ABSTRACT
Sporulation is an important feature of the clostridial life cycle, facilitating survival of these bacteria in harsh environments, contributing to disease transmission for pathogenic species, and sharing common early steps that are also involved in regulating industrially important solvent production by some non-pathogenic species. Initial genomics studies suggested that Clostridia lack the classical phosphorelay that phosphorylates Spo0A and initiates sporulation in Bacillus, leading to the hypothesis that sporulation in Clostridia universally begins when Spo0A is phosphorylated by orphan histidine kinases (OHKs). However, components of the classical Bacillus phosphorelay were recently identified in some Clostridia. Similar Bacillus phosphorelay components have not yet been found in the pathogenic Clostridia or the solventogenic Clostridia of industrial importance. For some of those Clostridia lacking a classical phosphorelay, the involvement of OHKs in sporulation initiation has received support from genetic studies demonstrating the involvement of several apparent OHKs in their sporulation. In addition, several clostridial OHKs directly phosphorylate Spo0A in vitro. Interestingly, there is considerable protein domain diversity among the sporulation-associated OHKs in Clostridia. Further adding to the emergent complexity of sporulation initiation in Clostridia, several candidate OHK phosphotransfer proteins that were OHK candidates were shown to function as phosphatases that reduce sporulation in some Clostridia. The mounting evidence indicates that no single pathway explains sporulation initiation in all Clostridia and supports the need for further study to fully understand the unexpected and biologically fascinating mechanistic diversity of this important process among these medically and industrially important bacteria.
KEYWORDS: Clostridia, Spo0A, sporulation, histidine kinases, phosphatases
INTRODUCTION
The Clostridia, a polyphyletic class of Bacillota (synonym Firmicutes), do not grow under aerobic conditions and most species stain Gram-positive. They have a widespread environmental distribution, including sewage and soil.
The Clostridia are of considerable interest for several reasons. Some Clostridia, such as Clostridium leptum, Clostridium coccoides, and Clostridium scindens, are important members of the normal human intestinal microbiota that promote health (1). By producing potent toxins, other Clostridia such as Clostridioides difficile, Clostridium perfringens, Clostridium botulinum, and Clostridium tetani are important human and animal pathogens. C. difficile is classified as an urgent public health threat by the CDC (https://www.cdc.gov/drugresistance/biggest-threats.html#cdiff), causing nearly half a million cases of C. difficile infection every year in the United States, resulting in ~15,000 annual deaths (2). In humans and other animals, C. perfringens is an important cause of histotoxic infections, including gas gangrene and intestinal infections (3). C. perfringens is also responsible for causing nearly 1 million cases of food poisoning annually in the United States, ranking it as the second most common cause of bacterial foodborne disease (4, 5). C. botulinum and C. tetani cause lethal flaccid or spastic paralysis, respectively, in many animal species, including humans (6). Lastly, several nonpathogenic Clostridia, for example, Clostridium acetobutylicum and Acetivibrio thermocellus, have biotechnology importance (7, 8) because they produce industrially useful solvents such as acetone, ethanol, and butanol, some of which can be used as biofuels (7–9).
THE IMPORTANCE OF SPORULATION FOR THE CLOSTRIDIA
A key characteristic of most Clostridia is their ability to form spores. Since these spores are metabolically inert, spore production permits sustained survival of most Clostridia in nutrient-poor environments (10). Upon encountering an environment with improved nutrient availability, these spores can germinate back into vegetative cells (10).
In addition, all clostridial spores possess resistance against factors such as heat, cold, radiation, and chemicals (including disinfectants and food preservatives), which facilitate survival of these bacteria in harsh conditions. By improving survival in the environment, spore resistance similarly facilitates the transmission of Clostridia causing many diseases, including botulism, tetanus, C. difficile-associated disease, C. perfringens-induced gas gangrene, and C. perfringens type F food poisoning (10). Interestingly, the degree of spore resistance properties can sometimes vary among spores produced by different isolates of the same clostridial species. For example, spores made by many C. perfringens type F food poisoning strains are dramatically more resistant to heat and other food environment stresses than the spores produced by other C. perfringens strains (11), which enhances their survival in improperly cooked foods and thereby should assist foodborne disease transmission. Sporulation is also necessary for the production of C. perfringens enterotoxin, which mediates the pathogenesis of food poisoning and nonfoodborne gastrointestinal disease caused by C. perfringens type F (12). As will be discussed in more detail later, sporulation and solvent production are often closely associated with the industrially important Clostridia. For example, phosphorylation of the master transcriptional regulator Spo0A is required for both sporulation and solvent production by C. acetobutylicum (13).
Clostridial sporulation involves a relatively conserved sequence of gene expression that governs spore development (8, 10). In all endospore-forming species, including Clostridia, the process of sporulation begins with the activation of Spo0A, the master regulator of sporulation (14). However, the genes and mechanisms that control clostridial Spo0A activation and, thereby, the initiation of sporulation are incredibly diverse. This minireview will address recent advances in understanding the initiation of sporulation in different Clostridia, with a focus on the roles of phosphotransfer proteins in this process.
COMPARISON OF SPORULATION INITIATION BETWEEN CLOSTRIDIA AND BACILLI
As mentioned, all endospore-forming bacteria initiate sporulation through the highly conserved, essential transcriptional regulator, Spo0A (14). Spo0A functions as a response regulator and the DNA-binding domain is activated by phosphorylation at a conserved aspartate residue in the N-terminal receiver domain (15, 16). Upon phosphorylation, SpooA~P dimerizes and directly binds to specific promoter regions containing “0A boxes” to regulate sporulation-specific genes, along with additional stationary phase-associated genes (17). The decision to trigger spore formation requires input from multiple factors to coordinate environmental and metabolic cues that are reflected in the Spo0A phosphorylation state (18). spo0A mutants of spore-forming bacteria fail to activate sporulation gene programming and, as a result, are asporogenous.
Bacilli, including the extensively studied model organism Bacillus subtilis, govern Spo0A phosphorylation through an expanded two-component system, known as a phosphorelay (Fig. 1), that controls the flux of phosphate (15). B. subtilis encodes five orphan histidine kinases (OHKs), KinA-E, which influence spore formation, along with additional Spo0A-dependent stationary phase processes. The moniker “orphan” refers to histidine kinases encoded by genes not located with genes encoding a cognate response regulator. Upon activation, presumably in response to intracellular and extracellular signals, these histidine kinases autophosphorylate and transfer the phosphoryl group to an intermediate response regulator, Spo0F (19, 20). This phosphoryl group is subsequently relayed to Spo0A through the phosphotransfer protein, Spo0B (15, 21). These consecutive interactions between factors in the phosphorelay are conserved; Spo0F shares significant similarity to a phosphorylatable response regulator receiver domain, and Spo0B is reminiscent of the histidine phosphotransfer domain of histidine kinases (22–24).
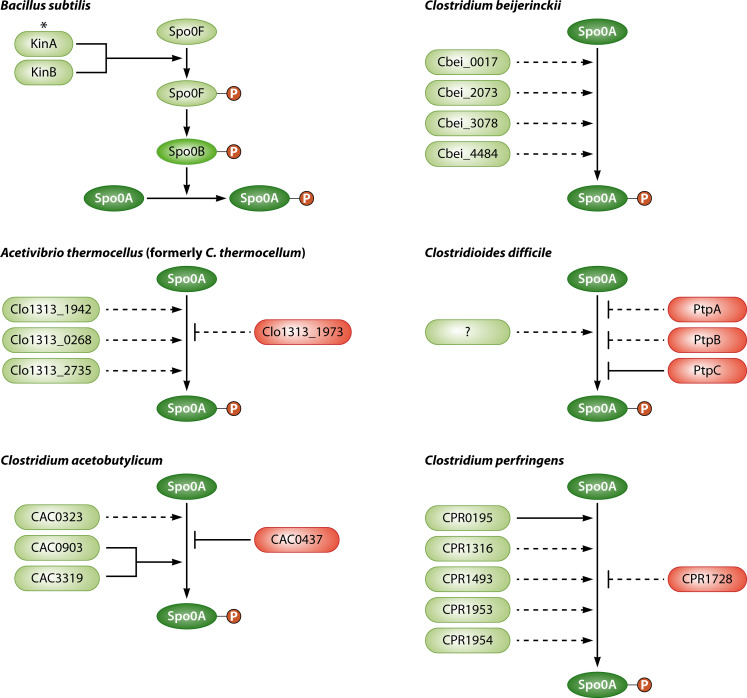
Fig 1.
Phosphotransfer proteins affecting the phosphorylation state of the master transcriptional regulator Spo0A in B. subtilis, A. thermocellus, C. acetobutylicum, C. beijerinckii, C. difficile, and C. perfringens. B. subtilis encodes five OHKs, KinA-E, but KinA and/or KinB are the major initiators of sporulation in B. subtilis by activating a phosphorelay that controls phosphate flux and leads to Spo0A phosphorylation. *, OHKs KinC-E in B. subtilis have minor roles in sporulation under specific genetic conditions and therefore are not shown in this figure. Comprehensive genome sequence analyses initially indicated that Clostridia lack a phosphorelay. More recent studies identified a phosphorelay in some Clostridia (see text), but for the Clostridial species shown in this figure, OHKs (light green) are directly or indirectly implicated in Spo0A phosphorylation. In addition, some OHKs (pink) have been shown to possess dephosphorylation activity and can remove a phosphoryl group from Spo0A-P (see text). Solid lines indicate direct interactions demonstrated in vitro while dashed lines represent putative direct interactions that are untested. This figure is updated and modified from reference 8.
Upon starvation, B. subtilis initiates sporulation through the histidine kinases KinA and KinB, which are cytosolic and membrane proteins, respectively (19, 25). KinA possesses three PAS (Per-Arnt-Sim) domains, which generally function as molecular sensors. These three KinA PAS domains are important for KinA oligomerization and autophosphorylation (26, 27). Although initially hypothesized to respond to various extracellular and intracellular signals, no known ligand has been identified and verified for these PAS domains. Instead, KinA kinase activity hinges on accumulation to a threshold level within the cell during slower cell growth, often triggered by nutrient deprivation, to contribute to Spo0A phosphorylation (28–30). This is supported by the evidence that KinA activity is regulated by its intracellular concentration (30). Little is known about KinB activation and regulation; however, KinA and KinB contributions to sporulation initiation vary depending upon the growth medium used (31). KinC plays a minor role in sporulation in unique genetic contexts by directly phosphorylating Spo0A (32, 33) but has since been shown to control cannibalism and biofilm formation through Spo0A via the phosphorelay (34, 35), functioning as either a kinase or phosphatase at different growth rates (36). Although an N-terminal PAS domain is necessary for KinC autophosphorylation activity, an activating signal of KinC has yet to be identified (37). KinD is also a bifunctional histidine kinase that promotes biofilm formation (20, 38, 39), similar to KinC. KinD delays sporulation during biofilm formation (38). Interestingly, osmotic pressure from the forming matrix polymer, glycerol, and manganese has been identified as signals to activate KinD activity through a conserved CACHE domain (39–41). Little is known about KinE’s contribution to B. subtilis sporulation. Similar to KinC and KinD, KinE appears to play a minor role in sporulation (20). While often generically grouped as the five OHKs required to activate sporulation in B. subtilis, KinA-E functions are not redundant, and they integrate diverse growth and environmental signals to influence Spo0A phosphorylation. This complex regulatory pathway calibrates Spo0A activity to control multiple physiological processes. This level of signal input diversity likely exists in other spore-forming bacteria as well.
Although the B. subtilis kinases directly interact with Spo0F, which passes the phosphoryl group to Spo0A via Spo0B, orthologs to Spo0F and Spo0B are notably missing in many Clostridial genomes (23, 42, 43). Historically, none of the class Clostridia species were thought to possess a phosphorelay architecture. However, a recent genome neighborhood conservation analysis discovered that many Clostridia encode predicted Spo0F and Spo0B proteins (44). A functional phosphorelay from a class Clostridia member, Desulfotomaculum acetoxidans, was experimentally verified, indicating that some Clostridia initiate sporulation through a four-protein phosphorelay similar to Bacilli (44). Yet, the absence of the phosphorelay in many other Clostridia, including the Families Clostridiaceae, Peptostreptococcaceae, and Ruminococcaceae, which contain many pathogenic or non-pathogenic solventogenic species of industrial importance, suggests that Clostridia employ different signaling pathways to initiate sporulation. Several mechanisms for Spo0A activation in the absence of the phosphorelay have been proposed; however, the most likely mechanism is that sporulation-associated sensor OHKs directly phosphorylate Spo0A without intermediate phosphotransfer proteins. Indeed, as detailed below, direct phosphorylation of Spo0A by OHKs has been demonstrated in several Clostridial species (45–48). Still, the possibility remains that a novel phosphorelay, perhaps between multiple histidine kinases, exists to control Spo0A activation in these species.
Finally, the additional early sporulation factors that influence sporulation initiation vary significantly between Bacilli and Clostridia as well (10, 42, 49, 50). In Bacilli, the flux of phosphate to Spo0A is further regulated by two classes of phosphatases that target either Spo0F or Spo0A and anti-kinases. Orthologs to many of these early sporulation factors are encoded in Clostridial species, and unsurprisingly, often exhibit different regulatory functions and mechanisms, likely adapting to the absence of a phosphorelay. While not the focus of this review, it is important to note the significant differences in the ecological niches between and within these two classes. The divergent functions of the early sporulation regulators likely reflect the diversity of environmental cues that trigger sporulation in different species. Supporting this notion, there is a poultry gut-adapted B. subtilis strain that does not encode two early sporulation factors, resulting in earlier and higher rates of sporulation (51).
OVERVIEW OF CLOSTRIDIAL PHOSPHOTRANSFER PROTEINS INVOLVED IN SPORULATION INITIATION
Clostridial phosphotransfer proteins (Table 1) have an assortment of structural architectures that perform a variety of functions in the Spo0A activation pathways of different species. Unfortunately, there are no apparent features, such as the presence of membrane-spanning segments or PAS domains, that allude to the roles of the individual Clostridial phosphotransfer proteins in Spo0A regulation. However, advances in the prediction of specificity residues for histidine kinase-response regulator interactions and function can provide clues to the operation of these proteins as phosphatases or kinases of Spo0A. The clostridial Spo0A proteins share 57%–76% amino acid identity, significantly more so with each other than they do with B. subtilis (52). However, analysis of these sporulation phosphotransfer proteins among different Clostridia reveals little structural or sequence similarity for those suspected of directly interacting with Spo0A to regulate sporulation.
TABLE 1.
Comparison of confirmed and potential sporulation phosphotransfer proteins
| Protein | Mutant sporulationa | Size | Protein structural domainsb,c | Reference(s) for mutant phenotypes |
|---|---|---|---|---|
| B. subtilis | ||||
| KinA | ↓ | 606 aa | 3 PAS – HisKA – HATPase | (19, 31) |
| KinB | ↓ | 428 aa | 5 TM – HisKA – HATPase | (25, 31) |
| KinC | No change | 428 aa | 2 TM – PAS – HisKA – HATPase | (32, 33) |
| KinD | ↑ | 506 aa | TM – dCache – HisKA – HATPase | (38, 53) |
| KinE | ND | 738 aa | 4 PAS – HisKA – HATPase | |
| A. thermocellus | ||||
| Clo1313_0268d | Spo- | 239 aa | HisKA – HATPase | (47) |
| Clo1313_0495 | ND | 604 aa | dCache – TM – HisKA – HATPase | |
| Clo1313_0496 | ND | 616 aa | TM – dCache – HisKA – HATPase | |
| Clo1313_1711 | ND | 501 aa | HisKA – HATPase | |
| Clo1313_1942 | Spo- | 424 aa | REC – HisKA – HATPase | (47) |
| Clo1313_1973 | ↑ | 712 aa | PPBP – HisKA – HATPase | (47) |
| Clo1313_2735 | Spo- | 387 aa | 2 TM – HisKA – HATPase | (47) |
| C. acetobutylicum | ||||
| CAC0323 | ↓ | 654 aa | 7 TM – PAS – HisKA – HATPase | (46, 54) |
| CAC0437 | ↑ | 637 aa | 2 PAS – HisKA – HATPase | (46, 55) |
| CAC0903 | ↓ | 683 aa | 7 TM – PAS – HisKA – HATPase | (46, 54) |
| CAC3319 | Spo- | 445 aa | HisKA – HATPase | (46, 54) |
| C. beijerinckii | ||||
| Cbei_0017 | ↓ | 301 aa | HisKA – HATPase | (56) |
| Cbei_0807 | ND | 636 aa | 7 TM – PAS – HisKA – HATPase | |
| Cbei_0808 | ND | 671 aa | 7 TM – PAS – HisKA – HATPase | |
| Cbei_2073 | ↓ | 444 aa | HisKA – HATPase | (57) |
| Cbei_2504 | ND | 480 aa | HisKA – HATPase | |
| Cbei_2732 | ND | 479 aa | HisKA – HATPase | |
| Cbei_3078 | ↓ | 754 aa | 2 PAS – HiskA – HATPase – REC | (56) |
| Cbei_3079 | ND | 1024 aa | 3 PAS – HisKA – HATPase – REC | |
| Cbei_4484 | ↓ | 568 aa | REC – 2 PAS – HisKA – HATPase | (57) |
| C. botulinum | ||||
| CBO0336 | ND | 615 aa | 5 TM – PAS – HisKA – HATPase | |
| CBO0340 | ND | 617 aa | 5 TM – PAS – HisKA – HATPase | |
| CBO0780 | ND | 301 aa | TM – HisKA – HATPase | |
| CBO1120 | ND | 477 aa | TM – HisKA – HATPase | |
| CBO2762 | ND | 702 aa | 2 PAS – HisKA – HATPase | |
| C. difficile | ||||
| PtpA | ↑ | 915 aa | 8 TM – 3 PAS – HisKA – HATPase | (58) |
| PtpB | ↑ | 912 aa | 8 TM – 2 PAS – HisKA – HATPase | (59) |
| PtpC | ↑ | 618 aa | PAS – HisKA – HATPase | (59) |
| C. perfringense | ||||
| CPR0195 | ↓ | 791 aa | 8 TM – PAS – HisKA – HATPase | (48, 60) |
| CPR1055 | No change | 558 aa | PAS – HisKA – HATPase | (48, 60) |
| CPR1316 | ↓ | 787 aa | 7 TM – PAS – HisKA – HATPase | (60) |
| CPR1493 | ↓ | 1086 aa | 9 BP/Y-Y-Y – TM – HisKA – HATPase | (60) |
| CPR1728 | ↑ | 571 aa | TM – sCache – HAMP – PAS – HisKA – HATPase | (60) |
| CPR1953 | ↓ | 678 aa | 8 TM – HisKA – HATPase | (60) |
| CPR1954 | ↓ | 624 aa | 7 TM – PAS – HisKA – HATPase | (60) |
ND: not determined.
BP/Y-Y-Y: β-propeller-associated domains; Cache: small molecule recognition; HAMP: histidine kinase, adenyl cyclase, methyl-binding, phosphatase domain; HATPase: histidine kinase-like ATPase; HisKA: His kinase A; PAS: Per-Arnt-Sim sensor; REC: receiver domain; TM: transmembrane domain.
Domains identified during preparation of this review using GenBank sequences of each protein and SMART (Simple Modular Architecture Research Tool) (https://smart.embl.de/).
Truncated product.
Sporulation phenotype of mutants in MDS.
The majority of experimental information on phosphotransfer protein contributions to the initiation of sporulation has been obtained from the Clostridiaceae family, which includes the species tetani, acetobutylicum, perfringens, beijerinckii, and botulinum. This review will now discuss recent progress in understanding the contributions of phosphotransfer proteins to sporulation initiation for different Clostridia. However, it is worthwhile emphasizing that there are dozens of Families within the Clostridia class for which sporulation, much less the pathways that regulate Spo0A activity, remains completely uncharacterized.
CONTRIBUTIONS OF OHKs TO SPORULATION INITIATION BY C. PERFRINGENS
Since C. perfringens lacks an identifiable phosphorelay (48, 61), bioinformatic analyses of the C. perfringens type F strain SM101 genome (62) were performed (48), which revealed that SM101 carries seven chromosomal genes encoding putative OHKs. Those putative OHK genes are designated as cpr0195, cpr1055, cpr1316, cpr1493, cpr1728, cpr1953, and cpr1954. Automated computational prediction using the PSORTb program suggested that the cpr0195, cpr1316, cpr1493, cpr1728, cpr1953, and cpr1954 genes encode putative OHKs with a membrane localization, while CPR1055 is predicted to be cytoplasmic (48, 60). Bioinformatic analyses using the SMART and Interpro programs indicate that all seven of these OHKs possess a histidine kinase-like ATPase (HATPase) domain and a histidine kinase A (HisKA) phosphoacceptor domain, with all but CPR1493 and CPR1953 also possessing a recognizable PAS domain.
Genetic analyses determined that cpr1953 and cpr1954 are overlapping genes, sharing 20 nucleotides in common, with the same orientation (60). While these two genes can be co-transcribed as an operon (60), the cpr1953 null mutant still expresses cpr1954, and the cpr1954 null mutant still expresses cpr1953 (60). Those observations suggest that cpr1953 and cpr1954 can also be expressed from independent promoters.
BLAST analysis (48, 60) indicated that the genes encoding these seven putative OHKs are present in nearly all other genome-sequenced C. perfringens strains, except type C strain JGS1495, which apparently lacks the genes encoding CPR1055 and CPR1316. Furthermore, those BLAST analyses suggested these C. perfringens OHKs are not encoded by most other Clostridia, including C. difficile. A BLAST search had initially indicated Clostridium novyi strain NCTC13108 carries genes encoding proteins with high similarity to CPR0195, CPR1493, CPR1728, and CPR1953 but this strain has now been reclassified as C. perfringens (https://www.culturecollections.org.uk/products/bacteria/detail.jsp?refId=NCTC+13108&collection=nctc).
Considerable progress was recently achieved in understanding the contributions of these putative OHKs to regulating sporulation and enterotoxin (CPE) production, which is sporulation dependent, by C. perfringens type F strain SM101. In 2019, Freedman et al. (48) showed that, in a modified Duncan-Strong sporulation medium (MDS), a cpr0195 null mutant of SM101 exhibited a ~1,000-fold reduction in sporulation, along with significantly reduced CPE production. In contrast, a cpr1055 null mutant of SM101 still showed wild-type sporulation and CPE production levels when cultured in MDS. These results indicated that some, but not all, putative OHKs are important for sporulation and CPE production by SM101 under this culture condition. It was also shown that, in vitro, the predicted kinase domain of CPR0195 can phosphorylate purified Spo0A. This in vitro evidence not only confirms that CPR0195 is a kinase but also supports the hypothesis that some C. perfringens OHKs can directly phosphorylate Spo0A, which is the critical first step in initiating sporulation.
A follow-up study (60) then evaluated the contributions of CPR0195 and CPR1055 for regulating sporulation and CPE production by SM101 in a more pathophysiologically relevant incubation condition than MDS. For this purpose, an ex vivo model using diluted mouse small intestinal contents (MIC) was developed and shown to support sporulation and CPE production by SM101 (60). Similar to the MDS results, no differences in the levels of sporulation or CPE production were detected between wild-type SM101 and the cpr1055 null mutant when cultured in MIC. Surprisingly, the cpr0195 null mutant, which exhibits reduced sporulation and CPE production in MDS, still sporulated and produced CPE at the same levels as wild-type SM101 when cultured in this new MIC model. This finding revealed that environmental conditions profoundly impact the importance of individual C. perfringens OHKs for sporulation and CPE production.
Therefore, seven SM101 mutants, each unable to produce a different putative OHK, were compared for their ability to sporulate and produce CPE in MIC vs MDS (60). The results revealed three phenotypes. The cpr1055 and cpr1728 null mutants still sporulated and produced CPE at approximately the same levels as wild-type SM101 in both MDS and MIC conditions. In contrast, the cpr0195, cpr1316, and cpr1493 mutants showed reduced sporulation and CPE production when cultured in MDS medium but were able to sporulate and produce CPE similarly to SM101 when cultured in MIC. Interestingly, the cpr1953 and cpr1954 mutants exhibited negligible sporulation and no CPE production in either MDS or MIC.
SM101 produced ~107 spores/mL when cultured in MDS but ~100-fold fewer spores/mL when incubated in MIC (60). While the SM101 mutants unable to produce CPR0195, CPR1316, or CPR1493 made the same number of spores as wild-type SM101 when cultured in MIC, these mutants showed a 102- to 104-fold reduction in sporulation compared to MDS cultures of wild-type SM101. However, the cpr1953 and cpr1954 null mutants exhibited a much greater sporulation defect, producing essentially no (i.e., only ~10/mL) spores whether cultured in MDS or MIC. These results indicated that the CPR1953 and CPR1954 OHKs are virtually essential for sporulation when SM101 is cultured in either MIC or MDS but CPR0195, CPR1316, and CPR1493 OHKs boost sporulation above those MIC sporulation levels when SM101 is cultured in MDS (Fig. 1). The intricate details of these OHK contributions in different incubation conditions require further study.
Bioinformatic analyses (60) detected the presence of a classical DHp histidine phosphotransfer motif in the translated open reading frame sequences encoding all seven putative OHKs. Therefore, an alanine was substituted for the key histidine residue in this phosphotransfer motif for CPR1316, CPR1493, CPR1953, or CPR1954. When plasmids encoding these alanine-substituted OHKs were transformed into their corresponding mutant, there was no increase in sporulation or CPE production, supporting these proteins as histidine kinases.
Using a spoIIA operon promoter-driven reporter plasmid, CPR0195, CPR1316, CPR1493, CPR1953, and CPR1954 were shown to function early in sporulation, that is, prior to the production of sporulation-associated sigma factors (60). This result is consistent with the involvement of these four OHKs in Spo0A production and Spo0A phosphorylation. Supporting this contention, Spo0A western blot analyses demonstrated that the cpr0195, cpr1316, and cpr1493 null mutants produced less Spo0A protein compared to wild-type SM101 when cultured for 3 h in MDS (60). However, under that same incubation condition, the cpr1953 and cpr1954 null mutants, which are almost completely unable to sporulate, made even less Spo0A than the cpr0195, cpr1316 or cpr1493 OHK mutants. If the incubation period was extended to 5 h in MDS, all mutants produced wild-type levels of Spo0A, except the cpr1953 null mutant, which still made reduced amounts of Spo0A.
As already mentioned, it was shown (48) that the predicted kinase domain of CPR0195 can directly phosphorylate Spo0A. Similar studies have not yet been performed with the CPR1316, CPR1493, CPR1953, or CPR1954 OHKs. However, studies (60) using Phos-Tag gels indicated that the cpr1954 kinase mutant has no detectable phosphorylation of Spo0A. Whether CPR1954 directly phosphorylates Spo0A or affects Spo0A phosphorylation through an intermediate remains to be determined, as does the ability of CPR1953 to phosphorylate Spo0A. Collectively, the reduced Spo0A production by the cpr1953 and cpr1954 mutants, and the lack of Spo0A phosphorylation for the cpr1954 mutant, can explain the profound defects in sporulation and CPE production by these mutants. The reduction in Spo0A production and phosphorylation for the cpr1954 mutant may be linked since Spo0A phosphorylation in Bacillus spp. increases Spo0A production (63).
Conceivably, CPR1055 or CPR1728 could be phosphatases that, under certain environmental conditions, affect Spo0A phosphorylation levels and thereby modulate (inhibit) sporulation, rather than acting as OHKs to promote sporulation (Fig. 1). Offering limited support for that possibility, the cpr1728 null mutant sporulated slightly better than SM101 in MDS, although that effect did not reach statistical significance. This mutant also produced slightly more CPE in MDS as assessed by western blotting, but that effect was not quantified.
CONTRIBUTION OF OHKs TO SPORULATION INITIATION BY C. DIFFICILE
C. difficile is a gastrointestinal pathogen and is the primary catalyst of antibiotic-associated diarrhea. The symptoms of C. difficile infection (CDI) are mediated by two large exotoxins, TcdA and TcdB (64), and range from mild diarrhea and abdominal pain to potentially lethal pseudomembranous colitis. Spores are critical to the C. difficile life cycle as they are essential for transmission; therefore, the dormant spore is the infectious form of this bacterium (65). Spores are resistant to many disinfectants used in healthcare settings, which provide persistence in the environment and are often impervious to antibiotic treatment, promoting the reoccurrence of CDI (65–67).
The Spo0A transcriptional regulator in C. difficile shares significant similarity to the B. subtilis Spo0A amino acid sequence and structure (52, 68). As in B. subtilis, the conserved aspartate residue in the N-terminal receiver domain is critical for C. difficile Spo0A phosphorylation and dimerization (52). Because C. difficile does not possess an identified phosphorelay, it is presumed that any activating histidine kinase would directly bind to and phosphorylate Spo0A, whereas sporulation-associated histidine kinases in B. subtilis directly interact with Spo0F. Comparative studies between the receiver domains of C. difficile Spo0A and B. subtilis Spo0A and Spo0F, coupled with extensive site-directed mutagenesis of conserved residues, revealed that C. difficile Spo0A utilizes functionally conserved regions to facilitate interactions with both positive and negative regulators (52).
C. difficile encodes several OHKs, three of which share significant homology to KinA and KinB of B. subtilis (45): PtpA (CD630_14920), PtpB (CD630_24920), and PtpC (CD630_15790), all named as phosphotransfer proteins (Ptp) for their function in sporulation. PtpA and PtpB are large, transmembrane proteins with three and two predicted PAS domains, respectively. These PAS domains are located intracellularly, although their function remains unknown. PtpC is a cytosolic protein containing a degenerate PAS domain. An early study briefly characterized two of these orphan kinases; they found that a ptpB mutant exhibited decreased sporulation, although this mutant was never complemented (45). They also demonstrated that PtpC directly transferred a phosphoryl group to Spo0A in vitro (45), suggesting that Spo0A phosphorylation and activation are directly controlled by orphan histidine kinases in C. difficile. However, the regulatory roles of PtpB and PtpC were not fully elucidated in this study.
A subsequent study revealed that PtpA inhibits C. difficile sporulation, as a ptpA mutant hypersporulated (58). Furthermore, the conserved histidine residue required for autophosphorylation and phosphoryl group transfer was critical for PtpA function. Interestingly, additional work revealed that a ptpB mutant exhibits increased sporulation in several lab conditions (59), similar to a ptpA mutant and in contrast to the initial study (45). Contrary to the initial hypotheses, PtpA and PtpB appear to function primarily as phosphatases that inhibit Spo0A activity and subsequently inhibit spore formation and thus are referred to as phosphotransfer proteins rather than sensor kinases.
PtpA and PtpB phenocopy each other and exhibit identical changes in gene expression (59). A ptpA ptpB double mutant displays the same hypersporulation phenotype as the single mutants, suggesting that PtpA and PtpB function in the same regulatory pathway to repress spore formation (59). Neither protein can replace the function of the other, indicating that PtpA and PtpB are nonredundant (59). Surprisingly, unlike PtpA, the conserved histidine residue of PtpB is not necessary for its function (59). It appears that PtpA and PtpB function together, not stepwise, to repress sporulation, suggesting that PtpA and PtpB may only be active as phosphatases when paired as hetero-oligomers. No direct evidence has yet demonstrated that PtpA and PtpB directly bind Spo0A; it remains possible that PtpA and PtpB serve as an endpoint in a serial dephosphorylation pathway. An alternative hypothesis is that PtpA and/or PtpB interact with an intermediate factor(s) to facilitate the dephosphorylation of Spo0A.
PtpA and PtpB affect additional virulence-associated physiological processes in C. difficile. Mutants of ptpA and ptpB produce less TcdA toxin, and a ptpA mutant exhibits an attenuated virulence phenotype in the hamster model of C. difficile infection (58, 59). PtpA and PtpB also promote motility gene expression, and the ptpA mutant is less motile than the parent (58, 59). The PtpA and PtpB regulatory pathway is linked with RstA, a multifunctional regulator that indirectly promotes sporulation and directly represses the expression of motility and toxin genes (58, 59, 69, 70). RstA inhibits the function of Spo0E, a small protein that directly binds to Spo0A and prevents its activation (71). PtpA/B and RstA reciprocally regulate sporulation, toxin production, and motility, which suggests that the activities of the proteins converge on a shared regulatory pathway. However, the regulatory relationship between PtpA/PtpB and RstA remains unclear.
The function of PtpC in early sporulation events has been difficult to discern. While PtpC was shown to directly transfer a phosphoryl group to Spo0A in vitro, a ptpC mutant exhibits variably increased sporulation on sporulation agar (45, 59). The ptpC mutant was complemented with ptpC alleles containing site-directed mutations in the predicated residues required for kinase and phosphatase activities, indicating PtpC may not be active on sporulation agar (59). Interestingly, overexpression of ptpC in the parent strain resulted in increased spore formation, which was dependent on the conserved histidine residue (59), indicating that at higher intracellular concentrations, PtpC functions to promote Spo0A phosphorylation. Altogether, PtpC appears to function as a dual kinase/phosphatase in response to unknown signals. It is possible that the strong phosphatase activity of PtpA and PtpB masks the effects of PtpC and/or that PtpC functions differently in the host. Additional studies are needed to better understand the contributions of PtpC to Spo0A activation.
There are additional OHKs encoded in C. difficile; however, not all of these have roles in regulating early sporulation events. The OHK CD630_13490 (CprK) was found to function as the sensor kinase for the cationic antimicrobial peptide-responsive CprABC system (72). The function of another OHK, CD630_19490, is unknown but does not impact spore formation (59). Another OHK, CD630_05760, now known as RgaS, has recently been identified as the cognate sensor kinase to the orphan response regulator, RgaR (73). RgaR directly activates the transcription of several operons, including agrB1D1, which encodes the gene products necessary to produce the AgrD1 quorum-sensing peptide, and spoZ, encoding a regulatory small RNA (73, 74). The AgrD1 quorum-sensing peptide accumulates extracellularly and promotes early-stage sporulation through an unknown regulatory pathway (75). SpoZ promotes later-stage sporulation through inhibiting the accumulation of a small protein (73). While RgaS does not directly influence Spo0A activation, the RgaSR two-component system functions at multiple points within the sporulation pathway, including during early sporulation events, to trigger C. difficile spore formation.
The presence of multiple, identified Spo0A inactivating factors suggests that mechanisms for deliberate Spo0A phosphorylation occur in C. difficile (Fig. 1). However, the mystery of which factor(s) in C. difficile are primarily responsible for Spo0A phosphorylation remains unsolved. It seems likely that Spo0A phosphorylation is directly mediated by still unidentified kinases. These potential kinases are traditionally difficult to predict beyond the status of an OHK. Further unraveling how the quorum-sensing peptide, AgrD1, promotes early sporulation may lead to the identification of an activating Spo0A factor. Further delineating the complex genetic pathways and molecular mechanisms by which Spo0A activity is controlled will provide greater insight into the environmental signals that trigger spore formation within the host.
CONTRIBUTIONS OF OHKs TO SPORULATION INITIATION BY CLOSTRIDIUM BOTULINUM AND RELATED SPECIES
C. botulinum is a diverse collection of species that has been historically clustered into four genetically distinct groups based on physiological traits (76). All C. botulinum produce the characteristic botulinum neurotoxins; however, each of the four groups has a phylogenetically related partner species that is non-toxigenic (77–79). The neurotoxigenic isolates are referred to as C. botulinum (Groups I-II) and C. argentinense (Group IV). The non-neurotoxigenic counterparts include C. sporogenes (Group I), C. taeniosporum (Group II), C. novyi (Group III), and C. argentinense, C. subterminale, or C. hastiforme (Group IV). In addition, some C. baratii and C. butyricum isolates can also make botulinum toxin but are not denoted as C. botulinum (80). Of all of these, the only information published on Spo0A post-transcriptional regulation is from the Group I C. botulinum, strain ATCC 3502.
In 2006, Wörner et al. scanned the genome in search of OHKs that could serve as C. botulinum Spo0A activators (80). This analysis led to the identification of five OHKs: CBO0336, CBO0340, CBO0780, CBO1120, and CBO2762. The investigators cloned and expressed CBO1120 and spo0A from C. botulinum in Bacillus subtilis, and assessed sporulation outcomes. Heterologous expression of spo0AC.b. alone could not complement the sporulation of a B. subtilis spo0A mutant but Spo0AC.b. was able to repress expression of the B. subtilis Spo0A-regulated gene, abrB. Furthermore, they observed that heterologous co-expression of CBO1120 and spo0A was lethal to B. subtilis. However, expression of CBO1120 alone, or a combination of CBO1120 and an inactive spo0A variant, had no effect. From this, they concluded that CBO1120 is likely a direct activator of Spo0A. However, no further studies have been performed to verify the interactions or functions of CBO1120 or the other OHKs. The limited information available on the factors and pathways that regulate Spo0A in the C. botulinum groups is likely due to the restrictions on experimentation with neurotoxin producers; however, exploration of these mechanisms in the non-pathogenic relatives or modified C. botulinum strains lacking the neurotoxin genes represents an opportunity to advance this field (81).
CONTRIBUTIONS OF OHKs TO SPORULATION INITIATION BY NON-PATHOGENIC CLOSTRIDIA
There are dozens of families under the order Eubacteriales of class Clostridia, most of which are non-pathogenic species. Compared to the pathogenic Clostridia, there is significantly less known about the post-transcriptional regulation of Spo0A or sporulation in non-pathogenic Clostridia. Only three non-pathogenic species of Clostridia have sporulation kinases or phosphatases that have been characterized genetically or biochemically. These include Clostridium acetobutylicum, Clostridium beijerinckii, and Acetivibrio thermocellus (previously Clostridium thermocellum). The studied non-pathogenic Clostridia are important biofuel/solvent generators that were historically employed for the production of the acetone-butanol-ethanol (ABE) solvents. Accordingly, most of what is known about the function of their genes is in the context of their use as industrial producers of compounds. In this section, we describe the current state of research for the three species with experimental evidence for Spo0A regulation: C. acetobutylicum, C. beijerinckii, and A. thermocellus.
Clostridium acetobutylicum
C. acetobutylicum has been employed for industrial solvent production for over a century. Consequently, there has been significant progress in understanding how C. acetobutylicum generates solvents and how to improve the solventogenesis process. It was understood more than 40 years ago that solvent production is closely tied to the activation of sporulation in Clostridial producer species (82–84). Research later verified that Spo0A is an important regulator of both sporulation and solvent production in Clostridia, including C. acetobutylicum (85–87). Given the importance of Spo0A in solventogenesis, investigators pursued the identification of factors that directly control Spo0A activity, including phosphotransfer proteins.
The genome of C. acetobutylicum (ATCC 824) encodes five OHK/phosphotransfer proteins, four of which affect sporulation (CAC0903, CAC3319, CAC0323, and CAC0437) and one that does not (CAC2730) (46). Of the four influential factors, CAC323, CAC0903, and CAC3319 promote sporulation, as null mutants of these genes demonstrated modest reductions in sporulation (46, 54). However, a CAC0437 null mutant had increased spore formation, suggesting that it functions as a phosphatase that deactivates Spo0A, rather than as a Spo0A kinase (46, 55). CAC0437, CAC0903, and CAC3319 demonstrated the ability to transfer phosphoryl groups with Spo0A in vitro, providing additional support for their roles as direct regulators of Spo0A activity (44, 46). Based on the phenotypes of selected double mutants and the evidence for their direct interaction with Spo0A, Steiner et al. proposed a model for Spo0A regulation that inferred three pathways for Spo0A regulation: (i) CAC0903-CAC3319 phosphorylation of Spo0A, (ii) CAC0323 phosphorylation of Spo0A, and (iii) CAC0437 dephosphorylation of Spo0A (46). However, a subsequent study found that a CAC3319 null mutant was unable to form spores (54), which disrupts the prior model. Investigators were unable to purify active CAC0323, so its ability to directly impact Spo0A phosphorylation remains uncertain. Some of these phosphotransfer proteins may act together in pathways to regulate Spo0A (Fig. 1); however, additional experimentation is required to sort out their specific roles and epistatic hierarchies.
Clostridium beijerinckii
C. beijerinckii is an important solventogenic species of interest as a biofuel producer. Because phenotypic traits were initially used to classify species, many C. beijerinckii were historically misclassified as C. acetobutylicum, and it was initially thought that regulation of sporulation and solventogenesis were similar between these species. However, genomic comparisons revealed extensive differences in the genomes of these two Clostridium, while transcriptional analyses have shown significant differences in their regulation (88–90). The C. beijerinckii (strain NCBI 8052) chromosome encodes more than a dozen OHKs, though only a subset was recently examined for a role in sporulation. In 2020, Xin et al. evaluated several C. beijerinckii OHKs for similarity to the C. acetobutylicum Spo0A kinases (57). Based on sequence alignment profiles, they investigated six OHKs: Cbei1553, Cbei2073, Cbei2087, Cbei2435, Cbei4484, and Cbei4925. They reported that null mutants in only two of these genes, Cbei2073 and Cbei4484, formed fewer heat-resistant spores, while deletions in the other four genes had no significant phenotypes. Cbei2073 is noted as having a similar structure and phenotype to the CAC3319 kinase of C. acetobutylicum but Cbei4484 is unlike any characterized sporulation phosphotransfer protein. Cbei4484 is predicted to contain the HisKA and HAPTase domains typical of sporulation kinases, but in addition, it includes a receiver domain that is typical of response regulators. The hybrid HK-RR structure of Cbei4484 suggests that this protein may be able to send and receive phosphoryl groups in a complex regulatory arrangement that could include a phosphorelay.
In 2023, Humphreys and colleagues found that sub-culturing mutants for solvent production selected for variants with sporulation defects (56). Through genome sequence analyses, they identified mutagenic “hot spots” in the chromosomes of affected isolates occurring within spo0A and the predicted OHKs Cbei0017 and Cbei3078. When assessed for sporulation, the null mutants in both Cbei0017 and Cbei3078 demonstrated several log decreases in spores formed, suggesting that they are positive effectors of Spo0A activity. Cbei0017 appears to be a conventional histidine kinase, similar to Cbei2073. But like Cbei4484, Cbei3078 encodes both kinase and receiver domains, suggesting that it may function as a hybrid sensor-receiver, with a more complex signaling role than noted in previously characterized Clostridia. In addition, C. beijerinckii encodes at least six other predicted OHKs with similarity to sporulation kinases that have not been examined for sporulation regulatory functions: Cbei0807, Cbei0808, Cbei2160 (hybrid HK-receiver), Cbei2504, Cbei2732, and Cbei3079. Further experimentation, including epistasis analyses and in vitro phosphotransfer studies, is necessary to determine the pathways and order of operations for signaling through these factors.
Acetivibrio thermocellus (formerly Clostridium thermocellum)
A. thermocellus is a soil-dwelling member of the Oscillospiraceae family that is best characterized by its ability to generate bioethanol from cellulose (91). What is known about the kinases or phosphatases that influence A. thermocellus sporulation stems from a 2014 study of putative OHKs of strain DSMZ 1313 (47). Using a combination of domain predictions and homology to the C. difficile kinase CD2492 (PtpB), the authors identified six predicted sporulation OHKs: Clo1313_268, Clo1313_0495, Clo1313_1711, Clo1313_1942, Clo1313_1973, and Clo1313_2735. They were able to generate null mutations in each of these genes except Clo1313_0495 and Clo1313_1711. Sporulation tests revealed that Clo1313_268, Clo1313_1942, and Clos1313_2735 mutants produced no detectable heat-resistant spores, while the Clo1313_1973 mutant generated more spores than the wild-type. Double mutants in Clo1313_1973 paired with mutants for each of the other genes resulted in wild-type sporulation, suggesting that the function of Clo1313_1973 is dominant to the other factors. Also, the over-expression of Clo1313_0268, Clo1313_1942, or Clo1313_2735 could complement sporulation in any of these mutants, indicating some redundancy in their functions.
Like C. beijerinckii, the sporulation phosphotransfer proteins of A. thermocellus have diverse structural domains including a hybrid HK-receiver (Clo1313_1942), a predicted periplasmic binding domain (Clo1313_1973), and conventional histidine kinases (Clo1313_0268 and Clo1313_2735); however, none of the examined factors contain defined PAS domains, which are often found in sporulation-associated histidine kinases/phosphotransfer proteins. In addition to the unexplored predicted kinases Clo1313_0495 and Clo1313_1711, the kinase Clo1313_0496 is predicted to be in an operon with Clo1313_0495 and may also contribute to sporulation. Given the number of uncharacterized putative sporulation kinases in A. thermocellus, it is possible that multiple sporulation initiation pathways or complex regulatory circuits remain to be discovered in this organism.
SUMMARY
Recent research has revealed great diversity in Spo0A activation pathways used by different Clostridia. Contrary to previous dogma, it is now apparent that some Clostridia encode functional components of the Bacillus phosphorelay, but components of that classical phosphorelay have not yet been identified in the pathogenic Clostridia or the nonpathogenic, industrially important solventogenic species. The identification of kinases with receiver domains in some Clostridia opens the possibility that those bacteria may possess a novel phosphorelay for Spo0A phosphorylation. However, the previous hypothesis that Spo0A is directly phosphorylated by OHKs remains the most viable explanation for most of the pathogenic Clostridia and at least some Clostridia with biotechnology importance.
Considering these points, the diversity in Spo0A activation pathways, the variability in kinase/phosphatase use, the structural dissimilarities of the phosphotransfer proteins, and the diversity of the ecological niches inhabited by members of the Clostridia, it is apparent that incredible variability in Spo0A phosphoregulation exists among this class of bacteria. Consequently, no one model can explain Clostridial sporulation initiation; the remarkable diversity in factors that regulate Spo0A among different Clostridia make it difficult to even predict the proteins that directly interact with Spo0A or integrate signals to regulate the onset of sporulation. Therefore, despite much recent progress, further research is needed to continue addressing mechanisms of Clostridial Spo0A phosphoregulation that initiates sporulation, a topic that retains significant relevance for Clostridial pathogenesis and exploitation of Clostridia for industrial purposes.
ACKNOWLEDGMENTS
This work was generously supported by grants AI-019844 (BM), AI116933 (SM), and AI156052 (SM) from the National Institute of Allergy and Infectious Disease. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biographies

Iman Mehdizadeh Gohari, D.V.M., M.Sc., P.hD., is a research instructor faculty at the University of Pittsburgh School of Medicine. His primary research interest is pathogenesis of C. perfringens infections in humans and animals, with a particular emphasis on understanding the role of orphan histidine kinases in sporulation, genomics, and action of toxins, especially enterotoxin (CPE), produced by this bacterium. His research in the field of clostridial has been published in many peer-reviewed scientific journals (PLoS Pathogens, mBio, Virulence, Journal of Bacteriology, and Infection and Immunity, etc.). He presented his work in numerous international and national conferences. He serves as a reviewer for numerous peer-reviewed journals, such as Frontiers in Microbiology, Scientific Report Nature, Plasmid, Veterinary Microbiology, BMC Veterinary Research, Anaerobe, and others. He is a member of the American Society for Microbiology.

Dr. Adrianne N. Edwards received her B.S. in genetics at the University of Georgia in 2004 and earned her Ph.D. in microbiology and molecular genetics at Emory University in 2010, conducting her dissertation studies in Dr. Tony Romeo’s lab. She trained as a postdoctoral fellow in the field of synthetic biology in the Department of Chemistry at Emory University and then joined Dr. Shonna McBride’s lab in the Department of Microbiology and Immunology at Emory University as a postdoctoral fellow in 2012. She was promoted to instructor in 2016. Dr. Edwards’s research career has focused on the investigation and dissection of global regulatory networks and transcriptional and post-transcriptional regulatory mechanisms that impact bacterial physiology and pathogenicity. Since 2012, Dr. Edwards’s work has focused on identifying and unraveling the genetic pathways and molecular mechanisms controlling Clostridioides difficile sporulation.

Shonna M. McBride, Ph.D., is an associate professor in the Department of Microbiology and Immunology at Emory University School of Medicine. She received her Ph.D. in microbiology and immunology from the University of Texas Health Science Center at San Antonio and a B.S. from McNeese State University. She trained as a postdoctoral fellow in the field of bacterial pathogenesis at the Schepens Eye Research Institute of Harvard Medical School and the Tufts University School of Medicine. For almost 20 years, her research interests have focused on Clostridioides difficile, to identify and characterize the molecular mechanisms that control sporulation, antimicrobial resistance, and toxin production.

Bruce A. McClane, Ph.D., is a professor in the Department of Microbiology and Molecular Genetics at the University of Pittsburgh School of Medicine. His research has provided important insights into the genetics, structure-function relationships, action, and role in the pathogenesis of Clostridium perfringens toxins involved in enteric disease. Dr. McClane's research has also focused on understanding other aspects of C. perfringens pathogenicity, including quorum sensing, sporulation, and sialidases. He is a member of the American Society for Microbiology and a Fellow of the American Academy of Microbiology. Dr. McClane has received a Merit Award from the National Institutes of Health. He is a founder of the Clostpath International Conference on clostridial pathogens and an editor of mBio and PLoS Pathogens.
Contributor Information
Shonna M. McBride, Email: shonna.mcbride@emory.edu.
Bruce A. McClane, Email: bamcc@pitt.edu.
Jacob Yount, Ohio State University, Columbus, Ohio, USA.
Daniel Paredes-Sabja, Texas A&M University, College Station, Texas, USA.
REFERENCES
- 1. Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, Corthier G, Furet JP. 2009. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9:123. doi: 10.1186/1471-2180-9-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mada PK, Alam MU. 2023. Clostridioides difficile infection. In Statpearls (Internet) treasure Island. StatPearls Publishing, FL. [PubMed] [Google Scholar]
- 3. Mehdizadeh Gohari I, A Navarro M, Li J, Shrestha A, Uzal F, A McClane B. 2021. Pathogenicity and virulence of Clostridium perfringens. Virulence 12:723–753. doi: 10.1080/21505594.2021.1886777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McClane BA, Robertson SL, Li J. 2013. Clostridium perfringens, p 465–489. In Doyle MP, Buchanan RL (ed), Food microbiology: fundamentals and frontiers, 4th ed. ASM press, Washington D.C. [Google Scholar]
- 5. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.p11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pirazzini M, Montecucco C, Rossetto O. 2022. Toxicology and pharmacology of botulinum and tetanus neurotoxins: an update. Arch Toxicol 96:1521–1539. doi: 10.1007/s00204-022-03271-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poehlein A, Solano JDM, Flitsch SK, Krabben P, Winzer K, Reid SJ, Jones DT, Green E, Minton NP, Daniel R, Dürre P. 2017. Microbial solvent formation revisited by comparative genome analysis. Biotechnol Biofuels 10:58. doi: 10.1186/s13068-017-0742-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al-Hinai MA, Jones SW, Papoutsakis ET. 2015. The Clostridium sporulation programs: diversity and preservation of endospore differentiation. Microbiol Mol Biol Rev 79:19–37. doi: 10.1128/MMBR.00025-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fernández-Blanco C, Robles-Iglesias R, Naveira-Pazos C, Veiga MC, Kennes C. 2023. Production of biofuels from C1-gases with Clostridium and related bacteria-recent advances. Microb Biotechnol 16:726–741. doi: 10.1111/1751-7915.14220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen A, Edwards AN, Sarker MR, Paredes-Sabja D. 2019. Sporulation and germination in clostridia pathogens. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0017-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J, McClane BA. 2008. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathog 4:e1000056. doi: 10.1371/journal.ppat.1000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J, Paredes-Sabja D, Sarker MR, McClane BA. 2016. Clostridium perfringens sporulation and sporulation-associated toxin production. Microbiol Spectr 4. doi: 10.1128/microbiolspec.TBS-0022-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diallo M, Kengen SWM, López-Contreras AM. 2021. Sporulation in solventogenic and acetogenic clostridia. Appl Microbiol Biotechnol 105:3533–3557. doi: 10.1007/s00253-021-11289-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferrari FA, Trach K, LeCoq D, Spence J, Ferrari E, Hoch JA. 1985. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A 82:2647–2651. doi: 10.1073/pnas.82.9.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552. doi: 10.1016/0092-8674(91)90238-t [DOI] [PubMed] [Google Scholar]
- 16. Bird TH, Grimsley JK, Hoch JA, Spiegelman GB. 1993. Phosphorylation of Spo0A activates its stimulation of in vitro transcription from the Bacillus subtilis spoIIG operon. Mol Microbiol 9:741–749. doi: 10.1111/j.1365-2958.1993.tb01734.x [DOI] [PubMed] [Google Scholar]
- 17. Baldus JM, Green BD, Youngman P, Moran CP Jr. 1994. Phosphorylation of Bacillus subtilis transcription factor Spo0A stimulates transcription from the spoIIG promoter by enhancing binding to weak 0A boxes. J Bacteriol 176:296–306. doi: 10.1128/jb.176.2.296-306.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strauch MA, Hoch JA. 1993. Signal transduction in Bacillus subtilis sporulation. Curr Opin Genet Dev 3:203–212. doi: 10.1016/0959-437x(93)90024-j [DOI] [PubMed] [Google Scholar]
- 19. Perego M, Cole SP, Burbulys D, Trach K, Hoch JA. 1989. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol 171:6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang M, Shao W, Perego M, Hoch JA. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol 38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x [DOI] [PubMed] [Google Scholar]
- 21. Trach K, Burbulys D, Strauch M, Wu JJ, Dhillon N, Jonas R, Hanstein C, Kallio P, Perego M, Bird T. 1991. Control of the initiation of sporulation in Bacillus subtilis by a phosphorelay. Res Microbiol 142:815–823. doi: 10.1016/0923-2508(91)90060-n [DOI] [PubMed] [Google Scholar]
- 22. Trach KA, Chapman JW, Piggot PJ, Hoch JA. 1985. Deduced product of the stage 0 sporulation gene Spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc Natl Acad Sci U S A 82:7260–7264. doi: 10.1073/pnas.82.21.7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stephenson K, Hoch JA. 2002. Evolution of signalling in the sporulation phosphorelay. Mol Microbiol 46:297–304. doi: 10.1046/j.1365-2958.2002.03186.x [DOI] [PubMed] [Google Scholar]
- 24. Stephenson K, Lewis RJ. 2005. Molecular insights into the initiation of sporulation in gram-positive bacteria: new technologies for an old phenomenon. FEMS Microbiol Rev 29:281–301. doi: 10.1016/j.femsre.2004.10.003 [DOI] [PubMed] [Google Scholar]
- 25. Trach KA, Hoch JA. 1993. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol Microbiol 8:69–79. doi: 10.1111/j.1365-2958.1993.tb01204.x [DOI] [PubMed] [Google Scholar]
- 26. Wang L, Fabret C, Kanamaru K, Stephenson K, Dartois V, Perego M, Hoch JA. 2001. Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis. J Bacteriol 183:2795–2802. doi: 10.1128/JB.183.9.2795-2802.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kiehler B, Haggett L, Fujita M. 2017. The PAS domains of the major sporulation kinase in Bacillus subtilis play a role in tetramer formation that is essential for the autokinase activity. Microbiologyopen 6:e00481. doi: 10.1002/mbo3.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fujita M, Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev 19:2236–2244. doi: 10.1101/gad.1335705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eswaramoorthy P, Duan D, Dinh J, Dravis A, Devi SN, Fujita M. 2010. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. J Bacteriol 192:3870–3882. doi: 10.1128/JB.00466-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Narula J, Kuchina A, Zhang F, Fujita M, Süel GM, Igoshin OA. 2016. Slowdown of growth controls cellular differentiation. Mol Syst Biol 12:871. doi: 10.15252/msb.20156691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. LeDeaux JR, Yu N, Grossman AD. 1995. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol 177:861–863. doi: 10.1128/jb.177.3.861-863.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kobayashi K, Shoji K, Shimizu T, Nakano K, Sato T, Kobayashi Y. 1995. Analysis of a suppressor mutation ssb (kinC) of sur0B20 (spo0A) mutation in Bacillus subtilis reveals that kinC encodes a histidine protein kinase. J Bacteriol 177:176–182. doi: 10.1128/jb.177.1.176-182.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. LeDeaux JR, Grossman AD. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol 177:166–175. doi: 10.1128/jb.177.1.166-175.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. López D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106:280–285. doi: 10.1073/pnas.0810940106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. López D, Vlamakis H, Losick R, Kolter R. 2009. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol 74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Z, Srivastava P, Zarazúa-Osorio B, Marathe A, Fujita M, Igoshin OA. 2022. Bacillus subtilis histidine kinase KinC activates biofilm formation by controlling heterogeneity of single-cell responses. mBio 13:e0169421. doi: 10.1128/mbio.01694-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Devi SN, Vishnoi M, Kiehler B, Haggett L, Fujita M. 2015. In vivo functional characterization of the transmembrane histidine kinase KinC in Bacillus subtilis. Microbiology 161:1092–1104. doi: 10.1099/mic.0.000054 [DOI] [PubMed] [Google Scholar]
- 38. Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. 2010. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio 1:e00035–00010. doi: 10.1128/mBio.00035-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo J, Losick R. 2012. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol 85:418–430. doi: 10.1111/j.1365-2958.2012.08109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rubinstein SM, Kolodkin-Gal I, McLoon A, Chai L, Kolter R, Losick R, Weitz DA. 2012. Osmotic pressure can regulate matrix gene expression in Bacillus subtilis. Mol Microbiol 86:426–436. doi: 10.1111/j.1365-2958.2012.08201.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shemesh M, Chai Y. 2013. A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J Bacteriol 195:2747–2754. doi: 10.1128/JB.00028-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paredes CJ, Alsaker KV, Papoutsakis ET. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol 3:969–978. doi: 10.1038/nrmicro1288 [DOI] [PubMed] [Google Scholar]
- 43. Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, Rigden DJ. 2012. Genomic determinants of sporulation in bacilli and clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol 14:2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davidson P, Eutsey R, Redler B, Hiller NL, Laub MT, Durand D. 2018. Flexibility and constraint: evolutionary remodeling of the sporulation initiation pathway in firmicutes. PLoS Genet 14:e1007470. doi: 10.1371/journal.pgen.1007470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Underwood S, Guan S, Vijayasubhash V, Baines SD, Graham L, Lewis RJ, Wilcox MH, Stephenson K. 2009. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J Bacteriol 191:7296–7305. doi: 10.1128/JB.00882-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Steiner E, Dago AE, Young DI, Heap JT, Minton NP, Hoch JA, Young M. 2011. Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol Microbiol 80:641–654. doi: 10.1111/j.1365-2958.2011.07608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mearls EB, Lynd LR. 2014. The identification of four histidine kinases that influence sporulation in Clostridium thermocellum. Anaerobe 28:109–119. doi: 10.1016/j.anaerobe.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 48. Freedman JC, Li J, Mi E, McClane BA. 2019. Identification of an important orphan histidine kinase for the initiation of sporulation and enterotoxin production by Clostridium perfringens type F strain SM101. mBio 10:e02674–02618. doi: 10.1128/mBio.02674-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Edwards AN, McBride SM. 2014. Initiation of sporulation in Clostridium difficile: a twist on the classic model . FEMS Microbiol Lett 358:110–118. doi: 10.1111/1574-6968.12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee CD, Rizvi A, Edwards AN, DiCandia MA, Vargas Cuebas GG, Monteiro MP, McBride SM. 2022. Genetic mechanisms governing sporulation initiation in Clostridioides difficile. Curr Opin Microbiol 66:32–38. doi: 10.1016/j.mib.2021.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Serra CR, Earl AM, Barbosa TM, Kolter R, Henriques AO. 2014. Sporulation during growth in a gut isolate of Bacillus subtilis. J Bacteriol 196:4184–4196. doi: 10.1128/JB.01993-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. DiCandia MA, Edwards AN, Jones JB, Swaim GL, Mills BD, McBride SM. 2022. Identification of functional Spo0A residues critical for sporulation in Clostridioides difficile. J Mol Biol 434:167641. doi: 10.1016/j.jmb.2022.167641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Banse AV, Hobbs EC, Losick R. 2011. Phosphorylation of Spo0A by the histidine kinase KinD requires the lipoprotein MED in Bacillus subtilis. J Bacteriol 193:3949–3955. doi: 10.1128/JB.05199-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Du G, Zhu C, Wu Y, Kang W, Xu M, Yang ST, Xue C. 2022. Effects of orphan histidine kinases on clostridial sporulation progression and metabolism. Biotechnol Bioeng 119:226–235. doi: 10.1002/bit.27968 [DOI] [PubMed] [Google Scholar]
- 55. Du G, Zhu C, Xu M, Wang L, Yang S-T, Xue C. 2021. Energy-efficient butanol production by Clostridium acetobutylicum with histidine kinase knockouts to improve strain tolerance and process robustness. Green Chem 23:2155–2168. doi: 10.1039/D0GC03993D [DOI] [Google Scholar]
- 56. Humphreys JR, Debebe BJ, Diggle SP, Winzer K. 2022. Clostridium beijerinckii strain degeneration is driven by the loss of Spo0A activity. Front Microbiol 13:1075609. doi: 10.3389/fmicb.2022.1075609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xin X, Cheng C, Du G, Chen L, Xue C. 2020. Metabolic engineering of histidine kinases in Clostridium beijerinckii for enhanced butanol production. Front Bioeng Biotechnol 8:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Childress KO, Edwards AN, Nawrocki KL, Anderson SE, Woods EC, McBride SM. 2016. The phosphotransfer protein CD1492 represses sporulation initiation in Clostridium difficile. Infect Immun 84:3434–3444. doi: 10.1128/IAI.00735-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Edwards AN, Wetzel D, DiCandia MA, McBride SM. 2022. Three orphan histidine kinases inhibit Clostridioides difficile sporulation. J Bacteriol 204:e0010622. doi: 10.1128/jb.00106-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mehdizadeh Gohari I, Li J, Navarro MA, Mendonça FS, Uzal FA, McClane BA. 2023. Identification of orphan histidine kinases that impact sporulation and enterotoxin production by Clostridium perfringens type F strain SM101 in a pathophysiologically-relevant ex vivo mouse intestinal contents model. PLoS Pathog 19:e1011429. doi: 10.1371/journal.ppat.1011429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci U S A 99:996–1001. doi: 10.1073/pnas.022493799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Myers GSA, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, et al. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res 16:1031–1040. doi: 10.1101/gr.5238106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fujita M, Sadaie Y. 1998. Feedback loops involving Spo0A and AbrB in in vitro transcription of the genes involved in the initiation of sporulation in Bacillus subtilis. J Biochem 124:98–104. doi: 10.1093/oxfordjournals.jbchem.a022103 [DOI] [PubMed] [Google Scholar]
- 64. Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. doi: 10.1038/nature09397 [DOI] [PubMed] [Google Scholar]
- 65. Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile Spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. doi: 10.1128/IAI.00147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lawley TD, Clare S, Deakin LJ, Goulding D, Yen JL, Raisen C, Brandt C, Lovell J, Cooke F, Clark TG, Dougan G. 2010. Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl Environ Microbiol 76:6895–6900. doi: 10.1128/AEM.00718-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Edwards AN, Karim ST, Pascual RA, Jowhar LM, Anderson SE, McBride SM. 2016. Chemical and stress resistances of Clostridium difficile spores and vegetative cells. Front Microbiol 7:1698. doi: 10.3389/fmicb.2016.01698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK. 2012. C. difficile 630Δerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS One 7:e48608. doi: 10.1371/journal.pone.0048608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Edwards AN, Tamayo R, McBride SM. 2016. A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol Microbiol 100:954–971. doi: 10.1111/mmi.13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Edwards AN, Anjuwon-Foster BR, McBride SM. 2019. RstA is a major regulator of Clostridioides difficile toxin production and motility. mBio 10:e01991–01918. doi: 10.1128/mBio.01991-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. DiCandia MA, Edwards AN, Lee CD, Monteiro MP, Cuebas GNV, Bagchi P, McBride SM. 2023. A conserved switch controls virulence, Sporulation, and motility in C. difficile. bioRxiv:2023.03.28.534590. doi: 10.1101/2023.03.28.534590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Suárez JM, Edwards AN, McBride SM. 2013. The Clostridium difficile cpr locus is regulated by a noncontiguous two-component system in response to type A and B lantibiotics. J Bacteriol 195:2621–2631. doi: 10.1128/JB.00166-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Edwards AN, McBride SM. 2023. The RgaS-RgaR two-component system promotes Clostridioides difficile sporulation through a small RNA and the Agr1 system. PLoS Genet 19:e1010841. doi: 10.1371/journal.pgen.1010841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. O’Connor JR, Lyras D, Farrow KA, Adams V, Powell DR, Hinds J, Cheung JK, Rood JI. 2006. Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol 61:1335–1351. doi: 10.1111/j.1365-2958.2006.05315.x [DOI] [PubMed] [Google Scholar]
- 75. Ahmed UKB, Shadid TM, Larabee JL, Ballard JD. 2020. Combined and distinct roles of AGR proteins in Clostridioides difficile 630 sporulation, motility, and toxin production. mBio 11:e03190–03120. doi: 10.1128/mBio.03190-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, Svensson RT, Brown JL, Johnson EA, Smith LA, Okinaka RT, Jackson PJ, Marks JD. 2007. Genetic diversity among botulinum neurotoxin-producing clostridial strains. J Bacteriol 189:818–832. doi: 10.1128/JB.01180-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hutson RA, Thompson DE, Collins MD. 1993. Genetic interrelationships of saccharolytic Clostridium botulinum types B, E and F and related clostridia as revealed by small-subunit rRNA gene sequences. FEMS Microbiol Lett 108:103–110. doi: 10.1111/j.1574-6968.1993.tb06081.x [DOI] [PubMed] [Google Scholar]
- 78. Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MTG, Mitchell WJ, Carter AT, Bentley SD, Mason DR, Crossman L, et al. 2007. Genome sequence of a proteolytic (group I) Clostridium botulinum strain hall A and comparative analysis of the clostridial genomes. Genome Res 17:1082–1092. doi: 10.1101/gr.6282807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Smith TJ, Hill KK, Raphael BH. 2015. Historical and current perspectives on Clostridium botulinum diversity. Res Microbiol 166:290–302. doi: 10.1016/j.resmic.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wörner K, Szurmant H, Chiang C, Hoch JA. 2006. Phosphorylation and functional analysis of the sporulation initiation factor Spo0A from Clostridium botulinum. Mol Microbiol 59:1000–1012. doi: 10.1111/j.1365-2958.2005.04988.x [DOI] [PubMed] [Google Scholar]
- 81. Nowakowska MB, Selby K, Przykopanski A, Krüger M, Krez N, Dorner BG, Dorner MB, Jin R, Minton NP, Rummel A, Lindström M. 2022. Construction and validation of safe Clostridium botulinum group II surrogate strain producing inactive botulinum neurotoxin type E toxoid. Sci Rep 12:1790. doi: 10.1038/s41598-022-05008-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jones DT, van der Westhuizen A, Long S, Allcock ER, Reid SJ, Woods DR. 1982. Solvent production and morphological changes in Clostridium acetobutylicum. Appl Environ Microbiol 43:1434–1439. doi: 10.1128/aem.43.6.1434-1439.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Long S, Jones DT, Woods DR. 1984. The relationship between sporulation and solvent production in Clostridium acetobutylicum P262. Biotechnol Lett 6:529–534. doi: 10.1007/BF00139997 [DOI] [Google Scholar]
- 84. Long S, Jones D, Woods D. 1984. Initiation of solvent production, clostridial stage and endospore formation in Clostridium acetobutylicum P262. Appl Microbiol Biotechnol 20:256–261. doi: 10.1007/BF00250635 [DOI] [Google Scholar]
- 85. Ravagnani A, Jennert KCB, Steiner E, Grünberg R, Jefferies JR, Wilkinson SR, Young DI, Tidswell EC, Brown DP, Youngman P, Morris JG, Young M. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol Microbiol 37:1172–1185. doi: 10.1046/j.1365-2958.2000.02071.x [DOI] [PubMed] [Google Scholar]
- 86. Harris LM, Welker NE, Papoutsakis ET. 2002. Northern, morphological, and fermentation analysis of Spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J Bacteriol 184:3586–3597. doi: 10.1128/JB.184.13.3586-3597.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Atmadjaja AN, Holby V, Harding AJ, Krabben P, Smith HK, Jenkinson ER. 2019. CRISPR-Cas, a highly effective tool for genome editing in Clostridium saccharoperbutylacetonicum N1-4(HMT). FEMS Microbiol Lett 366:fnz059. doi: 10.1093/femsle/fnz059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Johnson JL, Toth J, Santiwatanakul S, Chen JS. 1997. Cultures of "Clostridium acetobutylicum" from various collections comprise Clostridium acetobutylicum, Clostridium beijerinckii, and two other distinct types based on DNA-DNA reassociation. Int J Syst Bacteriol 47:420–424. doi: 10.1099/00207713-47-2-420 [DOI] [PubMed] [Google Scholar]
- 89. Patakova P, Branska B, Vasylkivska M, Jureckova K, Musilova J, Provaznik I, Sedlar K. 2022. Transcriptomic studies of solventogenic clostridia, Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol Adv 58:107889. doi: 10.1016/j.biotechadv.2021.107889 [DOI] [PubMed] [Google Scholar]
- 90. Patakova P, Branska B, Sedlar K, Vasylkivska M, Jureckova K, Kolek J, Koscova P, Provaznik I. 2019. Acidogenesis, solventogenesis, metabolic stress response and life cycle changes in Clostridium beijerinckii NRRL B-598 at the transcriptomic level. Sci Rep 9:1371. doi: 10.1038/s41598-018-37679-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577, doi: 10.1128/MMBR.66.3.506-577.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]