Abstract
Hepatitis C virus (HCV) glycoprotein E2 binds to human cells by interacting with the CD81 molecule, which has been proposed to be the viral receptor. A correlation between binding to CD81 and species permissiveness to HCV infection has also been reported. We have determined the sequence of CD81 from the tamarin, a primate species known to be refractory to HCV infection. Tamarin CD81 (t-CD81) differs from the human molecule at 5 amino acid positions (155, 163, 169, 180, and 196) within the large extracellular loop (LEL), where the binding site for E2 has been located. Soluble recombinant forms of human CD81 (h-CD81), t-CD81, and African green monkey CD81 (agm-CD81) LEL molecules were analyzed by enzyme-linked immunosorbent assay for binding to E2 glycoprotein. Both h-CD81 and t-CD81 molecules were able to bind E2. Competition experiments showed that the two receptors cross-compete and that the t-CD81 binds with stronger affinity than the human molecule. Recently, h-CD81 residue 186 has been characterized as the critical residue involved in the interaction with E2. Recombinant CD81 mutant proteins were expressed to test whether human and tamarin receptors interacted with E2 in a comparable manner. Mutation of residue 186 (F186L) dramatically reduced the binding capability of t-CD81, a result that has already been demonstrated for the human receptor, whereas the opposite mutation (L186F) in agm-CD81 resulted in a neat gain of binding activity. Finally, the in vitro data were confirmed by detection of E2 binding to cotton-top tamarin (Saguinus oedipus) cell line B95-8 expressing endogenous CD81. These results indicate that the binding of E2 to CD81 is not predictive of an infection-producing interaction between HCV and host cells.
Hepatitis C virus (HCV) is the major etiologic agent of non-A non-B hepatitis, infecting an estimated 400 million people worldwide (3). A striking feature of HCV infection is the tendency towards a chronic status leading to liver diseases such as chronic hepatitis, cirrhosis, and hepatocellular carcinoma (24). Moreover, HCV infection is also associated with mixed cryoglobulinemia, a B-lymphocyte proliferative disorder (2). HCV is an enveloped virus classified as a Flavivirus (20); its genome is a positive-strand RNA of 9.5 kb, containing a single open reading frame encoding a large polyprotein precursor (5, 6), whose cleavage results in mature structural and nonstructural viral proteins (18). A basic polypeptide (core) and two envelope glycoproteins (E1 and E2) are the putative HCV structural proteins (30). Because of the lack of a cell culture system for virus growth, most of the studies aimed at understanding the biological activity of viral envelope proteins have been performed on recombinant proteins (15, 16, 27, 28). A truncated soluble recombinant form of HCV E2 glycoprotein has been reported to bind the surface of human cells (22) by interacting with the CD81 molecule (9, 19).
CD81 is a member of the tetraspanin family, membrane proteins containing four transmembrane domains, a short intracellular domain, and two extracellular loops (13, 14). CD81 is a widely expressed cell-surface protein which is conserved among different mammalian species. CD81 forms molecular complexes with cell-surface proteins, and these complexes vary in cell types of different lineages (23). In B cells CD81 is a component of supramolecular complexes involved in B-cell activation (26, 29). Engagement of CD81 with recombinant E2 has been reported to mimic natural ligand and activate biological functions on a B-cell-derived cell line (9). The binding site for E2 has been mapped to the large extracellular loop (LEL) domain (19). A soluble recombinant CD81 LEL molecule was shown to capture HCV viral particles (19), suggesting that the interaction between CD81 and E2 plays a role in HCV infection. The CD81 LEL domain is highly conserved in primates. Humans and chimpanzee are the only two species permissive to HCV, and their cells are both able to bind HCV E2 in a CD81-dependent manner (19). The African green monkey is not susceptible to HCV infection (1), and the African green monkey CD81 (agm-CD81), differing at four residues from the human molecule (h-CD81), is not capable of binding HCV E2 protein (9, 12).
Here, we report the sequence and the binding activity of HCV E2 glycoprotein to CD81 from a further primate, the tamarin. Surprisingly, though tamarins, New World monkeys of the genus Saguinus, are not permissive to HCV infection (11), E2 recombinant protein interacts with cell-surface-expressed tamarin CD81 (t-CD81). Moreover, binding experiments performed with soluble t-CD81 LEL show that the t-CD81 variant binds E2 with greater relative affinity than the human protein. Finally, mutagenesis of t-CD81 LEL suggests that the residue critical for interacting with E2 is shared between human and tamarin receptors.
MATERIALS AND METHODS
Animals and cells.
A captive, outbred Saguinus oedipus tamarin (B234) was housed at the Biomedical Primate Research Center, Rijswik, The Netherlands, and maintained under conditions that fulfilled all the ethical and scientific requirements for animal use. Saguinus labiatus primary hepatocytes were kindly supplied by Ralph Laufer. S. oedipus lymphoblast cell line B95-8 was obtained from the American Type Culture Collection (ATCC) (CRL-1612). The Molt-4 (human T-cell leukemia) line was obtained from the Medical Research Council ADP Repository. The 293 (human embryonic kidney) cell line was obtained from ATCC (CRL-1573), as was EL4 (mouse lymphoma) (TIB-39).
RNA preparation.
A portion (360 mg) of resected S. oedipus tamarin liver was used to extract total RNA using the Ultraspec II RNA isolation system (Biotecx), following the manufacturer's instructions. Pellets corresponding to 5 × 106 cells of both S. labiatus primary hepatocytes and S. oedipus B95-8 cultured cells were used to prepare total RNA using the system described above.
Amplification of t-CD81 sequences.
Total RNA (2.5 μg) was used as a template for first-strand cDNA synthesis in a 20-μl reaction mixture. The RNA was mixed with 10 pmol of antisense primer 98184 (5′-TCAGTACACGGAGCTGTTCCGGATG-3′) in a volume of 11 μl, denatured for 5 min at 90°C, chilled on ice, and centrifuged at 4°C. The following reagents were then added to the reaction mixture in amounts suitable to reach the indicated concentrations: 2.5% dimethyl sulfoxide, 10 U of RNasin (Promega) per ml, 1× Superscript buffer (Gibco-BRL), 10 mM ditheothreitol, and a 1.25 mM concentration of each deoxynucleoside triphosphate (dNTP). The reaction was performed by preincubation of the mixture at 42°C for 2 min, followed by incubation with 1 μl of Superscript II reverse transcriptase (Gibco-BRL) at 42°C for 50 min. The reaction was stopped by incubation at 70°C for 10 min. PCR was performed using 2.5-μl aliquots of first-strand cDNA synthesis reaction mixtures. Elongase B buffer (Gibco-BRL), dNTPs (a 250 μM concentration of each), sense primer 98183 (5′-ATGGGAGTGGAGGGCTGCACCAAGT-3′) and antisense primer 98184 (a 200 nM concentration of each), and 1 μl of Elongase mix (Gibco-BRL) were added to the cDNA in a total volume of 50 μl. PCRs were performed in a Perkin-Elmer 9600 or 2400 thermocycler by using the following conditions: 94°C for 7 min with a hot start, then 35 cycles of incubation at 94°C for 1 min, 50°C for 2 min, and 68°C for 2 min, followed by an extensive elongation step of 10 min at 68°C.
Construction of plasmids encoding CD81.
t-CD81-ORF/pCR2.1 was obtained by direct cloning of PCR products. The purified PCR products were ligated into the 3′ T-overhang pCR2.1 vector using the Original TA cloning kit (Invitrogen). The t-CD81-LEL/pGEX-2T plasmid was constructed by cloning the 264-bp HindII-RsaI fragment of t-CD81-ORF/pCR2.1 encoding the CD81 LEL domain into the filled-in EcoRI site of the pGEX-2T vector (Amersham Pharmacia Biotech). Construction of h-CD81 and agm-CD81 corresponding plasmids was as previously reported (9). Mutants (t-CD81-F186L and agm-CD81-L186F) were constructed as follows. Parental plasmids were digested with BamHI and AatII. Overlapping mutated fragments were generated and then assembled by PCR. The first fragment was generated using the sense primer (5′-GATCTGGTTCCGCGTGGATCC-3′) complementary to the vector sequence for both constructs in combination with the antisense primer 5′-aAgGAGGTTGGAAATGATACTGCTGCC-3′ for t-CD81-F186L or the antisense primer 5′-aAaGAGGTTGCTGATGATGTTGCTGCC-3′ for agm-CD81-L186F, where the mutated sequence is shown in lowercase. The second fragment was generated with the sense primer 5′-GGCAGCAGTATCATTTCCAACCTCcTt-3′ for t-CD81-F186L or the sense primer 5′-GGCAGCAACATCATCAGCAACCTCtTt-3′ for agm-CD81-L186F, where the mutated sequence is shown in lowercase, used in combination with the antisense primer 5′-CCACCTGACGTCTAAGAAACC-3′, complementary to the vector sequence for both constructs. The full-length final fragment was digested with BamHI and AatII and ligated into the parental digested vectors.
Sequencing and sequence analysis.
DNA sequencing of PCR products and individual clones was performed with the Big Dye Terminator Cycle Sequencing kit with AmpliTaq (Applied Biosystems) and run with an Applied Biosystems model 373A sequencer. Sequences were analyzed by the EditView and Autoassembler programs (Applied Biosystems). Sequence homology analysis was performed using the FASTA program, Wisconsin Package Version 9.1 (Genetics Computer Group, Madison, Wis.).
Expression and purification of CD81 recombinant proteins.
DNA of plasmids encoding CD81 LEL domains fused to glutathione-S-transferase (GST) protein was used to transform Escherichia coli DH5α competent cells, and cells were plated on Luria-Bertani (LB) agarose plates containing 100 μg of ampicillin per ml and 1% glucose. Single ampicillin-resistant colonies were inoculated in LB medium containing 100 μg of ampicillin per ml and 1% glucose and grown at 37°C with shaking. Overnight cultures were diluted 1:100 in LB-ampicillin and grown at 37°C till the optical density at 600 nm (OD600) was 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the culture was incubated for 3 h at room temperature. The bacterial pellet corresponding to 500 ml of induced culture was resuspended in 30 ml of phosphate-buffered saline (PBS) containing protease inhibitor cocktail (Boehringer) at 4°C. The cells were lysed with a microfluidizer (model 110-S; Microfluidics International Corporation), and the extract was centrifuged for 30 min at 4°C at 38,000 × g. Glutathione-Sepharose matrix (Pharmacia) was added to the bacterial lysate and incubated overnight at 4°C with gentle agitation. After extensive washing with cold PBS, the matrix-bound GST fusion protein was eluted with reduced glutathione (Sigma) at a final concentration of 10 mM in 50 mM Tris HCl (pH 8.0). Proteins were supplemented with protease inhibitor cocktail tablets (1 tablet/50 ml; Boehringer Mannheim) and 0.05% NaN3, and the concentration was determined by protein assay (Bio-Rad). Finally, proteins were checked by Western blotting analysis (25), using both anti-GST polyclonal antibody (Amersham Pharmacia Biotech) and anti-CD81 mouse monoclonal antibody (MAb) (1.3.3.22; Santa Cruz Biotechnology), and kept at −80°C in the presence of 10% glycerol.
Binding of E2 to recombinant CD81 proteins.
Enzyme-linked immunosorbent assay (ELISA) plates were coated with 1 μg of recombinant GST-CD81 proteins (t-CD81, h-CD81, agm-CD81, t-CD81-F186L, and agm-CD81-L186F, with GST as the control) diluted in PBS. After an overnight incubation at 4°C, plates were washed with washing buffer (0.05% Tween 20, PBS) and nonspecific binding sites were blocked with milk buffer (5% nonfat dry milk–0.05% Tween 20–0.05% NaN3 in PBS) for 1 h at 37°C. Serial dilutions of crude cell extract containing E2 proteins were performed in milk buffer supplemented with 50 μg of GST per ml, and the dilutions were preincubated 1 h at room temperature and then added to the CD81-coated plates for an overnight incubation at 4°C. After extensive washing, anti-His tag mouse MAb (Qiagen) diluted 1/400 in 2.5% bovine serum albumin–PBS was added and incubated for 3 h at 4°C. Alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (IgG) (Sigma) diluted 1/2000 in milk buffer was used as secondary antibody. Plates were finally washed as described above, and alkaline phosphatase was revealed by incubation at 37°C with a 1-mg/ml solution of p-nitrophenyl phosphate in ELISA substrate buffer (10% diethanolamine buffer, 0.5 mM MgCl2 [pH 9.8]). Results were expressed as the difference between the OD405 and the OD620, as determined with an automated ELISA reader (Labsystems Multiskan Bichromatic, Helsinki, Finland). Competition of the binding to t-CD81 was performed by preincubation of a nonsaturating amount of E2 protein with recombinant CD81 molecules.
Cloning and expression of E2 protein.
E2 protein representative of genotype 1a (strain H) was cloned into a VIJns-TPA plasmid (8). E2 (H-661) was amplified by PCR using the sense primer 5′-GGAGCAGTCTTCGTTTCGCCCGAAACCCACGTCACCGGGGGA-3′ and the antisense primer 5′-AGGCACAGCAGATCTTTAGTGGTGGTGGTGGTGGTGTGGCAGGGTCGTGAAAGAACACGG-3′, where the HCV-specific sequence is indicated in boldface and the sequence encoding the C-terminal histidine tag is italicized. The TPA fragment was optimized to ensure optimal cleavage and was PCR amplified using the sense primer 5′-CATGGGTCTTTTCTGCAGTCACCGTCCTTAGAT-3′ and the antisense primer 5′-TCCCCCGGTGACGTGGGTTTCGGGCGAAACGAAGACTGCTCC-3′. The E2 fragment and the TPA fragment were assembled by PCR, and the resulting product was purified, digested with BglII and PstI, and ligated to the vector. 293 cells were transfected with the plasmid by the calcium phosphate method (25). Culture supernatant was harvested 48 h later, concentrated 10 times using filter devices (Millipore Centricon Plus-80) and supplemented with protease inhibitor cocktail tablets (1 tablet/50 ml; Boehringer Mannheim) and 0.05% NaN3. Crude cell extracts were prepared in 1% Triton–20 mM Tris HCl (pH 7.5)–150 mM NaCl–1 mM EDTA supplemented by protease inhibitor cocktail tablets. Both cell extracts and concentrated supernatants were kept at −80°C in the presence of 10% glycerol. The amount of E2 in the extracts and in the supernatants was monitored by Western blotting by using an anti-His mouse MAb (Qiagen).
Binding of E2 proteins to cell lines.
Binding of E2 to the cell surface was analyzed using a fluorescence-activated cell sorting (FACS)-based assay. Cells were washed twice in phosphate buffered saline–2% fetal calf serum–0.05% NaN3 (washing buffer). Then, 2 × 105 cells were allowed to bind with E2 concentrated supernatants at room temperature for 1 h. After one wash in washing buffer, an anti-His mouse MAb (Qiagen) was added at the concentration of 2 μg/ml for 1 h at room temperature. As an isotype control, an anti-Flag mouse MAb (Kodak) was used to determine background fluorescence values. Finally, cell-bound MAb was visualized with anti-mouse IgG1-phycoerythrin conjugate (Serotec). For competition of E2 binding the supernatant was incubated for 1 h with the recombinant t-CD81 GST or GST alone (as the control) before incubating with cells. Detection of bound E2 was performed as described above. Competition of E2 binding was also obtained by incubation of cells with anti-CD81 (1.3.3.22; Santa Cruz Biotechnology) at a concentration of 10 μg/ml for 30 min before being tested for the ability to bind E2. Bound E2 was visualized by addition of anti-His conjugate with Alexa 488 dye. The conjugate was prepared by using the Alexa protein labeling kit (Molecular Probes). Flow cytometry data acquisition was performed by using a Becton Dickinson FACS Vantage flow cytometer. Dead cells were detected by Sytox (Molecular Probes) staining and were excluded from analyses.
Nucleotide sequence accession number.
The nucleotide sequence of the t-CD81 open reading frame from which the amino acidic sequence of the t-CD81 LEL domain has been deduced will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AJ250197.
RESULTS
Cloning of t-CD81 sequence.
To investigate the relationship between permissiveness to HCV viral infection and CD81, we sequenced the CD81 molecule from a primate genus (Saguinus) nonsusceptible to the infection (11). RNA was prepared from the S. oedipus-derived B95-8 lymphoblast cell line, from the liver of S. oedipus, and from hepatocytes of S. labiatus tamarins. Reverse transcription-PCR amplification of the CD81 open reading frame was performed using primers complementary to the h-CD81 sequence, and the product was cloned. The sequence of all three specimens was identical, indicating that there is no variability either between the two species examined or between primary cells and the cell line B95-8. The deduced amino acid sequence differs from that of the h-CD81 sequence in five amino acid positions, 155, 163, 169, 180, and 196, all located within the LEL domain, as shown in Fig. 1.
FIG. 1.
Alignment of CD81 LEL amino acid sequences of h-CD81 (SWISS-PROT accession no. P18582), chimpanzee CD81 (c-CD81; GenBank accession no. AF116600), agm-CD81 (GenBank accession no. AF116599) and t-CD81 (EMBL accession no. AJ250197). ∗, residue 186 mutagenized in agm-CD81 (L186F) and in t-CD81 (F186L).
Interaction of soluble CD81 LEL proteins with HCV E2 glycoprotein.
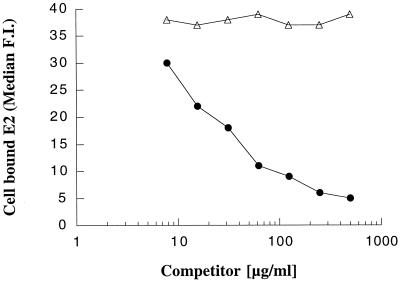
The binding site for HCV E2 glycoprotein has been mapped to the LEL domain of the CD81 molecule (9, 19). In general, fusions between tetraspanin large extracellular domains and GST have been described as correctly folded and functionally active (4). Moreover, a recombinant bacterially expressed h-CD81 LEL fused to GST has been reported to be able to bind HCV E2 envelope protein (9). We have analyzed the binding of E2 derived from genotype 1a (H-661) to recombinant CD81 LEL-GST fusion proteins from different species. All of the recombinant CD81 proteins were produced with comparable yields. Similarly, the concentrations of purified CD81 variant proteins, as measured by their reactivity by Western blotting with anti-CD81 antibodies, were comparable (data not shown). ELISA plates were coated with various CD81 molecules, and E2 binding was measured. agm-CD81 was unable to bind E2, whereas both human and tamarin proteins bound E2 (Fig. 2A). Indeed, E2 appeared to bind with higher relative affinity to t-CD81 than to h-CD81. To evaluate this further we compared the ability of soluble h-CD81 and t-CD81 proteins to compete for E2 interaction with t-CD81 bound to a solid support. Both human and tamarin proteins inhibited the interaction; however, a 10-fold-higher concentration of h-CD81 was required for the same level of inhibition (Fig. 2B). These results support the data shown in Fig. 2A, suggesting that E2 binds with higher affinity to t-CD81.
FIG. 2.
(A) Binding of E2 protein to recombinant CD81 molecules. ELISA plates were coated with the CD81 recombinant molecules, and 1:3 serial dilutions of crude lysate containing E2 were tested for binding. (B) Competition of E2 binding to t-CD81. t-CD81 was used to coat an ELISA plate. Competition was performed by preincubation of E2 extract with serial dilutions of recombinant CD81 molecules. ●, t-CD81; ⧫, h-CD81; ■, agm-CD81; ▵, GST (control).
Binding of HCV E2 to cell lines.
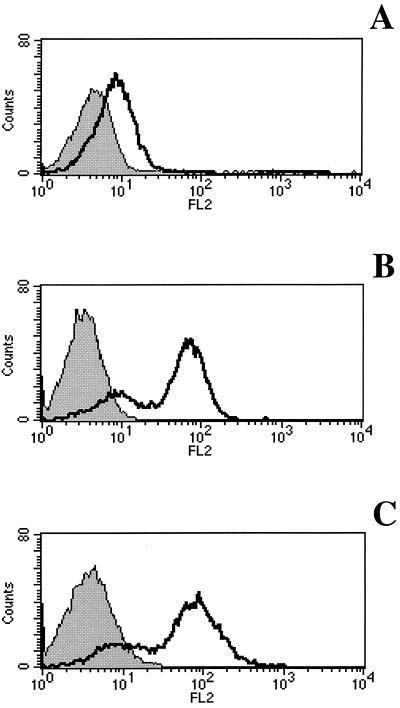
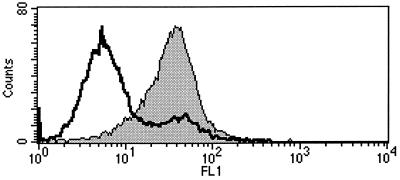
E2 ectodomain derived from HCV genotype 1a (H-661) binds to a variety of cell lines of human origin interacting with the CD81 receptor molecule (9, 19). We analyzed the binding of E2 to the cotton-top tamarin B95-8 cell line from which the t-CD81 cDNA was cloned for expression of the protein tested in the in vitro experiments described above. The binding was performed in parallel on the human cell line Molt-4 and on the mouse cell line EL4. The presence of the CD81 molecule on the cell surface was checked by FACS analysis using a fluorescein conjugate anti-h-CD81 monoclonal antibody. The antibody was cross-reactive with the t-CD81 and stained Molt-4 and B95-8 cells in a comparable manner (data not shown). E2 bound both human and tamarin cells, as revealed by the addition of a mouse MAb against the His tag of the recombinant protein followed by an anti-mouse IgG1-phycoerythrin conjugate (Fig. 3). The E2 interaction to B95-8 occurred in a CD81-dependent manner since nonsaturating levels of E2 could be competed by preincubation of E2 with soluble recombinant t-CD81 (Fig. 4). Moreover, preincubation of cells with anti-CD81 mouse MAb completely prevents E2 binding (Fig. 5). In this experiment directly labeled anti-His MAb (Alexa 488) was used to reveal the bound antigen.
FIG. 3.
FACS analysis of E2 binding to cell surface of EL4 (A), Molt-4 (B), and B95-8 (C) cell lines. Open curves, binding of E2 revealed by an MAb against the His tag; gray curves, binding of the isotype control. For panels A, B, and C, the median F.I. values were 7.8, 66.7, and 77.0, respectively.
FIG. 4.
FACS analysis of E2 binding to B95-8 cell surface in the presence of recombinant t-CD81. Competition was performed by preincubation of subsaturating amounts of E2 with increasing concentrations of t-CD81-GST (●) or GST (▵). The median F.I. for binding of E2 in the absence of the competitor was 38.8, and the median F.I. for binding of the mock antigen was 2.8.
FIG. 5.
FACS analysis of inhibition of E2 binding to B95-8 cells by anti-CD81 MAb. B95-8 cells were incubated with the monoclonal antibody at a concentration known to saturate the cell surface before the incubation with E2. Gray curve, E2 binding (median F.I., 34.2); open curve, binding competition by anti-CD81 MAb (median F.I., 5.3). The median F.I. for binding of the mock antigen to B95-8 was 2.7.
Defining the E2 binding region of t-CD81.
To gain further insight into the functional equivalence of t-CD81 and h-CD81 regions interacting with E2, we constructed a mutant t-CD81, introducing an amino acid substitution known to be crucial for the interaction of h-CD81 with E2 (12). The binding capability of h-CD81 is abrogated by replacing the phenylalanine at position 186 with a leucine, the corresponding residue present in agm-CD81. Hence, we mutated phenylalanine residue 186 to a leucine also in t-CD81. The mutant protein t-CD81-F186L was approximately 30-fold less active in binding E2 than the parental protein (Fig. 6). To obtain direct evidence for the requirement of a phenylalanine at position 186 for CD81-E2 interactions, the mutation L186F was introduced in agm-CD81 with the expectation that this mutation would have promoted E2 binding. Indeed, the effect of this mutation was striking, as it converted the inactive agm-CD81 to a high-affinity molecule having the ability to bind E2 with an apparent affinity threefold higher than that of the tamarin receptor (Fig. 6).
FIG. 6.
(A) Binding of E2 protein to recombinant, mutated CD81 molecules. ELISA plates were coated with the CD81 recombinant molecules, and 1:3 serial dilutions of crude lysate containing E2 were tested for binding. (B) Competition of E2 binding to t-CD81. t-CD81 was used to coat an ELISA plate. Competition was performed by preincubation of E2 extract with serial dilutions of recombinant CD81 molecules. ●, t-CD81; ○, t-CD81-F186L; ■, agm-CD81; □, agm-CD81-L186F.
DISCUSSION
CD81 has been identified as a putative receptor for HCV envelope glycoprotein E2. The binding region for E2 was mapped to the LEL of the CD81 molecule (10, 19). The LEL is highly conserved between humans and chimpanzees, the only two species known to be susceptible to HCV infection. Moreover, CD81 from nonsusceptible species, including the African green monkey (1), whose sequence differs from that of h-CD81 at only four residues, was shown to be unable to bind E2 (12, 19). To gain further information about the association between permissiveness to HCV infection and CD81 receptors we sequenced the CD81 gene from monkeys of the genus Saguinus, commonly called tamarins. Unsuccessful attempts to experimentally infect tamarins have led to the conclusion that these monkeys are resistant to HCV (11).
Cloning of the cDNA encoding CD81 from the S. oedipus cell line B95-8 allowed the characterization of this species' molecule, which is very similar to the h-CD81. We confirmed that t-CD81 obtained from the B95-8 cell line corresponded exactly to the molecule present in primary cells by sequencing the cDNA directly from two independent sources of hepatocytes belonging to the species S. oedipus and S. labiatus. The t-CD81 is identical to h-CD81 in the transmembrane domains, in the cytoplasmic domains, and in the extracellular loop 1, and it has only five amino acid differences, 155, 163, 169, 180, and 196, all located within the LEL region (Fig. 1). It is interesting to note that two of the five changes are conservative (T163S and D196E). Moreover, phenylalanine 186, which has been recently shown to be crucial for the interaction with E2 (12), is present in human and tamarin receptors and it is changed to leucine in the agm-CD81.
Production of soluble CD81 fusion proteins was achieved to perform comparative analysis of the interaction of E2 with CD81 molecules from different species. E2 protein from HCV interacts with soluble t-CD81 (Fig. 2). The comparative analysis of direct binding to E2 suggested that t-CD81 binds even better than the human receptor. This indication was confirmed by cross-competition experiments where a 10-fold-higher concentration of the human receptor than of the t-CD81 was required to obtain comparable levels of inhibition of E2 binding to t-CD81 (Fig. 2B).
Binding of E2 to S. oedipus-derived cells (B95-8) was performed with the aim of verifying that the interaction of the HCV envelope protein with the recombinant t-CD81 molecule reflected the ability of E2 to bind cell-surface-expressed t-CD81. B95-8 cells were indeed able to bind E2 in a CD81-dependent manner (Fig. 4 and 5), as already described for human cells (9).
Finally, we were interested in clarifying whether the LEL region involved in the interaction was the same for both human and tamarin receptors. A loss of function consequent to the mutation of h-CD81 residue 186 (F186L) has already been described (12). Here we show that the same mutation in the tamarin receptor clearly impairs the binding to E2 (Fig. 6), confirming that this amino acid residue is crucial for E2 binding of both h-CD81 and t-CD81. Even more interestingly, the opposite mutation in the corresponding residue of the inactive agm-CD81 (L186F) was sufficient to transform the agm-CD81 molecule to a strong binder, supporting the notion that the F in position 186 is absolutely required. The gain of function obtained in this case was very dramatic: we created a molecule capable of binding E2 with higher affinity than the best natural CD81 tested, t-CD81 (Fig. 6). Previous experiments had shown that the binding of the human receptor could be improved by mutagenesis of residue 163 (T163A), where alanine is the natural amino acid present in the agm-CD81 molecule (12). Probably the presence of alanine in position 163 confers high affinity binding on agm-CD81 when it is copresent with phenylalanine in position 186, as in the mutant agm-CD81-L186F. The construction of a series of mutants to acquire a complete picture of the residues responsible for the high binding activity of t-CD81 compared to that of its human counterpart is in progress.
The experiments presented in this study show that t-CD81 is able to bind HCV E2 glycoprotein, possibly through the same contacts of h-CD81-E2 interaction, in spite of the fact that HCV is unable to produce an infection in these animals. Thus, binding of HCV E2 glycoprotein to CD81 does not correlate with species permissiveness to HCV infection.
We could not assess whether t-CD81 is able to bind E2 displayed on HCV virions, since attempts to reproduce experiments on binding of HCV particles to CD81, as published by Pileri et al. (19), were unsuccessful with both the h-CD81 and the t-CD81 molecules. A possible explanation of this failure might be related to the availability of a convenient source of virus. We used high-titer HCV-infected human sera (data not shown), whereas Pileri et al. used experimentally infected chimpanzee sera, possibly obtained in a very early stage of infection, preceding the occurrence of antibodies complexing the viral particles.
Whether CD81 is the key molecule for HCV attachment to cells is an as-yet-unanswered question (21). We cannot exclude the possibility that HCV could gain entry into tamarin cells via CD81 and that infection would be blocked at a subsequent stage of the replicative cycle. On the other hand, CD81 may not be the sole receptor molecule involved in the interaction of the virus with the cells. We favor the hypothesis that a molecule exclusive for permissive species may be cooperative with CD81. One possibility is that cell entry of HCV requires, as does that of human immunodeficiency virus (7, 17), a coreceptor that would strengthen the interaction with CD81 and/or determine the viral tropism for the liver, which could not be achieved only by interacting with the almost ubiquitous CD81 molecule.
ACKNOWLEDGMENTS
We thank Giovanni Galfrè for helpful suggestions, Ralph Laufer and Tony Conley for S. labiatus tamarin primary hepatocytes, Ernst Verschoor for help with S. oedipus tamarin liver samples, and Mike Flint, Catherine Maidens, and Louise Wilson for reagents and technical expertise.
J.M. acknowledges financial support from The Wellcome Trust. S.L. was supported by Public Health Service grant P01 CA34233 from the National Institutes of Health.
REFERENCES
- 1.Abe K, Kurata T, Teramoto Y, Shiga J, Shikata T. Lack of susceptibility of various primates and woodchucks to hepatitis C virus. J Med Primatol. 1993;22:433–434. [PubMed] [Google Scholar]
- 2.Agnello V, Chung R T, Kaplan L M. A role for hepatitis C virus infection in type II cryoglobulinemia. N Engl J Med. 1992;327:1490–1495. doi: 10.1056/NEJM199211193272104. [DOI] [PubMed] [Google Scholar]
- 3.Alter M J. Epidemiology of hepatitis C. Hepatology. 1997;26:62S–65S. doi: 10.1002/hep.510260711. [DOI] [PubMed] [Google Scholar]
- 4.Azorsa D O, Moog S, Cazenave J, Lanza F. A general approach to the generation of monoclonal antibodies against members of the tetraspanin superfamily using recombinant GST fusion proteins. J Immunol Methods. 1999;229:35–48. doi: 10.1016/s0022-1759(99)00102-7. [DOI] [PubMed] [Google Scholar]
- 5.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 6.Choo Q L, Weiner A J, Overby L R, Kuo G, Houghton M, Bradley D W. Hepatitis C virus: the major causative agent of viral non-A, non-B hepatitis. Br Med Bull. 1990;46:423–441. doi: 10.1093/oxfordjournals.bmb.a072408. [DOI] [PubMed] [Google Scholar]
- 7.Doms R W, Peiper S C. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly J J, Martinez D, Jansen K U, Ellis R W, Montgomery D L, Liu M A. Protection against papillomavirus with a polynucleotide vaccine. J Infect Dis. 1996;173:314–320. doi: 10.1093/infdis/173.2.314. [DOI] [PubMed] [Google Scholar]
- 9.Flint M, Maidens C, Loomis-Price L D, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating J A. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint M, Thomas J M, Maidens C M, Shotton C, Levy S, Barclay W S, McKeating J A. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J Virol. 1999;73:6782–6790. doi: 10.1128/jvi.73.8.6782-6790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garson J A, Whitby K, Watkins P, Morgan A J. Lack of susceptibility of the cottontop tamarin to hepatitis C infection. J Med Virol. 1997;52:286–288. [PubMed] [Google Scholar]
- 12.Higginbottom A, Quinn E R, Kuo C C, Flint M, Wilson L, Bianchi E, Nicosia A, Monk P N, McKeating J, Levy S. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein. J Virol. 2000;74:3642–3649. doi: 10.1128/jvi.74.8.3642-3649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy S, Todd S C, Maecker H T. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol. 1998;16:89–109. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 14.Maecker H T, Todd S C, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 15.Matsuura Y, Suzuki T, Suzuki R, Sato M, Aizaki H, Saito I, Miyamura T. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology. 1994;205:141–150. doi: 10.1006/viro.1994.1629. [DOI] [PubMed] [Google Scholar]
- 16.Michalak J P, Wychowski C, Choukhi A, Meunier J C, Ung S, Rice C M, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 17.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 18.Neddermann P, Tomei L, Steinkuhler C, Gallinari P, Tramontano A, De Francesco R. The nonstructural proteins of the hepatitis C virus: structure and functions. Biol Chem. 1997;378:469–476. [PubMed] [Google Scholar]
- 19.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 20.Rice C M. Flaviviridae: the virus and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 21.Rice C M. Is CD81 the key to hepatitis C virus entry? Hepatology. 1999;29:990–992. doi: 10.1002/hep.510290356. [DOI] [PubMed] [Google Scholar]
- 22.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A J, Lau J Y, Choo Q L, Chien D, Pileri P, Houghton M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubinstein E, Le Naour F, Lagaudriere-Gesbert C, Billard M, Conjeaud H, Boucheix C. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur J Immunol. 1996;26:2657–2665. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- 24.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sato S, Miller A S, Howard M C, Tedder T F. Regulation of B lymphocyte development and activation by the CD19/CD21/CD81/Leu 13 complex requires the cytoplasmic domain of CD19. J Immunol. 1997;159:3278–3287. [PubMed] [Google Scholar]
- 27.Selby M J, Glazer E, Masiarz F, Houghton M. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- 28.Spaete R R, Alexander D, Rugroden M E, Choo Q L, Berger K, Crawford K, Kuo C, Leng S, Lee C, Ralston R, et al. Characterization of the hepatitis C virus E2/NS1 gene product expressed in mammalian cells. Virology. 1992;188:819–830. doi: 10.1016/0042-6822(92)90537-y. [DOI] [PubMed] [Google Scholar]
- 29.Szollosi J, Horejsi V, Bene L, Angelisova P, Damjanovich S. Supramolecular complexes of MHC class I, MHC class II, CD20, and tetraspan molecules (CD53, CD81, and CD82) at the surface of a B cell line JY. J Immunol. 1996;157:2939–2946. [PubMed] [Google Scholar]
- 30.van Doorn L J. Review: molecular biology of the hepatitis C virus. J Med Virol. 1994;43:345–356. doi: 10.1002/jmv.1890430406. [DOI] [PubMed] [Google Scholar]