Abstract
Organic nanoparticles are used in nanomedicine, including for cancer treatment and some types of COVID-19 vaccines. Here, we demonstrate the scalable, rapid, reproducible, and cost-effective synthesis of three model organic nanoparticle formulations relevant to nanomedicine applications. We employed a custom-made, low-cost fluid mixer device constructed from a commercially available three-dimensional printer. We investigated how systematically changing aqueous and organic volumetric flow rate ratios determined liposome, polymer nanoparticle, and solid lipid nanoparticle sizes, size distributions, and payload encapsulation efficiencies. By manipulating inlet volumes, we synthesized organic nanoparticles with encapsulation efficiencies approaching 100% for RNA-based payloads. The synthesized organic nanoparticles were safe and effective at the cell culture level, as demonstrated by various assays. Such cost-effective synthesis approaches could potentially increase the accessibility to clinically relevant organic nanoparticle formulations for personalized nanomedicine applications at the point of care, especially in nonhospital and low-resource settings.
Keywords: nanoparticles, liposomes, nanomedicine, lipid nanoparticles, RNA delivery, point of care
Graphical Abstract

Organic nanoparticles, such as liposomes, polymers, and lipid nanoparticles (LNPs), have attracted significant attention in the clinic due to their ability to encapsulate and deliver various payloads safely and effectively, such as small molecule drugs and nucleic acids.1–3 However, the storage, distribution, and accessibility of these nanoparticles can be challenging.4–6
To enable the future production of personalized nanomedicines, there is a need to decentralize the synthesis of safe and effective nanoparticle formulations. Ideally, personalized nanomedicines could be produced on demand and at the point of care.
Toward this vision, we sought to develop an accessible, low-cost synthesis platform to produce high-quality nanoparticle formulations. We hope that such synthesis platforms may enable clinicians and physicians in the future to produce and administer personalized nanoparticle formulations to patients on demand and at the point of care, thereby addressing the challenges associated with nanoparticle storage, distribution, and accessibility.7
Current state-of-the-art industry devices use a T-junction mixer to maintain a high throughput of organic nanoparticle formulations.8–10 This T-junction allows a mixture of lipids (or polymer) in an organic solvent to combine with an aqueous stream at certain flow rate ratios (FRRs). The controlled addition and mixing of the two liquid phases induce solvent polarity changes, leading to the formation of lipid (or polymer) nanoparticles. This synthesis approach creates uniform and monodisperse nanoparticles whose average size can be controlled by manipulating the FRR (aqueous:organic).11
Our vision was to adopt the industrial process of organic nanoparticle manufacturing and simplify it to a lab scale. Ideally, this system would contain a fluidic device controlled by a single unit. The device would need to synthesize various organic nanoparticles for downstream biomedical applications. Finally, we sought to keep the cost of this device as low as possible, as commercially available equipment can create organic nanoparticles. However, this equipment is expensive or requires more than one control panel to operate (Table S1).
Inspired by Saggiomo et al.,12 we designed a fluidic device by repurposing an Ender3 three-dimensional (3D) printer. Our demonstrated fluidic setup may enable a broad user base to synthesize organic nanoparticle formulations with a financial investment orders of magnitude below the cost of most commercial products (Tables S2 and S3). While the Ender3 T-mixing method is different from some of the benchmarked designs, the device is simpler, is more cost-effective, and produces nanoparticle quality comparable to that of the benchmarked products (Table S4). We provide a detailed blueprint containing optimized setup instructions and 3D printer files for the Ender3 syringe pumps in the Supporting Information. Additionally, using the Ender3 setup, we synthesized liposomes, poly(lactic-co-glycolic acid) (PLGA) nanoparticles, and LNPs and demonstrated their safety and efficacy at the cell culture level.
We transformed an Ender3 3D printer into a set of three programmable syringe pumps (labeled as X, Y, and Z) following the design of Saggiomo et al. (Figure 1a,b).12 Figure 1 provides a simplified schematic of how to synthesize organic nanoparticle formulations using the Ender3 syringe pumps. The Ender3 is a cost-efficient device that enables simultaneous and controlled fluid flow by manipulating simple coding parameters (Figure 1c). This level of control provides a scalable fluidic approach from microliters per minute to milliliters per minute, making the Ender3 setup suitable for reproducible synthesis techniques that require variable volumes. Additionally, the Ender3 is highly modular. Many existing mixing devices are compatible with Ender3 syringe pumps, including T-mixers, cross-mixers, staggered herringbone mixers, and flow-focused channels. We explain how to code the Ender3 and to calibrate 10 mL syringes to a particular step size in the Supporting Information. We confirmed that the calibrated step size is accurate for dispensing volumes of >1 mL (Figure S1a). Finally, we optimized the code to preserve the target flow rate when multiple syringe pumps were moving (Figure S1b).
Figure 1.
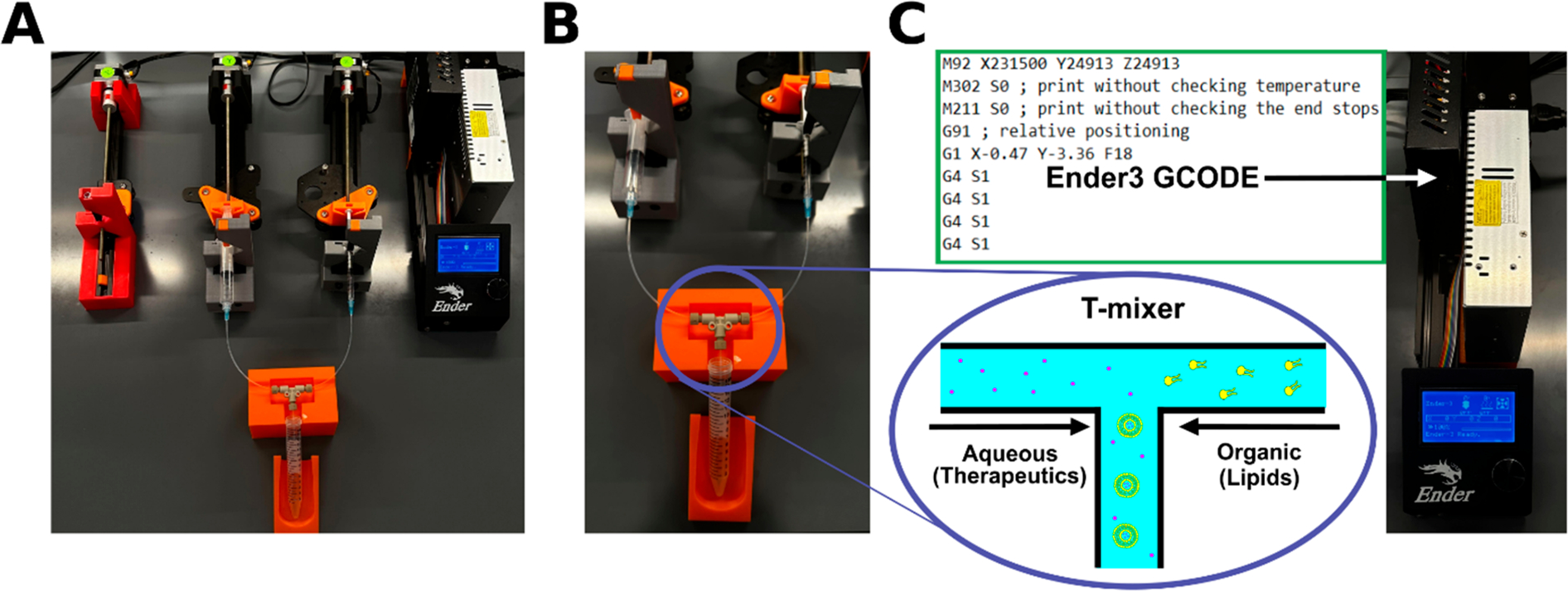
Ender3 3D printer-based fluidic setup for synthesizing organic nanoparticle formulations. (A) The Ender3 3D printer provides a set of three syringe pumps. The photograph shows the setup for liposome synthesis using two syringe pumps connected to a T-mixer. (B) Photograph highlighting the T-mixer fluidics. (C) Images of the GCODE used to program the syringe pumps. The GCODE is saved on a microUSB card and inserted into the receptacle highlighted by the white arrow. The bottom panel is a simplified schematic of the T-mixer, showing how payloads (e.g., therapeutics represented as dots) and organic components can be mixed to form payload-encapsulating nanoparticle formulations.
The maintenance expenses for the Ender3 are notably more favorable than those for alternate commercially available fluidic devices that often recommend using high-cost single-use fluidic cartridges. We present a cost analysis breakdown comparing the expenses of other fluidic devices with our Ender3 system (Table S3).
Next, we tested the Ender3 syringe pumps to manufacture liposomes. If the introduction of aqueous and organic fluids can be controlled, then monodisperse, high-quality liposomes can be produced without the need for resizing steps, ensuring the payload %ee remains relatively high.13–15 We first dissolved DSPC, cholesterol, and DSPE-PEG lipids in 100% ethanol at molar ratios listed in Table S5. Next, we ran the organic fluid stream of lipids against an aqueous solution of 1× phosphate-buffered saline (PBS) at different FRRs through a poly(ether ether ketone) (PEEK) T-mixer, which contained two inlet streams and one outlet stream. We observed that systematically increasing the FRR from 3 to 15 decreased the liposome hydrodynamic diameter (HDD) as determined by dynamic light scattering (DLS) measurements (Figure 2a). Lower FRRs resulted in the greatest shift in HDDs, and higher FRRs resulted in the best precision in liposome synthesis. This result confirmed previous findings that increasing the FRR decreases the liposome size.16
Figure 2.

Liposome characterization using dynamic light scattering (DLS). (A) Comparison of the liposome hydrodynamic diameter (HDD) synthesized at various flow rate ratios (FRRs) as measured by DLS. Red marks represent the mean values ± standard deviations (SD) (n = 3). The trend line is a mathematical fit of the obtained data points, modeled by the equation HDD = 124.3 × exp(−0.239 × FRR) + 61.96 × exp(−0.00153 × FRR). (B) Comparison of the liposome polydispersity index (PDI) at various FRRs as measured by DLS. Data represent average values ± SD (n = 3). A gray dashed line indicating the PDI benchmark has been included for the sake of convenience. (C) DLS data reporting the HDD and PDI of liposomes at various total flow rates (TFRs). Data represent average values ± SD (n = 3). (D) DLS data reporting the HDD and PDI of liposomes with different lipid bilayer membrane compositions (Table S8). Data represent average values ± SD (n = 3).
While liposomes synthesized with FRRs of ≥5 had polydispersity indices (PDIs) of <0.2 (Figure 2b), we found that the more commonly used FRR of 3 produced liposomes with PDIs of >0.2 (Table S6). Additionally, we observed that the total flow rate (TFR) affected the liposome HDD minimally but increasing the TFR increased nanoparticle monodispersity and synthesis consistency (Figure 2c and Table S6).
Next, we showed additional applications for synthesizing liposomes. First, we ensured that the Ender3-based synthesis approach is scalable (Figure S2 and Table S7). We then quantified the Ender3’s ability to synthesize liposomes of different lipid compositions. We selected three additional lipid compositions that offered varying levels of lipid bilayer rigidity and created liposomes at an FRR of 7 (Table S8), as liposomes made with an FRR of 7 produced more consistent results than FRRs of 5 and 3 (Figure 2a). Changing the lipid composition affected the resulting liposome size (Figure 2d and Table S6).
To demonstrate that therapeutics can be encapsulated within liposomes synthesized by the Ender3, we compared the %ee of dextran-encapsulating liposomes (TRITC-conjugated, molecular weight of 70 kDa) made via the fluidic and traditional extrusion methods.17 We found that the fluidic method produced liposomes with a %ee significantly higher than that of the traditional method (Figure S3 and Table S9).18
Polymeric nanoparticles are a second type of clinically relevant nanomedicines. The polymer poly[lactic-co-glycolic-acid] (PLGA) is a hydrophobic and biocompatible compound that breaks down to lactic acid and glycolic acid when hydrolyzed in the body.19 PLGA nanoparticles (PLGA-NPs) can efficiently encapsulate an average of 50–70% of hydrophobic payloads through microfluidic synthesis methods.19,20
With this in mind, we tested the Ender3’s potential to produce PLGA-NPs (Figure 3). Due to the unique formulation of polymeric nanoparticles (see the Supporting Information), we used a PEEK crossflow mixer with three syringe pumps, one for each inlet. We dissolved a PLGA copolymer in dimethyl sulfoxide (DMSO) and ran it against 1× PBS. The emulsifier PVA was used in the aqueous phase to assist in nanoparticle formation. We systematically varied the FRR and TFR to evaluate how these parameters affected the PLGA-NP synthesis.
Figure 3.

Characterization of PLGA nanoparticles. (A) Dynamic light scattering (DLS) data reporting the hydrodynamic diameter (HDD) and polydispersity index (PDI) of PLGA nanoparticles synthesized at different total flow rates (TFRs). The TFRs were calculated retroactively by combining the individual volumetric flow rates obtained from each syringe. Data represent average values ± standard deviations (SD) (n = 3). (B) DLS data reporting the HDD and PDI of PLGA nanoparticles synthesized at different flow rate ratios (FRRs). Data represent average values ± SD (n = 3). (C) DLS data reporting the HDD and PDI of PLGA-PEG NPs. Data represent average values ± SD (n = 3). (D) DLS data reporting the HDD and PDI of PLGA-NPs encapsulating rosiglitazone. Data represent average values ± SD (n = 3). (E) Encapsulation efficiency (%ee) of rosiglitazone within the PLGA nanoparticles. Data represent average values ± SD (n = 3).
Interestingly, we noted that TFRs between 0.1 and 1 mL/min did not change the PLGA-NP sizes (Figure 3a). As the TFR further increased to 10 mL/min, the PLGA-NPs had reduced time to equilibrate and grow, leading to smaller particles (Figure 3a and Table S9). Adjusting the FRR from 3 to 7 led to a slight decrease in PLGA-NP size and increased the PDI of the nanoparticles (Figure 3b and Table S10). However, we saw a weaker influence of FRR on PLGA-NP size compared with the liposome size. The trends described above mirrored other research on fluidic PLGA-NP manufacturing.20–22 We compared PLGA-NPs prepared using fluidics to PLGA-NPs prepared using a probe sonication technique and found that both methods produced the same quality of nanoparticles (Figure S4 and Table S10). However, the fluidic method is much faster to perform and does not require additional machinery, thus affirming that the Ender3 synthesis method possesses important advantages over the batch synthesis method.
To broaden the applicability of our synthesis method, we selected another type of PLGA polymer to synthesize nanoparticles. We chose to further study a PLGA-PEG polymer, because poly(ethylene glycol) (PEG) has been shown to increase the colloidal stability of clinically used nanoparticles and to reduce protein adsorption in vivo.23 The PLGA-PEG compound was dissolved in DMSO solvent at a concentration of 2.5 mg/mL and processed at an FRR of 5. Using DLS, we confirmed that the Ender3 setup created PLGA-PEG-based nanoparticles (Figure 3c and Table S10).
Next, we tested whether we could encapsulate model payloads in the PLGA nanoparticles. We selected the hydrophobic drug rosiglitazone as our model payload. Rosiglitazone reduces insulin resistance and may be useful for diabetic patients. To limit off-target effects, encapsulation of rosiglitazone into polymeric nanoparticles may be beneficial.24 We synthesized PLGA-NPs encapsulating rosiglitazone at an FRR of 7 and a TFR of 0.1 (Figure 3d), centrifuged them, and lysed them to reveal their encapsulated cargo, which was then measured using ultraviolet–visible (UV–vis) spectrophotometry. The resulting %ee was 14 ± 3% (Figure 3e and Table S9). This result is similar to the results of other microfluidic syntheses, because the use of DMSO does not offer a “droplet-based” method of drug entrapment, and the dissolved rosiglitazone is not inclined toward encapsulation.25,26 Other studies showed that combining dichloromethane and DMSO increases the level of drug encapsulation, which will be an appropriate target for a future study.20
In summary, our Ender3 setup can create PLGA nanoparticles with different polymer types. The nanoparticle size distributions can be tuned and optimized, and the resulting nanoparticles encapsulate hydrophobic drugs.
To further demonstrate the broad applicability of our Ender3-based fluidic mixing setup, we additionally synthesized different lipid nanoparticles (LNPs). LNPs have shown potential as a therapeutic delivery device, with prominent examples such as some COVID-19 vaccines and the Patisiran (Onpattro) pharmaceutical, which is used in hereditary transthyretin-mediated amyloidosis.27 Additionally, certain lipids, when implemented into LNP formulations, have shown to greatly increase the rate of uptake of LNPs in target organs, such as the spleen or lungs.28 LNPs often carry nucleic acids, ranging from large nucleic acids such as DNA and mRNA (mRNA) to smaller nucleic acids such as small interfering RNA (siRNA). While these nucleic acid molecules are promising therapeutics, they degrade quickly upon in vivo administration.29 When encapsulated into LNPs, the nanoparticles offer an efficient way to protect the nucleic acid payloads from degradation and deliver them safely and effectively to organs and cells.
We used the Ender3 syringe pumps to synthesize LNPs and encapsulated siRNA as a model nucleic acid payload. We adjusted FRRs from 5 to 9 and TFRs from 12 to 18 mL/min to optimize siRNA-LNP (siRNA-encapsulating LNPs) synthesis. We found that an FRR of 7 and a TFR of 18 mL/min produced the LNPs with the most consistent HDD and lowest PDIs (Figure 4a, Figure S5, and Table S11). While the previously described trend of FRR affecting liposome size was less prominent in this context due to the variations in the nitrogen:phosphate (nitrogen on lipids:phosphate groups on RNA) ratio,30 we established that changes in FRR influence LNP size with all tested FRRs producing LNPs with high %ee (>90%) (Figure 4b). Moreover, increasing the TFR from 12 to 18 mL/min resulted in more consistent synthesis results, aligning with prior studies focusing on controlling LNP size by manipulating the TFR (Figure S5a and Table S11).31,32 Notably, all TFR values led to LNPs with similarly high %ee values (Figure S5b). Future research will optimize LNP production for different FRRs and TFRs.
Figure 4.

Characterization of lipid nanoparticles (LNPs). (A) Dynamic light scattering (DLS) data reporting the hydrodynamic diameter (HDD) and polydispersity index (PDI) of siRNA-LNPs as a function of the flow rate ratio (FRR). Data represent average values ± standard deviations (SD) (n = 3). (B) Encapsulation efficiency (%ee) of siRNA-LNPs as a function of the FRR. Data represent average values ± SD (n = 3). (C) DLS data reporting the HDD and PDI of siRNA-LNPs before and after dialysis. Data represent average values ± SD. A t test was conducted against synthesized siRNA-LNPs and dialyzed siRNA-LNPs to compare changes in HDD and PDI (n = 3). No statistical test revealed any significance between the compared values. (D) %ee of siRNA-LNPs before and after dialysis. Data represent average values ± SD. A t test was conducted against synthesized siRNA-LNPs and dialyzed siRNA-LNPs to compare changes in %ee (n = 3). No statistical test revealed any significance between the compared values. (E) DLS data reporting the HDD and PDI of siRNA-LNPs at various time points during storage at 4 °C. Data represent average values ± SD. An analysis of variance (ANOVA) test was conducted on the siRNA-LNP HDD and PDI at different time points (n = 3). No statistical test revealed any significance between the compared values. (F) %ee of the siRNA-LNPs at various time points during storage at 4 °C. Data represent average values ± SD. An ANOVA test was conducted on the siRNA-LNP %ee at different time points (n = 3). No statistical test revealed any significance between the compared values.
We further confirmed that dialysis did not affect the LNP size or PDI values (Figure 4c). To determine the siRNA %ee, we used a fluorescence-based RiboGreen assay (Figure S6). Finally, we confirmed that %ee did not significantly change after dialysis (Figure 4d and Table S9). We compared the HDD, PDI, and %ee of LNPs produced via a hand mixing batch method to those of LNPs produced at an optimized FRR of 7. We found that the Ender3-synthesized LNPs had significantly lower PDI values, affirming that the Ender3 can effectively synthesize high-quality LNPs (Figure S7a,b and Tables S9 and S11). We additionally measured the HDD, PDI, %ee, and ζ potential of Ender3-synthesized LNPs under various storage conditions. We found that the HDD, %ee, and ζ potential values did not significantly change while stored at 4 °C for 3 weeks (p > 0.05), suggesting that the nanoparticles were colloidally stable (Figure 4e,f, Figure S8, and Table S12). Finally, while the ζ potential of the LNP significantly changed when it was stored at room temperature for 48 h (p < 0.05), the LNP HDD did not significantly change (p > 0.05), suggesting that the particles are relatively stable for short time periods at room temperature (Table S13).
Because siRNA is a relatively small nucleic acid molecule, we demonstrated the generalizability of our method for synthesizing LNPs with different nucleic acid payloads. We encapsulated a luciferase–mRNA cargo (~2,000 nucleotides) and demonstrated a consistent HDD, a low PDI (<0.2), and a high %ee for mRNA-encapsulating LNPs (Figure S9 and Tables S9 and S11).
Next, we demonstrated that the organic nanoparticle formulations synthesized by the Ender3 setup could be used in cell culture experiments. To assess the safety and efficacy of our nanoparticles, we elected to further study the LNPs, as they contained an siRNA payload that has demonstrated efficacy against prostate cancer cells.33 siUG is an siRNA known to inhibit the growth and viability of prostate cancer cells through RNA interference of androgen receptor coregulatory and essential gene networks. As a control, we used LNPs containing a nontarget siRNA (siNT). The siNT differs from the siUG in seven nucleotides corresponding to nucleotides 2–8 (the seed sequence of the siRNA) and does not affect cell growth or viability.
We selected 22Rv1 human prostate cancer cells as our model cell line. The 22Rv1 cells were seeded in 96-well plates and allowed to adhere overnight. The siNT-LNPs stained with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD) at a concentration of 50 μg/mL were prepared using the Ender3-based fluidic method. The encapsulated siRNA concentration was quantified, and LNPs, which contained 0–45 nM encapsulated siRNA, were introduced into the 22Rv1 cells. Following a variable incubation period, we observed an increasing quantity of DiD fluorescence with an increasing LNP concentration and uptake time. These results demonstrate concentration- and time-dependent LNP interactions with the 22Rv1 cells. We noted that higher concentrations (>30 nM siRNA) induced a slight reduction in cell viability, as shown by the reduced fluorescence value of the 45 nM group (Figure 5a,b and Table S14). Figure S10 provides confocal laser scanning microscopy images of the LNP-loaded 22Rv1 cells to confirm the LNP uptake within these cells.
Figure 5.

Characterization of siRNA-LNP uptake and efficacy. (A) Fluorescently labeled siRNA-LNPs were prepared and administered to 22Rv1 prostate cancer cells at the target concentrations. After a 24 h incubation period, the fluorescence intensity within these cells was measured using a Biotek Synergy Neo2 plate reader (644 nm excitation and 665 nm emission) and displayed as a function of siRNA concentration delivered by LNPs. Data represent average values ± standard deviations (SD). An analysis of variance (ANOVA) test was conducted to determine significant differences in cellular uptake (n = 5). The siRNA concentration of 15 nM showed significantly higher uptake than the siRNA concentration of 7.5 nM. *p < 0.05; ns, no significance. (B) Fluorescently labeled siRNA LNPs were prepared and administered to 22Rv1 prostate cancer cells at 30 nM siRNA. After the target time period, the fluorescence intensity within these cells was measured with the same parameters as in panel a and displayed as a function of incubation time. Data represent average values ± SD. An ANOVA test was conducted to determine significant differences in cellular uptake (n = 5). The 12 h time point displayed significantly higher LNP uptake than the 4 h time point (p = 0.014), and the 24 h time point displayed significantly higher LNP uptake than the 12 h time point (p = 0.009). (C) 22Rv1 cell growth in response to transfection with two different experimental groups: siRNA-LNPs loaded with siNT siRNA and siRNA-LNPs loaded with siUG siRNA. A cell-only group was used as a control. The relative confluency, as measured by the InCucyte, was used as a surrogate for cell growth. Points represent relative confluences of each well sample, and a line is provided for the sake of convenience. An ANOVA test was used to compare the siNT samples to the siUG and cell-only samples (n = 3). The cells dosed with siUG-LNPs had significantly lower confluences than those dosed with siNT-LNPs (*p < 0.005; **p < 0.0005). (D) Trypan blue cell viability data collected 156 h after the 22Rv1 transfections using a Nexcelom Cellometer Auto T4. Data are relative to the cell-only confluency and represent average values ± SD (n = 3). A t test was conducted to determine differences in cell viability. The cells dosed with siUG-LNPs showed significantly lower cell viability (p < 0.01) compared to those of the siNT-LNP and cell-only groups. (E) XTT cell viability data were collected from RAW 264.7 macrophages treated with the listed materials and incubated for 24 h. A cell-only group was used as a negative control. Data represent average values ± SD (n = 6). siNT = siNT siRNA, and siUG = siUGUUUGC siRNA. An ANOVA test was conducted to determine differences in cell viability. No groups had cell viability values statistically lower than those of the cell-only control group
Next, we studied the efficacy of the LNPs to deliver functional siRNA into cells grown in culture. We have previously demonstrated that 22Rv1 prostate cancer cells transfected with the siUGUUUGC siRNA, using commercially available transfection reagents, showed a decreased proliferation rate and a decreased viability.33
Following a similar protocol, we treated 22Rv1 cells with comparable amounts of LNPs loaded with 15 nM siUGUUUGC (siUG-LNPs) or with 15 nM siNT control (siNT-LNPs) and determined the cell growth rate and viability (Figure 5c,d). We observed that 22Rv1 cells dosed with siUG-LNPs showed significantly lower confluency, used as a proxy for growth rate, than the cells dosed with siNT-LNPs (p < 0.05), indicating that the LNPs can deliver functional siRNAs to the cells (Figure 5c, Table S15, and Figure S11). Additionally, we found that 22Rv1 cells dosed with siUG-LNPs exhibited significantly lower confluency than 22Rv1 cells transfected with siUGUUUGC using a commercially available Dharmacon reagent (Figure S12). These results indicate that the LNPs can deliver functional siRNA into 22Rv1 cells more effectively than other transfection methods.
It is noteworthy that we observed that untreated cells showed a small but significantly enhanced growth rate than cells dosed with the control siNT-LNPs, suggesting that treatment with the LNPs may have some effect on cell proliferation (Figure 5c and Table S15). We used Trypan blue to determine the effect of LNP treatment on the 22Rv1 cell viability (Figure 5d). The 22Rv1 cells treated with the siNT-LNPs showed the same viability as the untreated cells, indicating that 22Rv1 viability is unaffected by the LNPs. However, the cells dosed with siUG-LNPs exhibited viability significantly lower than that of the siNT-LNP or the untreated cells (p < 0.05) (Figure 5d and Table S16), confirming the toxicity of the siUGUUUGC for prostate cancer cells. These results demonstrate that LNPs synthesized with the Ender3 device can safely and efficiently deliver functional siRNA into prostate cancer cells.
Next, we wanted to assess the potential off-target cell toxicity of the Ender3-synthesized LNPs using murine macrophage cell RAW 264.7 as a model cell line. As these cells are not prostate cancer cells and are insensitive to the selected toxic siRNA, we expected a nonsignificant change in cell viability after dosing with siUG-LNPs. We ran an XTT-based cell viability assay following a 24 h incubation with 30 nM siUG- and siNT-encapsulating LNPs. Free siRNA and blank LNPs were used as controls. The concentration of the siRNA encapsulated into LNPs was selected on the basis of our published data using the Dharmacon transfection protocol33 and through our previous experiments determining LNP uptake. We noted that our siRNA-encapsulating LNPs had no significant effect on the viability in the selected macrophage cell line (p > 0.05) (Figure 5e and Table S17). With this, we confirmed that the siRNA-LNPs created by the Ender3 had limited toxicity at siRNA concentrations similar to those used in the Dharmacon transfection protocol.
To demonstrate the broad applicability of the Ender3-synthesized LNPs in cell transfection, we used our previously synthesized mRNA-LNPs containing luciferase and performed a luciferase bioluminescence assay. Briefly, HepG2 human hepatoma cells were dosed with LNPs loaded with 50–200 nM luciferase mRNA. After being incubated for 6 h, the mRNA-LNP sample was removed, and the transfection efficiency was evaluated using the luciferase bioluminescence assay. We observed that all doses of mRNA LNP produced luminescence intensities significantly greater than that of the control group (Figure S13). Additionally, we confirmed that the mRNA-LNPs did not significantly affect the HepG2 cell viability at the specified concentrations and time frame (p > 0.05) (Figure S14). These results demonstrate that LNPs synthesized through our Ender3 platform appear to transfect HepG2 cells safely and effectively under the tested parameters.
We showed that an Ender3 3D printer-based synthesis setup consisting of a synchronous, accurate, programmable syringe pump can be used to prepare a library of clinically relevant nanoparticle types, such as liposomes, PLGA nanoparticles, and LNPs. Our syringe pump system fulfilled the constraints of the outlined requirements. (i) It provides an exceptionally cost-effective platform for enabling the synthesis of diverse organic nanoparticle types. (ii) It exhibits synthesis trends comparable to established industry standards. (iii) It allows synchronous control of the syringe pumps with one control unit. The nanoparticle physicochemical properties did not significantly change during a storage period of 3 weeks, suggesting that the synthesized nanoparticles are colloidally stable and of high quality. We showed applications of siRNA and mRNA-encapsulating LNPs in cell culture, demonstrating that they appear to be safe and effective under the tested parameters. We created a cost-effective organic nanoparticle synthesis platform to prepare libraries of clinically relevant nanoparticle formulations. This synthesis platform has the potential to be broadly used at the point of care. We envision implementing our fluidics device into a benchtop point-of-care nanoparticle manufacturing system that monitors formulation physicochemical quality parameters to make safe and effective nanomedicines more broadly accessible, including potential personalized treatments and vaccines against infectious diseases and cancer.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the assistance of Drs. John Clegg, Paul DeAngelis, and Cecilia Pham. Additionally, the University of Oklahoma (OU) Samuel Roberts Noble Microscopy Laboratory (SRNML), the OU Mass Spectrometry, Proteomics & Metabolomics (MSPM) Core, the Oklahoma Medical Research Foundation (OMRF) Imaging Core Facility, and the Oklahoma COBRE in Cancer Imaging Research (Oklahoma Center of Medical Imaging for Translational Cancer Research). This work was supported in part by National Institutes of Health (NIH) MIRA R35 (1R35GM150758), NIH COBRE (P20GM135009), National Cancer Institute Cancer Center Support Grant (P30CA225520, COBRE P20GM103639), National Science Foundation CAREER (Grant 2048130), OCAST HR20–106, and the Oklahoma Tobacco Settlement Endowment Trust contract awarded to the University of Oklahoma Stephenson Cancer Center, and the Tissue Pathology and the Molecular Biology and Cytometry Research Shared Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Oklahoma Tobacco Settlement Endowment Trust.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.3c04171.
Tables comparing mixing techniques, device components, and device costs; physicochemical characterization data of nanoparticles; images and results from cell culture experiments; and experimental methods and materials (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.nanolett.3c04171
Contributor Information
Hamilton Young, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States.
Yuxin He, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States.
Bryan Joo, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States.
Sam Ferguson, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States.
Amberlynn Demko, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States.
Sarah K. Butterfield, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States
James Lowe, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States.
Nathan F. Mjema, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States
Vinit Sheth, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States.
Luke Whitehead, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States.
Maria J. Ruiz-Echevarria, Department of Pathology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma 73104, United States; Stephenson Cancer Center, Oklahoma City, Oklahoma 73104, United States
Stefan Wilhelm, Stephenson School of Biomedical Engineering, University of Oklahoma, Norman, Oklahoma 73019, United States; Institute for Biomedical Engineering, Science, and Technology (IBEST), Norman, Oklahoma 73019, United States; Stephenson Cancer Center, Oklahoma City, Oklahoma 73104, United States.
REFERENCES
- (1).Hou X; Zaks T; Langer R; Dong Y Lipid Nanoparticles for mRNA Delivery. Nat. Rev. Mater. 2021, 6 (12), 1078–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Tenchov R; Bird R; Curtze AE; Zhou Q Lipid Nanoparticles - From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15 (11), 16982–17015. [DOI] [PubMed] [Google Scholar]
- (3).Mitchell MJ; Billingsley MM; Haley RM; Wechsler ME; Peppas NA; Langer R Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Discov 2021, 20 (2), 101–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Barbier AJ; Jiang AY; Zhang P; Wooster R; Anderson DG The Clinical Progress of mRNA Vaccines and Immunotherapies. Nat. Biotechnol. 2022, 40 (6), 840–854. [DOI] [PubMed] [Google Scholar]
- (5).Jakubek ZJ; Chen S; Zaifman J; Tam YYC; Zou S Lipid Nanoparticle and Liposome Reference Materials: Assessment of Size Homogeneity and Long-Term – 70 and 4 °C Storage Stability. Langmuir 2023, 39 (7), 2509–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Crommelin DJA; Anchordoquy TJ; Volkin DB; Jiskoot W; Mastrobattista E Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2021, 110 (3), 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cheng Q; Wei T; Farbiak L; Johnson LT; Dilliard SA; Siegwart DJ Selective Organ Targeting (SORT) Nanoparticles for Tissue-Specific mRNA Delivery and CRISPR–Cas Gene Editing. Nat. Nanotechnol. 2020, 15 (4), 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lyon S Our COVID vaccines would not exist without this unsung Princeton technology. https://cbe.princeton.edu/news/our-covid-vaccines-would-not-exist-without-unsung-princeton-technology (accessed 2024-01-05). [Google Scholar]
- (9).Thorn CR; Sharma D; Combs R; Bhujbal S; Romine J; Zheng X; Sunasara K; Badkar A The Journey of a Lifetime — Development of Pfizer’s COVID-19 Vaccine. Curr. Opin Biotechnol 2022, 78, No. 102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Gindy ME; Leone AM; Cunningham JJ Challenges in the Pharmaceutical Development of Lipid-Based Short Interfering Ribonucleic Acid Therapeutics. Expert Opinion on Drug Delivery 2012, 9 (2), 171–182. [DOI] [PubMed] [Google Scholar]
- (11).Vogelaar A; Marcotte S; Cheng J; Oluoch B; Zaro J Use of Microfluidics to Prepare Lipid-Based Nanocarriers. Pharmaceutics 2023, 15 (4), 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Baas S; Saggiomo V Ender3 3D Printer Kit Transformed into Open, Programmable Syringe Pump Set. HardwareX 2021, 10, No. e00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Carugo D; Bottaro E; Owen J; Stride E; Nastruzzi C Liposome Production by Microfluidics: Potential and Limiting Factors. Sci. Rep 2016, 6 (1), No. 25876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yu B; Lee RJ; Lee LJ Microfluidic Methods for Production of Liposomes. Methods Enzymol 2009, 465, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhang G; Sun J Lipid in Chips: A Brief Review of Liposomes Formation by Microfluidics. IJN 2021, 16, 7391–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Vogelaar A; Marcotte S; Cheng J; Oluoch B; Zaro J Use of Microfluidics to Prepare Lipid-Based Nanocarriers. Pharmaceutics 2023, 15 (4), 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Berger N; Sachse A; Bender J; Schubert R; Brandl M Filter Extrusion of Liposomes Using Different Devices: Comparison of Liposome Size, Encapsulation Efficiency, and Process Characteristics. International journal of pharmaceutics 2001, 223, 55–68. [DOI] [PubMed] [Google Scholar]
- (18).Donahue ND; Vance EA; Sheth V; Francek ER; Wilhelm S Synthesis, Characterization, and Acute Cytotoxicity Evaluation of Chloroquine Encapsulating Liposomes. Micro 2023, 3 (1), 51–59. [Google Scholar]
- (19).Rezvantalab S; Keshavarz Moraveji M Microfluidic Assisted Synthesis of PLGA Drug Delivery Systems. RSC Adv. 2019, 9 (4), 2055–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Xu J; Zhang S; Machado A; Lecommandoux S; Sandre O; Gu F; Colin A Controllable Microfluidic Production of Drug-Loaded PLGA Nanoparticles Using Partially Water-Miscible Mixed Solvent Microdroplets as a Precursor. Sci. Rep 2017, 7 (1), 4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Precigenome LLC. PLGA Nanoparticles Synthesis System|Microfluidic Method, PreciGenome. https://www.precigenome.com/plga-nanoparticle-synthesis (accessed 2023-09-15).
- (22).Li X; Jiang X Microfluidics for Producing Poly (Lactic-Co-Glycolic Acid)-Based Pharmaceutical Nanoparticles. Adv. Drug Delivery Rev. 2018, 128, 101–114. [DOI] [PubMed] [Google Scholar]
- (23).Suk JS; Xu Q; Kim N; Hanes J; Ensign LM PEGylation as a Strategy for Improving Nanoparticle-Based Drug and Gene Delivery. Adv. Drug Delivery Rev. 2016, 99 (Part A), 28–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Quintanilla Rodriguez BS; Correa R Rosiglitazone. In StatPearls; StatPearls Publishing: Treasure Island, FL, 2023. [Google Scholar]
- (25).Bao Y; Maeki M; Ishida A; Tani H; Tokeshi M Effect of Organic Solvents on a Production of PLGA-Based Drug-Loaded Nanoparticles Using a Microfluidic Device. ACS Omega 2022, 7 (37), 33079–33086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Groner J; Tognazzi M; Walter M; Fleischmann D; Mietzner R; Ziegler CE; Goepferich AM; Breunig M Encapsulation of Pioglitazone into Polymer-Nanoparticles for Potential Treatment of Atherosclerotic Diseases. ACS Appl. Bio Mater. 2023, 6 (6), 2111–2121. [DOI] [PubMed] [Google Scholar]
- (27).Luigetti M; Romano A; Di Paolantonio A; Bisogni G; Sabatelli M Diagnosis and Treatment of Hereditary Transthyretin Amyloidosis (hATTR) Polyneuropathy: Current Perspectives on Improving Patient Care. Ther Clin Risk Manag 2020, 16, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wang X; Liu S; Sun Y; Yu X; Lee SM; Cheng Q; Wei T; Gong J; Robinson J; Zhang D; Lian X; Basak P; Siegwart DJ Preparation of Selective Organ-Targeting (SORT) Lipid Nanoparticles (LNPs) Using Multiple Technical Methods for Tissue-Specific mRNA Delivery. Nat. Protoc. 2023, 18, 265–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hattab D; Gazzali AM; Bakhtiar A Clinical Advances of siRNA-Based Nanotherapeutics for Cancer Treatment. Pharmaceutics 2021, 13 (7), 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hartl N; Adams F; Costabile G; Isert L; Doeblinger M; Xiao X; Liu R; Merkel O The Impact of Nylon-3 Copolymer Composition on the Efficiency of siRNA Delivery to Glioblastoma Cells. Nanomaterials 2019, 9, 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Yanar F; Mosayyebi A; Nastruzzi C; Carugo D; Zhang X Continuous-Flow Production of Liposomes with a Millireactor under Varying Fluidic Conditions. Pharmaceutics 2020, 12, 1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Erfle P; Riewe J; Bunjes H; Dietzel A Stabilized Production of Lipid Nanoparticles of Tunable Size in Taylor Flow Glass Devices with High-Surface-Quality 3D Microchannels. Micromachines (Basel) 2019, 10 (4), 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Corbin JM; Georgescu C; Wang L; Wren JD; Bieniasz M; Xu C; Asch AS; Ruiz Echevarría MJ An Unbiased Seed-Based RNAi Selection Screen Identifies Small RNAs That Inhibit Androgen Signaling and Prostate Cancer Cell Growth. Mol. Ther.– Nucleic Acids 2023, 33, 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


