ABSTRACT
Macroautophagy/autophagy is a complex degradation process with a dual role in cell death that is influenced by the cell types that are involved and the stressors they are exposed to. Ferroptosis is an iron-dependent oxidative form of cell death characterized by unrestricted lipid peroxidation in the context of heterogeneous and plastic mechanisms. Recent studies have shed light on the involvement of specific types of autophagy (e.g. ferritinophagy, lipophagy, and clockophagy) in initiating or executing ferroptotic cell death through the selective degradation of anti-injury proteins or organelles. Conversely, other forms of selective autophagy (e.g. reticulophagy and lysophagy) enhance the cellular defense against ferroptotic damage. Dysregulated autophagy-dependent ferroptosis has implications for a diverse range of pathological conditions. This review aims to present an updated definition of autophagy-dependent ferroptosis, discuss influential substrates and receptors, outline experimental methods, and propose guidelines for interpreting the results.
Abbreviation: 3-MA:3-methyladenine; 4HNE: 4-hydroxynonenal; ACD: accidentalcell death; ADF: autophagy-dependentferroptosis; ARE: antioxidant response element; BH2:dihydrobiopterin; BH4: tetrahydrobiopterin; BMDMs: bonemarrow-derived macrophages; CMA: chaperone-mediated autophagy; CQ:chloroquine; DAMPs: danger/damage-associated molecular patterns; EMT,epithelial-mesenchymal transition; EPR: electronparamagnetic resonance; ER, endoplasmic reticulum; FRET: Försterresonance energy transfer; GFP: green fluorescent protein;GSH: glutathione;IF: immunofluorescence; IHC: immunohistochemistry; IOP, intraocularpressure; IRI: ischemia-reperfusion injury; LAA: linoleamide alkyne;MDA: malondialdehyde; PGSK: Phen Green™ SK;RCD: regulatedcell death; PUFAs: polyunsaturated fatty acids; RFP: red fluorescentprotein;ROS: reactive oxygen species; TBA: thiobarbituricacid; TBARS: thiobarbituric acid reactive substances; TEM:transmission electron microscopy.
KEYWORDS: Cell death, ferritinophagy, iron, lipid peroxidation, lipophagy, lysosome
Introduction
Maintaining a delicate balance between cell survival and cell death is essential for the normal biological functions of living organisms. The disruption in this equilibrium can be pathogenic. For example, impaired cell death mechanisms can result in uncontrolled cell proliferation and tumorigenesis, while excessive cell death may disrupt normal tissue function and contribute to inflammation-related diseases. Cell death can be broadly categorized into accidental cell death (ACD) and regulated cell death (RCD) [1]. Unlike ACD, RCD is a dynamic process precisely regulated by signal transduction pathways and molecular networks. Whereas apoptosis has been extensively studied for many years [2], recent attention has shifted toward investigating the underlying mechanisms of non-apoptotic forms of RCD and their potential therapeutic applications [3–9].
Among the non-apoptotic types of RCD, ferroptosis was defined in 2012 as an iron-dependent cell death pathway characterized by the production of reactive oxygen species (ROS) through the Fenton reaction, resulting in unrestricted lipid peroxidation [5]. The core mechanism of ferroptosis bears a striking resemblance to the early notion of “oxytosis”, which described a glutamine-induced cell death involving glutathione (GSH) depletion and oxidative damage in neural cells [10]. Initially, ferroptosis was observed to selectively target cancer cells harboring oncogenic RAS mutations [11,12]. However, further research has revealed that ferroptosis can also occur in non-cancer cells or normal tissues independently of RAS mutation [13–16], highlighting its broad implication in various physiological and pathological conditions. The pharmacological induction of ferroptosis (e.g., using analogs of erastin or RSL3) holds great promise as a prospective strategy for cancer therapy, while the application of ferroptosis inhibitors (e.g., using ferrostatin-1 and liproxstatin-1) offers new opportunities for suppressing lipid peroxidation-related tissue damage and diseases in preclinical models [17–19].
Autophagy is a lysosome-dependent degradation process that plays a fundamental role in determining cell fate under various stress conditions [20–22]. Autophagy is often considered a pro-survival mechanism. It helps cells survive under stress conditions, such as nutrient deprivation, by recycling cellular components and providing energy and building blocks. Autophagy can remove damaged organelles and proteins, protecting cells from accumulating harmful materials that could trigger cell death. Although autophagy was initially viewed in part as a form of self-cannibalistic cell death that can occur during development [23], subsequent research suggested that the term “autophagic cell death” often used to describe the escalated autophagy process accompanying cell death, may be better characterized as an exaggerated autophagic activity during cell demise [24]. Nevertheless, there are validated contexts in which autophagy promotes cell death, either directly or indirectly through mitochondrial depletion [25–27]. Historically, ferroptosis was initially described as an autophagy-independent form of cell death. This interpretation was supported by studies demonstrating the ineffectiveness of the widely used autophagy inhibitor chloroquine (CQ) in blocking erastin-induced cell death in HT-1080, BJeLR, and Calu-1 cells [5]. However, emerging genetic evidence from various cell lines deficient in autophagy-related genes (e.g., ATG5 [autophagy related 5] and BECN1/Vps30/Atg6 [beclin 1]) suggests that ferroptosis can indeed be an autophagy-dependent form of cell death [28–31]. Although the signal and mechanism of autophagy-dependent ferroptosis (ADF) are still under investigation, it is suggested that impaired ADF is involved in the pathogenesis of various diseases.
In this review, our aim is to provide a comprehensive overview of the intricate relationship between autophagy and ferroptosis. Additionally, we propose guidelines for the definition, detection, and interpretation of ADF.
A brief overview of the ferroptotic machinery
Ferroptosis can occur through two major pathways: the extrinsic or transporter-dependent pathway, involving decreased cystine uptake and increased iron uptake, and the intrinsic or enzyme-regulated pathway, which includes the inhibition of GPX4 (glutathione peroxidase 4) (Figure 1). Although the ferroptotic machinery exhibits heterogeneity and plasticity [32], it appears that the SLC7A11 (solute carrier family 7 member 11)-GSH-GPX4 axis plays a major role in inhibiting ferroptosis [33]. Notably, classical ferroptosis inducers, such as erastin and RSL3, target SLC7A11 and GPX4, respectively [5,34]. SLC7A11 is a functional component of system xc−, which acts along with SLC3A2 as a plasma membrane amino acid antiporter that imports cystine from the extracellular space into the intracellular space, leading to subsequent cysteine production and synthesis of the antioxidant GSH. GSH serves as a cofactor for many enzymes and is vital for the activity of GPX4, the most important antioxidant enzyme responsible for reducing lipid peroxides/PLOOH to lipid alcohols [35]. Treatment with erastin or RSL3 induces peroxidation of polyunsaturated fatty acids (PUFAs), which are predominantly localized in cell membranes. This process involves ACSL4 (acyl-CoA synthetase long chain family member 4)-mediated production of PUFA-derived acyl-CoAs, which are utilized for the synthesis of various biological components, including phospholipids [36–38]. The initiation of lipid peroxidation is mediated by iron-dependent enzymes of the ALOX (arachidonate lipoxygenase) family or POR (cytochrome p450 oxidoreductase) enzymes [39–41]. These enzymes produce toxic lipid metabolites (such as 4-hydroxynonenal [4HNE] and 15-hydroperoxy-arachidonoyl-phosphatidylethanolamine [15-HpETE-PE]), leading to plasma membrane rupture and the release of danger/damage-associated molecular patterns (DAMPs) into the extracellular space. Enzymes, such as the Ca2+-independent PLA2G6 (phospholipase A2 group VI) and ALDH1B1 (aldehyde dehydrogenase 1 family member B1), which have a preference for hydrolyzing peroxidized phospholipids, play a critical role in eliminating signals that trigger ferroptotic cell death [42,43]. As a final line of defense, the assembly and activation of the endosomal sorting complex required for transport III/ESCRT-III complex are involved in repairing small membrane wounds caused by ferroptotic damage [44,45].
Figure 1.
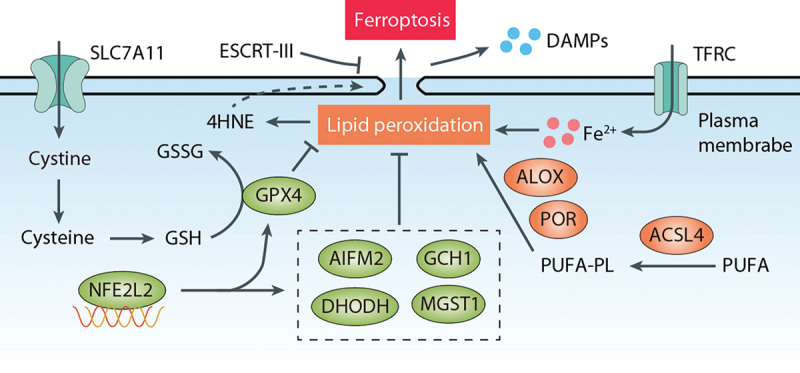
Overview of ferroptosis. The SLC7A11-GSH-GPX4 pathway and GPX4-independent pathways (such as AIFM2, GCH, DHODH, and MGST1) inhibit lipid peroxidation, whereas ACSL4, ALOX, and POR promote peroxidation of PUFA-containing phospholipids (PUFA-PL). Cystine is transported into cells by SLC7A11 and rapidly reduced to cysteine, which is utilized for GSH synthesis. GPX4 inhibits lipid peroxidation, the primary driver of ferroptosis, by converting GSH to GSSG. The transcription factor NFE2L2 serves as a key antioxidant system by upregulating genes involved in both GPX4-dependent and GPX4-independent pathways. Lipid peroxidation generates 4HNE, which can induce membrane damage and release DAMPs. Conversely, the endosomal sorting complex required for transport (ESCRT-III) machinery acts as a protective mechanism that delays membrane damage. TFRC increases intracellular iron levels, thereby promoting lipid peroxidation and ferroptosis.
Antioxidant enzymes are vital in catalyzing the transformation of ROS and their by-products into stable and nontoxic molecules, serving as a critical defense mechanism against cellular damage caused by oxidative stress [46]. Whereas GPX4 is well-known, several GPX4-independent antioxidant enzymes, such as AIFM2/FSP1 (apoptosis inducing factor mitochondria associated 2), DHODH (dihydroorotate dehydrogenase (quinone)), GCH1 (GTP cyclohydrolase 1), and MGST1 (microsomal glutathione S-transferase 1), exhibit alternative or parallel roles in GPX4-deficient or GPX4 expressing cells, although ongoing debates surround their specific functions [47–55]. Alongside these enzymes, the transcription factor NFE2L2/NRF2 (NFE2 like bZIP transcription factor 2) plays a multifaceted role in maintaining cellular redox homeostasis and inhibiting ferroptosis by upregulating genes involved in both GPX4-dependent and GPX4-independent pathways [56–60]. The identification of additional antioxidants, such as hydropersulfides, capable of blocking ferroptosis further emphasizes the existence of multiple defense systems within our cells and body [61,62]. Beyond the influence of GPX4, the inhibition of ferroptosis can also occur through the reconfiguration of the cellular phospholipid profile. This intricate process is orchestrated by phospholipid-modifying enzymes, namely MBOAT1 (membrane bound O-acyltransferase domain containing 1) and MBOAT2 [63].
A brief overview of autophagic machinery
Autophagy, a highly conserved cellular process, can be classified into three main types: microautophagy, macroautophagy, and chaperone-mediated autophagy (CMA) [64,65]. Microautophagy involves the direct invagination and fission of the vacuolar/lysosomal membrane to degrade cytoplasmic cargo [66], whereas CMA utilizes chaperones to deliver unfolded, individual substrate proteins across the lysosomal membrane for degradation [67]. Macroautophagy (hereafter referred to as autophagy) involves the formation of transient double-membrane compartments, phagophores, which engulf cellular components and close to form autophagosomes that subsequently fuse with lysosomes for degradation [68]. Macroautophagy is the most extensively studied form of autophagy in mammalian cells and diseases [69–72].
Understanding the complex process of autophagy provides insights into its importance for maintaining cellular homeostasis and adapting to various stress conditions. The process of classical autophagy involves several key steps: 1) Induction: Autophagy can be triggered by various cellular stressors, such as nutrient deprivation, oxidative stress, or the accumulation of damaged proteins or organelles. These stressors activate specific signaling pathways, including the MTOR (mechanistic target of rapamycin kinase) pathway, AMP-activated protein kinase (AMPK) pathway, or oxidative stress response pathways, which converge on the activation of ATG (autophagy related) proteins. 2) Nucleation: The initiation of autophagy involves the formation of the double-membraned phagophore, which is the dynamic sequestering compartment. Two key complexes, on the one hand, the ULK complex (including ULK1 [unc-51 like autophagy activating kinase 1], ATG13 [autophagy related 13], ATG101 [autophagy related 101], and RB1CC1/FIP200 [RB1 inducible coiled-coil 1]) and, on the other hand, the phosphatidylinositol 3-kinase/PtdIns3K complex (including PIK3C3/Vps34 [phosphatidylinositol 3-kinase catalytic subunit type 3], PIK3R4/Vps15 [phosphoinositide-3-kinase regulatory subunit 4], ATG14 [autophagy related 14], NRBF2 [nuclear receptor binding factor 2] and BECN1), play essential roles in phagophore assembly. 3) Expansion and elongation: The phagophore expands and elongates, forming a complete autophagosome, which is a double-membraned vesicle that contains the cargo targeted for degradation. This step requires the recruitment and activity of the ATG12 (autophagy related 12) – ATG5-ATG16L1 (autophagy related 16 like 1) complex. This complex acts as a ubiquitin-like conjugating system and facilitates the covalent attachment of phosphatidylethanolamine to Atg8-protein family members, especially MAP1LC3 (microtubule associated protein 1 light chain 3) in mammals. The lipidation of MAP1LC3 proteins directs their localization to the growing phagophore membrane, contributing to phagophore expansion and maturation [73]. 4) Cargo recognition and sequestration: The autophagosome recognizes and sequesters specific cargo, such as damaged organelles, protein aggregates, or invading pathogens. Autophagy receptors mediate this recognition by interacting with both the cargo and MAP1LC3 on the phagophore membrane, thus determining the selectivity of autophagic degradation [74]. 5) Fusion with a lysosome: The mature autophagosome fuses with a lysosome, forming an autolysosome; this may be preceded by fusion with an endosome to form an intermediate amphisome. This fusion process involves soluble N-ethylmaleimide-sensitive factor attachment protein receptors/SNARE proteins and enables the degradation of the sequestered cargo by lysosomal hydrolases. 6) Cargo degradation and recycling: The cargo inside the autolysosome is degraded by lysosomal enzymes, and the resulting breakdown products are released into the cytoplasm for recycling and reuse by the cell. 7) Termination: Once the cargo is degraded and recycled, the autolysosome undergoes autophagic lysosome reformation/ALR to regenerate a functional lysosome.
Concept of autophagy-dependent ferroptosis
Within the normal cellular life cycle, macromolecules fulfill their intended functions but must also undergo degradation or turnover to maintain tight regulation. Additionally, cellular components, such as proteins and organelles, can suffer damage, necessitating mechanisms to eliminate these damaged entities and ensure cell viability [75]. Autophagy emerges as a critical player in cellular responses to stress, capable of promoting cell survival or triggering cell death, depending on the intensity and duration of the stress.
Autophagy-dependent cell death refers to a form of RCD that mechanistically relies on the autophagic machinery or its components, as defined by the Nomenclature Committee on Cell Death [1]. In a similar vein, we define ADF as a form of ferroptotic cell death that mechanistically depends on the autophagic machinery or its components. In contrast, cases where ferroptosis occurs in the presence of protective autophagy, or when autophagy does not induce ferroptosis or any other form of cell death, fall under the category of “autophagy-independent ferroptosis” (Figure 2). To avoid ambiguity, we refrain from using terms such as “autophagy-mediated ferroptosis” or “autophagy-induced ferroptosis” as autophagy can also mediate or enhance apoptosis and necroptosis [76]. Therefore, it is essential to inhibit autophagy genetically or pharmacologically to strictly demonstrate the dependence of ferroptosis on autophagy. Combining both genetic and chemical approaches is recommended to establish the crucial role of autophagy in the ferroptosis process, considering the potential off-target effects of compounds and the autophagy-independent functions of certain autophagy-related genes and proteins.
Figure 2.
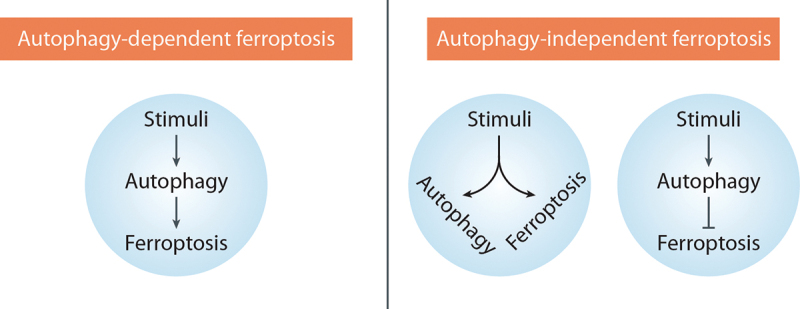
Classification of ferroptosis based on autophagic response. Autophagy-dependent ferroptosis: this process relies on the autophagic machinery to induce ferroptosis. Autophagy-independent ferroptosis: In this case, autophagy occurs alongside ferroptosis but does not directly induce ferroptosis or may even have a protective role in the context of ferroptosis.
Furthermore, it is essential to highlight that the occurrence and outcomes of ADF are context-dependent and can vary across different cell types and experimental models [77]. While an increased autophagy response correlates with enhanced sensitivity to ferroptosis in many cases, it is important to note that not all instances of ferroptosis depend on autophagy. In certain scenarios, autophagy can actually exert a protective role against ferroptosis [78], as discussed in the section on “The Protective Roles of Autophagy in Ferroptosis”. Hence, to fully comprehend the complex mechanisms underlying the interplay between autophagy and ferroptosis, it is crucial to consider the specific context in which these processes occur. An array of diverse factors, such as cell types, experimental conditions, and the presence of modulatory circumstances, can influence the relationship between autophagy and ferroptosis, leading to diverse outcomes.
Mechanisms of autophagy-dependent ferroptosis
Non-selective autophagy, also known as bulk autophagy, apparently involves the nondiscriminatory engulfment of a portion of the cell’s cytoplasm. In contrast, selective autophagy is a more precise process that specifically targets and eliminates distinct cellular components [79,80]. The impact of selective autophagy on cell fate determination can vary depending on the specific context. Selective autophagy relies on autophagy receptors that are specific to the cargo being targeted [81]. These receptors recognize and interact with the cargo, facilitating its sequestration by a phagophore for subsequent degradation. In the context of ferroptosis, various forms of selective autophagy have been implicated in promoting this form of cell death, usually through indirect mechanisms in which selective autophagy favors biochemical pathways that facilitate pro-ferroptotic oxidative reactions (see below). The activation of ADF is further regulated by multiple stress signal pathways, either directly or indirectly. Indeed, protein-protein interactions or sensors of stress signals play a crucial role in controlling the activation of ADF. Phase separation is an important process that promotes autophagy function through facilitating the assembly of autophagosome and degradation of protein cargos [82]. While the involvement of phase separation in ADF remains unclear, elucidation of the role of phase separation in ADF will provide insights into the molecular mechanism underlying ferroptosis.
Ferritinophagy
Ferritin is a cytosolic protein involved in the storage of iron and is composed of two subunits, FTH1 (ferritin heavy chain 1) and FTL (ferritin light chain). NCOA4 (nuclear receptor coactivator 4) acts as a receptor for the autophagic degradation of ferritin, a process known as ferritinophagy, and plays a crucial role in maintaining iron homeostasis [83]. The relationship between autophagy, specifically ferritinophagy, and ferroptosis was first elucidated by two research groups in 2016 [28,29]. They discovered that ATG genes, including ATG3 (autophagy related 3), ATG5, ATG7 (autophagy related 7), ATG13, and ULK1, serve as positive regulators of ferroptosis in both normal and tumor cells. Classical autophagy inhibitors, such as CQ, bafilomycin A1, and 3-methyladenine (3-MA), prevent lipid peroxidation and ferroptosis induced by erastin or RSL3 in mouse embryonic fibroblasts/MEFs, PANC1, and HT-1080 cells [28,29,84]. The activation of NCOA4-mediated degradation of ferritin leads to an increase in intracellular Fe2+ levels, ultimately resulting in the induction of ferroptosis. Conversely, knockdown of NCOA4 prevents the accumulation of Fe2+ and lipid peroxidation induced by ferroptosis inducers [28,29]. These findings establish a genetic link between autophagy and ferroptosis.
As a key regulator of ferritinophagy, the expression and function of NCOA4 are controlled by both transcriptional and post-transcriptional mechanisms (Figure 3). Hypoxia decreases NCOA4 transcription and inhibits ferritinophagy-dependent ferroptosis in macrophages [85], while PTBP1 (polypyrimidine tract binding protein 1) promotes ferroptosis in liver cancer cells by regulating NCOA4 translation [86]. TRIM7 (tripartite motif containing 7), an E3 ubiquitin ligase, directly interacts with NCOA4 and mediates its K48-linkage ubiquitination, thereby reducing NCOA4-mediated ferritinophagy and ferroptosis in human glioblastoma cells [87]. NFE2L2-mediated transcription of HERC2 (HECT and RLD domain containing E3 ubiquitin protein ligase 2), an E3 ubiquitin ligase for NCOA4, leads to a simultaneous decrease in ferritin and NCOA4 and the inhibition of ferritinophagy [58]. ATM (ATM serine/threonine kinase) phosphorylates NCOA4, facilitating the interaction between NCOA4 and ferritin and thereby sustaining ferritinophagy-dependent ferroptosis [88].
Figure 3.
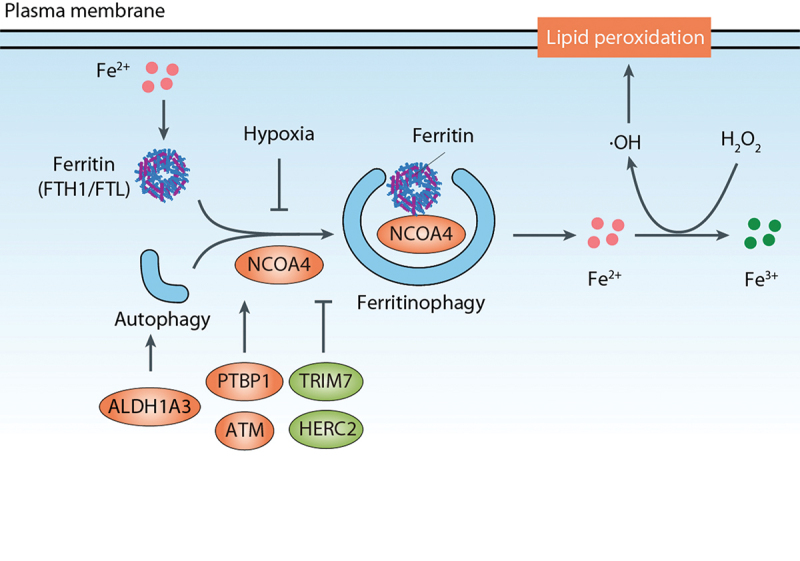
Ferritinophagy in ferroptosis. Ferritin is a cytosolic protein involved in iron storage, and NCOA4 serves as a receptor for the autophagic degradation of ferritin, a process referred to as ferritinophagy. Conditions and factors such as hypoxia, TRIM7, and HERC2 decrease NCOA4 expression, thereby inhibiting ferritinophagy-dependent ferroptosis. Conversely, PTBP1 and ATM promote NCOA4-mediated ferritinophagy. Additionally, ALDH1A3 can bind to MAP1LC3 to enhance ferritinophagy-dependent ferroptosis.
In addition, ALDH1A3 (aldehyde dehydrogenase 1 family member A3), a member of the ALDH (aldehyde dehydrogenase) family, can bind to MAP1LC3 to enhance ferritinophagy-dependent ferroptosis [89]. Interestingly, other ALDH family enzymes, including ALDH1B1, ALDH2 (aldehyde dehydrogenase 2 family member), and ALDH3A2 (aldehyde dehydrogenase 3 family member A2), negatively regulate ferroptotic cell death [43,90,91]. Thus, the function of ALDH in autophagy and ferroptosis is context-dependent.
Besides classical ferroptosis inducers like erastin, certain anticancer agents, such as chrysin, found in honey and propolis, can trigger ferritinophagy-dependent ferroptotic death to suppress tumor growth. Chrysin enhances chemosensitivity to gemcitabine by directly targeting CBR1 (carbonyl reductase 1) in pancreatic cancer cells, resulting in increased tumor suppression [92]. Because ferritinophagy likely interacts with other cellular processes, such as iron metabolism, redox regulation, and other forms of autophagy [93], investigating the crosstalk between ferritinophagy and these processes will provide a more comprehensive understanding of its role in cellular physiology and disease.
Lipophagy
Lipophagy refers to the process of autophagic clearance of lipid droplets, which serve as storage organelles for neutral lipids [94]. These lipid droplets, found in various organisms from bacteria to humans, consist of a hydrophobic core of neutral lipids (such as triglycerides and sterol esters) surrounded by a polar lipid monolayer and specific proteins. The biogenesis and degradation of lipid droplets are tightly regulated by various signals and pathophysiological conditions [95].
Lipophagy plays a role in promoting ferroptotic cell death [96]. In ferroptotic cells, aggregation of lipid droplets is observed at the early stages, potentially acting as a protective response against oxidative damage [96]. Likewise, the formation of lipid droplets mediated by PLTP (phospholipid transfer protein) antagonizes the occurrence of ferroptosis [97]. However, as cells progress and pro-ferroptotic signals intensify, lipophagy degrades the lipid droplets. This releases free fatty acids that serve as substrates for lipid peroxidation, ultimately culminating in ferroptosis [96]. This lipophagy-dependent ferroptosis is positively regulated by PGRMC1 (progesterone receptor membrane component 1)-induced tubulin detyrosination, although PGRMC1 has also a direct role in promoting autophagy by binding with MAP1LC3 and UVRAG (UV radiation resistance associated) [98,99].
Manipulation of lipid droplet dynamics has also been linked to ferroptosis regulation (Figure 4). Depletion of TPD52 (tumor protein D52), a resident protein in lipid droplets involved in lipid storage, increases ferroptosis induced by RSL3 in HepG2 cells [96]. The small guanosine triphosphatase (GTPase) RAB7A (RAB7A, member RAS oncogene family), which is associated with lipid droplets, drives lipophagy, and its knockdown significantly prevents RSL3-induced lipid peroxidation and subsequent ferroptosis [96]. Additionally, the heme-containing membrane protein PGRMC1 triggers lipophagy-dependent ferroptosis in cancer cells [98].
Figure 4.
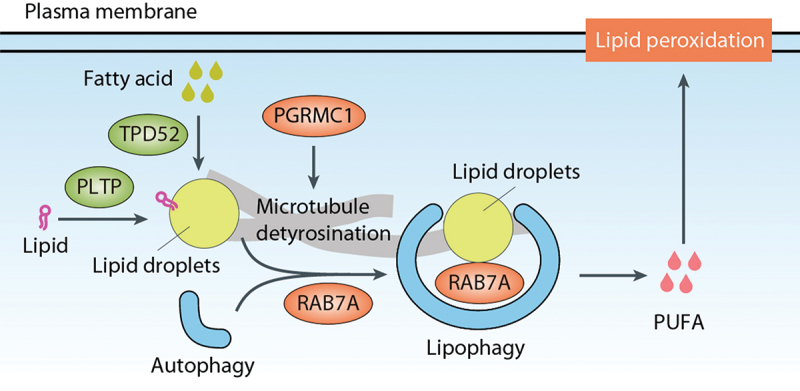
Lipophagy in ferroptosis. Lipophagy refers to the autophagic clearance of lipid droplets, which function as storage organelles for neutral lipids. RAB7A, associated with lipid droplets, drives lipophagy and promotes subsequent ferroptosis. The formation of lipid droplets, mediated by TPD52 and PLTP, antagonizes lipophagy-dependent ferroptosis. In contrast, PGRMC1 triggers lipophagy-dependent ferroptosis. PUFA, polyunsaturated fatty acid.
Altogether, these findings highlight the role of lipophagy-dependent ferroptosis as a potential approach for targeting and eradicating cancer cells. The interplay between lipophagy and ferroptosis offers new insights into the complex mechanisms underlying lipid metabolism and cell death pathways.
Mitophagy
Mitophagy, the selective autophagic clearance of damaged, dysfunctional or superfluous mitochondria, is a conserved process from yeast to humans [100,101]. While the role of mitochondria in ferroptosis is still debated [5,102], emerging evidence suggests that mitophagy may play a role in ferroptosis induction [103]. Several observations support this link between mitophagy and ferroptosis (Figure 5):
Key effectors of mitophagy, including PINK1 (PTEN induced kinase 1), DNM1L/DRP1 (dynamin 1 like), and FUNDC1 (FUN14 domain containing 1), positively regulate ferroptosis. The PINK1-PRKN (parkin RBR E3 ubiquitin protein ligase) pathway is a well-studied mitophagy-stimulatory pathway, where PINK1 phosphorylates ubiquitin on the outer mitochondrial membrane (OMM) and recruits PRKN, an E3 ubiquitin ligase. PRKN then ubiquitinates OMM proteins, facilitating their recognition by mitophagy receptors such as FUNDC1 and subsequent degradation [100].
Inhibition or knockdown of mitophagy effectors like PINK1 or DNM1L can impede mitophagy, leading to decreased ROS generation and cell death in response to ferroptotic stimuli [104].
Agents that activate mitophagy can promote ADF. For example, in melanoma cells, the inhibition of mitochondrial complex I triggers mitophagy-dependent ROS induction and cell death, including ferroptosis [104]. Similarly, deficiency of the mitochondrial protein FTMT (ferritin mitochondrial) or treatment of osteoblasts with the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone/CCCP promotes ROS production and ferroptosis [105].
Various compounds, such as WJ460, TDCPP, BAY 11-7085, and rapamycin, both trigger mitophagy and promote ferroptotic cell death [106–109].
O-GlcNAcylation, the primary nutrient sensor of glucose flux, orchestrates both ferritinophagy and mitophagy, rendering cells more susceptible to ferroptosis [110].
Figure 5.
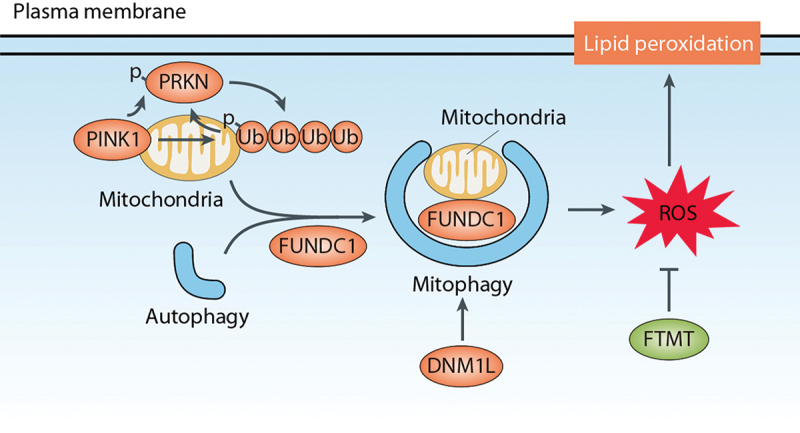
Mitophagy in ferroptosis. Mitophagy is a selective process that involves the autophagic clearance of damaged or dysfunctional mitochondria. The PINK1-PRKN pathway is a well-studied regulatory pathway for mitophagy. In this pathway, PINK1 recruits PRKN, which facilitates the degradation of mitochondria through mitophagy receptors such as FUNDC1. Mitochondrial fission induced by DNM1L/Drp1 can promote mitophagy-dependent ferroptosis. Additionally, the mitochondrial protein FTMT inhibits ROS production and ferroptosis.
Whereas mitophagy appears to contribute to ferroptosis, the specific mechanisms by which mitophagy influences the duration and intensity of lipid peroxidation in ferroptosis remain to be fully understood. Furthermore, the involvement of MFN1 (mitofusin 1)- and MFN2 (mitofusin 2)-mediated mitochondrial fusion in promoting erastin-induced ferroptosis in certain contexts highlights the complexity of the interplay between mitophagy and ferroptosis [111].
Clockophagy
BMAL1/ARNTL (basic helix-loop-helix ARNT like 1) is a transcription factor that plays a crucial role in the circadian clock by driving the rhythmic expression of genes, including those encoding its repressors PER (period circadian regulator) and CRY (cryptochrome circadian regulator) [112,113]. This creates a feedback loop that periodically suppresses its own transcription. The degradation of BMAL1 is implicated in the process of ferroptosis. In ferroptosis induced by GPX4 inhibitors (such as RSL3 and FIN56), BMAL1 is selectively degraded through autophagy, a process referred to as clockophagy [114]. This degradation of BMAL1 is not observed in response to SLC7A11 inhibitors (such as erastin, sulfasalazine, and sorafenib). The autophagy receptor SQSTM1/p62 (sequestosome 1) is involved in the recognition and degradation of BMAL1 during clockophagy [114].
The levels of BMAL1 directly affect the occurrence of lipid peroxidation and ferroptosis (Figure 6). The overexpression of BMAL1 reduces lipid peroxidation and ferroptosis, whereas the knockdown of BMAL1 enhances these processes [114]. The degradation of BMAL1 leads to the upregulation of EGLN2 (egl-9 family hypoxia inducible factor 2), a target gene of BMAL1. EGLN2, in turn, inhibits the function of HIF1A (hypoxia-inducible factor 1 subunit alpha). HIF1A acts as a ferroptosis suppressor by inducing the expression of two key target genes, FABP3 (fatty acid binding protein 3) and FABP7 (fatty acid binding protein 7) [114]. The corresponding proteins regulate fatty acid uptake and lipid storage, thereby having an impact upon the susceptibility to ferroptosis.
Figure 6.
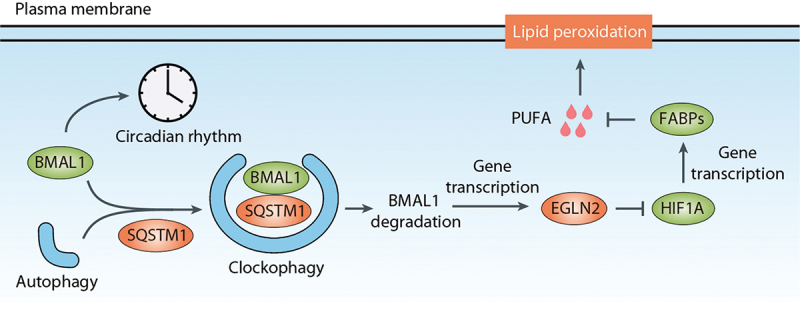
Clockophagy in ferroptosis. BMAL1/ARNTL, a transcription factor involved in the circadian clock, undergoes selective degradation through autophagy, a process known as clockophagy. The autophagy receptor SQSTM1/p62 plays a role in recognizing and degrading BMAL1 during clockophagy. Degradation of BMAL1 results in the upregulation of EGLN2, a target gene of BMAL1. EGLN2, in turn, inhibits the function of HIF1A, which acts as a suppressor of ferroptosis by inducing the expression of FABP3 and FABP7. These proteins regulate fatty acid uptake and lipid storage, thereby limiting the availability of polyunsaturated fatty acid (PUFA) and influencing the susceptibility to ferroptosis.
Given the protective role of BMAL1 in ferroptotic cancer cells, targeting clockophagy may hold potential as an anticancer strategy. Additionally, clockophagy-dependent ferroptosis is implicated in inflammation-related injury. Mice with specific depletion of pancreatic Bmal1 are more susceptible to acute pancreatitis induced by L-arginine compared to control mice [115]. Understanding the complex interplay between BMAL1, clockophagy, and ferroptosis provides insights into the regulation of lipid peroxidation and the potential development of therapeutic strategies targeting these pathways in cancer and inflammation.
CMA- and TAX1BP1-mediated GPX4 degradation
Accumulating studies have provided insights into the mechanisms underlying GPX4 degradation during ferroptosis and its connection to autophagy. One pathway involved in GPX4 degradation is CMA (Figure 7). GPX4 contains potential KEFRQ-like motifs that are recognized by CMA [116]. Treatment with ferroptosis inducers, such as erastin, activates CMA and increases the punctate pattern signals of an mCherry-tagged CMA-targeting substrate [116]. The lysosomal receptor LAMP2 (lysosomal associated membrane protein 2) plays a crucial role in CMA by serving as a subunit of the translocation complex responsible for the transport of specific cytosolic substrates across the lysosomal membrane for degradation [117]. HSPA8/HSC70 (heat shock protein family A (Hsp70) member 8) interacts with CMA substrates and LAMP2, thus facilitating CMA [118,119]. Along these lines, increased interactions between GPX4 and HSPA8 or LAMP2 are observed in 661W or HT-22 cells during ferroptosis [116].
Figure 7.
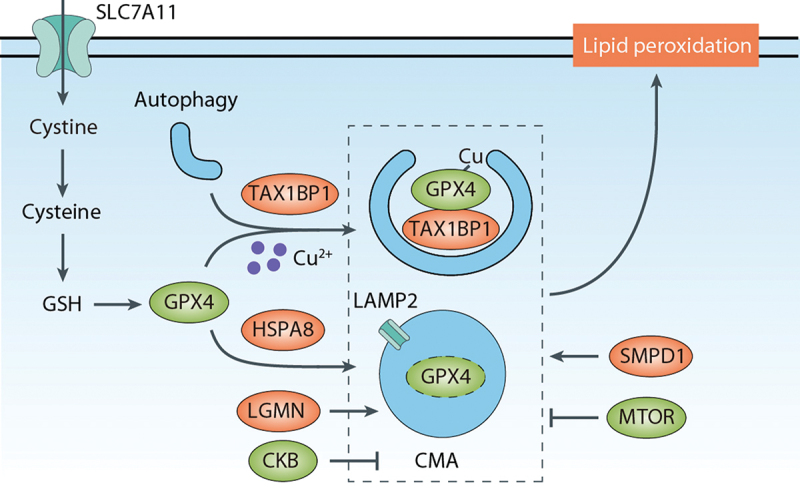
CMA- or TAX1BP1-mediated GPX4 degradation in ferroptosis. CMA serves as a pathway for GPX4 protein degradation during ferroptosis. LAMP2 plays a key role in transporting GPX4 across the lysosomal membrane, and HSPA8/HSC70 interacts with GPX4 and LAMP2, facilitating CMA. CMA-mediated degradation of GPX4 can be inhibited by CKB-mediated GPX4 phosphorylation. LGMN assists in CMA-mediated GPX4 degradation. Furthermore, macroautophagy/autophagy also contributes to GPX4 degradation during ferroptosis. SMPD1/ASM promotes the autophagic degradation of GPX4, whereas MTOR inhibits GPX4 protein degradation and subsequent ferroptosis. Copper directly binds to GPX4, leading to its autophagic degradation, and, in this process, TAX1BP1 acts as an autophagic receptor.
CMA-mediated GPX4 degradation can be inhibited by active, phosphorylated CKB (creatine kinase B), leading to GPX4 phosphorylation at S104 in cancer cells [120]. In PC12 cells (rat adrenal pheochromocytoma), GPX4 degradation via CMA contributes to antimony-induced ferroptosis [121]. Additionally, a conserved asparaginyl endopeptidase called LGMN (legumain) has been identified as a promoter of tubular ferroptosis in acute kidney injury by facilitating CMA-mediated GPX4 degradation [122]. Enhancing autophagy by HSPA8 inhibition contributes to ferroptosis in rifampicin-induced hepatotoxicity [123]. However, whether there is a role for impaired CMA-mediated GPX4 degradation in rifampicin-induced hepatotoxicity when HSPA8 is inhibited remains unclear.
Autophagy may also contribute to GPX4 degradation during ferroptosis (Figure 7). The ferroptosis inducer FIN56 promotes GPX4 protein degradation in an ACACA-dependent manner [124]. Inhibition of autophagy by bafilomycin A1 or the knockdown of ULK1 or ATG3 attenuates FIN56-induced GPX4 degradation and ferroptosis in 253J and T24 bladder cancer cells [125]. SMPD1/ASM (sphingomyelin phosphodiesterase 1), an enzyme involved in sphingolipid metabolism, is required for the autophagic degradation of GPX4 in both HT-1080 and Calu-1 cells [126]. Rapamycin, a well-known MTOR inhibitor and autophagy activator, can induce GPX4 protein degradation and ferroptosis in human pancreatic cancer cells [127]. However, the role of MTOR in ferroptosis is highly context-dependent [128], meaning that the impact of MTOR on ferroptosis can vary depending on specific conditions or cellular contexts. Additionally, copper, an essential metal in biology, promotes ADF by inducing GPX4 degradation in pancreatic cancer cells [129]. Copper directly binds to GPX4, leading to its ubiquitination and formation of aggregates, while TAX1BP1 (Tax1 binding protein 1) acts as an autophagic receptor for GPX4 degradation during copper-induced ferroptosis [129]. In addtion, N6F11, a small molecule compound, can selectively induce ferroptosis by targeting TRIM25-mediated GPX4 degradation via proteasome in cancer cells while sparing immune cells [130]. Further understanding the connection between proteasome and autophagy-dependent GPX4 degradation may provide a framework for the identification of biomarkers and predictive indicators of ferroptosis sensitivity in cancer.
SQSTM1-mediated KEAP1 degradation
NFE2L2/NRF2 is a transcription factor that plays a pivotal role in preventing oxidative stress and ferroptotic responses. Under normal, unstressed conditions, NFE2L2 levels are kept low due to KEAP1 (kelch like ECH associated protein 1)-mediated proteasomal degradation. However, during cellular stress, such as oxidative stress or the accumulation of misfolded proteins, the upregulation of SQSTM1 can occur. SQSTM1 interacts with ubiquitinated proteins, leading to the sequestration of KEAP1 [131,132]. This interaction prevents the degradation of NFE2L2 protein, enabling its translocation into the nucleus and subsequent activation of antioxidant response element (ARE)-dependent genes. These genes encode a range of cytoprotective proteins, including antioxidant enzymes and phase II detoxification enzymes, which help mitigate the effects of oxidative stress and maintain cellular homeostasis during ferroptosis.
Increased autophagy flux can promote the degradation of SQSTM1 protein, leading to enhanced KEAP1 stability and subsequent KEAP1-dependent degradation of NFE2L2 [57]. In contrast, autophagy deficiency can upregulate endogenous SQSTM1 levels, inhibiting KEAP1-dependent degradation of NFE2L2 and sustaining NFE2L2-dependent gene transcription [57]. Notably, SQSTM1 itself is a target gene of NFE2L2 [133], highlighting a feedback control mechanism involved in regulating KEAP1 degradation as well as autophagy levels. SQSTM1 can also be released into the extracellular space during ferroptosis [134]. Once released, extracellular SQSTM1 facilitates the AGER (advanced glycosylation end-product specific receptor)-dependent gene expression of ACSL4, which modulates cellular lipid composition to enhance autophagosome formation and promote ferroptosis in a model of pancreatitis [134].
SQSTM1-mediated SLC40A1 degradation
SLC40A1/ferroportin (solute carrier family 40 member 1) is an iron exporter. The stability of SLC40A1 protein is tightly regulated by cellular processes, such as the ubiquitin-proteasome system and autophagy [135,136]. In normal physiological conditions, the hormone HAMP (hepcidin antimicrobial peptide) controls iron homeostasis by inducing the internalization of SLC40A1 into cells and its subsequent degradation [137]. SLC40A1 acts as a key anti-ferroptotic regulator by reducing intracellular Fe2+ levels in various disease states, including neoplasia [138,139], Alzheimer disease [140], endotoxemia [141], liver fibrosis [142], and intervertebral disc degeneration [143]. Notably, ferroptosis can trigger the degradation of SLC40A1 through autophagy in specific cancer cells such as HT-1080 or PANC1 [144].
Interestingly, the autophagy receptor SQSTM1, but not other receptors such as NBR1 (NBR1 autophagy cargo receptor), CALCOCO2/NDP52 (calcium binding and coiled-coil domain 2), OPTN (optineurin), or NCOA4, is required for the autophagic degradation of SLC40A1 in cancer cells [144]. This suggests that the selective autophagy pathway involving SQSTM1 plays a specific role in regulating SLC40A1 degradation and potentially influencing cellular susceptibility to ferroptosis. These findings shed light on the intricate relationship between iron metabolism, autophagy, and ferroptosis. Understanding the mechanisms underlying the regulation of SLC40A1 and its degradation through autophagy in different disease contexts will contribute to the development of novel therapeutic strategies targeting iron dysregulation and ferroptosis-related disorders.
HPCAL1-mediated CDH2 degradation
HPCAL1/VILIP-3 (hippocalcin like 1) is a member of the visinin-like protein superfamily and primarily functions as a neuronal calcium sensor protein [145]. HPCAL1 plays diverse roles in regulating signaling pathways in specific cell types [146–148]. Intriguingly, HPCAL1 can act as an autophagy receptor in ADF, distinct from classical autophagy induced by starvation or the MTOR inhibitor rapamycin [149]. Specifically, HPCAL1 acts as a mediator of the autophagic degradation of CDH2 (cadherin 2) protein in HT-1080 and pancreatic cancer cells, leading to a decrease in membrane tension and subsequent induction of ferroptosis [149]. The degradation of CDH2 is dependent on a specific MAP1LC3/LC3-interacting region/LIR motif, DEFFKKI (amino acids 46–51), within HPCAL1. Activation of HPCAL1 by PRKCQ (protein kinase C theta) induces phosphorylation of HPCAL1 at Thr149, which is necessary for its role in mediating CDH2 degradation and promoting ferroptosis [149]. These findings provide insights into the multifaceted functions of HPCAL1 and its involvement in ADF, highlighting the complexity of the molecular mechanisms underlying this regulated cell death pathway.
Autophagic degradation of DHFR
Tetrahydrobiopterin (BH4) is an important intracellular antioxidant that plays a crucial role in protecting cells from lipid peroxidation and ferroptosis [51,52]. BH4 can be oxidized to dihydrobiopterin (BH2), but BH2 is regenerated back to BH4 by the enzyme DHFR (dihydrofolate reductase). As a result, DHFR acts as a suppressor of ferroptotic cell death [52]. However, CD38 (CD38 molecule), a membrane-bound enzyme involved in calcium signaling and NAD metabolism, mediates the production of ROS and ferroptosis susceptibility [150]. The autophagic degradation of DHFR enhances the susceptibility of bone marrow-derived macrophages (BMDMs) to ferroptosis [150]. Understanding the precise mechanisms and identifying the autophagy receptors involved in targeting DHFR for autophagic degradation could provide valuable insights into the regulation of DHFR levels and its role in cellular processes such as ferroptosis. Further research is needed to unravel the molecular details of DHFR autophagic degradation and its significance in cellular homeostasis and disease conditions.
BECN1-mediated SLC7A11 inhibition
BECN1 is a core component of the autophagy machinery, forming a complex with PIK3C3 [151]. Knockdown of BECN1 disrupts erastin-triggered ferroptosis through the inhibition of autophagy [152]. The expression of the BECN1 gene or protein in ADF is regulated by multiple factors. For instance, the BECN1 gene is epigenetically activated by m6A modification, leading to ADF in hepatic stellate cells [153]. The RNA-binding protein ELAVL1/HuR (ELAV like RNA binding protein 1) directly binds to and stabilizes BECN1 mRNA, promoting ADF [154]. Another RNA-binding protein, CIRBP (cold inducible RNA binding protein), interacts with ELAVL1 and activates ferritinophagy during ferroptosis-induced renal ischemia-reperfusion injury [155]. In contrast, the RNA-binding protein PCBP1 (poly(rC) binding protein 1) destabilizes BECN1 mRNA, whereas ZFP36 (ZFP36 ring finger protein) destabilizes ATG16L1 mRNA, thereby suppressing autophagy flux (ADF) in head and neck cancer cells or hepatic stellate cells [156,157]. Additionally, USP11 (ubiquitin specific peptidase 11) deubiquitinates and stabilizes BECN1 protein, promoting BECN1-mediated ADF in spinal cord ischemia-reperfusion injury [158].
Apart from its role in autophagic degradation, BECN1 also plays a role in inducing ferroptosis by directly binding to and inhibiting SLC7A11 [31]. The phosphorylation of BECN1 by AMPK enhances the formation of the BECN1-SLC7A11 complex, promoting ferroptosis [31,159]. Conversely, the expression of DUSP1 (dual specificity phosphatase 1), which is induced by erastin and RSL3, acts as a negative regulator of ADF by inhibiting the phosphorylation of ULK1 and BECN1 [160,161]. Furthermore, exosomes containing BECN1, secreted by human umbilical cord mesenchymal stem cells, can downregulate the expression of SLC7A11 and GPX4 [162]. This downregulation leads to the induction of ferroptosis in LX-2 hepatic stellate cells [162]. Collectively, these findings indicate that both endogenous and exogenous BECN1 can inhibit SLC7A11 activity and expression, thereby contributing to the promotion of ferroptosis.
In a separate mechanism, the transmembrane protein tyrosine kinase ERBB2 (erb-b2 receptor tyrosine kinase 2) interacts with BECN1 and inhibits autophagy initiation [163]. This interaction acts as a blockade for autophagy. Furthermore, the phosphatase CDC25A (cell division cycle 25A) inhibits ADF by dephosphorylating nuclear PKM (pyruvate kinase M1/2), which subsequently upregulates ERBB2 expression in cervical cancer cells [164]. It will be interesting to investigate whether an energy metabolism mechanism exists that controls ERBB2 expression and, subsequently, the function of BECN1 in regulating SLC7A11 activity.
PIR-mediated nuclear DNA sensing
PIR (pirin) was initially identified as a nuclear protein involved in regulating gene expression [165]. In addition to its role in gene transcription, PIR functions as a nuclear redox sensor in human pancreatic cancer cells. PIR is upregulated in response to erastin- or RSL3-induced DNA damage to limit oxidative damage to DNA [166]. The depletion of PIR leads to increased nuclear DNA oxidative damage, resulting in the translocation and release of nuclear DAMPs, including DNA and HMGB1 (high mobility group box 1) [166]. The translocation of HMGB1 from the nucleus to the cytosol is associated with HMGB1 acetylation, a process regulated by the activity of HDAC (histone deacetylase) [167]. Cytosolic HMGB1 promotes autophagy by binding to BECN1, whereas extracellular HMGB1 acts as a mediator of ferroptotic damage-induced cytokine release by activating the AGER pathway in macrophages [168–170]. Targeting the PIR-HMGB1 pathway holds promise for enhancing ferroptosis-mediated tumor suppression or preventing sterile inflammation induced by ferroptotic damage.
STING1-mediated cytosolic DNA sensing
STING1 (stimulator of interferon response cGAMP interactor 1) is a well-known adaptor protein involved in sensing cGAMP in the innate immune system. It also plays a significant role in mediating the activation of autophagy [171]. The anti-HIV drug, zalcitabine, induces mitochondrial DNA stress in pancreatic cancer cells, resulting in the activation of STING1-mediated autophagy and ADF [172]. This process involves the degradation of TFAM (transcription factor A, mitochondrial), leading to the release of mitochondrial DNA into the cytosol, oxidative DNA damage, and activation of the CGAS (cyclic GMP-AMP synthase)-STING1 DNA sensor pathway [172]. Moreover, STING1 interacts with NCOA4 to trigger ferritinophagy-dependent ferroptosis in macrophages [173]. In addition to promoting active autophagy, STING1 enhances ferroptosis in pancreatic cancer cells by increasing MFN1- or MFN2-dependent mitochondrial fusion, independent of mitophagy [111]. These findings highlight the broad role of STING1 in mediating both autophagy and cell death processes. In contrast, mitochondrial CGAS plays a STING1-independent role in inhibiting ferroptosis in liver cancer cells [174], highlighting the location-dependent role of CGAS in regulating immune response and cell death.
TMEM164-mediated phagophore membrane dynamics
The expansion of the phagophore membrane in the early stages of starvation-induced autophagy involves the multispanning membrane protein ATG9A (autophagy related 9A), which functions as a scramblase, facilitating the flipping of phospholipids between the two membrane leaflets present on phagophores and the endoplasmic reticulum (ER) [175]. Interestingly, ATG9A is dispensable for autophagy induced by erastin and RSL3 [176]. In contrast, TMEM164 (transmembrane protein 164), an ER-sessile transmembrane protein, is a crucial component required for phagophore membrane expansion during ferroptosis [176]. In pancreatic cancer cells, the loss of TMEM164 diminishes the binding between ATG5 and ATG16L1 in response to RSL3, while not affecting the stimulation caused by HBSS-induced starvation [176]. These findings unveil a mechanism to discern the early signals and molecular players involved in autophagy induction by starvation and ferroptosis activators. Additionally, TMEM164 exhibits acyltransferase activity and promotes ferroptosis by increasing polyunsaturated ether phospholipids [177]. However, whether this acyltransferase function is necessary for ADF remains unclear.
The protective role of autophagy in ferroptosis
In certain circumstances, selective autophagy can serve as a pro-survival mechanism during ferroptosis by selectively removing damaged or dysfunctional cellular components. This process aids in limiting lipid peroxidation and maintaining cellular homeostasis, as elaborated below.
Reticulophagy
Reticulophagy, also known as ER-phagy, is a selective form of autophagy that specifically targets and degrades portions of the ER [178]. Among the ER-resident receptors facilitating ER degradation through autophagy is RETREG1/FAM134B (reticulophagy regulator 1), which interacts with MAP1LC3 to promote ER degradation [179]. In the context of ferroptosis, the ferroptosis inducer sorafenib effectively activates RETREG1-mediated reticulophagy, thereby protecting hepatocellular carcinoma cells from undergoing ferroptosis [180]. Conversely, when RETREG1 is knocked down, reticulophagy is blocked, resulting in increased sensitivity of hepatocellular carcinoma cells to ferroptosis [180]. Furthermore, the RNA-binding protein PABPC1 (poly(A) binding protein cytoplasmic 1) can interact with RETREG1 mRNA to enhance its translation, ultimately inhibiting ferroptosis through the induction of RETREG1-mediated reticulophagy [180]. This clarifies the role of reticulophagy, its regulation by RETREG1, and its impact on ferroptosis in hepatocellular carcinoma cells.
Lysophagy
Lysophagy is a selective autophagy process responsible for clearing damaged lysosomes [181]. G3BP1 (G3BP stress granule assembly factor 1), a multifunctional binding protein, plays diverse roles in biological functions such as cell proliferation, cell differentiation, and cell death. In the context of intervertebral disc degeneration, G3BP1 forms a complex with TSC2 (TSC complex subunit 2) to regulate lysophagy activity, thereby protecting cells against compression-induced ferroptosis [182]. In contrast, inhibiting the G3BP1-TSC2 complex accelerates lysosomal dysfunction and promotes ferroptosis [182]. Lysophagy contributes to the regulation of cellular metabolism by promoting the turnover of lysosomal components under various stressful conditions. Understanding the interplay between lysophagy and other autophagic pathways would provide valuable insights into their collective regulation of ferroptosis determination.
CMA-mediated ACSL4 degradation
ACSL4 contributes to formation of lipid substrates for lipid peroxidation during ferroptosis [36–38]. ACSL4 protein is a substrate of CMA, likely because it contains six KFERQ-like motifs [183]. While CMA-mediated degradation of GPX4 promotes ferroptosis, CMA-mediated degradation of ACSL4 could inhibit this process. For example, GMFB (glia maturation factor beta), a growth and differentiation factor, plays a significant role in the pathogenesis of diabetes. In ARPE19 (human retinal pigment epithelium) cells, high glucose-induced GMFB secretion induces ferroptosis by inhibiting CMA-mediated ACSL4 degradation through impairing lysosomal function [183]. In contrast, treatment with GMFB antibody or CMA activator Q×77 facilitates ACSL4 degradation via CMA, thus preventing early diabetic retinopathy [183]. These findings underscore the potential of therapeutically inducing CMA for the suppression of ferroptotic cell death in the context of diabetes.
Role of lysosomes in ferroptosis
Lysosomes are essential intracellular organelles with an acidic internal pH that plays a pivotal role in digesting autophagic substrates. Consequently, the extensive investigation into the involvement of lysosomes in promoting ADF has shed light on the regulatory role of lysosomal activities in this process. For example, the inhibition of lysosomal function using compounds, such as bafilomycin A1, ammonium chloride, or PepAMe, protects cells from erastin- or RSL3-induced ferroptosis in HT-1080 and Calu-1 cells [184]. Furthermore, increased lysosomal membrane permeabilization is observed in ferroptotic glioblastoma cells. TFEB (transcription factor EB), a master regulator of autophagy and lysosomal biogenesis, favors ROS generation in response to ferroptosis inducers [185,186]. Specifically, TFEB drives ferroptosis by promoting lysosomal degradation of ferritin in breast cancer cells [187]. Lysosomal function heavily relies on an array of proteases, including cathepsins, which, under specific conditions, can be released into the cytoplasm and catalyze the proteolytic cleavage of substrates. CTSB (cathepsin B), a cysteine protease belonging to the cathepsin family, promotes ferroptosis through histone H3 cleavage or DNA damage induction [188,189]. Signaling via STAT3 (signal transducer and activator of transcription 3) is crucial for CTSB induction and ferroptosis in human pancreatic cancer cell lines [190]. Additionally, lysosomal nitric oxide mediates ferroptosis induced by plasma-activated Ringer’s lactate during cancer therapy [191].
While most studies support the pro-ferroptotic role of lysosomes, specific lysosomal proteins or degradation products, such as cysteine and selenium, can also exert inhibitory effects on ferroptosis. GRN (granulin precursor), derived from microglial lysosomes, inhibits ferroptosis-mediated neuronal injury by upregulating the expression of GPX4, NFE2L2, and SLC7A11 [192]. In contrast, the knockdown of LAMP2A, a receptor for CMA, induces ferroptosis in retinal pigment epithelial cells by depleting cytosolic cysteine and GSH levels [193]. The knockdown of PSAP (prosaposin) leads to impaired lysosomal function and autophagic flux [30]. This leads to the accumulation of iron within lipofuscin-like lysosomes, rendering neurons uniquely susceptible to ferroptosis, unlike several other cell types [30]. Interestingly, lysosomal degradation of extracellular ALB (albumin) increases cysteine levels and replenishes GSH content, thus suppressing ferroptosis induced by cystine deprivation [194]. Moreover, selenium utilization by GPX4 is necessary to prevent ferroptosis [195]. The cellular selenium levels regulated by LRP8/ApoER2 (LDL receptor related protein 8) influence ferroptosis. LRP8 acts as a receptor for SELENOP/SEPP1 (selenoprotein P), which undergoes lysosomal breakdown to release selenium [196]. Cellular selenium levels regulated by LRP8 protect cells from ferroptosis by enhancing the translation of the selenoprotein GPX4 [196].
In summary, the implication of lysosomes in ferroptosis is intricate and multifaceted. While lysosomes predominantly exhibit pro-ferroptotic functions, specific lysosomal proteins or degradation products can exert inhibitory effects on ferroptosis. Understanding the complex interplay between lysosomes and ferroptosis may shed light on novel therapeutic strategies targeting this cell death pathway.
Autophagy-dependent ferroptosis in diseases
ADF is implicated in multiple diseases, including both cancer and non-cancer diseases. In this section, we provide a summary of the significant advancements in understanding the pathological role of ADF in various diseases (Figure 8).
Figure 8.
Autophagy-dependent ferroptosis in diseases. Autophagy-dependent ferroptosis plays a role in various diseases, encompassing both cancer and non-cancerous conditions.
Cancer
ADF plays a multifaceted role in tumor initiation, development, and therapy, akin to other forms of cell death. On the one hand, persistent tissue damage resulting from ADF can contribute to tumorigenesis. For example, in the context of ferroptosis, dying cells release the oncogenic KRASG12D protein through secretory autophagy, an unconventional mechanism for protein secretion [197]. The uptake of KRASG12D protein from ferroptotic cells induces polarization of surrounding macrophages via the AGER receptor, fostering an immune-suppressed tumor microenvironment that facilitates tumor growth [197]. Ferroptotic macrophages engulfing asbestos fibers secrete holo-ferritin-loaded exosomes, which leads to oxidative DNA damage in the carcinogenesis-target mesothelial cells [198]. On the other hand, pharmacological induction of ADF holds promise as a therapeutic approach for various cancer types [199]. It can be employed either as a monotherapy to eliminate cancer cells or in combination with other agents to overcome drug resistance (Table 1). For instance, the induction of ADF using 4-octyl itaconate, a cell-permeable derivative of the metabolite itaconate, has demonstrated effectiveness in targeting acquired multi-drug-resistant/MDR retinoblastoma cells that have developed resistance due to prolonged carboplatin treatment [200]. Additionally, the loss of COPZ1 (COPI coat complex subunit zeta 1) induces ferritinophagy and ferroptosis in glioblastoma cells [225], whereas the loss of lncRNA EGFR-AS1 (EGFR antisense RNA 1) promotes ADF in cervical cancer [226].
Table 1.
Autophagy-dependent ferroptosis inducers in cancer treatment.
| Agents | Mechanism of action | Cancer types | Ref |
|---|---|---|---|
| 4-octyl itaconate | Activates ferritinophagy | Retinoblastoma | [200] |
| 6-Gingerol | Inhibits USP14 | Lung cancer | [201] |
| d-Borneol | Activates ferritinophagy | Lung cancer | [202] |
| Allicin | Activates AMPK-MTOR | Esophageal squamous | [203] |
| Amentoflavone | Activates AMPK-MTOR | Glioma | [204] |
| ATPR | Increases ROS | Acute myeloid leukemia | [205] |
| Chrysin | Increases ROS | Pancreatic cancer | [92] |
| Curcumin | Increases BECN1 and MAP1LC3-II | Lung cancer | [206] |
| Dihydroartemisinin | Activates ferritinophagy | Acute myeloid leukemia | [207] |
| Erastin | Activates ferritinophagy | Cervical cancer | [208] |
| FIN56 | Induces GPX4 degradation and activates ferritinophagy | Bladder cancer, lung cancer | [114,125] |
| Formosanin C | Activates ferritinophagy | Liver cancer | [209] |
| FTY720 | Activates AMPK | Multiple myeloma | [210] |
| Itaconic acid | Activates ferritinophagy | Pancreatic cancer | [211] |
| JQ1 | Activates ferritinophagy | Lung cancer, breast cancer | [212] |
| MMRi62 | Activates ferritinophagy | Pancreatic cancer | [213] |
| Newcastle disease virus | Activates ferritinophagy | Glioma | [214] |
| Nanoparticle: CPNs-ART | Increases ROS | Lewis lung carcinoma | [215] |
| Nanoparticle: FPBC@SN | Activates ferritinophagy | Breast cancer | [216] |
| Nanoparticle: HMCMs | Activates autophagy | Breast cancer | [217] |
| Nanoparticle: IONP@PTX | Increases BECN1 | Glioblastoma | [218] |
| Nanoparticle: NFER | Activates autophagy | Breast cancer | [219] |
| Nanoparticle: TFPs | Activates autophagy | Breast cancer | [220] |
| Nanoparticle: TreMMM | Activates ferritinophagy | Pancreatic cancer, breast cancer | [221] |
| Plasma-activated lactate | Activates lysosomal NO production and activates autophagy | Mesothelioma | [191] |
| Quercetin | Activates TFEB | Breast cancer | [187] |
| Rapamycin | Induces GPX4 degradation | Pancreatic cancer | [127] |
| RSL3 | Activates clockophagy | Lung cancer | [114] |
| Shikonin | Activates ferritinophagy | Multiple myeloma | [222] |
| Typhaneoside | Activates AMPK | Acute myeloid leukemia | [223] |
| Vitamin C | Activates ferritinophagy | Anaplastic thyroid cancer | [224] |
| Zalcitabine | Activates mitochondrial DNA stress | Pancreatic cancer | [172] |
Of note, triggering ferroptosis can serve as an alternative approach to eliminate apoptosis-resistant cancer cells [18]. Cancer cells undergoing the epithelial-mesenchymal transition (EMT) often exhibit resistance to apoptosis inducers, but they become vulnerable to ferroptosis inducers due to EMT-associated upregulation of ferroptosis-related genes [227]. Given that direct induction of ferroptosis may have immune side effects by causing cell death in immune cells [13,14], it is imperative to conduct additional research to authenticate and establish whether the initiation of ADF (or alternative approaches) could serve as a potential solution to circumvent this constraint in forthcoming investigations [228].
ADF is not only implicated in disease processes but also in chemotherapy-induced side effects on normal tissues. For example, recent studies have shed light on the involvement of ADF as a molecular mechanism underlying cisplatin-induced hearing loss [229]. Inhibition of autophagy using CQ alleviates cisplatin-induced ferroptosis in auditory cells [229]. Additionally, the administration of ferrostatin-1 has demonstrated attenuation of cisplatin-induced hearing loss in animal models [229]. These findings highlight the potential of targeting ADF as a therapeutic strategy to mitigate chemotherapy-related side effects.
Ischemia-reperfusion injury diseases
Ischemia-reperfusion injury (IRI) is a pathological condition characterized by tissue damage resulting from the interruption and subsequent restoration of blood supply to an organ or tissue. This phenomenon is observed in various diseases, including myocardial infarctions, stroke, and acute kidney injury, where severe cell damage or death occurs. Extensive evidence supports a direct link between ferroptosis and IRI. ADF plays a significant role in the tissue damage induced by IRI in the brain [230], spinal cord [158], kidney [155], and myocardium [231]. For example, the inhibition of autophagy using 3-MA or knockdown of ATG5 suppresses myocardial hypoxia-reperfusion-induced ferroptosis and cell injury [154,231]. Additionally, ferritinophagy-dependent ferroptosis contributes to myocardial IRI during the onset of type 2 diabetes mellitus/T2DM, but this process can be inhibited by SR9009, a specific synthetic agonist of circadian rhythm modulation activity, or by the DNMT1 (DNA methyltransferase 1) inhibitor 5-aza-2’-deoxycytidine [232,233]. Moreover, oxygen-glucose deprivation-reoxygenation promotes myocardial IRI by inducing BECN1-mediated autophagy, which can be inhibited by the polyphenol compound resveratrol [234]. Based on these findings, inhibiting ADF represents a potential therapeutic approach for the treatment of ischemia-reperfusion injury.
Brain diseases
Neurodegenerative diseases, such as Alzheimer, Parkinson, and Huntington diseases, are characterized by abnormal iron accumulation in specific brain regions. This iron overload contributes to oxidative stress and neuronal damage, playing a significant role in the development and progression of these disorders. Allelic variations in the APOE gene represent the most significant risk factor for sporadic Alzheimer disease, although the underlying mechanisms remain elusive. The APOE protein, irrespective of its isoform, exerts a neuroprotective effect by impeding ferroptosis in neuronal cells through PI3K/AKT-mediated inhibition of ferritinophagy [235]. The APOE ε4 risk allele might enhance susceptibility to ferroptosis due to reduced protein levels of APOE and elevated iron levels observed in individuals carrying this particular variant [235,236]. In Parkinson disease models, ferritinophagy activation is observed, and inhibiting autophagy can suppress ferritinophagy-dependent ferroptosis [237]. Similarly, paraquat, a neurotoxicant linked to Parkinson disease, induces ferroptosis through the activation of ferritinophagy in neuroblastoma cells [238].
Additionally, ADF is implicated in acute brain damage, such as subarachnoid hemorrhage. Inhibition of autophagy can alleviate neurological deficits and cell death in experimental models, while flavonoids and targeted inhibition of microglial S100A8 (S100 calcium binding protein A8) suppress ADF and improve neural function [239]. Moreover, polystyrene microplastics can trigger ADF, leading to nervous system damage in chickens [240]. These findings highlight the involvement of ADF in various neurological conditions and provide potential targets for therapeutic interventions.
Liver diseases
Liver fibrosis is a condition characterized by the excessive accumulation of scar tissue (fibrosis) in the liver, which occurs as a response to chronic liver damage and inflammation. The pathogenesis of liver fibrosis involves hepatic stellate cells, which play a crucial role in the development and progression of fibrosis. These cells undergo transdifferentiation into matrix-producing myofibroblasts, contributing to the excessive production of extracellular matrix components. Notably, studies have identified the involvement of RNA-binding proteins in the regulation of ADF signaling pathways within hepatic stellate cells. For instance, the RNA-binding protein ZFP36 (ZFP36 ring finger protein) inhibits ferroptosis in hepatic stellate cells by downregulating autophagy signaling, thereby promoting liver fibrosis in mice [157]. Conversely, the RNA-binding protein ELAVL1, along with the key m6A reader protein YTHDF1 (YTH N6-methyladenosine RNA binding protein F1), promotes BECN1 mRNA stability, leading to ADF in hepatic stellate cells [153,241]. The antimalarial drug artesunate also exerts its anti-fibrotic effects through the activation of ferritinophagy-dependent ferroptosis in hepatic stellate cells [242]. Understanding the intricate mechanisms involving these RNA-binding proteins and the interplay between ferroptosis, autophagy, and fibrosis holds potential for the development of therapeutic approaches targeting hepatic stellate cells to mitigate liver fibrosis.
Furthermore, ferroptosis appears to play a crucial role in liver injury associated with various pathological conditions. For instance, alcohol-induced liver injury is associated with ferroptosis in hepatocytes, which can be reduced by the administration of ferrostatin-1 [243]. In contrast, inhibitors of autophagy, such as CQ and 3-MA, reportedly attenuate alcohol-induced ferroptosis by activating the NFE2L2 pathway [243]. This finding suggests that ADF contributes to alcohol-induced liver injury. Additionally, in a mouse model of concanavalin A-induced acute liver injury, disulfiram, an FDA-approved alcohol-aversive drug, ameliorates liver injury by suppressing CMA-mediated degradation of GPX4 [244].
Pancreatic diseases
At physiological conditions, the pancreas does not serve as a major site for iron storage or accumulation compared to organs like the liver or spleen. However, excessive iron buildup in the pancreas can lead to oxidative stress and tissue damage, potentially contributing to the development of pancreatic diseases, particularly acute pancreatitis. Experimental studies have shown that high-iron diets or the conditional knockout of Gpx4 can accelerate the development of acute pancreatitis induced by cerulein or L-arginine in mice [245]. This process is further enhanced by trypsin, an enzyme secreted by the pancreas as trypsinogen, which is activated by enterokinase in the duodenum [246].
ADF plays a crucial role in the pathogenesis of acute pancreatitis. HPCAL1 acts as an inducer of pancreatic ferroptosis by promoting the autophagic degradation of CDH2 [149]. Pharmacological inhibition of HPCAL1 using a small compound, termed iHPCAL1, protects against ferroptosis-associated acute pancreatitis induced by cerulein in mice [149]. Moreover, interventions, such as SQSTM1-neutralizing antibody or the conditional knockout of Ager in the pancreas, prevent tissue damage and inflammatory responses induced by ADF in experimental acute pancreatitis in mice [134]. These findings highlight the significance of iron metabolism and ADF in the development and progression of acute pancreatitis.
Heart diseases
Autophagy contributes to the bioenergetics of the cardiovascular system [247], however the occurrence of ferroptosis in myocytes (heart muscle cells) may play a significant role in the pathophysiology of heart failure. Integrated bioinformatics analysis has identified the upregulation of TLR4 (toll like receptor 4) and NOX4 (NADPH oxidase 4) during heart failure, suggesting their involvement in the disease process [248]. Mechanistically, TLR4 and NOX4 can activate ADF in cardiomyocytes of rats with heart failure [248]. In contrast, the knockdown of TLR4 or NOX4 in cardiomyocytes decreases ferroptosis, resulting in an improvement of cardiac function [248].
Furthermore, ferritinophagy-dependent ferroptosis plays a role in sepsis-induced cardiac injury. Inhibition of ferritinophagy through methods like 3-MA treatment, ATG5 knockdown, or NCOA4 knockdown blocks ferroptosis and cellular injury in H9c2 myofibroblasts in vitro [249]. Moreover, inhibiting ferroptosis using compounds like ferrostatin-1, the iron chelator dexrazoxane, or the mitochondrial iron chelator deferiprone improves survival and cardiac function in mice injected with bacterial lipopolysaccharide [249]. Thus, targeting ADF holds promise for potential therapeutic interventions aimed at preserving cardiac function during bacterial infection.
Kidney diseases
ADF is considered a critical mediator in the pathogenesis of acute kidney injury. Studies have demonstrated its involvement in various forms of acute kidney injury induced by different toxins [248]. For instance, mercuric chloride induces ferroptosis in chicken embryo kidney cells by triggering ferritinophagy [248], and acute exposure to cadmium promotes ferroptosis in renal tubular epithelial cells through the activation of ER stress-mediated ferritinophagy [250]. Bisphenol A, a common environmental contaminant, also induces ferroptosis in renal tubular epithelial cells by activating ferritinophagy mediated by the AMPK-MTOR-ULK1 pathway [251]. Patulin, a common mycotoxin, induces acute kidney injury in mice through the activation of ADF [252]. In contrast, the administration of the ferroptosis inhibitor ferrostatin-1 and a natural bioactive compound, isoliquiritigenin, attenuate septic acute kidney injury by inhibiting ferritinophagy and ferroptosis [253,254]. Furthermore, the impact of ADF on the progression and outcomes of chronic kidney diseases, such as diabetic nephropathy or chronic kidney failure, remains an area of active research. Elucidating the role of ADF in the chronic stages of kidney disease could have significant implications for the development of targeted therapies.
Respiratory diseases
In asthma, the airway epithelium undergoes significant changes that compromise lung function. Increased cell death of airway epithelial cells, through apoptosis and non-apoptotic mechanisms, disrupts the integrity of the epithelial barrier. A recent study found that ferritinophagy contributes to the development of house dust mite-induced ferroptosis in human bronchial epithelial cells and a mouse asthma model [255]. Accordingly, ferroptosis inhibitors like ferrostatin-1 or iron chelators reduce airway inflammation induced by house dust mites [255]. Thus, by targeting ferroptotic pathways, it might be possible to intervene in the pathogenesis of asthma.
Cigarette smoke exposure also promotes ferritinophagy-dependent ferroptosis in lung epithelial cells, thus contributing to the pathogenesis of chronic obstructive pulmonary disease/COPD [256]. CMA-mediated GPX4 degradation promotes ferroptosis in a radiation-induced lung injury model, and this process can be limited using NVP-AUY922, a resorcinylic isoxazole amide drug [257]. Another study suggests the involvement of ferritinophagy-dependent ferroptosis in the pathogenesis of sepsis-induced acute lung injury [258]. The transcriptional regulator YAP1 (Yes1 associated transcriptional regulator) acts as a negative regulator of ferritinophagy and inhibits sepsis-induced acute lung injury [258]. Pending further confirmation, these studies suggest that the inhibition of ADF might result in the preservation of lung function.
Eye diseases
Glaucoma is an ocular disease characterized by retinal ganglion cell death resulting from an elevated intraocular pressure (IOP). Increased levels of Fe2+ resulting from elevated IOP can induce ferroptosis in retinal ganglion cells through ferritinophagy [259]. The depletion of NCOA4 can lower the Fe2+ levels in the retina, and oral administration of the iron chelator deferiprone inhibits ferroptosis in retinal ganglion cells and protects visual function [259].
Diabetic retinopathy involves damage to the blood vessels in the retina. Research has indicated that high glucose levels can induce mitophagy and ferritinophagy in ARPE-19 cells, which are retinal pigment epithelial cells [260]. The use of ferroptosis inhibitors, such as ferrostatin-1 and iron chelator, reduce ADF in these cells [260]. However, further in vivo experimentation is necessary to demonstrate the contribution of ADF to diabetes-induced blindness.
Age-related macular degeneration (AMD) stands as a leading cause of blindness among the elderly [261]. The early and atrophic (referred to as “dry”) manifestation of AMD represents its most prevalent form, currently lacking effective therapeutic interventions [262,263]. Recent research has unveiled that escalated levels of LCN2 (lipocalin 2) within retinal pigmented epithelial (RPE) cells contribute to the exacerbation of dry AMD’s pathogenesis. This is accomplished through the inhibition of autophagy flux and the disruption of iron homeostasis, consequently triggering inflammasome activation, oxidative stress, and ferroptosis within the RPE cells [264]. Moreover, an innovative monoclonal antibody designed to counteract LCN2 has demonstrated the capability to restore autophagy and mitigate ferroptosis in a well-characterized murine model of dry AMD, namely the Cryba1 cKO mice [264]. While evidence has been building that ferroptosis triggers numerous human diseases, this is the first study linking ADF to dry AMD.
Periodontal diseases
Periodontitis, a chronic inflammatory condition affecting the gums and supporting structures of the teeth, is associated with various forms of cell death. One critical process in the development of periodontitis is the loss of periodontal ligament fibroblasts [265]. Butyrate, a metabolite produced by commensal bacteria at levels found in periodontitis, kills periodontal ligament fibroblasts by inducing multiple types of cell death, including ferroptosis [266]. Specifically, butyrate induces ferritinophagy and ferroptosis in periodontal ligament fibroblasts, partly through the activation of the HIF1A-MAPK14/p38 (mitogen-activated protein kinase 14) pathway [266]. Theoretically, these processes may contribute to the pathogenesis of periodontitis.
Bone diseases
The loss of bone mass that characterizes osteoporosis involves an imbalance between osteoblasts and osteoclasts in favor of the latter. In the context of osteoclastogenesis triggered by TNFSF11/RANKL (TNF superfamily member 11) stimulation under normoxia, ferritinophagy-dependent ferroptosis is activated [267]. Conversely, the presence of HIF1A induced by hypoxia inhibits ferritinophagy and protects osteoclasts from ferroptosis [267]. In studies conducted on ovariectomized mice, the knockdown of HIF1A or the use of a HIF1A inhibitor called 2-methoxyestradiol promotes iron accumulation and induces ferroptosis in osteoclasts, thereby providing protection against osteoporosis [267]. Additionally, exosomes derived from vascular endothelial cells counteract glucocorticoid-induced osteoporosis by inhibiting ferritinophagy-dependent ferroptosis in osteoblasts [268]. In contrast, mitophagy induced by carbonyl cyanide m-chlorophenylhydrazone/CCCP increases ferroptosis in osteoblasts, potentially contributing to the pathogenesis of type 2 diabetic osteoporosis [105]. However, a significant challenge for osteoporosis treatment in the future lies in developing specific strategies to regulate the ferroptosis of osteoblasts.
Other diseases
Increased ADF is implicated in iron overload-induced toxicity in mouse thymus tissue [269]. L-citrulline, an endogenous metabolite in the urea cycle, restrains ferroptosis-induced thymus damage and preserves immune function in mice [269]. The cytotoxic effects induced by various agents such as nanoparticles [270,271], arsenic [272,273], antimony [121], PM2.5 [274,275], cadmium telluride [276], and ionizing radiation [277] can also be inhibited through genetic or pharmacological inhibition of ADF. These findings suggest that suppressing ADF might mitigate the toxic effects of these agents.
Methods for monitoring autophagy-dependent ferroptosis
Detection of a ferroptotic response
Lipid peroxidation
Lipid peroxidation is a crucial characteristic of ferroptotic cell death. Various techniques are commonly employed to assay lipid peroxidation in cells or tissues. Here, we provide a succinct list of these methods:
Mass spectrometry allows for direct quantification and identification of specific lipid peroxidation products. Because polyunsaturated phospholipids, particularly phosphatidyl-ethanolamines (PE), are the major substrates of ferroptosis-induced peroxidation, employment of liquid chromatography-mass spectrometry (LC-MS) is the preferred methodology for the detection and identification of peroxized ferroptosis biomarkers [36]. Combination of MS-based lipidomics with proteomics can be used for characterization of ferroptotic covalent adducts of oxidatively-truncated electrophilic PE with target proteins [278]. Recently, imaging mass-spectrometry has been applied for the spatial characterization of intracellular localization of peroxidized phospholipids [279]. Gas chromatography-mass spectrometry found limited applications in studies of ferroptosis as it is mostly effective in the analysis of oxidatively-modified free fatty acids [280], rather than oxygenated phospholipids. While mass spectrometry based protocols provide exceptional sensitivity, specificity, and the ability to detect and quantify a wide range of lipid species involved in ferroptosis [281], they require specialized equipment, expertise, and sample preparation. The latest breakthroughs employing high-resolution mass spectrometry in conjunction with ion mobility and cutting-edge tandem mass spectrometry techniques have additionally enabled the direct identification of intact glycerophospholipids that bear oxidative labeling [282,283]. These advancements provide specific insights into the precise localization of these oxidative modifications [282,283].
Thiobarbituric acid reactive substances (TBARS) assay: The TBARS assay, which can be performed using commercial kits, is used to measure the levels of malondialdehyde (MDA), a product of lipid peroxidation [284]. In this assay, MDA reacts with thiobarbituric acid (TBA) to form a colored complex that can be quantified spectrophotometrically (Figure 9). The TBARS assay offers a convenient and rapid method for assessing lipid peroxidation. However, it is important to note that the TBARS assay lacks specificity because other reactive aldehydes can also react with TBA, potentially confounding the measurement of lipid peroxidation.
Fluorescent probes: BODIPY dyes, such as BODIPY-581/591-C11 and BODIPY-665/676, are commonly used for the detection of lipid peroxidation [285]. BODIPY-581/591-C11 undergoes a fluorescence shift from red to green when its unsaturated diene bridge is oxidatively disrupted (Figure 10A). The Click-iT linoleamide alkyne (LAA) assay utilizes LAA, which inserts into cell membranes and is oxidized to produce lipid peroxide-derived aldehydes that can bind to protein nucleophiles (Figure 10B) [286]. Liperfluo, a perylene derivative, reacts with lipid hydroperoxide to generate a green fluorescent signal (Figure 10C) [287]. These probes provide valuable insights into lipid peroxidation but have limitations regarding specificity, potential interference by cellular autofluorescence, and the need for careful calibration and controls. For example, while BODIPY dyes can detect peroxidized lipids induced by pharmaceutical agents, they frequently struggle to identify ferroptosis under natural conditions. Therefore, it is advisable to exercise caution when interpreting negative data derived from BODIPY-based assays.
Immunohistochemistry (IHC) or immunofluorescence (IF): IHC or IF can be employed to detect the formation of specific lipid peroxidation products, such as 4HNE [288] or 8-hydroxy-2’-deoxyguanosine/8-OHdG [289], in tissues or cells [290]. Antibodies specific for these markers are used to visualize and quantify lipid peroxidation. However, IHC and IF assays require careful optimization, validation, and appropriate controls to ensure specificity and reliable results.
Figure 9.
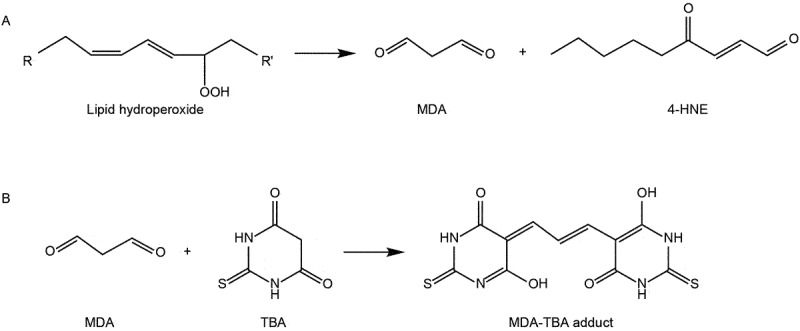
Reaction between MDA and TBA to form the MDA-TBA adduct.
Figure 10.
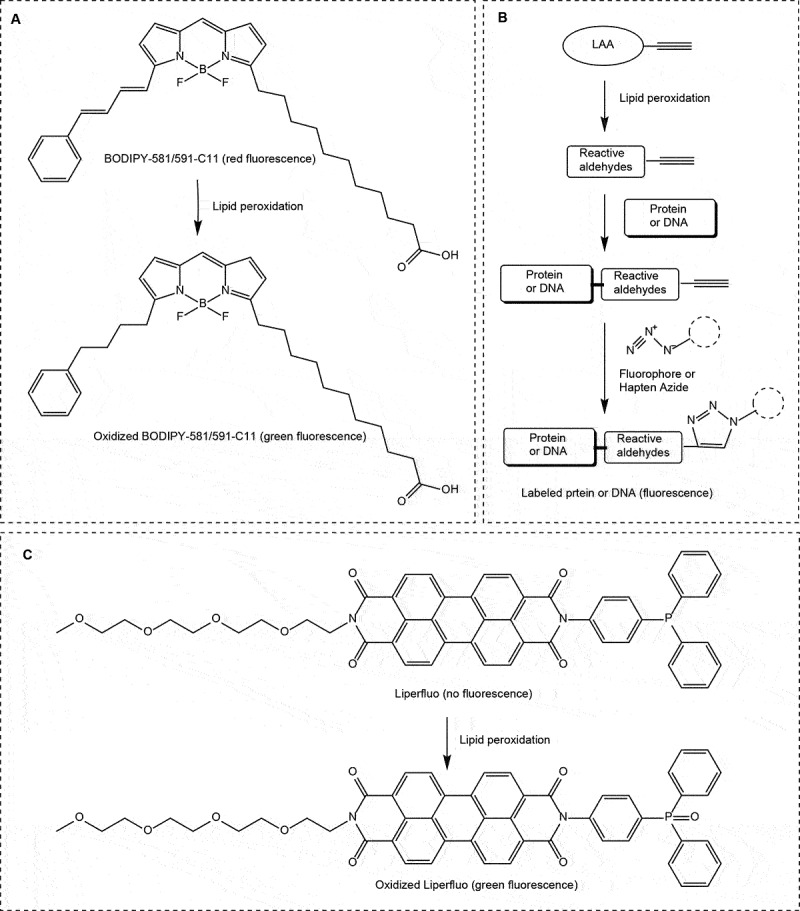
Fluorescent probe-based assays for monitoring lipid peroxidation. (A) BODIPY − 581/591-C11 assay; (B) click-iT LAA assay; (C) LiperFluo assay.
Iron redox forms
Several methods can be used to measure iron redox forms in cells and/or tissues. Here are some commonly employed techniques:
Ferrous/ferric ion assays: Spectrophotometric assays, such as the ferrozine assay or the bathophenanthroline sulfonate/BPS assay, are used to measure the levels of ferrous (Fe2+) and ferric (Fe3+) ions [291]. These assays rely on colorimetric reactions where specific reagents form colored complexes with either Fe2+ or Fe3+. Quantification is achieved through spectrophotometry. Commercial iron colorimetric assay kits are available based on this principle. However, these assays may lack specificity and can be influenced by interfering substances or other redox-active species in the sample.
Redox-sensitive fluorescent probes: Fluorescent probes such as FerroOrange (or RhoNox-4) or Phen Green™ SK (PGSK) can be used to track intracellular Fe2+. FerroOrange undergoes a fluorogenic reaction upon Fe2+-induced deoxygenation of N-oxide, resulting in an orange fluorescence (Figure 11) [292]. PGSK can permeate cell membranes and reacts with various metal ions, including Fe2+ (Figure 12) [293]. Binding of PGSK to cellular iron leads to fluorescence quenching. These changes in fluorescence can be detected using techniques such as flow cytometry or confocal microscopy. However, these probes may have limitations in terms of specificity and potential cytotoxicity, and require careful calibration and control experiments.
Electron paramagnetic resonance (EPR) spectroscopy: EPR spectroscopy is a powerful technique for studying paramagnetic species, including iron ions [294]. This method can provide direct measurements of the relative amounts of Fe2+ and Fe3+ in cells or tissues. EPR spectroscopy requires specialized equipment and expertise, making it less accessible for routine use. Additionally, sample preparation and handling for EPR analysis can be challenging, particularly when dealing with biological samples.
Mössbauer spectroscopy: Mössbauer spectroscopy is a technique used to study the electronic and magnetic properties of iron [295]. The method can provide detailed information on the oxidation state and coordination environment of iron ions. Mössbauer spectroscopy is highly sensitive and specific for iron redox forms and is particularly useful for investigating iron in complex biological samples. However, it requires specialized instrumentation and expertise, limiting its widespread use.
Figure 11.
Assessing Fe2+ with the FerroOrange assay.
Figure 12.
Assessing Fe2+ with the phen green SK assay.
Key ferroptosis-related genes or proteins
The levels of gene and protein markers associated with ferroptosis can be assessed using various techniques, including real-time reverse transcription polymerase chain reaction/RT-PCR, western blotting, immunofluorescence, and immunohistochemistry. These markers encompass key regulators of ferroptosis, such as SLC7A11, GPX4, AIFM2, DHODH, NFE2L2, ACSL4, and TFRC (transferrin receptor), as well as other ferroptosis-related proteins such as PTGS2 (prostaglandin-endoperoxide synthase 2) [296].
Changes in the protein levels of SLC7A11, GPX4, or ACSL4 can reflect the susceptibility or resistance of cells to ferroptotic cell death [297]. The degradation of GPX4 protein is often associated with increased susceptibility to ferroptosis [298], although other GPX4-independent antioxidant systems may also play a role. Elevated levels of TFRC or ACSL4 proteins, as well as increased expression of PTGS2 genes, can serve as markers for ferroptosis in cell cultures and tissue samples [34,299].
However, assessing the expression levels of ferroptosis-related genes can be challenging due to their dynamic and context-dependent regulation. These genes are often subject to complex transcriptional regulation and can be influenced by various factors, including cellular stress, signaling pathways, and genetic backgrounds. Obtaining accurate and reliable measurements of gene expression requires careful experimental design and the inclusion of appropriate controls. Additionally, the availability and specificity of antibodies used for protein analyses may be a limiting factor.
Ultrastructure of subcellular organelles
Ferroptotic cell death is characterized by distinct morphological changes, including necrotic cell morphology with organelle swelling, plasma membrane rupture, and cell lysis. The morphological alterations observed in ferroptotic cells depend on the stage of induction, and various organelles contribute to the regulation of ferroptosis sensitivity. Notably, mitochondria undergo remarkable changes during ferroptosis, and monitoring their morphological features is commonly achieved using transmission electron microscopy (TEM). TEM enables the visualization of condensed mitochondrial membrane densities, reduced mitochondrial volume, diminished mitochondrial cristae, and ruptured outer membranes [5], providing valuable insights into the ultrastructure of mitochondria in ferroptotic cells.
In addition to mitochondrial changes, ferroptotic cells often exhibit an increase in autophagic vesicles, indicative of altered autophagy activity [144]. The presence of autophagic vesicles can be observed alongside mitochondrial alterations using TEM. However, it is crucial to recognize that the ultrastructural changes observed during ferroptosis can also occur in other types of cell death or cellular stress conditions. Therefore, additional confirmatory assays are often necessary to distinguish ferroptosis from other cell death mechanisms. Measurement of lipid peroxidation or the use of specific molecular markers associated with ferroptosis can help confirm the occurrence of ferroptosis in conjunction with TEM observations.
Cell viability and cell death assays
Various methods are employed to measure cell viability, including the MTS, CCK-8, and Alamar Blue assays. These assays are relatively simple and widely used for monitoring ferroptosis in kinetic experiments and dose-response studies. For enhanced specificity, it may be worthwhile to conduct these assays both in the absence and presence of ferroptosis inhibitors, such as ferrostatin-1. As a comparative control, apoptosis inhibitors like Z-VAD-FMK could be employed. However, it is important to consider their limitations. For example, cell viability assays such as the MTS assay rely on changes in optical density/OD, which can be directly influenced by certain agents (e.g., antioxidants) or their color [300]. This can lead to potential interference and affect the accuracy of the viability measurements. Furthermore, a reduction in cell viability, as indicated by these assays, may not always signify cell death but could instead be attributed to growth inhibition or other non-lethal effects.
To accurately assess the reduction in ferroptosis-related cytotoxicity, it is necessary to complement cell viability assays with cell death assays. Several methods are available for evaluating cell death [301]. These methods include propidium iodide staining, SYTOX Green staining, and LDH release assay. These assays allow for the detection of different aspects of cell death, such as membrane integrity loss or the release of cytoplasmic contents. Additionally, the release of DAMPs, such as HMGB1 [167], ATP [302], SQSTM1 [134], and DCN (decorin) [303], can be monitored using ELISA both in vitro and in vivo. Of particular note, the release of DCN into the extracellular space serves as a relatively specific biomarker for early-stage ferroptotic damage, distinguishing it from other forms of cell death [303]. By combining cell viability assays with cell death assays and specific biomarker measurements, researchers can obtain a more comprehensive understanding of the effects of ferroptosis and distinguish it from other forms of cell death.
Genetically engineered mouse model
The Gpx4 conditional knockout mouse model is one of the most extensively studied genetically engineered models of ferroptosis. However, it is important to note that the depletion of Gpx4 in specific cells or tissues in mice does not always result in spontaneous ferroptotic damage. One of the limitations of the Gpx4 knockout model is the existence of several GPX4-independent regulators of ferroptosis. Initial studies indicated that conditional knockout of Gpx4 in the kidney leads to neonatal lethality, which can be rescued by the addition of ferroptosis inhibitors [15]. However, subsequent studies have not consistently observed lethality upon conditional Gpx4 knockout in the kidney, although some tissue damage has been observed in a sex-dependent manner [304].
Furthermore, conditional knockout of Gpx4 in murine myeloid cells or erythroid precursors induces pyroptosis or necroptosis instead of ferroptosis [305,306]. Similarly, depletion of mitochondrial Gpx4 in mice leads to a cone-rod dystrophy-like phenotype characterized by apoptosis, rather than ferroptosis [307]. These findings highlight the complexity of GPX4 depletion-mediated oxidative stress and its ability to induce or enhance cell death sensitivity through multiple pathways.
Another genetically engineered mouse model is a CRISPR-Cas9 genome edited mouse with homozygous expression of an R748W mutation in the mouse Pnpla9 gene, which corresponds to the human R747W mutation linked to Parkinson disease and other human neurodegenerative diseases [42]. These mice exhibit motor deficits accompanied by elevated levels of 4-HNE and 15-HpETE-PE with recued levels of GSH.
Detection of autophagy
Autophagic vesicles
Autophagic vesicles play a vital role in the autophagy process, and their ultrastructural features can be observed using TEM. TEM provides a high-resolution view of autophagic structures in the nanometer range. Autophagosomes, the initial autophagic vacuoles, typically exhibit a double membrane, while autolysosomes, the late/degradative autophagic vacuoles, usually have a single limiting membrane containing visible cytoplasmic cargos [308]. It is important to note that the cargo composition differs between selective and nonselective autophagy.
Nonetheless, TEM has some inherent challenges. Autophagy is a dynamic process with rapid vesicle turnover, while TEM analysis provides only a static snapshot at a specific time point. This limitation makes it challenging to capture the dynamics of autophagy and accurately assess vesicle formation and degradation rates. Additionally, counting and quantifying autophagic vesicles in TEM images can be subjective and time-consuming. The random distribution of vesicles within cells, along with potential obscuration by cellular structures, introduces variability and limits the statistical robustness of the analysis. Despite these challenges, TEM remains the gold standard technique for visualizing autophagic vesicles with their ultrastructural characteristics.
MAP1LC3 protein
During autophagy, MAP1LC3 undergoes a series of modifications and turnover that can be indicative of autophagic activity. These modifications involve the conversion of MAP1LC3 from its cytosolic form (MAP1LC3-I) to a lipidated form (MAP1LC3-II), which is associated with phagophore and autophagosomal membranes [309]. The levels of MAP1LC3-II correlate with the number/volume of autophagosomes in cells [310]. The MAP1LC3-II needs to be related to a loading control like actin (and not to MAP1LC3-I), because after autophagosome-lysosome fusion the MAP1LC3-II associated with the outer autophagosome membrane is converted back to MAP1LC3-I, while the MAP1LC3-II in the inner membrane is degraded. MAP1LC3-II levels can be determined using western blotting. As MAP1LC3-II levels reflect autophagosome load in the cells, an increase can either reflect an increase in synthesis (compared to degradation rates), or impaired autophagosome/MAP1LC3-II degradation. To infer autophagosome formation rates (or autophagic flux if one is in steady-state conditions), one can clamp autophagosome/MAP1LC3-II degradation using lysosomal inhibitors, such as bafilomycin A1, CQ or protease inhibitors such as E64d plus leupeptin, and monitoring the resulting changes in MAP1LC3-II levels over time. Overall, monitoring MAP1LC3-II formation rates provides valuable insights into the regulation and dynamics of autophagy especially when viewed in the context of overall MAP1LC3-II levels, allowing researchers to study the modulation of autophagic activity under different conditions or in response to various stimuli, such as erastin or RSL3. One final note is that it is generally helpful to monitor an autophagic substrate along with MAP1LC3-II to help in the interpretation of the results as discussed below. While SQSTM1 is often used as an autophagy substrate care needs to be taken especially in the context of ferroptosis, as its transcription is ROS-dependent (hence the need for this protein to measure its mRNA levels, or to transfect in exogenous protein driven by a promoter that is independent of ROS levels).
Autophagic flux
Autophagic flux refers to the entire process of autophagy, which involves the formation, maturation, and degradation of autophagosomes within cells [311]. Autophagy is a dynamic mechanism that ensures the proper turnover of cellular components and the maintenance of cellular homeostasis. To measure autophagic flux, several probes and techniques are commonly used. Here are some widely employed methods:
MAP1LC3-II protein turnover assay: See 9.2.2 and note that this does not directly measure turnover but assesses MAP1LC3-II formation rates. In mammals, the Atg8-protein family encompasses various members, including MAP1LC3A (microtubule associated protein 1 light chain 3 alpha), MAP1LC3B (microtubule associated protein 1 light chain 3 beta), MAP1LC3B2, MAP1LC3C (microtubule associated protein 1 light chain 3 gamma), GABARAP (GABA type A receptor-associated protein), GABARAPL1 (GABA type A receptor associated protein like 1), and GABARAPL2 (GABA type A receptor associated protein like 2) [312]. Whereas MAP1LC3B has been extensively studied, examining additional members of the Atg8-protein family may provide a more comprehensive understanding of autophagosomal action and yield broader insights into the process. However, it is important to consider that some of the Atg8 family members act at different stages of the pathway, thus it is simplest to start with MAP1LC3B.
MAP1LC3-binding substrate degradation assay: Autophagic flux can be assessed by monitoring the degradation of specific autophagy substrates, such as SQSTM1 [313], which serves as an autophagy receptor and is selectively incorporated into phagophores through its binding to MAP1LC3. To measure autophagic flux, the degradation of SQSTM1 can be monitored using techniques like western blotting or immunofluorescence analysis. This analysis can be performed in the presence or absence of autophagy inhibitors like CQ, bafilomycin A1, or lysosomal protease inhibitors, which help in distinguishing changes in SQSTM1 degradation specifically due to autophagy. It is important to note that when using the levels of a MAP1LC3-binding substrate as an indicator of autophagic activity, it is necessary to measure the mRNA levels of the substrate to confirm that changes in protein levels are not solely due to transcriptional induction. By assessing both protein degradation and mRNA levels, researchers can obtain a more comprehensive understanding of autophagic flux and its underlying mechanisms.
Lysosomal pH-sensitive probes: pH-sensitive dyes such as LysoTracker or LysoSensor are commonly used to monitor changes in lysosomal pH, providing valuable insights into autophagic flux [314]. An increase in lysosomal acidity is generally associated with enhanced autophagic activity. However, it is important to note that alterations in lysosomal pH alone may not always directly correlate with autophagic activity. Other factors, including lysosomal enzyme activity and lysosomal membrane integrity, also contribute to the overall autophagic flux. To obtain a comprehensive understanding of autophagy dynamics, it is crucial to interpret changes in lysosomal pH in conjunction with complementary assays and assessments. Several probes or methods are available to assay overall lysosomal integrity, such as lysosomal-METRIQ, which measures the overall integrity of lysosomes [315]. Additionally, LGALS3 (galectin 3) can be used as a marker for assessing lysosomal membrane damage [316], whereas MagicRed is a probe that specifically detects cathepsin enzyme activity within lysosomes [317].
Detection of lysosomal integrity and function in vivo presents additional challenges, as many of the above tools require preservation of lysosomal pH and/or expression of exogenous reporters. Some of the pH-sensitive probes and assays for lysosomal enzyme activity have been adapted to flow cytometry detection in isolated mammalian cells [318]. Alternatively, evaluating lysosomal activity can be achieved using freshly purified tissue lysosomes. Furthermore, the integrity of lysosomal membranes can be gauged in tissue sections through immunofluorescence, involving a comparison of the cellular distribution of lysosomal membrane proteins (e.g., LAMP1 [lysosomal associated membrane protein 1]) and soluble lysosomal enzymes (e.g., cathepsins) [319].
(4) mRFP-GFP-LC3 tandem probe: This fluorescent probe allows simultaneous visualization of autophagosomes and autolysosomes [320], and takes advantage of the differential pH stability of green fluorescent protein (GFP) and red fluorescent protein (RFP). Autophagosomes labeled with both GFP and RFP emit yellow fluorescence, while autolysosomes exhibit only RFP fluorescence due to GFP quenching in the acidic environment. The ratio of yellow (autophagosomes) to red (autolysosomes) puncta can indicate autophagic flux. The combination of GFP and RFP/mCherry in tandem reporters may not be ideal due to the occurrence of Förster resonance energy transfer (FRET) from GFP to RFP/mCherry [320]. This FRET process leads to a weakening of the RFP/mCherry signal (as it acts as the FRET acceptor) following the degradation of GFP (serving as the FRET donor). Furthermore, methodologies like single-cell analysis approaches offer the capability to meticulously dissect and quantify the entire intracellular reservoir of autophagosomes, autolysosomes, and lysosomes. These techniques also facilitate the assessment of autophagosome flux, denoting the count of autophagosomes per cell or per hour [321,322]. The significance lies not only in gauging the level of basal or induced autophagic activity but also in differentiating between autophagic and proteinaceous cargo flux. This distinction is growing in importance as it directly influences the onset of proteotoxicity and cellular demise [323].
(5) GFP-LC3-RFP(-LC3ΔG) reporter: This reporter is a specialized fluorescent probe extensively employed for assessing autophagic flux in cells [324]. The reporter is an adapted version of the mRFP-GFP-LC3 tandem probe. Within cells, the GFP-LC3-RFP-LC3ΔG fusion protein undergoes processing by ATG4, resulting in the generation of GFP-LC3, which indicates autophagy activity, and “free” RFP-LC3ΔG, lacking the essential C-terminal glycine required for membrane conjugation. The latter component functions as a control for reporter expression and cellular viability. The ratio of GFP-LC3 to RFP-LC3ΔG is inversely proportional to cumulative autophagic degradation activity. Importantly, this method offers the advantage of not relying on microscopy-based imaging of MAP1LC3 puncta, as it can be conveniently utilized with flow cytometers or fluorescence microplate readers.
(6) HaloTag reporter: A recent addition to the methodology for monitoring autophagy flux relies on the HaloTag (Halo) protein, which gains resistance to proteolysis when attached to its ligand. Comparison of HaloLigand-LC3 or -GFP following pulse labeling with ligand allows detection of an increase in free HaloLigand by immunoblot, in-gel fluorescence imaging or microscopy upon autophagy induction [325,326].
(7)SRAI-LC3B assay: This ratiometric assay that generates a positive signal when there is lysosomal degradation of an autophagic substrate and provides a direct readout of autophagic flux. The assay exploits the SRAI reporter, a tandem construct consisting of TOLLES (a blue fluorescent protein resistant to acid-denaturation and proteolysis) and YPet (a yellow fluorescent protein, which undergoes acid-denaturation and proteolysis in lysosomes). Delivery of the SRAI reporter to lysosomes causes degradation of YPet, which leads to a detectable shift in fluorescence of the tandem construct as the FRET-associated quenching of the TOLLES signal is relieved after YPet degradation. To adapt this tool for autophagy, the SRAI reporter is fused to the N-terminus of MAP1LC3B, which markedly increases the sensitivity of the reporter as a macroautophagy assay, as the ratio of blue to yellow fluorescence reflects the proportion of MAP1LC3B that is undergoing lysosomal degradation [327].
In summary, each method mentioned above has its strengths and limitations, and researchers often employ a combination of techniques to gain a comprehensive understanding of autophagy dynamics in specific cells or tissues of interest.
Selective autophagy
Multiple forms of selective autophagy are involved in the regulation of ferroptosis sensitivity. The assay of selective cargoes, such as mitochondria, lipid droplets, ER, ferritin protein, and GPX4 protein, can be performed using various imaging and labeling methods to assess their colocalization with autophagosomes or lysosomes. Recently, specific flux reporters have been developed to monitor the dynamics of selective autophagy for mitochondria (e.g., mito-SRAI [328], mito-QC [329], or pSU-Halo-mGFP [325], and the ER (e.g., ssGFP-RFP-KDEL [330], Halo-mGFP-KDEL [325], mCherry-GFP-SERP1/RAMP4 [331], or mCherry-GFP-REEP5 [332] in real-time. To gain further insights into the dynamics of these cargo degradation processes, the combined use of autophagy inhibitors such as CQ, 3-MA, spautin-1, bafilomycin A1, or lysosomal protease inhibitors, or the knockout of key autophagy genes such as ATG5 and ATG7, can be employed. These approaches can help elucidate the interplay between selective autophagy pathways and ferroptosis sensitivity.
Genetically engineered mouse model
Transgenic mice expressing tandem fluorescent-tagged LC3/tfLC3 or mice transfected with mCherry-GFP-LC3 via intraventricular injection of adeno-associated viruses have been developed as valuable tools to measure autophagic activity in vivo, without the need for lysosomal inhibitors [333–335]. These models allow for the visualization and quantification of autophagosomes and autolysosomes in specific cells or tissues. Furthermore, conditional knockout or overexpression of core autophagy genes, such as ATG5, ATG7, BECN1, or SQSTM1, in specific cells or tissues of mice can be employed to investigate whether a particular phenotype is driven by autophagy. While ferroptosis has been investigated in lower model organisms (e.g., Caenorhabditis elegans [336]), the exploration of ADF remains absent in these organisms.
Interpretation of autophagy-dependent ferroptosis
Most of the available assays for assessing the relationship between autophagy and ferroptosis fail to consider the complex nature of injury and the anti-injury response, which is highly context-dependent and influenced by various factors such as the initiating stimulus, stage, and cell types involved. Interpreting assays related to ADF faces several challenges that need to be carefully considered.
First, there is a significant variation in the literature regarding the doses of ferroptosis inducers, such as erastin and RSL3, with reported ranges of 0.5–50 µM for erastin [337,338] and 0.1–20 µM for RSL3 [266,339]. This variation in dosage can have implications for the type of stress and selective autophagy induced, as distinct inducers may have different thresholds for activation. Moreover, there is an ongoing debate surrounding the direct targets of these inducers. While erastin was initially identified as a VDAC (voltage dependent anion channel) activator [340], recent studies suggest its inhibition of system xc− as the primary mechanism [341]. Similarly, the direct inhibition of GPX4 by RSL3 is still under scrutiny, with evidence suggesting that it may instead target TXNRD1 (thioredoxin reductase 1) [63]. As a result, the diverse doses of ferroptosis inducers can activate distinct pathways, both targeted and untargeted, leading to varying levels of autophagy induction. Thus, when evaluating the autophagic response during ferroptosis stimulation, it is crucial to consider the IC50 values of erastin or RSL3 specific to the cell type being studied.
Second, it is important to recognize that both lipid peroxidation and autophagy are dynamic processes. Under physiological conditions, moderate levels of lipid peroxidation and autophagy can actually promote cell survival [342]. However, excessive or uncontrolled lipid peroxidation and autophagy can lead to cell death [343]. Therefore, in addition to considering the degradation of autophagic substrates, it is crucial to take into account the stage of ferroptosis and how it influences the function of associated autophagic responses. Conducting time-dependent dynamic analyses can provide valuable insights into the sequence of events during which changes in autophagy and core markers of ferroptosis occur. Furthermore, it may be relevant to inhibit specific autophagy receptors, rather than core autophagy regulators, to evaluate the impact of selective autophagy on ferroptosis. This approach could help elucidate the specific contribution of selective autophagy processes, rather than bulk autophagy, in the context of ferroptosis. By focusing on the selective autophagy pathways that are directly involved in the degradation of specific cargo, we can gain a more precise understanding of their impact on ferroptotic cell death.
Third, different cell types possess distinct genetic backgrounds, resulting in variations in molecular networks and basal levels of core autophagy or ferroptosis regulators. Cell lines, such as HT-1080 (human fibrosarcoma) and PANC1 (human PDAC), are commonly used in ADF studies due to their high sensitivity to erastin and RSL3. However, it should be noted that these cell lines exhibit relatively high expression of MAP1LC3-II [144,344,345], perhaps indicating a higher baseline autophagy activity compared to other cell lines with lower MAP1LC3-II expression. As a result, under ferroptosis stimulation and oxidative stress, these cell lines may induce excessive autophagy, which can lead to autophagy-dependent cell death. The response of cells may be also influenced by the number of generations of cell culture they undergo in different laboratories. To fully investigate the role of inducible autophagy in ferroptosis, it is crucial to include primary normal cells alongside these cell lines to provide a comparative analysis and to better understand the specific contribution of autophagy in the context of ferroptosis.
Fourth, it is important to acknowledge that in vivo models related to ADF are influenced by various factors. Many studies investigating ADF in vivo rely on murine tumor xenograft models or the administration of ferroptosis inhibitors (e.g., liproxstatin-1) or autophagy inhibitors (e.g., CQ) [15]. The in vivo drug metabolism and tissue distribution can significantly influence the interpretation of experimental results. The impact of infiltrating immune cells and the release of DAMPs on the outcomes of ADF also requires further investigation. In models of cell death-related diseases, the early response often involves inflammation-mediated wound healing processes, which involve the participation of diverse immune cells [346]. Therefore, when assessing ADF, it is essential to consider the influence of immune cells and their responses on the overall dynamics and outcomes of ADF. By investigating the interaction between autophagy, ferroptosis, and immune cells, a more comprehensive understanding of the complex interplay between these processes can be achieved.
In summary, the assessment of ADF necessitates a comprehensive evaluation using a range of in vitro and in vivo assays. These assays should collectively determine the circumstances under which autophagy promotes either a pro-survival or pro-death phenotype. It is crucial to tightly regulate autophagy within a narrow window to harness its beneficial effects while minimizing any potential negative consequences, particularly in the context of disease conditions.
Conclusions and perspectives
In conclusion, the relationship between autophagy and ferroptosis is complex and highly dependent on the specific context. Current evidence suggests that autophagy can have both promoting and protective effects on ferroptotic cell death, but the underlying mechanisms and checkpoints regulating these opposing effects are still poorly understood. The interplay between autophagy and lipid peroxidation further complicates the understanding of the sequence of events and causality.
As the field progresses, it is crucial to establish guidelines and standards that ensure consistency in experimental approaches. These guidelines should be adaptable to accommodate the diverse research questions and experimental systems employed by different researchers. Additionally, the development of novel techniques for monitoring autophagy and ferroptosis will greatly enhance our understanding of ADF and facilitate the unraveling of its intricate mechanisms.
While ADF has been extensively studied in vitro, its contribution to ferroptosis in vivo and its relevance to various diseases require further investigation. Assessing ADF in vivo remains challenging due to the lack of specific markers, often necessitating the use of general autophagy markers such as MAP1LC3. Therefore, it is imperative to develop specific and reliable methods for detecting autophagy in vivo.
In the future, addressing these knowledge gaps and expanding our understanding of ADF in both physiological and pathological contexts will be crucial. Advances in this field have the potential to uncover novel therapeutic targets and strategies for the treatment of diseases associated with dysregulated autophagy and ferroptosis. By elucidating the intricate interplay between autophagy and ferroptosis, we can pave the way for the development of precision therapies that harness the therapeutic potential of these interconnected processes.
Funding Statement
The work was supported by the National Natural Science Foundation of China [82272660]. X.C. was supported by the Plan on Enhancing Scientific Research in GMU [02-410-2302289XM] and the National Natural Science Foundation of China [82272660]. D.T. was supported by grants from the National Institutes of Health [NIH] of USA [R01CA160417, R01CA229275, and R01CA211070]. D.J.K. was supported by NIH grant GM131919. G.K. is supported by the Ligue contre le Cancer [équipe labellisée]; Agence National de la Recherche [ANR] – Projets blancs; AMMICa US23/CNRS UMS3655; Association pour la recherche sur le cancer [ARC]; Cancéropôle Ile-de-France; Fondation pour la Recherche Médicale [FRM]; a donation by Elior; Equipex Onco-Pheno-Screen; European Joint Programme on Rare Diseases (EJPRD); European Research Council Advanced Investigator Award [ERC-2021-ADG, ICD-Cancer, Grant No. 101052444], European Union Horizon 2020 Projects Oncobiome, Prevalung [grant No. 101095604] and Crimson; Fondation Carrefour; Institut National du Cancer [INCa]; Institut Universitaire de France; LabEx Immuno-Oncology [ANR-18-IDEX-0001]; a Cancer Research ASPIRE Award from the Mark Foundation; the RHU Immunolife; Seerave Foundation; SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination [SOCRATE]; and SIRIC Cancer Research and Personalized Medicine [CARPEM]. This study contributes to the IdEx Université de Paris ANR-18-IDEX-0001.
Disclosure statement
L.G. is/has been holding research contracts with Lytix Biopharma, Promontory and Onxeo, has received consulting/advisory honoraria from Boehringer Ingelheim, AstraZeneca, OmniSEQ, Onxeo, The Longevity Labs, Inzen, Imvax, Sotio, Promontory, Noxopharm, EduCom, and the Luke Heller TECPR2 Foundation, and holds Promontory stock options. D.C.R. is a consultant for Aladdin Healthcare Technologies Ltd., Mindrank AI, Nido Biosciences, Drishti Discoveries, Retro Biosciences and PAQ Therapeutics. D.I. G. is an employee and shareholder of AstraZeneca. M. K. serves on the Scientific Advisory Boards of Engine Biosciences, Casma Therapeutics, Cajal Neuroscience, Alector, and Montara Therapeutics, and is an advisor to Modulo Bio and Recursion Therapeutics. X.J. holds inventorship of patents related to autophagy and cell death, and holds equity as well as consults for Exarta Therapeutics and Lime Therapeutics. G.K. has been holding research contracts with Daiichi Sankyo, Eleor, Kaleido, Lytix Pharma, PharmaMar, Osasuna Therapeutics, Samsara Therapeutics, Sanofi, Tollys, and Vascage. G.K. is on the Board of Directors of the Bristol Myers Squibb Foundation France. G.K. is a scientific co-founder of everImmune, Osasuna Therapeutics, Samsara Therapeutics and Therafast Bio. G.K. is in the scientific advisory boards of Hevolution, Institut Servier and Longevity Vision Funds. G.K. is the inventor of patents covering therapeutic targeting of aging, cancer, cystic fibrosis and metabolic disorders. G.K.’wife, Laurence Zitvogel, has held research contracts with Glaxo Smyth Kline, Incyte, Lytix, Kaleido, Innovate Pharma, Daiichi Sankyo, Pilege, Merus, Transgene, 9 m, Tusk and Roche, was on the on the Board of Directors of Transgene, is a cofounder of everImmune, and holds patents covering the treatment of cancer and the therapeutic manipulation of the microbiota. G.K.’s brother, Romano Kroemer, was an employee of Sanofi and now consults for Boehringer-Ingelheim. The other authors declare no conflicts of interest or financial interests. The funders had no role in the writing of the manuscript.
References
- [1].Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vitale I, Pietrocola F, Guilbaud E, et al. Apoptotic cell death in disease—Current understanding of the NCCD 2023. Cell Death Differ. 2023;30(5):1097–1154. doi: 10.1038/s41418-023-01153-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cookson BT, Brennan MA.. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9(3):113–114. doi: 10.1016/S0966-842X(00)01936-3 [DOI] [PubMed] [Google Scholar]
- [4].Degterev A, Huang Z, Boyce M, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1(2):112–119. doi: 10.1038/nchembio711 [DOI] [PubMed] [Google Scholar]
- [5].Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Song X, Zhu S, Xie Y, et al. JTC801 induces pH-dependent death specifically in cancer cells and slows growth of tumors in mice. Gastroenterology. 2018;154(5):1480–1493. doi: 10.1053/j.gastro.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375(6586):1254–1261. doi: 10.1126/science.abf0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu X, Nie L, Zhang Y, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol. 2023;25(3):404–414. doi: 10.1038/s41556-023-01091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Galluzzi L, Kepp O, Chan F-M, et al. Necroptosis: mechanisms and relevance to disease. Annu Rev Pathol Mech Dis. 2017;12(1):103–130. doi: 10.1146/annurev-pathol-052016-100247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tan S, Schubert D, Maher P. Oxytosis: a novel form of programmed cell death. Curr Top Med Chem. 2001;1(6):497–506. doi: 10.2174/1568026013394741 [DOI] [PubMed] [Google Scholar]
- [11].Dolma S, Lessnick SL, Hahn WC, et al. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3(3):285–296. doi: 10.1016/S1535-6108(03)00050-3 [DOI] [PubMed] [Google Scholar]
- [12].Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15(3):234–245. doi: 10.1016/j.chembiol.2008.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim R, Hashimoto A, Markosyan N, et al. Ferroptosis of tumour neutrophils causes immune suppression in cancer. Nature. 2022;612(7939):338–346. doi: 10.1038/s41586-022-05443-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yao Y, Chen Z, Zhang H, et al. Selenium–GPX4 axis protects follicular helper T cells from ferroptosis. Nat Immunol. 2021;22(9):1127–1139. doi: 10.1038/s41590-021-00996-0 [DOI] [PubMed] [Google Scholar]
- [15].Friedmann Angeli JP, Schneider M, Proneth B, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Matsushita M, Freigang S, Schneider C, et al. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J Exp Med. 2015;212(4):555–568. doi: 10.1084/jem.20140857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266–282. doi: 10.1038/s41580-020-00324-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen X, Kang R, Kroemer G, et al. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18(5):280–296. doi: 10.1038/s41571-020-00462-0 [DOI] [PubMed] [Google Scholar]
- [19].Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23(3):369–379. doi: 10.1038/cdd.2015.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Galluzzi L, Yamazaki T, Kroemer G. Linking cellular stress responses to systemic homeostasis. Nat Rev Mol Cell Biol. 2018;19(11):731–745. doi: 10.1038/s41580-018-0068-0 [DOI] [PubMed] [Google Scholar]
- [23].Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131(6):1137–48. doi: 10.1016/j.cell.2007.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9(12):1004–10. doi: 10.1038/nrm2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhu J-H, Horbinski C, Guo F, et al. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol. 2007;170(1):75–86. doi: 10.2353/ajpath.2007.060524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhu J, Gusdon A, Cimen H, et al. Impaired mitochondrial biogenesis contributes to depletion of functional mitochondria in chronic MPP+ toxicity: dual roles for ERK1/2. Cell Death Dis. 2012;3(5):e312–e. doi: 10.1038/cddis.2012.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu Y, Shoji-Kawata S, Sumpter RM Jr, et al. Autosis is a Na + ,K + -ATPase–regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia–ischemia. Proc Natl Acad Sci, USA. 2013;110(51):20364–20371. doi: 10.1073/pnas.1319661110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hou W, Xie Y, Song X, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12(8):1425–8. doi: 10.1080/15548627.2016.1187366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gao M, Monian P, Pan Q, et al. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26(9):1021–32. doi: 10.1038/cr.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tian R, Abarientos A, Hong J, et al. Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nat Neurosci. 2021;24(7):1020–1034. doi: 10.1038/s41593-021-00862-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Song X, Zhu S, Chen P, et al. AMPK-Mediated BECN1 phosphorylation promotes ferroptosis by directly blocking system xc– activity. Curr Biol. 2018;28(15):2388–2399.e5. doi: 10.1016/j.cub.2018.05.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dai E, et al. A guideline on the molecular ecosystem regulating ferroptosis. Nat Cell Biol. 2024; doi: 10.1038/s41556-024-01360-8 [DOI] [PubMed] [Google Scholar]
- [33].Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–85. doi: 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–31. doi: 10.1016/j.cell.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xie Y, Kang R, Klionsky DJ, et al. GPX4 in cell death, autophagy, and disease. Autophagy. 2023;19(10):1–18. doi: 10.1080/15548627.2023.2218764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kagan VE, Mao G, Qu F, et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol. 2017;13(1):81–90. doi: 10.1038/nchembio.2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Doll S, Proneth B, Tyurina YY, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13(1):91–8. doi: 10.1038/nchembio.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yuan H, Li X, Zhang X, et al. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun. 2016;478(3):1338–1343. doi: 10.1016/j.bbrc.2016.08.124 [DOI] [PubMed] [Google Scholar]
- [39].Yang WS, Kim KJ, Gaschler MM, et al. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci U S A. 2016;113(34):E4966–75. doi: 10.1073/pnas.1603244113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yan B, Ai Y, Sun Q, et al. Membrane damage during Ferroptosis is caused by oxidation of phospholipids catalyzed by the oxidoreductases POR and CYB5R1. Mol Cell. 2020;81(2):355–369.e10. doi: 10.1016/j.molcel.2020.11.024 [DOI] [PubMed] [Google Scholar]
- [41].Zou Y, Li H, Graham ET, et al. Cytochrome P450 oxidoreductase contributes to phospholipid peroxidation in ferroptosis. Nat Chem Biol. 2020;16(3):302–9. doi: 10.1038/s41589-020-0472-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sun W-Y, Tyurin VA, Mikulska-Ruminska K, et al. Phospholipase iPla2β averts ferroptosis by eliminating a redox lipid death signal. Nat Chem Biol. 2021;17(4):465–476. doi: 10.1038/s41589-020-00734-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen X, Huang J, Yu C, et al. A noncanonical function of EIF4E limits ALDH1B1 activity and increases susceptibility to ferroptosis. Nat Commun. 2022;13(1):6318. doi: 10.1038/s41467-022-34096-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pedrera L, Espiritu RA, Ros U, et al. Ferroptotic pores induce Ca(2+) fluxes and ESCRT-III activation to modulate cell death kinetics. Cell Death Differ. 2020;28(5):1644–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dai E, Meng L, Kang R, et al. ESCRT-III–dependent membrane repair blocks ferroptosis. Biochem Biophys Res Commun. 2020;522(2):415–421. doi: 10.1016/j.bbrc.2019.11.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pisoschi AM, Pop A, Iordache F, et al. Oxidative stress mitigation by antioxidants-an overview on their chemistry and influences on health status. Eur J Med Chem. 2021;209:112891. doi: 10.1016/j.ejmech.2020.112891 [DOI] [PubMed] [Google Scholar]
- [47].Doll S, Freitas FP, Shah R, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–8. doi: 10.1038/s41586-019-1707-0 [DOI] [PubMed] [Google Scholar]
- [48].Bersuker K, Hendricks JM, Li Z, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–92. doi: 10.1038/s41586-019-1705-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dai E, Zhang W, Cong D, et al. AIFM2 blocks ferroptosis independent of ubiquinol metabolism. Biochem Biophys Res Commun. 2020;523(4):966–971. doi: 10.1016/j.bbrc.2020.01.066 [DOI] [PubMed] [Google Scholar]
- [50].Mao C, Liu X, Zhang Y, et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593(7860):586–90. doi: 10.1038/s41586-021-03539-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kraft VAN, Bezjian CT, Pfeiffer S, et al. GTP cyclohydrolase 1/Tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent Sci. 2020;6(1):41–53. doi: 10.1021/acscentsci.9b01063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Soula M, Weber RA, Zilka O, et al. Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat Chem Biol. 2020;16(12):1351–60. doi: 10.1038/s41589-020-0613-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kuang F, Liu J, Xie Y, et al. MGST1 is a redox-sensitive repressor of ferroptosis in pancreatic cancer cells. Cell Chem Biol. 2021;28(6):765–775.e5. doi: 10.1016/j.chembiol.2021.01.006 [DOI] [PubMed] [Google Scholar]
- [54].Mao C, Liu X, Yan Y, et al. Reply to: DHODH inhibitors sensitize to ferroptosis by FSP1 inhibition. Nature. 2023;619(7968):E19–E23. doi: 10.1038/s41586-023-06270-7 [DOI] [PubMed] [Google Scholar]
- [55].Mishima E, Nakamura T, Zheng J, et al. DHODH inhibitors sensitize to ferroptosis by FSP1 inhibition. Nature. 2023;619(7968):E9–E18. doi: 10.1038/s41586-023-06269-0 [DOI] [PubMed] [Google Scholar]
- [56].Anandhan A, Dodson M, Schmidlin CJ, et al. Breakdown of an ironclad defense system: the critical role of NRF2 in mediating ferroptosis. Cell Chem Biol. 2020;27(4):436–447. doi: 10.1016/j.chembiol.2020.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sun X, Ou Z, Chen R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63(1):173–84. doi: 10.1002/hep.28251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Anandhan A, Dodson M, Shakya A, et al. NRF2 controls iron homeostasis and ferroptosis through HERC2 and VAMP8. Sci Adv. 2023;9(5):eade9585. doi: 10.1126/sciadv.ade9585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lane DJR, Alves F, Ayton S, et al. Striking a NRF2: the rusty and rancid vulnerabilities toward ferroptosis in Alzheimer’s disease. Antioxid Redox Signal. 2023. doi: 10.1089/ars.2023.0318 [DOI] [PubMed] [Google Scholar]
- [60].Wu Y, Lim Y-W, Stroud DA, et al. Caveolae sense oxidative stress through membrane lipid peroxidation and cytosolic release of CAVIN1 to regulate NRF2. Dev Cell. 2023;58(5):376–97. e4. doi: 10.1016/j.devcel.2023.02.004 [DOI] [PubMed] [Google Scholar]
- [61].Barayeu U, Schilling D, Eid M, et al. Hydropersulfides inhibit lipid peroxidation and ferroptosis by scavenging radicals. Nat Chem Biol. 2023;19(1):28–37. doi: 10.1038/s41589-022-01145-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wu Z, Khodade VS, Chauvin JR, et al. Hydropersulfides inhibit lipid peroxidation and protect cells from ferroptosis. J Am Chem Soc. 2022;144(34):15825–15837. doi: 10.1021/jacs.2c06804 [DOI] [PubMed] [Google Scholar]
- [63].Cheff DM, Huang C, Scholzen KC, et al. The ferroptosis inducing compounds RSL3 and ML162 are not direct inhibitors of GPX4 but of TXNRD1. Redox Biol. 2023;62 :102703. doi: 10.1016/j.redox.2023.102703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Debnath J, Gammoh N, Ryan KM. Autophagy and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol. 2023;24(8):1–16. doi: 10.1038/s41580-023-00585-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yamamoto H, Zhang S, Mizushima N. Autophagy genes in biology and disease. Nat Rev Genet. 2023;24(6):382–400. doi: 10.1038/s41576-022-00562-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang L, Klionsky DJ, Shen HM. The emerging mechanisms and functions of microautophagy. Nat Rev Mol Cell Biol. 2023;24(3):186–203. doi: 10.1038/s41580-022-00529-z [DOI] [PubMed] [Google Scholar]
- [67].Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19(6):365–81. doi: 10.1038/s41580-018-0001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Xie Y, Li J, Kang R, et al. Interplay between lipid metabolism and autophagy. Front Cell Dev Biol. 2020;8:431. doi: 10.3389/fcell.2020.00431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi: 10.1016/j.cell.2018.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19(6):349–64. doi: 10.1038/s41580-018-0003-4 [DOI] [PubMed] [Google Scholar]
- [71].Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12(9):814–822. doi: 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rybstein MD, Pedro JM B-S, Kroemer G, et al. The autophagic network and cancer. Nat Cell Biol. 2018;20(3):243–251. doi: 10.1038/s41556-018-0042-2 [DOI] [PubMed] [Google Scholar]
- [73].Weidberg H, Shvets E, Shpilka T, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29(11):1792–802. doi: 10.1038/emboj.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Abdrakhmanov A, Gogvadze V, Zhivotovsky B. To eat or to die: deciphering selective forms of autophagy. Trends Biochem Sci. 2020;45(4):347–64. doi: 10.1016/j.tibs.2019.11.006 [DOI] [PubMed] [Google Scholar]
- [75].Lopez-Otin C, Kroemer G. Hallmarks of health. Cell. 2021;184(7):1929–39. doi: 10.1016/j.cell.2021.03.033 [DOI] [PubMed] [Google Scholar]
- [76].Denton D, Kumar S. Autophagy-dependent cell death. Cell Death Differ. 2019;26(4):605–16. doi: 10.1038/s41418-018-0252-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Liu J, Kuang F, Kroemer G, et al. Autophagy-Dependent Ferroptosis: Machinery and Regulation. Cell Chem Biol. 2020;27(4):420–435. doi: 10.1016/j.chembiol.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Li W, Luo LX, Zhou QQ, et al. Phospholipid peroxidation inhibits autophagy via stimulating the delipidation of oxidized LC3-PE. Redox Biol. 2022;55:102421. doi: 10.1016/j.redox.2022.102421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Vargas JNS, Hamasaki M, Kawabata T, et al. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Biol. 2023;24(3):167–85. doi: 10.1038/s41580-022-00542-2 [DOI] [PubMed] [Google Scholar]
- [80].Sica V, Galluzzi L, Pedro JM B-S, et al. Organelle-specific initiation of autophagy. Molecular Cell. 2015;59(4):522–539. doi: 10.1016/j.molcel.2015.07.021 [DOI] [PubMed] [Google Scholar]
- [81].Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20(3):233–42. doi: 10.1038/s41556-018-0037-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Noda NN, Wang Z, Zhang H. Liquid–liquid phase separation in autophagy. J Cell Bio. 2020;219(8):e202004062. doi: 10.1083/jcb.202004062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Mancias JD, Wang X, Gygi SP, et al. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–9. doi: 10.1038/nature13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hanke N, Rami A. Inhibition of autophagy rescues HT22 hippocampal neurons from erastin-induced ferroptosis. Neural Regen Res. 2023;18(7):1548–52. doi: 10.4103/1673-5374.360246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fuhrmann DC, Mondorf A, Beifuss J, et al. Hypoxia inhibits ferritinophagy, increases mitochondrial ferritin, and protects from ferroptosis. Redox Biol. 2020;36:101670. doi: 10.1016/j.redox.2020.101670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yang H, Sun W, Bi T, et al. The PTBP1‑NCOA4 axis promotes ferroptosis in liver cancer cells. Oncol Rep. 2023;49(2):49. doi: 10.3892/or.2023.8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Li K, Chen B, Xu A, et al. TRIM7 modulates NCOA4-mediated ferritinophagy and ferroptosis in glioblastoma cells. Redox Biol. 2022;56:102451. doi: 10.1016/j.redox.2022.102451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wu H, Liu Q, Shan X, et al. ATM orchestrates ferritinophagy and ferroptosis by phosphorylating NCOA4. Autophagy. 2023;19(7):1–16. doi: 10.1080/15548627.2023.2170960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wu Y, Kram H, Gempt J, et al. ALDH1-Mediated Autophagy Sensitizes Glioblastoma Cells to Ferroptosis. Cells. 2022;11(24):11. doi: 10.3390/cells11244015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhu ZY, Liu YD, Gong Y, et al. Mitochondrial aldehyde dehydrogenase (ALDH2) rescues cardiac contractile dysfunction in an APP/PS1 murine model of Alzheimer’s disease via inhibition of ACSL4-dependent ferroptosis. Acta Pharmacol Sin. 2022;43(1):39–49. doi: 10.1038/s41401-021-00635-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Yusuf RZ, Saez B, Sharda A, et al. Aldehyde dehydrogenase 3a2 protects AML cells from oxidative death and the synthetic lethality of ferroptosis inducers. Blood. 2020;136(11):1303–16. doi: 10.1182/blood.2019001808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zhou L, Yang C, Zhong W, et al. Chrysin induces autophagy-dependent ferroptosis to increase chemosensitivity to gemcitabine by targeting CBR1 in pancreatic cancer cells. Biochem Pharmacol. 2021;193:114813. doi: 10.1016/j.bcp.2021.114813 [DOI] [PubMed] [Google Scholar]
- [93].Ajoolabady A, Aslkhodapasandhokmabad H, Libby P, et al. Ferritinophagy and ferroptosis in the management of metabolic diseases. Trends Endocrinol Metab. 2021;32(7):444–462. doi: 10.1016/j.tem.2021.04.010 [DOI] [PubMed] [Google Scholar]
- [94].Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–5. doi: 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20(3):137–55. doi: 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bai Y, Meng L, Han L, et al. Lipid storage and lipophagy regulates ferroptosis. Biochem Biophys Res Commun. 2019;508(4):997–1003. doi: 10.1016/j.bbrc.2018.12.039 [DOI] [PubMed] [Google Scholar]
- [97].Gnanapradeepan K, Indeglia A, Stieg DC, et al. PLTP is a p53 target gene with roles in cancer growth suppression and ferroptosis. J Biol Chem. 2022;298(12):102637. doi: 10.1016/j.jbc.2022.102637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].You JH, Lee J, Roh JL. PGRMC1-dependent lipophagy promotes ferroptosis in paclitaxel-tolerant persister cancer cells. J Exp Clin Cancer Res. 2021;40(1):350. doi: 10.1186/s13046-021-02168-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Mir SU, Schwarze SR, Jin L, et al. Progesterone receptor membrane component 1/Sigma-2 receptor associates with MAP1LC3B and promotes autophagy. Autophagy. 2013;9(10):1566–78. doi: 10.4161/auto.25889 [DOI] [PubMed] [Google Scholar]
- [100].Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20(9):1013–22. doi: 10.1038/s41556-018-0176-2 [DOI] [PubMed] [Google Scholar]
- [101].Xie Y, Liu J, Kang R, et al. Mitophagy Receptors in Tumor Biology. Front Cell Dev Biol. 2020;8(10):594203. doi: 10.3389/fcell.2020.594203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Gao M, Yi J, Zhu J, et al. Role of mitochondria in Ferroptosis. Mol Cell. 2019;73(2):354–363.e3. doi: 10.1016/j.molcel.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chen X, Kang R, Kroemer G, et al. Organelle-specific regulation of ferroptosis. Cell Death Differ. 2021;28(10):2843–56. doi: 10.1038/s41418-021-00859-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Basit F, van Oppen LM, Schockel L, et al. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. 2017;8(3):e2716. doi: 10.1038/cddis.2017.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Wang X, Ma H, Sun J, et al. Mitochondrial Ferritin Deficiency Promotes Osteoblastic Ferroptosis Via Mitophagy in Type 2 Diabetic Osteoporosis. Biol Trace Elem Res. 2022;200(1):298–307. doi: 10.1007/s12011-021-02627-z [DOI] [PubMed] [Google Scholar]
- [106].Rademaker G, Boumahd Y, Peiffer R, et al. Myoferlin targeting triggers mitophagy and primes ferroptosis in pancreatic cancer cells. Redox Biol. 2022;53:102324. doi: 10.1016/j.redox.2022.102324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Qian B, Jiang RJ, Song JL, et al. Organophosphorus flame retardant TDCPP induces neurotoxicity via mitophagy-related ferroptosis in vivo and in vitro. Chemosphere. 2022;308:136345. doi: 10.1016/j.chemosphere.2022.136345 [DOI] [PubMed] [Google Scholar]
- [108].Chang LC, Chiang SK, Chen SE, et al. Heme oxygenase-1 mediates BAY 11-7085 induced ferroptosis. Cancer Lett. 2018;416:124–137. doi: 10.1016/j.canlet.2017.12.025 [DOI] [PubMed] [Google Scholar]
- [109].Feng Y, Xu J, Shi M, et al. COX7A1 enhances the sensitivity of human NSCLC cells to cystine deprivation-induced ferroptosis via regulating mitochondrial metabolism. Cell Death Dis. 2022;13(11):988. doi: 10.1038/s41419-022-05430-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yu F, Zhang Q, Liu H, et al. Dynamic O-GlcNAcylation coordinates ferritinophagy and mitophagy to activate ferroptosis. Cell Discov. 2022;8(1):40. doi: 10.1038/s41421-022-00390-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Li C, Liu J, Hou W, et al. STING1 promotes ferroptosis through MFN1/2-dependent mitochondrial fusion. Front Cell Dev Biol. 2021;9:698679. doi: 10.3389/fcell.2021.698679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Marcheva B, Ramsey KM, Buhr ED, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–31. doi: 10.1038/nature09253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164–79. doi: 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Yang M, Chen P, Liu J, et al. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci Adv. 2019;5(7):eaaw2238. doi: 10.1126/sciadv.aaw2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Liu Y, Wang Y, Liu J, et al. The circadian clock protects against ferroptosis-induced sterile inflammation. Biochem Biophys Res Commun. 2020;525(3):620–625. doi: 10.1016/j.bbrc.2020.02.142 [DOI] [PubMed] [Google Scholar]
- [116].Wu Z, Geng Y, Lu X, et al. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc Natl Acad Sci U S A. 2019;116(8):2996–3005. doi: 10.1073/pnas.1819728116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273(5274):501–3. doi: 10.1126/science.273.5274.501 [DOI] [PubMed] [Google Scholar]
- [118].Chiang HL, Terlecky SR, Plant CP, et al. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246(4928):382–5. doi: 10.1126/science.2799391 [DOI] [PubMed] [Google Scholar]
- [119].Bandyopadhyay U, Kaushik S, Varticovski L, et al. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28(18):5747–5763. doi: 10.1128/MCB.02070-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Wu K, Yan M, Liu T, et al. Creatine kinase B suppresses ferroptosis by phosphorylating GPX4 through a moonlighting function. Nat Cell Biol. 2023;25(5):714–25. doi: 10.1038/s41556-023-01133-9 [DOI] [PubMed] [Google Scholar]
- [121].Yu S, Li Z, Zhang Q, et al. GPX4 degradation via chaperone-mediated autophagy contributes to antimony-triggered neuronal ferroptosis. Ecotoxicol Environ Saf. 2022;234:113413. doi: 10.1016/j.ecoenv.2022.113413 [DOI] [PubMed] [Google Scholar]
- [122].Chen C, Wang D, Yu Y, et al. Legumain promotes tubular ferroptosis by facilitating chaperone-mediated autophagy of GPX4 in AKI. Cell Death Dis. 2021;12(1):65. doi: 10.1038/s41419-020-03362-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zhou J, Tan Y, Hu L, et al. Inhibition of HSPA8 by rifampicin contributes to ferroptosis via enhancing autophagy. Liver Int. 2022;42(12):2889–2899. doi: 10.1111/liv.15459 [DOI] [PubMed] [Google Scholar]
- [124].Shimada K, Skouta R, Kaplan A, et al. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat Chem Biol. 2016;12(7):497–503. doi: 10.1038/nchembio.2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Sun Y, Berleth N, Wu W, et al. Fin56-induced ferroptosis is supported by autophagy-mediated GPX4 degradation and functions synergistically with mTOR inhibition to kill bladder cancer cells. Cell Death Dis. 2021;12(11):1028. doi: 10.1038/s41419-021-04306-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Thayyullathil F, Cheratta AR, Alakkal A, et al. Acid sphingomyelinase-dependent autophagic degradation of GPX4 is critical for the execution of ferroptosis. Cell Death Dis. 2021;12(1):26. doi: 10.1038/s41419-020-03297-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Liu Y, Wang Y, Liu J, et al. Interplay between MTOR and GPX4 signaling modulates autophagy-dependent ferroptotic cancer cell death. Cancer Gene Ther. 2021;28(1–2):55–63. doi: 10.1038/s41417-020-0182-y [DOI] [PubMed] [Google Scholar]
- [128].Lei G, Zhuang L, Gan B. mTORC1 and ferroptosis: regulatory mechanisms and therapeutic potential. BioEssays. 2021;43(8):e2100093. doi: 10.1002/bies.202100093 [DOI] [PubMed] [Google Scholar]
- [129].Xue Q, Yan D, Chen X, et al. Copper-dependent autophagic degradation of GPX4 drives ferroptosis. Autophagy. 2023;19(7):1–15. doi: 10.1080/15548627.2023.2165323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Li J, Liu J, Zhou Z. et al, et al (2023). Tumor-specific GPX4 degradation enhances ferroptosis-initiated antitumor immune response in mouse models of pancreatic cancer. Sci. Transl. Med., 15((720) eadg3049.), 10.1126/scitranslmed.adg3049 [DOI] [PubMed] [Google Scholar]
- [131].Ichimura Y, Waguri S, Sou YS, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51(5):618–631. doi: 10.1016/j.molcel.2013.08.003 [DOI] [PubMed] [Google Scholar]
- [132].Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–23. doi: 10.1038/ncb2021 [DOI] [PubMed] [Google Scholar]
- [133].Jain A, Lamark T, Sjottem E, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285(29):22576–22591. doi: 10.1074/jbc.M110.118976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Yang L, Ye F, Liu J, et al. Extracellular SQSTM1 exacerbates acute pancreatitis by activating autophagy-dependent ferroptosis. Autophagy. 2022;19(6):1–12. doi: 10.1080/15548627.2022.2152209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Tang Z, Jiang W, Mao M, et al. Deubiquitinase USP35 modulates ferroptosis in lung cancer via targeting ferroportin. Clin Transl Med. 2021;11(4):e390. doi: 10.1002/ctm2.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Jiang L, Wang J, Wang K, et al. RNF217 regulates iron homeostasis through its E3 ubiquitin ligase activity by modulating ferroportin degradation. Blood. 2021;138(8):689–705. doi: 10.1182/blood.2020008986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Qiao B, Sugianto P, Fung E, et al. Hepcidin-induced endocytosis of ferroportin is dependent on ferroportin ubiquitination. Cell Metab. 2012;15(6):918–924. doi: 10.1016/j.cmet.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Geng N, Shi BJ, Li SL, et al. Knockdown of ferroportin accelerates erastin-induced ferroptosis in neuroblastoma cells. Eur Rev Med Pharmacol Sci. 2018;22(12):3826–36. doi: 10.26355/eurrev_201806_15267 [DOI] [PubMed] [Google Scholar]
- [139].Wei D, Ke YQ, Duan P, et al. MicroRNA-302a-3p induces ferroptosis of non-small cell lung cancer cells via targeting ferroportin. Vol. 55. Free Radic Res; 2021. pp. 821–830. [DOI] [PubMed] [Google Scholar]
- [140].Bao WD, Pang P, Zhou XT, et al. Loss of ferroportin induces memory impairment by promoting ferroptosis in Alzheimer’s disease. Cell Death Differ. 2021;28(5):1548–62. doi: 10.1038/s41418-020-00685-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Fang J, Kong B, Shuai W, et al. Ferroportin-mediated ferroptosis involved in new-onset atrial fibrillation with LPS-induced endotoxemia. Eur J Pharmacol. 2021;913:174622. doi: 10.1016/j.ejphar.2021.174622 [DOI] [PubMed] [Google Scholar]
- [142].Li L, Wang K, Jia R, et al. Ferroportin-dependent ferroptosis induced by ellagic acid retards liver fibrosis by impairing the SNARE complexes formation. Redox Biol. 2022;56:102435. doi: 10.1016/j.redox.2022.102435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Lu S, Song Y, Luo R, et al. Ferroportin-dependent iron homeostasis protects against oxidative stress-induced nucleus pulposus cell ferroptosis and ameliorates intervertebral disc degeneration in vivo. Oxid Med Cell Longev. 2021;2021:6670497. doi: 10.1155/2021/6670497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Li J, Liu J, Xu Y, et al. Tumor heterogeneity in autophagy-dependent ferroptosis. Autophagy. 2021;17(11):3361–74. doi: 10.1080/15548627.2021.1872241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Braunewell KH, Klein-Szanto AJ. Visinin-like proteins (VSNLs): interaction partners and emerging functions in signal transduction of a subfamily of neuronal Ca2+ -sensor proteins. Cell Tissue Res. 2009;335(2):301–316. doi: 10.1007/s00441-008-0716-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Wang X, Xie X, Zhang Y, et al. Hippocalcin-like 1 is a key regulator of LDHA activation that promotes the growth of non-small cell lung carcinoma. Cell Oncol. 2022;45(1):179–191. doi: 10.1007/s13402-022-00661-0 [DOI] [PubMed] [Google Scholar]
- [147].Zhang D, Liu X, Xu X, et al. HPCAL 1 promotes glioblastoma proliferation via activation of Wnt/β-catenin signalling pathway. J Cell Mol Med. 2019;23(5):3108–3117. doi: 10.1111/jcmm.14083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Wang W, Zhong Q, Teng L, et al. Mutations that disrupt PHOXB interaction with the neuronal calcium sensor HPCAL1 impede cellular differentiation in neuroblastoma. Oncogene. 2014;33(25):3316–24. doi: 10.1038/onc.2013.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Chen X, Song X, Li J, et al. Identification of HPCAL1 as a specific autophagy receptor involved in ferroptosis. Autophagy. 2022:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Ma Y, Yi M, Wang W, et al. Oxidative degradation of dihydrofolate reductase increases CD38-mediated ferroptosis susceptibility. Cell Death Dis. 2022;13(11):944. doi: 10.1038/s41419-022-05383-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Kihara A, Kabeya Y, Ohsumi Y, et al. Beclin–phosphatidylinositol 3-kinase complex functions at the trans -golgi network. EMBO Rep. 2001;2(4):330–335. doi: 10.1093/embo-reports/kve061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Park E, Chung SW. ROS-mediated autophagy increases intracellular iron levels and ferroptosis by ferritin and transferrin receptor regulation. Cell Death Dis. 2019;10(11):822. doi: 10.1038/s41419-019-2064-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Shen M, Li Y, Wang Y, et al. N(6)-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 2021;47:102151. doi: 10.1016/j.redox.2021.102151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Chen HY, Xiao ZZ, Ling X, et al. ELAVL1 is transcriptionally activated by FOXC1 and promotes ferroptosis in myocardial ischemia/reperfusion injury by regulating autophagy. Mol Med. 2021;27(1):14. doi: 10.1186/s10020-021-00271-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Sui M, Xu D, Zhao W, et al. CIRBP promotes ferroptosis by interacting with ELAVL1 and activating ferritinophagy during renal ischaemia-reperfusion injury. J Cell Mol Med. 2021;25(13):6203–6216. doi: 10.1111/jcmm.16567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Lee J, You JH, Roh JL. Poly(rC)-binding protein 1 represses ferritinophagy-mediated ferroptosis in head and neck cancer. Redox Biol. 2022;51:102276. doi: 10.1016/j.redox.2022.102276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Zhang Z, Guo M, Li Y, et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2020;16(8):1482–505. doi: 10.1080/15548627.2019.1687985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Rong Y, Fan J, Ji C, et al. USP11 regulates autophagy-dependent ferroptosis after spinal cord ischemia-reperfusion injury by deubiquitinating beclin 1. Cell Death Differ. 2022;29(6):1164–75. doi: 10.1038/s41418-021-00907-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Huang P, Zhao H, Pan X, et al. SIRT3-mediated autophagy contributes to ferroptosis-induced anticancer by inducing the formation of BECN1-SLC7A11 complex. Biochem Pharmacol. 2023;213:115592. doi: 10.1016/j.bcp.2023.115592 [DOI] [PubMed] [Google Scholar]
- [160].Wang J, Zhou JY, Kho D, et al. Role for DUSP1 (dual-specificity protein phosphatase 1) in the regulation of autophagy. Autophagy. 2016;12(10):1791–803. doi: 10.1080/15548627.2016.1203483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Xie Y, Kuang F, Liu J, et al. DUSP1 blocks autophagy-dependent ferroptosis in pancreatic cancer. Journal Of Pancreatology. 2020;3(3):154–60. doi: 10.1097/JP9.0000000000000054 [DOI] [Google Scholar]
- [162].Tan Y, Huang Y, Mei R, et al. HucMSC-derived exosomes delivered BECN1 induces ferroptosis of hepatic stellate cells via regulating the xCT/GPX4 axis. Cell Death Dis. 2022;13(4):319. doi: 10.1038/s41419-022-04764-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Wang BJ, Her GM, Hu MK, et al. ErbB2 regulates autophagic flux to modulate the proteostasis of APP-CTFs in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2017;114(15):E3129–E38. doi: 10.1073/pnas.1618804114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Wang C, Zeng J, Li LJ, et al. Cdc25A inhibits autophagy-mediated ferroptosis by upregulating ErbB2 through PKM2 dephosphorylation in cervical cancer cells. Cell Death Dis. 2021;12(11):1055. doi: 10.1038/s41419-021-04342-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Wendler WM, Kremmer E, Forster R, et al. Identification of pirin, a novel highly conserved nuclear protein. J Biol Chem. 1997;272(13):8482–8489. doi: 10.1074/jbc.272.13.8482 [DOI] [PubMed] [Google Scholar]
- [166].Hu N, Bai L, Dai E, et al. Pirin is a nuclear redox-sensitive modulator of autophagy-dependent ferroptosis. Biochem Biophys Res Commun. 2021;536:100–106. doi: 10.1016/j.bbrc.2020.12.066 [DOI] [PubMed] [Google Scholar]
- [167].Wen Q, Liu J, Kang R, et al. The release and activity of HMGB1 in ferroptosis. Biochem Biophys Res Commun. 2019;510(2):278–283. doi: 10.1016/j.bbrc.2019.01.090 [DOI] [PubMed] [Google Scholar]
- [168].Ye F, Chai W, Xie M, et al. HMGB1 regulates erastin-induced ferroptosis via RAS-JNK/p38 signaling in HL-60/NRASQ61L cells. Am J Cancer Res. 2019;9(4):730–739. [PMC free article] [PubMed] [Google Scholar]
- [169].Wu Y, Zhao Y, Yang HZ, et al. HMGB1 regulates ferroptosis through Nrf2 pathway in mesangial cells in response to high glucose. Biosci Rep. 2021;41(2):41. doi: 10.1042/BSR20202924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Tang D, Kang R, Livesey KM, et al. Endogenous HMGB1 regulates autophagy. J Cell Bio. 2010;190(5):881–892. doi: 10.1083/jcb.200911078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Gui X, Yang H, Li T, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567(7747):262–6. doi: 10.1038/s41586-019-1006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Li C, Zhang Y, Liu J, et al. Mitochondrial DNA stress triggers autophagy-dependent ferroptotic death. Autophagy. 2021;17(4):948–60. doi: 10.1080/15548627.2020.1739447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Wu J, Liu Q, Zhang X, et al. The interaction between STING and NCOA4 exacerbates lethal sepsis by orchestrating ferroptosis and inflammatory responses in macrophages. Cell Death Dis. 2022;13(7):653. doi: 10.1038/s41419-022-05115-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Qiu S, Zhong X, Meng X, et al. Mitochondria-localized cGAS suppresses ferroptosis to promote cancer progression. Cell Res. 2023;33(4):299–311. doi: 10.1038/s41422-023-00788-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Imai K, Hao F, Fujita N, et al. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J Cell Sci. 2016;129:3781–3791. doi: 10.1242/jcs.196196 [DOI] [PubMed] [Google Scholar]
- [176].Liu J, Liu Y, Wang Y, et al. TMEM164 is a new determinant of autophagy-dependent ferroptosis. Autophagy. 2023;19(3):945–56. doi: 10.1080/15548627.2022.2111635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Reed A, Ware T, Li H, et al. TMEM164 is an acyltransferase that forms ferroptotic C20: 4 ether phospholipids. Nat Chem Biol. 2023;19(3):378–88. doi: 10.1038/s41589-022-01253-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Bernales S, KL M, Walter P, et al. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4(12):e423. doi: 10.1371/journal.pbio.0040423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Khaminets A, Heinrich T, Mari M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522(7556):354–8. doi: 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- [180].Liu Z, Ma C, Wang Q, et al. Targeting FAM134B-mediated reticulophagy activates sorafenib-induced ferroptosis in hepatocellular carcinoma. Biochem Biophys Res Commun. 2022;589:247–253. doi: 10.1016/j.bbrc.2021.12.019 [DOI] [PubMed] [Google Scholar]
- [181].Chae CW, Yoon JH, Lim JR, et al. TRIM16-mediated lysophagy suppresses high-glucose-accumulated neuronal Aβ. Autophagy. 2023;19(10):1–17. doi: 10.1080/15548627.2023.2229659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Li S, Liao Z, Yin H, et al. G3BP1 coordinates lysophagy activity to protect against compression-induced cell ferroptosis during intervertebral disc degeneration. Cell Prolif. 2022;e13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Liu C, Sun W, Zhu T, et al. Glia maturation factor-beta induces ferroptosis by impairing chaperone-mediated autophagic degradation of ACSL4 in early diabetic retinopathy. Redox Biol. 2022;52:102292. doi: 10.1016/j.redox.2022.102292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Torii S, Shintoku R, Kubota C, et al. An essential role for functional lysosomes in ferroptosis of cancer cells. Biochem J. 2016;473(6):769–777. doi: 10.1042/BJ20150658 [DOI] [PubMed] [Google Scholar]
- [185].Zhang X, Guo Y, Li H, et al. FIN56, a novel ferroptosis inducer, triggers lysosomal membrane permeabilization in a TFEB-dependent manner in glioblastoma. J Cancer. 2021;12(22):6610–9. doi: 10.7150/jca.58500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Wang ZX, Ma J, Li XY, et al. Quercetin induces p53-independent cancer cell death through lysosome activation by the transcription factor EB and reactive oxygen species-dependent ferroptosis. Br J Pharmacol. 2021;178(5):1133–1148. doi: 10.1111/bph.15350 [DOI] [PubMed] [Google Scholar]
- [187].An S, Hu M. Quercetin promotes TFEB nuclear translocation and activates Lysosomal degradation of Ferritin to induce ferroptosis in breast cancer cells. Comput Intell Neurosci. 2022;2022:5299218. doi: 10.1155/2022/5299218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Nagakannan P, Islam MI, Conrad M, et al. Cathepsin B is an executioner of ferroptosis. Biochim Biophys Acta, Mol Cell Res. 2021;1868(3):118928. doi: 10.1016/j.bbamcr.2020.118928 [DOI] [PubMed] [Google Scholar]
- [189].Kuang F, Liu J, Li C, et al. Cathepsin B is a mediator of organelle-specific initiation of ferroptosis. Biochem Biophys Res Commun. 2020;533(4):1464–1469. doi: 10.1016/j.bbrc.2020.10.035 [DOI] [PubMed] [Google Scholar]
- [190].Gao H, Bai Y, Jia Y, et al. Ferroptosis is a lysosomal cell death process. Biochem Biophys Res Commun. 2018;503(3):1550–1556. doi: 10.1016/j.bbrc.2018.07.078 [DOI] [PubMed] [Google Scholar]
- [191].Jiang L, Zheng H, Lyu Q, et al. Lysosomal nitric oxide determines transition from autophagy to ferroptosis after exposure to plasma-activated Ringer’s lactate. Redox Biol. 2021;43:101989. doi: 10.1016/j.redox.2021.101989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Chen T, Shi R, Suo Q, et al. Progranulin released from microglial lysosomes reduces neuronal ferroptosis after cerebral ischemia in mice. J Cereb Blood Flow Metab. 2023;43(4):505–517. doi: 10.1177/0271678X221145090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Lee JJ, Ishihara K, Notomi S, et al. Lysosome-associated membrane protein-2 deficiency increases the risk of reactive oxygen species-induced ferroptosis in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2020;521(2):414–419. doi: 10.1016/j.bbrc.2019.10.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Armenta DA, Laqtom NN, Alchemy G, et al. Ferroptosis inhibition by lysosome-dependent catabolism of extracellular protein. Cell Chem Biol. 2022;29(11):1588–600 e7. doi: 10.1016/j.chembiol.2022.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Ingold I, Berndt C, Schmitt S, et al. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell. 2018;172(3):409–22. e21. doi: 10.1016/j.cell.2017.11.048 [DOI] [PubMed] [Google Scholar]
- [196].Li Z, Ferguson L, Deol KK, et al. Ribosome stalling during selenoprotein translation exposes a ferroptosis vulnerability. Nat Chem Biol. 2022;18(7):751–61. doi: 10.1038/s41589-022-01033-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Dai E, Han L, Liu J, et al. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy. 2020;16(11):2069–83. doi: 10.1080/15548627.2020.1714209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Ito F, Kato K, Yanatori I, et al. Ferroptosis-dependent extracellular vesicles from macrophage contribute to asbestos-induced mesothelial carcinogenesis through loading ferritin. Redox Biol. 2021;47:102174. doi: 10.1016/j.redox.2021.102174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Chen F, Cai X, Kang R, et al. Autophagy-dependent ferroptosis in cancer. Antioxid Redox Signaling. 2023;39(1–3):79–101. doi: 10.1089/ars.2022.0202 [DOI] [PubMed] [Google Scholar]
- [200].Liu K, Huang J, Liu J, et al. Induction of autophagy-dependent ferroptosis to eliminate drug-tolerant human retinoblastoma cells. Cell Death Dis. 2022;13(6):521. doi: 10.1038/s41419-022-04974-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Tsai Y, Xia C, Sun Z. The Inhibitory Effect of 6-Gingerol on Ubiquitin-Specific Peptidase 14 Enhances Autophagy-Dependent Ferroptosis and Anti-Tumor in vivo and in vitro. Front Pharmacol. 2020;11:598555. doi: 10.3389/fphar.2020.598555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Li J, Yuan J, Li Y, et al. D-Borneol enhances cisplatin sensitivity via autophagy dependent EMT signaling and NCOA4-mediated ferritinophagy. Phytomedicine. 2022;106:154411. doi: 10.1016/j.phymed.2022.154411 [DOI] [PubMed] [Google Scholar]
- [203].Guo Z, Zhang Y. RETRACTED: Allicin promotes autophagy and ferroptosis in esophageal squamous cell carcinoma by activating AMPK/mTOR signaling. Heliyon. 2022;8(10):e11005. doi: 10.1016/j.heliyon.2022.e11005 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [204].Chen Y, Li N, Wang H, et al. Amentoflavone suppresses cell proliferation and induces cell death through triggering autophagy-dependent ferroptosis in human glioma. Life Sci. 2020;247:117425. doi: 10.1016/j.lfs.2020.117425 [DOI] [PubMed] [Google Scholar]
- [205].Du Y, Bao J, Zhang MJ, et al. Targeting ferroptosis contributes to ATPR-induced AML differentiation via ROS-autophagy-lysosomal pathway. Gene. 2020;755:144889. doi: 10.1016/j.gene.2020.144889 [DOI] [PubMed] [Google Scholar]
- [206].Tang X, Ding H, Liang M, et al. Curcumin induces ferroptosis in non-small-cell lung cancer via activating autophagy. Thorac Cancer. 2021;12(8):1219–1230. doi: 10.1111/1759-7714.13904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Du J, Wang T, Li Y, et al. DHA inhibits proliferation and induces ferroptosis of leukemia cells through autophagy dependent degradation of ferritin. Free Radic Biol Med. 2019;131:356–369. doi: 10.1016/j.freeradbiomed.2018.12.011 [DOI] [PubMed] [Google Scholar]
- [208].Gryzik M, Asperti M, Denardo A, et al. NCOA4-mediated ferritinophagy promotes ferroptosis induced by erastin, but not by RSL3 in HeLa cells. Biochim Biophys Acta, Mol Cell Res. 2021;1868(2):118913. doi: 10.1016/j.bbamcr.2020.118913 [DOI] [PubMed] [Google Scholar]
- [209].Lin PL, Tang HH, Wu SY, et al. Saponin Formosanin C-induced Ferritinophagy and ferroptosis in human hepatocellular carcinoma cells. Antioxidants. 2020;9(8):682. doi: 10.3390/antiox9080682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Zhong Y, Tian F, Ma H, et al. FTY720 induces ferroptosis and autophagy via PP2A/AMPK pathway in multiple myeloma cells. Life Sci. 2020;260:118077. doi: 10.1016/j.lfs.2020.118077 [DOI] [PubMed] [Google Scholar]
- [211].Qu C, Dai E, Lai T, et al. Itaconic acid induces ferroptosis by activating ferritinophagy. Biochem Biophys Res Commun. 2021;583:56–62. doi: 10.1016/j.bbrc.2021.10.054 [DOI] [PubMed] [Google Scholar]
- [212].Sui S, Zhang J, Xu S, et al. Ferritinophagy is required for the induction of ferroptosis by the bromodomain protein BRD4 inhibitor (+)-JQ1 in cancer cells. Cell Death Dis. 2019;10(5):331. doi: 10.1038/s41419-019-1564-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Li J, Lama R, Galster SL, et al. Small-Molecule MMRi62 induces Ferroptosis and inhibits metastasis in pancreatic cancer via degradation of ferritin heavy chain and mutant p53. Mol Cancer Ther. 2022;21(4):535–545. doi: 10.1158/1535-7163.MCT-21-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Kan X, Yin Y, Song C, et al. Newcastle-disease-virus-induced ferroptosis through nutrient deprivation and ferritinophagy in tumor cells. iScience. 2021;24(8):102837. doi: 10.1016/j.isci.2021.102837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Li Z, Wang C, Dai C, et al. Engineering dual catalytic nanomedicine for autophagy-augmented and ferroptosis-involved cancer nanotherapy. Biomaterials. 2022;287:121668. doi: 10.1016/j.biomaterials.2022.121668 [DOI] [PubMed] [Google Scholar]
- [216].Zuo T, Fang T, Zhang J, et al. pH-sensitive molecular-switch-containing polymer nanoparticle for breast cancer therapy with ferritinophagy-cascade ferroptosis and tumor immune activation. Adv Healthc Mater. 2021;10(21):e2100683. doi: 10.1002/adhm.202100683 [DOI] [PubMed] [Google Scholar]
- [217].An P, Gao Z, Sun K, et al. Photothermal-enhanced inactivation of glutathione peroxidase for ferroptosis sensitized by an autophagy promotor. ACS Appl Mater Interfaces. 2019;11(46):42988–42997. doi: 10.1021/acsami.9b16124 [DOI] [PubMed] [Google Scholar]
- [218].Chen H, Wen J. Iron oxide nanoparticles loaded with paclitaxel inhibits glioblastoma by enhancing autophagy-dependent ferroptosis pathway. Eur J Pharmacol. 2022;921:174860. doi: 10.1016/j.ejphar.2022.174860 [DOI] [PubMed] [Google Scholar]
- [219].Li Y, Wang X, Yan J, et al. Nanoparticle ferritin-bound erastin and rapamycin: a nanodrug combining autophagy and ferroptosis for anticancer therapy. Biomater Sci. 2019;7(9):3779–3787. doi: 10.1039/C9BM00653B [DOI] [PubMed] [Google Scholar]
- [220].Hu R, Dai C, Dai X, et al. Topology regulation of nanomedicine for autophagy-augmented ferroptosis and cancer immunotherapy. Sci Bull (Beijing). 2023;68(1):77–94. doi: 10.1016/j.scib.2022.12.030 [DOI] [PubMed] [Google Scholar]
- [221].Yang J, Ding L, Yu L, et al. Nanomedicine enables autophagy-enhanced cancer-cell ferroptosis. Sci Bull. 2021;66(5):464–77. doi: 10.1016/j.scib.2020.10.021 [DOI] [PubMed] [Google Scholar]
- [222].Li W, Fu H, Fang L, et al. Shikonin induces ferroptosis in multiple myeloma via GOT1-mediated ferritinophagy. Front Oncol. 2022;12:1025067. doi: 10.3389/fonc.2022.1025067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Zhu HY, Huang ZX, Chen GQ, et al. Typhaneoside prevents acute myeloid leukemia (AML) through suppressing proliferation and inducing ferroptosis associated with autophagy. Biochem Biophys Res Commun. 2019;516(4):1265–1271. doi: 10.1016/j.bbrc.2019.06.070 [DOI] [PubMed] [Google Scholar]
- [224].Wang X, Xu S, Zhang L, et al. Vitamin C induces ferroptosis in anaplastic thyroid cancer cells by ferritinophagy activation. Biochem Biophys Res Commun. 2021;551:46–53. doi: 10.1016/j.bbrc.2021.02.126 [DOI] [PubMed] [Google Scholar]
- [225].Zhang Y, Kong Y, Ma Y, et al. Loss of COPZ1 induces NCOA4 mediated autophagy and ferroptosis in glioblastoma cell lines. Oncogene. 2021;40(8):1425–39. doi: 10.1038/s41388-020-01622-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Dong C, Dong R, Song J, et al. Knockdown of lncRNA EGFR-AS1 promotes autophagy-mediated ferroptosis in cervical cancer via regulating EGFR expression through miR-133b. Mol Cell Toxicol. 2023. [Google Scholar]
- [227].Viswanathan VS, Ryan MJ, Dhruv HD, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547(7664):453–7. doi: 10.1038/nature23007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Liu J, Kang R and Tang D. (2024). S2405–8033. Adverse effects of ferroptotic therapy: mechanisms and management. Trends in Cancer, 10.1016/j.trecan.2024.01.002 [DOI] [PubMed] [Google Scholar]
- [229].Jian B, Pang J, Xiong H, et al. Autophagy-dependent ferroptosis contributes to cisplatin-induced hearing loss. Toxicol Lett. 2021;350:249–260. doi: 10.1016/j.toxlet.2021.07.010 [DOI] [PubMed] [Google Scholar]
- [230].Zhu H, Zhong Y, Chen R, et al. ATG5 knockdown attenuates ischemia‒reperfusion injury by reducing excessive autophagy-induced ferroptosis. Transl Stroke Res. 2022;15(1):153–164. doi: 10.1007/s12975-022-01118-0 [DOI] [PubMed] [Google Scholar]
- [231].Tang S, Wang Y, Ma T, et al. MiR-30d inhibits cardiomyocytes autophagy promoting ferroptosis after myocardial infarction. Panminerva Med. 2020. doi: 10.23736/S0031-0808.20.03979-8. [DOI] [PubMed] [Google Scholar]
- [232].Huang Q, Tian L, Zhao X, et al. Rev-erbs agonist SR9009 alleviates ischemia-reperfusion injury by heightening endogenous cardioprotection at onset of type-2 diabetes in rats: down-regulating ferritinophagy/ferroptosis signaling. Biomed Pharmacother. 2022;154:113595. doi: 10.1016/j.biopha.2022.113595 [DOI] [PubMed] [Google Scholar]
- [233].Li W, Li W, Wang Y, et al. Inhibition of DNMT-1 alleviates ferroptosis through NCOA4 mediated ferritinophagy during diabetes myocardial ischemia/reperfusion injury. Cell Death Discov. 2021;7(1):267. doi: 10.1038/s41420-021-00656-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Li T, Tan Y, Ouyang S, et al. Resveratrol protects against myocardial ischemia-reperfusion injury via attenuating ferroptosis. Gene. 2022;808:145968. doi: 10.1016/j.gene.2021.145968 [DOI] [PubMed] [Google Scholar]
- [235].Belaidi AA, Masaldan S, Southon A, et al. Apolipoprotein E potently inhibits ferroptosis by blocking ferritinophagy. Mol Psychiatry. 2022. doi: 10.1038/s41380-022-01568-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Ayton S, Faux NG, Bush AI. Ferritin levels in the cerebrospinal fluid predict Alzheimer’s disease outcomes and are regulated by APOE. Nat Commun. 2015;6(1):6760. doi: 10.1038/ncomms7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Tian Y, Lu J, Hao X, et al. FTH1 inhibits ferroptosis through ferritinophagy in the 6-OHDA model of Parkinson’s disease. Neurotherapeutics. 2020;17:1796–812. doi: 10.1007/s13311-020-00929-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Zuo Y, Xie J, Li X, et al. Ferritinophagy-mediated ferroptosis involved in paraquat-induced neurotoxicity of dopaminergic neurons: implication for neurotoxicity in PD. Oxid Med Cell Longev. 2021;2021:9961628. doi: 10.1155/2021/9961628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Tao Q, Qiu X, Li C, et al. S100A8 regulates autophagy-dependent ferroptosis in microglia after experimental subarachnoid hemorrhage. Exp Neurol. 2022;357:114171. doi: 10.1016/j.expneurol.2022.114171 [DOI] [PubMed] [Google Scholar]
- [240].Yin K, Wang D, Zhao H, et al. Polystyrene microplastics up-regulates liver glutamine and glutamate synthesis and promotes autophagy-dependent ferroptosis and apoptosis in the cerebellum through the liver-brain axis. Environ Pollut. 2022;307:119449. doi: 10.1016/j.envpol.2022.119449 [DOI] [PubMed] [Google Scholar]
- [241].Zhang Z, Yao Z, Wang L, et al. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14(12):2083–103. doi: 10.1080/15548627.2018.1503146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Kong Z, Liu R, Cheng Y. Artesunate alleviates liver fibrosis by regulating ferroptosis signaling pathway. Biomed Pharmacother. 2019;109:2043–2053. doi: 10.1016/j.biopha.2018.11.030 [DOI] [PubMed] [Google Scholar]
- [243].Zhao Y, Lu J, Mao A, et al. Autophagy inhibition plays a protective role in ferroptosis induced by alcohol via the p62–Keap1–Nrf2 pathway. J Agric Food Chem. 2021;69(33):9671–9683. doi: 10.1021/acs.jafc.1c03751 [DOI] [PubMed] [Google Scholar]
- [244].Liu JP, Cen SY, Xue Z, et al. A class of disulfide compounds suppresses ferroptosis by stabilizing GPX4. ACS Chem Biol. 2022;17(12):3389–3406. doi: 10.1021/acschembio.2c00445 [DOI] [PubMed] [Google Scholar]
- [245].Dai E, Han L, Liu J, et al. Ferroptotic damage promotes pancreatic tumorigenesis through a TMEM173/STING-dependent DNA sensor pathway. Nat Commun. 2020;11(1):6339. doi: 10.1038/s41467-020-20154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Liu K, Liu J, Zou B, et al. Trypsin-mediated sensitization to ferroptosis increases the severity of pancreatitis in mice. Cell Mol Gastroenterol Hepatol. 2022;13(2):483–500. doi: 10.1016/j.jcmgh.2021.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Bravo-San Pedro JM, Kroemer G, Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ Res. 2017;120(11):1812–1824. doi: 10.1161/CIRCRESAHA.117.311082 [DOI] [PubMed] [Google Scholar]
- [248].Chen X, Xu S, Zhao C, et al. Role of TLR4/NADPH oxidase 4 pathway in promoting cell death through autophagy and ferroptosis during heart failure. Biochem Biophys Res Commun. 2019;516(1):37–43. doi: 10.1016/j.bbrc.2019.06.015 [DOI] [PubMed] [Google Scholar]
- [249].Li N, Wang W, Zhou H, et al. Ferritinophagy-mediated ferroptosis is involved in sepsis-induced cardiac injury. Free Radic Biol Med. 2020;160:303–318. doi: 10.1016/j.freeradbiomed.2020.08.009 [DOI] [PubMed] [Google Scholar]
- [250].Zhao C, Yu D, He Z, et al. Endoplasmic reticulum stress-mediated autophagy activation is involved in cadmium-induced ferroptosis of renal tubular epithelial cells. Free Radic Biol Med. 2021;175:236–248. doi: 10.1016/j.freeradbiomed.2021.09.008 [DOI] [PubMed] [Google Scholar]
- [251].Bao L, Zhao C, Feng L, et al. Ferritinophagy is involved in bisphenol A-induced ferroptosis of renal tubular epithelial cells through the activation of the AMPK-mTOR-ULK1 pathway. Food Chem Toxicol. 2022;163:112909. doi: 10.1016/j.fct.2022.112909 [DOI] [PubMed] [Google Scholar]
- [252].Hou Y, Wang S, Jiang L, et al. Patulin induces acute kidney injury in mice through autophagy–ferroptosis pathway. J Agric Food Chem. 2022;70(20):6213–6223. doi: 10.1021/acs.jafc.1c08349 [DOI] [PubMed] [Google Scholar]
- [253].Tang Y, Luo H, Xiao Q, et al. Isoliquiritigenin attenuates septic acute kidney injury by regulating ferritinophagy-mediated ferroptosis. Ren Fail. 2021;43(1):1551–1560. doi: 10.1080/0886022X.2021.2003208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Linkermann A, Skouta R, Himmerkus N, et al. Synchronized renal tubular cell death involves ferroptosis. Proc Natl Acad Sci U S A. 2014;111(47):16836–16841. doi: 10.1073/pnas.1415518111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Zeng Z, Huang H, Zhang J, et al. HDM induce airway epithelial cell ferroptosis and promote inflammation by activating ferritinophagy in asthma. FASEB J. 2022;36(6):e22359. doi: 10.1096/fj.202101977RR [DOI] [PubMed] [Google Scholar]
- [256].Yoshida M, Minagawa S, Araya J, et al. Involvement of cigarette smoke-induced epithelial cell ferroptosis in COPD pathogenesis. Nat Commun. 2019;10(1):3145. doi: 10.1038/s41467-019-10991-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Li L, Wu D, Deng S, et al. NVP-AUY922 alleviates radiation-induced lung injury via inhibition of autophagy-dependent ferroptosis. Cell Death Discov. 2022;8(1):86. doi: 10.1038/s41420-022-00887-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Zhang J, Zheng Y, Wang Y, et al. YAP1 alleviates sepsis-induced acute lung injury via inhibiting ferritinophagy-mediated ferroptosis. Front Immunol. 2022;13:884362. doi: 10.3389/fimmu.2022.884362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Yao F, Peng J, Zhang E, et al. Pathologically high intraocular pressure disturbs normal iron homeostasis and leads to retinal ganglion cell ferroptosis in glaucoma. Cell Death Differ. 2022;30(1):69–81. doi: 10.1038/s41418-022-01046-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Singh LP, Yumnamcha T, S Devi DT. Mitophagy, Ferritinophagy and ferroptosis in retinal pigment epithelial cells under high glucose conditions: implications for diabetic retinopathy and age-related retinal diseases. JOJ Ophthalmol. 2021;8(5):77–85. doi: 10.19080/JOJO.2021.08.555748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Wang J, Zibetti C, Shang P, et al. ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nat Commun. 2018;9(1):1364. doi: 10.1038/s41467-018-03856-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Whitcup SM, Sodhi A, Atkinson JP, et al. The role of the immune response in age-related macular degeneration. Int J Inflamm. 2013;2013:1–10. doi: 10.1155/2013/348092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Wong WL, Su X, Li X, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2(2):e106–e116. doi: 10.1016/S2214-109X(13)70145-1 [DOI] [PubMed] [Google Scholar]
- [264].Gupta U, Ghosh S, Wallace CT, et al. Increased LCN2 (lipocalin 2) in the RPE decreases autophagy and activates inflammasome-ferroptosis processes in a mouse model of dry AMD. Autophagy. 2023;19(1):92–111. doi: 10.1080/15548627.2022.2062887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Golz L, Memmert S, Rath-Deschner B, et al. LPS from P. gingivalis and hypoxia increases oxidative stress in periodontal ligament fibroblasts and contributes to periodontitis. Mediators Inflamm. 2014;2014:986264. doi: 10.1155/2014/986264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Zhao Y, Li J, Guo W, et al. Periodontitis-level butyrate-induced ferroptosis in periodontal ligament fibroblasts by activation of ferritinophagy. Cell Death Discov. 2020;6(1):119. doi: 10.1038/s41420-020-00356-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Ni S, Yuan Y, Qian Z, et al. Hypoxia inhibits RANKL-induced ferritinophagy and protects osteoclasts from ferroptosis. Free Radic Biol Med. 2021;169:271–282. doi: 10.1016/j.freeradbiomed.2021.04.027 [DOI] [PubMed] [Google Scholar]
- [268].Yang RZ, Xu WN, Zheng HL, et al. Exosomes derived from vascular endothelial cells antagonize glucocorticoid-induced osteoporosis by inhibiting ferritinophagy with resultant limited ferroptosis of osteoblasts. J Cell Physiol. 2021;236(9):6691–6705. doi: 10.1002/jcp.30331 [DOI] [PubMed] [Google Scholar]
- [269].Ba T, Zhao D, Chen Y, et al. L-Citrulline supplementation restrains ferritinophagy-mediated ferroptosis to alleviate iron overload-induced thymus oxidative damage and immune dysfunction. Nutrients. 2022;14(21):14. doi: 10.3390/nu14214549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Qin X, Zhang J, Wang B, et al. Ferritinophagy is involved in the zinc oxide nanoparticles-induced ferroptosis of vascular endothelial cells. Autophagy. 2021;17(12):4266–85. doi: 10.1080/15548627.2021.1911016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Liang Q, Ma Y, Wang F, et al. Silica nanoparticles induce hepatocyte ferroptosis and liver injury via ferritinophagy. Environ Sci Nano. 2022;9(8):3014–3029. doi: 10.1039/D2EN00116K [DOI] [Google Scholar]
- [272].Wei S, Qiu T, Yao X, et al. Arsenic induces pancreatic dysfunction and ferroptosis via mitochondrial ROS-autophagy-lysosomal pathway. J Hazard Mater. 2020;384:121390. doi: 10.1016/j.jhazmat.2019.121390 [DOI] [PubMed] [Google Scholar]
- [273].Xiao J, Zhang S, Tu B, et al. Arsenite induces ferroptosis in the neuronal cells via activation of ferritinophagy. Food Chem Toxicol. 2021;151:112114. doi: 10.1016/j.fct.2021.112114 [DOI] [PubMed] [Google Scholar]
- [274].Guo CC, Xu ZM, Lyu Y, et al. PM 2.5 induces autophagy-dependent ferroptosis by endoplasmic reticulum stress in SH-SY5Y cells. J Appl Toxicol. 2023;43(7):1013–1025. doi: 10.1002/jat.4439 [DOI] [PubMed] [Google Scholar]
- [275].Hu H, Li L, Zhang H, et al. Mechanism of YY1 mediating autophagy dependent ferroptosis in PM2.5 induced cardiac fibrosis. Chemosphere. 2023;315:137749. doi: 10.1016/j.chemosphere.2023.137749 [DOI] [PubMed] [Google Scholar]
- [276].Liu N, Liang Y, Wei T, et al. The role of ferroptosis mediated by NRF2/ERK-regulated ferritinophagy in CdTe QDs-induced inflammation in macrophage. J Hazard Mater. 2022;436:129043. doi: 10.1016/j.jhazmat.2022.129043 [DOI] [PubMed] [Google Scholar]
- [277].Zhou H, Zhou YL, Mao JA, et al. NCOA4-mediated ferritinophagy is involved in ionizing radiation-induced ferroptosis of intestinal epithelial cells. Redox Biol. 2022;55:102413. doi: 10.1016/j.redox.2022.102413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Amoscato A, Anthonymuthu T, Kapralov O, et al. Formation of protein adducts with hydroperoxy-PE electrophilic cleavage products during ferroptosis. Redox Biol. 2023;63:102758. doi: 10.1016/j.redox.2023.102758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Tian H, Sparvero LJ, Anthonymuthu TS, et al. Successive high-resolution (H2O) n-GCIB and C60-SIMS imaging integrates multi-omics in different cell types in breast cancer tissue. Anal Chem. 2021;93(23):8143–8151. doi: 10.1021/acs.analchem.0c05311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Liu J, Yeo HC, Doniger SJ, et al. Assay of aldehydes from lipid peroxidation: gas chromatography–mass spectrometry compared to thiobarbituric acid. Anal Biochem. 1997;245(2):161–166. doi: 10.1006/abio.1996.9990 [DOI] [PubMed] [Google Scholar]
- [281].Kawai Y, Takeda S, Terao J. Lipidomic analysis for lipid peroxidation-derived aldehydes using gas chromatography-mass spectrometry. Chem Res Toxicol. 2007;20(1):99–107. doi: 10.1021/tx060199e [DOI] [PubMed] [Google Scholar]
- [282].Morel Y, Jones JW. Utilization of LC–MS/MS and drift tube ion mobility for characterizing intact oxidized arachidonate-containing glycerophosphatidylethanolamine. J Am Soc Mass Spectrom. 2023;34(8):1609–1620. doi: 10.1021/jasms.3c00083 [DOI] [PubMed] [Google Scholar]
- [283].Lin Q, Jian R, Wang S, et al. Characterization of oxidized glycerophosphoethanolamines via radical-directed dissociation tandem mass spectrometry and the paternò–Büchi Derivatization. Anal Chem. 2023;95(25):9422–9427. doi: 10.1021/acs.analchem.3c00792 [DOI] [PubMed] [Google Scholar]
- [284].Tsikas D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: analytical and biological challenges. Anal Biochem. 2017;524:13–30. doi: 10.1016/j.ab.2016.10.021 [DOI] [PubMed] [Google Scholar]
- [285].Stockert JC. Lipid peroxidation assay using BODIPY-Phenylbutadiene probes: a methodological overview. Methods Mol Biol. 2021;2202:199–214. [DOI] [PubMed] [Google Scholar]
- [286].Mandavilli BS, Aggeler R. Abstract 512: cell-based analysis of oxidative stress, lipid peroxidation and lipid peroxidation-derived protein modifications using fluorescence microscopy. Cancer Res. 2014;74(19_Supplement):512–512. doi: 10.1158/1538-7445.AM2014-512 [DOI] [Google Scholar]
- [287].Liu Y, Wu L, Dai Y, et al. A novel fluorescent probe based on a triphenylamine derivative for the detection of HSO 3 − with high sensitivity and selectivity. Anal Methods. 2021;13(33):3667–3675. doi: 10.1039/D1AY00800E [DOI] [PubMed] [Google Scholar]
- [288].Comportl M. Lipid peroxidation and biogenic aldehydes: from the identification of 4-hydroxynonenal to further achievements in biopathology. Free Radic Res. 1998;28(6):623–35. doi: 10.3109/10715769809065818 [DOI] [PubMed] [Google Scholar]
- [289].Toyokuni S, Tanaka T, Hattori Y, et al. Quantitative immunohistochemical determination of 8-hydroxy-2’-deoxyguanosine by a monoclonal antibody N45. 1: its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab Invest. 1997;76:365–374. [PubMed] [Google Scholar]
- [290].Valavanidis A, Vlachogianni T, Fiotakis C. 8-hydroxy-2′-deoxyguanosine (8-OHdG): a critical biomarker of oxidative stress and carcinogenesis. Journal Of Environmental Science And Health Part C. 2009;27(2):120–139. doi: 10.1080/10590500902885684 [DOI] [PubMed] [Google Scholar]
- [291].Riemer J, Hoepken HH, Czerwinska H, et al. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem. 2004;331(2):370–5. doi: 10.1016/j.ab.2004.03.049 [DOI] [PubMed] [Google Scholar]
- [292].Hirayama T, Niwa M, Hirosawa S, et al. High-throughput screening for the discovery of iron homeostasis modulators using an extremely sensitive fluorescent probe. ACS Sens. 2020;5(9):2950–2958. doi: 10.1021/acssensors.0c01445 [DOI] [PubMed] [Google Scholar]
- [293].Petrat F, Rauen U, de Groot H. Determination of the chelatable iron pool of isolated rat hepatocytes by digital fluorescence microscopy using the fluorescent probe, phen green SK. Hepatology. 1999;29(4):1171–9. doi: 10.1002/hep.510290435 [DOI] [PubMed] [Google Scholar]
- [294].Cammack R, Cooper CE. Electron paramagnetic resonance spectroscopy of iron complexes and iron-containing proteins. Methods Enzymol. 1993;12;227:353–384. [DOI] [PubMed] [Google Scholar]
- [295].Schünemann V, Winkler H. Structure and dynamics of biomolecules studied by mössbauer spectroscopy. Rep Prog Phys. 2000;63(3):263. doi: 10.1088/0034-4885/63/3/202 [DOI] [Google Scholar]
- [296].Chen X, Comish PB, Tang D, et al. Characteristics and biomarkers of Ferroptosis. Front Cell Dev Biol. 2021;9:637162. doi: 10.3389/fcell.2021.637162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Müller T, Dewitz C, Schmitz J, et al. Necroptosis and ferroptosis are alternative cell death pathways that operate in acute kidney failure. Cell Mol Life Sci. 2017;74(19):3631–3645. doi: 10.1007/s00018-017-2547-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Wang C, Zheng C, Wang H, et al. Dual degradation mechanism of GPX4 degrader in induction of ferroptosis exerting anti-resistant tumor effect. Eur J Med Chem. 2023;247:115072. doi: 10.1016/j.ejmech.2022.115072 [DOI] [PubMed] [Google Scholar]
- [299].Feng H, Schorpp K, Jin J, et al. Transferrin Receptor Is a Specific Ferroptosis Marker. Cell Rep. 2020;30(10):3411–3423.e7. doi: 10.1016/j.celrep.2020.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Van Tonder A, Joubert AM, Cromarty AD. Limitations of the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) assay when compared to three commonly used cell enumeration assays. BMC Res Notes. 2015;8(1):1–10. doi: 10.1186/s13104-015-1000-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Galluzzi L, Aaronson SA, Abrams J, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009;16(8):1093–107. doi: 10.1038/cdd.2009.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Efimova I, Catanzaro E, Van der Meeren L, et al. Vaccination with early ferroptotic cancer cells induces efficient antitumor immunity. J Immunother Cancer. 2020;8(2):8. doi: 10.1136/jitc-2020-001369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Liu J, Zhu S, Zeng L, et al. DCN released from ferroptotic cells ignites AGER-dependent immune responses. Autophagy. 2022;18(9):2036–49. doi: 10.1080/15548627.2021.2008692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Ide S, Ide K, Abe K, et al. Sex differences in resilience to ferroptosis underlie sexual dimorphism in kidney injury and repair. Cell Rep. 2022;41(6):111610. doi: 10.1016/j.celrep.2022.111610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Kang R, Zeng L, Zhu S, et al. Lipid peroxidation drives gasdermin D-Mediated pyroptosis in lethal polymicrobial sepsis. Cell Host Microbe. 2018;24(1):97–108 e4. doi: 10.1016/j.chom.2018.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Canli O, Alankus YB, Grootjans S, et al. Glutathione peroxidase 4 prevents necroptosis in mouse erythroid precursors. Blood. 2016;127(1):139–148. doi: 10.1182/blood-2015-06-654194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Azuma K, Koumura T, Iwamoto R, et al. Mitochondrial glutathione peroxidase 4 is indispensable for photoreceptor development and survival in mice. J Biol Chem. 2022;298(4):101824. doi: 10.1016/j.jbc.2022.101824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy. 2021;17:1–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–92. doi: 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- [310].Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Mizushima N, Murphy LO. Autophagy assays for biological discovery and therapeutic development. Trends Biochem Sci. 2020;45(12):1080–1093. doi: 10.1016/j.tibs.2020.07.006 [DOI] [PubMed] [Google Scholar]
- [312].Shpilka T, Weidberg H, Pietrokovski S, et al. Atg8: an autophagy-related ubiquitin-like protein family. Genome Bio. 2011;12(7):1–11. doi: 10.1186/gb-2011-12-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Bjørkøy G, Lamark T, Pankiv S, et al. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. [DOI] [PubMed] [Google Scholar]
- [314].Zhou J, Tan S-H, Nicolas V, et al. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res. 2013;23(4):508–23. doi: 10.1038/cr.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Ishii S, Matsuura A, Itakura E. Identification of a factor controlling lysosomal homeostasis using a novel lysosomal trafficking probe. Sci Rep. 2019;9(1):11635. doi: 10.1038/s41598-019-48131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Papadopoulos C, Meyer H. Detection and clearance of damaged lysosomes by the endo-lysosomal damage response and lysophagy. Curr Biol. 2017;27(24):R1330–R1341. doi: 10.1016/j.cub.2017.11.012 [DOI] [PubMed] [Google Scholar]
- [317].Pryor PR. Analyzing lysosomes in live cells. In: Methods in enzymology. Elsevier; 2012. pp. 145–157. [DOI] [PubMed] [Google Scholar]
- [318].Ritzel RM, Li Y, Lei Z, et al. Functional and transcriptional profiling of microglial activation during the chronic phase of TBI identifies an age-related driver of poor outcome in old mice. Geroscience. 2022;44(3):1407–1440. doi: 10.1007/s11357-022-00562-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Sarkar C, Jones JW, Hegdekar N, et al. PLA2G4A/cPLA2-mediated lysosomal membrane damage leads to inhibition of autophagy and neurodegeneration after brain trauma. Autophagy. 2020;16(3):466–85. doi: 10.1080/15548627.2019.1628538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–60. doi: 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- [321].Loos B, Ad T, Hofmeyr J-H. Defining and measuring autophagosome flux—concept and reality. Autophagy. 2014;10(11):2087–2096. doi: 10.4161/15548627.2014.973338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].du Toit A, Hofmeyr J-H, Gniadek TJ, et al. Measuring autophagosome flux. Autophagy. 2018;14:1060–1071. doi: 10.1080/15548627.2018.1469590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Loos B, Engelbrecht A-M, Lockshin RA, et al. The variability of autophagy and cell death susceptibility: unanswered questions. Autophagy. 2013;9(9):1270–85. doi: 10.4161/auto.25560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Kaizuka T, Morishita H, Hama Y, et al. An autophagic flux probe that releases an internal control. Mol Cell. 2016;64(4):835–849. doi: 10.1016/j.molcel.2016.09.037 [DOI] [PubMed] [Google Scholar]
- [325].Yim WW, Yamamoto H, Mizushima N. A pulse-chasable reporter processing assay for mammalian autophagic flux with HaloTag. Elife. 2022;11:11. doi: 10.7554/eLife.78923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Wen-You Yim W, Yamamoto H, Mizushima N. A HaloTag-based reporter processing assay to monitor autophagic flux. Autophagy. 2023;19(4):1363–4. doi: 10.1080/15548627.2022.2123638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Wrobel L, Hill SM, Djajadikerta A, et al. Compounds activating VCP D1 ATPase enhance both autophagic and proteasomal neurotoxic protein clearance. Nat Commun. 2022;13(1):4146. doi: 10.1038/s41467-022-31905-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Katayama H, Hama H, Nagasawa K, et al. Visualizing and modulating mitophagy for therapeutic studies of neurodegeneration. Cell. 2020;181(5):1176–87. e16. doi: 10.1016/j.cell.2020.04.025 [DOI] [PubMed] [Google Scholar]
- [329].McWilliams TG, Prescott AR, Allen GF, et al. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Bio. 2016;214(3):333–345. doi: 10.1083/jcb.201603039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Chino H, Hatta T, Natsume T, et al. Intrinsically disordered protein TEX264 mediates ER-phagy. Molecular Cell. 2019;74(5):909–21. e6. doi: 10.1016/j.molcel.2019.03.033 [DOI] [PubMed] [Google Scholar]
- [331].Liang JR, Lingeman E, Ahmed S, et al. Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Bio. 2018;217(10):3354–67. doi: 10.1083/jcb.201804185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [332].Chen Q, Xiao Y, Chai P, et al. ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr Biol. 2019;29(5):846–55. e6. doi: 10.1016/j.cub.2019.01.041 [DOI] [PubMed] [Google Scholar]
- [333].Li L, Wang ZV, Hill JA, et al. New autophagy reporter mice reveal dynamics of proximal tubular autophagy. J Am Soc Nephrol. 2014;25(2):305. doi: 10.1681/ASN.2013040374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].Lee J-H, Rao MV, Yang D-S, et al. Transgenic expression of a ratiometric autophagy probe specifically in neurons enables the interrogation of brain autophagy in vivo. Autophagy. 2019;15(3):543–57. doi: 10.1080/15548627.2018.1528812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [335].Siddiqi FH, Menzies FM, Lopez A, et al. Felodipine induces autophagy in mouse brains with pharmacokinetics amenable to repurposing. Nat Commun. 2019;10(1):1817. doi: 10.1038/s41467-019-09494-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [336].Perez MA, Magtanong L, Dixon SJ, et al. Dietary lipids induce ferroptosis in caenorhabditiselegans and human cancer cells. Dev Cell. 2020;54(4):447–54 e4. doi: 10.1016/j.devcel.2020.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Zhang Y, Fan B-Y, Pang Y-L, et al. Neuroprotective effect of deferoxamine on erastin-induced ferroptosis in primary cortical neurons. Neural Regen Res. 2020;15(8):1539. doi: 10.4103/1673-5374.274344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].Xuan W, Lu X, Yang Z, et al. Propofol protects against erastin-induced ferroptosis in HT-22 cells. J Mol Neurosci. 2022;72(9):1797–1808. doi: 10.1007/s12031-022-02017-7 [DOI] [PubMed] [Google Scholar]
- [339].Wei Z, Hao C, Huangfu J, et al. Aging lens epithelium is susceptible to ferroptosis. Free Radic Biol Med. 2021;167:94–108. doi: 10.1016/j.freeradbiomed.2021.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Yagoda N, von Rechenberg M, Zaganjor E, et al. RAS–RAF–MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447(7146):865–869. doi: 10.1038/nature05859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [341].Yan R, Xie E, Li Y, et al. The structure of erastin-bound xCT–4F2hc complex reveals molecular mechanisms underlying erastin-induced ferroptosis. Cell Res. 2022;32(7):687–690. doi: 10.1038/s41422-022-00642-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [342].Hill BG, Haberzettl P, Ahmed Y, et al. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem J. 2008;410(3):525–34. doi: 10.1042/BJ20071063 [DOI] [PubMed] [Google Scholar]
- [343].Su L-J, Zhang J-H, Gomez H, et al. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid Med Cell Longevity. 2019;2019:1–13. doi: 10.1155/2019/5080843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [344].Leri M, Ramazzotti M, Vasarri M, et al. Bioactive compounds from Posidonia oceanica (L.) delile impair malignant cell migration through autophagy modulation. Mar Drug. 2018;16(4):137. doi: 10.3390/md16040137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [345].Liu J, Zheng L, Zhong J, et al. Oleanolic acid induces protective autophagy in cancer cells through the JNK and mTOR pathways. Oncol Rep. 2014;32(2):567–572. doi: 10.3892/or.2014.3239 [DOI] [PubMed] [Google Scholar]
- [346].Zitvogel L, Kepp O, Kroemer G. Decoding cell death signals in inflammation and immunity. Cell. 2010;140(6):798–804. doi: 10.1016/j.cell.2010.02.015 [DOI] [PubMed] [Google Scholar]


