Abstract
The majority of bacteriophage diversity remains uncharacterized, and new intriguing mechanisms of their biology are being continually described. Members of some phage lineages, such as the Crassvirales, repurpose stop codons to encode an amino acid by using alternate genetic codes. Here, we investigated the prevalence of stop codon reassignment in phage genomes and its subsequent impacts on functional annotation. We predicted 76 genomes within INPHARED and 712 vOTUs from the Unified Human Gut Virome Catalogue (UHGV) that repurpose a stop codon to encode an amino acid. We re-annotated these sequences with modified versions of Pharokka and Prokka, called Pharokka-gv and Prokka-gv, to automatically predict stop codon reassignment prior to annotation. Both tools significantly improved the quality of annotations, with Pharokka-gv performing best. For sequences predicted to repurpose TAG to glutamine (translation table 15), Pharokka-gv increased the median gene length (median of per genome median) from 287 to 481 bp for UHGV sequences (67.8% increase) and from 318 to 550 bp for INPHARED sequences (72.9% increase). The re-annotation increased median coding capacity from 66.8% to 90.0% and from 69.0% to 89.8% for UHGV and INPHARED sequences predicted to use translation table 15. Furthermore, the proportion of genes that could be assigned functional annotation increased, including an increase in the number of major capsid proteins that could be identified. We propose that automatic prediction of stop codon reassignment before annotation is beneficial to downstream viral genomic and metagenomic analyses.
Keywords: bacteriophages, stop codons, annotation, viromics, human gut, microbiome
Bacteriophages, hereafter phages, are increasingly recognized as a vital component of microbial communities in all environments where they have been studied in detail [1–3]. Phages are known to drive bacterial evolution and community composition through predator–prey dynamics and their potential as agents of horizontal gene transfer [4, 5]. The use of viral metagenomics, or viromics, has massively expanded our understanding of global viral diversity and shed light on the ecological roles that phages play [1–3].
Much of the study into viral communities has been conducted on the human gut. Here, viromics has uncovered ecologically important viruses that are difficult to bring into culture using standard laboratory techniques [6], shown the potential roles of viruses in disease states [3], and allowed for the recovery of enormous phage genomes larger than any brought into culture [7]. As the majority of phage diversity remains uncharacterized, new and enigmatic diversification mechanisms are being described continually, including the potential use of alternative translation tables.
Lineage-specific stop codon reassignment has been described previously in bacteriophages [8, 9], whereby a stop codon is repurposed to encode an amino acid. Notably, annotations of Lak “megaphages” assembled from metagenomes were observed to exhibit unusually low coding density (~70%) when genes are predicted using the standard bacterial, archaeal, and plant plastid genetic code (translation table 11) [7], much lower than the value observed for most cultured phages of ~90% [10]. The Lak megaphages were predicted to repurpose the TAG stop codon into an as-of-yet unknown amino acid [7]. More recently, uncultured members of Crassvirales have been predicted to repurpose TAG to glutamine (translation table 15) and TGA to tryptophan (translation table 4) [9], and since then, the use of translation table 15 has been experimentally validated in two phages belonging to Crassvirales [11]. Although the reasons for stop codon reassignment in viruses are not yet understood, it has been suggested that stop codon reassignment is involved in the regulation of lytic genes that are involved in late-stage infection [12].
As stop codon reassignment may be widespread in human gut viruses, we trained a fork of Prodigal [13], named prodigal-gv, to predict stop codon reassignment in phages [14] and implemented it in the pyrodigal-gv library to provide efficient Cython bindings to Prodigal-gv with pyrodigal [15]. Additionally, the virus discovery tool geNomad incorporates pyrodigal-gv to predict stop codon reassignment for viral sequences identified in metagenomes and viromes [14]. Similarly, others have developed a tool for the detection of stop codon reassignment named MgCod [16]. However, the detection of translation table 15 still has limited support in many tools, and the impacts of stop codon reassignment on functional annotation are rarely considered in viral genomics and metagenomics.
To assess the extent of stop codon reassignment in studied phage genomes and the impacts on functional annotation, we extracted phage genomes from INPHARED [10] and predicted those using alternative stop codons. We also added high-quality and complete vOTUs from the Unified Human Gut Virome Catalogue (UHGV; https://github.com/snayfach/UHGV) predicted to use alternative codons. The viral genomes were re-annotated using modified versions of the commonly used annotation pipelines Prokka [17] and Pharokka [18], implementing prodigal-gv and pyrodigal-gv for gene prediction (see Supplementary Methods). Hereafter, the modified versions are referred to as Prokka-gv and Pharokka-gv.
From INPHARED, 49 genomes (0.24%) were predicted to use translation table 15, and 27 (0.13%) were predicted to use translation table 4. From the UHGV, 666 vOTUs (1.2%) were predicted to use translation table 15, and 46 (0.08%) were predicted to use translation table 4. These genomes and vOTUs were not constrained to one particular clade of viruses, being predicted to occur on both dsDNA viruses of the realm Duplodnaviria and ssDNA viruses of the realm Monodnaviria. At the family level, we see clear lineages of viruses that conserve this feature, such as the Suoliviridae of Crassvirales; however, it also appears sporadically in other families that are not widely known to re-purpose stop codons, such as the Demerecviridae (Supplementary Table S1). The appearance of stop codon repurposing on distant lineages of viruses suggests this is a phenomenon that has arisen on multiple occasions. The lower frequency of these genomes in cultured isolates (INPHARED) versus human viromes (UHGV) may be due to culturing and sequencing biases, perhaps including modifications to DNA that are known to be recalcitrant to sequencing.
Although the mechanism for stop codon reassignment in phages is not fully understood, suppressor tRNAs are suggested to play a role [8, 19]. Consistent with previous findings, we found 375/715 (52.4%) phages predicted to use translation table 15 encoded at least one suppressor tRNA corresponding to the amber stop codon (Sup-CTA tRNA), and 11/73 (15.1%) of those predicted to use translation table 4 encoded at least one suppressor tRNA corresponding to the opal stop codon (Sup-TCA tRNA) [8, 19, 20]. Although fewer of those predicted to use translation table 4 encoded the relevant suppressor tRNA, 22/27 (81%) of the INPHARED phages predicted to use translation table 4 were viruses of Mycoplasma or Spiroplasma. As Mycoplasma and Sprioplasma are known to use translation table 4, many of the viruses predicted to use translation table 4 may be simply using the same translation table as their host.
Prediction of stop codon reassignment led to improved annotations for both Prokka and Pharokka, although the extent of this varied with the two datasets, translation tables, and annotation pipelines tested (Fig. 1; Supplementary Table S2; Supplementary Results). As Pharokka-gv outperformed Prokka-gv on all metrics tested, only Pharokka-gv is discussed further, and the equivalent results for Prokka-gv can be found in Supplementary Results. Despite using the same method for initially predicting ORFs, Prokka-gv filters more predicted ORFs than Pharokka-gv, which likely caused the difference in results.
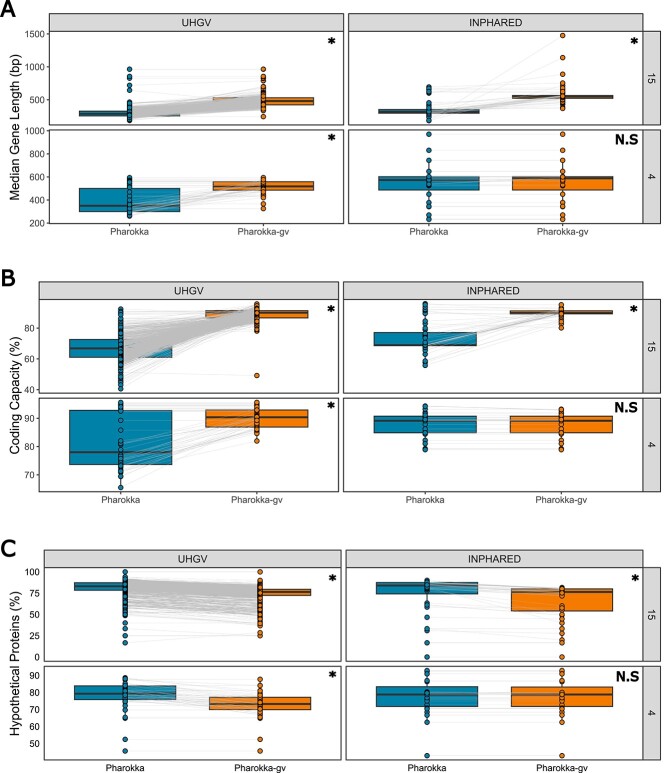
Figure 1.
Re-annotating with predicted stop codon reassignment increases the quality of annotations. Comparison of (A) median predicted gene length (bp), (B) coding capacity (%), and (C) proportion of unannotated “hypothetical” proteins for INPHARED genomes and UHGV vOTUs annotated with Pharokka (translation table 11 only) and Pharokka-gv (prediction of stop codon reassignment), grouped by dataset and predicted stop codon reassignment. Grey lines indicate pairing of the genomes across the two annotation strategies tested. Asterisk indicates significance at P ≤ 10e-10 with P determined by a paired sample T test and adjusted with the Benjamini–Hochberg procedure.
The largest improvements to annotations were observed for sequences predicted to use translation table 15, for which Pharokka-gv increased the median gene length (median of per genome medians) from 287 to 481 bp for UHGV sequences (67.8% increase) and from 318 to 550 bp for INPHARED sequences (72.9% increase; Fig. 1A). This was also reflected in an increase of median coding capacity from 66.8% to 90.0% for UHGV and 69.0% to 89.8% for INPHARED (Fig. 1B). Overall, these improved gene calls led to an increased gene length and a reduction in the number of predicted genes per kb (Supplementary Table S2). This was mirrored by an increase in the proportion of predicted proteins that could be assigned functions, with the median proportion of unannotated “hypothetical proteins” decreasing from 83.1% to 76.4% for UHGV and from 84% to 76.4% for INPHARED (Fig. 1C). As it is commonly used as a phylogenetic marker for bacteriophages, we investigated how commonly the major capsid protein (MCP) could be identified with and without predicted stop codon reassignment [21]. For those viruses we predicted to use translation table 15, annotation using the default translation table 11 only resulted in the MCP being identified in 407/715 (56.9%) of the genomes. In contrast, using translation table 15 with Pharokka-gv, we could identify the MCP in 475/715 (66.4%).
When investigating the sequences for which translation table 4 was predicted to be optimal, a substantial increase was also observed for UHGV sequences, with Pharokka-gv increasing median gene length (median of per genome medians) from 350 to 518 bp (a 48.0% increase in length; Fig. 1A), resulting in an increase of median coding capacity from 78.0% to 90.4% (Fig. 1B), and a decrease in the median proportion of unannotated hypothetical proteins from 79.3% to 73.2% (Fig. 1C). However, the same was not observed for the 27 INPHARED genomes predicted to use translation table 4. Reannotation resulted in a modest increase in median gene length (median of per genome medians) from 573 to 588 bp (a 2.6% increase in length; Fig. 1A). Median coding capacity was not increased, with both Pharokka and Pharokka-gv obtaining 89.1% (Fig. 1B). As the median gene length and coding capacity for INPHARED sequences predicted to use translation table 4 are in line with expected values, their prediction to use an alternate translation table may not be true. Similarly, many of these sequences belong to the viruses Mycoplasma and Sprioplasma, bacteria that are known to use translation table 4. Perhaps similarities of these viruses and their hosts have led to the prediction of translation table 4. Reassuringly, the prediction of translation table 4 has not hindered the quality of annotations for those genomes that have not observed a clear improvement in functional annotation.
The analysis of viral (meta)genomes relies on accurate protein predictions, with predicted ORFs being used in common analyses, including (pro)phage prediction, functional annotation, and phylogenetic analyses. The clear differences in protein predictions with/without predicted stop codon reassignment will likely have downstream impacts upon these analyses. However, this phenomenon is not yet widely considered in viral (meta)genomics. We have demonstrated the impacts of stop codon reassignment in the functional annotation of phages and provided tools for the automatic prediction and annotation of viral genomes that repurpose stop codons. Our analysis highlights the need for accurate viral ORF prediction and further experimental validation to elucidate the mechanisms of stop codon reassignment.
Supplementary Material
Contributor Information
Ryan Cook, Quadram Institute Bioscience, Norwich NR4 7UQ, United Kingdom.
Andrea Telatin, Quadram Institute Bioscience, Norwich NR4 7UQ, United Kingdom.
George Bouras, Faculty of Health and Medical Sciences, Adelaide Medical School, The University of Adelaide, Adelaide, SA 5070, Australia; Department of Surgery—Otolaryngology Head and Neck Surgery, University of Adelaide and the Basil Hetzel Institute for Translational Health Research, Central Adelaide Local Health Network, Adelaide, SA 5070, Australia.
Antonio Pedro Camargo, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, United States.
Martin Larralde, Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Meyerhofstraße 1, 69117 Heidelberg, Germany.
Robert A Edwards, Flinders Accelerator for Microbiome Exploration, College of Science and Engineering, Flinders University, Bedford Park, Adelaide, SA 5042, Australia.
Evelien M Adriaenssens, Quadram Institute Bioscience, Norwich NR4 7UQ, United Kingdom.
Conflicts of interest
The authors declare no conflicts of interest.
Funding
This research was supported by the BBSRC Institute Strategic Programme Food Microbiome and Health BB/X011054/1 and its constituent projects BBS/E/F/000PR13631 and BBS/E/F/000PR13633; and by the BBSRC Institute Strategic Programme Microbes and Food Safety BB/X011011/1 and its constituent projects BBS/E/F/000PR13634, BBS/E/F/000PR13635, and BBS/E/F/000PR13636. R.C. and E.M.A. were supported by the BBSRC grant Bacteriophages in Gut Health BB/W015706/1. This research was supported in part by the NBI Research Computing through the High-Performance Computing cluster. We gratefully acknowledge CLIMB-BIG-DATA infrastructure (MR/T030062/1) support for the provision of cloud resources. R.A.E. was supported by an award from the NIH NIDDK RC2DK116713 and an award from the Australian Research Council DP220102915. The work conducted by the US Department of Energy Joint Genome Institute (https://ror.org/04xm1d337) and the National Energy Research Scientific Computing Center (https://ror.org/05v3mvq14) is supported by the US Department of Energy Office of Science user facilities, operated under contract no. DE-AC02-05CH11231.
Data availability
The genomes used in this analysis are from two publicly available datasets; INPHARED (https://github.com/RyanCook94/inphared) and the Unified Human Gut Virome (UHGV; https://github.com/snayfach/UHGV). The details of included sequences are shown in Supplementary Table S1. The code for Prokka-gv is available on GitHub (https://github.com/telatin/metaprokka). The code for Pharokka is available on GitHub (https://github.com/gbouras13/pharokka). The code for Prodigal-gv is available on GitHub (https://github.com/apcamargo/prodigal-gv). The code for Pyrodigal-gv is available on GitHub (https://github.com/althonos/pyrodigal-gv).
References
- 1. Gregory AC, Zayed AA, Conceição-Neto Net al. Marine DNA viral macro- and microdiversity from pole to pole. Cell 2019;177:1109–1123.e14. 10.1016/j.cell.2019.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roux S, Emerson JB. Diversity in the soil virosphere: to infinity and beyond? Trends Microbiol 2022;30:1025–35. 10.1016/j.tim.2022.05.003 [DOI] [PubMed] [Google Scholar]
- 3. Clooney AG, Sutton TDS, Shkoporov ANet al. Whole-Virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 2019;26:764–778.e5. 10.1016/j.chom.2019.10.009 [DOI] [PubMed] [Google Scholar]
- 4. Borodovich T, Shkoporov AN, Ross RPet al. Phage-mediated horizontal gene transfer and its implications for the human gut microbiome. Gastroenterol Rep (Oxf) 2022;10:goac012. 10.1093/gastro/goac012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown TL, Charity OJ, Adriaenssens EM. Ecological and functional roles of bacteriophages in contrasting environments: marine, terrestrial and human gut. Curr Opin Microbiol 2022;70:102229. 10.1016/j.mib.2022.102229 [DOI] [PubMed] [Google Scholar]
- 6. Dutilh BE, Cassman N, McNair Ket al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 2014;5:4498. 10.1038/ncomms5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Devoto AE, Santini JM, Olm MRet al. Megaphages infect Prevotella and variants are widespread in gut microbiomes. Nat Microbiol 2019;4:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ivanova NN, Schwientek P, Tripp HJet al. Stop codon reassignments in the wild. Science 2014;344:909–13. 10.1126/science.1250691 [DOI] [PubMed] [Google Scholar]
- 9. Yutin N, Benler S, Shmakov SAet al. Analysis of metagenome-assembled viral genomes from the human gut reveals diverse putative CrAss-like phages with unique genomic features. Nat Commun 2021;12:1044. 10.1038/s41467-021-21350-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cook R, Brown N, Redgwell Tet al. INfrastructure for a PHAge REference database: identification of large-scale biases in the current collection of cultured phage genomes. Phage 2021;2:214–23Cold Spring Harbor Laboratory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peters SL, Borges AL, Giannone RJet al. Experimental validation that human microbiome phages use alternative genetic coding. Nat Commun 2022;13:5710. 10.1038/s41467-022-32979-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Borges AL, Lou YC, Sachdeva Ret al. Widespread stop-codon recoding in bacteriophages may regulate translation of lytic genes. Nat Microbiol 2022;7:918–27. 10.1038/s41564-022-01128-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hyatt D, Chen GL, LoCascio PFet al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010;11:119. BioMed Central. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Camargo AP, Roux S, Schulz Fet al. Identification of mobile genetic elements with geNomad. Nat Biotechnol 2023. 10.1038/s41587-023-01953-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larralde MP. Python bindings and interface to prodigal, an efficient method for gene prediction in prokaryotes. J Open Source Softw 2022;7:4296. 10.21105/joss.04296 [DOI] [Google Scholar]
- 16. Pfennig A, Lomsadze A, Borodovsky M. MgCod: gene prediction in phage genomes with multiple genetic codes. J Mol Biol 2023;435:168159. 10.1016/j.jmb.2023.168159 [DOI] [PubMed] [Google Scholar]
- 17. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068–9. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 18. Bouras G, Nepal R, Houtak Get al. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics 2023;39:btac776. 10.1093/bioinformatics/btac776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pfennig A, Lomsadze A, Borodovsky M. Annotation of phage genomes with multiple genetic codes. bioRxiv. 2022.2006.2029.4959982022. 10.1101/2022.06.29.495998 [DOI] [PubMed] [Google Scholar]
- 20. Chan PP, Lowe TM. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol 2019;1962:1–14. 10.1007/978-1-4939-9173-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simmonds P, Adriaenssens EM, Zerbini FMet al. Four principles to establish a universal virus taxonomy. PLoS Biol 2023;21:e3001922. 10.1371/journal.pbio.3001922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Telatin A, Fariselli P, Birolo G. SeqFu: a suite of Utilities for the Robust and Reproducible Manipulation of sequence files. Bioengineering 2021;8:59. 10.3390/bioengineering8050059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Terzian P, Olo Ndela E, Galiez Cet al. PHROG: families of prokaryotic virus proteins clustered using remote homology. NAR Genomics and Bioinformatics. Oxford Academic, 2021;3:lqab067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Team, R. C . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, 2018. [Google Scholar]
- 25. Benjamini Y, Hochberg Y. Journal of the Royal Statistical Society: Series B (Methodological), Vol. 57. John Wiley & Sons, Ltd, Hoboken, 1995, 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 26. Wickham H. Ggplot2: Elegant Graphics for Data Analysis, 2nd edn. Springer International Publishing, New York, 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomes used in this analysis are from two publicly available datasets; INPHARED (https://github.com/RyanCook94/inphared) and the Unified Human Gut Virome (UHGV; https://github.com/snayfach/UHGV). The details of included sequences are shown in Supplementary Table S1. The code for Prokka-gv is available on GitHub (https://github.com/telatin/metaprokka). The code for Pharokka is available on GitHub (https://github.com/gbouras13/pharokka). The code for Prodigal-gv is available on GitHub (https://github.com/apcamargo/prodigal-gv). The code for Pyrodigal-gv is available on GitHub (https://github.com/althonos/pyrodigal-gv).