Abstract
Despite substantial advancements in development of cancer treatments, lack of standardized and physiologically-relevant in vitro testing platforms limit the early screening of anticancer agents. A major barrier is the complex interplay between the tumor microenvironment and immune response. To tackle this, a dynamic-flow based 3D bioprinted multi-scale vascularized breast tumor model, responding to chemo and immunotherapeutics is developed. Heterotypic tumors are precisely bioprinted at pre-defined distances from a perfused vasculature, exhibit tumor angiogenesis and cancer cell invasion into the perfused vasculature. Bioprinted tumors treated with varying dosages of doxorubicin for 72 h portray a dose-dependent drug response behavior. More importantly, a cell based immune therapy approach is explored by perfusing HER2-targeting chimeric antigen receptor (CAR) modified CD8+ T cells for 24 or 72 h. Extensive CAR-T cell recruitment to the endothelium, substantial T cell activation and infiltration to the tumor site, resulted in up to ≈70% reduction in tumor volumes. The presented platform paves the way for a robust, precisely fabricated, and physiologically-relevant tumor model for future translation of anti-cancer therapies to personalized medicine.
Keywords: bioprinting, cancer, CAR-T immunotherapy, tumor models
1. Introduction
Cancer remains one of the leading causes of mortality worldwide, accounting for ≈10 million deaths in 2020 alone.[1] Despite remarkable advances in cancer treatment modalities, dearth of physiologically-relevant pre-clinical diagnostic platforms limits the successful clinical translation of anti-cancer therapeutics.[2] Genetic and epigenetic heterogeneity of a tumor microenvironment and the underlying immune-cancer interactions are critical aspects in determining effective therapeutic response.[3,4] Even though the field of immunotherapy has witnessed tremendous growth in checkpoint inhibitor-based immunotherapy against programmed cell death protein 1, programmed death-ligand 1 (PD-L1), and cytotoxic T-Lymphocyte associated protein 4 in melanoma[5] non-small cell lung cancer,[6] and renal cell carcinoma;[7] these treatments have shown promising results in only a subset of patients. Additionally, checkpoint inhibitor therapy for breast cancer is only limited to triple negative breast cancer patients expressing PD-L1 protein.[8] Similarly, the adoptive transfer of T cells expressing chimeric antigen receptors (CAR) against tumor associated antigens has produced benefits for hematological diseases but they are less effective for solid tumors.[9] Extensive antigen heterogeneity in solid tumors hinder detection of cancer cells by T cells and weakens CAR-T therapy.[10] Another major challenge involves CAR-T cell penetration to tumor site and subsequent survival of CAR-Ts in the tumor microenvironment.[9] CAR-Ts might be unable to penetrate the vasculature surrounding the tumor tissue and eventually return to the bloodstream or lymphatic system where they come in contact with blood tumor cells, which explains their superior efficacy in hematological cancers but not in solid tumors. Developing an effective CAR-T cell-based treatment specific to breast cancer, which can successfully navigate the immunosuppressive breast tumor microenvironment and generate potent anti-tumor response is a major challenge.[11]
Engineered in vitro platforms have emerged as powerful tools for assessing tumor immune interactions in the context of immunotherapy. Complex 3D models employing multicellular spheroids in conjunction with microfluidics have been employed in studying drug response of tumors under flow and endothelial regulation of drug transport in ovarian and lung cancer based tumor spheroids.[12,13] However, these studies have primarily been limited to microfluidic-based devices, which entails a fairly lengthy fabrication process with limited control on localization of tumors and no flexibility of altering their positions with respect to primary vasculature without changing the initial master mold design. Moreover, immune-cancer interactions have mostly been studied with monocytes (THP-1) as model immune cells, which fail to capture the cross talk of the CAR-T cells and the tumor microenvironment.[14,15] As CAR-Ts are programmed to kill cancer cells, inadequate understanding of the inhibitory or stimulatory receptor mediated immune cell activation is a deterrent to developing effective immunotherapy. The other microfluidic models present a reductionist view of the tumor microenvironment by portraying immune-cancer interactions in the absence of tumor vasculature, or continuous media perfusion.[16,17] Additionally, distance of the tumor from a perfused blood vessel plays a predominant role in tumor growth as it controls diffusion of oxygen and nutrients, which is critical to tumor survival.[18] This necessitates disseminating CAR-T-cancer interactions in a more physiologically-relevant microenvironment.
In an attempt to develop more representative platforms, for the first time, we studied CAR-T induced cytotoxicity in a vascularized and dynamic-flow based breast tumor microenvironment.[19] Employing aspiration-assisted bioprinting, heterotypic tumors were positioned in a biomimetic matrix with 50 μm positional accuracy.[20,21] Dynamic flow of media through the endothelialized vasculature and precise control over tumor location revealed higher invasion and intravasation of metastatic breast cancer (MDA-MB-231) cells when bioprinted proximal to the perfused vasculature. To understand if the perfused platform was capable of responding to anti-cancer therapeutics, doxorubicin, a chemotherapeutic drug, was perfused for 72 h resulting in a dose dependent reduction in tumor volume. After validating the system with the chemotherapeutic drug, CAR-T cells, engineered to recognize epidermal growth factor receptor 2 (HER2) on aggressive MDA-MB-231 cells, were perfused for 72 h to understand CAR-T infiltration to the tumor site and immune response generated as a result of CAR-T cell-based immunotherapy. The presented perfusable tumor model can be harnessed for disseminating the role of immune modulators, which will eventually lead to better clinical translations of CAR-T cell therapy.
2. Results
2.1. Device Fabrication
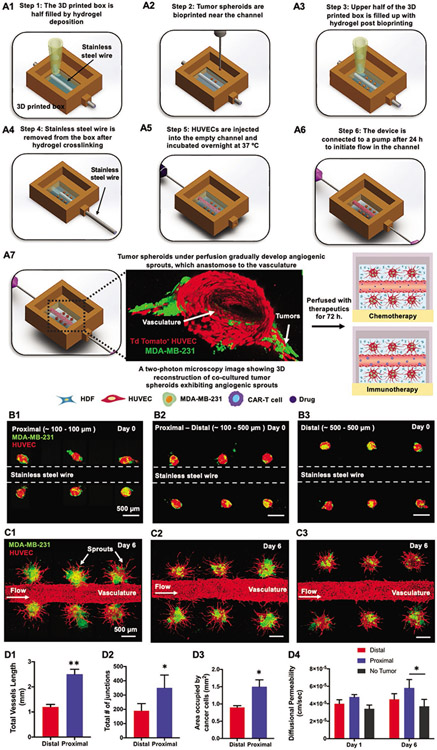
In this work, a bioprinted device was designed with a chamber to contain the biologics with perforations on opposite walls, which would serve as connection ports for perfusion. A stainless-steel wire was inserted through the wall perforations followed by hydrogel deposition to half-fill the device (Figure 1A1). A combination of tdTomato+ human umbilical vein endothelial cells (HUVECs), green fluorescent protein positive (GFP+) MDA-MB-231, and human dermal fibroblasts (HDFs), henceforth named “H231F,” was determined to be representative of a native tumor microenvironment as well as mechanically suitable for bioprinting. For development, optimization and characterization of H231F spheroids, the reader is referred to the Results Section in the Supporting Information and Figures S1 and S2 (Supporting Information). Employing aspiration-assisted bioprinting, heterotypic H231F tumor spheroids were precisely bioprinted at pre-defined distances from the stainless-steel wire within the first 5 min of the deposition of a composite collagen/fibrinogen (henceforth named “C2F3,” made of 2 mg mL−1 collagen and 3 mg mL−1 fibrinogen) hydrogel to prevent irreversible damage to the hydrogel integrity (Figure 1A2; Movie S1, Supporting Information). Changes in hydrogel rheological properties with the onset of cross-linking was determined to optimize the maximum allowable bioprinting time, which yielded a 50-μm positional accuracy for the bioprinting of H231F spheroids (see the Results Section in the Supporting Information and Figures S2 and S3, Supporting Information). After bioprinting, additional C2F3 was deposited to completely fill the device (Figure 1A3) and incubated at 37 °C for 30 min. On complete cross-linking, the stainless-steel wire was gently removed from the device (Figure 1A4), creating an open channel. Next, HUVECs were introduced into the channel at a concentration of 25–30 million cells mL−1 (Figure 1A5) and the following day, the device was connected to an external pump to initiate media flow through the endothelialized channel (Figure 1A6). The bioprinted tumor spheroids gradually developed angiogenic sprouts, where the extent of sprouting was dependent on the distance of the tumor from the perfused vasculature. These vascularized devices were further evaluated for their efficacy in chemo and immunotherapy, as presented later in the paper (Figure 1A7).
Figure 1.
A schematic representation of a 3D perfusable tumor model fabricated using aspiration-assisted bioprinting. A1–A6) Step 1-Step 6 enumerates the design and work flow followed for successful fabrication of the perfusable device. A7) A schematic representation along with a 3D reconstructed two-photon fluorescent image of the tumor angiogenesis observed after device was connected to a dynamic flow-based culture. After induction of tumor angiogenesis, the device was utilized for chemo and immunotherapy. B1–B3) Devices with tumor spheroids bioprinted at a proximal only (distance of ≈100 μm), proximal-distal combined (distance of ≈100 and ≈500 μm) and distal (≈500 μm) from the stainless-steel wire. C1–C3) Fluorescent images of bioprinted devices after six days of perfusion exhibiting tumor angiogenesis. D1–D3). Graphical representation of total vessels length, total number of junctions and area occupied by cancer cells as a function of distance from the central vasculature. D4) Graphical representation of diffusional permeability measured for distally and proximally bioprinted tumors at Days 1 and 6 (n = 3 for all, p*** < 0.001, p** < 0.01, p* < 0.05).
2.2. 3D Bioprinting of Tumor Spheroids at Varying Distances from the Perfusable Vasculature
Employing aspiration-assisted bioprinting, H231F spheroids, were bioprinted at pre-defined distances from the perfusable vasculature to analyze the effect of distance on tumor angiogenesis and cancer invasion. In the first set of devices, all spheroids were bioprinted at a distance of ≈100 μm on both sides of the vasculature (proximal, Figure 1B1). Next, spheroids were bioprinted at a distance of ≈100 μm on one side (proximal) and ≈500 μm (distal) on the other side of the vasculature, as shown in Figure 1B2. The third device included all spheroids bioprinted at ≈500 μm distance (distal, Figure 1B3). Tumor angiogenesis was induced in all these devices under perfusion, maintaining a perfusion speed of 0.7 μL min−1. Optimal sprout formation and maintenance of endothelial barrier was observed under this flow rate (see the Results Section in Supporting Information and Figure S5, Supporting Information). The angiogenic sprouts developed hollow capillaries (Figure S6A, Supporting Information) and vascularized the tumor spheroids (Figure 1C1-C3; Movie S2, Supporting Information). For the proximally bioprinted tumors, sprouts seemed to be originating from the heterotypic tumor and the central vasculature, which probably anastomosed and connected to the main vasculature, representative of in vivo physiology (Figure S6B, Supporting Information). Thus, the sprouting for proximally bioprinted tumors could be a constitutive effect of both sprouting events, which eventually led to cancer cell intravasation into the central vasculature (Figure S6C, Supporting Information). It was interesting to observe that the extent of tumor angiogenesis with regards to the total vessel length and vessel branching varied with distance of spheroids from the perfused vasculature. A higher sprouting density was observed for proximal tumors as compared to the distal spheroids (Figure 1C1-C3). The total vessel length for the proximal spheroids had an average value of ≈2.5 mm as compared to ≈1.2 mm for the distal spheroids (Figure 1D1). Additionally, the total number of junctions was approximately 2-fold higher for the proximal spheroid as compared to the distal ones (Figure 1D2). Moreover, the sprouts formed in the distal spheroids were unable to connect to the perfused vasculature. The distance of spheroids from the vasculature also affected cancer invasion. For proximal tumor spheroids, cancer cells proliferated and were observed to invade into the vasculature (Figure S6C and Movie S3, Supporting Information). Spatial location of tumor spheroids also affected cancer invasion. MDA-MB-231 cells invaded into its surrounding matrix for both the proximal and distal spheroids. However, the area occupied by proximal spheroids was ≈1.5-fold higher than the area occupied by distal spheroids (Figure 1D3).
Devices with tumors bioprinted proximal to the perfused vasculature exhibited higher diffusional permeability as compared to the distally-bioprinted tumors and no tumor group. However, this difference was not statistically significant on Day 1. On Day 6 of perfusion, the diffusional permeability of the proximally-bioprinted group was found to be significantly higher than the no tumor (control) group (Figure 1D4). Endothelialized channels with tumors proximal to the perfused vasculature had a 1.6-fold higher diffusional permeability as compared to the no tumor group and 1.3-fold higher than the distally-bioprinted tumor group. Devices with distally-bioprinted tumors exhibited a slightly higher diffusional permeability as compared to no tumor group but it was not statistically significant.
2.3. Effects of Chemotherapeutic Drug, Doxorubicin, on Tumor Growth under a Dynamic Flow-Based Culture
In order to validate the in vitro platform before the exposure of CAR-T cells, doxorubicin, an anthracycline based chemotherapeutic drug commonly used for treating breast cancer,[22] was utilized in our experiments.
First, free-standing and C2F3 encapsulated tumor spheroids were subjected to doxorubicin treatment under static culture conditions prior to perfusing it through 3D bioprinted devices. The goal was to understand if the heterotypic tumors exhibited drug response under static conditions to be able to later apply the drug under dynamic conditions. Doxorubicin induced dose dependent cytotoxicity in both free standing and C2F3 encapsulated static cultured spheroids with an IC50 value of ≈0.65 μm and ≈0.9 μm, respectively (see Figures S7 and S8 and the Results Section in Supporting Information).
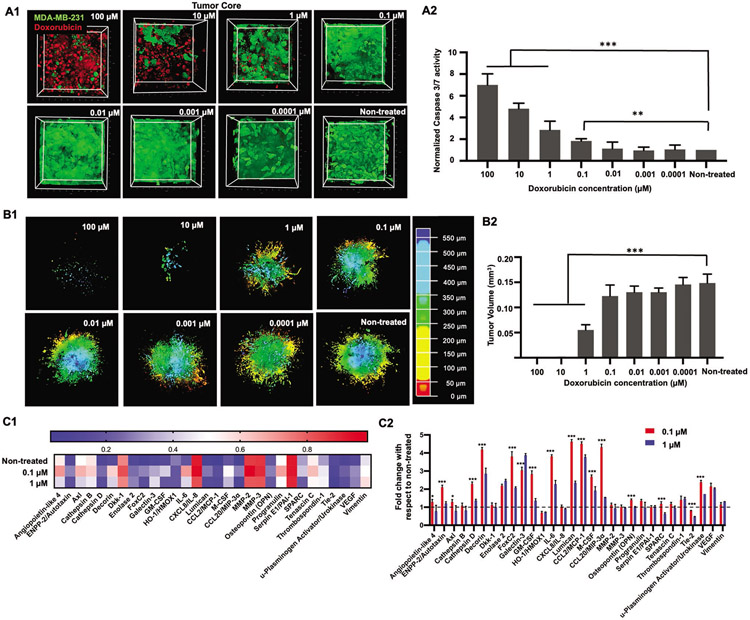
Having analyzed the effect of doxorubicin on static cultures, doxorubicin was then perfused through the devices at varying concentrations. First, H231F tumors were bioprinted at a distance of ≈100 μm (proximal) from the perfused vasculature. Tumors were cultured under flow for 3 days to induce angiogenic sprouts, which further anastomosed to the central vasculature. Doxorubicin, diluted in EGM-2MV media to the desired concentration (0–100 μm), was perfused through the central vasculature for 72 h. The tumor core was imaged to understand if the drug had diffused to tumor core. Doxorubicin molecule was excited at 780 nm to analyze doxorubicin uptake by cancer cells under perfusion and co-localization of the drug in cell nuclei.[23] There was an extensive uptake and localization of doxorubicin in the tumor core for both 10 and 100 μm treatments, identified by the doxorubicin auto-fluorescence signals (Figure 2A1). Widespread cellular debris in tumor core and significantly less density of GFP+ MDA-MB-231 cells were observed for these two above-mentioned concentrations. For the 1 μm treatment, doxorubicin was detected in tumor core but at a lower density as compared to 100 and 10 μm treatments. Post 1 μm treatment, presence of doxorubicin was not prominent in the tumor core for rest of the drug concentrations. As doxorubicin induces cellular apoptosis, we sought to understand Caspase 3/7, an early apoptosis marker activation after 24 h of doxorubicin perfusion in devices. As shown in Figure 2A2, Caspase 3/7 activity was observed to be ≈5–7 folds higher for 10 and 100 μm treatments, as compared to the non-treated group. Additionally, Caspase 3/7 activity was threefold and twofold higher than that for the non-treated group for the 1 and 0.1 μm treatment, respectively. For 0.01 to 0.0001 μm treatments, no significant difference in Caspase activity was observed as compared to the non-treated group. It is interesting to note that drug concentrations of 0.1 μm and lower were particularly resistant and showed little to no cytotoxicity in perfusion conditions. Whereas, in static cultures, drug concentration of 0.1 μm resulted in ≈40% cytotoxicity for free-standing spheroids (Figures S7 and S8, Supporting Information).
Figure 2.
A1) 3D Reconstruction of the tumor core showing GFP+ MDA-MB-231 cells and auto-fluorescent doxorubicin co-localized in cell nuclei. Doxorubicin concentration was varied from 100 to 0.0001 μm. A2) Graphical representation of normalized Caspase 3/7 activity after 24 h of the doxorubicin treatment. B1) 3D Reconstruction of the entire tumor remaining after 72 h of doxorubicin treatment. The tumors were depth coded to represent the entire tumor volume. B2) Graphical representation of tumor volumes after 72 h of doxorubicin perfusion. C1) Heatmap of selected proteins from the human oncology array performed on perfusates collected from devices treated with 1, 0.1, and 0 μm (non-treated) doxorubicin. C2) Graphical representation of fold-change in protein expression for treated devices as compared to non-treated devices. The dotted line at 1 through the bar graph helps identify protein expressions, which had values close or equal to the non-treated group. Thus, values higher than 1 represented an increase in the protein expression and values lower than 1 depicted decrease in expression as compared to the non-treated group (n = 3 for all, p*** < 0.001, p** < 0.01, p* < 0.05).
After perfusing doxorubicin for 72 h, GFP+ MDA-MB-231 cells remaining in the entire tumor volume was imaged for all concentrations. As shown in Figure 2B1, 10 and 100 μm treatments were extremely cytotoxic, resulting in only a few MDA-MB-231 cells remaining viable post treatment. This was also observed from our previous static cultures (Figures S7 and S8, Supporting Information). 1 μm treatment was also cytotoxic to MDA-MB-231 cells, resulting in fewer GFP+ cells remaining. Doxorubicin cytotoxicity was observed to be waning from 0.1 to 0.0001 μm. As compared to the non-treated group, where the tumor volume spanned a depth of ≈600 μm, a significantly smaller tumor was found for 10 and 100 μm treatments. The 1 μm treatment was also observed to be less dense as compared to other concentrations. On quantifying the tumor volumes, a significantly low volume (0.0004 mm3) was found for 10 and 100 μm treatments. For the 1 μm treatment, a mean tumor volume of ≈0.05 mm3 was obtained. For 0.0001– 0.1 μm, tumor volumes were determined to range from ≈0.12 to 0.15 mm3, which were close to the non-treated group (≈0.15 mm3) (Figure 2B2).
In order to understand the effect of doxorubicin on common cancer-related protein expressions, a proteome array containing 84 human oncoproteins was used to detect protein expressions in treated devices. The experimental groups chosen for this analysis included 0.1 μm, 1 μm, and non-treated (control) perfusion devices. As 10 and 100 μm treatments resulted in extensive cell death in static as well as perfusion cultures, 1 μm was chosen as the next “high” concentration to be analyzed. Additionally, as the 0.1 μm treatment resulted in comparable tumor volumes after 72 h of treatment with respect to the non-treated group, hence 0.1 μm would represent a “low” concentration. Figure S9 (Supporting Information) portrays a heatmap of all 84 oncoproteins from which a group of 29 proteins were identified having significantly high and variable expression among the three experimental groups. As shown in the heatmap in Figure 2C1, Angiopoietin-like 4, Axl, Capthepsin-B, Dickkopf-1 (Dkk-1), interleukin-8 (IL-8), matrix matalloproteinase-2 (MMP-2), MMP-3, Progranulin, Serpin E1, Tenascin C, and Vimentin were found to be comparatively higher in expression (> 0.5) as compared to other proteins, for all treated and non-treated groups. Overall, on comparing the two doxorubicin treated groups, the 0.1 μm treatment exhibited expression values higher than the non-treated group for some proteins. Thus, to further understand how the protein expression in the treated groups varied from the non-treated ones, fold-change of protein expression was calculated with respect to the non-treated group (Figure 2C2). We found comparable expressions of Angiopoietin-like 4, Axl, Capthepsin-B, Dkk-1, C-X-C motif chemokine ligand 8 (CXCL8)/IL-8, MMP-2, MMP-3, Progranulin, Serpin E1, Tenascin C, and Vimentin, for all the three groups, irrespective of doxorubicin treatment. There was a 2–4-fold increase in expression levels of Decorin, Enolase-2, Forkhead box protein C2 (FoxC2), Galectin-3, IL-6, Lumican, C-C motif chemokine ligand 2/monocyte chemoattractant protein-1 (CCL2/ MCP-1), macrophage colony-stimulating factor (M-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), Osteopontin (OPN), urokinase plasminogen activator (uPA), and vascular endothelial grow factor (VEGF) for both doxorubicin treated groups (0.1 and 1 μm), as compared to the non-treated group. Additionally, the 0.1 μm treatment exhibited increased expression of ectonucleotide pyrophosphate phosphodiesterase type II (ENPP-2)/Autotaxin, Cathepsin D, and CCL20, while all of these were similar in expression for the 1 μm treatment, when compared to the non-treated group. In contrast, a 0.5–0.7-fold decrease in the expression of heme oxygenase 1 (HMOX1) was observed for both 0.1 and 1 μm treated groups, and a decrease in secreted protein acidic and rich in cysteine (SPARC) and Tyrosine-protein kinase receptor (Tie-2) expression was observed for the 1 μm treatment as well.
2.4. Treating Tumors with Anti-HER2 CAR-T Cells in a Dynamic Flow-Based Culture
We next asked whether the engineered CAR-T cells would be effective against solid tumors when subjected to a dynamic flow-based culture. To understand the efficacy of the engineered CARs in identifying and eliminating HER+ MDA-MB-231 cells in 3D cultures, we first utilized free-standing tumors and evaluated their response against anti HER2 and anti CD19 CAR-T cells under static culture conditions without perfusion. Homotypic and heterotypic tumors treated with anti HER2 CAR-T cells exhibited ≈60–100% decrease in GFP intensity of MDA-MB-231 cells, over 3 days of culture, which indicated cancer cell death (Figures S11(A1-A3) and S12(A1-A3) Supporting Information). Whereas heterotypic H231F tumors exposed to anti CD19 CAR-T cells did not exhibit any significant decrease in GFP intensity (Figure S12(A2,A3) Supporting Information). In contrast, anti CD19 CAR-T cell treatment of homotypic MDA-MB-231 tumors resulted in ≈60–80% decrease in GFP intensity for treatment densities higher than 2xT (Figure S11(A2,A3) Supporting Information).
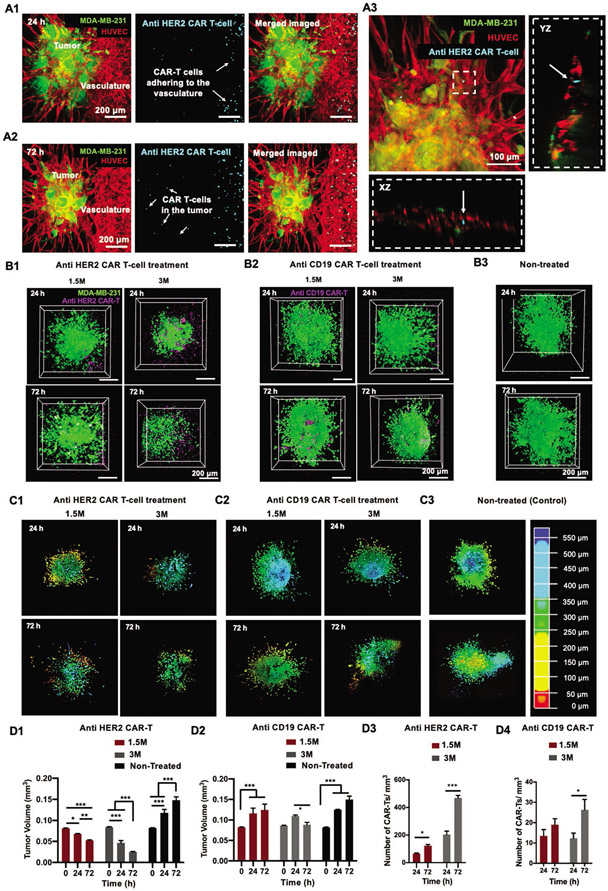
Following in vitro static cultures, CAR-T cells were perfused through the engineered vasculature to analyze the cancer-immune interaction in a dynamic-flow based culture (Movie S4, Supporting Information). After 24 h, CAR-T cells were found adhering to the central vasculature, through which they were perfused (Figure 3A1). The number of CAR-T cells adhering to the endothelium increased after 72 h of perfusion, which indicated an inflammatory microenvironment (Figure 3A2). Additionally, CAR-T cells were also found to have infiltrated to the tumor site, probably via the anastomosed capillaries and the porous C2F3 matrix. CAR-T cells were also found located near the sprouts as well as inside the tumor-associated capillaries (Figure 3A3).
Figure 3.
Culture of engineered CAR-T cells in a 3D bioprinted perfusable and vascularized tumor microenvironment. A1) Fluorescent images of devices perfused with anti HER2 CAR-T cells for 24 h. CAR-T cells were labeled with cell tracker violet (CTV) and observed to adhere to HUVECs in the perfused vasculature. A2) Fluorescent images of devices perfused with anti-HER2 CAR-T cells for 72 h. CAR-T cells were observed to have infiltrated to the tumor site. A3) Orthogonal projection of anti HER2 CAR-T cells inside a capillary (denoted by white arrows). 3D Reconstruction of tumor cores illustrating GFP+ MDA-MB-231 cells and CTV-labelled B1) anti-HER2 CAR-T cells and B2) anti-CD19 CAR-T cells. CAR-T cell treatment density was varied as 1.5 and 3 million active CAR-T cells. Perfusion was carried out for 24 h or 72 h for both concentrations. B3) 3D Reconstruction of tumor cores illustrating the GFP+ MDA-MB-231 cells for non-treated (control) groups. 3D Reconstruction of the entire tumor remaining after 24 or 72 h of C1) anti HER2 CAR-T cell and C2) anti CD19 CAR-T cell treatment and subsequent C3) non-treated (control) tumors. Tumors were depth coded to represent their entire volume in 3D. Graphical representation of tumor volumes after 24 or 72 h of D1) anti-HER2 CAR-T cell and D2) anti CD19 CAR-T cell perfusion. Graphical representation of average CAR-T cell numbers located near the tumor after D3) anti HER2 and D4) anti CD19 CAR-T treatment. (n = 3 for all, p*** < 0.001, p** < 0.01, p* < 0.05).
To better understand the effect of CAR-T cell treatment on the bioprinted tumor microenvironment, two treatment densities, comprising of 1.5 million (1.5 M) and 3 million (3 M) active CAR-T cells were perfused for a period of 24 or 72 h. For both treatment densities, CAR-T cells had infiltrated to the tumor site (Figure 3B1,B2). For the 3 M treatment density, presence of CAR-T cells in the tumor core was visibly higher after 72 h of perfusion for the anti HER2 treatment as compared to the anti CD19 treatment (Figure 3B1,B2). The number of anti HER2 CAR-T cells found near tumors for both 1.5 M and 3 M treatments was ≈2–2.5 folds higher after 72 h perfusion as compared to 24 h (Figure 3D3). Additionally, for both 24 h and 72 h anti HER2 CAR-T 3 M treatment, the number of CAR-T cells found in and around the tumor post treatment was ≈3–4 folds higher as compared to the 1.5 M treatment. In contrast, anti CD19 CAR-Ts exhibited lower CAR-T numbers irrespective of the treatment densities or treatment duration (Figure 3D4). For the 3 M treatment, after 72 h of anti CD19 CAR-T perfusion, there was a approximately twofold increase in CAR-T cell number around the tumor, however, it was still significantly lower than the anti HER2 treatment.
Under this dynamic flow-based culture, anti HER2 CAR-T cells suppressed the tumor growth as compared to the non-treated group (Figure 3B1-B3). 3D Reconstruction of tumor volumes after CAR-T cell treatment further revealed a reduction in tumor volume after 24 and 72 h of anti HER2 CAR-T treatment as compared to both anti CD19 and non-treated groups (Figure 3C1-C3). On comparing the tumor volume (≈0.081 mm3) prior to onset of treatment (0 h), tumors under 1.5 M anti HER2 CAR-T cell treatment for 24 h had a ≈16% reduction in volume (≈0.068 mm3) and a further ≈22% reduction (0.053 mm3) after 72 h treatment (Figures 3C1 and 4D****1****). Increasing the treatment density to 3 M resulted in a significant ≈44% (≈0.047 mm3) and a further ≈46% (≈0.025 mm3) decrease in tumor volume after 24 and 72 h CAR-T cell perfusion, respectively (Figure 3C1,D1). In contrast, as compared to the tumor volume before the treatment (0.082 mm3), perfusing 1.5 M anti CD19 CAR-T cells resulted in a ≈41% (0.116 mm3) increase in tumor volume after 24 h and a further increase of ≈7.5% (0.124 mm3) in the next two days (72 h) (Figure 3C2,D2). Increasing the anti-CD19 CAR-T cell treatment density to 3 M resulted in a ≈28% (0.11 mm3) increase in tumor volume after 24 h of CAR-T cell perfusion and a subsequent ≈20% decrease (0.088 mm3) in tumor growth after 72 h of perfusion (Figure 3C2,D2). Tumor volumes of non-treated tumors increased by ≈80% (≈0.15 mm3) in volume after 72 h of perfusion (Figure 3C3,D1,D2).
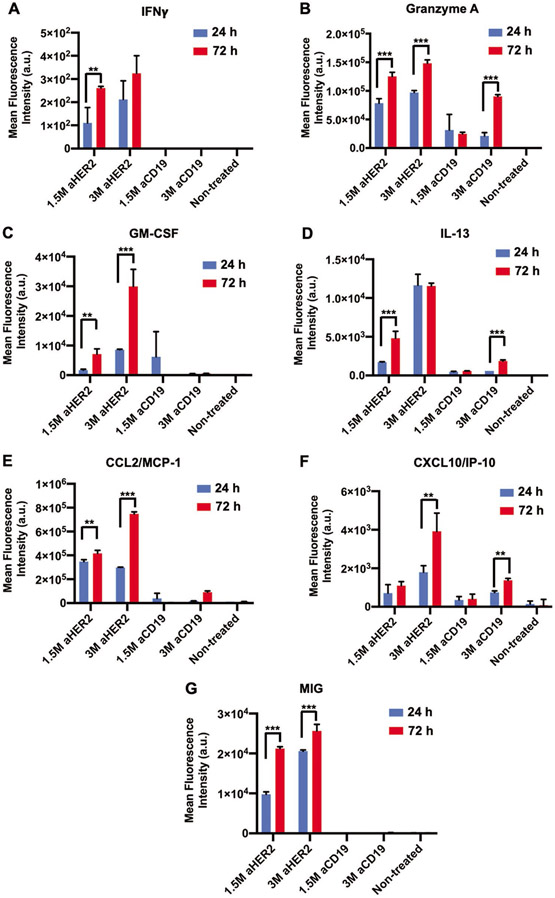
Figure 4.
Graphical representation of the mean fluorescence intensity of the cytokines and chemokines secreted after 24 or 72 h of CAR-T cell perfusion including A) IFNγ, B) Granzyme A, C) GM-CSF, D) IL-13, E) CCL2/ MCP-1, F) CXCL10/ IP-10, and G) MIG (n = 3, p* < 0.05, p** < 0.01, p*** < 0.001).
Cytokines and chemokines, secreted as an aftermath of the cancer-immune interactions, were assessed from the culture supernatants collected from the last 24 h of the entire 72-h-perfusion culture with CAR-T cells. Increased expression of interferon-gamma (IFNγ) was only observed after anti HER2 CAR-T cell perfusion, for both 1.5 M and 3 M treatments, which was indicative of HER2 specific CAR-T cell activation (Figure 4A). Subsequent cytotoxic Granzyme A secretion was also observed to be higher for the anti HER2 CAR-T cell treatment, as compared to anti CD19 treatment (Figure 4B). Additionally, cytokines GM-CSF and IL-13, and chemokines CCL2/MCP1, CXCL10 (also known as interferon γ induced protein 10 (IP-10)), and monokine induced by gamma interferon (MIG) were all detected in higher quantities in the culture perfusates of anti HER2 CAR-T cell treated tumors as compared to anti CD19 CAR-T cell treated counterparts (Figure 4C-G). Furthermore, expression of all cytokines and chemokines were higher after 72 h of CAR-T cell perfusion as compared to 24 h, which indicated enhanced immune activation and immune response over time. Enhanced secretion of these immune activators was also observed under in vitro static culture conditions for both homotypic MDA-MB-231 and heterotypic H231F tumors (Figures S11(B1-B7) and S12(B1-B7) Supporting Information). Specifically, expression of IFNγ, Granzyme A, GM-CSF, and IL-13 were higher with anti HER2 CAR-T cell treatment as compared to anti CD19 CAR T control condition (Figures S11(B1-B4) and S12(B1-B4), Supporting Information). In contrast, chemokines CCL2/MCP1, CXCL10/IP-10 and MIG were equally expressed in in vitro static cultures after both anti HER2 and anti CD19 CAR T-treatments, which indicated higher degree of non-specific cancer immune interactions in static cultures (Figures S11(B5-B7) and S12(B5-B7) Supporting Information).
3. Discussion
This study demonstrates the effect of chemo and immunotherapeutics on a 3D bioprinted vascularized breast tumor model. Employing aspiration-assisted 3D bioprinting, heterotypic tumors comprising of endothelial cells, cancer cells, and fibroblasts were precisely bioprinted at desired locations from a central vasculature in a composite collagen/fibrin based biomimetic matrix. The addition of endothelial cells with cancer cells pre-vascularizes the tumor and exhibits a suitable co-culture model for studying cancer-endothelial interaction in vitro. The co-cultured spheroids sprout capillaries from the tumor, which in an in vivo setting, anastomose with existing capillaries around the tumor.[19,24] Constant flow of media supplemented with growth factors, at low shear stress through the main vasculature, eventually induced tumor angiogenesis and aided cancer metastasis in tumors bioprinted proximal or distal to the vasculature. Low shear stress supported angiogenenic sprouting as high shear stresses have shown to be inhibitory to sprouting.[25] The incorporation of fibroblasts in spheroids not only rendered them mechanically suitable for bioprinting, it also helped to recapitulate the ‘reactive stroma’ of a native tumor microenvironment.[26] We had also previously observed specific upregulation of genes and activation of signaling pathways indicating the transformation of these fibroblasts to a ‘Cancer-associated fibroblast-like’ state. RNA-sequencing of a heterogeneous tumor microenvironment revealed genes upregulated in the composite system, which positively affected metastasis and angiogenesis in the presence of human dermal fibroblasts.[19]
Tumors bioprinted proximal to the central vasculature developed longer and denser vessels with a higher junction density probably due to enhanced diffusion of nutrients through the main vasculature and subsequent local accumulation of growth factors and cytokines, such as VEGF-A, basic fibroblast growth factor, and platelet-derived growth factor-BB.[27] This could also be attributed to the probable upregulation of local matrix metalloproteinases (MMPs) near the perfused vasculature, which contributes to greater matrix degradation, thus enabling HUVECs in the proximal tumors to develop longer vessels.[28] Moreover, these results also suggest that HUVECs associated with the tumor are capable of sensing metabolic and physiological changes in the surrounding stroma, which eventually activates the angiogenic signaling cascade.[29]
The efficacy of doxorubicin-based chemo and CAR-T cell-based immunotherapy were effectively studied in this vascularized and dynamic microenvironment. Doxorubicin, an anthracycline-based chemotherapy drug, primarily intercalates into the DNA and inhibits topoisomerase II (TOP2) in proliferating cancer cells, eventually leading to cancer cell death.[30] Perfusion of doxorubicin to treat proximal tumors resulted in extensive cancer cell death for 100, 10, and 1 μm dosages, which was evident from decreasing tumor volumes post-treatment. Overall, testing doxorubicin under a flow-based culture exhibited a dose dependent reduction in tumor growth with decreased doxorubicin cytotoxicity below 1 μm. Induction of apoptotic pathways are generally associated with the activation of a group of cysteine proteases, called caspases.[31] Specifically, Caspase 3 that catalyzes cleavage of key cellular proteins, DNA fragmentation, cell rupture, and formation of apoptotic bodies was significantly enhanced in 100–1 μm doxorubicin treated cultures.[32] Proteome array analysis also suggested chemo-resistance in tumors below 1 μm and protein expression profiles of both treated groups (1 and 0.1 μm) compared to the non-treated group, showed a substantial increase in the expression of several cytokines, chemokines, and growth factors, post-chemotherapy. Proteins such as decorin, enolase-2, foxC2, galectin-3, IL-6, lumican, CCL2/MCP-1, M-CSF, GM-CSF, OPN, uPA, and VEGF were all significantly higher in expression post chemotherapy. These cytokines and growth factors secreted by cancer cells play a major role in cancer cell proliferation and survival, and progression as well as formation of tumor stroma.[33] Thus, overexpression of these proteins could provide proliferative and anti-apoptotic signals aiding tumor survival and helping the tumor escape from drug-mediated apoptosis. Cytokines and chemokines, such as IL-6, IL-8, MCP-1, M-CSF, and GM-CSF, are all produced by cancer cells and heavily involved in a myriad of paracrine and autocrine functions.[34] IL-6 and IL-8 are both known to stimulate tumor cell proliferation as well as promote angiogenesis.[35,36] GM-CSF and M-CSF are both immune modulatory cytokines secreted by activated immune (macrophage, monocyte) cells, stromal (fibroblast, vascular endothelial cells), or even cancer cells, including, MDA-MB-231 cells, in response to various stimuli.[37,38] Increased expression of FoxC2 increases the promoter activity of adenosine triphosphate binding cassette transporters, which are known to be associated with multidrug resistance and epithelial–mesenchymal transition.[39,40] All of these proteins are involved in matrix remodeling, [41] cell migration,[42] cell metabolism, and their increased expression after the doxorubicin treatment exhibits the propensity of metastatic cancer cells to evade apoptosis.[43] Increased expression of all these factors enables the cancer cells to evade drug-induced death.[44,45]
Cellular engineering of immune cells as CAR-T cells has shown great therapeutic promise against hematological tumors but is not yet effective against solid tumors. An immune-suppressive microenvironment composed of a dense mass of tumor and stromal cells and abnormal vasculature restricts immune cell infiltration and thus suppresses immune function. [46,47] Our 3D vascularized tumor model consisting of different facets of the tumor microenvironment (dense tumor mass, stromal cells, and vasculature) itself might be suggestive of immune-suppressive microenvironment. Perfusing CAR T-cells through the central vasculature resulted in extensive CAR-T cell adhesion to vasculature wall and this phenomenon could be attributed to the presence of E- and P-selectins, which enable rolling of CAR-T cells on the endothelium. [48] Furthermore, activated T cells express ligands, which enable them to bind to intercellular adhesion molecule 1 (ICAM-1) and vascular cell adhesion molecule 1 (VCAM-1) present on endothelial cells.[49] It was also interesting to observe the anti HER2 CAR-T cells had a higher infiltration to the tumor site as compared to the anti CD19 CAR-T cells. Migration of CAR-T cells toward a tumor is dependent on the presence of various stimulatory chemokines. Increased expression of IFNγ post culture with anti-HER2 CAR-T cell versus B cell lymphoma specific anti-CD19 CAR-T cells, which served as controls, revealed antigen specific T cell activation leading to antitumor activity.[50] Higher expression of IFNγ along with cytotoxic granzymes, which are known to mediate cellular apoptosis by diffusing through perforin pores on the plasma membrane of target cells, possibly resulted in an antiproliferative, and pro-apoptotic tumor microenvironment in HER2 targeted CAR treatments.[51,52] Furthermore, higher secretion of cytokines GM-CSF and IL-13 in HER2 targeted CAR-T cell treated groups as compared to anti-CD19 CARs, also substantiated HER2 specific CAR-T cell activation. Chemokines, such as CCL2/MCP-1, and CXCL10/IP-10, and MIG/CXCL9, play a crucial role in immune cell stimulation and immune cell recruitment to intra-tumoral sites.[53] Their secretion is known to be induced by IFNγ are produced by a wide range of cell types, including endothelial cells, fibroblasts, monocytes, and neutrophils.[54-56] All of these cytokines play a role in cytokine release syndrome or cytokine storm, which is a major side effect of CAR-T cell based immunotherapy.[57] Higher expression of these factors after HER2 targeted CAR-T cell treatment as compared to CD19 CARs, further validated the 3D bioprinted perfusable tumor model as an effective platform for not only immunotherapy screening but also further eliciting immune responses post-treatment. It does not only advance the field but also sets a framework for modeling such 3D environments with patient-specific tumor, stromal, as well as immune cells. Even though we have studied the responsiveness of the bioprinted tumor model with regards to chemo and CAR-T cell-based immunotherapy, this model could potentially be utilized to study natural killer cell-based immunotherapy as well. Tumor microenvironments are often found to be hypoxic,[58] which could also be modeled in our system. Oxygen scavenging molecules, such as sodium sulphite, could be perfused through the central vasculature to create an oxygen gradient, prior to introducing anti-cancer therapeutics.[59] In the future, to enhance the clinical relevance and translatability of this 3D model, patient-derived organoids from different cancer types will be bioprinted in place of heterotypic tumor spheroids in this perfusable model.
4. Conclusions
In summary, we devised a method to study CAR-T interaction with solid tumors in a vascularized tumor microenvironment, thus taking a step toward understanding the intricacies needed to develop more targeted therapies. Precise control over the spatial location of tumors substantiated that distance of tumors from a perfused vasculature affects tumor angiogenesis and cancer invasion, two of the major hallmarks of cancer. Dose dependent reduction in tumor volumes as well as drug resistance beyond 0.1 μm of doxorubicin validated the responsiveness of tumor microenvironment against therapeutics. Most importantly, perfusing HER2 targeted CAR-T cells through the tumor vasculature, resulted in extensive T cell recruitment to the endothelium, suggestive of an inflammatory microenvironment. Perfusing HER2 targeted CARs through vasculature induced secretion of inflammatory cytokines and chemokines, activating CARs to generate antitumor response, which resulted in decreased tumor growth. Overall, fabrication of a physiologically-relevant in vitro 3D tumor model is not only essential for understanding critical cancer-stroma cross talk but also identifying novel therapeutic targets and developing targeted therapies against cancer.
5. Experimental Section
Cells and Reagents:
GFP+ MDA-MB-231 breast cancer cells were donated by Dr. Danny Welch, from University of Kansas (Kansas City, KS). They were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Corning Cellgro, Manassas, VA) supplemented with 5% fetal bovine serum (FBS) (Life Technologies, Grand Island, NY), 1 mm Glutamine (Life Technologies, Carlsbad, CA) 1 mm penicillin-streptomycin (Life Technologies, Carlsbad, CA). HUVECs and HDFs were procured from Lonza (Walkersville, MD). HUVECs were cultured in MCDB 131 media (Corning, NY) supplemented with 10% FBS, 1 mm Glutamine, 1 mm penicillin-streptomycin, 0.5 mm bovine brain extract (BBE, Lonza, Walkersville, MD), 1200 U mL−1 heparin (Sigma-Aldrich, St. Louis, MO) and 0.25 mm endothelial cell growth supplement (ECGS, Sigma-Aldrich). HUVECs were used at passages 2 through 7. HDFs were cultured in DMEM supplemented with 10% FBS, 1% glutamine, 1% sodium pyruvate, and 1% penicillin-streptomycin. HDFs were used at passages 2 through 8. Cells were maintained at 37 °C with 5% CO2 in an air-humidified atmosphere. Cell culture medium was changed every 2–3 days. Sub-confluent cultures were detached from cell culture flasks using a 0.25% trypsin-0.1% ethylenediaminetetraacetic acid (EDTA) solution (Life Technologies) and split to maintain cell growth. HUVECs were further transduced with tdTomato lentiviral vector to ease visualization for all experiments.
Transduction of HUVECs:
HUVECs (Lonza, Walkersville, MD) were transduced at passage 2 (≈50% confluency) with EF1 tdTomato lentiviral vector (Vectalys, Toulouse, France). A multiplicity of infection (MOI) of 20 was maintained for the transduction process. Briefly, a transduction mix was prepared by adding a measured amount of viral vector solution in complete culture media and 800 μg mL−1 polybrene (Sigma). This transduction mix was then transferred to a cell flask at ≈50% confluency and incubated for 8 h. The transduction mix was then discarded and the flask was rinsed with Dulbecco’s phosphate buffered saline (DPBS, 1X) and replenished with culture media. Cells were then allowed to grow in flasks for another 48 h before sorting them on a MoFlo Astrios sorter (Beckman Coulter, Pasadena, CA) for the brightest cells. The brightest cells were then collected for further cell culture.
Tumor Spheroid Formation for 3D Bioprinting:
Tumor (H231F) spheroids comprised of 7000 HUVECs, 1000 MDA-MB-231 cells, and 1000 HDFs. Cells were individually trypsinized and combined according to the above-mentioned ratio and seeded in a 96-well U-bottom cell repellant plate (Greiner Bio-One, Monroe, NC) in 75 μL of EGM-2MV media per well. Cells were cultured in the U-bottom plate for 24 h to form spheroids.
Immunostaining of Tumor Spheroids:
Tumor spheroids were fixed in 4% paraformaldehyde (Sigma-Aldrich) and rinsed in DPBS (1X). The spheroids were permeabilized using 0.1% Triton-X100 (Sigma–Aldrich, Burlington, MA) for 15 min and blocked with 10% normal goat serum (NGS, Thermo Fisher, Waltham, MA), 0.3 m glycine (Sigma-Aldrich), 0.1% Tween-20 (Sigma-Aldrich), and 1% BSA (Sigma–Aldrich) in DPBS for 1 h. Samples were then incubated with rabbit monoclonal anti-platelet-derived growth factor receptor (PDGFR) alpha antibody (1:200, Abcam, Waltham, MA) in the same blocking solution overnight at 4 °C. Afterward, they were washed twice with DPBS and incubated with goat anti-rabbit IgG (H + L)-Alexa Fluor 647 (1:1000, Invitrogen, Waltham, MA) for 1 h at room temperature, followed by incubation with Hoechst 33 258 (1:200, Sigma–Aldrich) for 30 min to visualize the cell nuclei. Images were taken using a Leica SP8 DIVE Multiphoton Microscope (Leica Microsystems, Germany).
Cellular Distribution Analysis:
H231F spheroids were fixed in 4% paraformaldehyde then rinsed in DPBS (1X). The spheroids were paraffin embedded and sectioned to obtain 8 μm-thick histological sections. The sections were then dewaxed using Leica Autostainer XL (Leica, Germany), then permeabilized using 0.1% Triton-X100 (Sigma–Aldrich, Burlington, MA) for 15 min and blocked with 10% NGS, 0.3 m glycine (Sigma-Aldrich), 0.1% Tween-20 (Sigma-Aldrich), and 1% BSA (Sigma–Aldrich) in DPBS for 1 h. Samples were then incubated with rabbit anti-PDGFR antibody (1:100, Invitrogen, Waltham, MA) in the same blocking solution overnight at 4 °C. Afterward, they were washed twice with DPBS and incubated with goat anti-mouse IgG (H + L)-Alexa Fluor 647 (1:1000, Invitrogen, Waltham, MA) for 1 h at room temperature, followed by incubation with Hoechst 33 258 (1:200, Sigma–Aldrich) for 30 min to visualize the cell nuclei. Images of the sections were taken with Zeiss LSM880 confocal microscope (Oberkochen, Germany), and plot profile of each image was obtained using Image J.
Immunostaining for α-Smooth Muscle Actin (αSMA):
Homotypic MDA-MB-231 only tumors, heterotypic H231, and H231F tumor spheroids were generated and kept cultured for 3 days. Spheroids were then fixed in 4% paraformaldehyde (Sigma-Aldrich) and rinsed in DPBS (1X). The samples were permeabilized using 0.1% Triton-X100 (Sigma Aldrich, Burlington, MA) for 15 min and blocked with 10%, 0.3 m glycine (Sigma–Aldrich), 0.1% Tween-20 (Sigma–Aldrich), and 1% BSA (Sigma Aldrich) in DPBS for 1 h. Samples were then incubated with mouse monoclonal anti-αSMA antibody (1:100, Invitrogen, Waltham, MA) in the same blocking solution overnight at 4 °C. Afterward, they were washed twice with DPBS and incubated with goat anti-mouse IgG (H + L)-Alexa Fluor 647 (1:1000, Invitrogen, Waltham, MA) for 1 h at room temperature, followed by incubation with Hoechst 33 258 (1:200, Sigma Aldrich) for 30 min to visualize the cell nuclei. Images were taken using a Zeiss LSM880 confocal microscope (Oberkochen, Germany).
Picrosirius Staining:
In order to study the collagen secretion of tumor spheroids, homotypic MDA-MB-231 only tumors, heterotypic H231 and H231F tumor spheroids were cultured for 3 days. After the fixation in 4% paraformaldehyde, spheroids were paraffin embedded and sectioned to obtain 8 μm histological sections. The sections were then dewaxed using Leica Autostainer XL, and stained with 2% picrosirius red solution (Thermo Fisher, Waltham, MA) for 1 h at room temperature. The dye was then removed, and samples were washed 10 times with DI water, followed by dehydration with ascending alcohol and clearing with xylene. All samples were mounted and imaged using the EVOS microscope. During the sectioning process of MDA-MB-231 cells-only spheroids, a part of the spheroids was lost due to their fragility.
Collagen Extraction:
Collagen type-I was extracted from rat tails according to a previously published protocol.[60] Collagen fibers were extracted from rat tail tendons, dissolved in 0.02 m acetic acid (Sigma Aldrich) and then subsequently freeze-dried to obtain collagen sponges. These sponges were again re-dissolved in acetic acid at a desired concentration, centrifuged to remove insoluble impurities, and then sterilized using Spectra/Por 1 dialysis tubing (6–8 kDa MWCO) (Spectrum Labs, Rancho Dominguez, CA).
Hydrogel Preparation for 3D Perfusable Devices:
Fibrinogen and thrombin were purchased from Sigma Aldrich (Burlington, MA). Equal volumes of 4 mg mL−1 type I collagen and 6 mg mL−1 fibrin was mixed to obtain a final concentration of 2 mg mL−1 collagen and 3 mg mL−1 fibrin.[19] This composite hydrogel is referred to as “C2F3”. Briefly, 5 μL DPBS (10X), 0.51 μL sodium hydroxide (1 n), 22.4 μL EGM-2MV media, 50 μL fibrinogen (6 mg mL−1), 22.2 μL collagen (9 mg mL−1) and 1.5 U mL−1 thrombin (50 U mL−1) were mixed in this specific order. All the above-mentioned solutions, except fibrinogen, were kept on ice prior to mixing. HDFs at a concentration of 0.5 million mL−1 were suspended in C2F3 and cast in the device prior to 3D bioprinting.
Ultrastructural Analysis of the Tumor Spheroids and Hydrogel:
Field emission scanning electron microscopy (SEM, Apreo, Thermo Fisher Scientific, Waltham, MA) was used to investigate the hydrogel architecture as well as to assess surface topography of tumor spheroids. Tumor spheroids were harvested after 24 h of culture in U-bottom well plates. Hydrogel samples comprised of 2 mg mL−1 collagen (C2), 3 mg mL−1 fibrin (F3) and the C2F3 composite hydrogel. All samples were fixed in 4% paraformaldehyde (Santa Cruz Biotechnology, Dallas, TX) overnight. Samples were then washed in DPBS (1X) to remove the fixative. Next, they were dehydrated using graded ethanol solutions (25%, 50%, 70%, 90%, 100%). To ensure complete removal of water, samples were further dried in a critical point dryer (CPD300, Leica EM, Wetzlar, Germany) for 4 h. On complete dehydration, they were sputter coated with iridium using a sputter coater (Leica) and observed at an accelerating voltage of 3–5 keV on the Apreos SEM (Thermo Fisher).
Rheological Studies on Collagen-Fibrin Composite Hydrogel:
Rheological properties of C2F3 were characterized using a rheometer (MCR 302, Anton Paar, Austria). All rheological measurements were performed in triplicates with a 25 mm and 50 mm cone plate with a 1-degree gap angle at room temperature (22 °C) and 37 °C controlled by a Peltier system. The time sweep test was carried out in an angular frequency range of 1 Hz. To monitor changes in rheological properties of C2F3 incubated at two different temperatures (22 °C to mimic bioprinting condition and 37 °C to mimic incubation conditions post-bioprinting), a time sweep test was carried out under constant dynamic-mechanical conditions at a fixed angular frequency of 1 Hz and a fixed shear strain of 0.1%, which was within the linear viscosity region. The storage modulus (G′) was logged from the amplitude sweep of all samples at a constant frequency of 1 Hz at a strain range from 0.01% to 2% after 40 min incubation time at 37 °C. The relative stiffness of C2, F3, and C2F3 was determined according to a published protocol.[61] The storage modulus of each sample at 1% strain was obtained and the elastic moduli (E) was determined using the following equation:
| (1) |
where , is Poisson’s ratio of hydrogel that is typically 0.5.
Measurement of Mechanical Strength of Tumor Spheroids:
A pulled micropipette with a final inner diameter of 70–85 μm, connected to vacuum, was used to measure the mechanical strength of MDA-MB-231, H231, and H231F spheroids. The micropipette was placed on an optical microscope (Motic, Schertz, TX) and the testing was performed in medium with a fixed aspiration pressure of 4 kPa to apply the force to spheroids for 10 min. The aspiration pressure was controlled by a pressure controller (Ultimus I, Nordson EFD, Westlake, OH). Videos were recorded for 10 min and images were taken every 1 min. The deformation of spheroids was calculated based on the change in length of aspiration in the micropipette tip from pre-deformed spheroids. The elastic modulus was calculated using a previously established equation, (Equation 2) under the assumption of the homogeneous half-space model.[62,63]
| (2) |
where is the elastic modulus, is the inner radius of the micropipette, is the length of aspiration, is the aspiration pressure, and is the geometry of pulled micropipette.
Device Fabrication and Aspiration-Assisted Bioprinting of Tumor Spheroids:
A poly-lactic acid (PLA)- based 3D printed structure was used as the perfusable chamber (Figure 1A1). This structure was designed using Autodesk Inventor (Autodesk, San Rafael, CA) and then 3D printed by Ultimaker 2 (Ultimaker, Germany). This 3D printed device was coated with 1 mg mL–1 of poly-D lysine hydrobromide (Sigma Aldrich) overnight. Devices were then thoroughly rinsed with sterile DI water to remove excess lysine and air dried prior to printing. H231F tumor spheroids were suspended in 3 mg mL–1 fibrinogen prior to bioprinting. Tumor spheroids were individually aspirated from fibrinogen using a 30G blunt nozzle, having a diameter of ≈150 μm, and vacuum pressure of ≈20 mmHg. They were then bioprinted into the semi-crosslinked C2F3 matrix at a speed of 5 mm s–1. The first tumor was bioprinted within 60 sec of C2F3 deposition. A total of six spheroids were bioprinted in C2F3 within 5 min. Spheroids were bioprinted at controlled distances of ≈100 μm (proximal) and ≈500 μm (distal) from the stainless-steel wire (later referred to as the “central vasculature”) as shown in Figure 1B1-B3. After device fabrication, it was transferred to a humidified incubator at 37 °C and 5% CO2 flow for complete hydrogel cross-linking. The stainless- steel wire was removed from the device leaving behind an open channel. The open channel was flushed with cell culture media a few times. HUVECs were injected into the channel, and the device was turned every 30 min to ensure homogenous cell attachment on both upper and lower channel walls. After 60 min, the device was submerged in EGM-2MV media and incubated at 37 °C and 5% CO2 flow, overnight, to allow HUVECs to completely adhere to the channel walls.
Measurement of Bioprinting Accuracy:
To evaluate the accuracy and precision of bioprinting of H231F spheroids in C2F3, spheroids were bioprinted at a predetermined target position on a micrometer calibration ruler. The calibration ruler was placed at the bottom of a Petri dish and recorded by a microscopic camera (Plugable USB Digital Microscope, Plugable Technologies, Redmond, WA) to monitor the target position. A total of 246 spheroids were bioprinted and analyzed by ImageJ (National Institutes of Health (NIH), MD, USA). Accuracy was represented by the root mean square error (RSME) and calculated using the following equation:
| (3) |
where, and are the and coordinates of the target position, respectively, and are the positions of bioprinted spheroid measured in and axes, respectively, and is the sample size. Precision was represented as the square root of the standard deviation.
Measuring Tumor Circularity after Bioprinting:
To represent the effect of bioprinting on the deformation of spheroids, H231F spheroids (n = 50) were bioprinted in C2F3 matrix. Free-standing spheroids in 96-well plates before bioprinting were used as a control group. To quantify the morphology of spheroids, circularity was calculated using ImageJ software according to the following equation:
| (4) |
where represents circularity, is the area, and is the perimeter of the bioprinted spheroid. The value “0” indicated an infinitely elongated polygon and “1” indicated a perfectly circular shape.
Perfusion, Imaging, and Quantitative Analysis of 3D Bioprinted Perfusion Device:
HUVECs were injected into the channel to form a uniform endothelial lining, as described under device fabrication. After HUVEC attachment to the channel walls, the device was connected to an external pump (Reglo Ismatec, MasterFlex, Radnor, PA) and perfused for a period of 6 days. The perfusion speed was maintained at 0.7 μL min−1. Formation of endothelium was confirmed using dextran perfusion and immunostaining. 3D Bioprinted devices were fixed with 4% formaldehyde (Santa Cruz, California, USA), overnight. Devices were then washed with DPBS (1X) and imaged using a 16x immersion lens on the Leica SP8 DIVE multiphoton microscope. For imaging the entire region including the six bioprinted spheroids, an imaging workflow was set up on the Leica software interface, where Z stacks were collected from individually defined regions and then stitched together using the ‘mosaic merge’ function. Quantitative analysis on tumor angiogenesis was performed using the Angiotool software.[64]
Dextran Perfusion for Diffusional Permeability:
Diffusional permeability was measured for the engineered vasculature by perfusing 20 μg mL−1 (Fluorescein isothiocyanate) FITC-conjugated 70 kDa dextran (Sigma Aldrich) in EGM-2MV media, for 40 min. Images were captured every 5 min using a fluorescence microscope (AxioObserver, Zeiss, NY). The diffusion of dextran and subsequent change in fluorescence intensity was measured using ImageJ. The diffusional permeability was then quantified based on the following equation:
| (5) |
Measurements were all performed on channels with and without endothelium (n = 3).
Drug Study:
Doxorubicin hydrochloride (Tocris Biosciences, Minneapolis, MN) was dissolved in dimethyl sulfoxide (DMSO) to prepare a stock concentration of 50 mm, aliquoted and stored at −30 °C. In this study, doxorubicin concentration was diluted 10-folds each time, starting from a very high concentration of 100 μm up to a very low concentration of 0.0001 μm. The non-treated control group contained only the vehicle (DMSO) diluted in EGM-2MV media (0.1%). For free-standing spheroids cultured in static conditions, spheroids in well plates were directly exposed to doxorubicin for a period of 72 h. Similarly, to understand the effect of doxorubicin on spheroids encapsulated in C2F3, spheroids were first cultured in C2F3 for a period of 3 days. Then, doxorubicin, diluted in culture media (100 – 0.0001 μm), was added to these cultures and incubated for 72 h. As the MDA-MB-231 cells used were GFP+, images were captured daily using a 4x lens on the EVOS fluorescent microscope to assess the changes in fluorescence intensity of the cancer cells over time. For imaging doxorubicin uptake after perfusion, devices were fixed overnight with 4% paraformaldehyde at 4 °C. Then, devices were rinsed well with 1x DPBS and imaged using a 16x immersion lens on SP8 multiphoton (Leica Microsystems, Morrisville, NC). To image doxorubicin, a 780 nm laser excitation was used and the emission was detected into the red/far-red spectrum following two-photon excitation.[23]
Viability Study after Drug Testing:
After incubating free-standing and hydrogel encapsulated tumor spheroids with doxorubicin for a 72-h culture period, Alamar Blue assay (Thermo Fisher) was performed according to the manufacturer’s protocol in order to quantify the cell viability after the drug treatment. Briefly, samples were incubated with Alamar Blue solution, which was diluted at a ratio of 1:10 in culture media, for a period of 4 h. The culture supernatant was then removed and fluorescence intensity of the resorufin produced was measured at an excitation between 530–560 and an emission at 590 nm, using a fluorescence-based microplate reader (Tecan, Morrisville, NC).
Caspase 3/7 Activity Assay:
Induction of apoptosis after 24 h of doxorubicin treatment was quantified by analyzing Caspase 3/7 activity using the Caspase-Glo 3/7 3D assay (Promega, Madison, WI). The assay protocol was followed from the manufacturer’s website. Briefly, the Caspase Glo 3/7 3D buffer was mixed with the Caspase Glo 3/7 3D substrate. This mixture constituted the Caspase Glo 3/7 3D reagent, which was then added to the perfusion devices. Devices were incubated at room temperature (RT) for 30 min. The solution was removed from the devices and luminescence was recorded on microplate reader (Tecan, Morrisville, NC).
Proteome Array:
In this study, Proteome Profiler Human XL Oncology Array (R&D Systems, Minneapolis, MN) was used to identify 84 different cancer-related antibodies. The experimental groups included a drug treated group (1 μm doxorubicin) and a non-treated group (0 μm doxorubicin). Cell culture supernatant was collected after 72 h drug perfusion, aliquoted and stored at −80 °C for later use. Approximately 300 μL of this supernatant was used for the proteome array for both drug treated and non-treated groups, and the manufacturer’s protocol was followed for this assay. ChemiDoc MP Imaging System (Bio Rad Laboratories, Hercules, CA, USA) was used to image the protein blots and the intensity of the protein blots was quantified using ImageJ.
Designing CAR Constructs:
Anti-CD19 CAR and anti-HER2 CAR encoding lentiviral vectors were produced and titrated as described below. CAR constructs consisted of CD8 alpha signal peptide, single chain variable fragment (scFv) of a CD19 or HER2 antibody, CD8 hinge domain, CD8 transmembrane domain, 4-1BB (CD137) intracellular domain, and CD3ζ domain that were designed with Snapgene and synthesized via Genscript. CD8a signal peptide, CD8 hinge, CD8 transmembrane domain, 4-1BB intracellular domain, and CD3ζ domain sequences were obtained from Ensembl Gene Browser and codon optimized with SnapGene by removing the restriction enzyme recognition sites that are necessary for subsequent molecular cloning steps while preserving the amino acid sequences. Anti-CD19 and anti-HER2 scFv amino acid sequences were obtained from Addgene plasmids #79 125 and #85 424, respectively, reverse translated to DNA sequences and codon optimized with Snapgene 5.2.4. The constructs were then cloned into a lentiviral expression vector with a multiple cloning site separated from GFP reporter via an Internal Ribosomal Entry Site (IRES.
Lentiviral Production and Titration:
Cloned lentiviral constructs including anti-CD19 CAR and anti-HER2 CAR encoding vectors were co-transfected with the packaging plasmids VSVG, pLP1 and pLP2 into 293 cells using Lipofectamine™ 3000 (Invitrogen) according to the manufacturer’s protocol. Viral supernatants were collected 24 – 48 h post-transfection, filtered through a 0.45 μm syringe filter (Millipore) to remove cellular debris, and concentrated with Lenti-X (Invitrogen) according to the manufacturer’s protocol. Lentivirus supernatant stocks were aliquoted and stored at −80°C. To measure viral titers, virus preps were serially diluted on Jurkat cells. 72 h after infection, GFP+ cells were counted using flow cytometry and the number of cells transduced with virus supernatant was calculated as infectious units/per mL. The cells were cultured in complete RPMI 1640 medium (RPMI 1640 supplemented with 10% FBS; Atlanta Biologicals, Lawrenceville, GA), 8% GlutaMAX (Life Technologies), 8% sodium pyruvate, 8% MEM vitamins, 8% MEM nonessential amino acid, and 1% penicillin/streptomycin (all from Corning Cellgro) for 72 h. Trypsin-0.53 mm EDTA(0.05%) (Corning Cellgro) was used to detach adherent cells.
Source and Purification of CD8+ T Cells:
Leukopaks from healthy adults were purchased from ALLCells (Alameda, CA) in deidentified form and processed to isolate Peripheral Blood Mononuclear Cells (PBMCs) using Ficoll-paque plus (GE Healthcare). CD8+ T cells were purified from PBMCs using Dynal CD8 Positive isolation kit (Invitrogen) according to the manufacturer’s instructions. Isolated CD8+ T cells were at >99% purity.
Engineering of CAR-T Cells:
CD8+ T cells were activated using anti-CD3/CD28 coated beads (Invitrogen) at a 1:2 ratio (beads:cells) and infected with anti-CD19 CAR, anti-HER2 CAR, or empty lentivectors with multiplicity of infection (MOI) of 3–10. Cells were then expanded in complete RPMI 1640 medium supplemented with 10% FBS (Atlanta Biologicals, Flowery Branch, GA), 1% penicillin/streptomycin (Corning Cellgro) and 20 ng mL−1 of IL-2 for 10–12 days and cultured at 37 °C and 5% CO2 supplemented incubators. Respective viruses were added 24 h after the activation. Cells were expanded for 12–20 days and cytotoxicity assays were performed following their expansion.
2D Cytotoxicity Assay:
Following the expansion of effector cells for 10–12 days, the cells were analyzed for their red fluorescent protein (RFP) and CAR expressions. CAR expression was determined with human HER2 / ErbB2 Protein, Fc Tag (Acro), or human CD19 (20-291) Protein, Fc Tag, low endotoxin (super affinity) (Acro) (data for anti-CD19 CAR expression is not shown) followed by a secondary staining with APC-conjugated anti-human IgG Fc antibody (Biolegend, San Diego, CA). HER2-expressing MDA-MB-231 that transduced with a GFP-encoding empty lentivector as a marker was used as target cell. Effector to target cell ratio (1:1) was calculated based on the number of CAR expressing cells. Effector and target cells were combined in U-bottom 96-well plates and incubated for 72 h. Cytotoxicity assay was analyzed with using flow cytometry at 72 h of co-incubation. The cells were first gated based on their average forward and size scatter densities, then target cells were identified with GFP expression and effector cells were identified with CD3 staining (APC/Cyanine7 anti-human CD3 antibody).
Anti HER2 CAR-T Cell Static Cultures:
Static cultures were conducted with aHER2 CAR-T cells using two types of tumor spheroids, similar to the chemotherapy experiments. Monocellular tumor spheroids, containing MDA-MB-231 cells only, and co-cultured tumor spheroids (H231F) were used. CAR-T to MDA-MB-231 cell ratio was varied as 10:1, 20:1, 40:1, and 80:1. Control group consisted of tumor spheroids without the presence of CAR-T cells. CAR-T cells were suspended in a 1:1 mixture of RPMI and EGM-2MV media, supplemented with IL-2 at 1:500 dilution. Tumor spheroids, cultured in 96 U-bottom well plates were then directly exposed to varying concentrations of CAR-T cells and cultured for 72 h. Fluorescent images were taken daily using the EVOS FL Auto microscope, at a constant light and exposure setting for all groups, to monitor the change in GFP intensity of the cancer cells over time. The change in GFP intensity was quantified using ImageJ.
Anti-HER2 CAR-T Cell Perfusion and Imaging:
To understand the efficacy of anti HER2 CAR-T cells in killing MDA-MB-231 cells, static cultures were conducted prior to perfusion studies. CAR-T cells were labeled with cell trace violet (Invitrogen, Waltham, MA) prior to using them for perfusion experiments, following manufacturer’s protocol. After staining, CAR-T cells were suspended in 1:1 mixture of RPMI and EGM-2MV media. This media combination was further supplemented with IL-2 at a 1:500 dilution. CAR-T cells, suspended in this media combination, were introduced in 3D bioprinted perfusion devices and perfused for 24 or 72 h. Perfusion devices were then fixed with 4% paraformaldehyde and imaged using a 16x immersion lens on the SP8 multiphoton microscope. Tumor volumes were quantified by imaging individual tumors post bioprinting.
Supernatant Analysis:
Qbeads immunoassay was used for analyzing cytokines and chemokines secreted after CAR-T cell perfusion for 24 or 72 h. Capture beads fluorescently tagged with a unique signature and coated with capture antibodies directed against a specific analyte were incubated with cell culture supernatants in a 96-well V-bottom plate. Once the analyte was bound by the capture beads, a fluorescent detection antibody was added to the reaction which then bound to the analyte already bound to the beads. To maximize analyte sensitivity and reduce fluorescence background, the bead/analyte/detection were washed. Data were acquired on Intellicyt iQue Screener Plus (Albuquerque, NM). The fluorescence signal was associated with the bead complex and the fluorescence intensity directly correlated to the quantity of bound analyte. Data were represented as mean fluorescence intensity. To assess the production of multiple secreted proteins and cytokines, including GM-CSF, Granzyme A, IFN-γ, IL-13, CCL2 (MCP1), MIG, and IP10, the assay was multiplexed.
Statistics:
All data were presented as the mean ± standard deviation and analyzed by Minitab 17.3 (Minitab Inc., State College, PA) using one-way analysis of variance (ANOVA) followed by the Posthoc Tukey’s multiple comparison test. When comparing multiple groups with a single control group, a Dunnett Multiple Comparisons test was used. Statistical differences were considered significant at *p < 0.05, **p <0.01, ***p < 0.001.
Supplementary Material
Acknowledgements
The authors are grateful to Dr. Danny Welch, from University of Kansas (Kansas City, KS) for providing GFP+ MDA-MB-231 cells. The authors also acknowledge the support from The Huck Institutes of Life Sciences and Materials Research Institute at Penn State (University Park, PA) and Jackson Laboratory (Farmington, CT) for providing facilities for characterization of experiments. This work was supported by NSF awards 1914885 (I.T.O.) and 1624515 (I.T.O.), H. G. Barsumian, M.D. Memorial Fund (I.T.O.), 2236 CoCirculation2 of TUBITAK award 121C359 (I.T.O.), NCI R21 CA224422 01A1 (I.T.O. and D.U.), and NCI P30 CA034196 (D.U.). The opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by NSF, TUBITAK, and NCI.
Footnotes
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/adfm.202203966.
Conflict of Interest
I.T.O. has an equity stake in Biolife4D and is a member of the scientific advisory board for Biolife4D and Brinter. Other authors confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Madhuri Dey, Department of Chemistry, Penn State University, University Park, PA 16802, USA; The Huck Institutes of the Life Sciences, Penn State University, University Park, PA 16802, USA.
Myoung Hwan Kim, The Huck Institutes of the Life Sciences, Penn State University, University Park, PA 16802, USA; Biomedical Engineering Department, Penn State University, University Park, PA 16802, USA.
Mikail Dogan, The Jackson Laboratory for Genomic Medicine, Farmington, CT 06032, USA.
Momoka Nagamine, Department of Chemistry, Penn State University, University Park, PA 16802, USA; The Huck Institutes of the Life Sciences, Penn State University, University Park, PA 16802, USA.
Lina Kozhaya, The Jackson Laboratory for Genomic Medicine, Farmington, CT 06032, USA.
Nazmiye Celik, The Huck Institutes of the Life Sciences, Penn State University, University Park, PA 16802, USA; Engineering Science and Mechanics Department, Penn State University, University Park, PA 16802, USA.
Derya Unutmaz, The Jackson Laboratory for Genomic Medicine, Farmington, CT 06032, USA; University of Connecticut Health Center, Farmington, CT 06032, USA.
Ibrahim T. Ozbolat, The Huck Institutes of the Life Sciences, Penn State University, University Park, PA 16802, USA; Biomedical Engineering Department, Penn State University, University Park, PA 16802, USA; Engineering Science and Mechanics Department, Penn State University, University Park, PA 16802, USA; Materials Research Institute, Penn State University, University Park, PA 16802, USA; Cancer Institute, Penn State University, Hershey, PA 17033, USA; Neurosurgery Department, Penn State University, Hershey, PA 17033, USA; Department of Medical Oncology, Cukurova University, Adana 01330, Turkey
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
References
- [1].Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, Bray F, Int. J. Cancer 2021, 149, 778. [DOI] [PubMed] [Google Scholar]
- [2].Adams DJ, Trends Pharmacol. Sci 2012, 33, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Guo M, Peng Y, Gao A, Du C, Herman JG, Biomark. Res 2019, 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gottlieb B, Trifiro M, Batist G, Trends in Cancer 2021, 7, 400. [DOI] [PubMed] [Google Scholar]
- [5].Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJM, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbé C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ, N. Engl J. Med 2010, 363, 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Doroshow DB, Sanmamed MF, Hastings K, Politi K, Rimm DL, Chen L, Melero I, Schalper KA, Herbst RS, Clin. Cancer Res 2019, 25, 4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ghatalia P, Zibelman M, Geynisman DM, Plimack ER, Curr. Treat. Options Oncol 2017, 18, 7. [DOI] [PubMed] [Google Scholar]
- [8].Polk A, Svane IM, Andersson M, Nielsen D, Cancer Treat. Rev 2018, 63, 122. [DOI] [PubMed] [Google Scholar]
- [9].Marofi F, Motavalli R, Safonov VA, Thangavelu L, Yumashev AV, Alexander M, Shomali N, Chartrand MS, Pathak Y, Jarahian M, Izadi S, Hassanzadeh A, Shirafkan N, Tahmasebi S, Khiavi FM, Stem Cell Res. Ther 2021, 12, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kailayangiri S, Altvater B, Wiebel M, Jamitzky S, Rossig C, Cancers (Basel) 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Toulouie S, Johanning G, Shi Y, J. Cancer 2021, 12, 1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nashimoto Y, Okada R, Hanada S, Arima Y, Nishiyama K, Miura T, Yokokawa R, Biomaterials 2020, 229, 119547. [DOI] [PubMed] [Google Scholar]
- [13].Haase K, Offeddu GS, Gillrie MR, Kamm RD, Haase K, Kamm RD, Offeddu GS, Gillrie MR, Adv. Funct. Mater 2020, 30, 2002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim BS, Cho W-W, Gao G, Ahn M, Kim J, Cho D-W, Kim BS, Cho W, Ahn M, Kim J, Cho D, Gao G, Small Methods 2021, 5, 2100072. [DOI] [PubMed] [Google Scholar]
- [15].Kim D, Seob Hwang K, Seo EU, Seo S, Chul Lee B, Choi N, Choi J, Nam Kim H, Kim D, Hwang KS, Seo EU, Seo S, Lee BC, Choi N, Kim HN, Adv. Healthcare Mater 2022, 11, 2102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pavesi A, Tan AT, Koh S, Chia A, Colombo M, Antonecchia E, Miccolis C, Ceccarello E, Adriani G, Raimondi MT, Kamm RD, Bertoletti A, JCI Insight 2017, 2, 89762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ando Y, Siegler EL, Ta HP, Cinay GE, Zhou H, Gorrell KA, Au H, Jarvis BM, Wang P, Shen K, Adv. Healthcare Mater 2019, 8, 1900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Forster J, Harriss-Phillips W, Douglass M, Bezak E, Hypoxia 2017, 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dey M, Ayan B, Yurieva M, Unutmaz D, Ozbolat IT, Adv. Biol 2021, 5, 2100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ayan B, Heo DN, Zhang Z, Dey M, Povilianskas A, Drapaca C, Ozbolat IT, Sci. Adv 2020, 6, eaaw5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Heo DN, Ayan B, Dey M, Banerjee D, Wee H, Lewis GS, Ozbolat IT, Biofabrication 2021, 13, 015013. [DOI] [PubMed] [Google Scholar]
- [22].Moo TA, Sanford R, Dang C, Morrow M, PET Clin 2018, 13, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Carlson M, Watson AL, Anderson L, Largaespada DA, Provenzano PP, J. Biomed. Opt 2017, 22, 116010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ehsan SM, Welch-reardon KM, Waterman ML, Christopher CW, George SC, Genetics M, Integr. Biol 2015, 6, 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ghaffari S, Leask RL, Jones EAV, Dev. 2015, 142, 4151. [DOI] [PubMed] [Google Scholar]
- [26].Kalluri R, Zeisberg M, Nat. Rev. Cancer 2006, 6, 392. [DOI] [PubMed] [Google Scholar]
- [27].Nagy JA, Chang S-H, Dvorak AM, Dvorak HF, Br. J. Cancer 2009, 100, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Galie PA, Nguyen DHT, Choi CK, Cohen DM, Janmey PA, Chen CS, Proc. Natl. Acad. Sci. USA 2014, 111, 7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hanahan D, Weinberg RA, Cell 2011, 144, 646. [DOI] [PubMed] [Google Scholar]
- [30].Bodley A, Liu LF, Israel M, Seshadri R, Koseki Y, Giuliani FC, Kirschenbaum S, Silber R, Potmesil M, Cancer Res. 1989, 49, 5969. [PubMed] [Google Scholar]
- [31].Fan TJ, Han LH, Cong RS, Liang J, Acta Biochim. Biophys. Sin. (Shanghai) 2005, 37, 719. [DOI] [PubMed] [Google Scholar]
- [32].Porter AG, Jänicke RU, Cell Death Differ. 1999, 6, 99. [DOI] [PubMed] [Google Scholar]
- [33].Levina V, Su Y, Nolen B, Liu X, Gordin Y, Lee M, Lokshin A, Gorelik E, Int. J. Cancer 2008, 123, 2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dankbar B, Padró T, Leo R, Feldmann B, Kropff M, Mesters RM, Serve H, Berdel WE, Kienast J, Blood 2000, 95, 2630. [PubMed] [Google Scholar]
- [35].Nilsson MB, Langley RR, Fidler IJ, Cancer Res. 2005, 65, 10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Strieter RM, Burdick MD, Mestas J, Gomperts B, Keane MP, Belperio JA, Eur. J. Cancer 2006, 42, 768. [DOI] [PubMed] [Google Scholar]
- [37].Hong IS, Exp. Mol. Med 2016, 48, e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ahmad R, Thomas R, Al-Rashed F, Akhter N, Al-Mulla F, Biomol 2019, 9, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Saxena M, Stephens MA, Pathak H, Rangarajan A, Cell Death Dis. 2011, 2, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chen Y, Deng G, Fu Y, Han Y, Guo C, Yin L, Cai C, Shen H, Wu S, Zeng S, Onco. Targets. Ther 2020, 13, 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Neill T, Schaefer L, Iozzo RV, Adv. Drug Delivery Rev 2016, 97, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Marer NL, Hughes RC, J. Cell. Physiol 1996, 168, 51. [DOI] [PubMed] [Google Scholar]
- [43].Vizin T, Kos J, Radiol. Oncol 2015, 49, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ding Caijun Wu C, Tan X, Hu X, Zhou M, J Immunol Ref 2021, 204, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang M, Liu Q, Li L, Ning J, Tu J, Lei X, Mo Z, Tang S, Oncol. Rep 2019, 41, 1807. [DOI] [PubMed] [Google Scholar]
- [46].Zhao Z, Chen Y, Francisco NM, Zhang Y, Wu M, Acta Pharm. Sin. B 2018, 8, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Springuel L, Lonez C, Alexandre B, Van Cutsem E, Machiels JPH, Van Den Eynde M, Prenen H, Hendlisz A, Shaza L, Carrasco J, Canon JL, Opyrchal M, Odunsi K, Rottey S, Gilham DE, Flament A, Lehmann FF, BioDrugs 2019, 33, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nolz JC, Starbeck-Miller GR, Harty JT, Immunotherapy 2011, 3, 1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Slaney CY, Kershaw MH, Darcy PK, 2014. [Google Scholar]
- [50].Alizadeh D, Wong RA, Gholamin S, Maker M, Aftabizadeh M, Yang X, Pecoraro JR, Jeppson JD, Wang D, Aguilar B, Starr R, Larmonier CB, Larmonier N, Chen MH, Wu X, Ribas A, Badie B, Forman SJ, Brown CE, Cancer Discov 2021, 11, 2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ, Front. Immunol 2018, 9, 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Voskoboinik I, Whisstock JC, Trapani JA, Nat. Rev. Immunol 2015, 15, 388. [DOI] [PubMed] [Google Scholar]
- [53].Kohli K, Pillarisetty VG, Kim TS, Cancer Gene Ther. 2021, 29, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Deshmane SL, Kremlev S, Amini S, Sawaya BE, https://home.liebertpub.com/jir 2010, 29, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Liu M, Guo S, Stiles JK, Oncol. Lett 2011, 2, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, McSkane M, Baba H, Lenz HJ, Cancer Treat. Rev 2018, 63, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wang Z, Han W, Biomark. Res 2018, 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li Y, Zhao L, Li XF, Technol. Cancer Res. Treat 2021, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen YA, King AD, Shih HC, Peng CC, Wu CY, Liao WH, Tung YC, Lab Chip 2011, 11, 3626. [DOI] [PubMed] [Google Scholar]
- [60].Rajan N, Habermehl J, Coté M-F, Doillon CJ, Mantovani D, Nat. Protoc 2006, 1, 2753. [DOI] [PubMed] [Google Scholar]
- [61].Cox T, Madsen C, Bio-Protocol 2017, 7, 2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sato M, Theret DP, Wheeler LT, Ohshima N, Nerem RM, 1990. [DOI] [PubMed] [Google Scholar]
- [63].Richards DJ, Li Y, Kerr CM, Yao J, Beeson GC, Coyle RC, Chen X, Jia J, Damon B, Wilson R, Starr Hazard E, Hardiman G, Menick DR, Beeson CC, Yao H, Ye T, Mei Y, Nat. Biomed. Eng 2020, 4, 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zudaire E, Gambardella L, Kurcz C, Vermeren S, PLoS One 2011, 6, e27385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.