Abstract
Background
The sporadic nature of blood transfusion therapy coupled with the alteration of HAMP genes may exacerbate the risk of iron burden in sickle cell anaemia (SCA) patients. The study determined the polymorphic distribution of the HAMP promoter gene rs10421768 and hepcidin levels in SCA patients.
Method
Sixty participants aged ≥12years [45 SCA patients and 15 controls (HbA)] were recruited from 15th March, 2023 to 20th July, 2023 for a case-control study at Methodist Hospital Wenchi, Ghana. Complete blood count and hepcidin levels assessment were done using haematology analyzer and ELISA, respectively. Genomic DNA was extracted using the Qiagen Kit, and HAMP gene rs10421768 (c.-582 A>G) was sequenced using the MassARRAY method. Data were analysed using SPSS version 26.0.
Results
The frequencies of the HAMP promoter rs10421768 genotypes AA, AG, and GG were 64.4%, 33.3%, and 2.2% in SCA patients, and 86.7%, 13.3%, and 0% in the controls, respectively. Serum hepcidin levels were significantly higher among controls than cases [204.0 (154.1–219.3) vs 150.2 (108.1–195.6)μg/L, p<0.010]. Participants with HAMP rs10421768 homozygous A genotype had higher serum levels of hepcidin compared with those in the wild genotypes (AG/GG) group [(188.7 (130.9–226.9) vs 136.8 (109.7–157.8)μg/L, p<0.016]. Disease severity and blood cell parameters were not associated with the HAMP variants (p>0.05).
Conclusion
The HAMP promoter rs10421768 AA genotype has the highest frequency of distribution and the GG genotype with the least distribution. Participants with HAMP rs10421768 G allele (c.-582A>G) had reduced levels of hepcidin. HAMP rs10421768 genotypes had no association with blood cell parameters and disease severity. The HAMP rs10421768 genotypes may influence serum levels of hepcidin. Further study is required to elucidate the potential effect of the G allele on hepcidin transcription.
Introduction
Sickle cell anaemia (SCA) is a genetic condition involving the homozygous inheritance of haemoglobin S (HbS) with a wide spectrum of disorders [1]. It occurs as a result of a transversion mutation in the second nucleotide at the sixth position (GAG to GTG) of the beta-globin gene resulting in abnormal HbS, which has poor solubility when deoxygenated [2]. Under hypoxic conditions, haemoglobin S polymerises to form tactoids with increased red blood cell adherence to the vascular endothelium [3]. Polymerised haemoglobin S is known to be central to vaso-occlusive crisis in SCA patients and leads to secondary processes such as inflammation, haemolysis, anaemia, vasculopathy and oxidative stress affecting many organs [4,5].
The burden in sub-Saharan Africa is predicted to be 64% of the global 400,000 children born with sickle cell disease (SCD) annually with an increased death rate [6]. In Ghana, almost 2% of newborn babies have SCD, with approximately 56% having sickle cell anaemia [7].
Hydroxyurea, penicillin V and folic acid therapy have been used as the primary management protocols for SCA [8]. Blood transfusion plays a significant role in the management of patients with SCA by reducing the proportion of HbS, to limit haemolysis and endothelial damage that results from high proportions of sickle polymer-containing red cells. The blood transfusion is usually used as supportive care for patients who experience severe form of the condition and are unresponsive to the first line of medication [9,10]. Although, transfusion may treat symptoms of anaemia or prevent complications of SCD-related vaso-occlusion such as stroke and severe acute chest syndrome, it is not without drawbacks. An average healthy person’s body has 4 g of iron, whereas people who receive multiple and frequent transfusions can accumulate 5 to 10 g annually. Long-term transfusion can lead to iron overload which may result in further complications including heart failure, growth retardation, endocrine problems and splenomegaly [11,12].
The body has no definite mechanism for eliminating excess iron; therefore, iron metabolism, storage and transport are tightly regulated through the hepcidin-ferroportin axis to avoid accumulation [13]. Hepcidin (major iron regulator) is a small antimicrobial peptide encoded by the hepcidin antimicrobial peptide (HAMP) gene located on chromosome 19q13. The gene is composed of three exons and has a length of 2637 bp. [14]. Research has shown that hepcidin mostly expressed by the hepatocytes is stimulated by iron overload (mediated through the bone morphogenetic protein (BMP) signalling pathway) and inflammation (by interleukin 6 (IL-6) and other cytokines through the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signalling pathway), whereas erythropoietic activity, anaemia or hypoxia suppress its synthesis [15,16]. Hepcidin and ferroportin form a complex, which is absorbed by these cells, where it causes ferroportin to degrade, inhibiting iron outflow and, as a result, reducing intestinal iron absorption and bioavailability [17]. Similar to other genes, various mutations in the HAMP gene have been reported.
It has been postulated that, the presence of G allele (c.-582A>G) may downregulate hepcidin transcription, which promotes iron absorption [16]. Previous studies reported an association between the hepcidin promoter c.-582 A>G (rs10421768) polymorphism and iron overload in patients with β-thalassaemia major [18,19]. The different transcriptional activation that happens through E-boxes inside the gene’s promoter region may be the cause of the association between the HAMP gene variation and iron metabolism [19]. Another study found reduced levels of hepcidin in severely anaemic children with SCD, independent of inflammation or markers of erythropoiesis [20]. Again, a previousstudy reported that SCA patients with history of multiple blood transfusions had elevated serum hepcidin levels compared to the control groups, and the anaemia of chronic inflammation was found to be a contributing factor to the anaemia of SCD patients [21].
There is paucity of data regarding on the distribution of hepcidin (rs10421768) promoter gene polymorphisms and its expression among SCA patients in Ghana, hence, the need for this study. Findings from this study would provide the possible relationship between the HAMP polymorphic variants and hepcidin protein expression in SCA patients.
Materials and methods
Study site/design
This was a hospital-based case-control study conducted at Methodist Hospital, Wenchi, a Christian faith-based hospital in the Bono Region of Ghana. This study was carried out from 15th March, 2023 to 20th July, 2023. The hospital, which has 250 beds, serves as a referral hub for 20 public and private healthcare facilities in the Wenchi municipality and some parts of the Savannah region. The hospital offers specialist surgical services, including orthopaedic, urology, and general surgery. The digital address of the hospital is BW-0005-0306. The population of the municipality according to the 2021 population and housing census stands at 124,758, with 60,960 males and 63,798 females with majority of the inhabitants been farmers [22].
Ethical consideration
All procedures were carried out in accordance with the ethical standards of the Institutional Review Board (IRB) of the University for Development Studies, Ghana (UDS/RB/006/23).
Permission was sought from the management of Methodist Hospital, Wenchi. Participants 18years and above provided written informed consent. The consents of the study participants below 18 years were obtained from guardians or parents.
Study population
The study included 45 SCA patients, and 15 healthy (HbA) controls aged ≥12 years. The control participants were people without sickle cell disease who reside in the same geographical area and of similar ethic group as the cases. Participants below the age of 12 years with haemoglobin variants other than HbS and co-morbidities such as diabetes mellitus, hypertension, human immunodeficiency virus (HIV), and hepatitis were excluded from the study. Pregnant women, lactating mothers, and those who refused to provide consent were also excluded.
Sample size determination
The necessary sample size was obtained by employing the Kelsey’s formula:
is the required sample size for the sickle cell anaemia group.
r is the ratio of sickle cell anaemia group to the group without SCA, which is 2:1 in this study.
represents the critical value of the normal dispersion at α/2 (for this study at confidence level of 95%, α is 0.05 and the critical value is 1.96).
Zβ represents the critical value of the normal distribution at β (this study used a power of 80%, β is 0.2 and the critical value is 0.84.
p1 represents the proportion of HAMP mutant allele in SCA group, 8.5% [23].
p2 is the proportion of HAMP mutant allele in the control group, 6.7% [23]
p1-p2 is the smallest difference in proportions that is clinically important.
The minimum number of participants with SCA patients required for this study was 30 with corresponding participants without SCA at 15.
However, this study employed 60 participants: 45 SCA patients and 15 controls (age-matched HbA healthy group).
Data collection techniques and tools
Demographic characteristics such as age and sex were obtained from the participants. The severity of the sickle cell disease was determined using a modified method as proposed by Hedo eta al. [24], based on the clinical presentation of the patients which included the number of blood transfusions received per year, number of vaso-occlusive crises per year, number of hospitalisations per year due to relapse or crisis that require medical attention, use of hydroxyurea, iron chelation therapy, presence of acute chest syndrome, osteomyelitis, renal failure, heart failure, avascular necrosis of the femoral hand, pneumonia, dehydration, anaemia, pigment gallstones and jaundice obtained from the clinical records. The total severity score was classified as mild SCA (Score <3), moderate SCA (Score >3 and ≤ 7) and severe SCA (Score > 7) with detailed description in previous study by Hedo et al. [24].
Categorization of blood cell transfusion was done as follows: multiple red cell transfusion (regular) when the subject received ≥ 3 units/year, rare transfusion when a client received <3 unit/year and nil transfusion when a patient received no transfusion per year.
Sample collection and processing
Five millilitres of venous blood samples from each patient were aseptically collected and divided into two tubes: 3 mL into ethylenediamintetraacetic acid (EDTA) and 2 mL into a gel tube. The EDTA sample was used for complete blood count estimation, sickling and Hb electrophoresis tests. Genomic DNA was extracted from whole blood using the spin protocol with commercial DNA extraction kit (Qiagen, Germany), and segments of DNA encompassing the HAMP rs10421768 (c.-582 A>G) promoter gene were sequenced (Inqaba BiotecTM West Africa Ltd, Ghana). The gel tube sample was allowed to clot and spun at 3500 rpm for five minutes. The resulting serum was transferred into Eppendorf tubes and stored at -20°C for hepcidin estimation using the enzyme-linked immunosorbent assay (ELISA) at University for Development Studies Laboratory.
Haematological and biochemical determinations
The EDTA whole blood samples were used for analysis of the complete blood count using an automated five-part haematology analyser (Sysmex XN-550, Japan).
Sickle slide test was done using sodium metabisulphite and haemoglobin variants determined using cellulose acetate electrophoresis method at an alkaline pH of 8.4.
Serum hepcidin levels were assayed by the sandwich ELISA method using commercially prepared ELISA kits (Biobase, China). Microtitter plate wells were coated with purified human hepcidin antibody forming a solid phase antibody. Patients/control sera were added to the wells, incubated and upon the addition of horseradish peroxidase (HRP) labeled antibody (conjugate), antibody-antigen-enzyme-labeled antibody complex was formed. After washing using WHYM201 microplate washer and the addition of TMB substrate solution, a blue coloured product was formed. Weak sulphuric acid solution was added to terminate the reaction and the optical density of the resulting colour change was measured using ELISA reader (Powean-Medical, China) at 450 nanometers.
DNA extraction
Genomic DNA was extracted from patients’ whole blood at the Kintampo Health Research Center using the QIAGEN kits (QIAGEN, Germany), according to the manufacturer’s instructions. In a 1.5 mL microcentrifuge tube, 200 μL of blood sample and 20 μL of protease K were combined. Two hundred (200) μL AL buffer was added, vortex for 15 seconds and incubated for 10 minutes at 56°C. Brief centrifugation was done at 8000 rpm to clear the drips from the lid’s interior. The sample was mixed thoroughly for 15 seconds, 200 μL of ethanol was added before brief centrifugation. The mixture was put into a QIAamp Mini spin column, centrifuged for 1 minute at 8000 rpm, 500 μL of AW 1 buffer was added, centrifuged for 1 minute and residue discarded. A 500 μL of AW2 buffer was subsequently added, centrifuged at 14000 rpm for 3 minutes to eliminate the residue. After dry centrifugation, 200 μL of AE buffer added, incubated for 1 minute at ambient temperature and centrifuged for 1 minute at 8000 rpm. The concentration of DNA (μg/μL) (OD 260 x 50 ng/μL) yielded from the extraction was evaluated using a NanoDrop spectrophotometer. The extracted DNA was stored at −70°C until analysis [25].
Identification of the HAMP gene polymorphisms
Genotyping was performed by real-time polymerase chain reaction (PCR) using the Agena MassARRAY method with iPLEX® PCR (Inqaba BiotecTM West Africa Ltd, Ghana).The SNP Genotyping assay consisted of an initial locus-specific PCR reaction, followed by a single-base extension using mass-modified dideoxynucleotide terminators of an oligonucleotide primer (Table 1), which annealed immediately upstream of the polymorphic site of interest. Using MALDI-TOF mass spectrometry, the distinct mass of the extended primer identifies the SNP allele as detailed in the technical manual [26]. The cluster plots for rs10421768 using the Agena MassARRAY System is shown in Fig 1.
Table 1. Primers used for the study.
| HAMP rs10421768 Nucleotides | |
|---|---|
| rs10421768_W1_F | ACGTTGGATGGTTTGAAGCTTTGGGCTACG |
| rs10421768_W1_R | ACGTTGGATGTGGAAACCCATGGAGTTCG |
| rs10421768_W1_E | TGTTCGTGTTCTATGAT |
Fig 1. Cluster plots for rs10421768 output.
Statistical analysis
Statistical Package for the Social Sciences (SPSS) software, version 26.0 (Armonk, NY, USA) was used for the statistical analysis. Shapiro-Wilk and one-sample Kolmogorov-Smirnov tests were used to assess the distribution of the continuous data. Parametric data were presented as mean±standard deviation whilst non-parametric data were presented as median (25th-75th percentiles). Categorical data were appropriately compared using Pearson Chi-square or Fisher’s exact test. Bivariate continuous data were compared using independent sample T-test (for parametric data) or Mann-Whitney test (for non-parametric data). A p< 0.05 was considered statistically significant.
Results
Demographic and allele frequency of HAMP rs10421768 gene
Table 2 shows the demographic and allele frequency of the HAMP polymorphism rs10421768 of the study participants. The 60 participants included in the study consisted of 45 (75.0%) sickle cell anaemia patients, and 15 (25.0%) healthy individuals. Of the 60 participants, 34 (56.7%) were females while 26 (43.3%) were males, with a median age of 19.0 (16.0–22.0) years. The majority (42/70.0%) of the participants had the common genotype (AA), 17 (28.3%) expressed the heterozygote genotype (AG), and 1 (1.7%) had the rare genotype (GG). Among those with SCA, the majority 29(64.4%) showed the AA genotype, 15 (33.3%) showed the AG genotype and just 1(2.2%) of the participants showed the GG genotype. In the control group, the majority 13(86.7%) expressed the common genotype (AA), with a few of them 2(13.3%) expressing the heterozygous type (AG). No significant difference was observed in the distribution of rs10421768 variants with the participants’ type (p = 0.326) and sex (p = 0.064).
Table 2. Demographic and allele frequency of HAMP rs10421768 gene.
| Variable | Category | HAMP rs10421768 SNP | Total (%) | P-value | ||
|---|---|---|---|---|---|---|
| AA (%) 42 (70.0) |
AG (%) 17 (28.3) |
GG (%) 1 (1.7) |
||||
| Participants | Controls (HbA) | 13 (86.7) | 2 (13.3) | 0(0.0) | 15 (25.0) | 0.326 |
| Cases (HbS) | 29 (64.4) | 15 (33.3) | 1(2.2) | 45 (75.0) | ||
| Sex | Male | 22 (84.6) | 4 (15.4) | 0 (0.0) | 26 (43.3) | 0.064 |
| Female | 20 (58.8) | 13 (38.2) | 1(2.9) | 34 (56.7) | ||
| Age | 19.0 (16.0–22.0) | |||||
HAMP: Hepcidin Antimicrobial Peptide; SNP: Single Nucleotide Polymorphism; SCA: Sickle cell anaemia; A: Adenine; G: Guanine; N: Number. Categorical data presented in frequencies with corresponding percentages in parenthesis. Pearson’s Chi-square and Fisher’s exact test were used to compare the association between two categorical data. Age was presented in median (25th-75th percentiles). P<0.05 was considered statistically significant.
Relationship between HAMP rs10421768 variants and disease severity
Table 3 displays the relationship between HAMP promoter gene variants and disease severity among the sickle cell anaemia patients. Cases with the mild form of the disease showed genotypic frequencies of AA, AG, and GG of 16 (64.0%), 8 (32.0%), and 1 (4.0%), respectively. Among cases with moderate disease, the allele frequencies of AA and AG were observed in 9 (56.2%) and 7 (43.8%) cases, respectively, while those with severe disease showed only the mutant genotype (AA) 4 (100%). There was no significant difference between HAMP promoter gene variants and disease severity.
Table 3. Relationship between HAMP rs10421768 variants and disease severity.
|
Disease Severity |
HAMP Genotypes |
Total N (%) |
P-value |
||
|---|---|---|---|---|---|
| AA (%) | AG (%) | GG (%) | |||
| Mild Disease | 16 (64.0%) | 8 (32.0%) | 1 (4.0%) | 25 (100.0%) | 0.448 |
| Moderately Disease | 9 (56.2%) | 7 (43.8%) | 0 (0.0) | 16 (100%) | |
| Severe Disease | 4 (100%) | 0 (0.0) | 0 (0.0) | 4 (100%) | |
| Total | 29 (64.4%) | 15 (33.3%) | 1 (2.2%) | 45(100%) | |
G: Guanine, A: Adenine, N: Number, Categorical information is displayed as frequencies with corresponding percentages in parentheses. Association was determined using Pearson’s Chi-square. Statistical significance was set at p<0.05.
Serum levels of hepcidin of study participants stratified by cases and controls
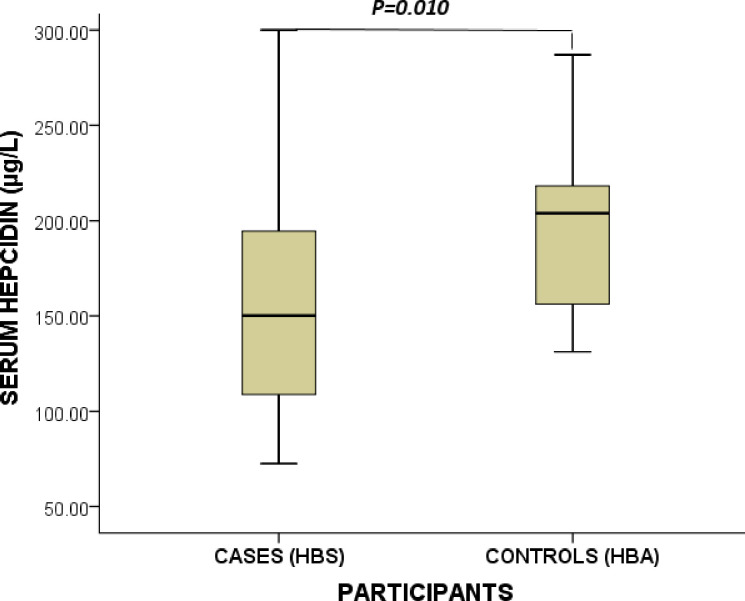
Fig 2 illustrates the hepcidin levels of the study participants stratified by cases and controls. The mean serum level of hepcidin among the study participants was 158.6 (127.8–214.8) μg/. Hepcidin levels were significantly higher among control subjects compared to their counterparts with SCA [204.0 (154.1–219.3) vs 150.2 (108.1–195.6)μg/L, p<0.010].
Fig 2. Serum levels of hepcidin of study participants stratified by cases and controls.
μg/L = microgram per litre. Data were compared using the Student T-Test. P< 0.05 was deemed statistically significant.
Serum levels of hepcidin of study participants stratified by HAMP rs10421768 variants
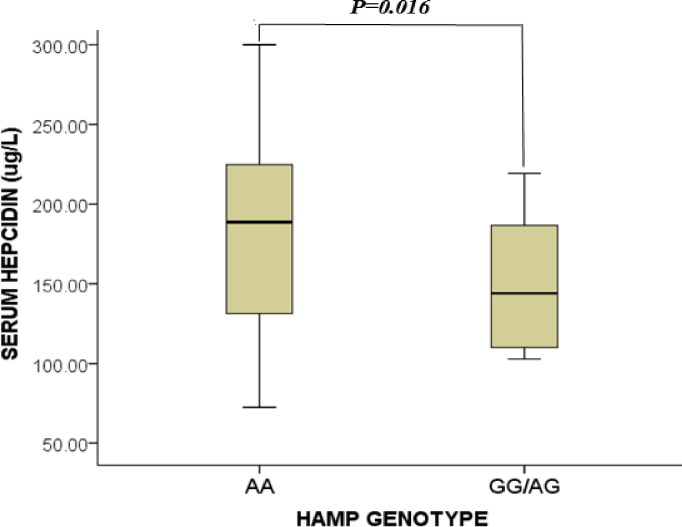
Fig 3 shows the serum levels of hepcidin of participants stratified by HAMP rs10421768 variants. The mean serum hepcidin levels of HAMP mutant genotype AA were significantly higher compared to the wild genotype AG/GG [(188.7 (130.9–226.9) vs 136.8 (109.7–157.8)μg/L, p<0.016].
Fig 3. Serum levels of hepcidin of study participants stratified by HAMP rs10421768 variants.
μg/L = microgram per litre. Data was compared using the Student T-Test. P<0.05 was deemed statistically significant.
Relationship between HAMP rs10421768 variants and blood cell parameters of the participants Table 4 shows the blood cell parameters of the study participants stratified by HAMP rs10421768 genotype variants. No statistically significant difference was observed in the blood cell parameters between the genotypes AA and AG/GG (p>0.05).
Table 4. Relationship between HAMP rs10421768 variants and blood cell parameters of the participants.
| Blood cell parameters | HAMP rs10421768 Genotypes | P-value | |
|---|---|---|---|
| AA N = (42) |
AG/GG N = (18) |
||
| RBC×103/μL | 3.3 (2.3–4.4) | 3.0 (2.4–4.3) | 0.859 |
| Hb (g/dL) | 9.9±3.0 | 9.1±1.9 | 0.262 |
| HCT% | 29.3±8.3 | 27.2 ±5.5 | 0.270 |
| MCV (fL) | 86.0 (80.0–91.8) | 87.3 (78.6–95.1) | 0.646 |
| MCH (pg) | 30.2 (27.2–32.7) | 28.7 (26.3–31.8) | 0.388 |
| MCHC (g/dL) | 34.7 (33.3–36.0) | 34.5 (33.9–35.9) | 0.840 |
| RDW-CV% | 15.5 (9.2–18.9) | 16.9 (14.0–22.1) | 0.217 |
| TWBC×103/uL | 7.6 (5.1–11.9) | 8.8 (7.1–11.2) | 0.287 |
| PLT.×103/uL | 269.0 (188.3–438.0) | 270.0 (199.3–358.0) | 0.834 |
μL = Microliter; fL = Femtolitre; g/dL = Gram per deciliter; μL = microgram per litre; pg = Picrogram; N = Number of participants; RBC = Absolute red blood cell count; Hb = Haemoglobin concentration; HCT = Haematocrit; MCV = Mean cell volume; MCH = Mean cell haemoglobin; MCHC = Mean cell haemoglobin concentration; RDW-CV = Red blood cell distribution width-coefficient of variation; TWBC = Total white blood cell count; PLT. = Platelet count; Parametric data presented as mean ± standard deviation was compared using Student T-test, and non-parametric data presented as median (25th-75thpercentiles) were compared using Mann- Whitney U-test. P<0.05 was considered statistically significant.
Discussion
Hepcidin is essential for keeping the body’s iron levels balanced, and its significance in sickle cell anaemia is vital for comprehending the intricate interplay between iron metabolism and this hereditary disease [27]. Previous studies have indicated that specific genetic variations in the HAMP promoter region can lead to diminished hepcidin expression, potentially leading to elevated serum iron levels [28–30] This study aimed to determine the genotypic variants in the hepcidin gene promoter (rs10421768) and the phenotypic levels of hepcidin protein in sickle cell anaemia patients.
In the present study, the polymorphic distribution of HAMP rs10421768 genotypes AA, AG and GG among SCA patients was 64.4%, 33.3%, and 2.2% respectively, whereas in the healthy controls, the AA, AG and GG genotypic frequencies were 86.7%,13.3% and 0% respectively. This finding corroborates with an earlier study conducted among multiple sclerosis patients that reported 75.9% frequency of A allele and 24.1% of G allele [16]. It also agrees with a study by Zarghamian et al. in β-thalassemia major patients that found the frequencies of AA, AG and GG genotypes of 53.9%, 40.2% and 5.9% respectively [19].
Conversely, a previous study conducted in northern Saudi Arabia reported frequencies of HAMP promoter rs10421768 genotypes AA, AG and GG to be 3.5%, 96.5% and 0% in the iron-deficient women and 12%, 88% and 0% in the healthy women, respectively [31]. The difference in the findings may be due to the variations in the study participants and geographical locations. Whiles the current study was conducted among SCA patients in Northern Ghana, the Al-amer & Alshari study was conducted among iron deficient women in northern part of Saudi Arabia.
Additionally, the A allele was more common among males (84.6%) compared to females (58.8%), whereas the heterozygous (AG) allele was more prevalent in females (38.2%) than in males (15.4%). The homozygous G allele was rare in both sexes with only (2.9%) in females. The data obtained from the study showed an increase in A allele homozygosity of the HAMP promoter (C-582 A>G) polymorphism in those without SCA (86.7%) compared with patients with SCA (64.4%) although not statistically significant. The slight difference in genotype distribution between sexes, as indicated, may warrant further investigation to explore the potential gender-specific effects of this polymorphism on blood cell parameters and SCA susceptibility. This is highlighted by an earlier study on the genetic variation within the HAMP gene and its potential role in the context of SCA [31].
This study explored the potential link between genetic variants of HAMP rs10421768 and the disease severity in individuals with sickle cell anaemia. Even though, previous studies postulated that genetic, environmental, or clinical factors could exert considerable influence over the clinical progression and disease susceptibility [31,32], the current study did not find any association between the 582 A/G gene polymorphisms and disease severity. Notably, the majority of SCA patients, regardless of their genotype, 62.5% had mild disease, 33.3% had moderate and 8.9% had the severe form of the disease. Severe disease was uncommon in individuals with the G allele. Findings from this study corroborate a study by Parajes et al. [33] that found no association between serum iron, serum transferrin, transferrin saturation or ferritin levels and the c.-582A > G HAMP promoter variant in a healthy population [33]. The study however contradicts previous studies on potential iron overload in β-thalassaemic patients [15,18]. The variation in the findings may be attributed to demographics and differences of the study subjects. This study recruited SCA patients whereas the earlier studies recruited β-thalassaemic patients.
This study found significantly lower hepcidin levels in SCA patients than the healthy controls. Previous studies have reported similar findings and this could be explained by the overriding effect of intense erythropoiesis or hypoxia that downregulates hepcidin synthesis as against the stimulatory effect of inflammatory cytokines (IL-6) on hepcidin experienced by SCA patients [17,26]. This contradicts an earlier study in Egypt that reported elevated serum hepcidin levels in sickle cell disease subjects with multiple blood transfusions [21]. The difference in the findings may be due to the variations in the study participants. Ismail et al. [21] recruited β-thalassaemia major patients who are transfusion dependent whilst the current study recruited SCA patients.
Additionally, this study found significantly reduced serum hepcidin levels in study participants with the G allele compared to those with the A allele. This finding corroborates with previous studies [18,19,30]. The reason for the finding could be attributed to the inhibitory effect of the G allele on hepcidin transcription. The HAMP c.-582 A>G variants located in the E-box 1 acts as a responsive element for upstream stimulatory factors 1 and 2 (USF 1&2) necessary for promoting adequate transcription and synthesis of hepcidin. The presence of G variants affects the binding of the transcription factor to the E-box which down-regulates the transcription of the HAMP gene and its expression [16].
The relationship between blood cell parameters and the HAMP rs10472168 (c.-582 A>G) were assessed. The study did not find any relationship between blood cell parameters and HAMP gene variants, which corroborates findings from earlier studies [31,34,35].
This study could not assess the iron status of the study participants.
Conclusion
The HAMP promoter rs10421768 AA genotype has the highest frequency of distribution and the GG genotype with the least distribution. Participants with HAMP rs10421768 G allele (c.-582A>G) had reduced levels of hepcidin. HAMP rs10421768 genotypes had no association with blood cell parameters and disease severity. The HAMP rs10421768 genotypes may influence serum levels of hepcidin. Further study with large sample size is required to elucidate the potential effect of the G allele on hepcidin transcription.
Supporting information
(SAV)
Acknowledgments
Authors appreciate the enormous contributions of management and staff of Kintampo Health Research Center for providing us with necessary resources for the DNA extraction. We also appreciate the immense support of University for Development Studies and the management of Methodist Hospital Wenchi, Ghana. Lastly, we thank all participants in the study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Pinto VM, Balocco M, Quintino S, Forni GL. Sickle cell disease: a review for the internist Sickle cell disease: a review for the internist. 2019;(August). [DOI] [PubMed] [Google Scholar]
- 2.Onimoe G, Rotz S. Sickle cell disease: A primary care update. Vol. 87, Cleveland Clinic Journal of Medicine. Cleveland Clinic Educational Foundation; 2020. p. 19–27. [DOI] [PubMed] [Google Scholar]
- 3.Amanor et al. Iron stores in steady‐state sickle cell disease children accessing care at a sickle cell disease clinic in Kumasi, Ghana: A cross‐sectional study. 2022;5(August):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houwing ME, Pagter D, Beers V, Draat V. Sickle cell disease Blood reviews Publisher’s PDF, also known as Version of record Publication date: Blood Reviews Sickle cell disease: Clinical presentation and management of a global health challenge. Blood Rev [Internet]. 2023;37:100580. Available from: 10.1016/j.blre.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 5.Steinberg M, Steinberg MH. Treating Sickle Cell Anemia: A New Era Dawns Treating sickle cell anemia: A new era dawns. 2020;(January). [Google Scholar]
- 6.Asare E V., Wilson I, Benneh-Akwasi Kuma AA, Dei-Adomakoh Y, Sey F, Olayemi E. Burden of Sickle Cell Disease in Ghana: The Korle-Bu Experience. Adv Hematol. 2018. Dec;2018:1–5. doi: 10.1155/2018/6161270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oppong M, Lamptey H, Kyei-Baafour E, Aculley B, Ofori EA, Tornyigah B, et al. Prevalence of sickle cell disorders and malaria infection in children aged 1–12 years in the Volta Region, Ghana: a community-based study. Malar J [Internet]. 2020;19(1):1–11. Available from: 10.1186/s12936-020-03500-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumayr LD, Hoppe CC, Brown C. Sickle Cell Disease: Current Treatment and Emerging Therapies. 2022;(March). [PubMed] [Google Scholar]
- 9.Ware RE, de Montalembert M, Tshilolo L, Abboud MR. Sickle cell disease. Lancet [Internet]. 2017;390(10091):311–23. Available from: doi: 10.1016/S0140-6736(17)30193-9 [DOI] [PubMed] [Google Scholar]
- 10.Tanhehco YC, Shi PA, Schwartz J. Transfusion therapy in sickle cell disease. 2022;(4):1–13. [Google Scholar]
- 11.Soliman AT, Corporation HM, Sanctis V De Hospital QP, Yassin MA, Alshurafa A, et al. Blood transfusion and iron overload in patients with Sickle Cell Disease (SCD): Personal experience and a short update of diabetes mellitus occurrence. 2022;(September). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkindi S, Panjwani V, Al-Rahbi S, Al-Saidi K, Pathare A V. Iron Overload in Patients With Heavily Transfused Sickle Cell Disease—Correlation of Serum Ferritin With Cardiac T2* MRI (CMRTools), Liver T2* MRI, and R2-MRI (Ferriscan®). Front Med. 2021;8(October):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andolfo I, Russo R. Novel Insights and Future Perspective in Iron Metabolism and Anemia. Metabolites. 2022;12(2):14–6. doi: 10.3390/metabo12020138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faraj and Al-Abedy. Serum Hepcidin Hormone Level and Its Genes Polymorphism. Genetic Variation. 2021. [Google Scholar]
- 15.Zarghamian P, Azarkeivan A, Arabkhazaeli A, Mardani A, Shahabi M. Hepcidin gene polymorphisms and iron overload in β-thalassemia major patients refractory to iron chelating therapy. BMC Med Genet. 2020;21(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stachowska L, Dorota Koziarska BK, Artur Kotwas, Anna Knyszy ´nska MF, Karolina Dec ES 1, Viktoria Hawryłkowicz 1, Monika Kulaszy ´nska 6 JSP 7 and ˙Zydecka KS. Transferrin Receptor 2 (rs7385804) Gene Polymorphism Might Be Associated with the Origin of Multiple Sclerosis. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichert CO, Marafon F, Levy D, Maselli LMF, Bagatini MD, Blatt SL, et al. Influence of Hepcidin in the Development of Anemia. Curr Top Anemia. 2018. [Google Scholar]
- 18.Andreani M, Radio FC, Testi M, De Bernardo C, Troiano M, Majore S, et al. Association of hepcidin promoter c.-582 A>G variant and iron overload in thalassemia major. Haematologica. 2009;94(9):1293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarghamian P, Azarkeivan A, Arabkhazaeli A, Mardani A, Shahabi M. Hepcidin gene polymorphisms and iron overload in β-thalassemia major patients refractory to iron chelating therapy. BMC Med Genet. 2020. Apr;21(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee N, Makani J, Tluway F, Makubi A, Armitage AE, Pasricha S rayn, et al. EBioMedicine Decreased Hepcidin Levels Are Associated with Low Steady-state Hemoglobin in Children With Sickle Cell Disease in Tanzania. EBioMedicine [Internet]. 2019;34(2018):158–64. Available from: 10.1016/j.ebiom.2018.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ismail et al. The Relation between Serum Hepcidin, Ferritin, Hepcidin: Ferritin Ratio, Hydroxyurea and Splenectomy in Children with β—Thalassemia. 2019;1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.GSS. Ghana 2021 population and housing census General report 3A. 2021. [Google Scholar]
- 23.Rahman HAA, Abou-elew HH, El-shorbagy RM, Fawzy R, Youssry I. Influence of Iron Regulating Genes Mutations on Iron Status in Egyptian Patients with Sickle Cell Disease. 2014;44(3):304–9. [PubMed] [Google Scholar]
- 24.Hedo CC, Aken’ova YA, Okpala IE, Durojaiye AO, Salimonu LS. Acute phase reactants and severity of homozygous sickle cell disease. J Intern Med. 1993. Jun;233(6):467–70. doi: 10.1111/j.1365-2796.1993.tb01000.x [DOI] [PubMed] [Google Scholar]
- 25.QIAamp DNA Mini Handbook. Blood Mini Handbook QIAGEN Sample and Assay Technologies. 2016;(May). [Google Scholar]
- 26.Chanté & Abera. Title: MassARRAY Single Nucleotide Polymorphism Genotyping Title: MassARRAY Single Nucleotide Polymorphism Genotyping. Vol. 2017. 2021. [Google Scholar]
- 27.Wood and Coates JC. HHS Public Access. 2018;177(5):703–16. [Google Scholar]
- 28.Piperno A, Pelucchi S, Mariani R. Inherited iron overload disorders. 2020;1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camaschella C, Nai A, Silvestri L. Iron metabolism and iron disorders revisited in the hepcidin era Correspondence: 2020;105(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Island ML, Jouanolle AM, Mosser A, Deugnier Y, David V, Brissot P, et al. A new mutation in the hepcidin promoter impairs its BMP response and contributes to a severe phenotype in HFE related hemochromatosis. Haematologica. 2009;94(5):720–4. doi: 10.3324/haematol.2008.001784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-amer O, Alsharif KF. Frequency of the HAMP (c. -582 A > G) Polymorphism in Iron Deficiency in Saudi Arabia. 2021. [DOI] [PubMed] [Google Scholar]
- 32.Liang et al. 2017. Association of Single-Nucleotide Polymorphism in the Hepcidin Promoter Gene with Susceptibility to Extrapulmonary Tuberculosis _ Genetic Testing and Molecular Biomarkers. 2017. [DOI] [PubMed] [Google Scholar]
- 33.Parajes S, González-quintela A, Campos J, Quinteiro C, Domínguez F. Genetic study of the hepcidin gene (HAMP) promoter and functional analysis of the c. -582A > G variant. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatem NL, Kamal MY, Younan DN, Ayad AM. Study of Association of Hepcidin Promotor C-582 A > G Variant and Iron Over Load in B-Thalassemia Major. 2023;6(7). [Google Scholar]
- 35.Abdelrhman AH, Abdelgadir EA, Khalid KM. Journal of DNA AND RNA research ISSN NO: 2575–7881 Sample Preparation and PCR Detection of HAMP DNA Polymorphisms. 2020;(2):23–30. [Google Scholar]