Abstract
We have characterized the human herpesvirus 6B (HHV-6B) rep gene, which is a homologue of the adeno-associated virus type 2 rep and is unique in the herpesvirus family. Three transcripts, 9.0, 5.0, and 2.7 kb (the major transcript), were detected by Northern blotting using an HHV-6B rep probe under late conditions. We investigated the expression kinetics of the rep gene using cycloheximide (CHX) and phosphonoformic acid (PFA), which are inhibitors of protein synthesis and viral DNA synthesis, respectively. The 5.2-kb transcript was mainly detected in the absence of protein biosynthesis upon infection, and none of the 9.0-, 5.0-, and 2.7-kb transcripts detected under the late conditions were detected in the presence of CHX and PFA. Sequences obtained from a cDNA library showed that the 5.0- and 2.7-kb transcripts were spliced from two and three exons, respectively, and the 2.7-kb transcript was more abundant. Immunohistochemistry using an antibody raised against the HHV-6 rep gene product (REP) revealed that REP was mainly present in the nucleus of MT-4 cells within 24 h after infection with HHV-6B. Using pull-down assays, coimmunoprecipitation, and a mammalian two hybrid system, we showed that HHV-6 REP binds to a transcription factor, human TATA-binding protein, through its N-terminal region.
Human herpesvirus 6 (HHV-6) is a recently isolated member of the herpesvirus family (44), which causes exanthem subitum as a primary infection (54). Thereafter, HHV-6 establishes a latent infection, but it can be reactivated during immunosuppression and has been recovered from immunodeficient individuals (10, 34, 44; H. Agut, D. Guetard, H. Collandre, C. Dauguet, L. Montagnier, J. M. Miclea, H. Baurmann, and A. Gessain, Letter, Lancet i:712, 1988; R. G. Downing, N. Sewankambo, D. Serwadda, R. Honess, D. Crawford, R. Jarrett, and B. E. Griffin, Letter, Lancet ii:390, 1987; R. S. Tedder, M. Briggs, C. H. Cameron, R. Honess, D. Robertson, and H. Whittle, Letter, Lancet ii:390–392, 1987).
HHV-6 is now classified into two variants: HHV-6A and HHV-6B (1). Both variants (HHV-6A strain U1102 [15] and HHV-6B strain HST [26a]) contain a linear double-stranded DNA genome of approximately 161 kbp with 112 potential open reading frames (ORFs). Nucleotide sequencing has shown that HHV-6A strain U1102 contains an ORF, U94, encoding a 490-amino-acid protein homologous to Rep78/68, a nonstructural protein from the human parvovirus adeno-associated virus type 2 (AAV-2) (15, 49). The HHV-6 rep gene product (REP) is closely related to AAV-2 REP, and the proteins share 24% identity over the entire length of the 490-amino-acid sequence (49). We also found a similar ORF in HHV-6B HST (54), situated proximal to the right-hand terminal repeat of the viral genome (26a). Interestingly, the AAV-2 rep gene homologue is unique to HHV-6 and is not present in other herpesviruses; thus, the role of HHV-6 REP in the life cycle of HHV-6 is of particular interest.
Although the function of HHV-6 REP is unknown, AAV-2 REP is known to possess several biological activities, including DNA-binding, site- and strand-specific endonuclease, helicase, and ATPase activities (25, 26). An attractive feature of AAV is that the viral DNA preferentially integrates within a defined region of the cellular genome (33), thus reducing the risk of insertional mutagenesis that is associated with vectors that integrate randomly. Targeted integration, therefore, involves AAV vectors containing an intact rep gene and terminal repeats incorporating into the integration locus (AAVS1) at high frequencies (14, 32). HHV-6 was recently reported to also integrate into the human genome (35, 36, 51; M. Daibata, T. Taguchi, M. Kamiska, I. Kubonishi, H. Taguichi, and I. Miyoshi, Letter, Leukemia 12:1002–1004). Conservation between HHV-6 rep and AAV-2 rep may, therefore, mean that HHV-6 REP possesses a similar range of functions advantageous to the survival of HHV-6 within the host.
Nonetheless, HHV-6 REP appears to affect gene expression differently than does AAV REP. For instance, HHV-6A REP activates the human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) promoter in fibroblast cell lines but not in T cells, whereas AAV REP actively inhibits the HIV-1 LTR promoter in both fibroblasts and T-cell lines (50). On the other hand, it has also been reported that HHV-6 REP suppresses transformation by H-ras and inhibits transcription from HIV-1 LTR in a T-cell line (3). Furthermore, HHV-6 REP may complement replication of a rep-deficient AAV-2 genome (50). Thus far, however, there is no direct evidence for the expression of HHV-6 rep during the life cycle of HHV-6 within infected cells.
In the present study, we identify the HHV-6B rep locus and localize HHV-6 REP in HHV-6-infected cells. We also identify a transcription factor, human TATA-binding protein (hTBP), as an interaction partner of HHV-6 REP. The interaction of the HHV-6 REP with hTBP establishes a link between the effects of HHV-6 REP on transcription and the genetic machinery of the host cell.
MATERIALS AND METHODS
Cells and viruses.
Umbilical cord blood mononuclear cells (CBMCs) were isolated by centrifugation over Ficoll-Conray gradients. The cells were then stimulated for 2 or 3 days with phytohemagglutinin (5 μg/ml) in RPMI 1640 medium containing 10% fetal calf serum (FCS). To prepare virus stocks, HHV-6B HST (54) and HHV-6A U1102 (Downing et al., Letter) were propagated in stimulated CBMCs as previously described (48). When more than 80% of the cells exhibited a cytopathic effect, the infected cells were lysed by freezing, thawed twice, and spun at 1,500 × g for 10 min. The supernatant was stored at −80°C as a cell-free virus stock (41). A human T-cell line, MT-4, was also used for experiments.
Isolation of RNA.
MT-4 cells were infected with strain HST or mock infected for 72 h; Poly(A)+ RNA was then extracted using the FastTrack TM 2.0 Kit (Invitrogen) according to the manufacturer's protocol. Phosphonoformic acid (PFA), which inhibits viral DNA synthesis, was used to determine whether the rep gene is an immediate-early (IE), early (E), or late (L) gene. For IE or E transcripts, MT-4 cells were infected with HHV-6, cultured with medium supplemented with PFA (300 μg/ml), and harvested at 48 h after infection. For IE transcriptional studies, MT-4 cells were maintained for 1 h in the culture medium supplemented with cycloheximide (CHX) (100 μg/ml; Sigma) prior to infection, and infected MT-4 cells were maintained with CHX for 10 h until harvest.
cDNA library construction.
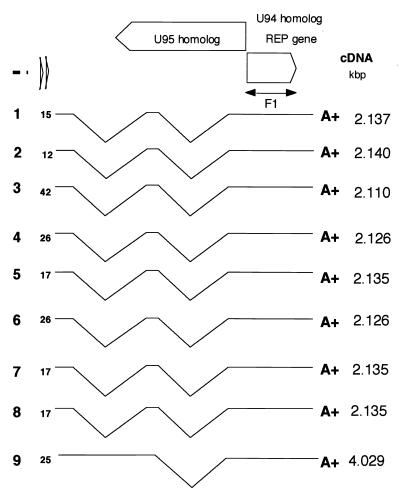
Poly(A)+ RNA (5 μg) was used as the template to construct a cDNA library. cDNA synthesis was primed using 1 μg of oligo (dT)12-18 and carried out with Superscript II reverse transcriptase and the Superscript Choice System for cDNA Synthesis (GIBCO, Grand Island, N.Y.). The cDNA was then inserted into a plasmid vector pGAD424 (Clontech Laboratories, Inc., Palo Alto, Calif.), introduced into Escherichia coli DH5α bacteria by electroporation, and cultivated. The cDNA clones were screened using 32P-labeled PCR products as F1 probes (Fig. 1).
FIG. 1.
Schematic interpretation of the cDNA clones isolated from the cDNA library. The PCR-amplified products of the full-length ORF U94 (F1) served as hybridization probes. cDNA clone numbers are indicated in boldface type on the left; the sizes of cDNA molecules are noted on the right. The locations of the 5′ ends of cDNAs hybridized by the F1 probe are indicated in front of each cDNA (positions of the first nucleotides are shown in Fig. 2). Each “A+” at the 3′ end of the cDNA indicates a poly(A)+ site.
Preparation of REP in insect cells using a recombinant baculovirus.
A DNA fragment comprising the full-length rep gene ORF was amplified by PCR and cloned, in frame, into the BamHI site of the baculovirus transfer vector, pAcG2T (PharMingen, San Diego, Calif.). The nucleotide sequences of the primers used for the PCR were as follows: sense, 5′-ggatccAATCTTGGAAGGCACAAACG-3′, and antisense, 5′-ggatccTCAATTCAGATCCTCTTCTGAGATGAGTTTTTGTTCTAAAATTTTTGGAACCGTGT-3′. The lowercase and underlined letters indicate additional restriction sites and c-myc epitope codons, respectively. The baculovirus, Autographa californica nuclear polyhedrosis virus, and a recombinant baculovirus were grown and assayed in Spodoptera frugiperda cells (Sf9 cells) in TC100 medium (GIBCO) supplemented with 0.26% Bacto Tryptose broth (Difco, Detroit, Mich.), kanamycin (100 μg/ml), and 10% FCS. A recombinant baculovirus, AcGST-REP, was generated by homologous recombination as described previously (38), and expression of the fusion protein was identified immunohistochemically using an anti-REP antibody (Ab) or anti-c-myc monoclonal Ab (MAb). Sf9 cells infected with AcGST-REP were harvested after 3 days of incubation, and the glutathione S-transferase (GST)-REP fusion protein was affinity purified.
Northern blot hybridization.
For Northern blot hybridization, 10 μg of the poly(A)+ RNA was subjected to electrophoresis on 1% agarose-formaldehyde gels, blotted onto a Hybond-N+ nylon membrane (Amersham Pharmacia, Little Chalfont Bucks, United Kingdom) and hybridized to oligonucleotide probes at 42°C in a 20 mM sodium phosphate buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× Denhardt's solution, 0.1% sodium dodecyl sulfate (SDS), 0.005 M EDTA, poly(A)n (10 μg/ml), sheared salmon sperm DNA (20 μg/ml), and yeast extract tRNA (20 μg/ml; 3 PRIME, Inc.). After hybridization, the membranes were washed in solutions of decreasing ionic strength (2× SSC–0.1% SDS, and 1× SSC–0.1% SDS) for 30 min each at 42°C. This was followed by exposure to Kodak XAR films with intensifying screens. A 0.24- to 9.5-kb RNA ladder marker was used in this study (GIBCO).
Radiolabeling of DNA probes.
The DNA probe used for hybridization was labeled with [α-32P]dCTP using a Multiprime DNA Labeling Systems kit (Amersham Pharmacia). Oligonucleotides were 5′ labeled with T4 polynucleotide kinase and [γ-32P]ATP using a Mega label kit (Takara, Kyoto, Japan). The sequence of the AR oligonucleotide was AGTTGATACTTATGTCTTTCCACCAC. The sequences of the oligonucleotides used for radiolabeling were as follows: ER, AAATGGGTGCTTCTGCATAATTACC; GR, ACCCGGATGGATGATT-GTCCTTGG; QR, CACTTGGATTTATTATGGAAAACATAC; HHV-6B IE1-1, CTGAGGCTGTACATACACAGTTAGGGCTTG; and HHV-6B IE1-2, ACAGTCATCTGACTCGCTGCTCGATTCAG.
5′ RACE.
Rapid amplification of cDNA 5′ ends (5′ RACE) was performed using the 5′ RACE System kit (version 2.0; GIBCO) according to the manufacturer's protocol. Primers used for 5′ RACE were as follows (Fig. 1): R1, GTACTAATCATTCAAGTTTACC; R2, TCTAGG-CAGGTCGGAGTCGAGGAAGA; and R3, TTAAGAACGACATAGTGATCACAGC.
Expression of the rep gene in E. coli and production of anti-REP antisera.
A DNA fragment spanning positions 1 to 705 of the ORF U94 (rep) and encoding the N-terminal portion of HHV-6 REP was amplified by PCR and cloned, in frame, into the pMALTM-c2 bacterial expression vector (New England Biolabs, Inc., Beverly, Mass.) at the BamHI and SalI sites. This vector also contained the maltose binding protein (MBP), and the resultant fusion protein (MBP-REPN1) was then expressed in E. coli DH5α. In addition, a DNA fragment comprising the entire amino acid coding region of the ORF U94 was amplified by PCR and cloned, in frame, into the pGEX-4T-1 bacterial expression vector (Amersham Pharmacia) at the BamHI and SalI sites. This vector contained GST, and the resultant GST-REP fusion protein was also expressed in E. coli DH5α. The expressed MBP-REPN1 (30) and GST-REP fusion proteins were affinity purified and used to raise Abs in rabbits. The rabbits were first immunized three times with GST-REP and then three times with MBP-REPN1. They were then bled, and the samples were used for subsequent immunological analysis. Polyclonal rabbit antiserum against HHV-6 REP, preimmune serum, anti-MBP rabbit serum (New England Biolabs, Inc.), and anti-GST rabbit serum were all used at a dilution of 1:700.
Immunohistochemical analysis of HST-infected MT-4 cells.
To examine REP expression patterns, immunohistochemical analysis was carried out using anti-REP rabbit serum as described previously (54). MT-4 cells were grown and maintained in RPMI 1640 supplemented with 10% FCS. After washing, cells were suspended in virus solution at a multiplicity of infection of 0.1 and incubated for 60 min at 37°C to adsorb virus, collected by centrifugation at 1,500 × g, and replated on cover glasses. The cells were fixed in cold acetone 3, 9, 12, 24, 36, 48, or 72 h after infection and then incubated for 30 min at room temperature with the respective primary antibodies (anti-REP serum and OHV-2, a MAb against HHV-6 nuclear antigen, which is expressed in the early stage). The cover glasses were then washed, rhodamine-conjugated goat anti-rabbit immunoglobulin secondary Ab (Chemicon) and Cy5-conjugated rabbit anti-mouse immunoglobulin secondary Ab (Chemicon) were added, and incubation was continued for an additional 15 min. Finally, the cells were incubated for 5 min with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride; Calbiochem, Nottingham, United Kingdom) to stain the DNA.
The distribution of the fluorescent label was visualized using the Delta Vision system (Applied Precision, Issaquah, Wash.), which is based on an inverted microscope (Carl Zeiss Co., Ltd., Jena, Germany) equipped with a Photometrics PXL-cooled charge-coupled device (CCD) camera. Sets of images of selected cells were collected while varying the plane of focus along the z axis. The resultant three-dimensional data sets were deconvolved and projected onto single planes. The images were then adjusted for contrast before exporting them as TIFF files to Adobe Photoshop.
Pull-down assay and immunoblotting.
Binding reactions were carried out by incubating 1-10 μg of either GST-REP or MBP-REPN1 fusion protein with human hTBP (Promega) in 100 μl of binding buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 2 mM dithiothreitol, 10% glycerol, 0.1% Tween 20, 1% bovine serum albumin [BSA]) for 1 h at 4°C on a rotating wheel. Proteins associated with GST or GST-REP were pulled down with glutathione-Sepharose beads, while those associated with MBP or MBP-REPN1 were pulled down with maltose-Sepharose beads. The beads were washed seven times with 0.5 ml of binding buffer without BSA. Bound proteins were then eluted by boiling in protein lysis buffer (10% glycerol, 50 mM Tris [pH 7.4], 1% 2-mercaptoethanol), separated by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS–12% PAGE), transferred to nylon membranes (Amersham Pharmacia), and reacted with the appropriate primary Abs (e.g., anti-hTBP MAb; Promega). Reactive bands were visualized using horseradish peroxidase-linked secondary conjugate and enhanced chemiluminescence detection reagents (Amersham Pharmacia).
Production of 35S-HHV-6 REP and coimmunoprecipitation with hTBP.
To generate 35S-labeled HHV-6 REP, the rep ORF was inserted into pcDNA3.1 (Invitrogen) downstream from the T7 promoter. The 35S-labeled HHV-6 REP was then produced by in vitro transcription-translation in rabbit reticulocyte lysates (TnT system; Promega), as directed by the kit's instructions. Equal aliquots (2,000 cpm) of 35S-labeled HHV-6 REP and 35S-labeled luciferase protein (control) were used for the immunoprecipitation assays. Initially, anti-hTBP Abs were incubated with protein A-Sepharose (Amersham Pharmacia) in 500 μl of binding buffer (25 mM Tris [pH 7.8], 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 150 mM NaCl, 5 mM MgCl2, 5 mM KCl, 1% BSA, and 0.1% Tween 20). The immune complex was then washed seven times in binding buffer without BSA and incubated with hTBP, followed by incubation with 35S-labeled HHV-6 REP or luciferase protein. Each incubation was carried out for 4 h at 4°C, and after each incubation the complexes were washed seven times with 1 ml of binding buffer. Finally, the products were boiled in sample buffer, subjected to SDS–10% PAGE, stained, and autoradiographed.
GAL4 mammalian two-hybrid analysis.
rep and hTBP expression vectors were constructed using standard techniques. The HHV6-B rep gene fragment was initially inserted into the SalI site of pBluescript II (Stratagene) followed by PCR amplification with a primer containing a SalI site at the end. The SalI fragment of the rep gene then was inserted into the SalI site of a pVP vector (Clontech) to make pVPrep. The rep gene was fused in frame with the herpes simplex virus VP16 transactivation domain ORF (pVP), to make pVP16rep. In the same way, the hTBP gene (23) was cloned into a yeast GAL 4 DNA binding domain (pM) vector, to make pMhTBP. To elucidate which region of the rep gene interacts with hTBP, the deletion mutant was constructed. For N-terminal deletion, the first 111 amino acids were deleted by excising EcoRI fragment from the pVP16Rep construct, referred to as pVP16RepΔEco. The end portion of EcoRI digestion was filled in with Klenow fragment and deoxynucleoside triphosphates to adjust the coding frame. For C-terminal deletion, the MluI fragment was excised, resulting in the deletion from the 244th to the last amino acid, referred to as pVP16 RepΔMlu. These plasmids were transfected into 293L cells (6, 13) in several combinations. Generally, 5.0 × 105 cells/well in six-well plates were seeded 24 h before transfection. A reporter plasmid, pG5 E1b Luc, which was generated by replacing the chloramphenicol acetyltransferase gene in pG5 E1b CAT (Clontech) with the firefly luciferase gene was transfected at 30 ng/well with either 0.3 μg of the pM-based or the pVP-based expression vector per well. These cells were harvested 24 h after transfection and lysed in lysis buffer (Picagene PGC50). The soluble fraction was collected, and the activity was normalized to the protein concentration using the Bradford method (8) with BSA as the standard and beta-galactosidase activity by cotransfecting pCMVβ (Clontech). Analyses were performed in triplicate, and relative luciferase activity was measured. The experiment was performed several times, and we obtained almost the same data each time.
RESULTS
Analysis of rep transcription.
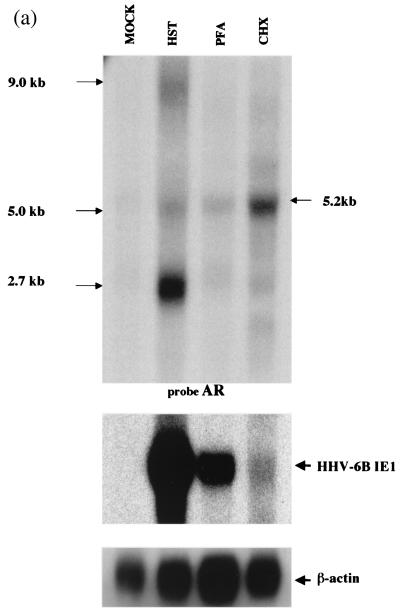
We initially attempted Northern blot analysis to determine the patterns of HHV-6B HST transcription. An antisense oligonucleotide (AR) from U94 served as the probe (Fig. 2). Analysis of poly(A)+ RNA from infected cells revealed three transcripts of approximately 9.0, 5.0, and 2.7 kb; of these, the 2.7-kb mRNA was the most abundant (Fig. 3a, lane HST).
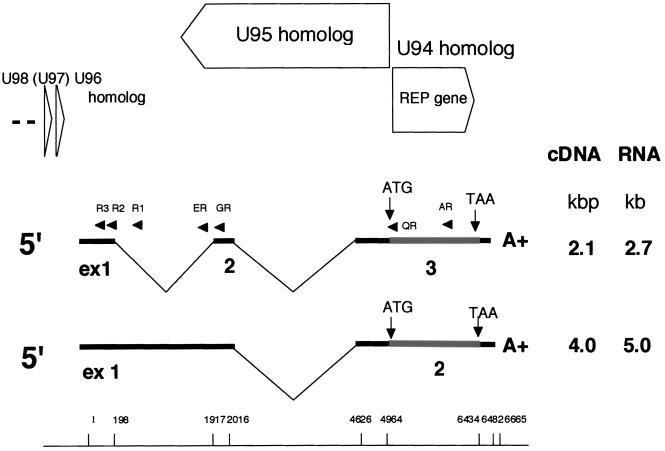
FIG. 2.
Organization and transcription pattern of the HHV-6B rep region. All exons and introns are drawn to scale. The drawing of the cDNAs is inverted relative to the HHV-6B genome. Numbers for splice sites apply to the last nucleotide of exon n and to the first nucleotide of exon n + 1; for special sequence motifs, the numbers apply to the first 5′ nucleotide. Thin lines represent introns; thick lines represent noncoding exons or parts of exons. Shaded boxes represent ORFs with ATG initiation codons and TAA termination codons. The single-stranded DNA probes (AR, ER, GR, and QR) used for Northern blot analyses and primers used for 5′ RACE (R1, R2, and R3) are indicated by closed arrows. The sizes of the cDNA molecules and corresponding mRNAs, identified by Northern blotting, are noted on the right. ORFs (HHV-6A U94, U95, U96, and U98 homologues) identified in the genomic sequence are symbolized by open arrows. The first nucleotide position is the 5′ end of both transcripts.
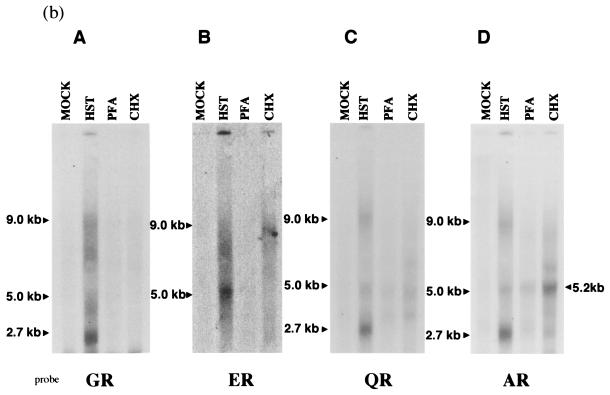
FIG. 3.
(a) Transcription of the putative rep region of HHV-6B HST under IE, E, and L conditions. Ten micrograms each of poly(A)+ RNA from mock-infected cells (lane MOCK), cells infected for 60 h in the presence of PFA (lane PFA), cells infected for 10 h in the presence of CHX (lane CHX), and cells infected for 60 h (HST) were subjected to Northern blot analyses. Sizes of mRNAs are indicated on the right. AR, an antisense, single-stranded DNA probe derived from an ORF U94 homologue (Fig. 2), detected three transcripts, 9.0, 5.0, and 2.7 kb in size. In the presence of CHX and PFA, the AR probe detected the only a 5.2-kb mRNA. The HHV-6B IE1 probe was used as the IE control. The β-actin was used as an internal control for the detection of mRNA. (b) Transcriptional patterns of the putative rep region of HHV-6B HST under IE, E, and L conditions, using several probes. ER (Fig. 2), an antisense probe derived from the first intron of the 2.7-kb mRNA detected only the 9.0- and 5.0-kb transcripts in mRNA from cells infected for 60 h. GR (Fig. 2), an antisense probe derived from the second exon of the 2.7-kb mRNA, detected all three of the transcripts (9.0, 5.0, and 2.7 kb). Moreover, the ER and GR probes detected nothing under the IE and E conditions. QR (Fig. 2), an antisense probe derived from the start site of the rep ORF, detected all three transcripts in mRNA from cells infected for 60 h but detected nothing under the IE and E conditions.
To investigate the kinetics of HHV-6 rep expression in HST-infected cells, Northern blot analyses were performed using samples treated with CHX or PHA (Fig. 3a). HHV-6B-infected MT-4 cells were maintained for 10 h in culture medium supplemented with CHX (100 μg/ml). E genes were defined as lytic-cycle transcripts whose expressions are not blocked by PFA, an inhibitor of viral DNA polymerase. Therefore, HHV-6B-infected MT-4 cells were maintained for 60 h in the culture medium either with or without PFA (300 μg/ml). Afterwards, each mRNA was harvested and subjected to Northern blot analysis. The AR probe recognized the 9.0-, 5.0-, and 2.7-kb (major) transcripts in mRNA originating from cells infected for 60 h. A 5.2-kb transcript was detected under CHX treatment (Fig. 1, lane CHX), and this transcript was more abundant than that of HHV-6 IE1 (Fig. 3a, HHV-6B IE1), which was used as a control. Moreover, the 5.2-kb transcript was detected faintly after PFA treatment (Fig. 3a, lane PFA). In contrast, the ER probe (Fig. 2), an antisense oligonucleotide derived from the first intron of the 2.7-kb mRNA, detected only the 5.0-kb transcript in mRNA from cells that had been infected for 60 h (Fig. 3b, panel B), and the GR probe (Fig. 2), an antisense oligonucleotide derived from the second exon of the 2.7-kb mRNA, detected both the 5.0- and 2.7-kb transcripts (Fig. 3b, panel A). Moreover, the ER and GR probes did not detect anything under the IE and E conditions (Fig. 3b, columns PHA and CHX). The QR probe (Fig. 2), an antisense oligonucleotide derived from the start site of the rep ORF, detected both the 5.0- and 2.7-kb transcripts in mRNA infected for 60 h but detected nothing under the IE and E conditions (Fig. 3b).
To map the mRNAs more precisely, a cDNA library was constructed from PBMCs infected with HHV-6B HST. After multiple screening steps, cDNA clones hybridizing in Southern blots to the radiolabeled, double-stranded F1 cDNA were selected using the full-length HHV-6 rep as a probe (Fig. 1). By screening approximately 106 clones, 11 positive clones were identified (Fig. 1). These clones were completely sequenced and all showed the same sense-strand orientation; no antisense clone was detected. Eight were approximately 2.1 kbp in length and had similar sequences; only one clone, approximately 4.0 kbp in length, was large, and the remaining two clones were comparatively short. The 2.1-kbp cDNA consisted of three exons, of which the first two appeared to be untranslated. The ATG sequence was located within the third exon. This cDNA is likely to correspond to the 2.7-kb transcript detected with Northern blotting. The 4.0-kbp cDNA consisted of two exons, and the ATG sequence was located within the second; in this case, only the first exon was untranslated. This cDNA is likely to correspond to the 5.0-kb transcript detected with Northern blotting. A stop codon (TAA) was found in all of the clones. The 2.7- and 5.0-kb transcripts had different termination sites, which is consistent with the sequences of the cDNAs. The 3′ untranslated region of the 4.0-kbp cDNA was 228 to 229 nucleotides long, while the 2.1-kbp cDNA had a 45-nucleotide 3′ untranslated region. No typical polyadenylation consensus signal was found in either cDNA. Both appeared to code for the same gene products. The mapping of these transcripts by cDNA sequencing is summarized in Fig. 2.
To confirm the start sites for transcription of the 2.7- and 5.0-kb mRNAs, 5′ RACE was carried out using the R1 primer in combination with either the R3 or R2 primer (Fig. 2). The start site of the 5.0-kb mRNA was determined using R1 and R3. Since R1 was derived from the intron between exons 1 and 2 of the 2.7-kb transcript and R2 was derived from exon 1, which was common to both transcripts, the use of R2 was expected to reveal the start sites of both transcripts. The RACE analysis showed that the start sites of the 5.0- and 2.7-kb transcripts were the same, and in each transcript a typical TATA box-like sequence, TATATTT, was found at a position 37 bp upstream of the cap site.
Expression of REP in MT-4 cells and CBMCs infected with strain HST.
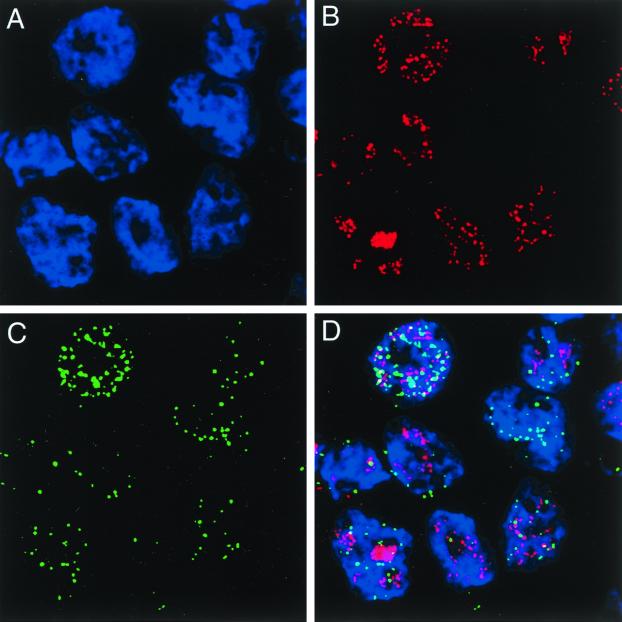
To verify HHV-6 rep expression in HST-infected cells and identify some of the properties of the transcripts, REP was localized within the infected cells. Immunofluorescence showing the distribution of anti-REP antibodies within MT-4 cells infected with HST for 48 h is shown in Fig. 4. It should be noted that there was significant overlap of the signals when infected cells were triple labeled (Fig. 4D) with anti-REP serum (Fig. 4C), DAPI (Fig. 4A), and OHV-2 (Fig. 4B), which indicates the presence of REP in the nucleus (Fig. 4C). REP was first detected in the nucleus 24 h after infection, appearing as localized spots. After 72 h, additional nuclear accumulation of REP was evident, as was the presence of REP in the cytoplasm (data not shown).
FIG. 4.
Cellular localization of HHV-6B REP in MT-4 cells infected with HST for 48 h. The optical sections traversing the nuclei of several HHV-6B-infected MT-4 cells labeled with DAPI (A); OHV-2, a MAb against HHV-6 nuclear antigen, which is expressed in the E stage (B); and anti-REP Ab (C) are shown. Note that REP is localized within the nucleus. (D) Overlaid images from panels A, B, and C.
In addition, no specific labeling was found in mock-infected cells or in infected cells incubated with preimmune serum, anti-MBP serum, or anti-GST serum (data not shown).
Interaction between a GST-HHV-6B REP fusion protein and hTBP in vitro.
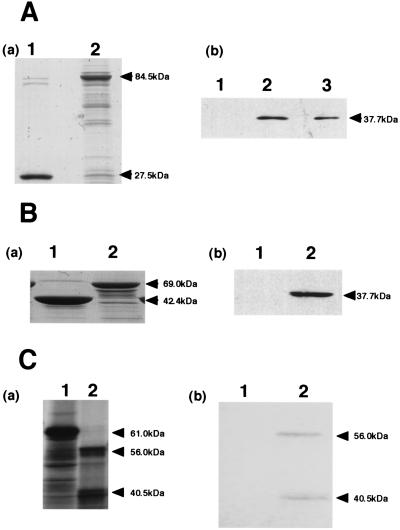
Hermonat et al. (22) showed that AAV-REP binds to hTBP, and we hypothesized that HHV-6 REP might do so as well. Direct interaction between HHV-6 REP and hTBP was assessed in vitro using pull-down assays. HHV-6B rep from baculovirus was expressed as a fusion protein with GST (GST-REP) and purified on glutathione-Sepharose beads. When the purified fusion proteins and GST were subjected to SDS-PAGE and stained with Coomassie blue, the quantity of protein in each sample was found to be about the same (Fig. 5A, panel a). When the purified samples were reacted with hTBP and then subjected to SDS-PAGE and analyzed by Western blotting using anti-hTBP Abs, GST itself did not associate with hTBP, but there was a strong interaction between anti-hTBP Abs and the GST-REP fusion protein, as indicated by the appearance of a band at 37.7 kDa (Fig. 5A, panel b, lane 2). When the GST-REP fusion protein was present on the blot in the absence of hTBP, the anti-hTBP Abs did not react (data not shown).
FIG. 5.
HHV-6B REP binds specifically to hTBP in vitro. (A) hTBP was retained on GST-REP beads. (a) Gel stained with Coomassie blue. Equal amounts of protein were applied as described for panel b. Lanes: 1, GST (27.5 kDa); 2, GST-REP (84.5 kDa). (b) Proteins bound to GST or GST-REP after incubation with 100 ng of hTBP were collected by centrifugation, washed, boiled in SDS sample buffer, and subjected to SDS-PAGE and Western blotting using an anti-hTBP MAb. Lanes: 1, GST; 2, GST-REP; 3, input hTBP. The input lane contains 10% of the amounts added to the pull-down reaction mixtures. (B) The N-terminal portion of REP is sufficient for REP-hTBP interaction. (a) Gel stained with Coomassie blue. Equal amounts of protein were applied as described for panel b. Lanes: 1, MBP (42.4 kDa); 2, MBP-REP (69.0 kDa). (b) Proteins bound to MBP or MBP-REPN1 after incubation with 100 ng of hTBP were visualized by immunoblotting using anti-hTBP MAb. Lanes: 1, MBP; 2, MBP-REPN1. (C) Coimmunoprecipitation of 35S-labeled REP with hTBP and anti-hTBP MAb. 35S-labeled REP was generated by in vitro transcription-translation. Equal aliquots of either 35S-labeled REP or 35S-labeled luciferase (control) were incubated with hTBP, anti-REP Ab, and protein A, as indicated, and then spun and subjected to SDS-PAGE and autoradiography. (a) Luciferase or REP produced by in vitro transcription and translation using reticulocyte lysate. Lanes: 1, 35S-labeled luciferase protein (61 kDa); 2, 35S-labeled REP protein (56.0 kDa). (b) 35S-labeled REP is coimmunoprecipitated with hTBP. Lanes: 1, 35S-labeled luciferase; 2, 35S-labeled REP.
The region of HHV-6 REP recognized by hTBP was then more specifically defined by carrying out pull-down assays using a fusion protein made of the N-terminal portion of REP and MBP (MBP-REPN1). We observed that MBP-REPN1 was bound by hTBP (Fig. 5B, panel b, lane 2), whereas an equivalent amount of MBP alone was not (Fig. 5B, panel b, lane 1).
Finally, the HHV-6 REP-hTBP interaction was further verified by a coimmunoprecipitation assay (Fig. 5C). The 35S-labeled HHV-6 REP was generated by in vitro transcription-translation and incubated with hTBP, anti-hTBP MAb, and protein A-Sepharose. hTBP coimmunoprecipitated with 35S-labeled HHV-6 REP, whereas hTBP did not bind to the radiolabeled luciferase that served as a control. The anti-hTBP Abs did not immunoprecipitate HHV-6 REP in the absence of hTBP. Thus, the results from two different assay systems clearly show that HHV-6 REP binds hTBP.
These results also indicate that only the N-terminal portion of HHV-6 REP was required for interaction with hTBP.
HHV-6B REP binds hTBP in vivo as determined using the GAL4 mammalian two-hybrid system.
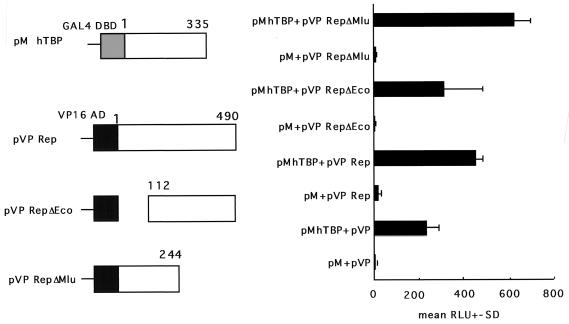
To extend the investigation of the REP-hTBP interaction in vivo, the GAL4 two-hybrid system was used (Fig. 6). hTBP ORF was fused in frame with the GAL4 DNA-binding domain (pM) ORF, and the Rep ORF was fused in frame with the HSV VP16 transactivation domain (pVP) ORF. These constructs were transfected with a reporter plasmid, pG5 E1b Luc, both alone and together. In this assay system, the level of luciferase activity from a GAL4-dependent reporter construct usually correlates with the strength of the in vivo interaction between the two proteins being examined (e.g., large T with p53 [Fig. 6]). These data are shown in Fig. 6. The positive control, pM53 + pVP16-T, was significantly elevated over all of the negative controls (pM + pVP16rep, pMhTBP + pVP16, or pM + pVP16). Importantly, the expression level of PMhTBP + pVP16rep was significantly greater than that of any of the negative controls. Furthermore, we constructed other plasmids, pVP16 RepΔEco and pVP16 RepΔMlu, to analyze which region of Rep was responsible for this interaction. pVP 16 RepΔEco showed higher luciferase activity than pVP16 Rep when cotransfected with pMhTBP. The data, however, vigorously fluctuated, which suggested that pVP16 RepΔEco could have the boundary region for the interaction. On the other hand, pVP16 RepΔMlu showed much higher activity than either pVP16 Rep or pVP16 RepΔEco. This suggested that the N-terminal part up to 244 amino acid should contain the element responsible for their interaction, which was coincident with the in vitro data. Thus, these data demonstrate a measurable in vivo interaction between REP and hTBP and support the validity of the in vitro studies.
FIG. 6.
REP binds hTBP in vivo as determined by GAL4 two-hybrid system. All of the negative controls resulted in relatively low signals (pG5 [reporter plasmid pG5 E1b Luc] + pM [GAL4 DNA binding domain-based expression plasmid] + pVP16 [HSV VP16 transactivation domain-based expression plasmid], pG5 + pMhTBP + pVP16, pG5 + pM + pVP16rep or pG5). In contrast, both the positive control (pG5 + pM53 [p53]; + pVP16-T [large T]) and pG5 + PMhTBP + pVP16rep gave significantly higher signals. Furthermore, pVP16 RepΔEco+pMhTBP showed higher luciferase activity than pVP16 Rep+pMhTBP. On the other hand, pVP RepΔMlu showed much higher activity than either pVP16 Rep or pVP16 RepΔEco. Thus, these data are consistent with a significant REP-TBP interaction in vivo. Data (means + standard deviations [error bars] of the relative luciferase activity) were calculated from triplicate cultures and are representative of three independent experiments.
DISCUSSION
HHV-6 ORF U94 encodes a protein that is homologous to the AAV-2 rep gene product and is unique among human herpesviruses (49). However, although the function of the AAV-2 rep gene is well documented (2, 4, 17–22, 26, 28–30) the function of the HHV-6 rep remains unknown. Interestingly, HHV-6 rep is able to complement replication of a rep-deficient AAV-2 genome (50), suggesting these genes may have similar biological functions.
In this study, we describe three transcripts of different size present in the poly(A)+ RNA of infected MT-4 cells (Fig. 3a). The 5.0- and 2.7-kb mRNAs were mapped within the rep gene region, while the 9.0-kb mRNA was not. In addition, the 5.0- and 2.7-kb (major) mRNAs, which were detected in L phase, appeared to use the same promoter and encode related polypeptides. In AAV-2, REP proteins are translated from four mRNA species whose expression is driven by two promoters, yielding four overlapping polypeptides with apparent molecular masses of 78 kDa (Rep78), 68 kDa (Rep68), 52 kDa (Rep52), and 40 kDa (Rep40) (5, 26, 39, 47). Of these, Rep68 and Rep78 have been shown to bind to AAV terminal hairpin DNA in a sequence-dependent manner, to bind to the origin of replication, and to exhibit ATP-dependent, site-specific endonuclease and DNA helicase activities (5, 26).
Herpesvirus genes have been classified as IE, E, and L (24). Expression of IE and E genes is independent of viral DNA replication; indeed, some IE and E genes are themselves involved in regulating gene expression and DNA replication. The L genes, in contrast, are dependent on viral DNA replication and mainly encode structural proteins (24). There has been one report that, based on reverse transcription-PCR analysis, HHV-6 rep is an IE gene (40). The Northern blot experiments described above indicated that the 2.7-kb transcript appeared abundantly at a later phase of the infectious cycle, although the 5.0-kb transcript (minor) was also detected (Fig. 3a). A 5.2-kb transcript was detected under IE conditions (Fig. 3a [CHX]) and detected faintly under E conditions (Fig. 3a [PFA]). This transcript was more abundant than that of HHV-6 IE1 under IE conditions (Fig. 3a [HHV-6B IE1]). The repression of HHV-6 IE1 by CHX is unusual for a herpesvirus IE gene but has been observed by another group (45). HHV-6 IE1 was expressed faintly with IE kinetics, but its expression increased and it accumulated with virus replication, indicating that the promoter might be activated by virus replication (Fig. 3a). The 5.2-kb transcript was not detected by Northern blotting using either the ER or GR probe (Fig. 3b) or by screening a cDNA library that was prepared from late-stage infected cells (72 h postinfection), indicating that the transcript may not be expressed in the late phase. Moreover, the QR probe (Fig. 2), an antisense oligonucleotide derived from the start site of the rep ORF, detected both the 5.0- and 2.7-kb transcripts in mRNA from cells infected for 60 h but detected no 5.2-kb transcript under the IE or E conditions (Fig. 3b). Therefore, it is not clear if the 5.2-kb transcript detected under the IE and E conditions codes for the REP protein.
In the present study, the HHV-6B REP protein was initially detected in the nucleus 24 h after infection and accumulated to high levels in the nucleus and was apparent in the cytoplasm within 72 h, but the REP protein was not detectable at any time in cells grown in the presence of PFA, CHX, and actinomycin D (data not shown). The reason for this discrepancy, in which the 5.2-kb transcript was detected under the IE condition but the REP protein was not, is not yet clear, but one possibility is that HHV-6 rep is transcribed at the IE stage and translated at the L stage. Alternatively, a low level of HHV-6 rep expression may make it difficult to detect at the IE stage. In addition, we have observed that transcription levels of the U95 gene homologue (Fig. 2), which is antisense with respect to rep, is very high (unpublished data). It is conceivable, therefore, that U95 transcription interferes with the rep transcript. We also tried to detect REP from cells infected with HST and grown in the presence of PFA or CHX for 72 h, by Western blotting and immunoprecipitation, but were not successful. It is possible that the expression level of the REP protein is too low to be detectable. The other possibility is that the 5.2-kb transcript detected under the IE condition may not code for the REP protein. We are currently investigating the nature of the 5.2-kb transcript. Finally, these results suggest that the 2.7-kb transcript codes for REP protein and is expressed at the L stage.
The level of HHV-6 rep transcription within infected cells was apparently small: only 11 cDNA clones were detected from among approximately 106 colonies making up the cDNA library. The reason for this tight control is not clear, but some evidence suggests that overexpression of this gene may be deleterious to the biological activity of HHV-6 (9).
Rotola et al. (43) used reverse transcription-PCR to detect expression of HHV-6 rep in PBMCs from healthy individuals who had latent HHV-6 infections, but the transcription of other genes, including IE genes, was not detectable. These investigators also reported being able to derive lymphoid cells in which HHV-6 REP was stably expressed but in which viral replication was restricted. They postulated that REP regulated viral gene expression in a manner that enabled the establishment or maintenance of latent infection in lymphoid cells.
In the present study, the 5.2-kb transcript was detected under the IE and E conditions by Northern blotting, but not under the L condition. Interestingly, the expression of the 5.2-kb transcript decreased with virus replication, indicating that this transcript might play a important role in the IE phase or in latency.
AAV2 Rep78 appears to bind to a variety of as yet uncharacterized cellular proteins (21) and to suppress heterologous promoters, at least in some cases, via Sp1 sites (20). We suspect that HHV-6 REP acts, at least in part, by interacting with cellular transcription factors. hTBP, which is critical for initiating transcription, seemed an obvious candidate. hTBP exists primarily as a subunit of several larger complexes: the Pol II-specific complex (TFIID) consists of hTBP and 10 to 12 hTBP-associated factors (TAFs), most of which have been highly conserved from Saccharomyces cerevisiae to humans (9, 16, 31, 42, 52) TAFs interact with other transcription factors, which in turn initiate transcription by binding to enhancer elements and to RNA polymerase II and its accessory proteins (19, 45).
In this study, HHV-6 REP binding to hTBP was demonstrated in vitro using pull-down assays (Fig. 5A) and coimmunoprecipitation (Fig. 5C) and in vivo using a mammalian two-hybrid assay (Fig. 6). Furthermore, we showed that the N-terminal portion of HHV-6 REP was sufficient for the interaction with hTBP in vitro and in vivo (Fig. 5B and 6). Two hybrid results suggested that the N-terminal part up to amino acid 244 should contain the element responsible for their interaction, which was coincident with the in vitro data. Taken together, the data still suggested that the N-terminal part should contain the interacting domain and that the coding region around amino acid 114 could be the boundary, although more precise analyses should be done. On the other hand, we could detect two bands in the coimmunoprecipitation assay (Fig. 5C). Although it is not clear that the small band (40.5 kDa) is specific, it may indicate the existence of N-terminal products of REP.
It appears, therefore, that by binding to hTBP, HHV-6 REP reduces the efficiency with which transcription is initiated. The mechanism of this inhibitory effect is as yet unknown. Perhaps HHV-6 REP contains a domain that, when bound to hTBP, actively interferes with some aspect of initiation; or, HHV-6 REP could block the binding of another, positively acting factor. For instance, HHV-6 REP binding to hTBP might significantly affect hTBP-TAF interactions and prevent certain TAFs from entering the TFIID complex. A similar mechanism has been suggested for p53-mediated inhibition of transcription (46). Another possibility is that HHV-6 REP itself may serve as a TAF, functioning as a connector or modulator in a manner analogous to SV40 large T (11, 37).
It has been reported that Rep78 binds to hTBP both in vitro and in vivo (22). It was also recently shown that Rep78 binds to CREB, thereby blocking activation of CREB-dependent transcription by protein kinase A (12) and that both Rep78 and Rep68 bind to transcription coactivator PC4 (53). Accordingly, we suggest that the function of the HHV-6 REP-hTBP interaction may be to modulate HHV-6 replication. This scenario supports a model wherein suppression by HHV-6 REP is involved in an autoregulatory loop controlling HHV-6 rep gene expression.
ACKNOWLEDGMENTS
This study was supported in a part by a grant-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Ablashi D, Agut H, Berneman Z, Campadelli-Fiume G, Carrigan D, Ceccerini-Nelli L, Chandran B, Chou S, Collandre H, Cone R, Dambaugh T, Dewhurst S, DiLuca D, Foa-Tomasi L, Fleckenstein B, Frenkel N, Gallo R, Gompels U, Hall C, Jones M, Lawrence G, Martin M, Montagnier L, Neipel F, Nicholas J, Pellet P, Razzaque A, Torrelli G, Thomson B, Salahuddin S, Wyatt L, Yamanishi K. Human herpesvirus-6 strain groups: a nomenclature. Arch Virol. 1993;129:363–366. doi: 10.1007/BF01316913. [DOI] [PubMed] [Google Scholar]
- 2.Antoni B A, Rabson A B, Miller I L, Trempe J P, Chejanovsky N, Carter B J. Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J Virol. 1991;65:396–404. doi: 10.1128/jvi.65.1.396-404.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araujo J C, Doniger J, Kashanchi F, Hermonat P L, Thompson J, Rosenthal L J. Human herpesvirus 6A ts suppresses both transformation by H-ras and transcription by the H-ras and human immunodeficiency virus type 1 promoters. J Virol. 1995;69:4933–4940. doi: 10.1128/jvi.69.8.4933-4940.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaton A, Palumbo P, Berns K I. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the rep protein. J Virol. 1989;63:4450–4454. doi: 10.1128/jvi.63.10.4450-4454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becerra S P, Koczot F, Fabisch P, Rose J A. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J Virol. 1988;62:2745–2754. doi: 10.1128/jvi.62.8.2745-2754.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blauvelt A, Herndier B G, Orenstein J M. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N Engl J Med. 1997;336:1837–1839. doi: 10.1056/NEJM199706193362517. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Braun D K, Dominguez G, Pellett P E. Human herpesvirus 6. Clin Microbiol Rev. 1997;10:521–567. doi: 10.1128/cmr.10.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 10.Carrigan D R, Knox K K, Tapper M A. Suppression of human immunodeficiency virus type 1 replication by human herpesvirus-6. J Infect Dis. 1990;162:844–851. doi: 10.1093/infdis/162.4.844. [DOI] [PubMed] [Google Scholar]
- 11.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 12.Di Pasquale G, Stacey S N. Adeno-associated virus Rep78 protein interacts with protein kinase A and its homolog PRKX and inhibits CREB-dependent transcriptional activation. J Virol. 1998;72:7916–7925. doi: 10.1128/jvi.72.10.7916-7925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foreman K E, Friborg J, Jr, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 14.Giraud C, Winocour E, Berns K I. Recombinant junctions formed by site-specific integration of adeno-associated virus into an episome. J Virol. 1995;69:6917–6924. doi: 10.1128/jvi.69.11.6917-6924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 16.Hahn S. The role of TAFs in RNA polymerase II transcription. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 17.Heilbronn R, Burkle A, Stephan S, zur Hausen H. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J Virol. 1990;64:3012–3018. doi: 10.1128/jvi.64.6.3012-3018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermonat P L. Down-regulation of the human c-fos and c-myc proto-oncogene promoters by adeno-associated virus Rep78. Cancer Lett. 1994;81:129–136. doi: 10.1016/0304-3835(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 19.Hermonat P L. Adeno-associated virus inhibits human papillomavirus type 16: a viral interaction implicated in cervical cancer. Cancer Res. 1994;54:2278–2281. [PubMed] [Google Scholar]
- 20.Hermonat P L, Santin A D, Batchu R B. The adeno-associated virus Rep78 major regulatory/transformation suppressor protein binds cellular Sp1 in vitro and evidence of a biological effect. Cancer Res. 1996;56:5299–5304. [PubMed] [Google Scholar]
- 21.Hermonat P L, Santin A D, Carter C A, Parham G P, Quirk J G. Multiple cellular proteins are recognized by the adeno-associated virus Rep78 major regulatory protein and the amino-half of Rep78 is required for many of these interactions. Biochem Mol Biol Int. 1997;43:409–420. doi: 10.1080/15216549700204201. [DOI] [PubMed] [Google Scholar]
- 22.Hermonat P L, Santin A D, Batchu R B, Zhan D. The adeno-associated virus Rep78 major regulatory protein binds the cellular TATA-binding protein in vitro and in vivo. Virology. 1998;245:120–127. doi: 10.1006/viro.1998.9144. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann A, Roeder R G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991;19:6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 26.Im D S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Isegawa Y, Mukai T, Nakano K, Kagawa M, Chen J, Mori Y, Sunagawa T, Kawanishi K, Sashihara J, Hata A, Zou P, Kosuge H, Yamanishi K. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J Virol. 1999;73:8053–8063. doi: 10.1128/jvi.73.10.8053-8063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellermann O K, Ferenci T. Maltose-binding protein from Escherichia coli. Methods Enzymol. 1982;90:459–463. doi: 10.1016/s0076-6879(82)90171-9. [DOI] [PubMed] [Google Scholar]
- 28.Kyostio S R, Wonderling R S, Owens R A. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J Virol. 1995;69:6787–6796. doi: 10.1128/jvi.69.11.6787-6796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labow M A, Hermonat P L, Berns K I. Positive and negative autoregulation of the adeno-associated virus type 2 genome. J Virol. 1986;60:251–258. doi: 10.1128/jvi.60.1.251-258.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labow M A, Graf L H, Jr, Berns K I. Adeno-associated virus gene expression inhibits cellular transformation by heterologous genes. Mol Cell Biol. 1987;7:1320–1325. doi: 10.1128/mcb.7.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee T I, Young R A. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 32.Linden R M, Winocour E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linden R M, Ward P, Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez C, Pellett P, Stewart J, Goldsmith C, Sanderlin K, Black J, Warfield D, Feorino P. Characteristics of human herpesvirus-6. J Infect Dis. 1988;157:1271–1273. doi: 10.1093/infdis/157.6.1271. [DOI] [PubMed] [Google Scholar]
- 35.Luppi M, Marasca R, Barozzi P, Ferrari S, Ceccherini-Nelli L, Batoni G, Merelli E, Torelli G. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol. 1993;40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 36.Luppi M, Barozzi P, Marasca R, Torelli G. Integration of human herpesvirus-6 (HHV-6) genome in chromosome 17 in two lymphoma patients. Leukemia. 1994;8(Suppl. 1):S41–S45. [PubMed] [Google Scholar]
- 37.Martin D W, Subler M A, Munoz R M, Brown D R, Deb S P, Deb S. p53 and SV40 T antigen bind to the same region overlapping the conserved domain of the TATA-binding protein. Biochem Biophys Res Commun. 1993;195:428–434. doi: 10.1006/bbrc.1993.2061. [DOI] [PubMed] [Google Scholar]
- 38.Matsuura Y, Possee R D, Overton H A, Bishop D H L. Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J Gen Virol. 1987;68:1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- 39.Mendelson E, Trempe J P, Carter B J. Identification of the trans-acting Rep proteins of adeno-associated virus by antibodies to a synthetic oligopeptide. J Virol. 1986;60:823–832. doi: 10.1128/jvi.60.3.823-832.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirandola P, Menegazzi P, Merighi S, Ravaioli T, Cassai E, Di Luca D. Temporal mapping of transcripts in herpesvirus 6 variants. J Virol. 1998;72:3837–3844. doi: 10.1128/jvi.72.5.3837-3844.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mori Y, Yagi H, Shimamoto T, Isegawa Y, Sunagawa T, Inagi R, Kondo K, Tano Y, Yamanishi K. Analysis of human herpesvirus 6 U3 gene, which is a positional homolog of human cytomegalovirus UL 24 gene. Virology. 1998;249:129–139. doi: 10.1006/viro.1998.9305. [DOI] [PubMed] [Google Scholar]
- 42.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 43.Rotola A, Ravaioli T, Gonelli A, Dewhurst S, Cassai E, Di Luca D. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc Natl Acad Sci USA. 1998;95:13911–13916. doi: 10.1073/pnas.95.23.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salahuddin S Z, Ablashi D V, Markham P D, Josephs S F, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 45.Schiewe U, Neipel F, Schreiner D, Fleckenstein B. Structure and transcription of an immediate-early region in the human herpesvirus 6 genome. J Virol. 1994;68:2978–2985. doi: 10.1128/jvi.68.5.2978-2985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seto E, Usheva A, Zambetti G P, Momand J, Horikoshi N, Weinmann R, Levine A J, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci USA. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeda K, Haque M, Sunagawa T, Okuno T, Isegawa Y, Yamanishi K. Identification of a variant B-specific neutralizing epitope on glycoprotein H of human herpesvirus-6. J Gen Virol. 1997;78:2171–2178. doi: 10.1099/0022-1317-78-9-2171. [DOI] [PubMed] [Google Scholar]
- 49.Thomson B J, Efstathiou S, Honess R W. Acquisition of the human adeno-associated virus type-2 rep gene by human herpesvirus type-6. Nature. 1991;351:78–80. doi: 10.1038/351078a0. [DOI] [PubMed] [Google Scholar]
- 50.Thomson B J, Weindler F W, Gray D, Schwaab V, Heilbronn R. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology. 1994;204:304–311. doi: 10.1006/viro.1994.1535. [DOI] [PubMed] [Google Scholar]
- 51.Torelli G, Barozzi P, Marasca R, Cocconcelli P, Merelli E, Ceccherini-Nelli L, Ferrari S, Luppi M. Targeted integration of human herpesvirus 6 in the p arm of chromosome 17 of human peripheral blood mononuclear cells in vivo. J Med Virol. 1995;46:178–188. doi: 10.1002/jmv.1890460303. [DOI] [PubMed] [Google Scholar]
- 52.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 53.Weger S, Wendland M, Kleinschmidt J A, Heilbronn R. The adeno-associated virus type 2 regulatory proteins rep78 and rep68 interact with the transcriptional coactivator PC4. J Virol. 1999;73:260–269. doi: 10.1128/jvi.73.1.260-269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;i:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]