Abstract
Numerous RNA viruses generate subgenomic mRNAs (sgRNAs) for expression of their 3′-proximal genes. A major step in control of viral gene expression is the regulation of sgRNA synthesis by specific promoter elements. We used barley yellow dwarf virus (BYDV) as a model system to study transcriptional control in a virus with multiple sgRNAs. BYDV generates three sgRNAs during infection. The sgRNA1 promoter has been mapped previously to a 98-nucleotide (nt) region which forms two stem-loop structures. It was determined that sgRNA1 is not required for BYDV RNA replication in oat protoplasts. In this study, we show that neither sgRNA2 nor sgRNA3 is required for BYDV RNA replication. The promoters for sgRNA2 and sgRNA3 synthesis were mapped by using deletion mutagenesis. The minimal sgRNA2 promoter is approximately 143 nt long (nt 4810 to 4952) and is located immediately downstream of the putative sgRNA2 start site (nt 4809). The minimal sgRNA3 core promoter is 44 nt long (nt 5345 to 5388), with most of the sequence located downstream of sgRNA3 start site (nt 5348). For both promoters, additional sequences upstream of the start site enhanced sgRNA promoter activity. These promoters contrast to the sgRNA1 promoter, in which almost all of the promoter is located upstream of the transcription initiation site. Comparison of RNA sequences and computer-predicted secondary structures revealed little or no homology between the three sgRNA promoter elements. Thus, a small RNA virus with multiple sgRNAs can have very different subgenomic promoters, which implies a complex system for promoter recognition and regulation of subgenomic RNA synthesis.
Synthesis of subgenomic mRNAs (sgRNAs) is a common strategy used by positive-sense RNA viruses for expression of their 3′-proximal genes. In combination with other strategies such as unconventional translational events and posttranslational proteolytic processing of precursor polyproteins, it allows efficient utilization of the viral genetic material. Synthesized later in infection, sgRNAs encode late viral genes whose products are required for pathogenesis and particle formation. Alphaviruses such as Sindbis virus and the alpha-like multipartite Bromoviridae produce one sgRNA for expression of the coat protein. Plant viruses that belong to such groups as potexvirus, tombusvirus, carmovirus, and tobamovirus, as well as some other alpha-like viruses, produce two or three sgRNAs for expression of the coat protein and movement proteins. RNA viruses with larger genomes, such as Closteroviridae (also alpha-like) and the Nidovirales, produce up to nine and seven sgRNAs, respectively (14, 23).
Several potential mechanisms for sgRNA synthesis have been proposed. However, only de novo internal initiation at a subgenomic promoter has been demonstrated unequivocally (36, 42, 46). A mechanism involving premature termination during minus-strand synthesis followed by replication of the sgRNA has been suggested in red clover necrotic mosaic dianthovirus (RCNMV) (40). Two different versions of a discontinuous transcription mechanism (leader priming and recombination during minus-strand synthesis) have been proposed for Coronaviridae and other members of the order Nidovirales (reviewed in reference 23).
Regardless of the actual mechanism of sgRNA synthesis, cis-acting elements required for transcription have been named subgenomic promoters. Subgenomic promoters have been mapped and characterized in several viruses (3, 5, 12, 17, 21, 28, 43, 45, 46, 51). The best-studied example is the promoter of RNA4 of brome mosaic virus (BMV) (1, 12, 39). The length of the subgenomic promoters, as mapped in vivo, ranges from 24 nucleotides (nt) in Sindbis virus (28) to over 100 nt in beet necrotic yellow vein virus (3). Almost all subgenomic promoters characterized to date, with the exception of that of beet necrotic yellow vein virus (3), are located largely upstream of the transcription start site. A combination of primary RNA sequence and secondary structural elements has been found to be required for sgRNA transcription in vivo (21, 45). In contrast, in vitro experiments with purified BMV replicase showed the importance of the primary RNA sequence, but not the secondary structure, for promoter activity (39).
Genomic locations of transcriptional control elements are not always confined to the areas colinear with the sgRNA 5′ ends, which suggests involvement of long-distance interactions in transcriptional regulation. Such long-distance interactions have been proposed in mouse hepatitis coronavirus (15, 29), potato virus X (19, 33), and tomato bushy stunt virus (52). One of the most unusual types of transcriptional control has been uncovered in the bipartite virus RCNMV where transcription of the sgRNA from RNA1 template is activated by base pairing between regulatory elements in RNAs 1 and 2 (40).
Considering RNA promoters are regions recognized by viral RNA polymerases or associated subunits, it is natural to expect conservation of certain features that determine specificity of this recognition. Indeed, viruses that have more than one sgRNA often contain short stretches of homologous sequence near their transcription initiation sites (reviewed in references 23 and 30). However, these short regions by themselves are insufficient for transcription and serve as parts of larger subgenomic promoters. Turnip crinkle virus (TCV), which has two sgRNAs, contains stable hairpins in both promoters in addition to the conserved GGG sequence at the initiation sites (46). Few data are available on mapping and detailed characterization of subgenomic promoters in other viruses containing multiple sgRNAs. However, based on the notion that the expression of gene products encoded by sgRNAs may be differentially regulated, it is reasonable to predict differences in the promoter structures within one virus.
In this study, we characterize transcriptional control of the three sgRNAs of barley yellow dwarf virus (BYDV). BYDV belongs to the genus Luteovirus of the family Luteoviridae (34). The virus has a 5.7-kb genomic RNA (gRNA) that encodes six open reading frames (ORFs) (Fig. 1A). Only ORFs 1 and 2, which encode viral replication proteins, are translated from the gRNA. SgRNA1 serves as the mRNA for ORFs 3 to 5. ORF3 encodes the 22-kDa coat protein. ORF4 encodes a 17-kDa protein required for plant systemic infection (7). It is translated by a leaky scanning mechanism (10). ORF5 is an extension of the coat protein gene, required for aphid transmission. It is translated by read-through of the ORF3 stop codon (9). ORF6 encodes a highly variable 6.7-kDa protein of unknown function which is expressed via sgRNA2. SgRNA3, at 0.3 kb, is the smallest sgRNA. It does not encode any protein, and its role in the viral life cycle is unclear.
FIG. 1.
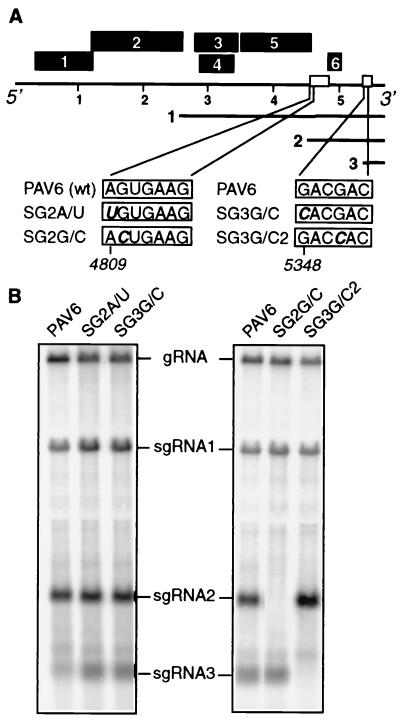
SgRNA2 and sgRNA3 knockout mutagenesis. (A) BYDV genome organization. Black boxes represent ORFs. Nucleotide positions of BYDV gRNA are shown in kilobases. Numbered solid lines represent sgRNAs. Nucleotides at the 5′ end of each sgRNA, according to Kelly et al. (18), are shown in boxes with mutations in boldface and italics. The name of each construct is to the left of each box. (B) Northern blot analysis of total RNA from oat protoplasts infected with the wild-type and mutant transcripts (∼24 hpi). A riboprobe complementary to the 3′ terminal 1.5 kb of BYDV gRNA was used. Left panel shows the wild-type viral RNA (PAV6) and the constructs whose mutations failed to abolish sgRNA synthesis (SG2A/U and SG3G/C). Right panel shows PAV6 and sgRNA2- and sgRNA3-deficient mutants (SG2G/C and SG3G/C2). Bands corresponding to gRNA and sgRNAs are indicated.
The family Luteoviridae contains two major genera, Luteovirus and Polerovirus (32). Members of both genera have high homology in the part of their genomes that contains ORFs 3 to 5. The 5′ halves, which contain RNA-dependent RNA polymerase (RdRp) genes, are as divergent as any known RdRp's: the Polerovirus RdRp belongs to supergroup 1, and the Luteovirus replicase belongs to supergroup 2 (22). Such genomic organization implies occurrence of a recombination event in the evolution of the family. The putative crossover sites are located at the regions corresponding to the 5′ ends of sgRNA1 and sgRNA2 of BYDV (35). Characterization of the promoters may contribute to the understanding of mechanisms of recombination.
We have previously characterized the sgRNA1 promoter and mapped it to a 98-nt region with the majority of the sequence (75 nt) located upstream of the transcription start site (21). We have also shown that the promoter folds into two stem-loops and that both RNA sequence and secondary structural elements are important for its activity.
We proposed recently that sgRNA2 plays a role in translational regulation of BYDV gene expression (47). Unlike the majority of eukaryotic mRNAs, BYDV genomic RNA is uncapped (2). To compensate for the lack of the 5′ cap, a cis element in a 3′ intergenic region mediates its cap-independent translation (48, 49). This 3′ translational enhancer (3′TE) located between nt 4810 and 4920 is indispensable for virus replication (2). In vitro, sgRNA2, which contains the 3′TE in its 5′ untranslated region (UTR), strongly inhibits translation of the gRNA in trans but only weakly inhibits translation of sgRNA1 (49). Thus, we suggest that sgRNA2, which can accumulate to a 20- to 40-fold molar excess over gRNA, mediates a switch from early (replicase) to late (coat protein) gene expression (47). Understanding sgRNA2 transcriptional regulation will help us test this model. Here, we report characterization of the sgRNA2 and sgRNA3 promoters of BYDV and compare them to the sgRNA1 promoter. We demonstrate that the three subgenomic promoters of BYDV have very limited sequence or structural resemblance.
MATERIALS AND METHODS
Plasmids.
The full-length infectious clone of BYDV-PAV, pPAV6 (8), was used to develop mutant constructs. All mutants were confirmed by sequencing. To make sgRNA2 knockout mutants (SG2A/U and SG2G/C), pPAV6 was PCR amplified by using downstream mutagenic primers (SG2A/T and SG2G/C, respectively) and the upstream primer CB0416 (Table 1). The product was digested with KpnI and BamHI. This was subcloned into pSG1 (47), containing a region corresponding to sgRNA1 of BYDV that had been cut with KpnI and BamHI. The resulting plasmids were digested with KpnI and SmaI. The fragments corresponding to the 3′ region of BYDV were purified by 0.8% low-melt agarose gel electrophoresis and were subcloned into pPAV6 cut with the same enzymes. SgRNA3 knockout mutants (SG3G/C and SG3G/C2) were constructed by two-step PCR (26). In the first step, a region of pPAV6 was amplified by using the upstream mutagenic primer (SG3G/C and SG3G/C2) and the downstream primer, 3′wt (Table 1). The gel-purified product was used as a downstream primer in PCR with the upstream primer CB0416. The resulting product was digested with KpnI and SmaI and was subcloned into pPAV6 cut with the same enzymes.
TABLE 1.
Primers used in this study
| Experiment | Primer name | Primer sequencea |
|---|---|---|
| SgRNA knockout | SG2A/T | CCCAGGATCCGATTTGTGCTAGTGGTGTTGTCTTCACAGGAATTGCGCCTTGTA |
| SG2G/C | CCCAGGATCCGATTTGTGCTAGTGGTGTTGTCTTCACTGGAATTGCGCCTTGTA | |
| SG3G/C | CCACGACCTGGTACAAGT | |
| SG3G/C2 | GAAGACGTTAAAACTCGACCACCTGGTACAAGTCGT | |
| 3′wt | ATACCCCGGGTTGCCGAA | |
| CB0416 | GGTCTAGATAATACGACTCACTATAGGGTACACAAACAAGCGAAT | |
| SgRNA2 promoter deletion | 4620D | CCTGAAGACGTACCTCCAAT_AAAGAAGAACCCCCA |
| 4686D | TCCACGCTACTCTATGAAAG_CTACTCCCACTGCC | |
| 4706D | GGCAACTTTTTATCCAGACT_GTGTCAACTACTTCAAACAT | |
| 4729D | CCAGACTTGTAGAAGCGA_AAGGGAGCAGCTCCG | |
| 4763D | ACTCCCACTGCCCCATCC_AATTCCAGCGGAATC | |
| 4790D | CAAACATGACAAGGGAGC_GCGTACAAGGCGCAA | |
| 4810D | CCGGGAGTACACTAGGATT_GTGAAGACAACACCAC | |
| 4811D | CCGGGAGTACACTAGGATT_TGAAGACAACACCACTA | |
| P018 | GCCTGTTTCCCAGGATCCTATAGTGAGTCGTATTACA | |
| Promoter duplication | 4620 | ATAGGTACCAAAGAAGAACCCCA |
| 4686 | ATAGGTACCCTACTCCCACTGCC | |
| 4810 | ATAGGTACCGTGAAGACAACACCACTA | |
| 4900 | ATAGGTACCACGGCGGTAGGTTG | |
| 4922 | ATAGGTACCCATCGGCCAAACACAATA | |
| 4952 | ATAGGTACCAATACAAACGGCGA | |
| 5249 | ATAGGTACCAAGGCTATCCCACC | |
| 5293 | ATAGGTACCAGTGGGTGACTTCG | |
| 5319 | ATAGGTACCGATCGTCAGGATTGA | |
| 5330 | ATAGGTACCTTGAAGACGTTAAACTC | |
| 5345 | ATAGGTACCCTCGACGACCTGGTACAA | |
| 5373 | ATAGGTACCAGTTTAACGACTTGTACCA | |
| 5388 | ATAGGTACCGTATCCACCCGAGTC | |
| 5402 | ATAGGTACCGGGCCGGGTGTGGTG | |
| 5432 | ATAGGTACCCGTTTCGTATCGTG |
Altered bases are in boldface, deletions are shown as underlined spaces, and KpnI recognition sites are underlined.
The PAVDSGP2-(1-8) deletion series was constructed by using two-step PCR (26). In the first step, a region of pPAV6 was amplified by a mutagenic upstream primer (4620D, 4686D, 4706D, 4729D, 4763D, 4790D, 4810D, or 4811D) and a downstream primer, P018 (Table 1). The product was gel purified and used as a downstream primer in the second-step PCR with the upstream primer CB0416. The product of the second-step PCR was digested with KpnI and BamHI, was gel purified, and was subcloned into pSG1 cut with the same enzymes. The resulting plasmid was cut with KpnI and SmaI, and the fragment containing the deletion mutation was gel purified and subcloned into pPAV6 cut with the same enzymes.
For sgRNA2 and sgRNA3 promoter mapping by duplication in the KpnI site of PAV6, promoter regions were PCR amplified with primers listed in Table 1, which contained flanking KpnI sites. The products were digested with KpnI and were subcloned into pPAV6 cut with KpnI except constructs SGP2BG/C, SGP2D, SGP2E, and SGP2E-BF, which were subcloned into the KpnI site of SG2G/C.
Protoplast infection and Northern blot analysis.
Oat protoplasts were prepared and electroporated with infectious transcripts essentially as previously described (10). Infectious transcripts were prepared by using the Megascript T7 RNA in vitro transcription system (Ambion, Austin, Tex.). Ten to fifteen micrograms of RNA was used for electroporation. Total RNA was extracted from protoplasts ∼24 h postinoculation (hpi) by using RNeasy plant RNA isolation kit (QIAGEN, Los Angeles, Calif.). RNA (5 to 10 μg) was analyzed by Northern blot hybridization essentially as previously described (38). A 32P-labeled riboprobe complementary to the 3′ terminus of BYDV was used to detect viral gRNA and sgRNAs. For positive-strand detection, the plasmid pSP10 (9) was linearized with HindIII and was used in an in vitro transcription reaction with T7 RNA polymerase. GeneScreen nylon membranes (Dupont) were hybridized with the probes and exposed to PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) screens for 5 to 24 h. Bands on blots were quantitated by using ImageQuant 4.2 (Molecular Dynamics). Identical rectangles were placed over each band and over a “band-free” region in the lane below each band. The latter was defined as background and subtracted from the counts obtained in the rectangle on the band of interest. After background subtraction, average counts per unit area were normalized for the length of the RNA being detected to obtain the values used to calculate molar ratios indicated in the figures.
RNA sequence and structure analysis.
Sequence alignments of BYDV isolates were performed with GCG software. RNA secondary structure predictions were carried out by using the MFOLD program, version 3.0, at the MFOLD website (http://mfold2.wustl.edu/∼mfold/rna/form1.cgi) (31, 53).
RESULTS
SgRNA2 and sgRNA3 are not required for virus replication in oat protoplasts.
In our previous study, we showed that sgRNA1, which encodes the coat protein, is not required for viral RNA replication in oat protoplasts (21). To determine the roles of sgRNA2 and sgRNA3 in BYDV replication, we developed mutants defective in their synthesis. In order to make sgRNA2- and sgRNA3-deficient mutants, we changed their initiation sites in the full-length viral infectious transcript PAV6 (8). The 5′ ends of sgRNAs 2 and 3 were previously mapped to nt 4809 and 5348, respectively (18). By using site-directed mutagenesis, we changed the sgRNA2 5′-terminal nucleotide A to a U (mutant SG2A/U) and the sgRNA3 5′-terminal nucleotide G to a C (mutant SG3G/C) (Fig. 1A). Northern blot analysis of the total RNA from infected protoplasts showed, surprisingly, that neither of these mutations had any effect on the accumulation of these sgRNAs (Fig. 1B).
SgRNAs 1 and 2, as well as the genomic RNA of BYDV, share a conserved hexanucleotide, GUGAAG, at their 5′ ends (18), and sgRNA1 starts at the first G of this hexanucleotide, whereas sgRNA2 start was mapped to an A 1 nt upstream of the conserved G (18). By analogy with sgRNA1, we mutated the first G of the conserved hexanucleotide at position 4810 to a C in the mutant SG2G/C (Fig. 1A). The sgRNA3 5′ terminus lacks the conserved hexanucleotide. In an attempt to make an sgRNA3-deficient mutant, we changed the nearest G, at position 5351, to a C in mutant SG3G/C2. Both mutants failed to produce their respective sgRNAs (Fig. 1B). It is possible that the essential G residues at nt 4810 and 5351 are the actual 5′ termini of sgRNAs 2 and 3, respectively. However, the mapping data published previously support 5′ ends at 4809 and 5348, respectively (18). None of the mutations that knocked out sgRNA synthesis negatively affected viral replication (Fig. 1B), indicating that neither sgRNA2 nor sgRNA3 is required for virus RNA replication in protoplasts.
Mapping the boundaries of the sgRNA2 promoter.
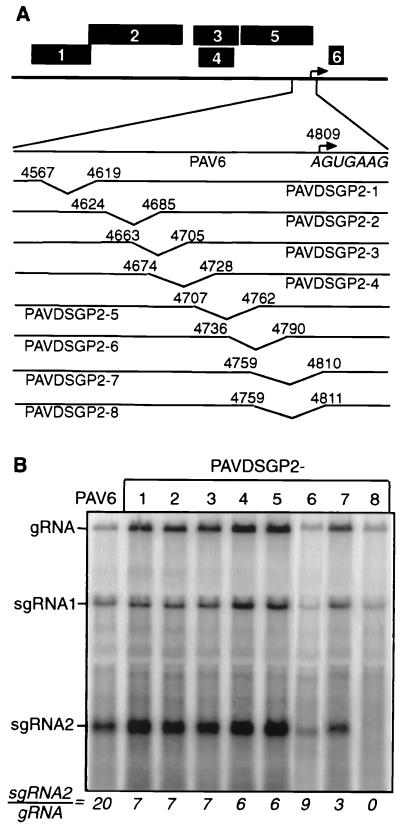
We began mapping the sgRNA2 promoter 5′ boundary by using deletion mutagenesis of the sequence upstream of the start site (Fig. 2A). Wild-type transcript (PAV6) gave an sgRNA2-gRNA ratio of 20:1. Surprisingly, each deletion, except PAVDSGP2-7 and PAVDSGP2-8, reduced the sgRNA2-gRNA ratio only two- to threefold (Fig. 2B). Mutant PAVDSGP2-7, in which A4809 is replaced with a U, reduced sgRNA2-gRNA to 3:1. Mutant PAVDSGP2-8, which has just one more base deleted (G4810 replaced by U) yielded no sgRNA2 (Fig. 2B). These data confirmed the results of the previous experiment, which indicated that the 5′-terminal base of sgRNA2 tolerates changes or that sgRNA2 does not initiate at position 4809 (Fig. 1B). The entire set of deletions indicates that no particular RNA sequence upstream of the sgRNA2 5′ end is essential for sgRNA2 synthesis.
FIG. 2.
Deletion mapping of the 5′ border of sgRNA2 promoter. (A) Maps of the constructs (PAVDSGP2-1 through PAVDSGP2-8) used in the experiment. Location of the sgRNA2 start site, as reported by Kelly et al. (18), is indicated by an arrow. The 5′-terminal sequence of sgRNA2 is shown in italics underneath. Sites of deletions are indicated by the angled lines between designated nucleotide positions at deletion boundaries. (B) Northern blot analysis of total RNA from oat protoplasts infected with the wild-type and deletion mutant transcripts (∼24 hpi). Bands were quantitated with ImageQuant 3.0, and the ratio of sgRNA2-gRNA is shown below each lane. The PAVDSGP2-6 lane is from a different experiment than the others.
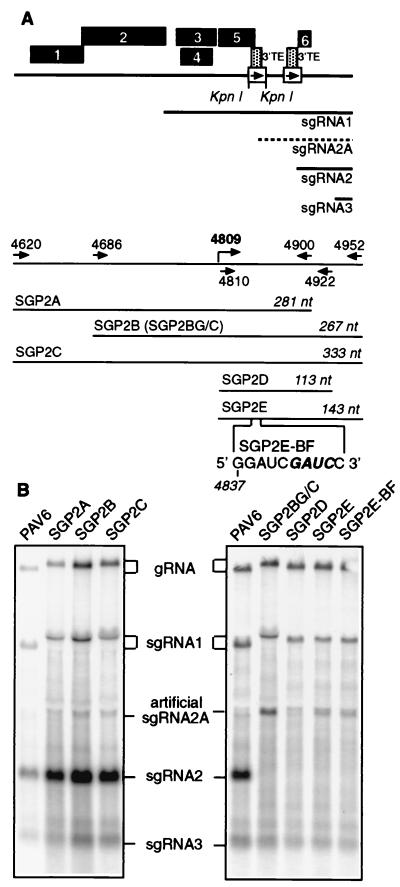
The 5′ UTR of sgRNA2 coincides with the 3′TE (nt 4809 to 4920); therefore, the promoter may overlap it. Thus, mutations in the subgenomic promoter may knock out 3′TE function, which would be lethal (2). To avoid this problem, we duplicated the sgRNA2 promoter in the unique KpnI site (nt 4154) of the BYDV genome in ORF5 (Fig. 3A), as we did previously for sgRNA1 promoter mapping (21). ORF5 is not required for virus replication in protoplasts (37). This ectopic expression of sgRNA has also been done in other viruses (3, 5, 12, 27, 43, 46). Mutants SGP2B and SGP2C, which contain 143 nt downstream of the transcription start site (nt 4810), duplicated in the KpnI site, produced small amounts of an artificial sgRNA of the expected size (sgRNA2A) (Fig. 3B). Mutant SGP2A, which contained only 91 nt downstream of the start site, gave no sgRNA (Fig. 3B).
FIG. 3.
Mapping sgRNA2 promoter by ectopic expression. (A) Map of the constructs that contain duplicated copies of the sgRNA2 promoter (box with arrow) in the KpnI site which is duplicated in the cloning process. The gray box represents the 3′TE. The artificial sgRNA2A is indicated by the dashed line. Arrows with nucleotide positions indicate PCR primers used to amplify promoter regions for ectopic expression. Lengths of the duplicated regions are shown in italics. SGP2E-BF contains the same sgRNA2 promoter region duplication as does SGP2E, but with the BamHI fill-in mutation (49) (boldface italics). (B) Northern blot analysis of total RNA from oat protoplasts (∼24 hpi) infected with the wild-type and mutant transcripts. Left panel shows PAV6 and the constructs with sgRNA2 promoter duplicated in the KpnI site of PAV6. Right panel shows PAV6 and the constructs with sgRNA2 promoter duplicated in the KpnI site of SG2G/C, a mutant deficient in sgRNA2 synthesis. SgRNA2A is not to be confused with an apparent faint band that migrates just below the 18S rRNA shadow. Ratios of sgRNA2A:gRNA are 3.5:1 for SGP2BG/C, 0:1 for SGP2D, and 1:1 for SGP2E and SGP2E-BF.
The amount of the artificial sgRNA2A synthesized by the mutants SGP2B and SGP2C was very low (Fig. 3B). Previous studies of the BMV subgenomic promoter (12) and the sgRNA1 promoter of BYDV (21) showed that downstream sgRNA promoters are preferentially used, and they reduce expression from those located upstream. To increase transcription from the ectopic (upstream) promoter, we subcloned it into the mutant SG2G/C that does not produce sgRNA2 in its natural location (Fig. 1A). Indeed, the same 267-nt region used in mutant SGP2B resulted in a much higher level of artificial sgRNA2A transcription in the context of SG2G/C (Fig. 3B, mutant SG2BG/C). To map the 3′ boundary of the promoter, we tested two additional mutants that contained the duplicated sgRNA2 promoter region extending from the essential G4810 to positions 4922 and 4952, respectively (Fig. 3A SGP2D and SGP2E). The ratios of sgRNA2A to gRNA revealed that SGP2E produced about one-third as much sgRNA2A as SGP2B/C, whereas SGP2D produced no detectable sgRNA2A (Fig. 3B, legend). This defines the 3′ promoter boundary to a region between nt 4922 and 4952 (Fig. 3B). Thus, we mapped the minimal sequence required for sgRNA2 synthesis to the 143 nt located between nt 4810 and 4952 of the BYDV genomic RNA. SGP2BG/C, which includes 120 nt upstream, appears to contain an enhancer sequence that provides the threefold increase in sgRNA2A accumulation relative to SGP2E. However, it is clear that the region from nt 4810 to 4952 is sufficient for transcription of significant levels of sgRNA. Moreover, this includes an essential sequence located between nt 4922 and 4952, over 100 nt downstream of the start site.
To test whether the 3′TE and sgRNA2 promoter are functionally connected, we introduced a 4-nt duplication (GAUC) into the BamHI site within the 3′TE (Fig. 3, SGP2E-BF), in the sgRNA2 promoter construct in the KpnI site. This mutation was shown previously to abolish the function of the 3′TE (49). This mutation had no effect on the synthesis of artificial sgRNA2A. This indicates that the sgRNA2 promoter does not require a functional 3′TE sequence, suggesting that they function independently of each other (Fig. 3B).
Mapping the sgRNA3 promoter.
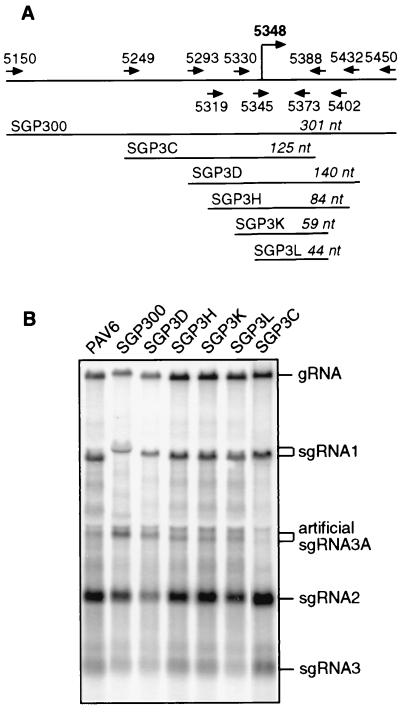
A mutant of PAV6 with a 100-nt deletion spanning the sgRNA3 start site failed to replicate in protoplasts (C. Paul and W. A. Miller, unpublished data). This made it difficult to map the sgRNA3 promoter in its original location by deletion mutagenesis. Therefore, as with sgRNA 1 and 2 promoters, we duplicated the sgRNA3 promoter and inserted it in the KpnI site in ORF5. Unfortunately, a band migrating just below the 18S rRNA front may confuse interpretation of the gel (Fig. 4B). However, the sgRNA3A bands are clearly discernible by the slight variations in mobility that correspond precisely to the size predicted by the insert size in the KpnI site. Construct SGP3L, which contains only 44 nt from the sgRNA3 promoter region (nt 5345 to 5388), yielded 15% as much sgRNA3A as SGP300, which contains a 300-nt insert, from bases 5150 to 5450 (Fig. 4B). The very weak band in the SGP3C lane is not significantly above background. Sequences upstream of the start site have a greater stimulatory effect than those upstream of the sgRNA2 promoter (Fig. 4B, compare SGP300 and SGP3L). Although the ends of the sgRNA2 and sgRNA3 promoters have not been mapped precisely, it is clear that the essential cores of both promoters, that are able to generate at least a low level of sgRNA, are located downstream of the transcription start site.
FIG. 4.
Mapping sgRNA3 promoter by ectopic expression. (A) Map of the promoter regions duplicated in the KpnI site of PAV6. Arrows with nucleotide positions indicate PCR primers used to amplify promoter regions for ectopic expression. Lengths of the duplicated regions are shown in italics. The position of the sgRNA3 start site, nt 5348, as reported by Kelly et al. (18), is shown in bold. (B) Northern blot analysis of total RNA from oat protoplasts (∼24 hpi) infected with the wild-type and mutant transcripts. The sgRNA3A bands were distinguished from the faint band that migrates just below the negatively labeled 18S rRNA band by the fact that the mobilities of the sgRNA3A bands varied exactly as expected, decreasing in size as the size of the insert (downstream of nt 5348) decreased. Ratios of sgRNA3A:gRNA are 1.2:1 for SGP300, 0.5:1 for SGP3D, 0.18:1 for SGP3H, 0.15:1 for SGP3K and SGP3L, and 0.06:1 for SGP3C.
BYDV sgRNA promoters have limited sequence homology.
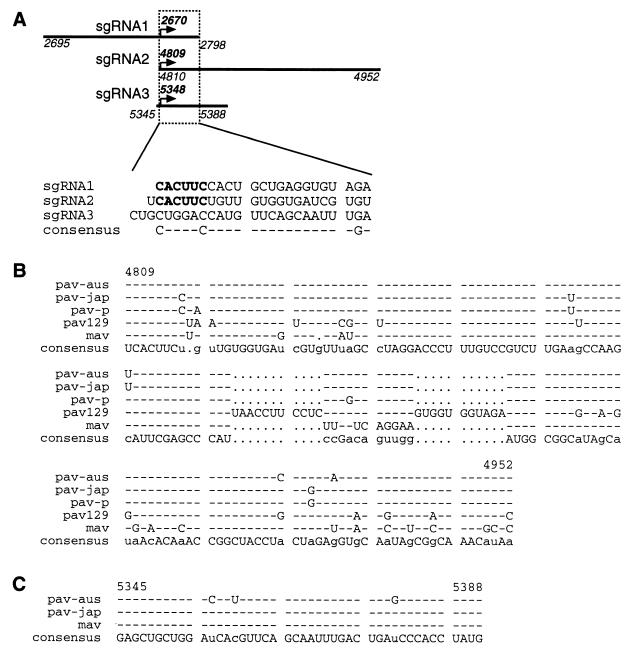
In an attempt to find common elements within sgRNA promoters of BYDV, we analyzed nucleotide sequences of the three subgenomic promoters. We chose sgRNA transcription initiation sites as the start points of our analysis. Because only the sgRNA2 and sgRNA3 promoters are located mostly downstream of the start site, proper alignment of the three promoters was difficult. Surprisingly, besides the conserved hexanucleotide (CUCAAC) in the minus strand of promoters for sgRNAs 1 and 2, no significant sequence homology was found between overlapping regions of the three subgenomic promoters (Fig. 5A).
FIG. 5.
Sequence comparisons of sgRNA promoters. All sequences are negative sense. Numbers that show nucleotide positions refer to the positive sense of BYDV-PAV-Australia gRNA, from which infectious clone PAV6 was derived. (A) Alignment of subgenomic promoter sequences of BYDV. Transcription initiation sites of the three subgenomic promoters are aligned, and only the overlapping regions are shown. The conserved hexanucleotide CACUUC is shown in bold. Dashes indicate the lack of consensus. (B) Alignment of sgRNA2 promoters in different strains of BYDV are as follow: PAV-Japan (pav-jap, accession no. D85783), PAV-Purdue (pav-p, accession no. D11032), PAV-Australia (pav-aus, accession no. X07653), PAV-129 (pav129, accession no. AF218798), and MAV (mav, accession no. D11028). In the consensus sequence, bases conserved in all strains are shown in upper case, those conserved in three or four strains are shown in lower case. Gaps are designated by dots. Bases identical to consensus are indicated by dashes. The 3′TE is located between nt 4810 and 4920. (C) Alignment of sgRNA3 promoters in three strains of BYDV. Strains are designated as in panel B. In the consensus sequence, bases conserved in all three strains are shown in upper case, those conserved in two strains are shown in lower case.
To explore the conservation of sgRNA promoters, we performed sequence alignments of the promoter regions in the negative strands of different strains of BYDV. Alignments of both sgRNA2 and sgRNA3 promoter regions showed significant sequence conservation among various strains of the virus (Fig. 5B and C). Sequences of BYDV-PAV129 and BYDV-MAV sgRNA2 promoters are most divergent, containing stretches of nonhomologous regions. The conservation of the sgRNA2 promoter sequence could be due to conservation of the essential 3′TE function that is contained in its complement (plus strand), rather than (or in addition to) conservation of promoter function.
Interestingly, BYDV-PAV129 lacks any sequence resembling the sgRNA3 promoter. Thus, we were unable to align BYDV-PAV129 sequence with the sgRNA3 promoter regions. It is not known whether BYDV-PAV129 produces sgRNA3.
BYDV sgRNA promoters have different secondary structures.
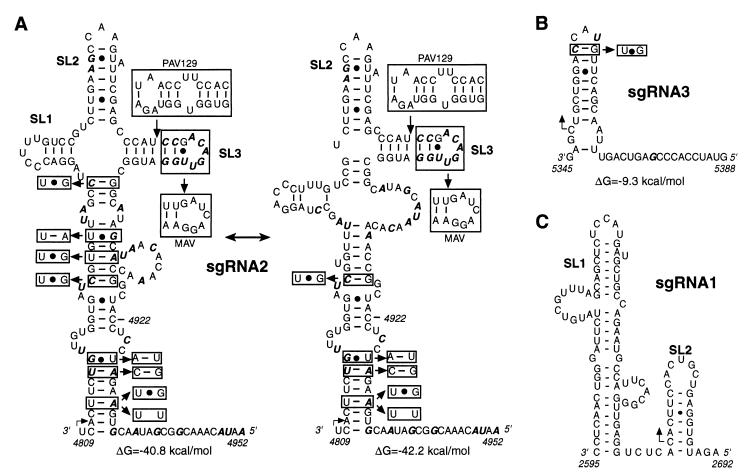
We have shown previously that both RNA primary and secondary structures are required for sgRNA1 synthesis (21). Therefore, we analyzed sgRNA2 and sgRNA3 promoter regions for potential secondary structure. Figure 6A presents optimal (ΔG = −42.2 kcal/mol) and suboptimal (ΔG = −40.8 kcal/mol) conformations of sgRNA2 promoter predicted using MFOLD (31, 53). The two conformations share identical stems at the base and two stem-loops (SL2 and SL3) at the top of the structure. The suboptimal structure has a different midsection and an additional stem-loop (SL1) which creates a four-way junction. The conservation of the SL3 structure, in spite of the sequence differences in BYDV-MAV and insertions that extend SL3 in BYDV-PAV129 (Fig. 5B), suggests its potential importance. The covariation found in the middle helix of the suboptimal structure favors formation of that structure and not of the theoretically most energetically stable one. The G-U pairs that covary with Watson-Crick pairs indicate selection for conservation of base-paired regions that would not form in the positive strand. The sgRNA3 promoter sequence is predicted to fold into a hairpin structure and a single-stranded region. Comparison of the secondary structure of the sgRNA1 promoter and the structures predicted for the other two promoters showed no common elements other than downstream stem-loops of various shapes and sizes (Fig. 6C), indicating that BYDV sgRNA transcription is controlled by very divergent cis elements.
FIG. 6.
Prediction of secondary structures of sgRNA promoters of BYDV RNA (negative sense). Angled arrows indicate sgRNA initiation sites. (A) Potential secondary structures of sgRNA2 promoter predicted by MFOLD. The optimal and suboptimal conformations are shown with the calculated free energy (ΔG) indicated for each structure. Nucleotides that vary in different strains of BYDV are in boldface italics. Base pair covariations that preserve proposed secondary structure are boxed. Extra bases in PAV129 extend the stem of SL3 (boxed). The variable distal end of SL3 in MAV is also boxed. (B) An MFOLD secondary structure prediction for sgRNA3 promoter. (C) Secondary structure of sgRNA1 promoter supported by MFOLD analysis, phylogenetic evidence, and nuclease probing (21).
DISCUSSION
One of the surprising findings of this study was the fact that mutations of sgRNA2 and sgRNA3 start sites as mapped by Kelly et al. (18) did not abolish sgRNA synthesis. They mapped both sgRNAs by using primer extension, assuming that sgRNAs are colinear with the gRNA. However, this is not true for all viruses (23). To our knowledge, no data exist on direct sequencing of isolated sgRNAs of BYDV. Potentially, there may be nontemplated addition of 5′-terminal nucleotides that would complicate precise mapping of their 5′ ends. Furthermore, small variations in the precise start sites of BYDV sgRNAs have been reported (L. Domier, personal communication). It is also possible that the sgRNA 5′ ends are exactly as determined by Kelly et al. (18) and that the sgRNA promoters tolerate variations at the first base of the sgRNA. In fact, several bases in the sgRNA promoters of the Bromoviridae and Alphaviridae are conserved, but the first nucleotide of the sgRNA is not (1, 12, 39).
Roles of BYDV sgRNAs.
Our experiments show that sgRNAs 2 and 3 are not required for viral RNA replication in protoplasts. We demonstrated previously that sgRNA1 is also dispensable for viral RNA replication. It serves as the mRNA for the viral coat protein which is usually not required for replication of positive-sense RNA viruses, with the exception of alfamoviruses and ilarviruses (reviewed in reference 16). The lack of requirement for sgRNA2 supports a previous observation that the ORF6 product encoded by sgRNA2 is not required for replication (37). Nevertheless, we expected that sgRNA2 may influence gRNA replication based on a novel regulatory role it may play in viral translation (47). This role is to preferentially inhibit translation of the viral replicase from the gRNA, thereby mediating the switch to the late gene expression (coat protein) from sgRNA1. Based on this model, it is not surprising that absence of sgRNA2 does not abolish virus replication, because none of the products of sgRNA1 are necessary for viral RNA replication in protoplasts.
While the lack of sgRNA2 transcription in SG2G/C did not adversely affect viral RNA replication, it might have deleterious consequences for other aspects of the viral life cycle that would not be detected by the assays used in this study. For example, the disruption of the stoichiometry of viral RNA and protein accumulation may result in inefficient encapsidation or movement of the virus. The product of ORF6, while dispensable for replication, may be needed for other processes. Another potential role for both sgRNAs 2 and 3 would be attenuation of virus replication, which could prevent premature death of the host and allow a larger window for transmission. Therefore, sgRNAs 2 and 3 may be a type of molecular parasite (much like defective interfering RNAs, but originating in cis) whose presence, however, created selective advantage for the virus. In planta infection experiments with mutants lacking sgRNAs will address these possibilities.
Recognition of divergent promoters.
The mapping experiments, sequence alignments, and RNA secondary structure predictions clearly demonstrate the lack of significant homology between the sgRNA promoters of BYDV. Location of the minimal core domains of the sgRNA2 and sgRNA3 promoters downstream of the transcription initiation sites, in contrast to the sgRNA1 promoter and the vast majority of subgenomic promoters mapped in other RNA viruses (5, 12, 17, 21, 28, 43, 45, 46, 51), is especially striking. This raises the question: how does the replicase complex recognize such different promoters? Either it has multiple RNA recognition domains or there are separate RNA-recognizing proteins for each promoter, and each of these interacts with the replicase. Various host proteins have been found associated with viral replication and transcription complexes (reviewed in reference 24). Different host factors control the specificity of bacteriophage Qβ replicase for the positive and negative strands of Qβ RNA (6). Another possibility is that there is one protein with a recognition domain that has different affinities for each promoter.
An RNA virus may evolve divergent promoters for differential temporal and quantitative regulation of sgRNA accumulation. Also, overlapping of subgenomic promoters and important ORFs and cis-acting elements in a small virus may result in evolution of dissimilar promoters. For example, the sgRNA1 promoter sequence has to accommodate an essential part of the replicase coding region and part of the 5′ UTR of sgRNA1 that controls translation (2). The region that contains the sgRNA2 promoter overlaps the 3′TE and part of ORF6, as well as the sgRNA2 5′ UTR. The sgRNA3 promoter is located in the region important for virus RNA replication (C. Paul and W. A. Miller, unpublished data). Thus, we propose that different sgRNA promoters arose independently within RNA sequences that have other functions. This versatility of promoter sequences facilitates small genome size by allowing overlapping functions on an RNA sequence.
More detailed characterization of sgRNA2 and sgRNA3 promoters is needed to elucidate RNA primary and secondary structural requirements for promoter activity. The negative-strand-specific conservation of the sgRNA2 promoter secondary structure suggests its potential importance for transcription. Studies with other RNA viruses have shown involvement of RNA secondary structure in various processes of the viral life cycle including RNA transcription and replication. The lack of secondary structural homology between BYDV subgenomic promoters is novel. This is in contrast to TCV, whose promoters have similar stable hairpins immediately upstream of the start site (46). The hairpin predicted to form within the sgRNA3 promoter structurally resembles SL2 of the sgRNA1 promoter (Fig. 6B versus C). However, not only is SL2 secondary structure not required for sgRNA1 transcription, it inhibits promoter activity (21). It will be interesting if future studies show that sgRNA3 promoter has no secondary structure requirement and that the hairpin has evolved to attenuate transcription, the role we suggested for SL2 of sgRNA1 promoter (21).
Mechanisms of sgRNA synthesis.
Based on the fact that internal initiation of sgRNA synthesis on the negative strand is the only unequivocally demonstrated mechanism of transcription in plant RNA viruses and that BYDV replicase belongs to the supergroup 2, the same supergroup as TCV, which employs internal initiation for its transcription, we expected internal initiation as the mechanism of BYDV sgRNA synthesis. However, as we have indicated (21), we do not exclude the possibility that premature termination with independent replication could be the mechanism, as has been suggested for RCNMV (40), which is very closely related to BYDV in the RdRp gene (22). For example, the region identified as the sgRNA1 promoter, which contains a helical structure indispensable for sgRNA synthesis, can form identical secondary structures in both the positive and negative strands (21). The overlapping of the 3′TE region with the sgRNA2 promoter indicates that the same RNA element has a dual function. Potentially, the 3′TE could function indirectly as an sgRNA2 promoter in the positive strand by serving as a terminator of the negative-strand sgRNA2 synthesis. Conceivably, binding of the factor(s) that mediate cap-independent translation to the 3′TE could create an obstacle for the viral RdRp synthesizing negative-strand RNA, causing termination and release of the negative-strand sgRNA2 which would serve as a template for the positive-strand sgRNA2. Although the BamHI fill-in mutation, which knocks out 3′TE function, did not affect synthesis of sgRNA (Fig. 3), it does not rule out the possibility that some TE-binding proteins may still bind. However, the phylogenetic conservation of the predicted secondary structure of sgRNA2 promoter in the negative strand (Fig. 6A, G-U pairs) argues that the negative strand also contains an important control signal. No such phylogenetic evidence has been found for the sgRNA1 promoter, and only one instance of such base pair covariation was found in the sgRNA3 promoter (Fig. 6B).
Mutations that resulted in the lack of sgRNA synthesis may have disrupted either the replicase recognition region in the negative strand or the termination element in the positive strand. Unfortunately, we know of no conclusive experimental data on the termination process in positive-sense RNA viruses which would allow us to compare sgRNA promoter regions with structures involved in termination of RNA synthesis. Involvement of RNA secondary structure has been suggested in RdRp pausing that leads to recombination in RNA viruses (11, 25, 50). Evidence from studies done with rhabdoviruses and retroviruses suggests that termination could be both sequence and secondary structure dependent (4, 13, 20, 41).
The increase of activity of the ectopic sgRNA2 promoter due to a knockout of its downstream endogenous copy is consistent with observations made in both the alpha-like BMV (12) and the coronaviruses (44). BMV uses internal initiation for sgRNA synthesis (36); however, consensus has not been reached on the coronavirus transcription model. Subgenomic promoter attenuation by other downstream promoters is also consistent with both transcription models (44). The attenuation effect could result either from viral RdRp dissociating from the plus-strand template when it encounters a subgenomic promoter (terminator) or from RdRp pausing and subsequently dissociating at the promoters on the minus strand where other transcription initiation complexes assemble. Either way, the largest sgRNA would experience the highest number of obstacles during its synthesis and therefore would be the least abundant, if all promoters were of equal strength. The diverse structures of the BYDV promoters may allow a more complex level of regulation than just genomic location.
ACKNOWLEDGMENTS
This work was supported by a grant from USDA-NRI.
We thank Theo Dreher for very useful suggestions. We also thank Randy Beckett for sequence of BYDV-PAV-129.
Footnotes
This is paper no. J-18710 of the Iowa Agriculture and Home Economics Experiment Station, project 3545.
REFERENCES
- 1.Adkins S, Siegel R W, Sun J H, Kao C C. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA. 1997;3:634–647. [PMC free article] [PubMed] [Google Scholar]
- 2.Allen E, Wang S, Miller W A. Barley yellow dwarf virus RNA requires a cap-independent translation sequence because it lacks a 5′ cap. Virology. 1999;253:139–144. doi: 10.1006/viro.1998.9507. [DOI] [PubMed] [Google Scholar]
- 3.Balmori E, Gilmer D, Richards K, Guilley H, Jonard G. Mapping the promoter for subgenomic RNA synthesis on beet necrotic yellow vein virus RNA 3. Biochimie (Paris) 1993;75:517–521. doi: 10.1016/0300-9084(93)90056-x. [DOI] [PubMed] [Google Scholar]
- 4.Barr J N, Whelan S P, Wertz G W. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boccard F, Baulcombe D. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology. 1993;193:563–578. doi: 10.1006/viro.1993.1165. [DOI] [PubMed] [Google Scholar]
- 6.Brown D, Gold L. RNA replication by Qβ replicase: a working model. Proc Natl Acad Sci USA. 1996;93:11558–11562. doi: 10.1073/pnas.93.21.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chay C A, Gunasinge U B, Dinesh-Kumar S P, Miller W A, Gray S M. Aphid transmission and systemic plant infection determinants of barley yellow dwarf luteovirus-PAV are contained in the coat protein readthrough domain and 17-kDa protein, respectively. Virology. 1996;219:57–65. doi: 10.1006/viro.1996.0222. [DOI] [PubMed] [Google Scholar]
- 8.Di R, Dinesh-Kumar S P, Miller W A. Translational frameshifting by barley yellow dwarf virus RNA (PAV serotype) in Escherichia coli and in eukaryotic cell-free extracts. Mol Plant-Microbe Interact. 1993;6:444–452. doi: 10.1094/mpmi-6-444. [DOI] [PubMed] [Google Scholar]
- 9.Dinesh-Kumar S P, Brault V, Miller W A. Precise mapping and in vitro translation of a trifunctional subgenomic RNA of barley yellow dwarf virus. Virology. 1992;187:711–722. doi: 10.1016/0042-6822(92)90474-4. [DOI] [PubMed] [Google Scholar]
- 10.Dinesh-Kumar S P, Miller W A. Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell. 1993;5:679–692. doi: 10.1105/tpc.5.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duggal R, Wimmer E. Genetic recombination of poliovirus in vitro and in vivo: temperature-dependent alteration of crossover sites. Virology. 1999;258:30–41. doi: 10.1006/viro.1999.9703. [DOI] [PubMed] [Google Scholar]
- 12.French R, Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison G P, Mayo M S, Hunter E, Lever A M. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res. 1998;26:3433–3442. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilf M E, Karasev A V, Pappu H R, Gumpf D J, Niblett C L, Garnsey S M. Characterization of citrus tristeza virus subgenomic RNAs in infected tissue. Virology. 1995;208:576–582. doi: 10.1006/viro.1995.1188. [DOI] [PubMed] [Google Scholar]
- 15.Huang P, Lai M M C. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3′ untranslated region, thereby altering RNA conformation. J Virol. 1999;73:9110–9116. doi: 10.1128/jvi.73.11.9110-9116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaspars E M J. Genome activation in alfamo- and ilarviruses. Arch Virol. 1999;144:843–863. doi: 10.1007/s007050050551. [DOI] [PubMed] [Google Scholar]
- 17.Johnston J C, Rochon D M. Deletion analysis of the promoter for the cucumber necrosis virus 0.9-kb subgenomic RNA. Virology. 1995;214:100–109. doi: 10.1006/viro.1995.9950. [DOI] [PubMed] [Google Scholar]
- 18.Kelly L, Gerlach W L, Waterhouse P M. Characterisation of the subgenomic RNAs of an Australian isolate of barley yellow dwarf luteovirus. Virology. 1994;202:565–573. doi: 10.1006/viro.1994.1378. [DOI] [PubMed] [Google Scholar]
- 19.Kim K H, Hemenway C L. Long-distance RNA-RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA. 1999;5:636–645. doi: 10.1017/s1355838299982006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klarmann G J, Schauber C A, Preston B D. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J Biol Chem. 1993;268:9793–9802. . (Erratum, 268:13764.) [PubMed] [Google Scholar]
- 21.Koev G, Mohan B R, Miller W A. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J Virol. 1999;73:2876–2885. doi: 10.1128/jvi.73.4.2876-2885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 23.Lai M M, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai M M C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 25.Lai M M C. Recombination in large RNA viruses: coronaviruses. Semin Virol. 1996;7:381–388. doi: 10.1006/smvy.1996.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landt O, Grunert H P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 27.Lehto K, Grantham G L, Dawson W O. Insertion of sequences containing the coat protein subgenomic RNA promoter and leader in front of the tobacco mosaic virus 30K ORF delays its expression and causes defective cell-to-cell movement. Virology. 1990;174:145–157. doi: 10.1016/0042-6822(90)90063-w. [DOI] [PubMed] [Google Scholar]
- 28.Levis R, Schlesinger S, Huang H V. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol. 1990;64:1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y-J, Zhang X, Wu R-C, Lai M M C. The 3′ untranslated region of coronavirus RNA is required for subgenomic mRNA transcription from a defective interfering RNA. J Virol. 1996;70:7236–7240. doi: 10.1128/jvi.70.10.7236-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maia I G, Seron K, Haenni A-L, Bernardi F. Gene expression from viral RNA genomes. Plant Mol Biol. 1996;32:367–391. doi: 10.1007/BF00039391. [DOI] [PubMed] [Google Scholar]
- 31.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 32.Mayo M A, D'Arcy C J. Family Luteoviridae: a reclassification of luteoviruses. In: Smith H G, Barker H, editors. The Luteoviridae. Wallingford, Oxon, England: CABI Publishing; 1999. pp. 15–22. [Google Scholar]
- 33.Miller E D, Plante C A, Kim K H, Brown J W, Hemenway C L. Stem-loop structure in the 5′ region of potato virus X genome required for plus-strand RNA accumulation. J Mol Biol. 1998;284:591–608. doi: 10.1006/jmbi.1998.2174. [DOI] [PubMed] [Google Scholar]
- 34.Miller W A. Luteoviridae. In: Webster R G, Granoff A, editors. Encyclopedia of virology. 2nd ed. London, England: Academic Press; 1999. pp. 901–908. [Google Scholar]
- 35.Miller W A, Dinesh-Kumar S P, Paul C P. Luteovirus gene expression. Crit Rev Plant Sci. 1995;14:179–211. [Google Scholar]
- 36.Miller W A, Dreher T W, Hall T C. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−) sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 37.Mohan B R, Dinesh-Kumar S P, Miller W A. Genes and cis-acting sequences involved in replication of barley yellow dwarf virus-PAV RNA. Virology. 1995;212:186–195. doi: 10.1006/viro.1995.1467. [DOI] [PubMed] [Google Scholar]
- 38.Seeley K A, Byrne D H, Colbert J T. Red light-independent instability of oat phytochrome mRNA in vivo. Plant Cell. 1992;4:29–38. doi: 10.1105/tpc.4.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel R W, Adkins S, Kao C C. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sit T L, Vaewhongs A A, Lommel S A. RNA-mediated transactivation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 41.Tuerk C, Gauss P, Thermes C, Groebe D R, Gayle M, Guild N, Stormo G, d'Aubenton-Carafa Y, Uhlenbeck O C, Tinoco I, Jr, et al. CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci USA. 1988;85:1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Kuyl A C, Langereis K, Houwing C J, Jaspars E M, Bol J F. cis-Acting elements involved in replication of alfalfa mosaic virus RNAs in vitro. Virology. 1990;253:327–336. doi: 10.1016/0042-6822(90)90004-b. [DOI] [PubMed] [Google Scholar]
- 43.van der Vossen E A, Notenboom T, Bol J F. Characterization of sequences controlling the synthesis of alfalfa mosaic virus subgenomic RNA in vivo. Virology. 1995;212:663–672. doi: 10.1006/viro.1995.1524. [DOI] [PubMed] [Google Scholar]
- 44.van Marle G, Luytjes W, van der Most R G, van der Straaten T, Spaan W J. Regulation of coronavirus mRNA transcription. J Virol. 1995;69:7851–7856. doi: 10.1128/jvi.69.12.7851-7856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Carpenter C D, Simon A E. Minimal sequence and structural requirements of a subgenomic RNA promoter for turnip crinkle virus. Virology. 1999;253:327–336. doi: 10.1006/viro.1998.9538. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Simon A E. Analysis of the two subgenomic RNA promoters for turnip crinkle virus in vivo and in vitro. Virology. 1997;232:174–186. doi: 10.1006/viro.1997.8550. [DOI] [PubMed] [Google Scholar]
- 47.Wang S, Guo L, Allen E, Miller W A. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans. RNA. 1999;5:728–738. doi: 10.1017/s1355838299981979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Miller W A. A sequence located 4.5 to 5 kilobases from the 5′ end of the barley yellow dwarf virus (PAV) genome strongly stimulates translation of uncapped mRNA. J Biol Chem. 1995;270:13446–13452. doi: 10.1074/jbc.270.22.13446. [DOI] [PubMed] [Google Scholar]
- 49.Wang S P, Browning K S, Miller W A. A viral sequence in the 3′ untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White K A, Morris T J. RNA determinants of junction site selection in RNA virus recombinants and defective interfering RNAs. RNA. 1995;1:1029–1040. [PMC free article] [PubMed] [Google Scholar]
- 51.Zavriev S K, Hickey C M, Lommel S A. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology. 1996;216:407–410. doi: 10.1006/viro.1996.0076. [DOI] [PubMed] [Google Scholar]
- 52.Zhang G, Slowinski V, White K A. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA. 1999;5:550–561. doi: 10.1017/s1355838299982080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]