Abstract
Reovirus nonstructural protein ςNS interacts with reovirus plus-strand RNAs in infected cells, but little is known about the nature of those interactions or their roles in viral replication. In this study, a recombinant form of ςNS was analyzed for in vitro binding to nucleic acids using gel mobility shift assays. Multiple units of ςNS bound to single-stranded RNA molecules with positive cooperativity and with each unit covering about 25 nucleotides at saturation. The ςNS protein did not bind preferentially to reovirus RNA over nonreovirus RNA in competition experiments but did bind preferentially to single-stranded over double-stranded nucleic acids and with a slight preference for RNA over DNA. In addition, ςNS bound to single-stranded RNA to which a 19-base DNA oligonucleotide was hybridized at either end or near the middle. When present in saturative amounts, ςNS displaced this oligonucleotide from the partial duplex. The strand displacement activity did not require ATP hydrolysis and was inhibited by MgCl2, distinguishing it from a classical ATP-dependent helicase. These properties of ςNS are similar to those of single-stranded DNA binding proteins that are known to participate in genomic DNA replication, suggesting a related role for ςNS in replication of the reovirus RNA genome.
Mammalian orthoreoviruses (reoviruses) encode three nonstructural proteins, μNS, ςNS, and ς1s, whose roles during viral infection remain poorly understood. This report concerns ςNS, which is known to be essential for reovirus replication based on the phenotype of a conditionally lethal (temperature-sensitive) mutant with its lesion in the ςNS-encoding S3 gene segment (9, 27). The ςNS protein comprises 366 amino acids and has a molecular mass of 41 kDa. Interaction of ςNS with the viral plus-strand RNAs in infected cells is well documented (3, 14). Moreover, when ςNS from infected cells is used to bind those RNAs in vitro, it protects 20- to 40-nucleotide fragments of the RNAs from RNase T1 digestion (34). Since the protected fragments include the 3′ ends of at least some of the plus-strand RNAs, it was proposed that ςNS binds specifically to those regions (34). In addition, ςNS and two other reovirus proteins, μNS and ς3, are found to associate with the viral plus-strand RNAs shortly after they are transcribed in infected cells and before minus-strand synthesis converts them into the double-stranded RNA (dsRNA) genome segments (3). These findings have led to a hypothesis that ςNS plays a role in selecting or condensing the viral plus-strand RNAs for packaging during early stages of particle morphogenesis (3, 14, 28, 34). A role for ςNS in translation of proteins from the reovirus plus-strand RNAs has also been suggested (10, 14).
Evidence for a direct role of ςNS in minus-strand synthesis is limited. The temperature-sensitive mutant whose lesion maps to the S3 gene segment (27) produces little or no dsRNA at restrictive temperatures (9, 15), but this indicates only that ςNS provides a required function at or before minus-strand synthesis in the replication cycle. Other previous work demonstrated that ςNS-containing ribonucleoprotein complexes isolated from reovirus-infected cells display a poly(C)-dependent poly(G) polymerase activity (12, 13). However, recombinant ςNS expressed in Escherichia coli or insect cells does not display this activity (10, 28), whereas recombinant reovirus protein λ3 obtained from mammalian or insect cells does (35; D. L. Farsetta and M. L. Nibert, unpublished data). These findings suggest that the ςNS-containing complexes characterized by Gomatos et al. (12, 13) also contained λ3, which is a known component of the reovirus RNA polymerase (22, 35). Thus, despite not having polymerase activity itself, ςNS might play a role in minus-strand synthesis within these λ3-containing complexes.
In the present study, we performed additional characterizations of the RNA-binding properties of ςNS to learn more about its roles in reovirus replication. In initial experiments, we analyzed purified, baculovirus-expressed ςNS (10) for its capacity to bind single-stranded RNA (ssRNA) molecules in gel mobility shift assays. The results indicated that multiple units of ςNS bind to single molecules of ssRNA with positive cooperativity and in numbers dependent on RNA length such that each unit of ςNS covered approximately 25 nucleotides at saturation. Competition experiments with various nucleic acids addressed the specificity of ςNS binding. These studies revealed a preference for ςNS to bind single-stranded over double-stranded nucleic acids, with a slight preference for RNA over DNA, but they showed no preference for ςNS to bind reovirus over nonreovirus RNA. We also characterized the capacity of ςNS to bind ssRNA to which short DNA oligonucleotides were hybridized. Displacement of these oligonucleotides from the partial duplexes was observed, and further characterizations demonstrated that this strand displacement activity of ςNS is distinct from that of a classical helicase. The observed activities of ςNS are similar to those of several well-characterized single-stranded DNA (ssDNA) binding proteins, which are known to be involved in genomic double-stranded DNA (dsDNA) replication, suggesting that ςNS may play a related role in replication of the reovirus dsRNA genome.
MATERIALS AND METHODS
Overexpression and purification of ςNS.
ςNS was overexpressed in insect cells infected with a recombinant baculovirus containing the ςNS-encoding reovirus type 1 Lang S3 gene and purified as described previously (10). Although RNase A was used in the purification procedure, no nuclease activity was detected in the final purified preparations of ςNS. As noted in the previous study (10), the ςNS protein obtained by this protocol was ≥95% pure according to results with Coomassie-stained sodium dodecyl sulfate-polyacrylamide gels and appeared to be relatively homogeneous in that it migrated between 7- and 9S in 5 to 20% sucrose gradients in the absence of RNA. Nonetheless, whether these complexes represent one discrete type of ςNS oligomer (e.g., tetramer or hexamer) or a mixture and whether or not they can self-associate under some conditions to form higher-order multimers remain to be determined.
Construction of the plasmids pGEM-DS4 and pGEM-3′DS4.
The reovirus type 3 Dearing S4 gene was amplified from a linearized plasmid (19) by PCR (29) with Vent DNA polymerase (New England Biolabs, Beverly, Mass.) and primers (Integrated DNA Technologies, Coralville, Iowa) representing the 5′ and 3′ ends of S4 plus EcoRI and PstI sites, respectively. The purified PCR product was digested with EcoRI and PstI, again purified, and ligated into those sites in the pGEM-4 plasmid (Promega, Madison, Wis.). A PCR with other primers was used to amplify nucleotides 738 to 1196 of the S4 gene from the new plasmid as well as the majority of pGEM-4 sequences between the PstI and EcoRI sites. Another PCR was used to amplify nucleotides 1 to 536 of the S4 gene. A final PCR was used to join and amplify the purified products from the first two reactions (8). The purified product from the final reaction was digested with SpeI and XhoI and ligated to the purified SpeI-XhoI fragment of the cloned S4 gene to generate new plasmid pGEM-DS4, with the entire S4 gene positioned immediately 3′ to the SP6 RNA polymerase promoter such that the first nucleotide of the SP6 transcript was S4 nucleotide 1 (23).
For subcloning the 3′ end of S4 from pGEM-DS4, we performed PCR with two primers (Integrated DNA Technologies) and Deep Vent DNA polymerase (New England Biolabs). One primer represented the plus strand of pGEM-DS4 from S4 nucleotides 1061 to 1096, except for two nucleotide substitutions that generated a new EcoRI site at S4 nucleotides 1082 to 1087, and the second primer represented the minus strand of pGEM-DS4 in vector sequences 3′ to the S4 gene and an intervening HindIII site. The amplified product was digested with EcoRI and HindIII and ligated into those sites in the pGEM-4Z vector to generate new plasmid pGEM-3′DS4. The first 6 nucleotides of the SP6 transcript from this plasmid were from pGEM-4Z, the next 6 were the EcoRI site, and the next 109 were from the S4 3′ end, beginning with S4 nucleotide 1088 and ending with the final S4 nucleotide, 1196.
Generation of RNA probes and competitors by in vitro runoff transcription.
To synthesize linear DNA templates with the desired termini for in vitro runoff transcription, we performed PCR with different primer pairs (Integrated DNA Technologies) (Table 1) and Deep Vent DNA polymerase (New England Biolabs). For making either full-length or partial (5′S4-121, 5′S4-151, 5′S4-181, 5′S4-211, 5′S4-271, and 3′S4-121) S4 plus-strand transcripts, one primer represented the plus strand of pGEM-DS4 or pGEM-3′DS4 in vector sequences 5′ to the SP6 promoter and the second primer represented the minus strand of pGEM-DS4 or pGEM-3′DS4 in S4 sequences beginning either at the 3′ end (relative to the plus strand) or the indicated (Table 1) internal position of S4. For making the 121-nucleotide vector transcript (pGEM-121), one primer represented the plus strand of pGEM-4Z in sequences 5′ to the SP6 promoter and the second primer represented the minus strand of pGEM-4Z in the appropriate downstream sequences. The fragments were isolated from 1% agarose gels and purified using the Qiaex II kit (Qiagen, Valencia, Calif.).
TABLE 1.
DNA primers used for PCR to generate linear DNA templates for in vitro transcription
| Transcripta | Plasmidb | Primerc
|
|
|---|---|---|---|
| Upstream | Downstream | ||
| S4 full length | pGEM-DS4 | 2787-GGCTTGTACATATTGTCG-2804 | 1196-GATGAATGAAGCCTGTCC-1179d |
| 5′S4-121 | pGEM-DS4 | 2787-GGCTTGTACATATTGTCG-2804 | 121-TCTTGCGCGCTGTAGATTGATACACG-96d |
| 5′S4-151 | pGEM-DS4 | 2787-GGCTTGTACATATTGTCG-2804 | 151-GGCTGTGCTGAGATTGTTTTGTCC-128d |
| 5′S4-181 | pGEM-DS4 | 2787-GGCTTGTACATATTGTCG-2804 | 181-ACGACGGCGCCACCACATACC-161d |
| 5′S4-211 | pGEM-DS4 | 2787-GGCTTGTACATATTGTCG-2804 | 211-GATCCAACAACACCTAGACAATGC-188d |
| 5′S4-271 | pGEM-DS4 | 2787-GGCTTGTACATATTGTCG-2804 | 271-TGATGACGGATCTGTTGATTACATCTATGG-242d |
| 3′S4-121 | pGEM-3′DS4 | 2654-GCTGCAAGGCGATTAAGTTGG-2674 | 1196-GATGAATGAAGCCTGTCC-1179d |
| pGEM-121 | pGEM-4Z | 2654-GCTGCAAGGCGATTAAGTTGG-2674 | 121-AGGAAACAGCTATGACCATGATTACGCC-94 |
| T7-17 | pGEM-3′DS4 | 349-GTCAGTGAGCGAGGAAGCGGAAGAGC-324 | 54-ATGCAAGCTTGTCTCCC-70 |
| SP6-146 | pGEM-3′DS4 | 2654-GCTGCAAGGCGATTAAGTTGG-2674 | 71-AGGGAGACAAGCTTGCATGC-52 |
RNA transcripts generated by in vitro runoff transcription from PCR-amplified DNA fragments. Each was generated using SP6 RNA polymerase except for T7-17, which was generated using T7 RNA polymerase.
Plasmid DNA used as the template for PCR.
Sequence numbers are those designated for pGEM-4Z in the Promega Life Science catalog unless otherwise indicated (see footnote d). All sequences are written in 5′-to-3′ orientation. Sequence numbers that decrease from left to right represent minus strands between those numbers (SP6 transcripts are defined as plus strands). Upstream and downstream are defined relative to the promoter (SP6 or T7) used in generating each transcript.
Sequence numbers are those for the type 3 Dearing S4 plus strand.
To generate RNA transcripts from each amplified DNA fragment, 1 to 2 μg of the purified fragment was combined with transcription buffer (50 mM HEPES [pH 7.5], 16 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol); 3 mM ATP, CTP, and UTP; 1 mM GTP (Pharmacia Biotech, Piscataway, N.J.); 200 U of SP6 RNA polymerase (Epicentre, Madison, Wis.); 20 U of RNasin (Promega); and 0.1 U of inorganic pyrophosphatase (Sigma, St. Louis, Mo.). To radiolabel the RNA transcripts, 0.13 nmol of [α-32P]GTP (specific activity, 3,000 Ci/mmol) (NEN, Boston, Mass.) was also added. The reaction mixtures were incubated at 37°C for 90 min prior to the addition of another 130 U of SP6 RNA polymerase, 13 U of RNasin, and 0.067 U of inorganic pyrophosphatase. The reaction mixtures were then incubated for another 90 min, after which they were extracted with phenol-chloroform, and the RNA was precipitated with 1 volume of 7.5 M ammonium acetate and 2 volumes of 100% ethanol. The RNAs were gel isolated from a 5% polyacrylamide Tris-glycine native gel (25 mM Tris, 192 mM glycine) and eluted overnight at room temperature in elution buffer (0.1% sodium dodecyl sulfate, 0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA; filter sterilized). The eluted RNA was then extracted with phenol-chloroform and precipitated with 0.3 M sodium acetate and 2 volumes of 100% ethanol. The RNA concentration was determined by scintillation counting (11).
Gel mobility shift assays.
Various amounts of purified ςNS in phosphate-buffered saline (137 mM NaCl, 3 mM KCl, 8.4 mM Na2HPO4, 1.6 mM KH2PO4) were incubated with various amounts of 32P-labeled RNA in gel shift buffer (12 mM Tris-HCl [pH 7.5], 0.1 mg of bovine serum albumin/ml, 10 mM β-mercaptoethanol, 0.2% Tween 20, 10 U of RNasin) (32) for 15 min at room temperature in a total volume of 10 μl. The samples were mixed with gel loading buffer (25 mM Tris, 192 mM glycine, 0.25% bromophenol blue, 0.25% xylene cyanole FF, 30% glycerol) and subjected to electrophoresis through either a 1% agarose-Tris-acetate-EDTA gel (40 mM Tris-acetate, 1 mM EDTA) at 90 V or a 5% polyacrylamide Tris-glycine native gel (described above) at 10 mA. The times of electrophoresis varied depending on the size of the RNA probe used in the assay. The gels were dried onto filter paper under vacuum, and the samples were visualized by phosphorimaging (Molecular Dynamics, Sunnyvale, Calif.). The amount of radioactivity in the samples was quantitated using ImageQuant software (Molecular Dynamics). Concentrations of NaCl from 0 to 300 mM had little effect on the binding of ςNS to RNA. The standard binding reaction mixture used throughout this study contained less than 30 mM NaCl.
Immunoblots.
Proteins in the 5% Tris-glycine polyacrylamide gels were blotted onto a nylon membrane (Bio-Rad, Hercules, Calif.) at 30 V overnight at 4°C in transfer buffer (25 mM Tris, 192 mM glycine). The membrane was probed with the anti-ςNS antibody (10) at a dilution of 1/500 using the protocol supplied with the Bio-Rad color development reagent kit. The secondary antibody was visualized with color development reagents 5-bromo-4-chloro-3-indoylphosphate p-toluidine salt and p-nitroblue tetrazolium chloride (Bio-Rad).
Hill plots.
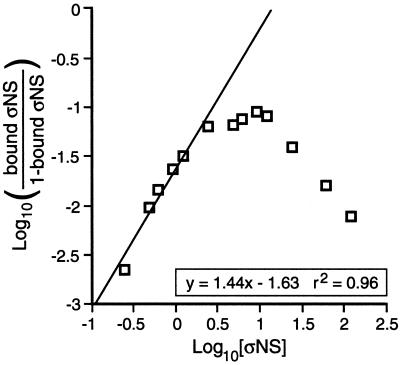
A Hill plot was generated for the 121-, 151-, and 181-nucleotide RNAs in Fig. 3 as well as for three other assays done with a 121-nucleotide RNA and ςNS amounts from 0.24 to 6.0 pmol (1 μg ≈ 24 pmol) (26). The amount of bound ςNS was calculated as follows. First the amount of radiolabeled RNA in each shifted band was quantitated and expressed as a percentage of total RNA in the reaction mixture. Next the percentage was multiplied by the number of ςNS units in each band (e.g., one ςNS unit in the fastest-migrating band and four in the slowest). Finally, the amounts of ςNS in all four bands were summed and the sum was divided by the total amount of protein added to each sample. The amount of unbound ςNS = 1 − the amount of bound ςNS. The log10 (bound ςNS/unbound ςNS) versus the log10 (ςNS concentration) was plotted, and the best-fit line to the linear portion of the graph was calculated using Excel (Microsoft, Redmond, Wash.). The slope of the line is the Hill coefficient (40) and was calculated for all six assays and averaged.
FIG. 3.
Hill plot of ςNS binding to ssRNA to determine cooperativity. A gel shift assay with 0.24 pmol of radiolabeled 5′S4-121 RNA was performed as described for Fig. 2A except that the concentrations of purified ςNS ranged from 0.24 to 6 pmol. The amount of ςNS bound at each concentration was calculated as described in Materials and Methods. Log10 (bound ςNS/1 − bound ςNS) was plotted relative to log10 (ςNS concentration), and a best-fit line was calculated for the linear portion of the graph (the equation and coefficient of determination for the line are shown in the box).
Competition assays.
A radiolabeled RNA probe (0.24 pmol each) was mixed with the unlabeled competitors in various relative amounts, after which 4.8 pmol of ςNS was added, and the samples were incubated for 15 min at room temperature. For one set of samples, ςNS was combined with radiolabeled RNA probe and incubated for 15 min as described above. The competitor was then added, and the samples were incubated for an additional 15 min or 2 h. Equivalent weights of RNA (concentration determined by A260 [30]) were used. The radiolabeled RNA probe used for both competition experiments was the 121-nucleotide fragment from the 5′ end of S4 in the pGEM-DS4 plasmid. The nucleic acid competitors used for competition between ss- and dsRNA and ss- and dsDNA were the following: ssRNA, full-length in vitro-transcribed S4 gene; dsRNA, reovirus type 3 Dearing genomic RNAs; ssDNA, circular M13 plasmid containing rhinovirus sequences; dsDNA, SmaI-linearized pGEM-4Z vector containing the reovirus type 1 Lang S3 gene (10). The unlabeled RNA competitors used for competitions were the following: the 5′ 121 nucleotides of S4 (same RNA used as the labeled probe), the 3′ 121 nucleotides of S4, and 121 nucleotides from the multiple-cloning region of the pGEM-4Z vector (Promega) that did not contain reovirus sequences. The amount of radiolabeled RNA bound was determined as the amount of RNA in the upper four shifted bands versus the total amount of RNA. All results are expressed relative to the sample containing no competitor.
RNA-DNA and RNA-RNA partial duplexes.
Nineteen-nucleotide DNA oligonucleotides (Integrated DNA Technologies) were designed to be complementary to the 3′ end, the middle, or the 5′ end of the 5′S4-121 RNA (Table 2). The oligonucleotides were 5′-end-labeled with polynucleotide kinase (New England Biolabs) and [γ-32P]ATP, extracted with phenol-chloroform, and precipitated with ethanol. The oligonucleotides were then hybridized to the 5′S4-121 RNA in hybridization buffer (100 mM NaCl, 50 mM Tris-HCl, 2 mM EDTA) at 90°C for 5 min and cooled to room temperature.
TABLE 2.
Oligonucleotides used in forming partial duplexes, RNA-DNA or RNA-RNA
| Type of oligonucleotide | Sequencea | Region of longer RNA strand to which oligonucleotide was hybridizedb | Tmc (°C) |
|---|---|---|---|
| DNA | 121-TCTTGCGCGCTGTAGATTG-103 | 3′ end | 43.1 |
| DNA | 68-CACGACCTGATGACCGTTG-50 | Middle | 44.1 |
| DNA | 19-GGGAAGAGGCAAAAATAGC-1 | 5′ end | 35.5 |
| RNA | 70-GGGAGACAAGCTTGCAT-54 | 3′ endd | 48.8 |
For DNA oligonucleotides, sequence numbers are those for the type 3 Dearing S4 plus strand. For the RNA oligonucleotide, sequence numbers are those for pGEM-4Z as designated in the Promega Life Science catalog. All sequences are written in 5′-to-3′ orientation. Sequence numbers decrease from left to right since the sequences represent minus strands (SP6 transcripts are defined as plus strands).
DNA oligonucleotides were individually hybridized to RNA 5′S4-121. The RNA oligonucleotide was hybridized to RNA SP6-146.
Melting temperatures calculated for RNA-DNA or RNA-RNA duplexes of the indicated sequences at 25 nM duplex and 25 mM salt using the algorithms of Sugimoto et al. (36) for RNA-DNA and Xia et al. (41) for RNA-RNA as implemented at http://bioweb.pasteur.fr/seqanal/interfaces/melting.html at the Pasteur Institute.
The 5′ end of the RNA oligonucleotide was hybridized to the penultimate 3′ nucleotide of the longer RNA.
To synthesize linear DNA templates for transcribing the 17-nucleotide RNA oligonucleotide (T7-17) (Table 2) and the 146-nucleotide RNA (SP6-146) to which T7-17 was later hybridized, we performed PCR with different primer pairs (Integrated DNA Technologies) (Table 1) and Deep Vent DNA polymerase (New England Biolabs). For making T7-17, one primer represented the minus strand of pGEM-3′DS4 in vector sequences 5′ to the T7 promoter and the second primer represented the plus strand of pGEM-3′DS4 in the appropriate downstream vector sequences. For making SP6-146, one primer represented the plus strand of pGEM-3′DS4 in vector sequences 5′ to the SP6 promoter and the second primer represented the minus strand of pGEM-3′DS4 in the appropriate downstream vector sequences. Both RNA transcripts were generated by runoff transcription in vitro as described above except that T7 RNA polymerase (Epicentre) and 12 mM MgCl2 were used in making T7-17. [α-32P]GTP was included in the reaction mixture to provide radiolabeling of T7-17, SP6-146, or both in different experiments. T7-17 was hybridized to the 3′ end of SP6-146 as described above for the RNA-DNA hybrids.
ATPase and displacement assays.
In the ςNS samples for the ATPase assay, 75 pmol of purified ςNS was incubated with 3 μCi of [α-32P]ATP (NEN) in gel shift buffer with or without EDTA (12.5 mM). In the core samples for the ATPase assay, 6 × 109 reovirus cores were incubated with 3 μCi of [α-32P]ATP (NEN) in nucleoside triphosphatase buffer (50 mM Tris-morpholinoethanesulfonic acid [MES] [pH 8.5], 5 mM MgCl2) (25) with or without EDTA (12.5 mM). The RNA-DNA hybrid (0.11 pmol) was added to all samples, which were incubated at room temperature or 35°C (cores) for 30 min in a total volume of 5 μl each. One microliter of each sample was then spotted onto a polyester-backed polyethyleneimine-cellulose thin-layer chromatography plate (Sigma) and developed with ascending solvent (1 M formic acid, 0.5 M LiCl). The reaction products were visualized by phosphorimaging (Molecular Dynamics). To assay for DNA oligonucleotide displacement activity in parallel with ATPase assays, samples were prepared in the same manner except that no [α-32P]ATP was added. The entire volume of each sample was then analyzed on a 5% polyacrylamide Tris-glycine native gel as described above. For other experiments in which DNA or RNA oligonucleotide displacement activity was measured, various amounts of purified ςNS and 0.24 to 0.26 pmol of RNA-DNA or RNA-RNA were added to gel shift buffer in a total volume of 10 μl and then the mixtures were incubated at room temperature for 15 min before running the entire sample on a 5% polyacrylamide Tris-glycine native gel.
RESULTS
Gel mobility shift assays for ςNS binding to 1,196- or 151-nucleotide ssRNAs.
The RNA-binding activity of purified ςNS was examined using an agarose gel mobility shift assay. In initial experiments, increasing amounts of ςNS were incubated with runoff transcripts representing full-length ssRNA plus strands of the reovirus type 3 Dearing S4 gene (data not shown). More than one band was seen in the mobility shift assay with this long RNA (1,196 nucleotides); however, most of the intermediate-size complexes migrated as a smear, making it difficult to distinguish the number of different complexes.
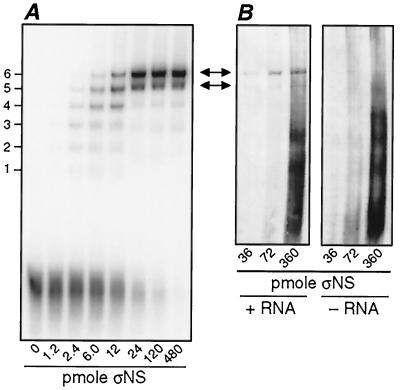
To improve resolution of the intermediate-size complexes, we decreased the length of the ssRNA and assayed for ςNS binding using a polyacrylamide gel mobility shift assay (Fig. 1A). For initial experiments with smaller ssRNA molecules, we used runoff transcripts representing the 5′ 151 nucleotides of the S4 plus strand (5′S4-151). As the amount of ςNS was increased, the mobility of the 5′S4-151 RNA was progressively retarded and clearly distinguishable, intermediate-size complexes were observed. At the larger amounts of ςNS (>12 pmol), a lowest-mobility complex was formed, accumulated, and was not further retarded with the addition of more protein. Based on the detection of more than one shifted RNA band over the range of ςNS amounts added, we concluded that more than one unit of ςNS can bind per molecule of 5′S4-151 plus-strand RNA. In addition, we concluded that the lowest-mobility complex represents RNA molecules that have each been saturatively bound by the same maximum number of ςNS units. Each binding unit of ςNS is hypothesized to be a small oligomer of that protein based on the sedimentation properties of the purified protein (10).
FIG. 1.
Gel mobility shift assay with a 151-nucleotide fragment of reovirus S4 plus strand and immunoblot analysis of a similar assay with anti-ςNS serum. (A) Increasing amounts of purified ςNS were incubated with 0.21 pmol of radiolabeled ssRNA comprising the 5′ 151 nucleotides of the reovirus S4 plus strand (5′S4-151). The samples were then subjected to electrophoresis in a 5% Tris-glycine polyacrylamide native gel until the free RNA was near the bottom of the gel. The radiolabeled RNA in the dried gel was visualized by phosphorimaging. ςNS-RNA complexes with six distinct mobilities are indicated at left. (B) Increasing amounts of ςNS were incubated with and without unlabeled 5′S4-151 RNA (0.8 pmol) and subjected to electrophoresis as described above. An immunoblot assay was performed to detect the protein. The amounts of RNA and protein in these samples were increased so that the ratios were similar to those in panel A but the protein was more easily detected. Arrows, comparable bands in panels A and B.
To confirm that the shifted bands contain ςNS, a gel from a mobility shift assay with unlabeled 5′S4-151 RNA was subjected to immunoblotting using a polyclonal antiserum for ςNS (10). The antiserum detected protein migrating in distinct bands (Fig. 1B, + RNA) with the same mobilities as the radiolabeled, protein-shifted RNA bands in Fig. 1A. The ςNS protein did not show this pattern of banding in the absence of ssRNA (Fig. 1B, − RNA), although ςNS did appear to concentrate in several distinct bands when excess amounts of the protein were present. Bands of similar mobility were visible in the presence of RNA as well (Fig. 1B, + RNA). The nature of these protein-specific bands is not known, but they may reflect a capacity of ςNS to self-associate into higher-order multimers in the absence of RNA under these conditions. Their appearance only with excess amounts of ςNS suggests that they hold limited significance for the RNA-binding results. The findings confirm that ςNS was present in the shifted RNA-protein complexes.
Gel mobility shift assay for ςNS binding to ssRNAs of other lengths.
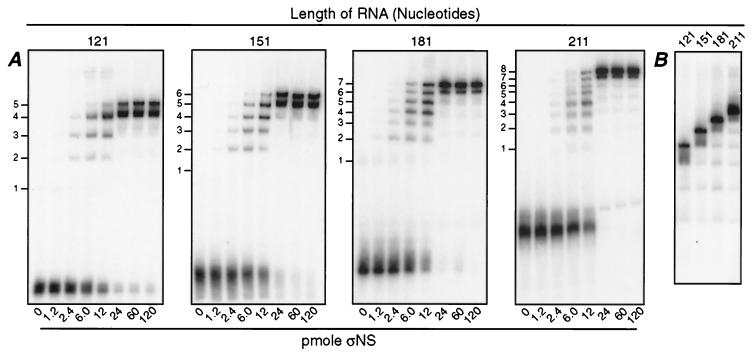
The 5′S4-151 RNA appeared to be shifted into six distinct bands over the range of amounts of ςNS (Fig. 1A). The presence of six shifted bands suggested that if the larger amounts of ςNS were fully saturating the RNA, then a maximum of six units of ςNS were binding to each RNA molecule. Moreover, since the RNA was 151 nucleotides long, each unit of ςNS must be covering about 25 nucleotides if the ςNS units are evenly distributed. To test this hypothesis, three additional plus-strand RNAs, differing in length by 30 nucleotides (5′S4-121, 5′S4-181, and 5′S4-211), were generated as runoff transcripts from the same S4 clone used to generate the 5′S4-151 RNA. The new RNAs were incubated with increasing amounts of ςNS and were analyzed in the polyacrylamide gel mobility shift assay. As predicted from initial findings with 5′S4-151 (Fig. 1A), 5′S4-121 was shifted into five distinguishable complexes, 5′S4-151 was shifted into six distinguishable complexes, 5′S4-181 was shifted into seven distinguishable complexes, and 5′S4-211 was shifted into eight distinguishable complexes with increasing amounts of ςNS (Fig. 2A). To compare the mobilities of the different complexes formed with each RNA, all four RNAs were incubated with the same amount of ςNS (18 pmol) and subjected to electrophoresis in adjacent lanes of the same gel (Fig. 2B). This analysis demonstrated that with each 30-nucleotide increase in RNA length over this range, one new lower-mobility complex was seen. In later experiments, 5′S4-271 RNA was also tested and found to be shifted into 11 distinguishable complexes with increasing amounts of ςNS (data not shown). Considering the size of each RNA and the number of distinguishable complexes formed with each, we concluded that the binding unit of ςNS covers a 24- to 27-nucleotide region of RNA and that the total number of ςNS units that bound per RNA molecule is determined by the overall length of the RNA.
FIG. 2.
Gel mobility shift assays with S4 plus-strand RNA fragments ranging in size from 121 to 211 nucleotides in 30-base increments. Radiolabeled ssRNA fragments comprising the following 5′ portions of the S4 plus strand were synthesized: 121 nucleotides, 5′S4-121; 151 nucleotides, 5′S4-151; 181 nucleotides, 5′S4-181; and 211 nucleotides, 5′S4-211. (A) Increasing amounts of purified ςNS were separately incubated with 0.21 to 0.22 pmol of each RNA, and the samples were analyzed as described for Fig. 1A. ςNS-RNA complexes with five to eight distinct mobilities are indicated to the left of each gel. (B) Purified ςNS (18 pmol) was incubated with each of the four RNAs as described for panel A (RNA sizes are indicated above the lanes) and subjected to electrophoresis as described for Fig. 1A. To provide better separation of the shifted complexes, the gel was run longer in this experiment such that the free RNA was run off the bottom.
Binding cooperativity.
Since multiple units of ςNS bound to a single RNA molecule, the binding could have exhibited positive, negative, or no cooperativity. To address these possibilities, the ratio of bound to total ςNS in each sample was calculated for the 121-, 151-, and 181-nucleotide RNAs from Fig. 2A. In addition, three other experiments using the 121-nucleotide RNA and smaller protein amounts were performed. Hill plots were generated for all six data sets (40). A representative plot for the 121-nucleotide RNA is shown (Fig. 3). The linear region of this curve has a slope >1 indicating that ςNS binds ssRNA with positive cooperativity. When the data from all six experiments were averaged and ςNS was assumed to bind ssRNA as a monomer, the Hill coefficient was calculated at 1.43 ± 0.10. Since the RNA-binding unit of ςNS is thought to be a small oligomer (10), the calculations were also done for ςNS binding as a dimer, trimer, and tetramer. The Hill coefficient increased by 0.02 to 0.03 unit with each step in oligomer order (data not shown), indicating that ςNS binding to ssRNA exhibited positive cooperativity regardless of the oligomeric nature of the binding unit.
Competition assays with single- and double-stranded nucleic acids.
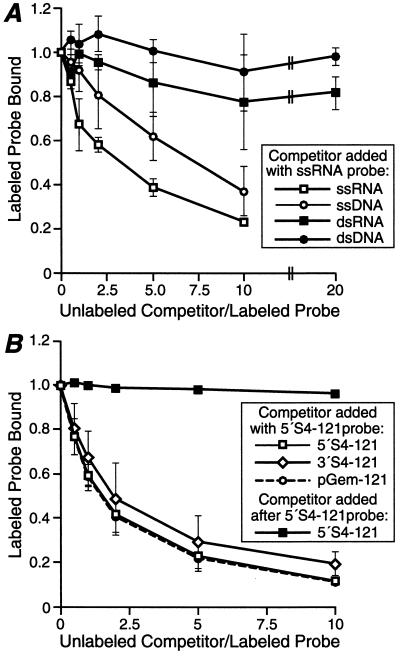
Previous work demonstrated that ςNS does not bind detectably to dsRNA or dsDNA in filter-binding or sedimentation assays (14, 28) but does bind to ssDNA in the filter-binding assay (28). To compare the affinities of ςNS for ssRNA and other nucleic acids in a more quantitative fashion, we performed competition assays. Radiolabeled 5′S4-121 RNA was combined with unlabeled competitor nucleic acids (reovirus ssRNA, reovirus dsRNA, ssDNA, or dsDNA) before the addition of ςNS. The RNA-protein complexes were then analyzed using the polyacrylamide gel mobility shift assay (Fig. 4A). Both ssRNA and ssDNA competed efficiently with the labeled ssRNA probe for ςNS binding, whereas dsRNA and dsDNA competed much less efficiently (Fig. 4A). For example, when 10-fold more ssRNA or ssDNA competitor was added only 23% ± 2% or 37% ± 11%, respectively, of the radiolabeled 5′S4-121 RNA was bound. In contrast, when 20-fold more dsRNA or dsDNA competitor was added, 82% ± 7% or 98% ± 4%, respectively, of the radiolabeled 5′S4-121 RNA was bound. In addition, with either single- or double-stranded competitors, RNA competed with only a slightly greater efficiency for binding to ςNS than did DNA (Fig. 4A). In summary, ςNS bound efficiently to single-stranded, but not double-stranded, nucleic acids and exhibited a slight preference for RNA over DNA.
FIG. 4.
Competition assays for ςNS binding to ssRNA using ssRNA, dsRNA, ssDNA, or dsDNA as the competitor. Each data point represents the mean from three experiments, and the error bars represent the standard deviation of the mean. (A) Increasing concentrations of unlabeled competitor ssRNA, dsRNA, dsDNA, or ssDNA (see Materials and Methods) were combined with 0.24 pmol of radiolabeled 5′S4-121 ssRNA. Purified ςNS (4.8 pmol) was added to each sample, and the sample was incubated. The samples were subjected to electrophoresis and visualized as described for Fig. 1A. The upper four shifted RNA bands were included in the quantitation of bound RNA. The ratios of competitor to probe RNA were calculated from weights rather than molar amounts of nucleic acid to reflect numbers of potential ςNS binding sites rather than numbers of nucleic acid molecules. (B) The assay was performed as described for panel A except that the unlabeled competitor ssRNAs were 121 nucleotides from the 5′ end of the S4 plus strand (5′S4-121), the 3′ end of the S4 plus strand (3′S4-121), or the vector sequence (pGEM-121). For one set of samples, ςNS was added to the labeled RNA prior to addition of the 5′S4-121 competitor.
Competition assays with reovirus and nonreovirus ssRNAs.
Previous work demonstrated ςNS binding to reovirus RNA in infected cells (3, 14) and to nonreovirus RNA in vitro (14, 28). However, no experiments addressed whether ςNS exhibits a preference for reovirus RNA (14, 28). Sequences at the 5′ and 3′ termini of all 10 reovirus RNAs are conserved (2) and therefore may be involved in distinguishing reovirus from nonreovirus RNA for binding by proteins such as ςNS during steps in reovirus replication. To test this hypothesis, we used a competition assay to compare the affinities of ςNS for RNAs representing the 5′ end of the reovirus S4 plus strand, the 3′ end of the reovirus S4 plus strand, and nonreovirus sequences derived from pGEM-4Z. Radiolabeled 5′S4-121 RNA was incubated with unlabeled competitor RNAs (5′S4-121, 3′S4-121, or pGEM-121) before addition of ςNS. The RNA-protein complexes were then analyzed using the polyacrylamide gel mobility shift assay (Fig. 4B). In this assay, all three competitor RNAs competed to approximately the same level with radiolabeled 5′S4-121 RNA for binding to ςNS. For example, when fivefold more 5′S4-121 competitor RNA was added, only 13% ± 3% of the radiolabeled 5′S4-121 RNA was bound by ςNS. When fivefold more 3′S4-121 competitor RNA was added, only 22% ± 12% of the radiolabeled 5′S4-121 RNA was bound. Last, when fivefold more pGEM-121 competitor RNA was added, only 13% ± 9% of the radiolabeled 5′S4-121 RNA was bound. Although these data suggested that ςNS does not preferentially bind reovirus sequences, it was possible that both ends of the RNA together might be required to confer specificity. However, the amount of 5′S4-121 RNA bound to ςNS in the presence of the full-length ssRNA competitor in Fig. 4A was similar to the amount bound in the presence of each of the competitor RNAs in Fig. 4B. Thus, all ssRNAs appeared to compete to about the same level with radiolabeled 5′S4-121 RNA for binding to ςNS. This suggests that ςNS does not distinguish between RNA sequences corresponding to the 5′ and 3′ ends of the S4 gene and does not exhibit preference for binding to reovirus sequences.
To investigate the stability of preformed RNA-protein complexes, radiolabeled 5′S4-121 RNA was incubated with ςNS prior to addition of competitor (Fig. 4B). Unlabeled 5′S4-121 RNA did not displace radiolabeled 5′S4-121 RNA that was already bound to ςNS, even when the competitor was added in 10-fold excess. The complex of 5′S4-121 RNA and ςNS was stable for at least 2 h after addition of this competitor (data not shown). These results suggest that, once ςNS bound to RNA, it formed a stable complex that could not be easily disrupted by subsequent addition of other RNA.
Gel mobility shift assays for ςNS binding to partially duplex RNA-DNA hybrids and evidence for a strand displacement activity.
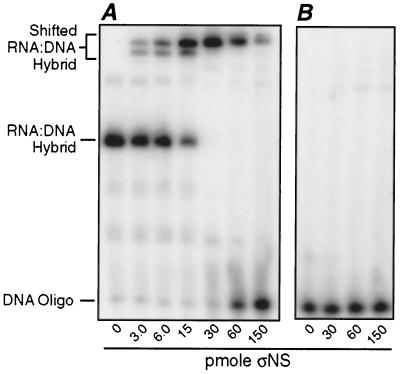
We hypothesized that ςNS may require a single-stranded 5′ or 3′ end of RNA to initiate binding. To test this hypothesis, an end-radiolabeled DNA oligonucleotide complementary to 19 nucleotides at the 3′ end of the 5′S4-121 RNA (Table 2) was hybridized to that RNA. The RNA-DNA hybrid was then incubated with increasing amounts of ςNS and analyzed using the polyacrylamide gel mobility shift assay (Fig. 5). The mobility of this RNA-DNA hybrid with a duplex region at the RNA 3′ end was retarded by the smaller amounts of ςNS (Fig. 5A). Moreover, the amount of ςNS that shifted the majority of this RNA-DNA hybrid (30 pmol of ςNS to shift 0.26 pmol of hybrid in Fig. 5A) was comparable to the amount that shifted the majority of the 121-nucleotide RNA (24 pmol of ςNS to shift 0.22 pmol of RNA in Fig. 2A). Similar results were observed when other end-radiolabeled DNA oligonucleotides (Table 2) were hybridized to either the middle or the 5′ end of the 5′S4-121 RNA (data not shown). We therefore concluded that ςNS does not require a large single-stranded region at either the 5′ or the 3′ end of RNA to initiate binding.
FIG. 5.
Gel shift assay with ssRNA having an RNA-DNA duplex region at its 3′ end. A 19-nucleotide DNA oligonucleotide that was exactly complementary to the 3′ end of the RNA was 32P-labeled at its 5′ end. (A) The oligonucleotide was hybridized to unlabeled ssRNA (5′S4-121) and purified. Differing amounts of purified ςNS were then mixed with 0.26 pmol of hybrid and assayed as described for Fig. 1A. (B) An unhybridized 32P-labeled oligonucleotide was incubated in the presence of increasing amounts of ςNS and also assayed as described for Fig. 1A. For both panels, the amounts of ςNS added to samples are indicated beneath the lanes, and the positions of the RNA-DNA hybrid and the DNA oligonucleotide are also indicated.
Interestingly, when larger amounts of ςNS were incubated with the RNA-DNA hybrid having a duplex region at the 3′ end, the shifted hybrids disappeared and increasing amounts of free oligonucleotide were observed (Fig. 5A). This suggests that, as the final unit of ςNS was binding to each molecule of the hybrid, it destabilized the duplex region and caused the DNA oligonucleotide to be released. The DNA oligonucleotides bound to the middle or the 5′ end of the RNA were also released when saturating amounts of ςNS were incubated with the respective hybrids (data not shown), suggesting that the displacement activity was not direction dependent.
Evidence that ςNS is not an ATP-dependent helicase.
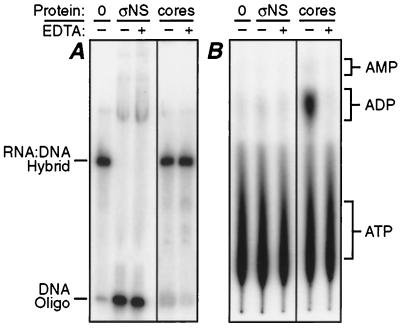
Since ςNS displaced DNA oligonucleotides from RNA-DNA hybrids, we recognized that the protein might possess helicase activity. If so, then ςNS like other RNA or DNA helicases, should require hydrolysis of ATP as a source of energy for the displacement activity (reviewed in reference 16). Although ATP was not added to the displacement assay mixtures described in the previous section, it is possible that there was contaminating ATP from the reaction mixture used to radiolabel the DNA oligonucleotides (see Materials and Methods). To investigate whether ATP hydrolysis was required for ςNS to displace the DNA oligonucleotides, the displacement assay was repeated in the absence or presence of EDTA, a known inhibitor of ATPase and RNA helicase functions through its chelation of divalent cations (16, 25). In addition, [α-32P]ATP was added to parallel samples, which were then analyzed by thin-layer chromatography to determine if ATP had been hydrolyzed. The ςNS protein displaced DNA from the RNA-DNA hybrid in the absence or presence of EDTA (Fig. 6A) but did not detectably hydrolyze ATP in either case (Fig. 6B). Reovirus cores, which were analyzed as a positive control for EDTA-sensitive ATPase activity (25), hydrolyzed ATP in the absence, but not in the presence, of EDTA (Fig. 6B). These data suggest that ςNS does not require hydrolysis of ATP during displacement of the DNA oligonucleotide from the RNA-DNA hybrid.
FIG. 6.
Analysis of ATP hydrolysis during strand displacement by ςNS. Either 12.5 mM EDTA (+) or H2O (−) was added to 75 pmol of purified ςNS (ςNS) or 6 × 109 reovirus core particles (cores) in each sample. Cores were included as a positive control for ATP hydrolysis. All samples were analyzed in duplicate. (A) The RNA-DNA hybrid described for Fig. 5 (0.11 pmol) was added to each sample in one set. These samples were then analyzed as described for Fig. 1A. The positions of the RNA-DNA hybrid and the DNA oligonucleotide are indicated. (B) Three microcuries of [α-32P]ATP and 0.11 pmol of the RNA-DNA hybrid were added to each sample in the other set. Products of the reaction with ATP were resolved by thin-layer chromatography and analyzed by phosphorimaging. The positions of unlabeled ATP, ADP, and AMP markers, as determined by UV absorption, are indicated.
To assess the effect of MgCl2 on the ςNS-associated strand displacement activity, the displacement assay was performed in the presence of increasing concentrations of MgCl2. The activity was progressively decreased with increasing MgCl2 (data not shown). At 10 mM MgCl2, for example, only 30% of the DNA oligonucleotide was displaced relative to the amount displaced in the absence of MgCl2 (data not shown), suggesting that Mg2+ inhibited rather than stimulated the displacement activity. This response to MgCl2 provided additional evidence that the strand displacement activity of ςNS is distinguishable from that of a classical helicase (16).
Test for strand displacement activity of ςNS using a partially duplex RNA molecule.
Regions of RNA-RNA duplex are generated during the normal course of reovirus replication, certainly in the genomic dsRNA segments and probably also in regions of secondary structure within the free plus-strand transcripts. The strand displacement activity that we demonstrated for ςNS using RNA-DNA hybrids may therefore reflect an essential activity of this protein at disrupting regions of RNA-RNA duplex during one or more steps in reovirus replication. The poor capacity of ςNS to bind to fully duplex RNA molecules (Fig. 4A) suggested that partial duplexes were more appropriate for further testing. In experiments with one such partially duplex RNA, we found that ςNS could not displace a fully complementary 17-nucleotide RNA oligonucleotide (Table 2) from the 3′ end of a longer RNA to which it was hybridized (data not shown). Notably, however, this RNA-RNA duplex had a higher calculated melting temperature than the RNA-DNA duplexes used in the preceding experiments (Table 2), which may have limited the capacity of ςNS to act on it. Further experiments are warranted before concluding that ςNS can or cannot disrupt dsRNA regions in partially duplex molecules.
DISCUSSION
ςNS binds in multiple copies to single ssRNA molecules, but there are unresolved questions about the nature of these complexes.
Results from the gel mobility shift assays indicate that multiple units of ςNS can bind to a single ssRNA molecule. Such molecules, bound by multiple units of ςNS, may represent the large nucleoprotein complexes obtained from both reovirus-infected mammalian cells (14) and insect cells infected with a recombinant baculovirus expressing ςNS (10). By binding in multiple units to the viral plus-strand RNAs (minus-strand RNAs are not released from template plus strands during reovirus replication [1, 31]), ςNS may provide multiple sites for interactions with other viral or cellular proteins that may play roles in translation, packaging, or minus-strand synthesis. Questions remaining to be answered include whether the multiple units of ςNS that bind to an RNA molecule stretch out like beads on a necklace or self-associate to form a more compact structure.
The positive cooperativity demonstrated for ςNS binding to ssRNA may arise either from changes in RNA conformation upon ςNS binding that promote subsequent ςNS units to bind more readily or from ςNS-ςNS interactions, similar to the case for poliovirus RNA polymerase (4, 26). Certain observations are consistent with each possibility. Data indicating that ςNS destabilizes short regions of a nucleic acid duplex and thereby alters the RNA conformation are consistent with an RNA-based explanation for cooperativity. On the other hand, evidence that ςNS oligomers might self-associate to form higher-order multimers under certain conditions (Fig. 1B) suggests that positive cooperativity may arise from such ςNS-ςNS interactions. Additional analyses of ςNS-RNA and ςNS-ςNS interactions are needed to establish the basis of the observed cooperativity.
One curious feature of the results is that saturative binding occurred at between 12 and 24 pmol of ςNS for each RNA, despite their different sizes (Fig. 2A). Assuming that ςNS was present in limiting amounts but not limiting concentrations in these experiments, we would expect saturation of the 211-nucleotide RNA to have required approximately twice the amount of ςNS as saturation of the 121-nucleotide RNA. However, since the interval between amounts of added ςNS in the saturation range was large (twofold), the experimental findings are reasonable. For example, if the 121-nucleotide RNA was saturatively bound by five units of ςNS when 15 pmol of ςNS was added in this assay, then the 211-nucleotide RNA should have been saturatively bound by eight units of ςNS when 24 pmol of ςNS was added. This is consistent with the data in Fig. 2A.
Another interesting feature of the results is that a faint ladder of higher-mobility complexes was seen even when saturative amounts of ςNS were added (Fig. 1A and 2). These complexes showed mobilities distinct from the major ones seen with smaller amounts of ςNS (Fig. 1A and 2A). Moreover, the higher-mobility complexes seen with saturative amounts of ςNS showed the same mobilities regardless of the size of added RNA and also comigrated with the saturated complexes of each smaller added RNA over the range at which those were analyzed (Fig. 2B). There are several possible explanations for these observations, but by far the most likely would seem to be that these higher-mobility complexes represent small amounts of partially degraded RNA molecules that were present in each sample and to which ςNS was also saturatively bound. Enumeration of these complexes in Fig. 2B lends support to the conclusion that each ςNS binding unit covers about 25 nucleotides of RNA.
What are the unit of ςNS that binds to ssRNA and the number of nucleotides of RNA that each unit contacts?
The purified ςNS protein migrates as 7- to 9S complexes in 5 to 20% sucrose gradients (10), consistent with oligomers containing three to six 41-kDa monomers of ςNS. As stated earlier, whether these complexes represent one discrete type of ςNS oligomer or a mixture and whether they can self-associate to form higher-order multimers in the absence of RNA remain unknown. We nonetheless hypothesize that one of these oligomers or multimers represents the unit of ςNS that binds to ssRNA in a number of copies dependent on RNA length. The molar ratio of ςNS to RNA at saturation in this study may indicate the number of ςNS subunits in the binding unit, that is, if the concentrations of ςNS are not limiting and if most or all of the added ςNS is competent for RNA binding. In this case, we can calculate that saturative binding was achieved with ςNS in 60- to 120-fold molar excess to each RNA in Fig. 2A (12 to 24 pmol of ςNS for about 0.2 pmol of RNA). Considering that only five to eight units of ςNS were bound to each RNA in the 121- to 211-nucleotide range at saturation, we can further calculate that each binding unit contained 8 to 24 subunits of ςNS. If the RNA-free, 7- to 9S ςNS oligomer is a tetramer, for example, this suggests that a multimer comprising two to six tetramers constitutes each binding unit. Given that some of the purified ςNS in our preparations might not have been competent for RNA binding, we recognize that these numbers indicate an upper limit for the size of the binding unit. It remains possible that each binding unit comprises one copy of a discrete type of ςNS oligomer that sediments in the 7- to 9S range in the absence of RNA (10).
The finding that each binding unit of ςNS covers about 25 nucleotides of RNA is consistent with previous work demonstrating that ςNS protects 20 to 40 nucleotides of reovirus ssRNAs from RNase T1 degradation (34). However, it is important to distinguish this 25-nucleotide region of ssRNA covered by ςNS from the number of nucleotides that constitutes the minimum or preferred size of binding site for each unit of ςNS. No experiments in this report directly address the latter, but further analyses of ςNS binding to different-size RNAs might be used to establish this number. For example, if a 140-nucleotide RNA binds five ςNS units covering 25 nucleotides each, are the 15 “extra” nucleotides enough to bind a sixth ςNS unit?
No evidence for sequence specificity in ςNS binding to ssRNA.
Since reovirus replication is cytoplasmic (reviewed in reference 24), there is no selective advantage for ςNS to have a strong preference for binding ssRNA over ssDNA. Accordingly, we found that ςNS binds to ssRNA with only slightly greater efficiency than to ssDNA. Our evidence that neither dsRNA nor dsDNA is an efficient competitor for ςNS binding to ssRNA is consistent with previous findings (28), but our further evidence for strand displacement from short duplex regions is novel (see below).
Although previous work suggested that ςNS can bind nonreovirus RNA (14, 28), this study provides the first evidence that nonreovirus RNA efficiently competes with reovirus RNA sequences for binding. Previous authors suggested that ςNS binds specifically to the 3′ end of the reovirus plus-strand RNAs (34). Our findings suggest that the reason reovirus 3′ RNA ends were protected from nuclease degradation in the previous study is that ςNS protects many regions along the length of the RNAs, including the 3′ ends. We have in fact demonstrated that ςNS binding to ssRNA partially protects it from in vitro digestion with RNase A (data not shown). Since our data indicate that ςNS does not bind specifically to the short conserved sequences at the 5′ and 3′ ends of all reovirus plus-strand RNAs (2), we hypothesize that ςNS is not involved in recognizing these sequences during packaging or minus-strand synthesis. Thus, these sequences are likely to be recognized by other viral or cellular proteins.
ςNS displaces DNA oligonucleotides from RNA-DNA hybrids but is not a classical helicase.
Since the strand displacement activity of ςNS bound to RNA-DNA hybrids occurred independently of ATP hydrolysis and was inhibited by MgCl2 but not by EDTA, the data strongly suggest that ςNS is not a classical, ATP-dependent helicase. Consistent with this conclusion is the fact that ςNS lacks the nucleoside triphosphate-binding motifs characteristic of those enzymes (16). Instead, it appears most likely that ςNS melts duplex regions as it completes its cooperative and saturative binding to a primarily ssRNA molecule, with the energy for melting provided by the binding event(s).
From the data in this study, we hypothesize that only short duplex regions, with low thermodynamic stability, are subject to melting by ςNS (Table 2). We favor this explanation for why unwinding activity was not demonstrated by ςNS on the one type of partially duplex RNA-RNA molecule that we examined in this study. Rather than concluding that ςNS is inactive at melting any RNA-RNA duplexes, we propose that the duplex we analyzed was simply too stable to be unwound by ςNS. In future work, we will more systematically test for the potential RNA-melting activity of ςNS by using molecules containing shorter duplex regions with lower predicted melting temperatures. Such duplexes may be more relevant to reovirus replication in any case since regions of continuous duplex shorter than 17 bp are much more likely to form within the reovirus plus strands according to RNA-folding predictions (J.-Y. Sgro and M. L. Nibert, unpublished data). We will also examine molecules having shorter duplex regions at the 5′ end, the middle, or the 3′ end of the longer RNA strand in order to increase our chances of seeing a specific type of strand displacement activity. If RNA-RNA melting activity can be shown for ςNS in vitro, then its role in reovirus-infected cells may be in melting short regions of intra- or intermolecular secondary structure within the viral plus-strand RNAs, regions which might otherwise interfere with the use of the RNAs in packaging and/or minus-strand synthesis during formation of progeny virions. ςNS almost certainly does not destabilize the reovirus dsRNA genome for transcription, since this process occurs within the inner capsid of assembled reovirus particles (6, 33), where ςNS is not packaged.
Similarities between ςNS and ssDNA binding proteins involved in dsDNA replication.
The adenovirus DNA binding protein Ad-DBP (21, 42), the herpes simplex virus type 1 protein ICP8 (5), and the Epstein-Barr virus ssDNA-binding protein BALF-2 (38) are all similar to ςNS in that they can destabilize regions of a nucleic acid duplex independently of ATP hydrolysis and in a manner that is inhibited by MgCl2 (5, 21, 38, 42). In addition to ATP-independent strand displacement activity, Ad-DBP and ICP8 share the following properties with ςNS: (i) binding in multiple units to a nucleic acid molecule in a sequence-independent manner; (ii) protection of the single-stranded nucleic acid from nuclease degradation (data for ςNS are not shown in this paper), (iii) strong preference for single-stranded over double-stranded nucleic acids; and (iv) positive cooperativity for nucleic acid binding (reviewed in references 7 and 20). These and other ssDNA binding proteins have been shown to be involved in dsDNA replication. For example, both in vitro and in vivo studies have shown that Ad-DBP is required for elongation of ssDNA by the adenovirus polymerase (7, 20), possibly involving the ATP-independent helix-destabilizing properties of Ad-DBP (21, 42). Two prokaryotic ssDNA binding proteins, E. coli SSB and bacteriophage T4 gene 32 protein (gp32), are also required for dsDNA replication and share nucleic acid-binding properties with the noted DNA animal virus proteins and ςNS but cannot destabilize DNA duplexes (7). Of course, a clear difference between ςNS and ssDNA binding proteins is the nucleic acid substrate for genome replication (RNA versus DNA). Nonetheless, considering the large number of similarities, we propose that ςNS plays a role in genome replication similar to that played by the ssDNA binding proteins. Specifically, ςNS may bind to the reovirus plus-strand RNAs and stabilize them in a conformation that allows the reovirus RNA polymerase and other potential cofactors to mediate minus-strand synthesis to produce the dsRNA gene segments. Some secondary structures in the plus-strand RNAs may be unwound directly by ςNS (similarly to Ad-DBP [21, 42]), whereas others may need to be unwound by another viral or cellular protein and then stabilized in the single-stranded state by ςNS binding (similarly to gp32 [7]). Studies to address interactions among ςNS, the polymerase, and other proteins should provide further insight into the potential role of ςNS in reovirus genome replication.
Similarities to the nonstructural proteins of other Reoviridae members.
Other members of the virus family Reoviridae, all of which have dsRNA genomes, also encode nonstructural proteins with ssRNA-binding activity. For instance, the bluetongue virus (orbivirus) nonstructural protein NS2 (41 kDa) binds ssRNA and forms 7S complexes (39). Similarly, the rotavirus nonstructural protein NSP2 (35 kDa) has sequence-independent ssRNA-binding activity (18), forms 10S multimers (17), and interacts with the rotavirus RNA polymerase VP1 (17). Recent data indicate that the binding activities of NSP2 are in fact very similar to those of ςNS shown in this study, including binding to single ssRNA molecules in multiple copies, with positive cooperativity, with little or no specificity for rotavirus sequences, and with substantially greater affinity for ssRNA than dsRNA (37). NSP2 has not yet been reported to destabilize nucleic acid duplexes, as was ςNS in this study, but was reported to have nucleoside triphosphatase and autophosphorylation activities (37). The latter have not been demonstrated for ςNS (Fig. 6). It remains to be determined whether reovirus ςNS, rotavirus NSP2, bluetongue virus NS2, and similar nonstructural proteins from other Reoviridae members play strictly analogous roles in the replication cycles of their respective viruses.
ACKNOWLEDGMENTS
This work was supported by NIH research grants AI-39533 (M.L.N.) and AI-32139 (L.A.S.), by ACS grant RPG-98-12701-MBC (L.A.S.), by a Hatch grant from USDA funds awarded to the College of Agricultural and Life Sciences (M.L.N.), and by a grant from the Lucille P. Markey Charitable Trust to the Institute for Molecular Virology (M.L.N.). A.L.G. received additional support as a Department of Biochemistry Steenbock Fellow. S.C.S. was supported by NIH Medical Scientist Training Program grant GM-08244 and by NIH Microbiology/Cancer Research training grant CA-09138. J.L. received additional support as a University of Wisconsin/Hilldale Undergraduate/Faculty Research Fellow. M.L.N. received additional support as a Shaw Scientist from the Milwaukee Foundation.
We thank T. Broering, D. Farsetta, and C. Luongo for useful discussions and comments on the manuscript. We especially acknowledge T. Baumstark and P. Ahlquist for advice on work with RNA, T. Broering for assistance in determining melting temperatures for the RNA-DNA and RNA-RNA duplexes, and S. Melcher for help with the Hill plots. Assistance in producing the S4 clone was provided by M. Moses and K. Thoemke. We also thank M. Chute, S. J. Harrison, J. Lugus, K. Thoemke, and X. Zhou for technical support and other members of our laboratories for helpful discussions.
REFERENCES
- 1.Acs G, Klett H, Schonberg M, Christman J, Levin D H, Silverstein S C. Mechanism of reovirus double-stranded ribonucleic acid synthesis in vivo and in vitro. J Virol. 1971;8:684–689. doi: 10.1128/jvi.8.5.684-689.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antczak J B, Chmelo R, Pickup D J, Joklik W K. Sequence at both termini of the 10 genes of reovirus serotype 3 (strain Dearing) Virology. 1982;121:307–319. doi: 10.1016/0042-6822(82)90170-2. [DOI] [PubMed] [Google Scholar]
- 3.Antczak J B, Joklik W K. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology. 1992;187:760–776. doi: 10.1016/0042-6822(92)90478-8. [DOI] [PubMed] [Google Scholar]
- 4.Beckman M T L, Kirkegaard K. Site size of cooperative single-stranded RNA binding by poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1998;273:6724–6739. doi: 10.1074/jbc.273.12.6724. [DOI] [PubMed] [Google Scholar]
- 5.Boehmer P E, Lehman I R. Herpes simplex virus type 1 ICP8: helix-destabilizing properties. J Virol. 1993;67:711–715. doi: 10.1128/jvi.67.2.711-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borsa J, Graham A F. Reovirus: RNA polymerase activity in purified virions. Biochem Biophys Res Commun. 1968;33:895–901. doi: 10.1016/0006-291x(68)90396-3. [DOI] [PubMed] [Google Scholar]
- 7.Chase J W, Williams K R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 8.Cormack B. Mutagenesis by the polymerase chain reaction. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1991. pp. 8.5.1–8.5.9. [Google Scholar]
- 9.Cross R K, Fields B N. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of viral RNA. Virology. 1972;50:799–809. doi: 10.1016/0042-6822(72)90434-5. [DOI] [PubMed] [Google Scholar]
- 10.Gillian A L, Nibert M L. Amino terminus of reovirus nonstructural protein ςNS is important for ssRNA binding and nucleoprotein complex formation. Virology. 1998;240:1–11. doi: 10.1006/viro.1997.8905. [DOI] [PubMed] [Google Scholar]
- 11.Gillian-Daniel D L, Gray N K, Astrom J, Barkoff A, Wickens M. Modifications of the 5′ cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol Cell Biol. 1998;18:6152–6163. doi: 10.1128/mcb.18.10.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomatos P J, Prakash O, Stamatos N M. Small reovirus particle composed solely of ςNS with specificity for binding different nucleic acids. J Virol. 1981;39:115–124. doi: 10.1128/jvi.39.1.115-124.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomatos P J, Stamatos N M, Sarkar N H. Small reovirus-specific particle with polycytidylate-dependent RNA polymerase activity. J Virol. 1980;36:556–565. doi: 10.1128/jvi.36.2.556-565.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huismans H, Joklik W K. Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology. 1976;70:411–424. doi: 10.1016/0042-6822(76)90282-8. [DOI] [PubMed] [Google Scholar]
- 15.Ito Y, Joklik W K. Temperature-sensitive mutants of reovirus. I. Patterns of gene expression by mutants of groups C, D, and E. Virology. 1972;50:189–201. doi: 10.1016/0042-6822(72)90359-5. [DOI] [PubMed] [Google Scholar]
- 16.Kadare G, Haenni A-L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kattoura M D, Chen X, Patton J T. The rotavirus RNA-binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral RNA polymerase. Virology. 1994;202:803–813. doi: 10.1006/viro.1994.1402. [DOI] [PubMed] [Google Scholar]
- 18.Kattoura M D, Clapp L L, Patton J T. The rotavirus non-structural protein, NS35, possesses RNA-binding activity in vitro and in vivo. Virology. 1992;191:698–708. doi: 10.1016/0042-6822(92)90245-k. [DOI] [PubMed] [Google Scholar]
- 19.Kedl R, Schmechel S, Schiff L. Comparative sequence analysis of the reovirus S4 genes from 13 serotype 1 and serotype 3 field isolates. J Virol. 1995;69:552–559. doi: 10.1128/jvi.69.1.552-559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1991. [Google Scholar]
- 21.Monaghan A, Webster A, Hay R T. Adenovirus DNA binding protein: helix destabilising properties. Nucleic Acids Res. 1994;22:742–748. doi: 10.1093/nar/22.5.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morozov S Y. A possible relationship of reovirus putative RNA polymerase to polymerases of positive-strand RNA viruses. Nucleic Acids Res. 1989;17:5394. doi: 10.1093/nar/17.13.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam S C, Kang C. Transcription initiation site selection and abortive initiation cycling of phage SP6 RNA polymerase. J Biol Chem. 1988;263:18123–18127. [PubMed] [Google Scholar]
- 24.Nibert M L, Schiff L A, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1557–1596. [Google Scholar]
- 25.Noble S, Nibert M L. Characterization of an ATPase activity in reovirus cores and its genetic association with core shell protein λ1. J Virol. 1997;71:2182–2191. doi: 10.1128/jvi.71.3.2182-2191.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pata J D, Schultz S C, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 27.Ramig R F, Mustoe T A, Sharpe A H, Fields B N. A genetic map of reovirus. II. Assignment of the double-stranded RNA-negative mutant groups C, D, and E to genome segments. Virology. 1978;85:531–534. doi: 10.1016/0042-6822(78)90459-2. [DOI] [PubMed] [Google Scholar]
- 28.Richardson M A, Furuichi Y. Synthesis in Escherichia coli of the reovirus nonstructural protein ςNS. J Virol. 1985;56:527–533. doi: 10.1128/jvi.56.2.527-533.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saiki R K, Gelfand D H, Stoffel S, Sharf S J, Hiuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Schonberg M, Silverstein S C, Levin D H, Acs G. Asynchronous synthesis of the complementary strands of the reovirus genome. Proc Natl Acad Sci USA. 1971;68:505–508. doi: 10.1073/pnas.68.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoumacher F, Erny C, Berna A, Godefroy-Colburn T, Stussi-Garaud C. Nucleic acid-binding properties of the alfalfa mosaic virus movement protein produced in yeast. Virology. 1992;188:896–899. doi: 10.1016/0042-6822(92)90549-5. [DOI] [PubMed] [Google Scholar]
- 33.Shatkin A J, Sipe J D. RNA polymerase activity in purified reoviruses. Proc Natl Acad Sci USA. 1968;61:1462–1469. doi: 10.1073/pnas.61.4.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatos N M, Gomatos P J. Binding to selected regions of reovirus mRNAs by a nonstructural reovirus protein. Proc Natl Acad Sci USA. 1982;79:3457–3461. doi: 10.1073/pnas.79.11.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starnes M C, Joklik W K. Reovirus protein lambda 3 is a poly(C)-dependent poly(G) polymerase. Virology. 1993;193:356–366. doi: 10.1006/viro.1993.1132. [DOI] [PubMed] [Google Scholar]
- 36.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 37.Taraporewala Z, Chen D, Patton J T. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J Virol. 1999;73:9934–9943. doi: 10.1128/jvi.73.12.9934-9943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsurumi T, Kishore J, Yokoyama N, Fujita M, Daikoku T, Yamada H, Yamashita Y, Nishiyama Y. Overexpression, purification and helix-destabilizing properties of Epstein-Barr virus ssDNA-binding protein. J Gen Virol. 1998;79:1257–1264. doi: 10.1099/0022-1317-79-5-1257. [DOI] [PubMed] [Google Scholar]
- 39.Uitenweerde J M, Theron J, Stoltz M A, Huismans H. The multimeric nonstructural NS2 proteins of Bluetongue virus, African horsesickness virus, and epizootic hemorrhagic disease virus differ in their single-stranded RNA-binding ability. Virology. 1995;209:624–632. doi: 10.1006/viro.1995.1294. [DOI] [PubMed] [Google Scholar]
- 40.van Holde K E. Physical biochemistry. 2nd ed. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1985. [Google Scholar]
- 41.Xia T, SantaLucia J, Jr, Burkard M E, Kierzek R, Schroeder S J, Jiao X, Cox C, Turner D H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 42.Zijderveld D C, van der Vliet P C. Helix-destabilizing properties of the adenovirus DNA-binding protein. J Virol. 1994;68:1158–1164. doi: 10.1128/jvi.68.2.1158-1164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]