Structure of the malaria blood-stage parasite’s main surface antigen reveals putative host cell–binding domains.
Abstract
The merozoite surface protein 1 (MSP-1) is the most abundant protein on the surface of the erythrocyte-invading Plasmodium merozoite, the causative agent of malaria. MSP-1 is essential for merozoite formation, entry into and escape from erythrocytes, and is a promising vaccine candidate. Here, we present monomeric and dimeric structures of full-length MSP-1. MSP-1 adopts an unusual fold with a large central cavity. Its fold includes several coiled-coils and shows structural homology to proteins associated with membrane and cytoskeleton interactions. MSP-1 formed dimers through these domains in a concentration-dependent manner. Dimerization is affected by the presence of the erythrocyte cytoskeleton protein spectrin, which may compete for the dimerization interface. Our work provides structural insights into the possible mode of interaction of MSP-1 with erythrocytes and establishes a framework for future investigations into the role of MSP-1 in Plasmodium infection and immunity.
INTRODUCTION
With 229 million reported cases and 409,000 deaths in 2019 worldwide and more than half of the world’s population at risk, malaria remains a great burden to global health (1). Earlier reductions in incident rates have leveled off between 2015 and 2019 (1), making the development of a vaccine as vital as ever. Merozoites are the erythrocyte-infecting form of the malaria-causing Plasmodium parasite, and merozoite surface proteins (MSPs) show potential as malaria vaccines [reviewed by Beeson et al. (2)]. After erythrocyte invasion, merozoites replicate within the parasitophorous vacuole (PV) for ~48 hours in the case of Plasmodium falciparum, until they egress by sequential rupture of the PV membrane (PVM) and host cell membrane and go on to invade new host cells within minutes (3). In this short time window, the merozoite surface is exposed to the immune system. Several integral and peripheral MSPs (4, 5) form a thick fibrillar coat on the merozoite surface, MSP-1 being the most abundant (5). Antibodies targeting MSP-1 are found in sera of individuals from malaria-endemic regions and have been shown to confer some immunity (6–8). Thus, MSP-1 is considered a prime candidate antigen for vaccine development (9). It consists of four subunits, p83, p30, p38, and p42, which are held together noncovalently (Fig. 1A). A glycosylphosphatidylinositol (GPI) anchor links the complex to the membrane via the p42 subunit. MSP-1 is produced by subtilisin-like protease 1 (SUB-1) cleavage of a ~190-kDa precursor protein (p190) just before erythrocyte egress. In a second processing step by SUB-2 during erythrocyte invasion, p42 is cleaved into p33 and p19; the latter remains attached to the parasite surface via its GPI anchor, while the rest of the complex is shed (10). The MSP-1 complex is known to interact with a range of other merozoite proteins, including MSP-3, MSP-6, MSP-7, and MSP-9, and MSP Duffy binding-like (MSPDBL)–1 and MSPDL-2 (11–13), forming various different complexes on the parasite surface (14–16). MSP-7 associates with MSP-1 immediately after translation (17), while other MSPs bind to the complex after maturation (11, 18, 19). Although the functional role of MSP-1 is not yet well understood, it is essential to Plasmodium development given that MSP-1 knockdown mutants could not be generated (20, 21). It is thought to play a role in early erythrocyte attachment and invasion (22); MSP-1 has been shown to bind the erythrocyte surface proteins glycophorin A (GPA) (23) and band 3 (15), the latter being essential in the merozoite invasion process (23). Other studies failed to detect direct binding of red blood cells (RBCs) by MSP-1 itself but reported interactions with other components of the MSP-1 complex, specifically MSPDBL-1, MSPDBL-2, MSP-6, and MSP-9, although the RBC host receptors of these components remain unknown, and conflicting data were reported for MSP-6 (12–15). MSP-1 also interacts with the erythrocyte cytoskeleton protein spectrin (24, 25) and plays a role in egress (25). Antibodies targeting all MSP-1 subunits (14, 26), as well as some associated MSPs (11, 13), have been shown to inhibit parasite growth in RBC cultures. Antibodies targeting the p19 subunit have received particular scrutiny (10, 20), although contradictory reports exist regarding their ability to prevent disease (6), and immunization trials using only p42 were unsuccessful in inducing significant protection against malaria (27). Given the additive inhibitory effect that was observed for antibodies targeting the different MSP-1 subunits in in vitro parasite growth assays (26), full-length MSP-1 may be a better vaccine antigen. A recent phase 1a clinical trial reported promising immunogenicity of a full-length recombinant MSP-1 “heterodimer” composed of two peptide chains corresponding to the N- and C-terminal “halves” of MSP-1, p83/30 and p38/42, respectively (hdMSP-1; Fig. 1A) (9), which was previously shown to assemble in the same manner as the native MSP-1 complex (28).
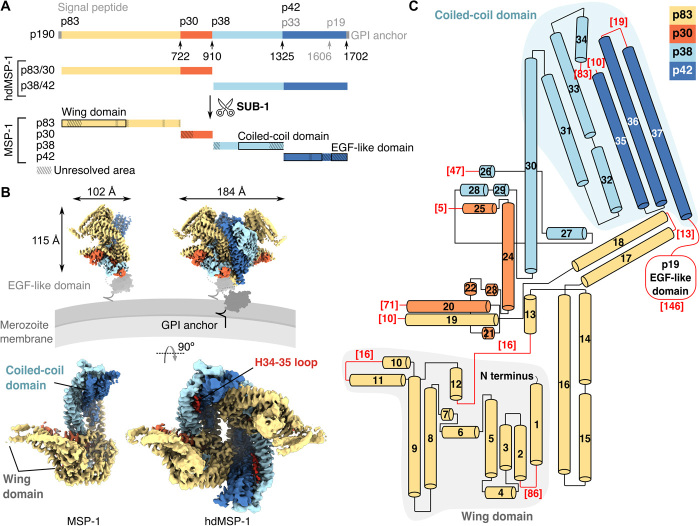
Fig. 1. Global architecture of MSP-1.
(A) Schematic of MSP-1 precursor protein (p190) and production of MSP-1 by SUB-1 cleavage. The canonical cleavage sites are indicated with arrows. The MSP-1 heterodimer (hdMSP-1) used here comprises the N- and C-terminal halves of MSP-1. The various domains are indicated by black boxes. Hatching indicates regions that were not resolved in the EM maps. (B) EM maps of the main conformations of MSP-1 (monomer, left) and hdMSP-1 (dimer, right). Top: Side view of presumed positioning with respect to the merozoite membrane. The EGF-like domain was not resolved here; a previously determined structure (29) is shown in gray surface representation for reference. Red density corresponds to the loop following H34 and was only resolved for hdMSP-1 (see also fig. S8A). (C) Topology of the main conformation of MSP-1. Red numbers correspond to the number of unresolved amino acids at the indicated loops/domain.
Despite its clinical relevance, structural information on MSP-1 is sparse and limited to the C-terminal epidermal growth factor (EGF)–like domain consisting of subunit p19 (29, 30) and global mapping of intersubunit interactions (28). As expected from secondary structure prediction, circular dichroism reports a largely α-helical structure with a significant portion of unstructured regions (25), which may explain the difficulty in obtaining well-diffracting crystals of MSP-1. More detailed knowledge about the structure of MSP-1 may aid in elucidating its essential role in Plasmodium infectivity.
RESULTS
Global architecture of MSP-1
Using single-particle cryo–electron microscopy (cryo-EM), we solved the structures of full-length MSP-1 from P. falciparum, the deadliest and most prevalent of the Plasmodium spp. infecting humans (1). MSP-1 was produced recombinantly as a heterodimer (hdMSP-1; Fig. 1A) in the absence of any of the other natively associated MSPs, as used in the aforementioned successful phase 1a clinical trial (9), using the previously described MSP-1D construct based on the P. falciparum strain 3D7 (28). Structural analysis was performed for both hdMSP-1 and its fully SUB-1–processed form (henceforth designated MSP-1; Fig. 1A and fig. S1). The data showed continuous flexibility (movie S1) and yielded several subtly different conformations (fig. S2 and table S1). Specifically, we solved six monomeric structures from the MSP-1 dataset, at nominal resolutions ranging from 3.1 to 3.6 Å and two dimeric structures from the hdMSP-1 dataset at nominal resolutions of 3.3 and 3.6 Å, which were used to build atomic models de novo (figs. S2 to S4). The highest-resolution monomeric structure was designated as the main conformation and is used for further analysis here.
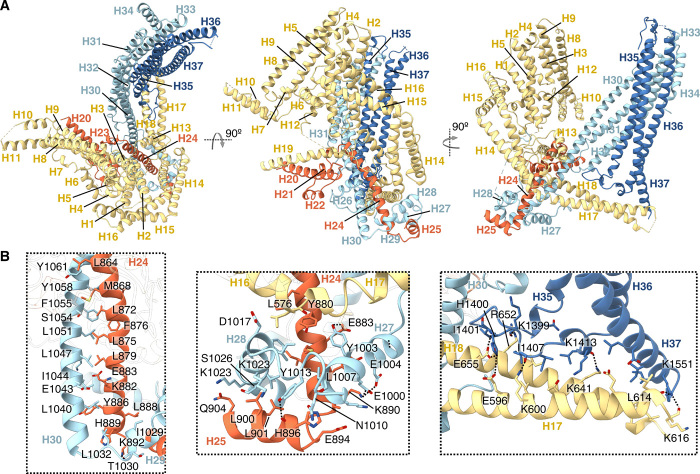
MSP-1 has no sequence orthologs outside of Plasmodium and adopts an unusual three-dimensional (3D) architecture with a central cavity, which is likely to face away from the parasite surface (Fig. 1B). The monomer consists of 37 α helices (Fig. 1C), which can be roughly divided into two interconnected domains found on either side of the cavity (Fig. 1, B and C). On one side, p38 and p42 form a helical bundle (H30 to H37) made up of left-handed coiled coils, which we term the coiled-coil domain (Figs. 1, B and C, and 2A). Flexible structure alignment (31) identified similarity to BAR/IMD domain-like and signal transducer and activator of transcription (STAT)–like t-snare folds, which are associated with membrane and cytoskeleton interactions (fig. S5). A flexible domain, which we term the wing domain, made up of the shorter N-terminal α helices of p83 (H1 to H12) is found on the opposite side of the central cavity (Figs. 1, B and C, and 2A). The EGF-like domain that makes up the C-terminal part of p42 (p19) was not resolved and is probably connected to the rest of the protein via a flexible linker including a region with no predicted secondary structure (Fig. 1, B and C, and figs. S6 and S7). The wing domain and the coiled-coil domain are held together via intersubunit interactions mediated by long helices extending outward from each domain and p30. The largest intersubunit contact surface is found between p30 and p38 (2900 ± 100 Å2, average of all MSP-1 conformations). The 67–amino acid–long H30 of p38 extends from the coiled-coil domain on one end of the central cavity to the opposite end, where it makes extensive contacts with H24 of p30, stabilized further by interactions with helices H26 to H29 that wrap around the H24/30 helix pair (Fig. 2). In addition, a long left-handed coiled coil made up of H14 to H18 of p83 kinks around the H24/30 pair and extends from the edge of the wing domain to the bottom end of the coiled-coil domain on the opposite side of cavity where H17 and H18 contact p42 at the H35-36 loop and H37 (Fig. 2; total buried surface 970 ± 70 Å2).
Fig. 2. Atomic model of MSP-1.
(A) Atomic model built for the main conformation of MSP-1. Different subunits are colored in the same manner as in Fig. 1. (B) Examples of intersubunit interactions holding the complex together. Hydrogen bonds and salt bridges are indicated by dotted lines.
Conformational flexibility
The highest degree of conformational flexibility was observed in the wing domain, and the solved conformations differed mainly in the position of this domain relative to the coiled-coil domain (Fig. 3, fig. S2, and movie S1). Consequently, the local resolution of the maps was lowest in this domain (fig. S3, E and F), and local refinement of the wing domain was performed to improve the map and aid model building of this region (figs. S2A, S3G, and S4A). In addition, cross-links between peptides from the wing domain observed by cross-linking mass spectrometry (XL-MS) were consistent with the model (fig. S4D). Nevertheless, side-chain density remained weak in certain regions (in particular, for H10 and H11), and the residual uncertainty in the model in this region is reflected in the relatively high local model B-factors (fig. S4C).
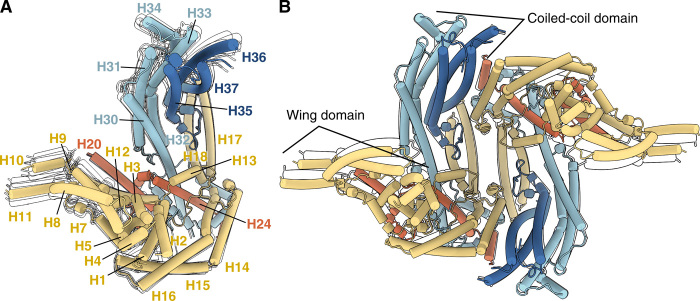
Fig. 3. Flexibility of MSP-1.
(A) Main conformation of MSP-1 and (B) conformation 1 of hdMSP-1 shown in colored cylinders with only silhouettes shown for alternative conformations 1 to 5 of MSP-1 and conformation 2 of hdMSP-1. Monomeric structures are aligned on H24 (residues 859 to 890) chosen for the low local B-factor (see fig. S4C). Superpositions of the corresponding EM maps are shown in fig. S2 (A and B).
Because of the continuous nature of the intrinsic flexibility of MSP-1, classification of the data into a limited number of classes led to slightly different resolved conformations for the MSP-1 and hdMSP-1 wing domains. No further notable differences were observed between the protomers of the dimeric hdMSP-1 and the monomeric MSP-1 forms. The only exception is found in the pocket between H30 and H35 (fig. S8A). Here, we observe density in the hdMSP-1 maps that most likely corresponds to the loop between H34 and H35 containing the p38/p42 SUB-1 processing site, although the density is not clear enough to allow unambiguous model building (Fig. 1B and fig. S8A). No clear density was observed in this pocket for the SUB-1–treated MSP-1 sample where this loop was cleaved, and thus likely to be less restrained than in the hdMSP-1 sample where it is not cleaved (Fig. 1B and fig. S8A). The p38/p42 processing has been shown to be critical for parasite viability (25). Thus, the absence of global (resolved) structural changes between the hdMSP-1 and fully processed MSP-1 samples suggests that the functional importance of processing of the H34-35 loop might be due to local differences. For example, the loop itself might interact with crucial erythrocyte or parasite targets that require this loop to be “released” from the H30/35 pocket; alternatively, the pocket itself might be an important binding site that is blocked by the H34-35 loop before its cleavage and becomes accessible once the loop is released upon processing. The other SUB-1 processing site that remains intact in hdMSP-1 (p83/p30) is located in an unstructured loop between H19 and H20 and was not resolved in any of the structures.
Conserved and variable regions of MSP-1
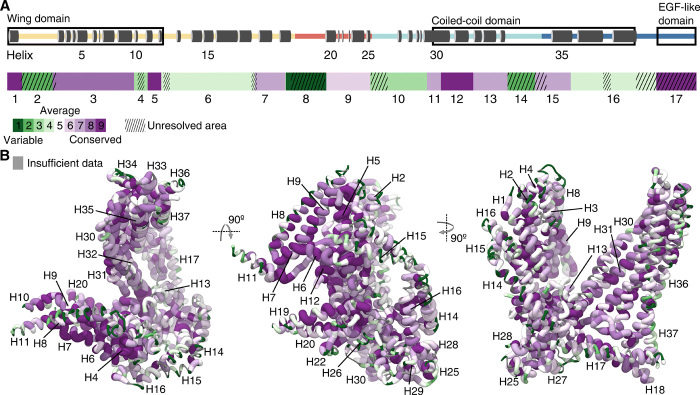
Because the MSP-1 sequence varies among P. falciparum strains, the question arises whether the structure of MSP-1 from other strains would differ from the structures presented here. The MSP-1 sequence can be divided into 17 blocks based on sequence variability as first described by Tanabe and co-workers (32) (Fig. 4A and fig. S6). They noted that the primary structure of P. falciparum MSP-1 has a dimorphic nature, and isoforms from different P. falciparum strains were broadly classified as belonging to one of two variants, termed K1 and MAD20 based on two representative isolates (32). Highest sequence homology was found for blocks 1 (comprising H1), 3 (H2 to H10), 5 (H11 and H12), 12 (H30 and H31), and 17 (the EGF-like domain; Fig. 4A). A similar pattern of MSP-1 sequence conservation is observed for MSP-1 isoforms from different Plasmodium spp. (Fig. 4B and fig. S7). The flexible p83 wing domain consists of a conserved core, composed of helices H5 to H7, H9, and H12 (Fig. 4, A and B). The core of the coiled-coil domain, made up of helices H31 to H33 and H35, also shows relatively high sequence homology across P. falciparum strains and Plasmodium spp. (Fig. 4, A and B). The most polymorphic region (block 2, 65 amino acids in MSP-1D) is located between H1 and H2 in the wing domain, is predicted to be unstructured (fig. S6), and was not resolved in the EM maps (Fig. 4A). Likewise, the second oligomorphic region (block 4) contains a predicted unstructured stretch of 16 amino acids between H10 and H11 at the edge of the wing domain that was also not resolved (Fig. 4A and figs. S6 and S7). This is echoed in blocks 6, 8, 10, 14, and 16, where unresolved (predicted unstructured) loops corresponded to the most variable regions (Fig. 4A and figs. S6 and S7). While the resolved parts of blocks 6 (H13 to H17), 10 (H26 to H30), and 16 (H35 to H37) also contain dimorphic regions, they show comparatively higher homology between different P. falciparum isolates and other Plasmodium spp. relative to the loop regions (Fig. 4A and figs. S6 and S7). Together, these observations suggest that the highest sequence variability is found in the flexible loop regions and that the global architecture of MSP-1 observed here may be conserved between different genotypes.
Fig. 4. Sequence variability of MSP-1.
(A) Graphical representation of the primary structure of MSP-1, indicating the resolved helices (top), and division of the sequence in 17 blocks based on sequence variability of the P. falciparum MSP-1 as determined by Tanabe et al. (32) (bottom). Hatching indicates regions that were not resolved in the EM maps. Green blocks correspond to high sequence variability, and purple blocks correspond to high sequence conservation. (B) MSP-1 sequence conservation is mapped onto the main conformation derived for MSP-1 using all 46 Plasmodium MSP-1 genes from various species available from PlasmoDB (63). Green thin tubes indicate high sequence variability, and thick purple tubes indicate high sequence conservation. Areas for which there are insufficient homologous sequence data are colored gray.
Monomer-dimer equilibrium
While MSP-1 is often depicted as monomeric (13, 25), dimeric forms of MSP-1 have previously been detected in detergent-resistant membranes of P. falciparum 3D7 schizonts by native polyacrylamide gel electrophoresis (PAGE) and chemical cross-linking (33). Here, both monomeric and dimeric forms were observed for MSP-1 and hdMSP-1, with 2D classification of different (control) EM datasets yielding varying fractions of dimer and monomer classes (fig. S8, B and C). The hdMSP-1 datasets used for high-resolution 3D reconstruction yielded a larger fraction of dimer classes (53 to 100% dimer; fig. S8C) compared to the dataset used for the SUB-1–processed MSP-1, which appeared to predominantly yield monomer classes (only 4% dimer; fig. S8B), supported by the apparent molecular weight of the samples used, gauged by size-exclusion chromatography (SEC; fig. S1A). Note that a smaller control dataset of a second MSP-1 sample yielded predominantly dimeric 2D class averages (89% dimer; fig. S8B) and a corresponding higher molecular weight estimate by SEC (fig. S1A), suggesting that SUB-1–processed MSP-1 is not monomeric per se. Microscale thermophoresis (MST) showed that in solution dimerization is concentration dependent with a dissociation constant (KD) of 60 ± 10 and 30 ± 20 nM for hdMSP-1 and MSP-1, respectively (Fig. 5A), suggesting that there is no significant difference in the dimerization propensity of the two MSP-1 forms, and the difference in monomer-to-dimer ratio observed in the EM data may have been introduced during sample or grid preparation. Given the observed low KD and high local concentration of MSP-1 on the merozoite surface, MSP-1 can be expected to form dimers on the parasite surface, as was observed for MSP-1 from P. falciparum 3D7 (33) on which the construct used here is based.
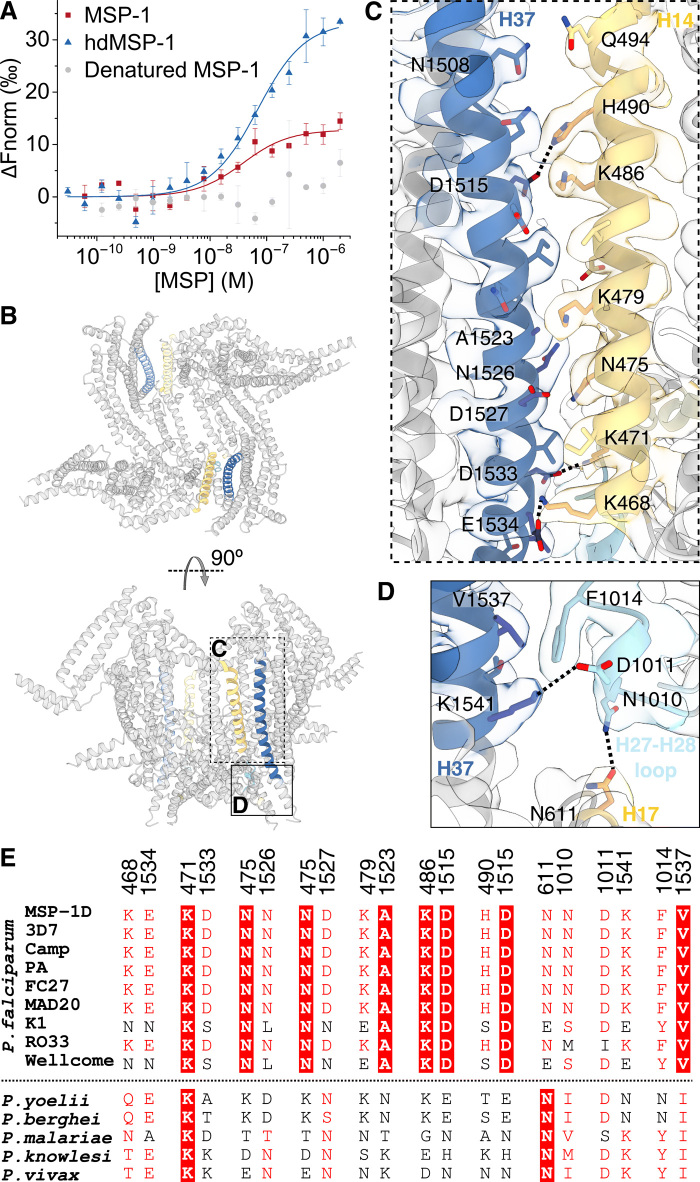
Fig. 5. Dimerization of MSP-1.
(A) Dimerization affinity for (hd)MSP-1 was determined by MST. KD values of 30 ± 20 nM (n = 3) and 60 ± 10 nM (n = 4) were found for MSP-1 and hdMSP-1, respectively. No significant dimerization was observed for heat-denatured MSP-1. Error bars represent SD. (B) Dimerization is mediated mainly through two symmetric interfaces (highlighted). (C and D) More detailed views of the dimerization interfaces. Salt bridges and H bonds are indicated by dashed lines. (E) Sequence alignment of residue pairs contributing to the dimerization interface is given for the MSP-1D sequence used here and other P. falciparum isolates, as well as other Plasmodium species. Full-sequence alignments can be found in figs. S6 and S7.
Whether or not MSP-1 forms dimers on the parasite surface is of interest because this could potentially shield the dimerization interface to some extent from immune detection. Dimerization is mediated mainly through interactions between p83 and p42, contributing 1100 ± 90 Å2 to the total dimerization interface, with contacts between p38 and p42 adding 250 ± 20 Å2 (average of both dimer conformations; Fig. 5, B to E). Most interprotomer contacts are found between helices H14 (p83) and H37 (p42), which show patches of complementary surface charge (Fig. 6). Salt bridges (H490-D1515, K468-E1534, and K471-D1533) and various polar (e.g., Q494-N1508, N475-D1527, and N475-N1526) and hydrophobic interactions (e.g., K479-A1523) hold the helices together (Fig. 5C). In addition, contacts are observed between the H27-28 loop of p38 and H37 (F1014-V1537 and salt-bridge D1011-K1541) and H17 (p83; N1010-N611; Fig. 5D). While all structural elements involved in dimerization fall within the dimorphic sequence blocks described by Tanabe et al. (32) (blocks 6, 10, and 16; Fig. 4A and fig. S6), a number of residues involved in the dimerization interface are conserved between different isolates (Fig. 5E). For most other residue pairs involved, the other genotypes could still allow favorable interprotomer contacts (Fig. 5E). Thus, dimerization may occur through the same interface in other P. falciparum genotypes. Other Plasmodium spp. show higher sequence variability (Figs. 4B and 5E and fig. S7), and it is unclear whether dimerization would be conserved across the genus.
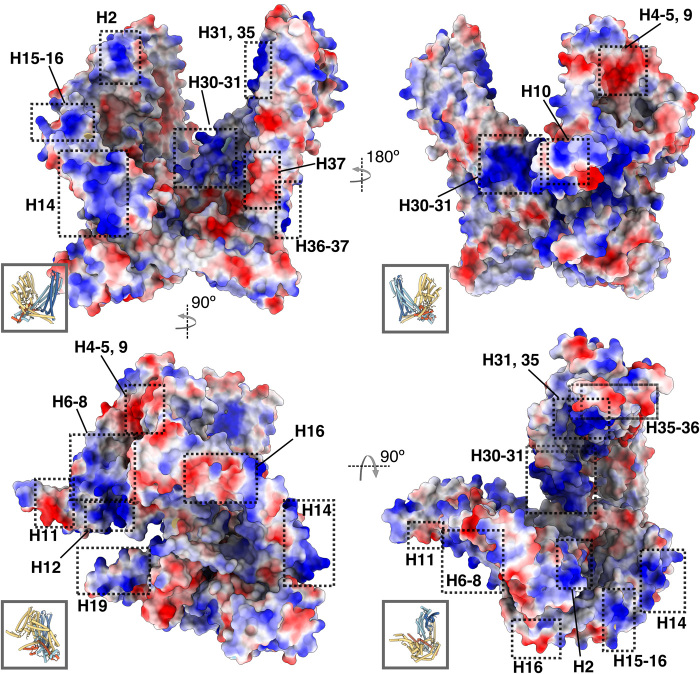
Fig. 6. Electrostatic surface potential.
MSP-1 model colored by electrostatic surface potential, where blue and red correspond to positive and negatively charged regions, respectively. Insets show same view of the model in cylinder representation for reference.
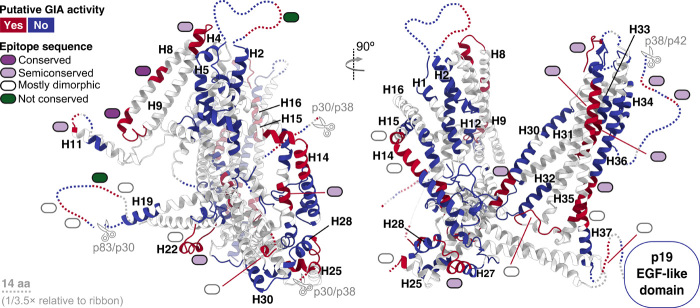
Antibodies directed against a region encompassing H11 to H16, which includes the dimerization interface H14, are associated with protection against P. falciparum malaria in adolescents from Mali (34). Furthermore, in the recent vaccine trial by Blank et al. (9) using hdMSP-1, epitopes were identified on H14, the H27-28 loop, and H37, which elicited immunoglobulin G (IgG) antibody responses putatively associated with inhibiting parasite growth in vitro (Fig. 7 and fig. S9). It is possible that these antibodies would destabilize the dimer, although whether this underlies their protective action remains an open question.
Fig. 7. Immunogenic epitopes.
Immunogenic B cell epitopes identified in an MSP-1 vaccine trial (9) are mapped onto MSP-1. On the basis of correlation between IgG responses and parasite growth inhibition assay (GIA) activity of the various investigated sera, Blank et al. (9) identified epitopes putatively associated with GIA activity. Blue, no role; red, possible role in inhibiting parasite growth. Colored ovals indicate the degree of sequence conservation for each epitope: Dark purple, light purple, white, and green correspond to conserved, semiconserved (>50% conserved residues, rest dimorphic), mostly dimorphic (<50% conserved residues, rest dimorphic), and nonconserved epitopes. Dotted lines represent unresolved loops (scaled 1/3.5 relative to ribbon). More details can be found in fig. S9. aa, amino acids.
MSP-1 domains involved in erythrocyte interactions and immunogenicity
As discussed in the introduction, MSP-1 has been shown to interact with erythrocyte components (15, 23–25), as well as a range of other merozoite proteins (11–13, 15, 17), forming various different complexes on the parasite surface (14–16). The molecular details of these interactions remain unclear. Given that the core of the flexible wing domain does not participate in the stabilization of the MSP-1 complex itself, its relatively high degree of conservation suggests that this domain may play a role in these interactions, as previously proposed by Kauth et al. (28). In support of this hypothesis, antibodies against the p83 subunit that makes up this domain were particularly effective in inhibiting infection of erythrocytes by malaria parasites in in vitro assays (26). The p83 subunit was furthermore identified as a major target of opsonizing antibodies in sera from semi-immune individuals from Burkina Faso (35). While p83 is indeed also known to bind other MSPs [such as MSP-7, MSPDBL-1, and MSPDBL-2 (11, 13, 17)], an antibody targeting p83 inhibited parasite growth without affecting complex formation with these MSPs (14), suggesting that p83 is important for infection not only as a binding site for other MSPs but also, in particular, through its direct interaction with erythrocytes. Notably, residues 232 to 278 (in MSP-1D numbering) of the p83 subunit, encompassing H6 to H8 (Fig. 8A), were shown to interact with a Glu-rich extracellular region of GPA on the erythrocyte surface (23). The positively charged patch found at H6 to H8 suggests that the interaction might be electrostatic (Fig. 6). Deletion of GPA reduced but did not completely abrogate parasitemia (23). Blank and co-workers (9) identified two conserved epitopes on H8, which elicited an IgG antibody response putatively associated with inhibiting parasite growth in vitro (Fig. 7).
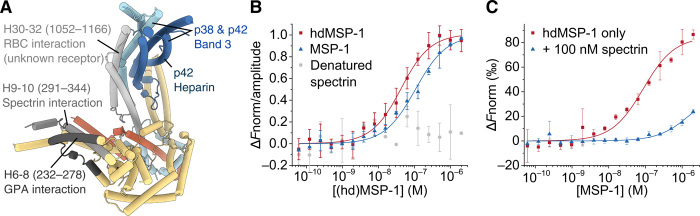
Fig. 8. Interaction surfaces.
(A) Various regions of MSP-1 that have been shown to interact with erythrocyte targets (15, 23–25) are indicated. Note that the MSP-1 regions were identified in studies using MSP-1 fragments, and other MSP-1 areas might also interact with the named targets. See main text for details. (B) MSP-1 was shown to bind spectrin using MST. KD values of 31 ± 5 nM (n = 4, red squares) and 100 ± 10 nM (n = 2, blue triangles) were found for the hdMSP-1–spectrin and MSP-1–spectrin complexes, respectively. No significant binding of MSP-1 to heat-denatured spectrin was observed (gray circles). (C) MST analysis of the dimerization affinity for hdMSP-1 in the absence (red squares) and presence (blue triangles) of 100 nM unlabeled spectrin yielded KD values of 80 ± 20 nM (n = 2) and 1.7 ± 0.9 μM (n = 2), respectively. Error bars represent SD.
Other MSP-1 subunits have also been implicated in interactions with RBCs. Residues 1052 to 1166 (in MSP-1D numbering) of the p38 subunit, encompassing H31 and part of H30 and H32 (Fig. 8A), were shown to specifically bind erythrocytes via an unknown mechanism independent of GPA and sialic acid, a negatively charged sugar moiety abundant on the erythrocyte surface (36). While the RBC target remained unclear (36), strongly basic patches present on H30 to H32 near the bottom of the MSP-1 cavity (Fig. 6) suggest that long-range electrostatic interactions with other erythrocyte components could play a role. When recombinantly produced, this part of the p38 domain was recognized by sera of semi-immune individuals (36). Notably, the recombinant p38 fragment and polyclonal antibodies against it could inhibit parasite invasion of RBCs in vitro by nearly 70 and 90%, respectively (36), suggesting that it plays an important role in Plasmodium infectivity. Furthermore, semiconserved B cell epitopes putatively associated with parasite growth inhibition in vitro were also identified on H30 and H32 in the study by Blank et al. (9) (Fig. 7). The p38 subunit, as well as the p42 subunit, was further shown to interact with specific peptides of the erythrocyte anion transporter band 3 (37). These band 3 peptides appeared to have higher affinity for p42 and, in particular, for the p19 domain than for p38 (37). Considering that band 3 is the most abundant protein on the erythrocyte surface and essential in the merozoite invasion process, these interactions may be highly important for parasite attachment, although a molecular mechanism or precise MSP-1 binding site remains unclear (37).
Besides its interaction with band 3 (37), the p42 subunit has been reported to bind highly negatively charged heparin-like polysaccharides, which are abundant on the erythrocyte surface (22). The resolved part of the p42 subunit (p33) contains two strongly basic patches on H35 and at the bottom of H36 to H37 (Fig. 6), which may play a role in heparin binding; the latter patch coincides with a semiconserved B cell epitope putatively associated with parasite growth inhibition (Fig. 7), supporting its functional importance (9).
In addition to these erythrocyte surface targets important for parasite invasion, MSP-1 has also been shown to play a role in egress through interaction with the erythrocyte cytoskeleton protein spectrin (24, 25). A previous study using a recombinant fragment of p83 (encompassing H2 to H10) identified residues 291 to 344 (in MSP-1D numbering), corresponding to H9 and H10 (Fig. 8A) as the binding site for spectrin (24). However, interactions with other subunits were not investigated (24), and the highly negatively charged spectrin could be expected to also interact with positively charged patches present elsewhere on the MSP-1 complex. As stated above, the resolved parts of p42 and p38 appear to adopt a BAR/IMD-like or STAT-like t-snare fold (fig. S5), which is associated with membrane and cytoskeleton interactions, and the most extensive positively charged surface region of MSP-1 is found on this domain (Fig. 6). The recombinant (hd)MSP-1 used here also bound spectrin (Fig. 8B), and in the presence of spectrin, hdMSP-1 appeared to dimerize less readily than in its absence (1.7 ± 0.9 μM versus 80 ± 20 nM, respectively; Fig. 8C), suggesting that spectrin may compete for the dimerization interface. This interface includes not only H37 of the p42 BAR/IMD-like coiled-coil domain but also H14 (p83), which is likewise part of a coiled-coil bundle (fig. S5) and contains a positively charged region (Fig. 6). MSP-1 has been linked to PVM rupture in the parasite egress pathway (24, 25, 38). Immuno-EM previously identified MSP-1 in high abundance on the PVM in regions where the parasite and RBC membrane were apparently in contact (24). Furthermore, inhibition of SUB-1 has been shown to prevent the MSP-1–spectrin interaction (25) and PVM rupture (38). Thus, the MSP-1–spectrin interaction may play a role in PVM rupture, and given their fold, it is possible that the coiled-coil regions of MSP-1 are involved in this mechanism. Note that Das and co-workers (25) previously reported that cleavage of the p38/p42 processing site was necessary for binding of MSP-1 to inside-out erythrocyte ghost vesicles, which was attributed to interactions with spectrin, while we observe binding of spectrin to hdMSP-1 in which this processing site is not cleaved. The reason for this discrepancy could lie in experimental differences but remains unclear.
While we can only speculate about the molecular mechanism of these interactions, the flexibility of MSP-1 and, in particular, its wing domain may be central to its role as a major interaction surface for these various erythrocyte proteins as well as other MSPs. That the previously reported B cell epitopes putatively associated with in vitro parasite growth inhibition can be mapped to various regions of the 3D structure (Fig. 7) suggests a multifaceted mode of interaction. As previously discussed, this ability of MSP-1 to bind multiple erythrocyte targets via seemingly different and redundant mechanisms might enhance the efficiency of erythrocyte attachment and allow evasion of host cell immune responses (14, 37).
DISCUSSION
In summary, this work defines the architecture of MSP-1, the most abundant antigen on the erythrocyte-invading malaria parasite surface. The protein appears to be flexible and adopts an unusual fold with a large central cavity, which may be fundamental to its proposed role as a major hub for interaction with other MSPs and erythrocyte surface proteins. Previously identified RBC interaction sites could be mapped onto the flexible domain, as well as a strongly basic region in the central cavity. This and other resolved basic patches could also play a role in initial weak adhesion to erythrocytes via long-range electrostatic interactions, e.g., with highly negatively charged heparin-like polysaccharides, which are abundant on the erythrocyte surface. Notably, the coiled-coil fold of the p38 and p42 subunits hints at a role in membrane remodeling or cytoskeleton interactions, which has indeed been proposed previously for MSP-1 (24, 25). Dimerization of MSP-1, which involves coiled-coil regions, is hampered by the presence of spectrin, supporting this hypothesis. The structures presented here only form a small piece of the puzzle, given that MSP-1 forms a complex with other MSPs on the parasite surface. Nevertheless, the high-resolution detail afforded by our models provides an important framework to guide future work to fully elucidate the role of the native MSP-1 complex in Plasmodium infectivity and antibody-mediated protection from malaria.
MATERIALS AND METHODS
MSP-1 preparation
MSP-1 was produced recombinantly in Escherichia coli and purified under GMP-compatible conditions by Biomeva GmbH (Heidelberg, Germany) using the P. falciparum MSP-1D construct described by Kauth et al. (28). Briefly, the coding sequence of MSP-1D is largely based on the P. falciparum strain 3D7, but the N-terminal signal peptide and the C-terminal GPI anchor have been removed. The coding sequence was split into two halves (p83/30 and p38/42); each was expressed individually in E. coli W3110-Z1 and recovered as inclusion bodies. The recombinant MSP-1D was reconstituted as a heterodimer (hdMSP-1) in a process called “pulse renaturation” (39). The protein was purified using ion exchange and hydroxyapatite chromatography followed by dialysis into phosphate-buffered saline (PBS) and lyophilization. MSP-1 produced in this way was previously shown to assemble in the same manner as the native MSP-1 complex (28) (density observed for a disulfide bond between C813 and C845 in our EM maps and the proximity of C211 and C216 in the maps are also indicative of native folding). Before use, lyophilized hdMSP-1 was reconstituted by adding ddH2O, filtered through a 0.22-μm pore centrifugal filter, and subjected to SEC at 4°C on a Superdex 200 Increase 10/300 column (GE Healthcare) equilibrated with gel filtration buffer [25 mM Hepes and 125 mM NaCl (pH 7.4)]. SEC peak fractions were pooled and concentrated to ~0.7 to 2.5 mg/ml using an Amicon Ultra centrifugal concentrator [molecular weight cutoff (MWCO) 100,000; Merck]. For the production of fully SUB-1–cleaved MSP-1, hdMSP-1 was processed in vitro following the procedure published by Koussis et al. (40). Lyophilized hdMSP-1 was reconstituted by adding ddH2O and dialyzed into SUB-1 buffer [50 mM tris and 15 mM CaCl (pH 7.6)]. After dialysis, precipitated protein was removed by centrifugation. Recombinant P. falciparum SUB-1 (SUB-1), provided by M. J. Blackman, was added to hdMSP-1 in SUB-1 buffer, and the sample was incubated at 16°C overnight. Nonspecific cleavage and protein degradation were prevented by adding the protease inhibitors E64, leupeptin, and Pefabloc. Successful processing was verified by SDS-PAGE. The processed sample was filtered through a 0.22-μm pore centrifugal filter and again subjected to SEC at 4°C on a Superdex 200 Increase 10/300 column equilibrated with gel filtration buffer. SEC peak fractions were pooled and concentrated to ~1.6 to 2.7 mg/ml using an Amicon Ultra centrifugal concentrator (MWCO 3000).
EM sample preparation and cryo-EM data collection
Concentrated hdMSP-1 and fully processed MSP-1 SEC peak fractions were diluted to ~0.02 mg/ml and examined by uranyl formate negative staining using a Tecnai Spirit BioTWIN electron microscope at 120 kV to confirm sample homogeneity. For cryo-EM, Quantifoil R2/2 and R1.2/1.3 holey carbon grids (copper and gold 300 mesh, respectively) were washed in chloroform overnight before use. Grids were prepared in the presence and absence of 0.125% (w/v) CHAPS (3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate) [0.25-fold CMC (critical micelle concentration)], which was added right before freezing. Grids were plasma-cleaned under an Ar/O2 atmosphere at a ratio of 95:5, at 35 W for 35 s (NanoClean, model 1070, Fischione Instruments), after which 3 μl of sample was applied to the grids. Grids were blotted for 4 to 5 s with a blot force of 20 at 100% humidity and 10°C and then plunge-frozen in liquid ethane using a using a Mark IV Vitrobot. The grids were screened, and data were collected at a pixel size of 1.077 Å (for hdMSP-1) or 1.073 Å (for MSP-1) on a Titan Krios electron microscope (Thermo Fisher Scientific) operated at 300 kV equipped with a Quantum energy filter and a K2 camera (Gatan) at the Max Planck Institute of Biophysics. For hdMSP-1, four datasets were collected with the same imaging conditions and combined (fig. S2B and table S1): two in the absence of CHAPS and two in the presence of CHAPS, one of which was collected at a fixed tilt angle of 15° (all from R2/2 holey carbon copper mesh grids). For fully processed MSP-1, one dataset was collected from an R1.2/1.3 holey carbon gold mesh grid prepared in the presence of CHAPS. Further data collection details are given in table S1.
Image processing
During data collection, datasets were preprocessed “on-the-fly” using Warp (41) for frame alignment, contrast transfer function (CTF) estimation, and particle picking; cryoSPARC (42) was used for 2D classification to monitor data quality including particle orientation and integrity. Of the 3978 original motion-corrected movies of the MSP-1 dataset, 3735 micrographs were used in processing, using Warp (41) to curate the dataset; micrographs containing signal at 3.9 Å or better, with estimated defocus values ranging from −0.5 to −3.0 μm, and average motion in the first one-third of the frames of no more than 1.5 Å per frame were retained. Subsequently, data were imported into RELION 3.0 (43). Motion correction and dose weighting were performed with MotionCor2 (44) using a 5 × 5 patch. CTF estimation was performed with CTFFIND 4.1 (45), after which ~1.6 million particles picked by Warp (41) were extracted using a box size of 288 pixels at 1.073 Å per pixel. The particle set was imported into cryoSPARC (42) and cleaned by 2D classification. A minority of the 2D classes (4.4% of particles) corresponded to dimeric MSP-1 and was also excluded. The cleaned particle set was subjected to 3D heterogeneous refinement (classification) using a 3D reference generated ab initio using cryoSPARC (42) from ~15,000 particles picked and preprocessed on-the-fly by Warp (41) during data collection. The resulting 3D classes (mostly defined by different conformations of the wing domain) were subjected to nonuniform (NU) refinement, followed by further cleaning by 2D and 3D classification. Particle star files were generated for the resulting particle sets using UCSF pyem (46), which were reimported into RELION 3.0 (43) and subjected to iterative CTF refinement, 3D refinement, and Bayesian polishing until no further improvements in the EM maps were observed. Particle sets were then imported into cryoSPARC (42), subjected to 2D classification (which identified residual junk particles in only two cases) and final NU 3D refinement, resulting in six maps with nominal resolutions ranging from 3.1 to 3.6 Å (auto-tightened masked applied). The highest-resolution map was designated the “main” conformation. The alternative conformations (Alt conf) were numbered according to the position of the wing domain, from furthest (1) to closest (5) to the coiled-coil domain (the main conformation represents an intermediate position; see fig. S2A). To improve the resolution in the wing domain to aid model building, local refinement was performed in cryoSPARC (42) using the particle set corresponding to the main conformation. To this end, soft masks encompassing either H1-12 (wing domain) or H13-37 (“rest”) were generated in RELION 3.0 (43) using a map generated from the initially built atomic model in UCSF chimera (47) filtered to 12 Å. For local refinement of the wing domain, placing the fulcrum at the tip of the wing domain at the end of helices H10 and H11 gave the best results. For local refinement of the rest of the map, density corresponding to the wing domain was subtracted, and the fulcrum for refinement was placed at the center of mass of the mask. A graphical representation of the processing strategy used for the MSP-1 dataset is provided in fig. S2A.
For hdMSP-1, the four datasets were initially processed individually. Collected movies were curated either manually based on resolution estimates from CTFFIND 4.1 (45) and visual inspection (dataset 1) or using Warp (41). For dataset 1, 1686 of 2272 micrographs containing signal at 5 Å or better were retained. For dataset 2, 3560 of 4725 micrographs were retained, containing signal at 5.2 Å or better, with estimated defocus values ranging from −0.3 to −3 μm, and average motion in the first one-third of the frames of no more than 2 Å per frame. For dataset 3, 5864 of 6610 micrographs were retained, containing signal at 3.9 Å or better, with estimated defocus values ranging from −0.3 to −2.5 μm, and average motion in the first one-third of the frames of no more than 1 Å oer frame. For dataset 4, 1391 of 1454 micrographs were retained, containing signal at 5 Å or better, with estimated defocus values ranging from −0.75 to −3 μm, and average motion in the first one-third of the frames of no more than 1 Å per frame. Subsequently, data were imported into RELION 3.0 (43). Motion correction and dose weighting were performed with MotionCor2 (44) using a 5 × 5 patch. CTF estimation was performed with CTFFIND 4.1 (45). Particle picking was performed using crYOLO (48) for dataset 1 (yielding ~219,000 particles) and Warp (41) for all other datasets (yielding ~696,000, ~1.2 million, and ~ 304,000 particles for datasets 2 to 4, respectively). Particles were extracted using a box size of 288 pixels at 1.077 Å per pixel. Various processing strategies were tested for all datasets, and those resulting in the best maps were eventually followed; a graphical representation of the final processing strategy is provided in fig. S2B. Datasets were either first cleaned by 2D classification using ISAC from the SPHIRE cryo-EM software suite (49) or subjected directly to 3D classification in RELION 3.0 (43) using either unbinned data (box size, 288 pixels) or 4× binned data (box size, 72 pixels). Further rounds of 3D classification in RELION 3.0 (43) were performed if necessary, followed by iterative CTF refinement, 3D refinement, and Bayesian polishing, until no further improvements to the maps were observed. Particles sets were then refined in 3D applying C2 symmetry (after having confirmed the symmetry in a 3D reconstruction without applying symmetry). No differences were observed in the final reconstructions of the four datasets at the obtained resolution beyond differences in the angular orientation distribution of the particles (fig. S2B), and thus, all particle sets were merged. Excluding any of the four datasets resulted in (slightly) worse final maps in subsequent processing steps. To better classify the conformational variability observed in the wing domain, the combined particle sets were subjected to symmetry expansion (50) and signal subtraction yielding a particle set corresponding to all monomers. The monomer mask used for signal subtraction was created in RELION 3.0 (43) using a monomer map generated from a dimer map in USCF Chimera (47) using the volume eraser tool. Further 3D classification without alignment into 10 classes using a monomer mask was performed. The best classes were refined individually in RELION 3.0 (43) yielding monomer maps with two distinct conformations of the wing domain. In a second approach, to get the best possible dimer maps, the best classes were also merged. Signal subtraction was reverted, and duplicates were removed. The resulting particle set was refined in 3D and subjected to 3D variability analysis in cryoSPARC (see movie S1) (51). Two extreme conformations were extracted from the trajectory of the first mode of the variability analysis and used as references for multi-referenced 3D classification in cryoSPARC (42). This resulted in two maps with wing domain conformations similar to those obtained from RELION (requesting further classes supplying more references did not yield any additional good maps). Local resolution estimation of the maps was performed using cryoSPARC (42), except for the symmetry expansion–derived maps for which RELION 3.0 (43) was used. UCSF pyem (46) was used to generate Euler angle distribution plots for 3D reconstructions generated in cryoSPARC (42). All reconstructed maps had the wrong handedness and were flipped for atomic model building.
Model building and analysis
Buccaneer (52) was used to start de novo model building followed by manual building in Coot (53) guided by PSIPRED 4.0 (54) secondary structure prediction, using either RELION 3.0 (43) with automatic B-factor estimation to sharpen the maps or the auto-sharpened refined maps resulting from NU refinement in cryoSPARC (42). DeepEMhancer was also used to postprocess the map corresponding to the main conformation of MSP-1 (55). The locally refined maps [auto-sharpened in cryoSPARC (42) or sharpened using DeepEMhancer (55)] were also used in model building of the main conformation of MSP-1. For hdMSP-1, the C2 symmetric dimer maps showed higher local resolution for most of the protein, while the monomer maps obtained from the symmetry-expanded particle set showed a better-resolved wing domain (figs. S2 to S4) and were used to build that part. PHENIX (56) was used for model refinement, and the model was scored with Molprobity (57). The refined model was then used to locally sharpen the refined EM map using LocScale (58). The LocScale-sharpened EM map was subsequently used in combination with the other maps for further iterative refinement in Coot (53), ISOLDE (59), and PHENIX (56). The best resolved conformation for each sample was built initially, and the resulting atomic model was fit into the maps of the other conformation(s) using ISOLDE (59). Interhelical interactions of the flexible wing domain were restrained until the model had settled into the density of the alternative conformation, after which restraints were released and further iterative refinement in Coot (53), ISOLDE (59), and PHENIX (56) was performed. After confirming that the independently built MSP-1 and hdMSP-1 models did not differ substantially, the wing domain built for the main conformation of MSP-1 was fit into the hdMSP-1 maps and refined in the same manner. Map-model FSC (Fourier shell correlation) curves were generated using PHENIX (56). The model was refined in the same manner against one of the independent half-maps and cross-validated against the both independent half-maps; the overlay of the resulting map-model FSCs indicated that no overrefinement took place. Further model quality statistics were determined using PHENIX (56) and can be found in table S1. Structural figures were prepared using USCF Chimera (47), USCF ChimeraX (60), and Inkscape. An initial topology cartoon was generated by Pro-Origami (61) and modified in Inkscape. The ConSurf webserver (62) and UCSF Chimera (47) were used to map the MSP-1 sequence conservation onto the derived structure using a sequence alignment of all Plasmodium MSP-1 genes available from PlasmoDB (63) generated by Clustal Omega 1.2.4 (64). The PBDsum webserver (65) and UCSF Chimera (47) were used to analyze the dimerization interface. The FATCAT webserver (31) was used to identify known structural motifs in the determined MSP-1 structure.
Cross-linking mass spectrometry
Cross-linking combined with mass spectrometry (MS) analysis was used to validate the MSP-1 model. Lyophilized hdMSP-1 (150 μg) was reconstituted by adding 150 μl of ddH2O, followed by buffer exchange into PBS. A stock solution of disuccinimidyl sulfoxide (DSSO; Thermo Fisher Scientific) was freshly prepared according to the manufacturer’s instructions. After optimizing the cross-linker concentration by analyzing cross-linked hdMSP-1 by SDS-PAGE, hdMSP-1 was incubated with a 20-fold molar excess of DSSO for 60 min at room temperature before stopping the reaction with quenching buffer. Samples were digested in urea lysis buffer [8 M urea, 100 mM NaCl, and 50 mM triethylamonium bicarbonate (TEAB) (pH 8.5)] using a two-stage Lys-C and trypsin digestion protocol. Samples were reduced and alkylated using 10 mM dithiothreitol (1 hour, 27°C) and 30 mM iodacetamide (30 min, room temperature, in the dark). Lys-C was added at a 1:100 enzyme-to-protein molar ratio, and the sample was incubated at 37°C for 4 hours. Afterward, the sample was diluted 1:5 using 50 mM TEAB. Trypsin was added at a 1:50 enzyme-to-protein molar ratio, and samples were incubated overnight at 37°C. Digestion was stopped by acidification [0.6% (v/v) trifluoroacetic acid (TFA)] before desalting using StageTips (66). Dried peptides were resuspended in reconstitution buffer (2.5% 1,1,1,3,3,3-hexafluoro-2-propanol and 0.1% TFA in water) before liquid chromatography (LC)–MS measurement, which was conducted using an ultra-performance liquid chromatography system (Ultimate 3000 UPLC) (Thermo Fisher Scientific) coupled to an Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific). During the LC separation, the peptides were first loaded onto a trapping cartridge (Acclaim PepMap300 C18, 5 μm, 300-Å-wide pore; Thermo Fisher Scientific) and washed for 3 min with 0.1% TFA in water. Analytical separation was performed using a nanoEase MZ Peptide analytical column (300 Å, 1.7 μm, 75 μm by 200 mm; Waters) and carried out for 90-min total analysis time. The 90-min LC method consisted of a multistep gradient going from 2 to 8% solvent B (80% acetonitrile and 20% water with 0.1% formic acid) in 5 min, 8 to 25% in 45 min, and 25 to 40% in 10 min followed by a washing and an equilibration step while solvent A was water and 0.1% formic acid. Eluting peptides were analyzed online by a coupled Q-Exactive-HF-X mass spectrometer running in data-dependent acquisition mode. Full scans were performed at 120K resolution on a mass range covering 375 to 1500 mass/charge ratio (m/z) with a maximum allowed injection time (maxIT) of 54 ms, followed by up to 8 tandem mass spectrometry (MS/MS) scans at 60K resolution with a maxIT of 100 ms for up to 1 × 105 ions [automatic gain control (AGC) target]. Precursors were isolated with a window of 1.6 m/z and fragmented with a collision energy of 30 NCE (normalized collision energy). Unassigned, singly and doubly charged features were excluded from fragmentation, and dynamic exclusion was set to 10 s. Raw files were converted into mzML files using default settings in MSConvert (v3.0.21048) from the ProteoWizard toolkit (67). mzML files were then analyzed using Merox (v2.0.1.4.) (68) for identification of cross-linked peptides using a single protein database of the MSP-1D protein. Overall, default search parameters were used with the following changes: Cross-linker was set to DSSO, lysines were set as cross-linking sites, consecutive peptides were ignored as cross-linkers, and analysis mode was set to “quadratic mode” as recommended for small fasta files. A 1% false discovery rate cutoff was used including manual inspection of lower-scoring hits. The resulting table was exported as csv and used for 3D structural analysis using R and ChimeraX (60).
Thermostability and MST characterization
The thermal stability and aggregation behavior of (hd)MSP-1 were investigated using nanoscale differential scanning fluorimetry (nanoDSF) on a Prometheus NT.48 instrument (NanoTemper Technologies). Samples obtained after SEC were concentrated to ~0.7 to 2.7 mg/ml and loaded into nanoDSF-grade standard capillaries (NanoTemper Technologies). Samples were heated from 15 to 95°C at 1°C/min, and the intrinsic fluorescence at λ = 330 nm after excitation at λ = 280 nm and scattering were used to monitor receptor denaturation and aggregation, respectively. Relatively high protein concentrations were needed because of the lack of Trp residues in MSP-1. The first derivative of the unfolding curves was used to determine the transition midpoint using PR.ThermControl software (NanoTemper Technologies).
The affinity of spectrin for MSP-1 was assessed by MST. Spectrin (Sigma-Aldrich) was labeled with NT-647-NHS dye (NanoTemper Technologies), as per the manufacturer’s instructions, and added to a dilution series of unlabeled MSP-1 (61 nM to 2 μM, final concentrations) in 25 mM Hepes and 125 mM NaCl (pH 7.4), supplemented with 0.05% (w/v) Tween-20 (final concentration) using LoBind tubes (Eppendorf) to prevent adsorption of the sample. The KD of the MSP-1 dimer was also assessed by MST. MSP-1 was labeled with NT-647-NHS dye (NanoTemper Technologies) as per the manufacturer’s instructions. Labeled MSP-1 (final concentration of 8 nM for fully processed MSP-1 and 10 nM for hdMSP-1) was added to a dilution series of unlabeled MSP-1 (61 pM to 2 μM), in the presence or absence of 100 nM spectrin (final concentrations) in 25 mM Hepes and 125 mM NaCl (pH 7.4), supplemented with 0.05% (w/v) Tween-20 (final concentration) using LoBind tubes (Eppendorf). Under these conditions, the fluorescence was found to be constant (within ±10%) for all points in the titration curves in all experiments. The samples were loaded into premium coated capillaries (NanoTemper Technologies). MST experiments were carried out on a blue/red Monolith NT.115 instrument (NanoTemper Technologies) using the red filter set, an excitation power of 100%, and an MST power of 40 or 60%; 60% MST power gave the best results (highest signal-to-noise ratio) in all cases except for the titration of hdMSP-1 against labeled spectrin, where both MST powers gave comparably good results and 40% was used. MST data were analyzed in MO.Affinity Analysis software (NanoTemper Technologies), using −1 to 0 s before the infrared (IR) laser was turned on as the cold region and 19 to 20 s after the IR laser was turned on as the hot region, except for the titration of hdMSP-1 against labeled spectrin, where 0.2 to 1.2 s after the IR laser was turned on was used as the hot region. Data were plotted using Origin Pro 2015 (Origin Lab).
Acknowledgments
We thank D. Mills and the EM staff at Max Planck Institute of Biophysics for expert technical assistance, J. F. C. Hernandez and Ö. Yildiz for computing support, M. Blackman for providing SUB-1, B. Hessling for contributions to the XL-MS analysis, and F. Frischknecht for critical reading of the manuscript. Funding: P.M.D., Y.Z., S.-Y.S.C., and M.K. were supported by the Sofja Kovalevskaja Award from the Alexander von Humboldt Foundation to M.K. Y.Z. was partially supported by an IMPRS international student scholarship. S.-Y.S.C. was partially supported by a GSSA student scholarship from the Taiwanese government. Author contributions: T.M. and H.B. provided the samples. T.M. and P.M.D. prepared the SUB-1–processed MSP-1. P.M.D. performed biophysical analyses, the optimization of EM grid preparation, EM data collection, processing, model building, and model analysis. Y.Z. contributed to the cryo-EM data collection and preliminary data analysis. S.-Y.S.C. contributed to the biophysical analyses. D.H. performed the XL-MS analysis. M.L. provided essential reagents. P.M.D. wrote the manuscript with input from the other authors. M.K. supervised the project and provided the project funding. Competing interests: H.B. is a shareholder of Sumaya GmbH & Co. KG. The other authors declare that they have no competing interests. Data and materials availability: The atomic coordinates of (hd)MSP-1 have been deposited at the Protein Data Bank (PDB) under accession codes 6ZBJ and 6ZBL for the dimeric hdMSP-1 conformations 1 and 2, and 6ZBC, 6ZBE, 6ZBD, 6ZBF, 6ZBG, and 6ZBH for the monomeric main MSP-1 conformation and alternative conformations 1 to 5, respectively. The corresponding cryo-EM density maps have been deposited at the Electron Microscopy Data Bank (EMDB) under accession codes EMD-11156, EMD-11157, EMD-11150, EMD-11152, EMD-11151, EMD-11153, EMD-11154, and EMD-11155, respectively. The original movies and refined particle datasets have been deposited at the electron microscopy public image archive (EMPIAR; accession code EMPIAR-10437). The XL-MS data have been deposited to the proteomeXchange Consortium via the PRIDE (proteomics identifications) partner repository with the dataset identifier PXD024749.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabg0465/DC1
REFERENCES AND NOTES
- 1.World Health Organization, “World malaria report 2020” (ISBN 978-92-4-001579-1, World Health Organization, 2020); www.who.int/publications/i/item/9789240015791.
- 2.Beeson J. G., Drew D. R., Boyle M. J., Feng G., Fowkes F. J. I., Richards J. S., Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria. FEMS Microbiol. Rev. 40, 343–372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hale V. L., Watermeyer J. M., Hackett F., Vizcay-Barrena G., van Ooij C., Thomas J. A., Spink M. C., Harkiolaki M., Duke E., Fleck R. A., Blackman M. J., Saibil H. R., Parasitophorous vacuole poration precedes its rupture and rapid host erythrocyte cytoskeleton collapse in Plasmodium falciparum egress. Proc. Natl. Acad. Sci. U.S.A. 114, 3439–3444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ladda R., Aikawa M., Sprinz H., Penetration of erythrocytes by merozoites of mammalian and avian malarial parasites. J. Parasitol. 55, 633–644 (1969). [PubMed] [Google Scholar]
- 5.Gilson P. R., Nebl T., Vukcevic D., Moritz R. L., Sargeant T., Speed T. P., Schofield L., Crabb B. S., Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteomics 5, 1286–1299 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Osier F. H. A., Fegan G., Polley S. D., Murungi L., Verra F., Tetteh K. K. A., Lowe B., Mwangi T., Bull P. C., Thomas A. W., Cavanagh D. R., McBride J. S., Lanar D. E., Mackinnon M. J., Conway D. J., Marsh K., Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infect. Immun. 76, 2240–2248 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fowkes F. J. I., Richards J. S., Simpson J. A., Beeson J. G., The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: A systematic review and meta-analysis. PLOS Med. 7, e1000218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards J. S., Arumugam T. U., Reiling L., Healer J., Hodder A. N., Fowkes F. J. I., Cross N., Langer C., Takeo S., Uboldi A. D., Thompson J. K., Gilson P. R., Coppel R. L., Siba P. M., King C. L., Torii M., Chitnis C. E., Narum D. L., Mueller I., Crabb B. S., Cowman A. F., Tsuboi T., Beeson J. G., Identification and prioritization of merozoite antigens as targets of protective human immunity to Plasmodium falciparum malaria for vaccine and biomarker development. J. Immunol. 191, 795–809 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blank A., Fürle K., Jäschke A., Mikus G., Lehmann M., Hüsing J., Heiss K., Giese T., Carter D., Böhnlein E., Lanzer M., Haefeli W. E., Bujard H., Immunization with full-length Plasmodium falciparum merozoite surface protein 1 is safe and elicits functional cytophilic antibodies in a randomized first-in-human trial. npj Vaccines. 5, 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackman M. J., Heidrich H.-G., Donachie S., McBride J. S., Holder A. A., A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J. Exp. Med. 172, 379–382 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kauth C. W., Woehlbier U., Kern M., Mekonnen Z., Lutz R., Mücke N., Langowski J., Bujard H., Interactions between merozoite surface proteins 1, 6, and 7 of the malaria parasite Plasmodium falciparum. J. Biol. Chem. 281, 31517–31527 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Paul G., Deshmukh A., Kumar Chourasia B., Kalamuddin M., Panda A., Kumar Singh S., Gupta P. K., Mohmmed A., Chauhan V. S., Theisen M., Malhotra P., Protein-protein interaction studies reveal the Plasmodium falciparum merozoite surface protein-1 region involved in a complex formation that binds to human erythrocytes. Biochem. J. 475, 1197–1209 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Lin C. S., Uboldi A. D., Marapana D., Czabotar P. E., Epp C., Bujard H., Taylor N. L., Perugini M. A., Hodder A. N., Cowman A. F., The merozoite surface protein 1 complex is a platform for binding to human erythrocytes by Plasmodium falciparum. J. Biol. Chem. 289, 25655–25669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C. S., Uboldi A. D., Epp C., Bujard H., Tsuboi T., Czabotar P. E., Cowman A. F., Multiple Plasmodium falciparum merozoite surface protein 1 complexes mediate merozoite binding to human erythrocytes. J. Biol. Chem. 291, 7703–7715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Chen H., Oo T. H., Daly T. M., Bergman L. W., Liu S.-C., Chishti A. H., Oh S. S., A co-ligand complex anchors Plasmodium falciparum merozoites to the erythrocyte invasion receptor band 3. J. Biol. Chem. 279, 5765–5771 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Ranjan R., Chugh M., Kumar S., Singh S., Kanodia S., Hossain M. J., Korde R., Grover A., Dhawan S., Chauhan V. S., Reddy V. S., Mohmmed A., Malhotra P., Proteome analysis reveals a large merozoite surface protein-1 associated complex on the Plasmodium falciparum merozoite surface. J. Proteome Res. 10, 680–691 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Pachebat J. A., Kadekoppala M., Grainger M., Dluzewski A. R., Gunaratne R. S., Scott-Finnigan T. J., Ogun S. A., Ling I. T., Bannister L. H., Taylor H. M., Mitchell G. H., Holder A. A., Extensive proteolytic processing of the malaria parasite merozoite surface protein 7 during biosynthesis and parasite release from erythrocytes. Mol. Biochem. Parasitol. 151, 59–69 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Trucco C., Fernandez-Reyes D., Howell S., Stafford W. H., Scott-Finnigan T. J., Grainger M., Ogun S. A., Taylor W. R., Holder A. A., The merozoite surface protein 6 gene codes for a 36 kDa protein associated with the Plasmodium falciparum merozoite surface protein-1 complex. Mol. Biochem. Parasitol. 112, 91–101 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Hodder A. N., Czabotar P. E., Uboldi A. D., Clarke O. B., Lin C. S., Healer J., Smith B. J., Cowman A. F., Insights into Duffy binding-like domains through the crystal structure and function of the merozoite surface protein MSPDBL2 from Plasmodium falciparum. J. Biol. Chem. 287, 32922–32939 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Donnell R., Saul A., Cowman A. F., Crabb B. S., Functional conservation of the malaria vaccine antigen MSP-119 across distantly related Plasmodium species. Nat. Med. 6, 91–95 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Combe A., Giovannini D., Carvalho T. G., Spath S., Boisson B., Loussert C., Thiberge S., Lacroix C., Gueirard P., Ménard R., Clonal conditional mutagenesis in malaria parasites. Cell Host Microbe 5, 386–396 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Boyle M. J., Richards J. S., Gilson P. R., Chai W., Beeson J. G., Interactions with heparin-like molecules during erythrocyte invasion by Plasmodium falciparum merozoites. Blood 115, 4559–4568 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Baldwin M. R., Li X., Hanada T., Liu S.-C., Chishti A. H., Merozoite surface protein 1 recognition of host glycophorin A mediates malaria parasite invasion of red blood cells. Blood 125, 2704–2711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrera S., Rudin W., Herrera M., Clavijo P., Mancilla L., de Plata C., Matile H., Certa U., A conserved region of the MSP-1 surface protein of Plasmodium falciparum contains a recognition sequence for erythrocyte spectrin. EMBO J. 12, 1607–1614 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das S., Hertrich N., Perrin A. J., Withers-Martinez C., Collins C. R., Jones M. L., Watermeyer J. M., Fobes E. T., Martin S. R., Saibil H. R., Wright G. J., Treeck M., Epp C., Blackman M. J., Processing of Plasmodium falciparum merozoite surface protein MSP1 activates a spectrin-binding function enabling parasite egress from RBCs. Cell Host Microbe 18, 433–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woehlbier U., Epp C., Kauth C. W., Lutz R., Long C. A., Coulibaly B., Kouyate B., Arevalo-Herrera M., Herrera S., Bujard H., Analysis of antibodies directed against merozoite surface protein 1 of the human malaria parasite Plasmodium falciparum. Infect. Immun. 74, 1313–1322 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogutu B. R., Apollo O. J., Kinney D. M., Okoth W., Siangla J., Dubovsky F., Tucker K., Waitumbi J. N., Diggs C., Wittes J., Malkin E., Leach A., Soisson L. A., Milman J. B., Otieno L., Holland C. A., Polhemus M., Remich S. A., Ockenhouse C. F., Cohen J., Ballou W. R., Martin S. K., Angov E., Stewart V. A., Lyon J. A., Gray Heppner D. Jr., Withers M. R.; MSP-1 Malaria Vaccine Working Group , Blood stage malaria vaccine eliciting high antigen-specific antibody concentrations confers no protection to young children in Western Kenya. PLOS ONE 4, e4708 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kauth C. W., Epp C., Bujard H., Lutz R., The merozoite surface protein 1 complex of human malaria parasite Plasmodium falciparum: Interactions and arrangements of subunits. J. Biol. Chem. 278, 22257–22264 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Morgan W. D., Birdsall B., Frenkiel T. A., Gradwell M. G., Burghaus P. A., Syed S. E. H., Uthaipibull C., Holder A. A., Feeney J., Solution structure of an EGF module pair from the Plasmodium falciparum merozoite surface protein 1. J. Mol. Biol. 289, 113–122 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Pizarro J. C., Chitarra V., Verger D., Holm I., Pêtres S., Dartevelle S., Nato F., Longacre S., Bentley G. A., Crystal structure of a Fab complex formed with PfMSP1-19, the C-terminal fragment of merozoite surface protein 1 from Plasmodium falciparum: A malaria vaccine candidate. J. Mol. Biol. 328, 1091–1103 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Ye Y., Godzik A., FATCAT: A web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 32, W582–W585 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanabe K., Mackay M., Goman M., Scaife J. G., Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 195, 273–287 (1987). [DOI] [PubMed] [Google Scholar]
- 33.Sanders P. R., Cantin G. T., Greenbaum D. C., Gilson P. R., Nebl T., Moritz R. L., Yates J. R. III, Hodder A. N., Crabb B. S., Identification of protein complexes in detergent-resistant membranes of Plasmodium falciparum schizonts. Mol. Biochem. Parasitol. 154, 148–157 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Tolle R., Früh K., Doumbo O., Koita O., N’Diaye M., Fischer A., Dietz K., Bujard H., A prospective study of the association between the human humoral immune response to Plasmodium falciparum blood stage antigen gp190 and control of malarial infections. Infect. Immun. 61, 40–47 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jäschke A., Coulibaly B., Remarque E. J., Bujard H., Epp C., Merozoite surface protein 1 from Plasmodium falciparum is a major target of opsonizing antibodies in individuals with acquired immunity against malaria. Clin. Vaccine Immunol. 24, e00155–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikodem D.-P., Davidson E.-A., Identification of a novel antigenic domain of Plasmodium falciparum merozoite surface protein-1 that specifically binds to human erythrocytes and inhibits parasite invasion, in vitro. Mol. Biochem. Parasitol. 108, 79–91 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Goel V. K., Li X., Chen H., Liu S.-C., Chishti A. H., Oh S. S., Band 3 is a host receptor binding merozoite surface protein 1 during the Plasmodium falciparum invasion of erythrocytes. Proc. Natl. Acad. Sci. U.S.A. 100, 5146–5169 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fierro M. A., Asady B., Brooks C. F., Cobb D. W., Villegas A., Moreno S. N. J., Muralidharan V., An endoplasmic reticulum CREC family protein regulates the egress proteolytic cascade in malaria parasites. MBio 11, e03078–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudolph R., Lilie H., In vitro folding of inclusion body proteins. FASEB J. 10, 49–56 (1996). [PubMed] [Google Scholar]
- 40.Koussis K., Withers-Martinez C., Yeoh S., Child M., Hackett F., Knuepfer E., Juliano L., Woehlbier U., Bujard H., Blackman M. J., A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 28, 725–735 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tegunov D., Cramer P., Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D. Asarnow, E. Palovcak, Y. Cheng, UCSF pyem v0.5 (2019).
- 47.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera — a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., Quentin D., Roderer D., Tacke S., Siebolds B., Schubert E., Shaikh T. R., Lill P., Gatsogiannis C., Raunser S., SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z., Fang J., Chittuluru J., Asturias F. J., Penczek P. A., Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20, 237–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou M., Li Y., Hu Q., Bai X.-c., Huang W., Yan C., Scheres S. H. W., Shi Y., Atomic structure of the apoptosome: Mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes Dev. 29, 2349–2361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Punjani A., Fleet D. J., 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Cowtan K., Fitting molecular fragments into electron density. Acta Crystallogr. Sec. D Struct. Biol. 64, 83–89 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P., Lohkamp B., Scott W., Cowtan K., Features and development of Coot. Acta Crystallogr. Sec. D Struct. Biol. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buchan D. W. A., Jones D. T., The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 47, W402–W407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez-Garcia R., Gomez-Blanco J., Cuervo A., Carazo J. M., Sorzano C. O. S., Vargas J., DeepEMhancer: A deep learning solution for cryo-EM volume post-processing. BioRxiv 2020.06.12.148296, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams P. D., Grosse-Kunstleve R. W., Hung L.-W., Iorger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C., PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. Sec. D Struct. Biol. 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jakobi A. J., Wilmanns M., Sachse C., Model-based local density sharpening of cryo-EM maps. eLife 6, e27131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps research papers. Acta Crystallogr. Sect. D Struct. Biol. 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stivala A., Wybrow M., Wirth A., Whisstock J. C., Stuckey P. J., Automatic generation of protein structure cartoons with pro-origami. Bioinformatics 27, 3315–3316 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N., ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aurrecoechea C., Brestelli J., Brunk B. P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Miller J. A., Nayak V., Pennington C., Pinney D. F., Roos D. S., Ross C., Stoeckert C. J. Jr., Treatman C., Wang H., PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laskowski R. A., Jabłońska J., Pravda L., Vařeková R. S., Thornton J. M., PDBsum: Structural summaries of PDB entries. Protein Sci. 27, 129–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rappsilber J., Mann M., Ishihama Y., Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M.-Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., Mallick P., A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Götze M., Pettelkau J., Fritzsche R., Ihling C. H., Schäfer M., Sinz A., Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J. Am. Soc. Mass Spectrom. 26, 83–97 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Kruschke J. K., Bayesian estimation supersedes the t test. J. Exp. Psychol. Gen. 142, 573–603 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Merkley E. D., Rysavy S., Kahraman A., Hafen R. P., Daggett V., Adkins J. N., Distance restraints from crosslinking mass spectrometry: Mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23, 747–759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller L. H., Roberts T., Shahabuddin M., McCutchan T. F., Analysis of sequence diversity in the Plasmodium falciparum merozoite surface protein-1 (MSP-1). Mol. Biochem. Parasitol. 59, 1–14 (1993). [DOI] [PubMed] [Google Scholar]
- 72.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabg0465/DC1