Abstract
Transcriptional transactivators (Tat) from many lentiviruses interact with their cognate transactivation response RNA structures (TAR) to increase rates of elongation rather than initiation of transcription. For several of them, the complex of Tat and a species-specific cyclin T1 must be formed before the binding to TAR can occur with high affinity and specificity. In sharp contrast, Tat from the bovine immunodeficiency virus (BIV) binds to its TAR without the help of the cyclin T1. This binding depends on the upper stem and 5′ bulge, but not the central loop in TAR. Moreover, cyclins T1 from different species can mediate effects of this Tat in cells. Unlike the situation with other lentiviruses, Tat transactivation can be rescued simply by linking a heterologous promoter to TAR in permissive cells. Thus, lentiviruses have evolved different strategies to recruit Tat and the positive transcription elongation factor b to their promoters, and interactions between Tat and TAR are independent from those between Tat and the cyclin T1 in BIV.
The bovine immunodeficiency virus (BIV) causes lymphocytosis, lymphadenopathy, progressive weakness, and central nervous system disorders in infected cattle (11, 19). It is a member of the genus Lentivirinae, which contains evolutionarily distinct retroviruses with a complex genomic organization. In this respect, BIV is very similar to primate lentiviruses, the human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2, respectively), as well as the simian immunodeficiency virus (SIV). Besides genes encoding obligate retrovirus Gag, Pol, and Env polyproteins, BIV codes for six accessory proteins, which are called Tat, Rev, Vif, Vpy, Vpw, and Tmx (18). These proteins play a critical role in the viral replicative cycle and contribute significantly to the pathogenesis of BIV (19).
The transcriptional transactivator Tat is essential for lentiviral replication (22). It increases levels of gene expression from the viral 5′ long terminal repeat (LTR). Based on their mechanism of action, Tat proteins can be categorized into two distinct groups. The first group contains visna virus (7, 8, 21), feline immunodeficiency virus (FIV) (9, 29), and caprine arthritis encephalitis virus (CAEV) (28). In these viruses, the transactivation response RNA structure (TAR) is absent from the 5′ end of viral transcripts. Therefore, these Tat proteins increase transcription in a TAR-independent manner. In sharp contrast, HIV-1, HIV-2, SIV (22), BIV (12, 24, 25), the Jembrana disease virus (JDV) (5), and equine infectious anemia virus (EIAV) (10) depend completely on promoter proximal TAR elements. That Tat proteins from this group affect gene expression via RNA makes them unique among other eukaryotic transcriptional activators, which act via DNA. Moreover, these Tat proteins increase rates of elongation rather than initiation of transcription by RNA polymerase II (22).
Recently, the cellular target for Tat from HIV-1 (hTat) was identified. It is called the positive transcription elongation factor b (P-TEFb) and consists of the cyclin-dependent kinase 9 (CDK9) and cyclin T1 (26, 32). The interaction of hTat with the human cyclin T1 (hCycT1) increases greatly the affinity and specificity of the binding between hTat and hTAR (2, 15, 16, 23). In this manner, hTat could position P-TEFb in closer proximity to the C-terminal domain (CTD) of RNA polymerase II. As a consequence, the phosphorylation of the CTD and possibly other targets leads to the efficient elongation of viral transcription (31). Much support for the notion that cyclins T1 are obligate binding partners of different Tat proteins came from studies of the species specificity of Tat transactivation, which had been observed with hTat and Tat from EIAV (eTat). For example, although hTat functions poorly in murine cells, its effects are rescued by the exogenous expression of the hCycT1 (2, 15, 16, 23). Similarly, the transactivation of the EIAV LTR (eLTR) by eTat is impaired and rescued by the exogenous expression of equine cyclin T1 (eCycT1) in human cells (1, 30). In summary, these findings suggest that the formation of a tripartite complex consisting of Tat, cyclin T1, and TAR, rather than the interaction between Tat and TAR alone, determines the host range of hTat or eTat transactivation (31).
In sharp contrast, Tat from BIV (bTat) can function in most cells. bTat transactivates strongly the BIV LTR (bLTR) in virally permissive canine (Cf2Th) cells and virally nonpermissive human (HeLa), monkey (CV1), and murine (3T3) cells (5, 13). Surprisingly, bTat functions poorly and independently of bTAR in virally permissive bovine (BLAC-20) and lapine (EREp) cells (13). Nevertheless, bTat should interact with P-TEFb. First, it shares similar sequence and domain organization with hTat (12, 24). Second, its longer arginine-rich RNA-binding motif (ARM) binds with high affinity and specificity to the upper stem and 5′ bulge in bTAR in vitro (6, 25, 27). Of note and unlike hTat, the central loop in bTAR is not essential for the function of bTat in vivo (3, 6). Thus, a combinatorial surface between bTat and a cyclin T1 might not be required for productive interactions between bTat and bTAR.
In this report, we asked several questions. Does bTat target the same host factors as other lentivirus Tat proteins for its role in viral transcription? Why can bTat transactivate the bLTR in cells from all species tested and yet not function in other cells? To these ends, we first demonstrated that bTat binds to cyclins T1 from different species. Using serum-starved canine cells, all of these cyclins T1 could cooperate with bTat. Next, the inability of bTat to function in lapine cells reflected low levels of transcription. Finally, bTat bound strongly to bTAR by itself, and hCycT1 did not increase this interaction. This binding required the upper stem and 5′ bulge, but not the central loop in bTAR. In summary, bTAR serves as a docking site for bTat, which recruits P-TEFb independently to elongate viral transcription.
MATERIALS AND METHODS
Cell culture.
HeLa and COS cells, lapine EREp, and canine Cf2Th cells were grown in Dulbecco's modified Eagle's medium (DMEM). Media were supplemented with 10% fetal bovine serum (FBS), 100 mM l-glutamine, and 50 μg each of penicillin and streptomycin per ml. For serum starvation, Cf2Th cells were maintained in DMEM without FBS.
Plasmid construction.
The plasmid targets pHIVSCAT and pBLTRCAT have been described previously (13, 14). To construct pHIVSCATbTAR, pHIVSCAT was cleaved at unique BglII and SacI sites, and bTAR sequence was inserted into pHIVSCAT by using two complementary bTAR oligonucleotides with BglII and SacI sites. The following oligonucleotide sequences were designed: 5′-GATCTGGCTCGTGTAGCTCATTAGCTCCGAGCCGAGCT-3′ and 5′-CGGCTCGGAGCTAATGAGCTACACGAGCCA-3′. For mammalian expression, bTat (amino acids [aa] 1 to 103) was amplified by PCR from BIV127 provirus. Subsequently, the fragment was subcloned into NcoI-EcoRI-cleaved modified pEFBOS in frame with an N-terminal Myc-epitope tag (pbTat). The resulting construct was named pbTat. Myc-tagged hCycT1 and eCycT1 (aa 1 to 300) were expressed from phCycT1-300 and peCycT1-300, respectively. N-terminally Myc-epitope-tagged mouse cyclin T1 (mCycT1) was expressed from pmCycT1-300. pGST-bTat for bacterial expression was made by using a PCR-amplified bTat gene (aa 1 to 103), which was ligated in-frame with the coding region of the glutathione S-transferase (GST) gene into SmaI-EcoRI-cleaved pGEX-2TK (Amersham Pharmacia Biotech, Piscataway, N.J.). Wild-type (nucleotides 1 to 28) or mutant TAR sequences derived from the sequence of BIV127 provirus were cloned into pGEM-3Z (EcoRI-HindIII; Promega, Madison, Wis.) with oligonucleotides containing EcoRI (5′) and HindIII (3′) linkers.
The oligonucleotide sequences were as follows: 1WT, 5′AATTCGGGTCTC TCTGGGGCTCGTGTAGCTCATTAGCTCCGAGCCCTAGGGAACCCA 3′; 2WT, 5′AGCTTGGGTTCCCTAGGGCTCGGAGCTAATGAGCTACACGAGCCCCAGAGAGACCCG 3′; 1ΔS, 5′AATTCGGGTCTCTCTGGGGCTCGTGTACCTCATTAGGTCCGAGCCCTAGGGAACCCA 3′; 2ΔS, 5′AGCTT GGGTTCCCTAGGGCTCGGACCTAATGAGGTACACGAGCCCCAGAG AGACCCG 3′; 1ΔL, 5′AATTCGGGTCTCTCTGGGGCTCGTGTAGCTTGCCAGCTCCGAGCCCTAGGGAACCCA 3′; 2ΔL, 5′AGCTTGGGTTCCCTAGGGCTCGGAGCTGGCAAGCTACACGAGCCCCAGAGAGACCCG3′.
pGEM3WT codes for the wild-type TAR RNA. pGEM3ΔS encodes TAR RNA with a nucleotide pair mutation G14:C23 to C14:G23 in the stem region of TAR RNA (6). pGEM3ΔL mutant TAR RNA has replaced the CAUU nucleotide sequence of the loop with UGCC sequence (6). All constructs used in this study were confirmed by DNA sequencing.
Transient transfection, CAT assays, and Western blotting.
All cell lines used in this study were transfected with Lipofectamine according to the manufacturer's instructions (Gibco-BRL, Rockville, Md.). For chloramphenicol acetyltransferase (CAT) enzymatic assays, Cf2Th, HeLa, and EREp cells were seeded into 50-mm-diameter petri dishes 12 h prior to transfections. Serum starvation of Cf2Th cells was performed 10 h before the transfection and continued until CAT assays. Cells were harvested 48 h posttransfection, and CAT activity was determined as described previously (14). Western blotting was done with a rabbit polyclonal anti-Myc antibody (c-Myc [A-14]; Santa Cruz Biotechnology, Santa Cruz, Calif.) followed by horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin secondary antibody (1:2,000; Amersham Life Science, Inc., Arlington Heights, Ill.). Blots were developed by chemiluminescence assay (NEN Life Science Products, Boston, Mass.).
In vitro transcription and translation.
The plasmids containing hCycT1-300, eCycT1-300, and mCycT1-300 were transcribed and translated in vitro with the TnT T7 coupled reticulocyte lysate system (Promega, Madison, Wis.) in the presence of 35S-labeled cysteine (NEN Life Science Products, Boston, Mass.).
Protein purification.
Hybrid GST-CycT and GST-bTat proteins were expressed in the BL21(DE3)pLysS strain of Escherichia coli (Novagen, Madison, Wis.) after a 4-h induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and purified from total cell lysates by using glutathione-Sepharose beads (Amersham-Pharmacia Biotech, Piscataway, N.J.). For the electrophoretic mobility shift assay (EMSA), purified GST-CycT proteins were eluted from glutathione-Sepharose beads by using 10 mM reduced glutathione in 50 mM Tris-HCl (pH 8.0) and subjected to dialysis against a buffer containing 30 mM Tris-HCl (pH 8.0), 70 mM KCl, and 1 mM dithiothreitol (DTT). The hybrid GST-bTat protein bound to glutathione-Sepharose beads was eluted with 50 mM reduced glutathione in 200 mM Tris-HCl (pH 8.0) and 500 mM NaCl. Dialysis and concentration were performed by using Centricon concentrators (cutoff, 10 kDa; Amicon, Inc., Beverly, Mass.). The purity of eluted proteins was determined by Coomassie blue staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the concentration was determined by a protein assay kit (Bio-Rad, Hercules, Calif.).
In vitro binding assays.
Binding assays between different cyclins T1, synthesized in the coupled transcription and translation system, and chimeric GST-bTat were performed as follows. Ten microliters of each cyclin T1 was incubated with 10 μg of the GST or GST-bTat protein, bound to glutathione-Sepharose beads, in 300 μl of binding buffer (20 mM HEPES [pH 7.8], 0.5% NP-40, 1% Triton X-100, 2 mM DTT, 0.1% bovine serum albumin, 0.05% SDS, 20 μM ZnCl2, 100 mM KCl) at 4°C for 4 h. After the incubation, GST-coupled beads were washed four times with the binding buffer. Bound proteins were eluted from beads by boiling in equal volumes of 2× SDS loading buffer, resolved by SDS-PAGE (10% polyacrylamide), and analyzed by autoradiography.
Preparation of TAR RNA and EMSA.
α-32P-labeled TAR and unlabeled TAR transcripts were prepared with in vitro-transcribing linearized plasmid templates (HindIII) with T7 RNA polymerase in the presence or absence of [α-32P]UTP by using the MEGAshortscript T7 kit (Ambion, Austin, Tex.). Reaction mixtures were incubated at 37°C for 1 h, and the DNA template was treated with 2 U of DNase I. TAR RNAs were purified with a phenol-chloroform-isoamyl alcohol mixture (Boehringer, Mannheim, Germany) and precipitated with ethanol. Prior to use, the RNA pellet was dissolved in 0.1 M NaCl and applied to a G-25 spin column (Boehringer). EMSA (16-μl final reaction volume) was carried out in the binding buffer (30 mM Tris-HCl [pH 8.0], 70 mM KCl, 0.01% NP-40, 5.5 mM MgCl2, 1 mM DTT, 12% glycerol) and contained 20,000 to 50,000 cpm of α-32P-labeled TAR RNA as well as 850 ng of poly(dI-dC) and 500 ng of poly(I-C) in the presence or absence of excessive amounts of unlabeled TAR transcripts. One microgram of purified GST, GST-hCycT, and/or 400 ng of GST-bTat (aa 1 to 103) was added to the reaction mixtures. The reaction mixtures were incubated for 10 min at 30°C, and RNA-protein complexes were separated on a prerun nondenaturing 6% Tris-glycine polyacrylamide gel in 1× Tris-glycine buffer (4 W, 3.5 h at 4°C). Gels were dried and then analyzed by autoradiography.
RESULTS
bTat binds equivalently to cyclins T1 from different species in vitro.
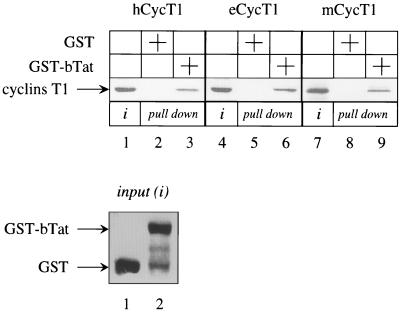
P-TEFb is the critical cellular target for hTat and eTat transactivation. Therefore, bTat might also use the same cellular machinery to perform its function. Moreover, the broad host range of bTat transactivation could reflect the binding of bTat to cyclins T1 from different species. To date, hCycT1, functionally indistinguishable canine T1 (cCycT1) and eCycT1, and mCycT1 have been isolated and characterized (31). To test our hypothesis, we performed GST pull down assays with the hybrid GST-bTat protein, which was expressed in E. coli, and hCycT1, eCycT1, and mCycT1, which were transcribed and translated in vitro with the rabbit reticulocyte lysate (Fig. 1). Since the N-terminal 300 residues in hCycT1 and eCycT1 support hTat and eTat transactivation, respectively (1, 2, 15, 16, 23, 30), proteins of this size were used in our binding experiments. Under stringent conditions, the hybrid GST-bTat protein bound to all of these cyclins T1 (Fig. 1, lanes 3, 6, and 9). This binding was specific, since no interaction between bacterially expressed GST and cyclins T1 was observed (Fig. 1, lanes 2, 5, and 8). The inputs of all cyclins T1 (Fig. 1, lanes 1, 4, and 7) and the hybrid GST-bTat protein and GST alone (Fig. 1, lower panel, lanes 1 and 2) were comparable. Thus, bTat binds to regulatory subunits of P-TEFb from different species in vitro.
FIG. 1.
The N-terminal 300 residues in hCycT1, eCycT1, and mCycT1 bind to bTat in vitro. 35S-labeled hCycT1, eCycT1, and mCycT1 were incubated with GST alone (lanes 2, 5, and 8) or with the hybrid GST-bTat protein (lanes 3, 6, and 9) and selected on glutathione-Sepharose beads. Bound cyclins T1 were separated on SDS-PAGE and subjected to autoradiography. The input of each cyclin T1 was equal in all reactions and represented 25% of the amount used for the binding assay (lanes 1, 4, and 7). The arrow to the left points to cyclins T1. Twenty-five percent of the input GST alone (lane 1) and the hybrid GST-bTat protein (lane 2) were comparable and are presented in the Coomassie blue-stained SDS-PAGE panel at the bottom. Arrows to the left indicate the presence of the hybrid proteins.
Given a stronger LTR, lapine cells support bTat transactivation.
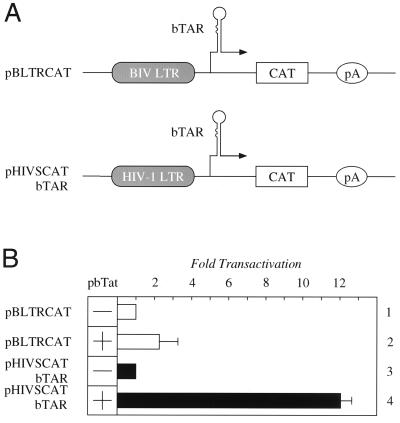
Having established that cyclins T1 from different species bind to bTat in vitro and knowing that they are conserved from Drosophila melanogaster to Homo sapiens, we were intrigued by low levels of bTat transactivation in virally permissive bovine BLAC-20 and lapine EREp cells (13). Although this effect of bTat was described as being independent of bTAR, we wondered if the absence of appropriate RNA tethering of bTat could play some role in these cells. The other possibility was that insufficient bTAR was synthesized so that bTat did not have enough target RNA for its effects. To distinguish between these possibilities, we used two different plasmid targets, which are presented schematically in Fig. 2A, and plasmid effectors that expressed bTat. When bTat was coexpressed with the pBLTRCAT in EREp cells, only a twofold increase in CAT enzymatic activity over basal levels was observed (Fig. 2B, lanes 1 and 2). In sharp contrast, when the U3 region from bLTR was replaced with that from HIV-1 so that bTAR remained (Fig. 2A, pHIVSCATbTAR), levels of transactivation increased 12-fold over basal values (Fig. 2B, lanes 3 and 4). When cyclins T1 from different species were coexpressed with pBLTRCAT and bTat, levels of bTat transactivation were not affected (data not shown). We conclude that the restriction to bTat transactivation does not map to the cyclin T1, but to the strength of the promoter in EREp cells. These data suggest that insufficient bTAR is synthesized as the substrate for bTat in these cells.
FIG. 2.
bTat transactivation in lapine cells is inhibited by low levels of basal transcription from bLTR. (A) Schematic representation of plasmid targets. pBLTRCAT contains the CAT reporter gene under the control of bLTR. pHIVSCATbTAR contains the hLTR linked to bTAR instead of hTAR. pA represents the polyadenylation signal. (B) bTat requires a strong promoter in lapine EREp cells. Cells were transfected with 0.3 μg of pBLTRCAT alone (lane 1, white bar) or together with 0.1 μg of pbTat (lane 2, white bar). pHIVSCATbTAR (0.1 μg) was expressed alone (lane 3, black bar) or together with 0.1 μg of pbTat (lane 4, black bar). Where no bTat was added, the amount of DNA was equilibrated with 0.1 μg of pEFBOS (lanes 1 and 3, white and black bars). The CAT enzymatic activity of pBLTRCAT or pHIVSCATbTAR alone was set to 1. Standard errors of the mean from three independent transfections are shown by error bars.
The exogenous expression of cyclins T1 from different species restores bTat transactivation in serum-starved Cf2Th cells.
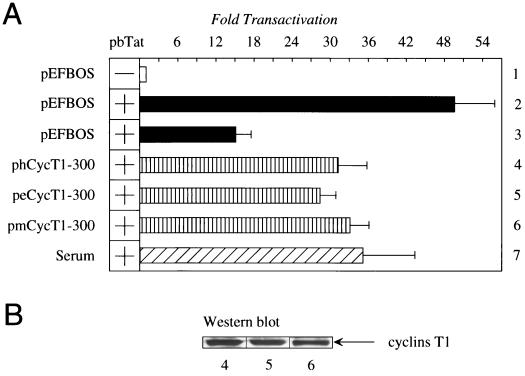
Next, we performed a functional assay to prove that targeting different cyclins T1 can support bTat transactivation in vivo. Since bTat can function in cells from many different species (Fig. 2), classical rescue experiments using different cyclins T1, like those for hTat or eTat (1, 2, 15, 16, 23, 30), were not possible. For example, the exogenous expression of hCycT1, eCycT1, and mCycT1 has neither positive nor negative effects on bTat transactivation in canine Cf2Th cells (data not shown). However, the state of activation and proliferation of cells dictate levels of cyclin T1 (17, 20). Consequently, blocking the progression of the cell cycle should reduce levels of intracellular cyclin T1. Therefore, experiments in which cyclins T1 were expressed exogenously were repeated with serum-starved Cf2Th cells (Fig. 3A). In this system, bTat transactivation dropped from 49-fold to 15-fold (Fig. 3A, compare lanes 2 and 3). Importantly, the exogenous expression of hCycT1, eCycT1, and mCycT1 restored bTat transactivation to levels observed with the addition of serum (Fig. 3A, compare lanes 4 to 6 with lane 7). All cyclins T1 were expressed at similar levels (Fig. 3B). In keeping with our binding studies and coexpression data from many laboratories (1, 2, 5, 13, 16, 23, 30), we conclude that different cyclins T1 can form productive complexes with bTat and bTAR.
FIG. 3.
Cyclins T1 from different species increase bTat transactivation in serum-starved Cf2Th cells. (A) bTat transactivates bLTR via different cyclins T1. Cells were serum starved before and after transfection (lanes 1, and lanes 3 to 6), before transfection only (lane 7), or grown in the medium with serum (lane 2). pBLTRCAT (0.3 μg) was expressed alone (lane 1, white bar) or together with 0.1 μg of pbTat (lanes 2 to 7). To bTat were added effector plasmids (1.0 μg) phCycT1, peCycT1, and pmCycT1 (lanes 4 to 6, striped bars). In all transfections, the amount of DNA was equilibrated with pEFBOS. Values are as in Fig. 2B. (B) Amounts of exogenously expressed cyclins T1 determined by Western blotting. Numbers under the Western blot correspond to lanes from the transient transfection assay (A).
The formation of the complex between bTat and bTAR, which occurs independently of the cyclin T1, explains the lack of species-specific restriction to bTat transactivation.
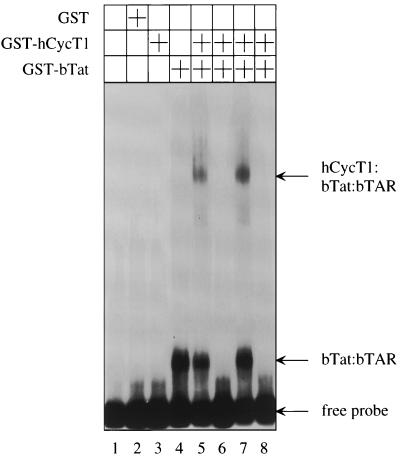
Previous studies of different lentivirus Tat proteins established that the binding of Tat to the cyclin T1 is required for the formation of the tripartite complex between Tat, cyclin T1, and TAR as well as efficient Tat transactivation (31). It is also known that the ARM peptide from bTat binds to bTAR with high affinity and specificity and that mutations in the central loop in bTAR do not affect bTat transactivation (3, 6, 25, 27). Since bTat binds to several different cyclins T1, the simplest scenario to explain the broad host range of bTat would have bTat bind to bTAR alone without the assistance of any cyclin T1. To prove this hypothesis, an EMSA was performed with bacterially expressed GST, as well as hybrid GST-hCycT1 and GST-bTat proteins, and in vitro-transcribed α-32P-labeled or unlabeled wild-type bTAR or mutated bTAR transcripts, which were changed in the stem or central loop regions, respectively (Fig. 4). Neither GST alone nor the hybrid GST-hCycT1 protein bound to the labeled probe (Fig. 4, lanes 2 and 3). In sharp contrast, the hybrid GST-bTat protein bound strongly to bTAR (Fig. 4, lane 4). The addition of the hybrid GST-hCycT1 protein did not improve the binding of the hybrid GST-bTat protein to bTAR, but resulted in a tripartite complex between the hybrid GST-CycT1 and GST-bTat proteins and bTAR (Fig. 4, lane 5). To demonstrate the specificity of this binding, competition experiments using unlabeled bTAR transcripts were performed. Excessive amounts of the unlabeled wild-type bTAR RNA completely blocked the formation of RNA-protein complexes (Fig. 4, lane 6). Importantly, the unlabeled mutant bTAR RNA, which changed the stem, did not compete with the labeled wild-type bTAR (Fig. 4, lane 7), confirming that this region is critical for the binding of bTat to bTAR (6, 25, 27). Moreover, the unlabeled bTAR RNA with mutations in the central loop had the same effect as the wild-type bTAR (Fig. 4, compare lanes 6 and 8), which agrees with previous functional studies (3, 6). Thus, these results demonstrate that bTat does not need the assistance of the cyclin T1 for its binding to bTAR, which explains the lack of species specificity in bTat transactivation.
FIG. 4.
bTat binds to the upper stem and 5′ bulge in bTAR without the help of the cyclin T1. Where indicated, bacterially expressed GST, hybrid GST-bTat, and GST-hCycT1 proteins were used in EMSA. α-32P-labeled wild-type bTAR was present in all reactions. For competition experiments, three different unlabeled competitor bTAR transcripts were used: bTARWT (lane 6), bTARΔS, which is mutated in the upper stem in bTAR (lane 7), and bTARΔL, which is mutated in the central loop in bTAR (lane 8). The resulting RNA-protein complexes were resolved on a 6% nondenaturing polyacrylamide gel and analyzed by autoradiography. Arrows to the right indicate the free bTAR probe and the presence of distinct RNA-protein complexes.
DISCUSSION
In this study, we demonstrated that the lack of species-specific restriction to bTat transactivation reflects the ability of bTat to bind efficiently to bTAR without the cyclin T1. Thus, bTat bound equally well to bTAR in the presence and absence of hCycT1. Moreover, this binding did not require the central loop and was specific to the upper stem and 5′ bulge in bTAR. Additionally, bTat bound indistinguishably to cyclins T1 from three different species, and they all restored levels of bTat transactivation in serum-starved canine Cf2Th cells. Finally, the lack of robust and bTAR-dependent effects could be blamed on the weakness of the bLTR in lapine EREp cells. Linking the hLTR to bTAR rescued the function of bTat in these cells. We conclude that the binding of bTat to bTAR is independent of the recruitment of a species-specific CycT1 and thus P-TEFb to the bLTR.
Since bTat functions in cells from many species, there was no need to isolate the bovine cyclin T1 (bCycT1). Moreover, comparisons between known cyclins T1 revealed that their sequences are 90% identical (1, 2, 15, 16, 23, 30). Because bTat contains an identical array of cysteines to that in hTat and only the 300 N-terminal residues of the cyclins T1 were required for our functional and binding studies, the binding of bTat to bCycT1 is expected to resemble that of hTat to hCycT1. In this tripartite complex, the ARM of bTat binds to the upper stem and 5′ bulge in bTAR. The activation domain of bTat then binds independently to the surface of bCycT1, which is located on the side opposite to where CDK9 binds. Thus, bTat brings P-TEFb to bTAR for the transcriptional needs of BIV. Moreover, we resolved the dilemma of why some cells that are permissive for BIV infection do not support bTat transactivation (13). In these cells, the bLTR is a weak promoter and probably does not synthesize sufficient amounts of bTAR transcripts. However, linking a heterologous promoter to bTAR rescued bTat transactivation. Thus, the inability of bTat to function in these cells is not due to the defective formation of the tripartite complex between bTat, P-TEFb, and bTAR. That these cells can be infected by BIV also suggests that virions or other virus proteins activate the integrated bLTR and facilitate the full replicative cycle of the virus.
JDV, which is another bovine lentivirus, is closely related to BIV and has a broad host range, and its Tat (jTat) has properties similar to those of bTat (4, 5). jTat not only activates its cognate LTR (jLTR), but also activates LTRs from other lentiviruses (5). Moreover, residues of the ARM from bTat and jTat are very similar, and neither protein requires the central loop in TAR for efficient transactivation (5). Taken together, these data lead us to speculate that jTat also binds to the upper stem and 5′ bulge in jTAR independently of bCycT1 and that these interactions between jTat and jTAR and jTat and bCycT1 occur independently. Thus, BIV and JDV are more closely related to each other than other lentiviruses and have adopted similar strategies for efficient replication in their primary host.
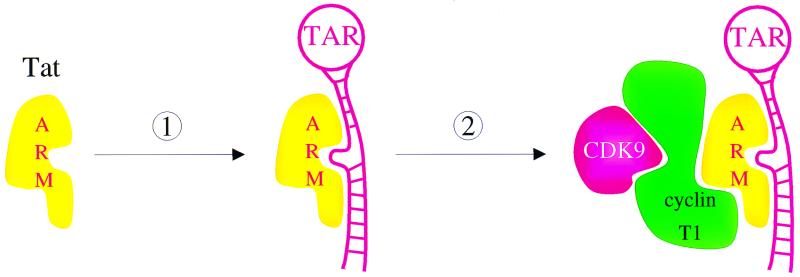
The model that emerges from this study is that unlike other Tat proteins, bTat binds to a stable structure and specific nucleotides in bTAR independently of the cyclin T1 (Fig. 5). This binding is of sufficient affinity and specificity to bring P-TEFb to the transcription complex. In this scenario, bCycT1 plays no role in the RNA tethering of bTat. This RNA-protein interaction is simpler than those that require a combinatorial surface between Tat and the cyclin T1. With HIV-1, hTat binds to the upper stem and 5′ bulge, and hCycT1 binds to the central loop in hTAR. Only the preassembled complex between hTat and P-TEFb interacts productively with hTAR (2, 15, 16, 23). With EIAV, no binding of eTat alone to RNA could be demonstrated. However, the complex of eTat and eCycT1 binds to the central loop in eTAR (1, 30). These combinatorial surfaces might have increased the specificity of the RNA tethering by different Tat proteins. Alternatively, they might have allowed for a more precise positioning of CDK9 with respect to its cellular targets on the transcription complex. Nevertheless, because both the N-terminal and C-terminal α-helices in the cyclin box of cyclins T1 are required for the binding of hTat and eTat and these three Tat proteins are so similar, we expect that the same binding surface is utilized by bTat (22). Future mutageneses and structural studies will reveal the validity of this model and suggest possible inhibitors of Tat transactivation.
FIG. 5.
Model for the formation of the tripartite complex between bTat, P-TEFb, and bTAR. The binding of bTat to bTAR (step 1) and the recruitment of P-TEFb (step 2) occur independently in BIV. The ARM from bTat interacts with the upper stem and 5′ bulge, but not the central loop, in bTAR. The activation domain from bTat binds to the cyclin T1. The binding of bTat to bTAR is of sufficient affinity and specificity to recruit P-TEFb from different species to bLTR for efficient elongation of transcription.
ACKNOWLEDGMENTS
We thank Paula Zupanc-Ecimovic and Michael Armanini for expert secretarial assistance; Steven E. Fong for providing pBLTRCAT, pBIV127, and Cf2Th and EREp cell lines; Nabila Jabrane-Ferrat and Christoph Turck for help with experiments; and other members of our laboratory for comments on the manuscript.
Koh Fujinaga is supported by a fellowship from the Universitywide AIDS Research Program. This work was supported by the Howard Hughes Medical Institute.
REFERENCES
- 1.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Highly divergent lentiviral Tat proteins activate viral gene expression by a common mechanism. Mol Cell Biol. 1999;19:4592–4599. doi: 10.1128/mcb.19.7.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter S, Nadin-Davis S A, Wannemuehler Y, Roth J A. Identification of transactivation-response sequences in the long terminal repeat of bovine immunodeficiency-like virus. J Virol. 1993;67:4399–4403. doi: 10.1128/jvi.67.7.4399-4403.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chadwick B J, Coelen R J, Wilcox G E, Sammels L M, Kertayadnya G. Nucleotide sequence analysis of Jembrana disease virus: a bovine lentivirus associated with an acute disease syndrome. J Gen Virol. 1995;76:1637–1650. doi: 10.1099/0022-1317-76-7-1637. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Wilcox G, Kertayadnya G, Wood C. Characterization of the Jembrana disease virus tat gene and the cis- and trans-regulatory elements in its long terminal repeats. J Virol. 1999;73:658–666. doi: 10.1128/jvi.73.1.658-666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Frankel A D. An RNA-binding peptide from bovine immunodeficiency virus Tat protein recognizes an unusual RNA structure. Biochemistry. 1994;33:2708–2715. doi: 10.1021/bi00175a046. [DOI] [PubMed] [Google Scholar]
- 7.Clements J E. Genetic regulation of the ungulate lentiviruses. AIDS. 1991;5:S15–S20. doi: 10.1097/00002030-199101001-00003. [DOI] [PubMed] [Google Scholar]
- 8.Davis J L, Clements J E. Characterization of a cDNA clone encoding the visna virus transactivating protein. Proc Natl Acad Sci USA. 1989;86:414–418. doi: 10.1073/pnas.86.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Parseval A, Elder J H. Demonstration that orf2 encodes the feline immunodeficiency virus transactivating (Tat) protein and characterization of a unique gene product with partial Rev activity. J Virol. 1999;73:608–617. doi: 10.1128/jvi.73.1.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn P L, Derse D. cis- and trans-acting regulation of gene expression of equine infectious anemia virus. J Virol. 1988;62:3522–3526. doi: 10.1128/jvi.62.9.3522-3526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egberink H, Horzinek M C. Animal immunodeficiency viruses. Vet Microbiol. 1992;33:311–331. doi: 10.1016/0378-1135(92)90059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fong S E, Greenwood J D, Williamson J C, Derse D, Pallansch L A, Copeland T, Rasmussen L, Mentzer A, Nagashima K, Tobin G, Gonda M A. Bovine immunodeficiency virus tat gene: cloning of two distinct cDNAs and identification, characterization, and immunolocalization of the tat gene products. Virology. 1997;233:339–357. doi: 10.1006/viro.1997.8589. [DOI] [PubMed] [Google Scholar]
- 13.Fong S E, Pallansch L A, Mikovits J A, Lackman-Smith C S, Ruscetti F W, Gonda M A. cis-acting regulatory elements in the bovine immunodeficiency virus long terminal repeat. Virology. 1995;209:604–614. doi: 10.1006/viro.1995.1292. [DOI] [PubMed] [Google Scholar]
- 14.Fujinaga K, Cujec T P, Peng J, Garriga J, Price D H, Graña X, Peterlin B M. The ability of positive transcription elongation factor b to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J Virol. 1998;72:7154–7159. doi: 10.1128/jvi.72.9.7154-7159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujinaga K, Taube R, Wimmer J, Cujec T P, Peterlin B M. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci USA. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garriga J, Peng J, Parreno M, Price D H, Henderson E E, Grana X. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene. 1998;17:3093–4002. doi: 10.1038/sj.onc.1202548. [DOI] [PubMed] [Google Scholar]
- 18.Garvey K J, Oberste M S, Elser J E, Braun M J, Gonda M A. Nucleotide sequence and genome organization of biologically active proviruses of the bovine immunodeficiency-like virus. Virology. 1990;175:391–409. doi: 10.1016/0042-6822(90)90424-p. [DOI] [PubMed] [Google Scholar]
- 19.Gonda M A, Luther D G, Fong S E, Tobin G J. Bovine immunodeficiency virus: molecular biology and virus-host interactions. Virus Res. 1994;32:155–181. doi: 10.1016/0168-1702(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann C H, Carroll R G, Wei P, Jones K A, Rice A P. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hess J L, Clements J E, Narayan O. cis- and trans-acting transcriptional regulation of visna virus. Science. 1985;229:482–485. doi: 10.1126/science.2990051. [DOI] [PubMed] [Google Scholar]
- 22.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 23.Kwak Y T, Ivanov D, Guo J, Nee E, Gaynor R B. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J Mol Biol. 1999;288:57–69. doi: 10.1006/jmbi.1999.2664. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z-Q, Sheridan D, Wood C. Identification and characterization of the bovine immunodeficiency-like virus tat gene. J Virol. 1992;66:5137–5140. doi: 10.1128/jvi.66.8.5137-5140.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallansch L A, Lackman-Smith C S, Gonda M A. Bovine immunodeficiency-like virus encodes factors which trans activate the long terminal repeat. J Virol. 1992;66:2647–2652. doi: 10.1128/jvi.66.5.2647-2652.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puglisi J D, Chen L, Blanchard S, Frankel A D. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 28.Saltarelli M J, Schoborg R, Gdovin S L, Clements J E. The CAEV tat gene trans-activates the viral LTR and is necessary for efficient viral replication. Virology. 1993;197:35–44. doi: 10.1006/viro.1993.1564. [DOI] [PubMed] [Google Scholar]
- 29.Sparger E E, Shacklett B L, Renshaw-Gegg L, Barry P A, Pedersen N C, Elder J H, Luciw P A. Regulation of gene expression directed by the long terminal repeat of the feline immunodeficiency virus. Virology. 1992;187:165–177. doi: 10.1016/0042-6822(92)90305-9. [DOI] [PubMed] [Google Scholar]
- 30.Taube R, Fujinaga K, Irwin D, Wimmer J, Geyer M, Peterlin B M. Interactions between equine cyclin T1, Tat, and TAR are disrupted by a leucine-to-valine substitution found in human cyclin T1. J Virol. 2000;74:892–898. doi: 10.1128/jvi.74.2.892-898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taube R, Fujinaga K, Wimmer J, Barboric M, Peterlin B M. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology. 1999;264:245–253. doi: 10.1006/viro.1999.9944. [DOI] [PubMed] [Google Scholar]
- 32.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]