ABSTRACT
Two bacteriophages (phages) of Klebsiella pneumoniae were isolated from sewage water collected from Dakar, Senegal. Phage vKpIN17 belongs to the Przondovirus genus within the Autographiviridae family, with double-stranded DNA genomes, whereas vKpIN18 belongs to the Webervirus genus of the Drexlerviridae family.
KEYWORDS: bacteriophages, Klebsiella pneumoniae, bacteriophage therapy, Healthcare associated infections
ANNOUNCEMENT
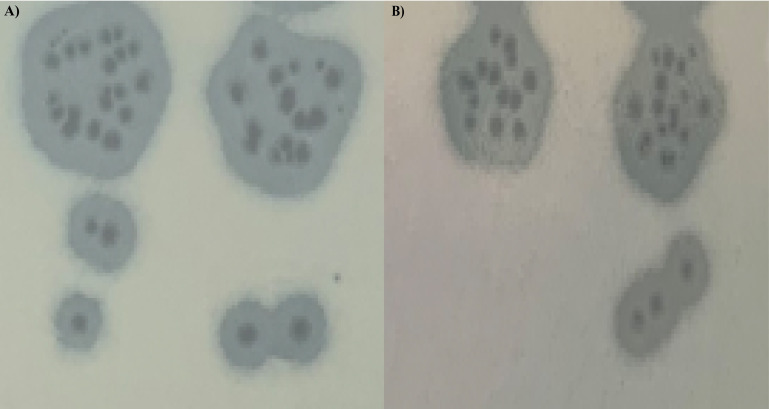
Using a clinical strain of Klebsiella pneumoniae KP26 isolated from healthcare-associated infection in a Children’s Hospital Center Albert Royer of Fann in Dakar, two lytic bacteriophages were isolated from two community sewage water (14.695121–17.455776; 14.685763–17.46923) in Dakar, Senegal. Twenty milliliters of sewage sample was centrifugated at 5,000 g for 10 min, and then the supernatants were filtered through a 0.22 µm pore size membrane filter. We assessed the filtered supernatants individually for phage presence using double layer agar method (1). Phage plaques were processed by three rounds of purification and amplified as previously described (2). The two phages produced clear plaques with a large halo (Fig. 1).
Fig 1.
Plaque morphology of phages (A) vKpIN17 and (B) vKpIN18.
Genomic DNA was isolated from high titer stocks (>109 PFU/mL). Briefly, 1 mL of phage lysate was treated with 10 µL of DNase I (20 U) and 4 µL of RNase A (20 mg/mL), incubated for 30 min at 37°C, followed by DNA extraction using phenol-chloroform method (2). Library preparations were performed using 1 ng of DNA and Nextera XT DNA library preparation kits (Illumina, San Diego, CA, USA) and executed according to the manufacturer’s protocol. Whole-genome sequencing was performed on Illumina iSeq100 sequencers utilizing the 300-cycle i1 Reagent V2 Kit (Illumina, San Diego, CA, USA).
A total of 299,580 and 323,244 (2 × 150 bp) paired-end reads were generated, respectively, for vKpIN17 and vKpIN18, and quality control was performed with FastQC v0.12.1 (3). Reads were trimmed using trim-galore v0.6.10 (4). The de novo assembly was performed using SPAdes v.3.15.5 (5) with careful parameters. Contig coverage and assembly validation were performed with BBMap v 35.85 (6). Furthermore, reads were sorted and indexed using Samtools v1.18 (7) and were submitted to assembly error corrections using Pilon v1.24 (8). Phage termini were identified with PhageTerm (9). Predicted coding sequences (CDS) were annotated using Pharokka v1.3.0 (10). For taxonomic classification, closely related genomes were obtained from the NCBI database.
The vKpIN17 and vKpIN18 genome sizes were 40,702 and 48,639 bp, respectively, with GC contents of 53.35% and 50.4%, and mean coverage of 1,674× and 1,481×. Both phages’ genomes are permutated and feature redundant ends. There are, respectively, 54 and 84 predicted CDS of which 24 (44%) and 48 (57%) are hypothetical proteins for vKpIN17 and vKpIN18. CDS with homology to other known genes encode, among others, structural elements, DNA, RNA and nucleotide metabolism, and host lysis. No genes associated with lysogeny (e.g., integrases), virulence, toxin, transfer RNAs, clustered regularly interspaced short palindromic repeats, or antibiotic resistance were detected within their genomes.
The closest phages of vKpIN17 and vKpIN18 were Klebsiella phage K11 (accession: NC_011043.1) (95.99%) and Klebsiella phage Kp8 (accession: NC_048700.1) (95.70%), respectively. The phage K11 belongs to the Przondovirus K11 species, Przondovirus genus, and Autographiviridae family, while Kp8 belongs to the Webervirus KLPPOU149 species, Webervirus genus, and Drexlerviridae family.
We have identified two phages, vKpIN17 and vKpIN18, that show significant promise as candidates for therapeutic applications. However, further characterization is needed to explore their putative therapeutic value.
ACKNOWLEDGMENTS
The authors thank Maïmouna Mbanne Diouf for technical assistance with the Sequencing and all the members of the Pole of Microbiology, Pasteur Institute of Dakar for their precious assistance.
This study was supported by GCDM at Institut de Recherche pour le Développement (IRD), France.
I.N. conceptualized the study, performed sampling, curated the data, performed formal analysis and investigation, designed the methodology, helped with software, wrote the original draft, and reviewed and edited the manuscript. L.D. reviewed and edited the manuscript and supervised the study. M.M.D. designed the methodology and reviewed and edited the manuscript. B.S.B. reviewed and edited the manuscript. O.S. helped with sampling and reviewed and edited the manuscript. A.C., C.F., Y.D., and N.D. reviewed and edited the manuscript. A.S. conceptualized the study, administrated the project, reviewed and edited the manuscript, and supervised the study. G.C.d.M. conceptualized the study, administrated the project, reviewed and edited the manuscript, acquired funding, and supervised the study.
Contributor Information
Issa Ndiaye, Email: seydina.indiaye14@gmail.com.
John J. Dennehy, Queens College Department of Biology, New York, USA
DATA AVAILABILITY
The data for the two phages are available in the European Nucleotide Archive (ENA) under the project number PRJEB68306 with accession numbers ERP153263 (vKpIN17) and ERP153263 (vKpIN18) for raw sequences reads and accession numbers GCA_963669465 (vKpIN17) and GCA_963669475 (vKpIN18) for genome sequences.
REFERENCES
- 1. Jofre J, Muniesa M. 2020. Bacteriophage isolation and characterization: phages of Escherichia coli. Methods Mol Biol:61–79. doi: 10.1007/978-1-4939-9877-7_4 [DOI] [PubMed] [Google Scholar]
- 2. Sambrook J, Russell DW. 2006. Identification of associated proteins by coimmunoprecipitation. CSH Protoc 2006:pdb.prot3898. doi: 10.1101/pdb.prot3898 [DOI] [PubMed] [Google Scholar]
- 3. Andrews S. 2010. Babraham bioinformatics-fastqc a quality control tool for high throughput sequence data. Available from: https://wwwbioinformaticsbabrahamacuk/projects/fastqc
- 4. Krueger F. 2015. Trim galore!: a wrapper around cutadapt and fastqc to consistently apply adapter and quality trimming to fastq files, with extra functionality for Rrbs data. Babraham Institute. [Google Scholar]
- 5. Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using spades de novo assembler. Curr Protoc Bioinformatics 70:e102. doi: 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- 6. Bushnell B. 2015. Bbmap short-read aligner, and other bioinformatics tools. University of California, Berkeley. [Google Scholar]
- 7. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of samtools and Bcftools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garneau JR, Depardieu F, Fortier L-C, Bikard D, Monot M. 2017. Phageterm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7:8292. doi: 10.1038/s41598-017-07910-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bouras G, Nepal R, Houtak G, Psaltis AJ, Wormald P-J, Vreugde S. 2023. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics 39:btac776. doi: 10.1093/bioinformatics/btac776 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data for the two phages are available in the European Nucleotide Archive (ENA) under the project number PRJEB68306 with accession numbers ERP153263 (vKpIN17) and ERP153263 (vKpIN18) for raw sequences reads and accession numbers GCA_963669465 (vKpIN17) and GCA_963669475 (vKpIN18) for genome sequences.