Directed differentiation of hPSCs and transition from 3D intestinal organoids into functional hIECs.
Abstract
Advanced technologies are required for generating human intestinal epithelial cells (hIECs) harboring cellular diversity and functionalities to predict oral drug absorption in humans and study normal intestinal epithelial physiology. We developed a reproducible two-step protocol to induce human pluripotent stem cells to differentiate into highly expandable hIEC progenitors and a functional hIEC monolayer exhibiting intestinal molecular features, cell type diversity, and high activities of intestinal transporters and metabolic enzymes such as cytochrome P450 3A4 (CYP3A4). Functional hIECs are more suitable for predicting compounds metabolized by CYP3A4 and absorbed in the intestine than Caco-2 cells. This system is a step toward the transition from three-dimensional (3D) intestinal organoids to 2D hIEC monolayers without compromising cellular diversity and function. A physiologically relevant hIEC model offers a novel platform for creating patient-specific assays and support translational applications, thereby bridging the gap between 3D and 2D culture models of the intestine.
INTRODUCTION
The human small intestine (hSI) is uniquely adapted for performing specialized functions, including initial and selective barrier activity, nutrient absorption, host-microbiome interactions, and regulation of host defense and immune responses (1). Drugs administered orally, the most common route of drug delivery, are absorbed and begin to get metabolized in the hSI. In the past decade, attempts have been made to recapitulate various aspects of the complex in vivo intestinal physiology and generate experimental models for evaluating drug absorption and metabolism in the small intestine using human colon adenocarcinoma cell lines, such as Caco-2 and HT-29, and artificial membrane systems. However, the existing models do not adequately mimic the functions of hSI, as these models do not have the four major intestinal epithelial subtypes, such as absorptive enterocytes, antimicrobial peptide-secreting Paneth cells, mucus-producing goblet cells, and hormone-secreting enteroendocrine cells (2), and do not show expression patterns for drug transporters, drug-metabolizing enzymes, and tight junctions observed in vivo (3). Furthermore, cytochrome P450 3A4 (CYP3A4) deficiency in these models, the most abundant drug-metabolizing P450 in hSI, renders them unsuitable for investigating intestinal first-pass metabolism and bioavailability (4). Primary human intestinal epithelial cells (hIECs) can act as alternatives for predicting absorption potential in humans, despite limitations regarding their number and accessibility (5). Hence, an evaluation system where a model can be used to predict intestinal absorption and CYP3A4-mediated first-pass metabolism using normal hIECs, without requiring repeated sampling of human tissue, is urgently required in the field of drug development.
Recently, notable advancements have been made in the development of hIEC models (6–8). Several groups have generated two-dimensional (2D) hSI monolayer models of epithelial-like cells from human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), although considerable room exists for improving the differentiation efficiency and achieving sufficient expression and activities of small intestinal drug-metabolizing enzymes and drug transporters (7, 9, 10). 3D human intestinal organoids (hIOs) with small intestine–like cell composition either derived from hPSCs or the dissociated crypts of intestinal tissues is another attractive model. We have previously reported a method for in vitro maturation of hPSC-derived hIOs that closely resemble adult human intestinal tissues at the molecular and functional levels (11). Although 3D hIOs represent an advanced culture system showing physiologically relevant phenotypes and integrate several microenvironmental features of hSI in vitro (12), they cannot be as easily used for classical intestinal assays as determined using 2D monolayer cultures. Generation of a monolayer hSI model derived from hIOs is challenging owing to the difficulty in growing intestinal epithelial cells as confluent monolayers due to the lack of the proliferative cell populations within hIOs (13). Therefore, an optimal differentiation and culture method that enables directed differentiation of hPSCs into hIEC monolayers and generates hIO-derived hIEC monolayers to mimic the in vivo–like intestinal cell composition and functional characteristics is required.
Wnt and Notch signaling are well-known signaling pathways; a cross-talk between these pathways regulates intestinal stem cell (ISC) differentiation (14, 15). Active Wnt and Notch signaling pathways are important for crypt maintenance via self-renewal of ISCs (16). Inhibition of Wnt signaling results in crypt loss due to ISC loss and destruction of intestinal epithelial barrier (17, 18). In the absence of Notch signaling, ISCs differentiate to secretory lineage cells leading to ISC depletion and ultimately secretory metaplasia (19). On the basis of these observations, several distinct in vitro differentiation protocols using Wnt and Notch modulators have been reported for the differentiation of hPSCs into intestinal lineages (20). Although current evidence suggests that absorptive enterocytes appear under conditions of Wnt inhibition and Notch activation (21), most of the studies have reported that hPSCs differentiate into intestinal absorptive enterocytes in the presence of Wnt activators such as R-spondin 1, Wnt3a, (20Z, 30E)-6-bromoindirubin-30-oxime (BIO), CHIR99021, and LY2090314 and Notch inhibitors such as N-[N-(3,5-difluorophenacetyl-l-alanyl)]-(2)-phenylglycine t-butyl ester (DAPT) (7, 8, 10, 22). Therefore, development and optimization of differentiation methods are required for generating functional hIECs comprising multiple intestinal cell types via fine-tuning of the Wnt and Notch signaling pathways and subsequent direct lineage differentiation into mature absorptive and secretory cells (16, 21).
To address these issues, we aimed to generate expandable and scalable hIEC progenitors from both hESCs and patient-specific iPSCs derived from intestinal biopsies, which can differentiate into functional hIECs in a Transwell system and exhibit in vivo–like intestinal cell type composition after lineage-specific modulation of key signaling pathways. Furthermore, functional hIECs may predict the fraction absorbed in human intestines based on intestinal absorption and first-pass metabolism. This hIEC culture system is a step toward transitioning from 3D hIOs to 2D monolayer culture, thereby bridging the gap between 3D organotypic culture and the 2D monolayer system of hIECs.
RESULTS
A novel differentiation method for generating hPSC-derived hIEC progenitors
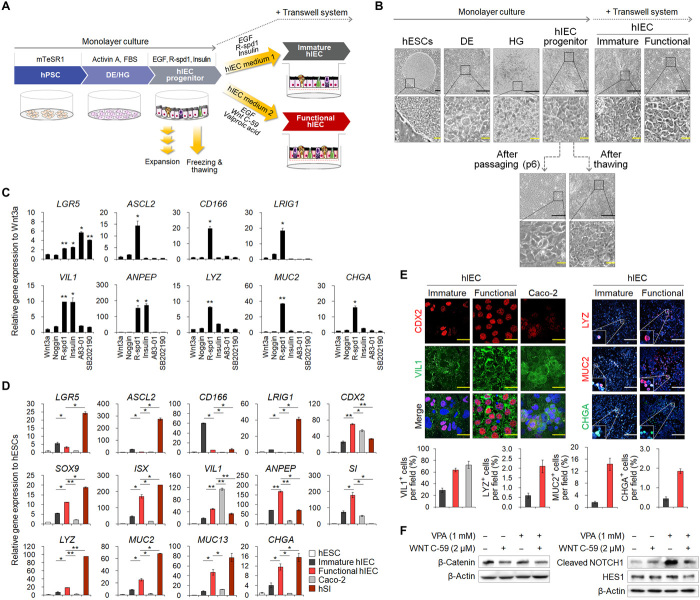
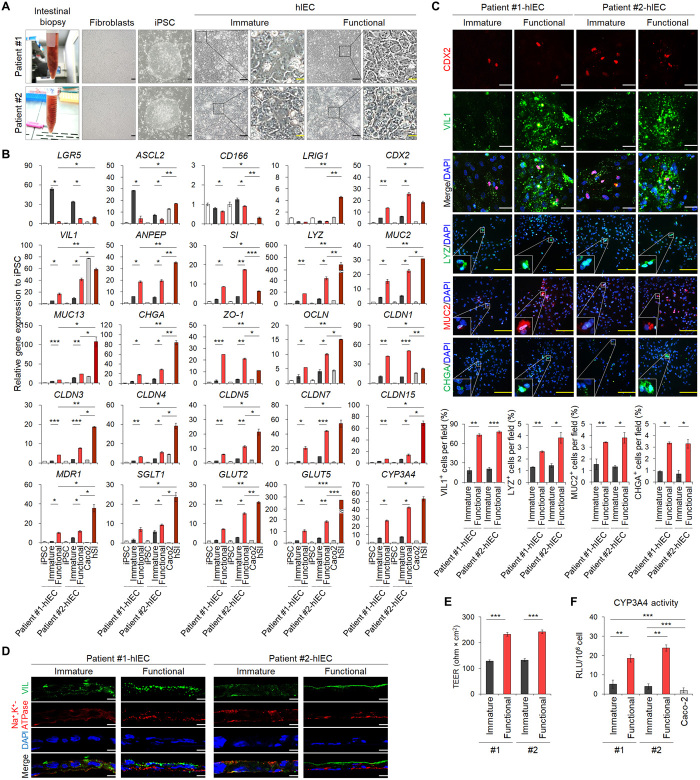
To generate a monolayer hSI model derived from hPSCs, we established a novel directed differentiation protocol, which can mimic in vivo intestinal developmental processes (Fig. 1A). As reported previously (8, 22, 23), hESCs were differentiated into definitive endoderm (DE) and hindgut (HG) via sequential treatment with growth factors such as activin A, fibroblast growth factor 4 (FGF4), and the Glycogen synthase kinase-3 beta (GSK3β) inhibitor CHIR99021; the cellular morphologies were similar to the previously reported phenotypes (Fig. 1B). Previous studies have reported that HG cells can be further differentiated into hIECs via activating various signaling pathways (20). To identify factors enhancing intestinal differentiation in 2D cultures, we tested selected small molecules that modulate signaling pathways such as Wnt, Notch, transforming growth factor–β, insulin, and mitogen-activated protein kinase. We observed that R-spondin 1, an agonist of Wnt signaling, induced the expression of markers of major intestinal cell types, including ISCs (LGR5, ASCL2, CD166, and LRIG1), enterocytes (VIL1 and ANPEP), and secretory lineage cells (LYZ for Paneth cells, MUC2 for goblet cells, and CHGA for enteroendocrine cells) (Fig. 1C). Insulin, which was selected for its role in different cell types in the intestinal epithelium via the insulin receptor (24), specifically increased differentiation into absorptive cells, as shown by the increase in VIL1 and ANPEP expression (Fig. 1C). Therefore, a newly developed hIEC medium 1 supplemented with R-spondin 1 and insulin is promising for enhancing hIEC differentiation and generation of the small intestinal cell types observed in vivo. hIECs were stably maintained after long-term passage without distinct changes in the expression of intestinal cell markers and tight junction molecules upon sequential freezing/thawing of the hIECs (>10 passages and >5 months in culture) (Fig. 1B and fig. S1, A and B; known as hIEC progenitors). Experiments regarding intestinal epithelial function are typically performed using a Transwell system (10). hIECs formed confluent monolayers in Transwell inserts and showed intestinal epithelial cell–like morphology (Fig. 1B) with transepithelial electrical resistance (TEER) values of approximately 144.39 ± 0.81 ohm × cm2 without cell viability impairment (fig. S1, C and D).
Fig. 1. Differentiation of hESCs into hIEC progenitors and functional hIECs.
(A) Schematic representation of the developed protocol. (B) Morphology of hESCs, DE, HG, hIECs progenitors, immature, functional, passaged, and thawed hIECs. Black scale bars, 200 μm. Yellow scale bars, 50 μm. (C) Relative gene expression of intestinal markers in the compound-treated hIECs. (D) Relative gene expression of intestinal markers in hESC, immature hIECs, functional hIECs, Caco-2 cells, and hSI. (E) Immunofluorescence analysis of the markers of enterocytes (CDX2 and VIL1), Paneth cells (LYZ), goblet cells (MUC2), and enteroendocrine cells (CHGA) in immature and functional hIECs (top). White scale bars, 200 μm. Yellow scale bars, 20 μm. Data represent means ± SEM (n = 4) of each marker positive cell per field (bottom). (F) Western blot analysis to verify expression levels of β-catenin, cleaved NOTCH1, and HES1 during differentiation into immature hIECs and functional hIECs. *P < 0.05 and **P < 0.01 using two-tailed t test.
Generation of functional hIECs with composition and distribution of intestinal cell types present in vivo
To further differentiate hIEC progenitors into mature and functional hIECs similar to their in vivo counterparts, we used the Wnt signaling inhibitor, Wnt-C59, and the Notch signaling agonist, valproic acid (VPA) (Fig. 1A), because Wnt inhibition and Notch activation contribute to differentiation into absorptive pre-epithelial cells in hSI (21). We cultured monolayers of hIEC progenitors with either hIEC medium 1 or hIEC medium 2 containing Wnt-C59 and VPA using the Transwell system. Morphological examination of the hIECs differentiated in hIEC medium 1 (immature hIECs) and hIEC medium 2 (functional hIECs) revealed that the functional hIECs were polygonal and had epithelial-like morphology (Fig. 1B). The functional hIECs showed significantly higher mRNA levels of intestinal transcription factors (CDX2, SOX9, and ISX) and intestinal cell type–specific markers than immature hIECs (Fig. 1D). In contrast, the expression level of all ISC markers was lower in the functional hIECs than in immature hIECs (Fig. 1D), implying that the hIEC progenitors differentiated into more specialized cells in hIEC medium 2. VIL1 expression in functional hIECs remained lower than that in Caco-2 cells, which undergo spontaneous differentiation into enterocytes when grown on Transwell inserts, while the expression levels of secretory cell type–specific markers (LYZ, MUC2, and CHGA) were higher than those in Caco-2 (Fig. 1D). Most VIL1-expressing enterocytes were observed in 28.80 ± 3.80% in immature hIECs, 68.81 ± 3.34% in functional hIECs, and 72.24 ± 6.17% in Caco-2 (Fig. 1E). There are 2.10 ± 0.33% LYZ+ Paneth cells, 14.35 ± 2.03% MUC2+ goblet cells, and 1.85 ± 0.12% CHGA+ enteroendocrine cells in functional hIECs, whereas only few cells were VIL1 positive in immature hIECs (Fig. 1E). Flow cytometry analysis revealed results similar to that obtained by immunostaining, indicating that cell populations undergo dynamic changes during differentiation (fig. S2).
Although we detected significantly more intestinal epithelial cell types in functional hIECs than in immature hIECs, reports indicate that inactivation of the Wnt pathway and activation of the Notch pathway suppress differentiation of ISCs into secretory cells (14, 15). To explain this discrepancy, we analyzed the expression level of proteins downstream of WNT and NOTCH, such as β-catenin, cleaved NOTCH1, and HES1. The expression of β-catenin was specifically inhibited by WNT C-59 (Fig. 1F). However, the expression levels of cleaved NOTCH1 and HES1, the downstream proteins of NOTCH pathway, significantly suppressed upon cotreatment of VPA with WNT C-59 even after notable enhancement of Notch signaling by single treatment with VPA (Fig. 1F). It is speculated that differentiation of hIEC progenitors into functional hIECs is triggered by the inhibition of WNT pathway, while the fate of differentiating into absorptive or secretory lineage cells depends on the activity of the NOTCH pathway. Together, in contrast with other small intestinal epithelial models comprising only absorptive enterocytes, our system allows study of in vivo–like intestinal tissue.
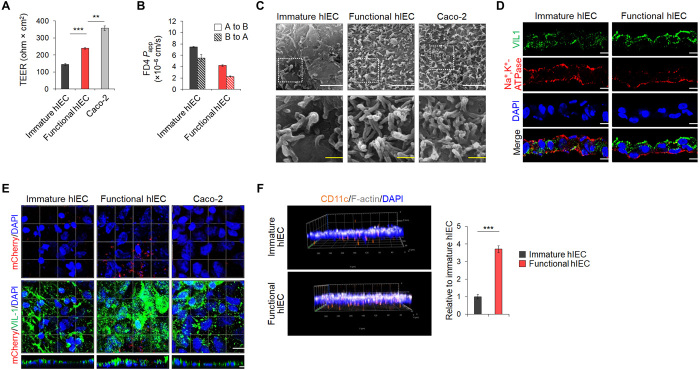
Functional hIEC monolayer displays pronounced polarity and barrier integrity
The TEER values, representing monolayer integrity, were 137.76 ± 4.77 ohm × cm2 and 238.56 ± 4.08 ohm × cm2 for the immature and functional hIEC monolayers, respectively, and were lower than that of the Caco-2 monolayer (357.28 ± 13.76 ohm × cm2) (Fig. 2A). The maximum TEER value, ranging from 203.28 ± 0.56 to 235.20 ± 5.60 ohm × cm2, was obtained after 14 days of culture on Transwell inserts when hIEC progenitors at each passage were further differentiated into functional hIECs (fig. S1E). The expression of tight junction genes and proteins in the functional hIECs was higher than in immature hIECs (fig. S3, A and B). In addition, structural proteins, such as ZO-1 and E-cadherin, were properly localized in functional hIECs (fig. S3C). The apparent permeability (Papp) values of fluorescein isothiocyanate (FITC)–dextran (4 kDa) across functional hIECs in both directions were significantly lower than those of immature hIECs (Fig. 2B). Compared to immature hIECs, functional hIECs formed a structurally polarized monolayer, as evident from the scanning electron microscopy (SEM) images and the polarized distribution of apical (VIL1) and basolateral cell surface proteins [Na+- and K+-dependent adenosine triphosphatase (Na+,K+-ATPase)] (Fig. 2, C and D). Although microvilli developed normally on the surface of both functional hIEC and Caco-2 cell monolayers, irregular blobs were specifically observed on the surface of the functional hIEC monolayer, which appeared like mucus extruded from goblet cells (25), as shown by high-resolution SEM images (Fig. 2C). Moreover, a significantly higher number of bacteria were in contact with the apical surface of the functional hIEC monolayers, as visualized using mCherry-expressing Lactobacillus plantarum, compared to immature hIECs and Caco-2 using immunofluorescence and quantitative analysis of the colony-forming units (Fig. 2E and fig. S4A). Consistently, the expression of mucins and mucin-related genes, which is critical for intestinal mucosal integrity as it allows sustained colonization by intestinal microbiota (26), was significantly increased in functional hIECs (fig. S4, B to D). Furthermore, a higher number of macrophages attached to the basolateral side of the functional hIEC monolayer and translocated to the upper side of the Transwell compared to that in immature hIECs (Fig. 2F), which may be attributed to elevated receptor expression of bacterial antigens (CD14 and TLR4) (fig. S5) (27).
Fig. 2. Functional characterization of hPSC-derived hIEC monolayers.
(A) TEER values of immature hIECs, functional hIEC, and Caco-2 cell monolayers. Data represent means ± SEM (n = 4). (B) The Papp coefficient of FITC-dextran 4 kDa in either the apical to basolateral (A to B) or the basolateral to apical (B to A) direction. Data represent means ± SEM (n = 4). (C) Representative SEM images of apical surface of the immature hIEC, functional hIEC, and Caco-2 cell monolayers. White scale bars, 2 μm. Yellow scale bars, 500 nm. (D) Confocal immunofluorescence analysis showing vertical sections of immature and functional hIECs stained for the markers of apical brush border (VIL1) and basolateral (Na+,K+-ATPase) side. Scale bars, 10 μm. (E) Representative confocal images of L. plantarum–mCherry (red) inoculated immature hIEC, functional hIEC, and Caco-2 cells at 2 hours after inoculation. XZ projection of a 3D confocal micrograph showing adhesion of L. plantarum–mCherry (red) to the apical side of immature hIEC, functional hIEC, and Caco-2 cells stained for the apical brush border (VIL1) marker. Scale bar, 10 μm. (F) Representative 3D confocal Z-stack images of macrophage migration across the immature hIECs or the functional hIECs after liposaccharide (LPS) treatment for 24 hours. F-actin, white; CD11c, orange; DAPI, blue; magnification ×20 (left). Data are means ± SEM (n = 3) of CD11c-positive cell per field (right). **P < 0.01 and ***P < 0.001 using two-tailed t test.
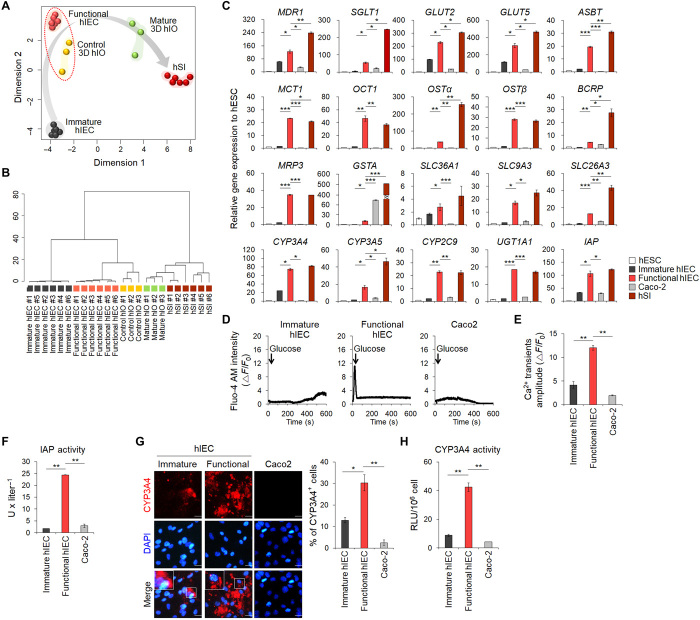
Improved intestinal transporter and enzyme activities in functional hIECs
To assess the differentiation status of functional hIECs based on global gene expression patterns, we conducted multidimensional scaling (MDS) (Fig. 3A) and hierarchical clustering of 85 small intestine markers (Fig. 3B and table S1) using our original and publicly available RNA sequencing (RNA-seq) datasets of 3D control and mature hIOs and hSI samples. The results demonstrated that functional hIECs were most closely related to control hIOs and became more similar to the hSI. Next, we analyzed the expression levels of various intestinal transporters and metabolic enzymes. Expression of 20 genes was up-regulated in functional hIECs compared to immature hIECs (Fig. 3C). In line with the higher expression of glucose transporter-encoding genes (SGLT, GLUT2, and GLUT5) (Fig. 3C), more calcium ions were released from intracellular organelles, including the endoplasmic reticulum, due to enhanced sensitivity to glucose stimulation in functional hIECs than in immature hIECs and Caco-2 cells (Fig. 3, D and E). This suggested that functional hIECs were more capable of incorporating nutrients such as glucose than immature hIECs and Caco-2 cells. Next, we assayed intestinal alkaline phosphatase (IAP) activity, which is essential for maintaining gut homeostasis as a crucial mucosal defense factor and is most commonly evaluated in functional assays of hIECs (28). Consistent with the mRNA expression levels (Fig. 3C), IAP activity was significantly higher in functional hIECs than in immature hIECs and Caco-2 cells (Fig. 3F). Orally administered drugs are metabolized primarily by cytochrome P450 not only in the liver but also in the small intestine (29). Although CYP3A4 is a dominant drug-metabolizing enzyme in the hIECs, CYP3A4 is rarely expressed in hPSC-derived enterocytes and Caco-2 cells (8). Consistent with the results of a previous report (8), CYP3A4 was negligibly expressed in Caco-2 cells (Fig. 3C). However, CYP3A4 expression was slightly more in immature hIECs than in Caco-2 cells and was considerably high in functional hIECs, with no significant difference between functional hIECs and hSI (Fig. 3C). The percentage of CYP3A4+ cells increased significantly in functional hIECs compared to immature hIECs and Caco-2 cells (Fig. 3G). CYP3A4 activity was also significantly higher in functional hIECs (Fig. 3H). Therefore, these results indicated that functional hIECs have drug transporter and drug-metabolizing enzyme activities, which are required for evaluating drug bioavailability in vitro (8).
Fig. 3. Improvement of functional properties of differentiated hIECs.
(A) MDS plot shows the pairwise distances between samples. Five homogeneous sample groups were observed: immature hIECs (gray, n = 6), functional hIECs (orange, n = 6), control 3D hIOs (yellow, n = 3), interleukin-2 (IL-2)–treated mature hIOs (green, n = 3), and adult hSI (dark red) (n = 6). (B) A dendrogram based on hierarchical clustering of the 85 small intestine marker gene set from the RNA-seq data using a maximum distance. (C) Quantitative polymerase chain reaction (qPCR) analysis of the expression of intestinal transporters and enzymes in hESCs, immature hIECs, functional hIECs, Caco-2 cells, and hSI. Data represent means ± SEM (n = 3). (D) Representative glucose-induced calcium fluctuations measured in immature (left), functional hIECs (middle), and Caco-2 cells (right) using Fluo-4 AM calcium indicator. (E) Mean value of delta calcium transient amplitude (n = 5 per group). (F) The enzyme activity of IAP was measured in hPSCs, immature hIECs, functional hIECs, and Caco-2 cells. Data represent means ± SEM (n = 4). (G) Immunofluorescence analysis for CYP3A4 (left) and the percentages of CYP3A4+ cells (right) in immature hIECs, functional hIECs, and Caco-2 cells. Scale bars, 20 μm. Data represent means ± SEM (n = 3). (H) Levels of CYP3A4 activity in immature, functional hIECs, and Caco-2 cells. RLU, relative luminescence unit. Data represent means ± SEM (n = 4). *P < 0.05, **P < 0.01, and ***P < 0.001 using two-tailed t test.
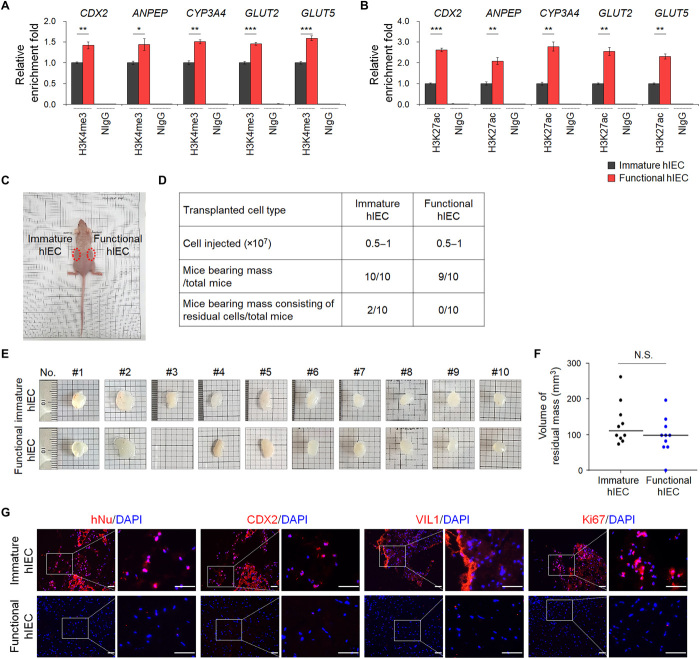
High active histone marks at intestine-specific gene promoters and low in vivo cell retention capacity of functional hIECs
To characterize the differentiated hIECs at an epigenetic level, chromatin immunoprecipitation (ChIP) assay was performed using antibodies against histone 3 lysine 4 trimethylation (H3K4me3) and histone 3 lysine 27 acetylation (H3K27ac), which are prominent histone modifications associated with active lineage-specific genes. The H3K4me3 and H3K27ac were significantly enriched at the promoter/enhancer regions of CDX2, ANPEP, CYP3A4, GLUT2, and GLUT5 in functional hIECs than in immature hIECs (Fig. 4, A and B), which was accompanied by an increase in their expression (Figs. 1D and 3C). This is in accordance with the results of previous reports showing that active methylation and acetylation at promoters and enhancers correlate with overexpression of lineage-specific genes during in vitro differentiation of hPSCs (30). These results suggested that the culture condition of functional hIECs induces the expression of intestine-specific markers via facilitating open chromatin structure due to epigenetic regulation, yielding more lineage-committed cells.
Fig. 4. The epigenetic status and in vivo cell retention capacity of functional hIECs.
The ChIP assay was performed using anti-H3K4me3 (A) and anti-H3K27ac antibodies (B). The result is shown as a relative enrichment fold compared to immature hIECs using a percentage of the input chromatin, n = 3. NIgG, normal IgG. *P < 0.05, **P < 0.01, and ***P < 0.001 by two-tailed t test. (C) Nude mice were injected subcutaneously with immature (left side) or functional hIECs (right side) on both sides, and each plug contained 0.5 to 1 × 107 cells within the Matrigel. (D) A summary of cell retention capacity of immature or functional hIECs. (E) Representative images of hIEC-Matrigel plugs recovered from mice after 6 to 10 weeks. (F) hIEC-Matrigel plugs were isolated 6 to 10 weeks after transplantation and volumes were measured. Data represent means ± SEM (n = 10). N.S. indicates nonsignificant by two-tailed t test. (G) Immunofluorescence analysis for human nuclei (hNu), intestinal markers (CDX2 and VIL1), and proliferation marker (Ki67) in hIEC-Matrigel plugs. Scale bars, 50 μm.
Highly differentiated cells exhibit reduced sustainability in vivo, as the proliferation and differentiation potentials of mature cells are highly restricted (31). To determine whether the immature and functional hIECs retained cell residual capacity in vivo, 0.5 to 1 × 107 cells were transplanted subcutaneously into the right and left flanks of recipient nude mice. After 6 to 10 weeks, all mice (10 of 10) injected with immature hIECs developed palpable masses, and 9 of 10 mice injected with functional hIECs developed subcutaneous masses (Fig. 4, C to E) with no significant difference in mass size between the immature and functional hIECs (Fig. 4F). We also assessed the presence of residual cells or further cellular differentiation after transplantation of hIECs using immunohistochemistry with human-specific antibodies. Two of 10 mice transplanted with only immature hIECs still contained hIEC-derived endodermal cells within the hIEC-Matrigel plugs, which expressed human-specific nuclear antigen (hNu), the intestinal transcription factor CDX2, intestinal protein VIL1, and the proliferation marker Ki-67 (Fig. 4G). In contrast, when functional hIECs were transplanted, none of the Matrigel plugs of mice contained any human cells even after prolonged in vivo cultivation (Fig. 4G), clearly demonstrating that functional hIECs did not leave any residual cells or differentiated further after in vivo transplantation.
Establishment of functional hIEC models from patient-specific iPSC lines
To reproducibly generate the patient-specific functional hIEC model, we used the developed protocol with iPSC lines independently established from two patients. Patient-specific fibroblasts obtained from the normal intestinal biopsy specimens were reprogrammed using a nonintegrating Sendai virus–based method (Fig. 5A). Two independent patient iPSC-derived cell lines were characterized for their morphological and molecular properties, karyotype, and differentiation potentials (fig. S6). The iPSCs efficiently differentiated into immature hIECs in hIEC medium 1 and further differentiated into functional hIECs in hIEC medium 2 (Fig. 5A). As expected, the expression level of ISC markers increased in iPSC-derived immature hIECs and decreased in functional hIECs (Fig. 5B). Accordingly, the iPSC-derived functional hIECs abundantly expressed the markers specific for intestinal cell types (Fig. 5B). The number of VIL+, LYZ+, MUC2+, and CHGA+ cells increased in the functional hIECs compared to immature hIECs (Fig. 5C). The functional hIECs formed well-defined polarized epithelial cell monolayers, as determined by their vertical cross sections (Fig. 5D). The TEER values of the functional hIECs differentiated from each iPSC line were significantly higher (232.68 ± 7.11 ohm × cm2 and 242.48 ± 7.12 ohm × cm2) than those of immature hIECs (128.52 ± 4.07 ohm × cm2 and 132.16 ± 5.31 ohm × cm2) (Fig. 5E). High expression of small intestinal drug transporters was also observed in functional hIECs (Fig. 5B). The expression and activity of CYP3A4 were higher in functional hIECs than in immature hIECs (Fig. 5, B and F). Thus, this newly developed culture system can support the generation of functional hIECs from patient-specific iPSCs and allow patient-tailored assays.
Fig. 5. Generation of functional hIECs derived from patient-specific iPSCs.
(A) Representative morphology of patient-derived intestinal samples from normal tissues, isolated fibroblasts, established iPSCs, and immature and functional hIECs differentiated from iPSCs. Black scale bars, 100 μm. Yellow scale bars, 20 μm. (B) Relative gene expression of intestinal markers in iPSC, immature hIECs, functional hIECs, Caco-2, and hSI. (C) Immunofluorescence analysis of the markers for enterocytes (CDX2 and VIL1), Paneth cells (LYZ), goblet cells (MUC2), and enteroendocrine cells (CHGA) in patient-specific immature hIECs and functional hIECs (top). White scale bars, 200 μm. Yellow scale bars, 50 μm. Data represent means ± SEM of each marker positive cell per field, n = 4 (bottom). (D) Confocal immunofluorescence analysis showing vertical cross sections of patient-specific immature and functional hIECs stained for the apical brush border marker (VIL1) and basolateral marker (Na+,K+-ATPase). Scale bars, 10 μm. (E) TEER values of patient-specific immature and functional hIEC monolayers. Data represent means ± SEM (n = 6). (F) Levels of CYP3A4 activity in patient-specific immature and functional hIECs and Caco-2 cells. Data are means ± SEM (n = 4). *P < 0.05, **P < 0.01, and ***P < 0.001 by two-tailed t test.
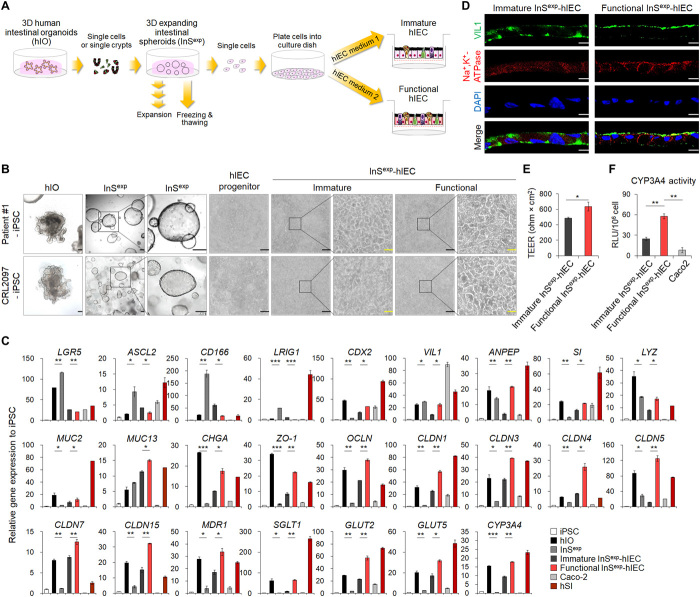
Development of 2D functional hIEC monolayer from 3D hIOs
3D hIOs have been widely used as an in vivo–like model system for the human intestinal epithelium (11, 32). However, 3D hIOs are not suitable for conventional assay systems, as the apical surface of hIOs is toward the inside of the 3D structure. Therefore, we attempted to apply our culture method for the transition of 3D hIOs to 2D hIEC monolayers (Fig. 6, A and B). To initiate the culture, we dissociated patient-specific iPSC-derived hIOs into single cell or single crypt and embedded them in a Matrigel dome to generate 3D expanding intestinal spheroids (InSexp). InSexp was enriched more in stem cells than hIOs, as was evident from the increased expression of ISC markers (Fig. 6C) and high proliferative activity (movie S1), which allowed further subculture and cryopreservation (fig. S7). For establishing 2D hIEC monolayer culture, the 3D InSexp was dissociated into single cells and seeded on the Matrigel-coated Transwell inserts. Monolayered InSexp-derived cells were maintained in InSexp culture medium for 7 days, followed by a change to hIEC medium 1 for differentiation into immature InSexp-derived hIECs (immature InSexp-hIECs) or to hIEC medium 2 for differentiation into functional InSexp-derived hIECs (functional InSexp-hIECs) (Fig. 6, A and B). After 7 days of differentiation, monolayer cells filled the inner chamber of the Transwell insert, and a more regular polygonal morphology was observed in functional InSexp-hIECs than in immature InSexp-hIECs (Fig. 6B). The expression of markers specific for absorptive and secretory cells was significantly higher in functional InSexp-hIECs than in immature InSexp-hIECs and was accompanied by decreased expression of ISC markers in functional InSexp-hIECs (Fig. 6C). Within 14 days of seeding, the functional InSexp-hIECs formed an apical-basolateral polarized monolayer, as evidenced by coimmunofluorescence for both apical (VIL1) and basolateral cell surface proteins (Na+,K+-ATPase) (Fig. 6D). Immature InSexp-hIEC and functional InSexp-hIEC monolayers exhibited an average TEER of 487.20 ± 13.86 and 635.41 ± 43.29 ohm × cm2, respectively, and enhanced expression of tight junction protein-coding genes was observed in functional InSexp-hIECs, indicating barrier integrity (Fig. 6, C and E). In addition, functional InSexp-hIECs displayed increased expression of intestinal transporters (Fig. 6C). The enzyme activity of CYP3A4 was also higher in functional InSexp-hIECs than immature InSexp-hIECs and Caco-2 cells (Fig. 6F). These data suggested that the newly developed hIEC culture system can be applied for 3D hIO-derived 2D monolayer hIEC culture. Moreover, immature InSexp-hIECs and functional InSexp-hIECs derived from control hIOs and in vitro–matured hIOs as reported in our previously work (11) maintained their maturation status as evident from the gene expression profile (fig. S8, A and B).
Fig. 6. Generation of 2D functional hIEC monolayer from 3D intestinal organoids.
(A) Schematic representation of the strategies for transitioning from hPSC-derived 3D hIO to 3D InSexp and 2D functional hIECs. (B) Morphological characterization of hPSC-derived hIOs, InSexp, hIECs progenitors, terminally differentiated immature hIECs, and functional hIECs. Black scale bars, 200 μm. Yellow scale bars, 40 μm. (C) Relative gene expression levels by qPCR of intestinal and tight junction markers in hPSC, hPSC-derived hIOs, InSexp, InSexp-derived immature and functional hIECs, Caco-2 cells, and hSI. (D) Immunofluorescence analysis showing vertical cross sections of patient-specific immature and functional hIECs stained for the apical brush border marker (VIL1) and basolateral marker (Na+,K+-ATPase). Nuclei were counterstained with DAPI (blue). Scale bars, 10 μm. (E) TEER values of patient-specific immature and functional hIEC monolayers. Data represent means ± SEM (n = 4). (F) Levels of CYP3A4 activity in patient-specific immature and functional hIECs and Caco-2 cells. Data are means ± SEM (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001 by two-tailed t test.
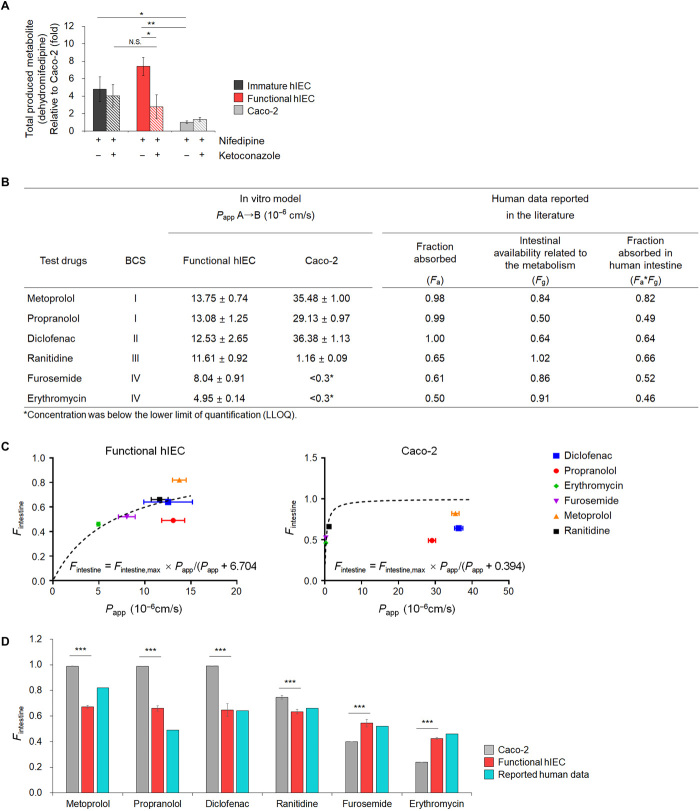
Functional hIECs as an in vitro model for predicting intestinal first-pass availability of drugs in humans
We studied the effect of the metabolic activity of CYP3A4 on the intestinal first-pass availability of nifedipine. CYP3A4-mediated metabolism of nifedipine was assayed and analyzed using liquid chromatography–tandem mass spectrometry (LC-MS/MS) to identify and quantify dehydro-nifedipine, the major active metabolite of nifedipine (33). Nifedipine was added to the monolayers of immature and functional hIECs and Caco-2 cells on a Transwell insert. After 2-hour incubation, the concentrations of dehydro-nifedipine increased by approximately 4.5-fold (P < 0.05) in the immature hIECs and by 7.4-fold (P < 0.01) in functional hIECs compared to that in Caco-2 cells (Fig. 7A). Upon pretreatment with 1 μM ketoconazole, a specific CYP3A4 inhibitor, CYP3A4 activity was sufficiently inhibited in immature as well as functional hIECs (fig. S9), and more than 62.5% CYP3A4-mediated metabolism of nifedipine was significantly inhibited in the functional hIECs (P < 0.01, Fig. 7A). In contrast, the concentrations of dehydro-nifedipine did not change significantly in immature hIECs and Caco-2 cells (Fig. 7A).
Fig. 7. Drug metabolizing activity of CYP3A4 using nifedipine and prediction of Fa × Fg in humans based on functional hIEC monolayers.
(A) Concentration of dehydro-nifedipine following 2 hours incubation in the absence or presence of ketoconazole, a specific CYP3A4 inhibitor, in immature hIECs, functional hIECs, and Caco-2 cells. Data represent means ± SD (n = 3). *P < 0.05 and **P < 0.01 by two-tailed t test. (B) Apical-to-basolateral permeability in functional hIECs and Caco-2 monolayers for six model drugs. Apparent permeability values (Papp) represent means ± SD (n = 3). Values for fraction absorbed by the human intestine (Fintestine) were obtained from literature. (C) The hyperbolic model fitted between the in vivo human Fintestine and observed Papp for model drugs in functional hIECs or Caco-2 cells. The dotted lines represent a fitted relationship curve, which was used to estimate Fintestine using the observed Papp. The equation for model structure was Fintestine = Fintestine,max × Papp(10−6 cm/s)/[Papp (10−6 cm/s) + Kd]; Kd represents the Papp at which Fintestine is 50% of Fintestine,max. (D) Comparison of predicted fraction absorbed by the human intestine [Fintestine; Fa (the fraction of orally administered drugs absorbed from the intestine) × Fg (the fraction of intestinal availability related to the metabolism)] of six model compounds in functional hIEC and Caco-2 cell system. Fintestine was calculated using the established model equation. The data represent means ± SD (n = 3). ***P < 0.001 by two-tailed t test.
Next, we evaluated the utility of the functional hIECs as a model for predicting human drug bioavailability by performing in vitro drug absorption assay using six test drugs. The Papp of metoprolol, propranolol, diclofenac, ranitidine, furosemide, and erythromycin was 13.75 ± 0.74, 13.08 ± 1.25, 12.53 ± 2.65, 11.61 ± 0.92, 8.04 ± 0.91, and 4.95 ± 0.14 (×10−6 cm/s), respectively, in functional hIECs, whereas it was 35.48 ± 1.00, 29.13 ± 0.97, 36.38 ± 1.13, 1.16 ± 0.09, <0.30, and <0.30 (×10−6 cm/s), respectively, in Caco-2 cells (Fig. 7B). Using the complete data, we determined the best fit model to estimate the fraction absorbed by the human intestine (Fintestine), expressed as Fa (fraction of orally administered drugs absorbed by the intestine) × Fg (fraction of intestinal availability related to the metabolism). Last, the hyperbolic model was used to determine the phenomenon for each system as Fintestine = Fintestine,max × Papp(10−6 cm/s)/[Papp (10−6 cm/s) + Kd]; where Kd represents the Papp at which Fintestine is 50% of Fintestine,max. The parameter (i.e., Kd) was estimated as 6.704 (% SE, 18.18%) and 0.394 (% SE, 72.63%) in functional hIECs and Caco-2 cells, respectively, whereas in vivo human Fintestine values were calculated from the reported values in the literature (34), and Fintestine,max was fixed as 1 (i.e., theoretical maximum Fintestine value) (Fig. 7C). Using the established corresponding model equation, the mean Fintestine values of metoprolol, propranolol, diclofenac, ranitidine, furosemide, and erythromycin were 0.67, 0.66, 0.65, 0.63, 0.55, and 0.42, respectively, in hIECs and 0.99, 0.99, 0.99, 0.75, 0.43, and 0.43, respectively, in Caco-2 cells (Fig. 7D). The functional hIECs correlated well with the published human in vivo Fintestine, suggesting that functional hIECs can better predict the absorption and extent of oral bioavailability in humans.
DISCUSSION
In this study, we aimed to develop a reliable and robust method for obtaining high-performance hIECs directly derived from hESCs and patient-specific iPSCs in a monolayer arrangement, which closely resembling the function and diversity of cell types observed in vivo. This method overcomes the limitations of the 3D structure of hIOs, which show limited accessibility to the apical epithelial surface, rendering them unsuitable for standard and high-throughput intestinal functional assays designed for monolayer formation.
We identified that treatment with a combination of R-spondin 1 and insulin promoted differentiation of hPSCs into hIEC progenitors and that their progenitor-like properties were stably maintained after continuous passaging and cryopreservation. R-spondin 1 promotes differentiation into hPSC-derived hIECs by strengthening the Wnt signaling pathway (8, 9). In addition, activation of Wnt signaling by R-spondin 1 increased the ISC population, as shown by the increase in the expression of ISC makers, and four major intestinal cell types (Fig. 1C). In contrast, insulin specifically increased enterocyte marker gene expression (Fig. 1C), which is possibly due to the selective differentiation from ISCs to enterocyte via insulin signaling. A previous report showed that reduced insulin signaling (23) in ISCs of the Drosophila adult midgut delayed the production of enterocytes, although the number of enteroendocrine cells from ISCs was not affected. Generation of lineage-committed expandable progenitors is an effective approach for ensuring sufficient quantity and quality of cell sources for various biomedical applications. Recent examples of this concept include generation of foregut stem cells (35) and multipotent endoderm progenitors (36) from hPSCs. The hIEC progenitors also contain proliferating progenitor-like cells that express ISC makers and are suitable for further intestinal differentiation.
The DAPT-mediated Notch pathway inhibition is mainly used to increase the efficiency of intestinal epithelial differentiation from hPSC in a 2D culture system (7, 8, 22). However, Notch inhibition is not an ideal strategy for producing high-performance hIEC from hPSCs as it inhibits self-renewal of ISCs and differentiation into absorptive cells, the main cell types of the intestinal epithelium (22). In contrast, Notch activators increase the ISC population when Wnt is turned on and promote differentiation into absorptive cells when Wnt is turned off (21). We treated cells with Wnt-C59 for Wnt inhibition and with VPA for Notch activation to maximize differentiation into absorptive cells (16, 21). Unexpectedly, when treated with Wnt-C59 and VPA in hIEC medium 2, hIEC progenitors showed simultaneously increased ability to differentiate into absorptive and secretory cell types that are present in vivo (Fig. 1, D and E, and fig. S2). This discrepancy may be due to the maintenance of basal activity of NOTCH pathway upon cotreatment of WNT C-59 and VPA (Fig. 1F). The activity of the NOTCH signaling pathway is probably regulated by the WNT signaling pathway. Although the underlying molecular mechanism of cross-talk between WNT and NOTCH pathway remains to be determined, one possible explanation is the suppression of NOTCH ligand production by WNT C-59 treatment. In colorectal cancer cells, the generation of NOTCH ligands such as DLL1 and JAG-1 is caused by activation of the WNT/β-catenin signaling pathway (37). Therefore, the inhibition of the WNT/β-catenin signaling pathway by WNT C-59 treatment may have reduced the production of the NOTCH ligands, thereby lowering the activity of the NOTCH pathway despite VPA treatment. In line with this, we propose that the stemness of hIEC progenitors is lost and differentiation begins when the WNT signaling pathway is inhibited by WNT C-59 treatment, and hIEC progenitors differentiate into absorptive and secretory lineage cells depending on the NOTCH pathway activity.
Approximately 90% of marketed drugs are administered orally, and therefore, it is important to predict their oral bioavailability in humans, which is regulated by absorption and gut metabolism. The functional hIECs developed in this study showed higher expression of drug transporters and metabolic enzymes (Figs. 3C, 5B, and 6C) compared to immature hIECs and Caco-2 cells. Functional hIEC monolayers generated via both directed differentiation and transition from 3D hIOs showed effective barrier function as evident by the increased expression level of tight junction molecules and polarized localization of the structural proteins (fig. S3, A to C). Although the functional hIECs differentiated closely to hSI compared to the immature hIECs and Caco-2 cells, the TEER value was slightly higher than that of hSI (40 to 100 ohm × cm2) (38). This appears to be attributed to low cellular diversity in functional hIECs (39). Therefore, there is a need to develop a differentiation method to differentiate functional hIECs into cells that resemble hSI in vivo.
Activity of the glucose transporter and metabolic enzymes, including IAP and CYP3A4, was high in the functional hIECs (Figs. 3, C to H, 5F, and 6F). Furthermore, the level of dehydro-nifedipine, formed when nifedipine is metabolized by CYP3A4, was significantly high in the functional hIEC monolayers (Fig. 7A). The functional hIEC system also showed better prediction of intestinal absorption fraction (Fintestine) than the Caco-2 system using model drugs with various physicochemical properties based on their solubility and permeability by Biopharmaceutical Classification System (BCS); metoprolol and propranolol (BCS I), diclofenac (BCS II), ranitidine (BCS III), and furosemide and erythromycin (BCS IV) (Fig. 7, C and D). In case of drugs with low permeability, it was difficult to determine the drug concentration transported by the Caco-2 monolayer because their concentration was very low, whereas no issues were faced in determining the drug concentration in functional hIECs (Fig. 7B), suggesting that hIECs are more appropriate for assaying drugs with low permeability in BCS IV. A large number of drugs showing Fg < 0.8 in the human intestine should be considered products of intestinal first-pass metabolism while predicting bioavailability and effective dose in drug discovery processes (34). The low expression level of CYP3A4 in Caco-2 cells leads to the difficulty in evaluating CYP3A4-mediated drug metabolism, an important factor in oral drug bioavailability; in addition, the permeability of hydrophilic drugs through the paracellular route is considerably low due to the absence of goblet cells (40). On the basis of these results, we propose that functional hIECs may be the most suitable in vitro small intestinal model system for evaluating drug bioavailability by overcoming the limitations of the Caco-2 monolayer, the gold standard for studying the absorption of new drugs.
Although the transcriptome profile of functional hIEC shifted extensively toward 3D hIOs, recapitulating the cellular diversity and specific functions of hSI, the whole transcriptome RNA-seq analysis revealed gaps between the functional hIECs and hSI. In case of hIOs, in vitro maturation by exposure to interleukin-2 (IL-2) enables the acquisition of intestinal traits, including more complex organ-like structures, high expression of mature intestinal markers, and increased intestine-specific functionalities (11). 3D hIOs contain the distinct closed luminal space in which intestinal epithelium exhibits polarity, thereby facilitating the quantification of the directional absorption of nutrients from the apical to the basolateral side, as it occurs in vivo. Therefore, further studies are required to build more accurate in vitro hSI monolayer models resembling the spatial organization of crypt-villi units and exhibiting distinctive gene expression patterns of the hSI. An in vitro hSI model system should be chosen according to the objective and purpose of the study considering its pros and cons. The functional hIECs cultured as 2D monolayers can be easily used for various conventional intestinal assays. From this perspective, this system is particularly useful for transitioning from 3D hPSC-derived hIOs to 2D hIEC monolayers, as well as directed differentiation of patient-derived iPSCs into hIEC monolayers, thereby enabling patient-tailored precise approaches (fig. S10).
In conclusion, we have successfully developed functional hIECs exhibiting in vivo–like cellular organization, which may be used as a platform for evaluating drug bioavailability and first-pass metabolism. The stability of the hIEC differentiation protocol was confirmed by the increase in the levels of H3K4me3 and H3K27ac and the absence of further differentiation following transplantation. Hence, we propose that the reliable and robust hIEC culture system described here, which leads to the generation of small intestinal epithelial monolayers with close to in vivo functionality, can be used to replace the current in vitro models and provide a versatile experimental platform.
MATERIALS AND METHODS
Isolation of human small intestinal fibroblasts, generation of iPSCs, and cell culture
The human small intestinal tissues were obtained from two adults during routine endoscopy, which was approved by the Institutional Review Board of Chungnam National University Hospital (IRB file no. CNUH 2016-03-018); informed consent was obtained from both patients. The tissue samples were digested using the collagenase type I (Thermo Fisher Scientific) for 3 hours in a 37°C shaking incubator and centrifuged. The pellets were washed, plated on a 0.2% gelatin-coated dish, and incubated in minimal essential medium (MEM; Thermo Fisher Scientific Inc.) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific Inc.), 1% penicillin and streptomycin (P/S; Thermo Fisher Scientific Inc.), and 1 mM nonessential amino acids (NEAAs; Thermo Fisher Scientific Inc.). The isolated fibroblasts were reprogrammed into iPSCs using the CytoTune-iPS 2.0 Sendai reprogramming kit and characterized as described previously (11). The H9 hESC line (WiCell Research Institute, Madison, WI, USA) and iPSCs were routinely cultured as described previously (11). Caco-2 cells (American Type Culture Collection, Manassas, VA, USA) were cultured using standard culture protocol in MEM supplemented with 10% FBS, 1% P/S, and 1% NEAA. For monolayer experiments, the Caco-2 cells (1.34 × 105 cells/cm2) were seeded on 5% Matrigel (Corning, NY, USA)–coated Transwell insert, and the medium was replaced on alternate days.
Differentiation of hPSCs into hIECs via hIEC progenitors
To induce formation of the DE, well-maintained hPSCs with more than 70% confluence were treated with activin A (100 ng/ml; R&D Systems, Minneapolis, MN, USA) for 3 days in RPMI 1640 medium (Thermo Fisher Scientific Inc.) containing 2 mM l-glutamine (Thermo Fisher Scientific Inc.), 1% P/S, and increasing concentration of 0, 0.2, or 2% FBS. The DE cells were almost confluent after 3 days of differentiation. The cells were further differentiated into HG by treating with FGF4 (250 ng/ml) (Peprotech, Rocky Hill, NJ, USA) and 1.2 μM CHIR99021 (Tocris Bioscience, Minneapolis, MN, USA) in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) (Thermo Fisher Scientific) supplemented with 2 mM l-glutamine, 1% P/S, and 2% FBS for 4 days. For differentiation into hIEC progenitors, the HG cells were prepared in single-cell suspension following treatment with trypsin-EDTA for 5 min at 37°C. Single HG cells were neutralized with DMEM/F12 containing 2% FBS and harvested. After centrifugation for 5 min at 1250 rpm, cell pellet was resuspended and reseeded onto 1% Matrigel-coated tissue culture plate in hIEC differentiation medium 1 (hIEC medium 1) containing DMEM/F12, epithelial growth factor (EGF) (100 ng/ml) (R&D Systems), R-spondin1 (100 ng/ml) (Peprotech), insulin (5 μg/ml) (Thermo Fisher Scientific Inc.), 2% FBS, 2% B27 supplement (Thermo Fisher Scientific Inc.), 1% N2 supplement (Thermo Fisher Scientific Inc.), 2 mM l-glutamine, 1% NEAA, and 15 mM Hepes buffer (Thermo Fisher Scientific Inc.). The hIEC medium 1 was replaced every other day. The hIEC progenitors were passaged after every 7 days. To passage, 7 days cultured hIEC progenitors were dissociated by treatment with trypsin-EDTA for 5 min at 37°C. Harvested hIEC progenitors were neutralized with DMEM/F12 containing 2% FBS before centrifugation for 5 min at 1250 rpm. Cell pellet was resuspended in hIEC medium 1 and reseeded onto 1% Matrigel-coated tissue culture plate in a 1:3 passage ratios. The hIEC progenitors were cultured for up to 10 passages without observing any significant differences in characteristics. For cryopreservation of the hIEC progenitors, the cells were digested with trypsin-EDTA and slowly frozen in CryoStor CS10 freezing medium until below −70°C for 24 hours. The frozen cells were stored at −196°C liquid nitrogen. To differentiate the hIEC progenitors into immature hIECs and functional hIECs, 1.34 × 105 cells/cm2 hIEC progenitors were reseeded onto 1% Matrigel-coated Transwell (Corning) inserts in hIEC medium 1 containing 10 μM Y-27632 (Tocris) and incubated until the cells were near confluence. Once confluent, the medium was replaced with either hIEC medium 1 or hIEC differentiation medium 2 (hIEC medium 2) containing DMEM/F12, EGF (100 ng/ml), 2 μM Wnt-C59 (Selleckchem, Huston, TX, USA), 1 mM VPA (Stemgent, Huston, TX, USA) acid, 2% FBS, 2% B27 supplement, 1% N2 supplement, 2 mM l-glutamine, 1% NEAA, and 15 mM Hepes buffer for differentiation into functional hIECs. The medium was changed every other day, and immature and functional hIECs were cultured for 10 to 14 days for further analysis. TEER values of immature and functional hIEC monolayers were determined after reaching complete confluence, and the immature and functional hIEC monolayers were maintained for up to 21 days with a survival rate of >90%.
Culture and differentiation of hIO-derived 3D InSexp into immature and functional hIECs
hPSC-derived hIOs were differentiated and matured in media containing 1 ng/ml of IL-2 (R&D Systems) as described by Jung et al. (11). The hIOs were incubated in trypsin-EDTA for 5 min and mechanically dissociated using 10 rounds of pipetting. The dissociated hIOs were resuspended in 10 ml of basal media and centrifuging at 1500 rpm, 4°C, for 5 min. The supernatant was discarded and the pellet was resuspended in Matrigel. The hIO-Matrigel mixture was plated onto a four-well culture plate and incubated at 37°C in a CO2 incubator for 10 min to solidify the Matrigel, followed by the addition of InSexp culture medium containing DMEM/F12, 2 mM l-glutamine, 15 mM Hepes buffer, 2% B27 supplement, 10 nM [Leu-15]-gastrin I (Sigma-Aldrich, St. Louis, MO, USA), human recombinant WNT3A (100 ng/ml) (R&D Systems), EGF (100 ng/ml), Noggin (100 ng/ml) (R&D Systems), R-spondin1 (100 ng/ml), 500 nM A-83-01 (Tocris), 500 μM SB202190 (Sigma-Aldrich), 10 nM prostaglandin E2 (Sigma-Aldrich), 1 mM N-acetylcysteine (Sigma-Aldrich), 10 mM nicotinamide (Sigma-Aldrich), 10 μl of Y-27632 (Tocris), and 1 μM Jagged-1 (AnaSpec, Fremont, CA, USA) for the first 2 days. Subsequently, the medium was replaced with the InS culture medium every 3 days. For differentiation of 3D InS into immature and functional hIECs, 3D InS were dissociated with trypsin-EDTA and seeded on 1% Matrigel-coated plates or Transwell inserts in InS culture medium supplemented with 10 μl of Y-27632 and 1 μM Jagged-1 for the first 2 days. The InS culture medium was replaced every 2 days until the cells reached near confluence, after which the InS culture medium was changed into either hIEC medium 1 or hIEC medium 2. The medium was replaced every other day.
Measurement of TEER
TEER measurement was performed using epithelial tissue volt/ohmmeter (EVOM2, WPI, Sarasota, FL, USA) according to the manufacturer’s instructions.
Transepithelial permeability assay
For permeability assays, immature, functional hIECs and Caco-2 cells grown on Transwell inserts for 14 days were used. The integrity of cell monolayers was evaluated by measuring TEER values on the day of the experiment. The TEER value ranges for immature hIEC, functional hIEC, and Caco-2 cell monolayers were 175 to 225 ohm × cm2, 195 to 265 ohm × cm2, and 230 to 330 ohm × cm2, respectively. A total of 500 μl of fluorescein isothiocyanate-dextran 4 kDa (FD4; 100 μg/ml; Sigma-Aldrich) or 40 kDa (FD40; Sigma-Aldrich) diluted in Hank’s balanced salt solution (HBSS) was applied to the apical side, and 1.5 ml of HBSS was added into basolateral side. The permeability assay was performed at 37°C for 120 min in the dark. After incubation, 100 μl of the supernatant was taken from the basolateral side and analyzed with a SpectraMax M3 microplate reader (Molecular Devices, Sunnyvale, CA, USA) at excitation and emission wavelength of 485 and 520 nm. The standard curve was obtained by evaluating serial dilutions of FD4 or FD40.
Western blot analysis
The IECs were lysed with radioimmunoprecipitation assay buffer (Sigma-Aldrich) supplemented with 1 mM protease inhibitor, 1× phenylmethylsulfonyl fluoride (Thermo Fisher Scientific), and 1× phosSTOP (Roche, Indianapolis, IN, USA). A total of 20 μg of protein was separated on precast gel (4 to 20% gradient; Bio-Rad Laboratories, Hercules, CA, USA) and transferred to methanol (Merck Millipore)–activated polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA, USA). Blocked membranes were incubated overnight with the appropriate primary antibodies diluted in Tris-buffered saline with 0.05% Tween ® 20 Detergent (TBST) buffer containing 4% bovine serum albumin (BSA). The membranes were washed and incubated with appropriate horseradish peroxidase–conjugated secondary antibody, and the protein band was analyzed using the LAS-3000 analyzer (Fuji Photo Film GMBH, Tokyo, Japan).
FACS analysis
Fluorescence-activated cell sorting (FACS) analysis was performed to determine the content of differentiated intestinal cell populations during hIEC differentiation. BD Pharmingen Transcription-Factor Buffer Set (BD Biosciences) was used to fix and permeabilize hIECs. The harvested immature and functional hIECs were fixed and permeabilized with Fix/Perm Buffer for 40 min at 4°C. Cells were then washed three times with Perm/Wash Buffer and blocked with blocking solution containing 0.5% BSA and 2% FBS for 40 min at 4°C. Subsequently, primary antibodies against the specific markers of each intestinal cell type (table S3) were diluted in blocking solution in recommended concentration, added to cells, and incubated for 1 hour at 4°C. After washing three times with Perm/Wash Buffer, cells were incubated with secondary antibodies for 1 hour at 4°C. After reaction, hIECs were washed three times with Perm/Wash Buffer, resuspended in phosphate-buffered saline (PBS), subjected to FACS on Accuri C6 Flow Cytometer (BD Biosciences), and analyzed with FlowJo V10 software (BD Biosciences).
Cell viability assay
To calculate the viability of the hIECs, immature and functional hIECs were dissociated to single cells using 0.25% trypsin-EDTA for 5 min at 37°C. Next, the single hIECs were neutralized with DMEM/F12 containing 2% FBS and centrifuged for 5 min at 1250 rpm. The hIECs were resuspended in DMEM/F12 containing 2% FBS and stained with trypan blue. EVE Automatic Cell Counter (Nanoenteck, Seoul, Korea) was used to calculate cell viability.
Scanning electron microscopy
Immature hIECs, functional hIECs, and Caco-2 cells cultured for 14 days in Transwell inserts were washed three times with PBS and fixed with 2.5% SEM grade glutaraldehyde (Sigma-Aldrich) for 2 hours at 4°C. After primary fixation, the cells were washed three times with PBS, and the secondary fixation was performed using osmium tetroxide (OsO4; Sigma-Aldrich) for 2 hours at 4°C. The used OsO4 was replaced with fresh OsO4, and fixed cells were further incubated for 1 hour at 4°C. The secondary fixed samples were washed twice with distilled water and dehydrated in increasing concentrations of ethanol (30, 50, 70, 80, 90, and 100%) for 5 min each at room temperature. Hexamethyldisilazane (Sigma-Aldrich) was added to cells, and samples were dried in a fume hood over night at room temperature. Next, the membranes were excised out of the cell inserts, fixed onto the carbon tape, and sputter-coated with platinum for 60 s each in the upright position and on both sides. Images of prepared samples were acquired using the field-emission SEM instrument (Inspect F50, FEI, USA).
Live calcium imaging with Fluo-4 AM
The hIECs and Caco-2 cells were seeded in glass bottom confocal dishes and treated with 5 μM Fluo-4 AM (Thermo Fisher Scientific Inc.) for 1 hour. The cells were washed thrice with Ca2+-free isotonic buffer (140 mM NaCl, 5 mM KCl, 10 mM Hepes, 5.5 mM d-glucose, and 2 mM MgCl2) and transferred to the stage of a confocal microscope (FV1000 Live, Olympus, Japan). The cells were stimulated with 50 mM glucose (Sigma-Aldrich) in Ca2+-free isotonic buffer and excited at 488 nm, and the signals were recorded at 505 to 530 nm. The fluorescence intensity of the region of interest was calculated using the FV1000 software (Olympus).
Alkaline phosphatase assay
Alkaline phosphatase activity was quantified using the alkaline phosphatase assay kit (ab83369; Abcam, Cambridge, UK) according to the manufacturer’s instructions. Briefly, the cell culture medium was harvested from 14-day-old cultured hIECs and Caco-2 cells and diluted (1:10) with assay buffer. Sample (80 μl) and 50 μl of 5 mM para-nitrophenyl phosphate solution were mixed and added to each well, and the plate was incubated at 25°C for 60 min in the dark. Next, 20 μl of stop solution was added to each well, and the absorbance of the plate was measured at 405 nm on a SpectraMax M3 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
CYP3A4 activity assay
CYP3A4 assay was conducted using the P450-Glo CYP3A4 assay kit (V9002; Promega, Madison, WI, USA) according to the manufacturer’s instructions. hIECs and Caco-2 cells cultured for 14 days were treated with 3 μM Luciferin-IPA at 37°C for 60 min. The harvested supernatant was transferred to a 96-well opaque white luminometer plate. An equal volume of luciferin detection reagent was added to each well and incubated at room temperature for 20 min. The luminescence was measured using a SpectraMax M3 microplate reader.
Quantitative reverse transcription-polymerase chain reaction
Total RNA was harvested, and cDNA was prepared using the RNeasy kit (Qiagen) and Superscript IV cDNA synthesis kit (Thermo Fisher Scientific Inc.), respectively. RNA extracted from adult hSI was purchased from Clontech (Fremont, CA, USA) and used as a positive control. Quantitative polymerase chain reaction (qPCR) was performed using a 7500 Fast Real-Time PCR system (Applied Biosystems, Foster city, CA, USA). Relative expression was calculated using the ΔΔCt method. The primers used in this study are listed in table S2.
Chromatin immunoprecipitation
ChIP was performed using the Magna ChIP A/G kit (Magna0013 and Magna0014; Millipore, Billerica, MA, USA), following the manufacturer’s instructions. hIECs were cross-linked with 1% formaldehyde (Sigma-Aldrich) for 10 min at room temperature, and the reaction was stopped with 1× glycine (Millipore) for 5 min at room temperature. Then, the cells were washed with cold 1× PBS (containing 1× protease inhibitor cocktail II). After nuclear extraction, the chromatin solution was sonicated using the Bioruptor Pico sonication device (B01060010; Diagenode, Belgium) with 20 cycles of 30 s on and 30 s off to obtain 200 to 1000 bp chromatin fragments. The sheared chromatin was incubated with 2 μg of anti-H3K4me3 (ab8580; Abcam, Cambridge, MA, USA) antibody, 2 μg of anti-H3K27ac (ab4729; Abcam) antibody, or 2 μg of normal rabbit immunoglobulin G (IgG) (2729S; Cell Signaling Technology Inc., Danvers, MA, USA) and 20-μl Magna ChIP A/G magnetic beads (Millipore) overnight at 4°C. The complexes were washed with washing buffer using a magnetic separation device and incubated with ChIP elution buffer and ribonuclease A mixture for 30 min at 37°C. Next, they were incubated with proteinase K for 120 min at 62°C. After DNA purification using spin columns, the samples were analyzed using qPCR. The primers used in this experiment are listed in table S2.
Immunofluorescence analysis
Cells were washed and fixed with 4% paraformaldehyde, cryopreserved in sucrose (10 to 30%), and embedded in the optimal cutting temperature (O.C.T.) compound (Sakura Finetek, Tokyo, Japan). For vertical sections, the frozen blocks were cut into 10-μm thickness using a cryostat microtome at −30°C. For immunofluorescence analysis, the cells were permeabilized with PBS containing 0.1% Triton X-100 and blocked with 4% BSA solution. The cells were incubated overnight with specific primary antibodies at 4°C. After incubation, the cells were washed with PBS containing 0.05% Tween 20 (Sigma-Aldrich) and incubated with the secondary antibody, followed by measurement of the fluorescent signal. The nucleus was labeled with 4′,6-diamidino-2-phenylindole (DAPI; 1 mg/ml, Thermo Fisher Scientific Inc.). The fluorescence was examined using a confocal microscope (LSM800, Carl Zeiss, Oberkochen, Germany) and a fluorescence microscope (IX51, Olympus, Japan). For the transplantation assay, the paraffin sections were deparaffinized and stained in a manner similar to that used for antigen detection in the frozen specimens. The transplant samples were observed using an EVOS microscope (FL Auto 2, Thermo Fisher Scientific Inc.). For the 3D image, the Transwell membrane were fixed with methanol (Merck Millipore) for 6 min at −20°C and blocked with 4% BSA solution. The membrane was incubated overnight with optimal primary antibodies at 4°C followed by incubation with fluorescence conjugated secondary antibody. The mounted cells were scanned with a LSM800 confocal microscope (Carl Zeiss, Gottingen) with Z-stack series capability. 3D images were assembled with ZEN software (Carl Zeiss). The list of primary antibodies used is provided in table S3.
Transplantation assay
Male BALB/c nude mice, aged 6 to 7 weeks, were purchased from the Jackson laboratory (Bar Harbor, ME, USA). Mice were housed in a standard animal maintenance facility under a 12-hour light:12-hour dark cycle. For subcutaneous injection, 5 to 10 × 106 immature or functional hIECs were mixed with 200 μl of Matrigel and implanted subcutaneously. The transplants were monitored over 6 to 10 weeks. The resulting hIEC-Matrigel plugs were surgically removed from the mice and fixed with 10% formaldehyde. The hIEC-Matrigel plugs were embedded in the O.C.T. compound, cut into 10-μm thickness using a cryostat microtome at −30°C, and stained with relevant primary antibodies. The list of primary antibodies used is provided in table S3. All animal studies were approved by the Institutional Animal Care and Use Committee of the Korea Research Institute of Bioscience and Biotechnology (approval no: KRIBB-AEC-19110).
Bacteria adhesion analysis
Bacteria adhesion analysis was performed using immature hIECs, functional hIECs, and Caco-2 cells on a Transwell system. mCherry-expressing L. plantarum were cultured in De Man, Rogosa, and Sharpe (MRS, Gibco) medium containing erythromycin (5 ng/ml) (Sigma-Aldrich) for 37°C under anaerobic conditions. For bacteria adhesion analysis, freshly cultured bacteria were harvested and diluted to 1 × 109 cells in culture medium and added to the IECs and Caco-2 cells. After 2 hours, the cells were washed three times with PBS to remove unbound bacteria, followed by treatment with 0.25% trypsin-EDTA (Gibco). Harvested cells were serially diluted and plated on MRS agar plates, and colony-forming units were enumerated. The images were captured using an LSM800 confocal microscope (Carl Zeiss, Gottingen).
Macrophage migration assay
hIECs were seeded on Transwell insert and differentiated for 14 days. The human THP-1 cell line was cultured in RPMI 1640 (Gibco) supplemented with 10% FBS (Gibco), 1% P/S (Gibco), and 1 × β-mercaptoethanol (Gibco). For macrophage differentiation, THP-1 cells (1.5 × 106 cells per well) were seeded in the 12-well plate (Corning) and incubated with culture medium containing phorbol 12-myristate 13-acetate (50 μg/ml) (Sigma-Aldrich) without β-mercaptoethanol. After 48 hours, the medium was replaced with THP-1 culture medium and incubated for 24 hours. Next, differentiated THP-1 were digested, reseeded (3 × 106 cells) onto the bottom surface of the Transwell inserts, and incubated for overnight at 37°C in 5% CO2 to allow cell adherence. The Transwell inserts were transferred into the original 12-well plate. For macrophage migration, the cells were stimulated with liposaccharide (LPS; 1 μg/ml; Sigma-Aldrich) for 24 hours.
Assay to assess CYP3A4-mediated metabolism
hIEC progenitors and Caco-2 cells (1.34 × 105 cells/cm2) were reseeded onto 1% Matrigel-coated Transwell (Corning) inserts in fresh culture media and cultured for 14 days. Before drug treatment, the TEER values were measured to assess cell condition, and only cells with TEER value > 200 ohm × cm2 were used. For CYP3A4 inhibition, the cells were incubated in culture medium containing 1 μM ketoconazole for 2 hours at 37°C before conducting the CYP3A4-mediated metabolism assay. The cells were washed three times with transport buffer containing 1× HBSS (Thermo Fisher Scientific Inc.), sodium bicarbonate (0.35 g/liter; Sigma-Aldrich), and 10 mM Hepes (Thermo Fisher Scientific Inc.). Five hundred microliters of the transport buffer containing 5 μM nifedipine (Sigma-Aldrich) was added to the apical side of the Transwell, and 1.5 ml of transport buffer was added to the basolateral side of the Transwell. After 2-hour incubation, the supernatants of the apical and basolateral sides were collected separately in fresh Eppendorf tubes. The concentrations of nifedipine and dehydro-nifedipine in each supernatant were quantitated using liquid chromatography–electrospray ionization/mass spectrometry analysis on a 4000 QTRAP LC-MS/MS system (Applied Biosystems) equipped with a Turbo VTM ion spray source and an Agilent 1200 series high-performance liquid chromatography system (Agilent Technologies, Palo Alto, CA, USA).
Measurement of Papp values
The cells were prepared as described in the previous section. The integrity of the hIEC progenitor and Caco-2 cells was evaluated by measuring TEER values on the day of the experiment, and TEER values ranged between 175 to 225 ohm × cm2, 195 to 265 ohm × cm2, and 230 to 330 ohm × cm2 for immature hIEC, functional hIEC, and Caco-2 cell monolayers, respectively. The hIEC progenitors and Caco-2 cells were washed three times with the transport buffer. For the permeability assay, test compounds 10 μM metoprolol (Sigma-Aldrich), propranolol (Sigma-Aldrich), diclofenac (Sigma-Aldrich), 20 μM ranitidine (Sigma-Aldrich), 20 μM furosemide (Sigma-Aldrich), or 20 μM erythromycin (Sigma-Aldrich) in 500 μl of transport buffer were added to the apical side of the Transwell, and 1.5 ml of transport buffer was added to the basolateral side of the Transwell. After 2-hour incubation, the supernatants from the apical and basolateral sides were collected separately in fresh Eppendorf tubes. The concentration of the test compounds in the samples was determined using LC-MS/MS. The apparent permeability coefficient was calculated according to the following equation
where dQ/dt, A, and C0 represent the rate of transport, the surface area of the insert, and the initial concentration of the compound in the donor compartment, respectively. Chromatographic quantification of the compounds was performed using an LC-MS/MS system equipped with a Shimadzu Prominence Ultra Performance Liquid Chromatography (UPLC) system (Shimadzu, Kyoto, Japan) and an API 2000 QTRAP mass spectrometer (Applied Biosystems, Foster City, CA, USA). Briefly, an aliquot (50 μl) of the samples was vortex-mixed with an acetonitrile solution containing an internal standard (50 ng/ml carbamazepine for metoprolol, ranitidine, and propranolol; 500 ng/ml 4-methylumbelliferone for diclofenac) and centrifuged at 3000g for 10 min at 4°C. Subsequently, an aliquot (10 μl) of the supernatant was directly injected into the LC-MS/MS system and separated using a gradient of 0.1% formic acid in acetonitrile and 0.1% formic acid in water (flow rate: 0.4 ml/min) using a Waters XTerra MS C18 column (2.1 mm by 50 mm, 5 μm; Milford, MA, USA). The following transitions were used for detecting the analytes: mass/charge ratio (m/z) 268.0 → 116.2 for metoprolol, m/z 260.00 → 56.00 for propranolol, m/z 294.00 → 250.10 for diclofenac, m/z 314.90 → 176.10 for ranitidine, m/z 329.06 → 204.80 for furosemide, m/z 734.46 → 576.3 for erythromycin, m/z 237.0 → 194.0 for carbamazepine, and m/z 175.0 → 119.0 for 4-methylumbelliferone.
RNA sequencing
RNA samples were analyzed on an Agilent 2100 Bioanalyzer system (Agilent Biotechnologies). Only high-quality RNA samples (RNA integrity number ≥ 7.5) were used for preparing the samples for sequencing. Libraries were prepared using Illumina TruSeq library preparation according to manufacturer specifications. RNA-seq was performed on Illumina HiSeq2500 (Illumina, San Diego, CA, USA) following the standard Illumina RNA-Seq protocol with a read length of 2 × 100 bases.
Bioinformatic analysis
The sequence data were evaluated using the NGSQCToolkit v.2.3.3 and the adapters were removed using Cutadapt v.1.18 with default settings; low-quality sequences were trimmed using Sickle v.1.33 with a Phred quality threshold score of 20. If the trimmed read contained any ambiguous character (such as N) or was less than 50 bp, it was excluded. After preprocessing of raw reads, the clean reads were mapped to the reference genome (GRCh38) using HISAT2 v.2.0.5 with default parameter settings and applying StringTie v.2.1.0 using the reference annotation file for estimating the expression levels of all genes and transcripts. Using all protein-coding genes, the MDS analysis was performed to cluster the samples according to their overall similarity of gene expression pattern to determine whether the gene expression pattern between the phenotype classes could be clearly distinguished. The log2 transformation values were used for this analysis, and rows with zero expression in all samples were eliminated. The MDS analysis of the pairwise distances of the sample was conducted using the function “dist” (maximum distance measure) in the R v.4.0.2 statistical programming language and plotted using the R package. In addition, hierarchical clustering was performed using the Maximum distance by the “hclust” function of the stats package v.3.6.2 in R. Adult small intestine RNA-seq data were downloaded from the public database under accession E-MTAB-1733.
Statistical analysis
All experiments were repeated at least three times, and results are presented as means ± SEM. Two-tailed Student’s t test was performed to determine the significance of the data statistically.
Acknowledgments
Funding: This work was supported by a grant from the Technology Innovation Program (nos. 20008777 and 20009774) funded by the Ministry of Trade, Industry and Energy (MOTIE, Korea); grant from the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning (NRF-2018M3A9H3023077); and the KRIBB Research Initiative Program. The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript. Author contributions: M.-Y.S., C.-R.J., and J.K. contributed to the study concept and design; O.K., K.B.J., K.-R.L., Y.S.S., H.L., J.-J.K., K.K., S.L., Y.-K.S., J.J., K.P., S.J.O., and H.C. performed experiments and acquired samples; M.-Y.S., K.-R.L., M.-O.L., T.-S.H., D.-S.K., H.-S.C., M.J.S., K.S.C., and S.-H.K. analyzed and interpreted the data; M.-Y.S., K.-R.L., and O.K. wrote the entire manuscript; all authors contributed to the draft manuscript and the critical revision of the manuscript and approved the final version. Competing interests: The other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabh1586/DC1
REFERENCES AND NOTES
- 1.Maloy K. J., Powrie F., Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Sinagoga K. L., Wells J. M., Generating human intestinal tissues from pluripotent stem cells to study development and disease. EMBO J. 34, 1149–1163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martignoni M., Groothuis G. M. M., de Kanter R., Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2, 875–894 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Kato M., Intestinal first-pass metabolism of CYP3A4 substrates. Drug Metab. Pharmacokinet. 23, 87–94 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Takenaka T., Harada N., Kuze J., Chiba M., Iwao T., Matsunaga T., Application of a human intestinal epithelial cell monolayer to the prediction of oral drug absorption in humans as a superior alternative to the Caco-2 cell monolayer. J. Pharm. Sci. 105, 915–924 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Jung P., Sato T., Merlos-Suárez A., Barriga F. M., Iglesias M., Rossell D., Auer H., Gallardo M., Blasco M. A., Sancho E., Clevers H., Batlle E., Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 17, 1225–1227 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Ogaki S., Shiraki N., Kume K., Kume S., Wnt and Notch signals guide embryonic stem cell differentiation into the intestinal lineages. Stem Cells 31, 1086–1096 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Ozawa T., Takayama K., Okamoto R., Negoro R., Sakurai F., Tachibana M., Kawabata K., Mizuguchi H., Generation of enterocyte-like cells from human induced pluripotent stem cells for drug absorption and metabolism studies in human small intestine. Sci. Rep. 5, 16479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogaki S., Morooka M., Otera K., Kume S., A cost-effective system for differentiation of intestinal epithelium from human induced pluripotent stem cells. Sci. Rep. 5, 17297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kauffman A. L., Gyurdieva A. V., Mabus J. R., Ferguson C., Yan Z., Hornby P. J., Alternative functional in vitro models of human intestinal epithelia. Front. Pharmacol. 4, 79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung K. B., Lee H., Son Y. S., Lee M.-O., Kim Y.-D., Oh S. J., Kwon O., Cho S., Cho H.-S., Kim D.-S., Oh J.-H., Zilbauer M., Min J.-K., Jung C.-R., Kim J., Son M.-Y., Interleukin-2 induces the in vitro maturation of human pluripotent stem cell-derived intestinal organoids. Nat. Commun. 9, 3039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Chen Y.-G., 2D- and 3D-based intestinal stem cell cultures for personalized medicine. Cell 7, 225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altay G., Larrañaga E., Tosi S., Barriga F. M., Batlle E., Fernández-Majada V., Martínez E., Self-organized intestinal epithelial monolayers in crypt and villus-like domains show effective barrier function. Sci. Rep. 9, 10140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collu G. M., Hidalgo-Sastre A., Brennan K., Wnt–Notch signalling crosstalk in development and disease. Cell. Mol. Life Sci. 71, 3553–3567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancho R., Cremona C. A., Behrens A., Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Rep. 16, 571–581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanDussen K. L., Carulli A. J., Keeley T. M., Patel S. R., Puthoff B. J., Magness S. T., Tran I. T., Maillard I., Siebel C., Kolterud Å., Grosse A. S., Gumucio D. L., Ernst S. A., Tsai Y.-H., Dempsey P. J., Samuelson L. C., Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 139, 488–497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fevr T., Robine S., Louvard D., Huelsken J., Wnt/β-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 27, 7551–7559 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto D., Gregorieff A., Begthel H., Clevers H., Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17, 1709–1713 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian H., Biehs B., Chiu C., Siebel C. W., Wu Y., Costa M., de Sauvage F. J., Klein O. D., Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 11, 33–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macedo M. H., Araújo F., Martínez E., Barrias C., Sarmento B., iPSC-derived enterocyte-like cells for drug absorption and metabolism studies. Trends Mol. Med. 24, 696–708 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Yin X., Farin H. F., van Es J. H., Clevers H., Langer R., Karp J. M., Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods 11, 106–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negoro R., Takayama K., Kawai K., Harada K., Sakurai F., Hirata K., Mizuguchi H., Efficient generation of small intestinal epithelial-like cells from human iPSCs for drug absorption and metabolism studies. Stem Cell Rep. 11, 1539–1550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi N. H., Lucchetta E., Ohlstein B., Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 18702–18707 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou W., Rowitz B. M., Dailey M. J., Insulin/IGF-1 enhances intestinal epithelial crypt proliferation through PI3K/Akt, and not ERK signaling in obese humans. Exp. Biol. Med. 243, 911–916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh M. N., Swift J. A., A study of the small intestinal mucosa using the scanning electron microscope. Gut 10, 940–949 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tailford L. E., Crost E. H., Kavanaugh D., Juge N., Mucin glycan foraging in the human gut microbiome. Front. Genet. 6, 81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciesielska A., Matyjek M., Kwiatkowska K., TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 78, 1233–1261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bilski J., Mazur-Bialy A., Wojcik D., Zahradnik-Bilska J., Brzozowski B., Magierowski M., Mach T., Magierowska K., Brzozowski T., The role of intestinal alkaline phosphatase in inflammatory disorders of gastrointestinal tract. Mediators Inflamm. 2017, 9074601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watkins P. B., Drug metabolism by cytochromes P450 in the liver and small bowel. Gastroenterol. Clin. North Am. 21, 511–526 (1992). [PubMed] [Google Scholar]
- 30.Vanhove J., Pistoni M., Welters M., Eggermont K., Vanslembrouck V., Helsen N., Boon R., Najimi M., Sokal E., Collas P., Voncken J. W., Verfaillie C. M., H3K27me3 does not orchestrate the expression of lineage-specific markers in hESC-derived hepatocytes in vitro. Stem Cell Rep. 7, 192–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel J., Donovan P., Khosrotehrani K., Concise review: Functional definition of endothelial progenitor cells: A molecular perspective. Stem Cells Transl. Med. 5, 1302–1306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kretzschmar K., Clevers H., Organoids: Modeling development and the stem cell niche in a dish. Dev. Cell 38, 590–600 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Watkins P. B., Non-invasive tests of CYP3A enzymes. Pharmacogenetics 4, 171–184 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Varma M. V. S., Obach R. S., Rotter C., Miller H. R., Chang G., Steyn S. J., El-Kattan A., Troutman M. D., Physicochemical space for optimum oral bioavailability: Contribution of human intestinal absorption and first-pass elimination. J. Med. Chem. 53, 1098–1108 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Hannan N. R. F., Fordham R. P., Syed Y. A., Moignard V., Berry A., Bautista R., Hanley N. A., Jensen K. B., Vallier L., Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Rep. 1, 293–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng X., Ying L., Lu L., Galvão A. M., Mills J. A., Lin H. C., Kotton D. N., Shen S. S., Nostro M. C., Choi J. K., Weiss M. J., French D. L., Gadue P., Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell 10, 371–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pannequin J., Bonnans C., Delaunay N., Ryan J., Bourgaux J.-F., Joubert D., Hollande F., The Wnt target jagged-1 mediates the activation of Notch signaling by progastrin in human colorectal cancer cells. Cancer Res. 69, 6065–6073 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Srinivasan B., Kolli A. R., Esch M. B., Abaci H. E., Shuler M. L., Hickman J. J., TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 20, 107–126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Araújo F., Sarmento B., Towards the characterization of an in vitro triple co-culture intestine cell model for permeability studies. Int. J. Pharm. 458, 128–134 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Artursson P., Ungell A.-L., Löfroth J.-E., Selective paracellular permeability in two models of intestinal absorption: Cultured monolayers of human intestinal epithelial cells and rat intestinal segments. Pharm. Res. 10, 1123–1129 (1993). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabh1586/DC1