-transformed lymphocytes. J Virol. 1988;62:1330–1338. doi: 10.1128/jvi.62.4.1330-1338.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radkov S A, Bain M, Farrell P J, West M, Rowe M, Allday M J. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J Virol. 1997;71:8552–8562. doi: 10.1128/jvi.71.11.8552-8562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radkov S A, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday M J. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73:5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rangan S R, Martin L N, Bozelka B E, Wang N, Gormus B J. Epstein-Barr virus-related herpesvirus from a rhesus monkey (Macaca mulatta) with malignant lymphoma. Int J Cancer. 1986;38:425–432. doi: 10.1002/ijc.2910380319. [DOI] [PubMed] [Google Scholar]
- 29.Rivailler P, Quink C, Wang F. Strong selective pressure for evolution of an Epstein-Barr virus LMP2B homologue in the rhesus lymphocryptovirus. J Virol. 1999;73:8867–8872. doi: 10.1128/jvi.73.10.8867-8872.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein J kappa. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson E S, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa) J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson E S, Ooka T, Kieff E D. Epstein-Barr virus vectors for gene delivery to B lymphocytes. Proc Natl Acad Sci USA. 1996;93:11334–11340. doi: 10.1073/pnas.93.21.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rooney C M, Smith C A, Ng C Y, Loftin S, Li C, Krance R A, Brenner M K, Heslop H E. Use of gene-modified virus-specific T lymphocytes to control Epstein-Barr-virus-related lymphoproliferation. Lancet. 1995;345:9–13. doi: 10.1016/s0140-6736(95)91150-2. [DOI] [PubMed] [Google Scholar]
- 34.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steven N M, Leese A M, Annels N E, Lee S P, Rickinson A B. Epitope focusing in the primary cytotoxic T cell response to Epstein-Barr virus and its relationship to T cell memory. J Exp Med. 1996;184:1801–1813. doi: 10.1084/jem.184.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomkinson B, Kieff E. Second-site homologous recombination in Epstein-Barr virus: insertion of type 1 EBNA 3 genes in place of type 2 has no effect on in vitro infection. J Virol. 1992;66:780–789. doi: 10.1128/jvi.66.2.780-789.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomkinson B, Kieff E. Use of second-site homologous recombination to demonstrate that Epstein-Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J Virol. 1992;66:2893–2903. doi: 10.1128/jvi.66.5.2893-2903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F, Gregory C, Sample C, Rowe M, Liebowitz D, Murray R, Rickinson A, Kieff E. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao Q Y, Tierney R J, Croom-Carter D, Cooper G M, Ellis C J, Rowe M, Rickinson A B. Isolation of intertypic recombinants of Epstein-Barr virus from T-cell-immunocompromised individuals. J Virol. 1996;70:4895–4903. doi: 10.1128/jvi.70.8.4895-4903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates J L, Camiolo S M, Ali S, Ying A. Comparison of the EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology. 1996;222:1–13. doi: 10.1006/viro.1996.0392. [DOI] [PubMed] [Google Scholar]
- 43.Zhao B, Marshall D R, Sample C E. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jkappa. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]