Abstract
Substrate positioning dynamics (SPD) orients the substrate to reactive conformations in the active site, accelerating enzymatic reactions. However, it remains unknown whether SPD effects originate primarily from electrostatic perturbation inside the enzyme or can independently mediate catalysis with a significant non-electrostatic component. Here we investigated how the non-electrostatic component of SPD affects transition state stabilization. Using high-throughput enzyme modeling, we selected Kemp eliminase variants with similar electrostatics inside the enzyme but significantly different SPD. The kinetic parameters of these selected mutants were experimentally characterized. We observed a valley-shaped, two-segment linear correlation between the TS stabilization free energy (converted from kinetic parameters) and an index used to quantify SPD. Favorable SPD was observed for a distal mutant R154W, leading to the lowest activation free energy among the mutants tested. R154W involves an increased proportion of reactive conformations. These results indicate the contribution of the non-electrostatic component of SPD to mediating enzyme catalytic efficiency.
Keywords: Protein Dynamics, Enzyme Kinetics, Mutation Effect, Electrostatic Effect, Kemp Eliminase
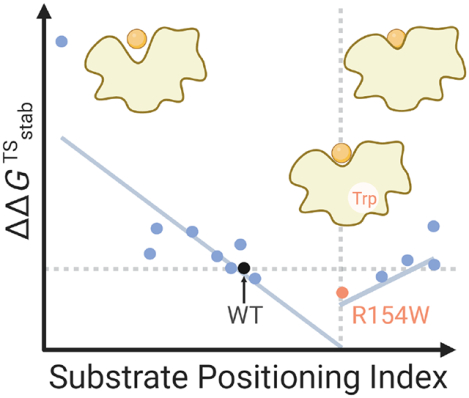
Graphical Abstract
Elucidating the catalytic origin of enzymes, a fundamental question in chemistry, guides the development of engineering strategies to create enzyme variants for chemical synthesis,1–3 waste degradation,4–7 fuel production,8–11 disease diagnosis, and treatment.12–14 Protein dynamics, which ranges over ten orders of magnitude,15–24 have been widely reported to mediate catalysis.17, 18, 25–28 For example, residue vibrations and collision have been proposed to facilitate transition state (TS) barrier crossing in the sub-picosecond time scale (e.g., lactate dehydrogenase, alcohol dehydrogenase, and purine nucleoside phosphorylase).15–17 Residue and loop motions have been proposed to facilitate the positioning of substrates to form reactive conformation (or near-attack conformation29) in the pico- to nanosecond time scale (e.g., dihydrofolate reductase, chitinase, β-lactamase, retro-aldolase, Kemp eliminase, glycoside hydrolase, Cytochrome P450, and soybean lipoxygenase).21–24, 30–37 Conformational change of loops and domains have been demonstrated to enable substrate binding, solvent shielding, or product releasing in the nanosecond to millisecond time scale (e.g., triosephosphate isomerase and adenylate kinase).18, 19, 38, 39
Substrate positioning dynamics (SPD) serves to orient the substrates for an energetically favorable barrier crossing and desired selectivity.33, 36, 40–52 Experimentally, the impact of SPD on catalysis has been investigated using mutagenesis, where mutating dynamically-important residues results in a significant rate reduction (e.g., Gly121 in dihydrofolate reductase53–56). Furthermore, rate-enhancing mutants have been created through optimizing SPD.49, 57 Broom et al. observed 700-fold rate acceleration in Kemp eliminase HG4 after multiple rounds of mutagenesis that turned out to rigidify the dynamic motion of active site residues.49 However, a major pitfall of this mutagenesis-based approach is that SPD is coupled to the electrostatics inside enzyme,25, 58 which is an established physical factor contributing to the high catalytic efficiency of enzymes based on theoretical,59 computational,34, 60–62 spectroscopy,63–65 kinetic and mutagenesis studies.58 Upon mutation, any change in SPD likely affects the projection of the enzyme electric field along the reacting bond. For instance, Wu et al. showed experimentally that the SPD mediates catalysis through tuning electrostatics in ketosteroid isomerase. This casts doubts on whether the correlation between the change of SPD and that of enzyme catalytic efficiency is confounded by electrostatics inside enzyme.63
We introduced eq. 1 and eq. 2 to quantitatively express the relationship between SPD and catalytic efficiency. Catalytic efficiency is represented by the transition state (TS) stabilization free energy,66–70 denoted as , which stands for the difference in free energy between the enzyme-catalyzed transition state and the transition state in water (eq. 1 and Scheme 1). This quantity is first introduced by Wolfenden using a thermodynamic box to describe the catalytic origin of enzyme.66, 68 The change of SPD affects through two components: electrostatic stabilization energy ΔGele(SPD) and a non-electrostatic component ΔGnonele(SPD) (eq. 2). To clarify, non-electrostatic component of SPD does not refer to non-electrostatic interactions between a substrate and enzyme residues but indicate the component of protein dynamics that mediates enzyme kinetics without perturbing interior electrostatics.
Scheme 1.
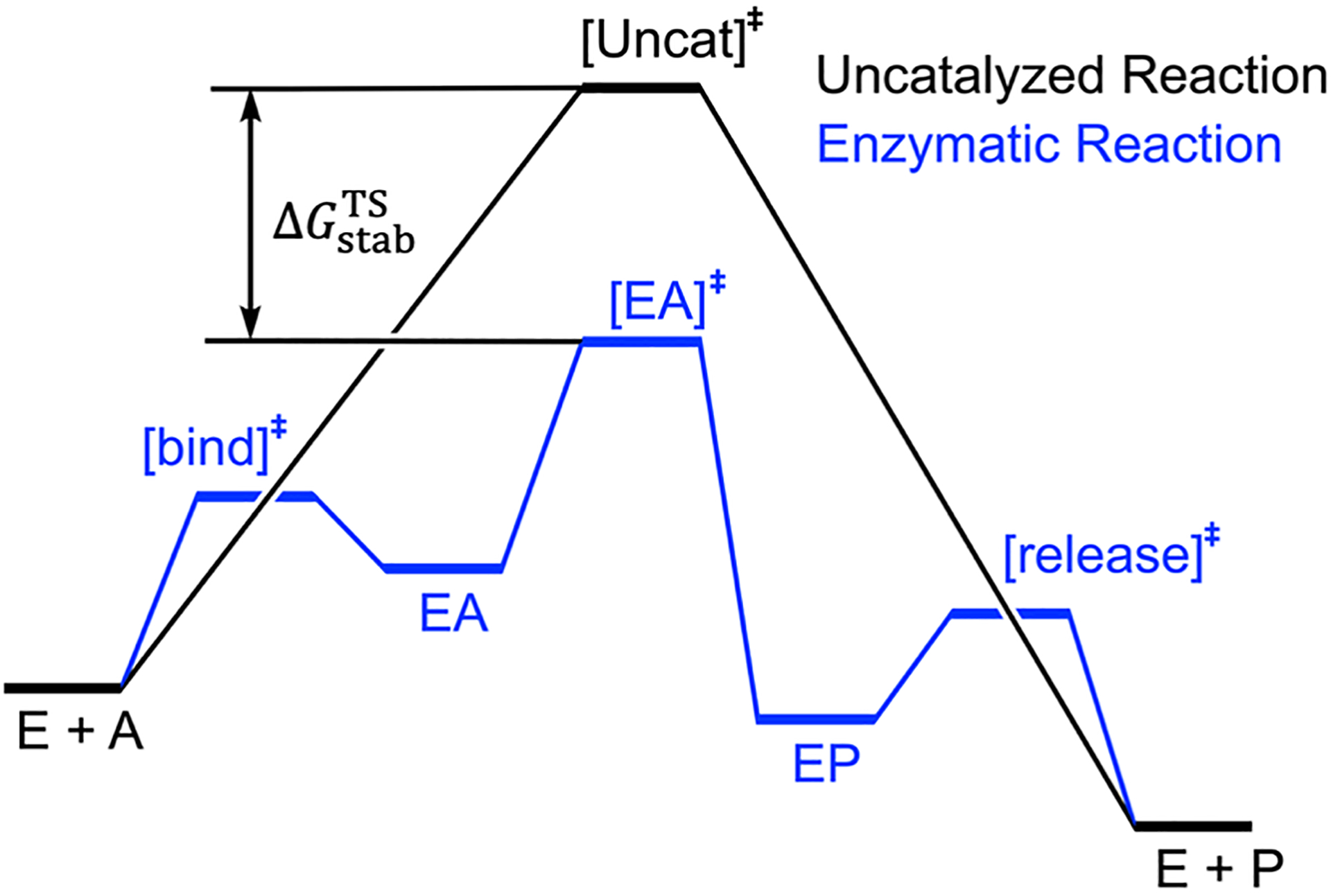
The definition of through a conceptual energy profile of enzymatic reaction (in blue) and uncatalyzed reaction (in black). [EA]‡ and [Uncat]‡ are transition states for the enzymatic reaction and uncatalyzed reaction, respectively. E, A, EA, EP, P are enzyme, substrate, enzyme-substrate complex, enzyme-product complex, and product, respectively. [bind]‡ and [release]‡ are transition states for the substrate binding and product release, respectively. is the transition state (TS) stabilization free energy.
| (1) |
| (2) |
Unless the impact of electrostatics is factored out in mutagenesis,27, 71–73 it will remain unknown whether SPD originates primarily from electrostatic perturbation inside enzyme (ΔGnonele(SPD) ≈ 0), or can independently mediate catalysis with a non-trivial ΔGnonele(SPD). The answer to the question will not only deepen our fundamental understanding of the catalytic origin of enzymes but also inform whether dynamics-related descriptors should be considered as a general and independent factor for the computational engineering of biocatalysts. In this work, we investigated how the non-electrostatic component of SPD, ΔGnonele(SPD), contributes to TS stabilization free energy, , using Kemp eliminase (KE07-R7–2) as a model enzyme.35, 74 Through in silico high-throughput screening by EnzyHTP,75 we identified single-point KE07-R7–2 mutants involving significantly different SPD but similar electrostatics inside enzyme (ΔGele(SPD) ≈ 0). We characterized the turnover rate and Michaelis constant of these mutants using biochemical assays. Based on these data, we investigated the correlation between the TS stabilization free energy and SPD to evaluate the contribution of ΔGnonele(SPD).
Model System: Kemp Eliminase.
We used Kemp eliminase (KE),34, 35, 39, 50, 61, 74, 76–79 the first de novo-designed enzyme, as the model enzyme in this study. The substrate 5-nitro-1,2-benzoxazole undergoes C–H deprotonation and ring opening catalyzed by a general base, generating 2-hydroxy-5-nitrobenzonitrile through a single transition state (Figure 1a top). In the KE07 series,74 the general acid-base mechanism is enabled by active site residues, including Ala9, Ile11, Ser48, Trp50, Glu101, Tyr128, His201, Arg202, and Lys222 (Figure 1a bottom). Specifically, Glu101 serves as the general base that deprotonates the substrate, Lys222 acts as the hydrogen bond donor to stabilize the phenoxide intermediate, and Trp50 acts as the π-stacking residue to stabilize the substrate binding and charge-separated transition state. Four polar residues (Ser48, Tyr128, His201, and Arg202) stabilize the substrate binding or transition state through electrostatic or polar interactions. The nonpolar residues Ala9 and Ile11 likely favor substrate binding via dispersion interactions.
Figure 1.
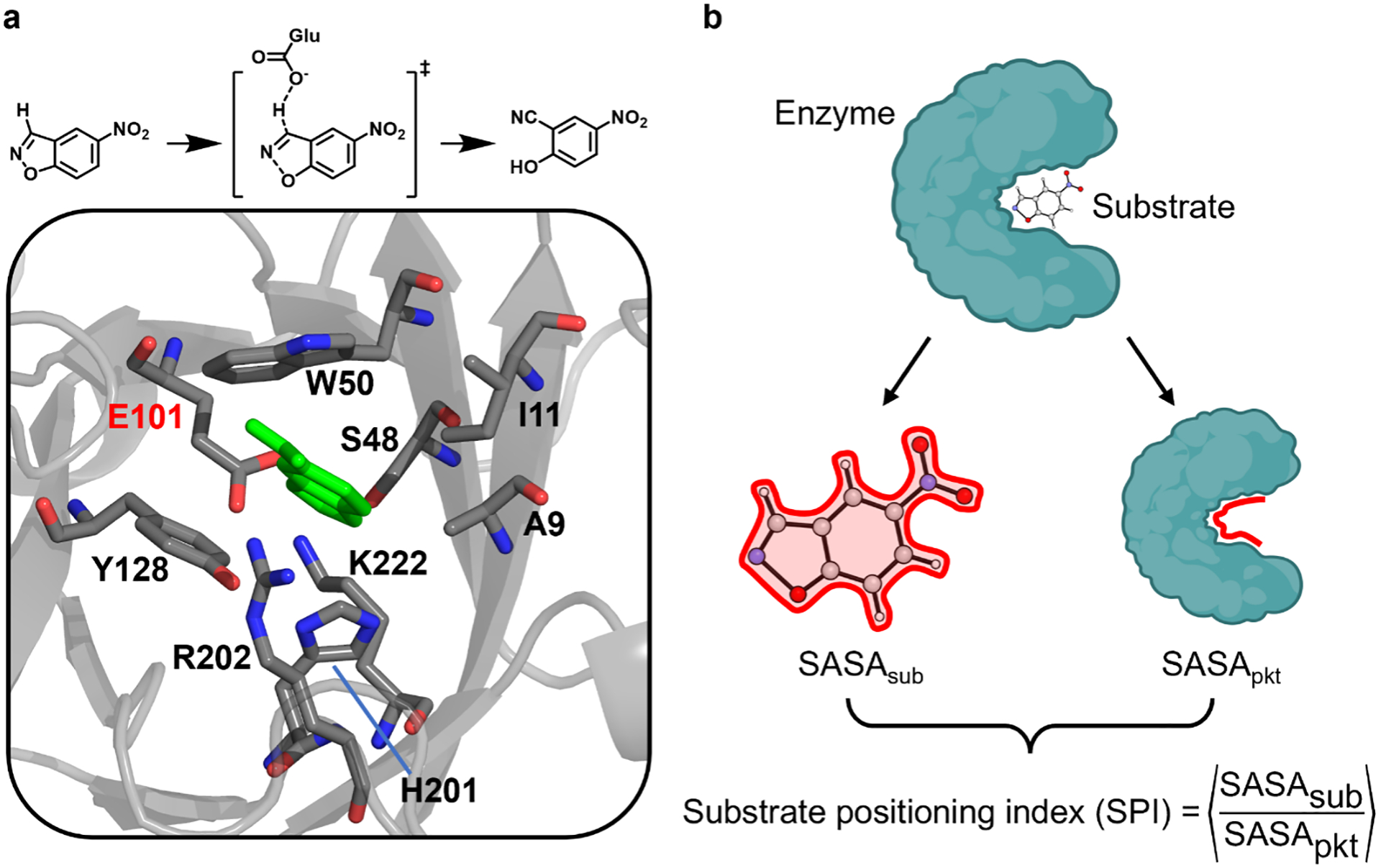
(a) The model enzyme, Kemp eliminase, used in the study and (b) the schematic definition of substrate positioning index, SPI. (a, top) A putative reaction mechanism. The carboxylic group of Glu101 deprotonates the C–H bond on the substrate, 5-nitro-1,2-benzoxazole. (a, bottom) The active site structure of Kemp eliminase, KE07-R7–2. The active site residues and the substrate are shown in stick. The substrate is shown in green, and the carbon, oxygen, and nitrogen of the residues are shown in gray, red, and blue, respectively. The catalytic base is labeled in red, and others are labeled in black. The structure is derived from the crystal structure with a PDB ID of 5D38.39 (b) The solvent-accessible surface area (SASA) of the substrate (SASAsub) and enzyme pocket (SASApkt) are evaluated separately by isolating these two parts from the complex. The SPI value is computed as an average over all snapshots sampled along the MD trajectories.
We employed KE07-R7–2, the KE variant out of seven rounds of directed evolution,35, 74 as the wild-type (WT) scaffold. Three reasons support this choice. First, computational benchmarks80 and biochemical assay protocols81 have been established for KE07-R7–2, ensuring accuracy and reproducibility. Second, kinetic parameters have been experimentally characterized by Bhowmick et al.35 for KE07-R7–2 mutants, providing references for this study. Third, both electrostatics inside the enzyme61 and protein dynamics35 are known to mediate catalytic efficiency of KE, making KE07-R7–2 a suitable model to investigate the electrostatic and non-electrostatic components of SPD.
Substrate Positioning Index.
To quantify the impact of protein dynamics on substrate positioning, we introduced a computational descriptor derived from molecular dynamics (MD) simulations. Existing descriptors for SPD, including mechanism-based bond parameters (e.g. length of a certain H-bond) and root-mean-square deviation (RMSD) of the active site, do not directly inform the dynamic response of substrate to the conformational fluctuation of the active site residues.44, 49 Instead, we defined a substrate positioning index (SPI) based on solvent-accessible surface area (SASA, and see Figure S5 for the solvent-exclusive surface area test). SPI was determined by averaging the SASA ratio values calculated from individual snapshots extracted from the trajectories. Specifically, the SASA ratio between the substrate and active site residues (listed in Figure 1a bottom, selected based on previous benchmark80) was first computed for each snapshot, with SASAsub and SASApkt calculated using isolated coordinates (as described in Text S1). Subsequently, these values were averaged across the conformational ensemble to derive the SPI value (i.e., <SASAsub/SASApkt>, Figure 1b).
When the same substrate binds to different enzyme mutants, a higher SPI value indicates that protein dynamics leads to a tighter positioning of the substrate in the active site. As such, SPI quantitatively describes SPD:
| (3) |
This descriptor was first introduced in our prior study of mutation effects in lactonase SsoPox,46 where a piecewise linear correlation was observed between the activation free energies and SPI values for various lactonase mutant-substrate pairs. An optimal range of SPI was identified that enables the non-native substrate to react as efficiently as the native substrate.
KE Mutant Selection.
We designed a high-throughput computational workflow to identify single amino acid mutations with significant variation in SPD (represented by SPI, eq. 3) but minimal change in electrostatics inside enzyme (Figure 2), therefore approximating ΔGele(SPD) to zero (eq. 2). Electrostatics inside enzyme is represented by the electric field (EF) change of the breaking C–H bond relative to the WT, ΔEFC–H (Text S1). Using EnzyHTP,75, 82 we constructed the high-throughput computational workflow to build structural models for 98 KE variants, identify thermally stable mutants using a folding stability test at room temperature (Table S1), and eventually perform a functional test to select mutations that perturb SPI significantly but electric field minimally (Figure 2a and Table S2).
Figure 2.
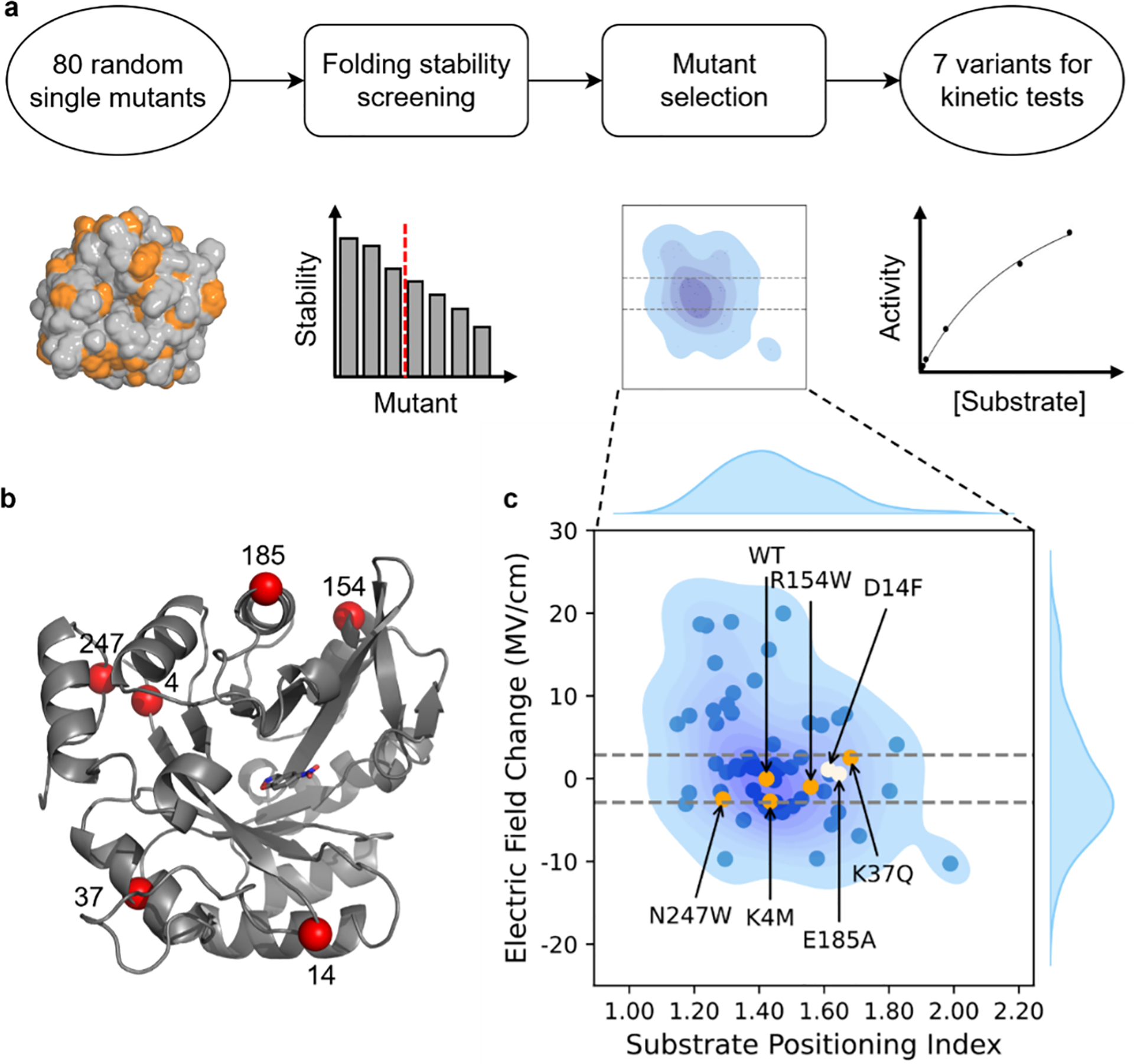
The computational protocol for the selection of KE variants for kinetic assessment. (a) Computational workflow to screen for mutants with a single amino acid substitution that significantly affects the substrate positioning dynamics but minimally the electric field strength on the breaking bond. (b) The structure of KE07 with the Cα atoms of mutation sites highlighted as red spheres. The structure is the KE07 design model by Röthlisberger et al.74 (c) Distribution of the electric field change, ΔEFC–H versus the substrate positioning index, SPI. The ΔEFC–H of WT is set to be the reference (0 MV/cm). The dashed lines show the EFC–H range cutoff from which mutants with a small electrostatic effect were selected. The dots include the data points for 63 KE variants, including 61 randomly generated variants that pass the folding stability test, E185A reported by Bhowmick et al.35, and the wild-type KE07-R7–2 enzyme. Orange and white dots are the mutants selected for the first and second rounds of kinetic characterization, respectively. Other mutants are shown in blue. The blue shade under the dots shows the data distribution in the 2D space and the curves with blue fill on the sides show the data distribution of EFC–H or SPI. The shade and curves are derived from fitting all 63 data points using Kernel Density Estimation (Text S1).
The 98 KE variants used in this study consist of 1 wild-type enzyme (KE07-R7–239, 74), 17 mutants reported by Bhowmick et al.35 and involved in our previous benchmark study,80 and 80 randomly-generated mutants using EnzyHTP.75 The 80 randomly generated mutants were first tested for folding stability using Rosetta cartesian_ddg83, 84 to minimize unexpressed and misfolded mutants. Through the stability test, 61 mutants were retained with their folding free energies of less than 10 Rosetta Energy Units. These mutants were used for the following MD simulations, in which the average SPI and ΔEFC–H were calculated using snapshots sampled from MD production trajectories. Structural constraints were applied throughout the MD simulations to enhance the sampling of enzyme conformations that stabilize pre-reaction complexes (Figure S1). To calculate ΔEFC2010033H, we first calculated the electric field strength projected along the breaking C–H bond of the substrate (EFC–H) in each MD snapshot. The EFC–H was summed at the middle point of the C–H bond from all partial charges of protein atoms. Then the relative electric field change, ΔEFC–H, was calculated as the difference between the average EFC–H of a mutant and the .
To factor out the impact of electrostatics, we selected the mutants whose averaged electric field strength was determined to be within ±2.88 MV/cm compared to the wild-type enzyme (KE07-R7–2). This range corresponds to the fluctuation of electrostatic stabilization energy (ΔGele) of ±0.1 kcal/mol, in which ΔGele was estimated by the projection of the electric field on the reacting dipole of C–H bond (Text S1). Within these variants, we further selected 5 KE variants for a kinetic assessment, including N247W, K4M, R154W, K37Q, and WT. These variants are evenly distributed across an SPI range from 1.30 to 1.70 (orange dots in Figure 2c) with an interval between 0.12 and 0.15.
After the first round of kinetic measurements, R154W was found to exhibit a 1.4-fold increase in kcat/KM compared to the WT (Tables S4 and S5). We further selected D14F (from our random mutants) and E185A (characterized by Bhowmick et al.35) for the second round of kinetic measurement (white dots in Figure 2c) because they contribute additional data points for mutants that possess an SPI value greater than that of R154W. These mutation sites are distant from the active site and from each other (Figure 2b). In addition to these 7 mutants, we identified another 7 mutants from the study of Bhowmick et al.,35 including H201A, M62A, N25S, K162A, K132M, H84Y, and L170A. Their projected electric field strengths also fall within ±2.88 MV/cm compared to KE07-R7–2. We thus obtained 14 variants for experimental characterization. Despite nearly identical averaged electric field strengths in the 14 mutants compared to the WT, a potential factor affecting the results may stem from multiple conformations, causing multimodal distribution of electric field strength. To test this possibility, we calculated the distributions of electric field strength for the 14 variants. Only single-peak EFC–H distribution is observed for all variants (Figure S2). This means that conformational diversity is unlikely to affect the results.
We measured the kinetic parameters for the 14 selected variants, and converted their turnover number kcat and Michaelis constant KM values to the change of TS stabilization free energy upon mutation85 (i.e.,) using eq. 4:
| (4) |
In this equation, KM approximates to be dissociation constant, kcat/Km approximates the apparent rate constant, and thus approximates the change of the apparent activation free energy upon mutation. According to Scheme 1, approximates to the change of upon mutation (i.e.,). Temperature T is set at 298 K and R represents the gas constant. Since ΔGnonele(SPD) ≈ 0, combining eq. 2–4 leads to:
| (5) |
This relation allows us to investigate the contribution of non-electrostatic component of substrate-positioning dynamics (ΔΔGnonele) directly from the change of TS stabilization free energy . Upon change of SPI, a trivial change in indicates that SPD originates primarily from electrostatic perturbation inside enzyme, while a significant change in indicates that SPD independent mediates catalysis with a substantial non-electrostatic component.
Non-electrostatic Component of Substrate Positioning Dynamics.
Based on eq. 5, we performed correlation analysis between and SPI to evaluate non-electrostatic component of SPD. A valley-shaped, two-segment piecewise linear correlation was observed between and SPI values for the 14 selected KE variants (Figure 3a). The first linear segment involves a gradual drop of from 1.7 kcal/mol (H201A) to −0.2 kcal/mol (R154W), which accompanies the increase of SPI value from 1.17 (H201A) to 1.56 (R154W). The second linear segment involves a gradual increase of from −0.2 kcal/mol (R154W) to 0.3 kcal/mol (K37Q), which accompanies the increase of SPI value from 1.56 (R154W) to 1.68 (K37Q). The Pearson correlation coefficients for the two linear segments are −0.83 and 0.83, respectively. R154W exhibits the most favorable value with an SPI of 1.56. The observed outcomes demonstrate a strong statistical significance, as the error bars of values show minimal overlap among the seven variants chosen from our computational mutagenesis (e.g., N247W, WT, K4M, R154W, D14F, E185A, and K37Q). The largest standard error is only 0.05 kcal/mol for N247W, which is half of the electrostatic stabilization energy selection window (i.e., 0.1 kcal/mol). Despite H201A appearing as an outlier, the removal of this data point does not disrupt the linear correlation on the left linear segment, which maintains a Pearson correlation coefficient of −0.82.
Figure 3.
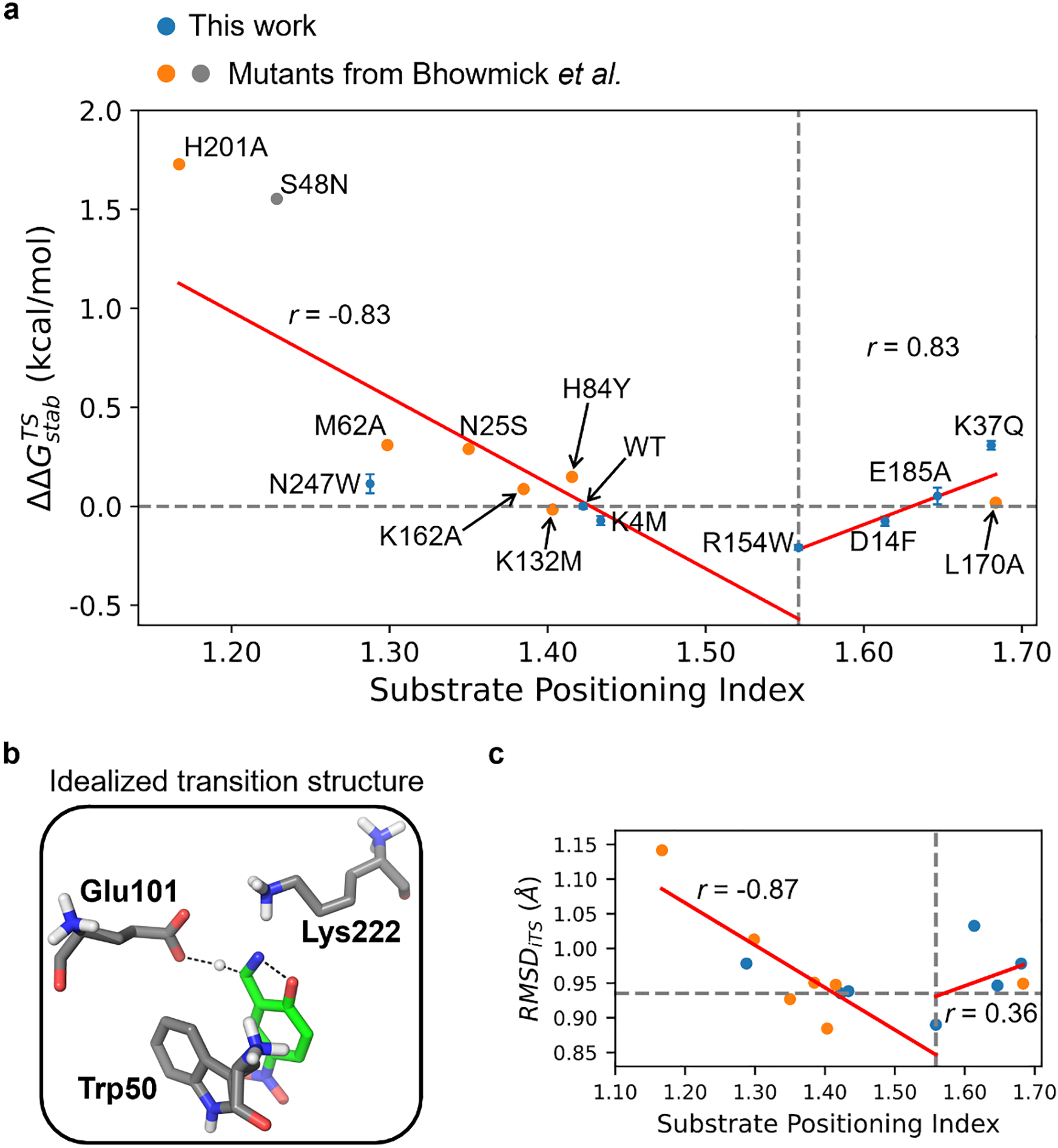
The impact of substrate positioning dynamics on the activation barrier and reaction conformation for KE07-R7–2 variants that are experimentally tested in this work (blue) and reported by Bhowmick et al.35 (orange and gray). (a) The correlation between change of activation free energy versus the substrate positioning index (SPI). For each data point tested in this work, the mean and standard error (shown as the vertical error bar) are derived from three independently repeated kinetic measurements. The horizontal dashed line indicates the position of . The vertical dashed line indicates the position of SPI = 1.56 where the beneficial mutant R154W is located. The vertical dashed line is also the boundary of the two-segment piecewise linear fitting. The fitting lines are shown in red and labeled with the corresponding Pearson correlation coefficient (r). The data point of R154W is included in both fitting lines. S48N is not included in the fitting because its electric field strength is outside the selection window of ±2.88 MV/cm. (b) Structure of the idealized transition state optimized from QM calculations.74 The dashed lines indicate the breaking or forming bonds. (c) Scatter plots for the root-mean-square deviation from the idealized transition state, i.e., RMSDiTS versus SPI of the selected KE07-R7–2 variants. The dashed and red lines are drawn similarly to (a). The horizontal dashed line indicates the value of WT RMSDiTS (0.94 Å).
The correlation shown in Figure 3a indicates the non-trivial contribution of non-electrostatic component of SPD in catalysis, with ΔΔGnonele changing from −0.2 to 1.7 kcal/mol upon variation of SPI. Although has been observed to change 3–7 kcal/mol upon variation of electric field inside enzyme through mutagenesis,64 an unfavorable SPD can substantially disrupt catalysis. As an example, we experimentally characterized the kinetic parameters for S48N (gray dot in Figure 3a). This mutant involves a favorable electrostatic environment (a ΔEFC–H of 5.68 MV/cm), but a small SPI value (1.23) that substantially deviates from the predicted optimal range (1.56). The value of S48N was measured to be +1.56 kcal/mol, which is >10-fold slower than the wild-type enzyme at room temperature. This negative impact of substrate-positioning dynamics is projected to be even worse if the favorable electrostatic contribution is factored out in S48N.
According to eq. 4, can be decomposed to contributions from and . To identify the key factor influencing the valley-shaped curve, we separately examined the correlation of SPI with and (Text S2 and Figure S6). The result shows that dominates the valley-shaped trend except for the outlier N247W, while exhibits minimal dependence on SPI. Notably, in our previous work for lactonase SsoPox, we observed a similar trend between and SPI.46 A recent study by Bååth et al.86 showed that the turnover of poly(ethylene terephthalate) hydrolases also follows a valley-shaped trend,87, 88 where the turnover rate is initially enhanced and then diminished as the monotonic decrease of enzyme-substrate binding affinity (Sabatier principle).
Substrate Positioning Dynamics Mediates the Sampling of Reactive Conformations.
The increase of SPI from 1.17 to 1.68 reflects the process in which mutations reshape protein dynamics, gradually positioning the substrate toward a more compact active-site conformation. As such, we hypothesized that the non-electrostatic component of SPD mediates enzyme kinetics by perturbing the population of reactive conformation. To validate this hypothesis, we calculated the mass-weighted RMSD relative to the active site of the idealized transition state (iTS),74 i.e., RMSDiTS, for each KE variant, and tested their correlation to SPI. RMSDiTS was calculated by considering all heavy substrate atoms, as well as Cα and side chains of Trp50, Glu101, and Lys222 amino acid residues, using the Röthlisberger et al. model as the reference structure74. The structural model of iTS is shown in Figure 3b. Using this structure as the reference, we calculated the mass-weighted RMSD of each MD snapshot as , where X represents the coordinate of an atom, i denotes the ith atom in this snapshot, and iTS denotes the corresponding atom in the reference iTS structure. mi is the mass of the ith atom. N is the total number of heavy atoms and M is the total mass. We have confirmed that RMSDiTS is an effective descriptor for reactive conformation population with a decent linear correlation (Pearson coefficient = 0.82, Figure S8).
As shown in Figure 3c, the correlation between the average RMSDiTS and SPI values also follows a valley-shaped, two-segment piecewise linear trend. The RMSDiTS decreases from 1.14 Å (H201A) to 0.88 Å (K132M) as SPI increases from 1.17 (H201A) to 1.56 (R154W) whereas RMSDiTS starts to increase beyond the SPI value of 1.56 and reaches a local maximum of 1.03 Å in D14F. The Pearson coefficients for the two fitting lines are −0.87 and 0.36. This result informs more physical details behind the valley-shaped correlation pattern. During the first linear segment, the increase of SPI leads to the reduction of active-site pocket space, which enhances the sampling of reactive conformations that resemble the active-site geometry of an idealized TS. However, when the pocket further shrinks and surpasses the optimal SPI range, the active site tends to populate in a non-reactive conformation that deviates significantly from the iTS. As such, the non-electrostatic component of SPD promotes enzyme kinetics by shifting the conformation ensemble towards TS-like geometries. This may help lower the conformational entropy cost during the transition from reactant to transition state. This two-segment piecewise linear correlation trend may exist universally in enzymes when electrostatic contributions are factored out, which is similar to the “volcano plot” or Sabatier principle broadly observed in catalysis89. The specific SPI value for optimal enzyme kinetics, however, is likely to be case-dependent.
To understand the molecular mechanism of how mutations mediate substrate positioning dynamics, we conducted conformational analyses on R154W (SPI = 1.56) and compared it against the results of WT (SPI = 1.42). As shown in Figure 4a, R154W is a remote mutation located on the surface of the enzyme and is spatially distant from the active site (i.e., ~19.4 Å away from the active site). Compared to the WT, the SASApkt of R154W decreases by 18.30 Å2. To identify which residue contributes the most to the change of active-site pocket, we decomposed the SASApkt into contributions of individual residues (Table S6). The decomposition shows that Trp50 contributes over 84% of the overall decrease. As shown in Figure 4b, the large reduction in SASApkt is driven by the shortening of spatial proximity between Trp50 and Ser144. This is supported by the downshift of distance distribution between Ser144 Oγ and Trp50 Hε upon mutation (average values: 4.45 Å in R154W; 6.87 Å in WT, Figure 4c left). Furthermore, the formation of hydrogen bond between Ser144 Oγ and Trp50 Hε is observed in R154W (around 2.72 Å) but is absent in WT.
Figure 4.
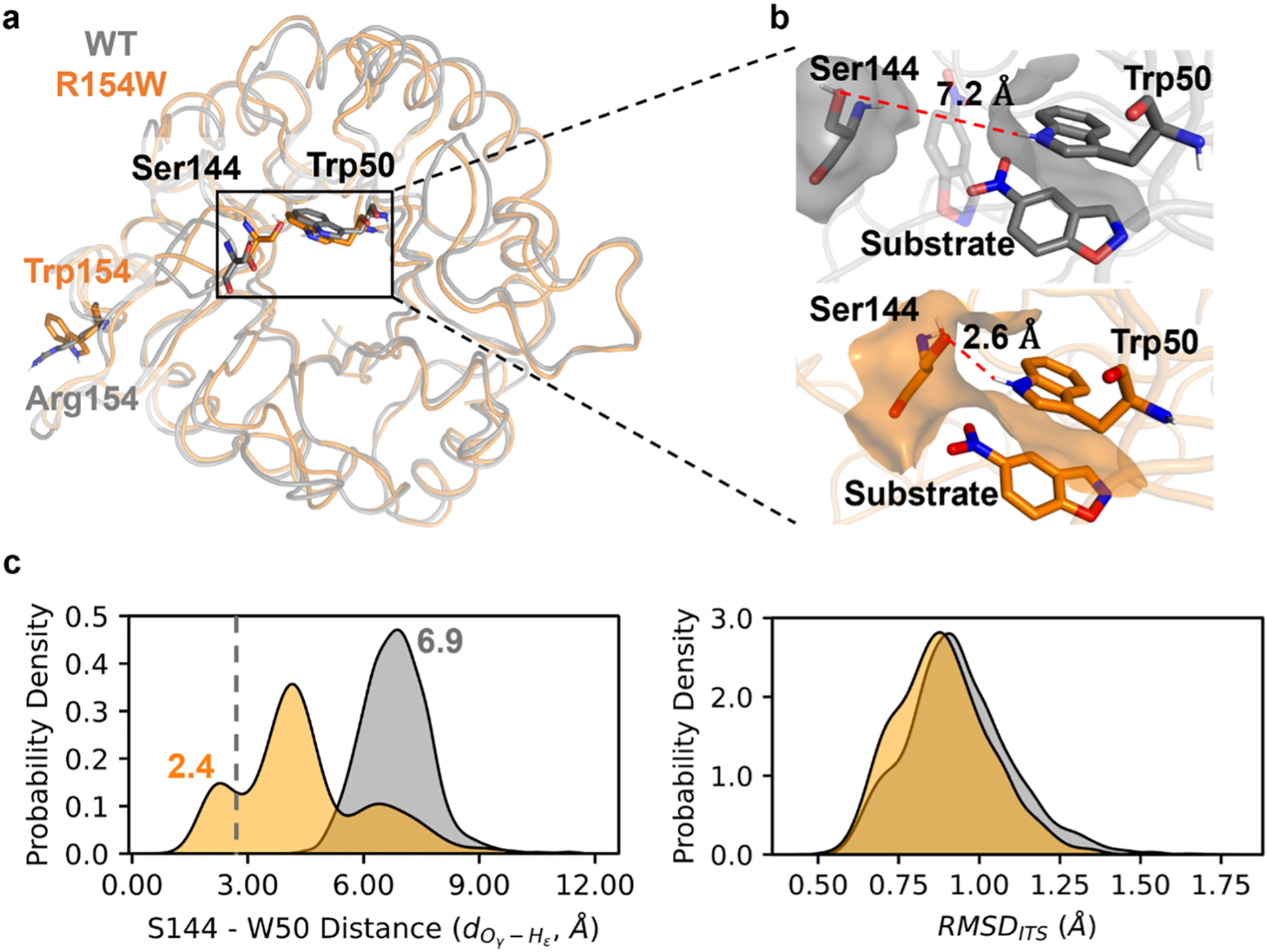
The molecular mechanism underlying the impact of R154W mutation on substrate positioning dynamics. (a) Structure overlay of the representative conformation of WT (gray) and R154W (orange) with the residues at the sites of 50, 144, and 154 shown in stick. The structures of the representative conformation are chosen from MD trajectories based on the S144 – W50 distance , that is, picked randomly from structures that have the distance in the range of 6.9 ± 0.3 and 2.4 ± 0.3 Å for WT and R154W, respectively. They are peak values of the distance distribution shown in Figure 4c left. (b) Surfaces of Trp50 and Ser144 in WT and R154W. The distance between Ser144 Oγ and Trp50 Hε, , is shown as the red dashed line. The opaque substrate indicates its favorable position in the active site, while the transparent substrate illustrates a potentially unfavorable position in the WT. This unfavorable position is a conceptual illustration since it is prohibited by the constraints applied during the MD simulation. (c) Distribution of the distance between Ser144 Oγ and Trp50 Hε, in the WT and R154W (left) and RMSD to the idealized transition station (right). In the distribution, the gray vertical dashed line represents the sum of van der Waals radii for oxygen and hydrogen (2.72 Å). The curves are made by fitting the corresponding data from all snapshots from MD using Kernel Density Estimation (Text S1).
Compared to WT, the close contact between Ser144 and Trp50 eliminates the accessible space of the substrate, forcing it to adopt a conformation that is parallel to the sidechain of Trp50 (Figure 4b). This conformation directs the breaking C–H bond towards the carboxylic group of Glu101. The RMSDiTS distribution of R154W shifts towards smaller values, generating more conformations that resemble the iTS (Figure 4c right). This likely reduces the activation entropy cost, which ultimately reduces the activation barrier. Notably, a similar phenomenon has been observed in HG3, another member of the Kemp eliminase family.57 Otten et al. showed that the evolved HG3 variants have more ordered side-chain orientations, leading to optimal positioning of the residues crucial to the chemical transformation and constraining of the ligand in the reactive pose.
In summary, we combined computational and experimental approaches to investigate the non-electrostatic component of substrate positioning dynamics (SPD) in mediating enzyme kinetics using Kemp eliminase (KE) as a model system. To quantitatively describe SPD, we introduced a molecular dynamic-derived descriptor, substrate positioning index (SPI), which is defined using the ratio of solvent-accessible surface area between the substrate and the enzyme active site residues. We designed a high-throughput computational workflow to identify stable KE variants that involve similar electrostatics inside enzyme but distinct SPI values.
The resulting KE variants were characterized using kinetic assays. The correlation between activation free energies and SPI values demonstrates a valley-shaped, two-segment piecewise linear relationship. The trend was validated using additional KE data reported by Bhowmick et al.35 The presence of an optimal SPI value was observed in R154W, which corresponds to the lowest activation free energy among the selected mutants. We further investigated the relationship between SPI and the root-mean-square deviation of each conformational ensemble from an idealized active-site transition state model. The results show that the non-electrostatic component of SPD promotes enzyme kinetics by shifting the conformation ensemble towards TS-like geometries. To understand the molecular details behind how mutation reshapes the SPD, we performed conformational analyses on R154W and compared the results against the WT. We found that this distal mutation has a significant impact on the conformational distribution at the active site, where the mutation enables a hydrogen bonding between Ser144 and Trp50, limiting the accessible space of the substrate and positioning the substrate towards chemical activation.
These results indicate the presence of a non-electrostatic component of SPD in mediating enzyme catalysis. To promote catalysis, SPD has to position the substrate in an optimal active-site cavity to favor barrier crossing. The study implies that SPD should be considered as an independent factor in developing strategies for pinpointing rate-enhancing mutants for biocatalysis. The study also highlights SPI as a descriptor that informs the impact of the mutation on substrate positioning dynamics. SPI can be easily calculated from molecular mechanical modeling and implemented in high-throughput computational workflows for computational enzyme engineering. On a separate note, statistical energy (EMaxEnt), which quantifies the fitness of a specific sequence in evolution, has been recently demonstrated to display a strong anti-correlation90 (correlation values are −0.88 and −0.89 for log(kcat/KM) and logkcat, respectively) to the corresponding activity (log(kcat/KM) or logkcat) in the KE variants reported by Bhowmick et al.35 Although EMaxEnt is different from SPI, further investigations into the relationship between substrate positioning dynamics and the evolutionary profile of sequences may inform the synergy between the free energy landscape and fitness landscape that mediates enzyme catalysis in evolution.57
Computational and Experimental Methods
Computational Methods.
We employed EnzyHTP, a software developed by our lab, to perform high-throughput computational screening of Kemp eliminase (KE) mutants.75 A job script was prepared that leveraged EnzyHTP functions to automate the process of enzyme structure preparation, random mutation generation,91 folding stability assessment,83, 84 molecular dynamics simulation using AMBER18,91 quantum mechanics calculation using Gaussian1692 and TeraChem,93, 94 and post-analysis of results using PyMol.95 The workflow starts from KE07-R7–2,39 the “wild-type” structure used in this study and then creates and simulates 98 random KE variants with single amino acid substitution. The configurations of the EnzyHTP functions are detailed in Text S1.
Experimental Methods and Characterization.
The enzymes were expressed in Escherichia coli BL21(DE3) using a pET-29b(+) vector (Novagen) and purified using Ni-NTA resin (Invitrogen). Kinetic parameters were determined using 5-nitro-1,2-benzoxazole as the substrate, with concentrations ranging from 5 to 1500 μM. The reactions were initiated by adding 50 μL of the enzyme (8 μM final concentration) to 150 μL of the substrate in a 96-well plate (Corning-Costa) at 25 °C in 25 mM HEPES (pH 7.25), 100 mM NaCl, 5% glycerol and 1.5% (v/v) acetonitrile.35 The formation of the product was monitored at 380 nm using a SpectraMax iD3 microplate reader (Molecular Device). Vmax and Km were calculated by nonlinear regression with the Michaelis-Menten model using GraphPad Prism software (Version 8).96 Three biologically independent replicates were used to calculate means and standard deviations. More details can be found in Text S1.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the startup grant from Vanderbilt University. Z. J. Yang, Y. Jiang, N. Ding, and Q. Shao are supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM146982. Z. J. Yang thanks the sponsorship from Rosetta Commons Seed Grant Award and the Dean’s Faculty Fellowship in the College of Arts and Science at Vanderbilt. S. L. Stull acknowledges financial support from the 24 Vanderbilt Undergraduate Summer Research Program and the Department of Computer Science. This work used SDSC Dell Cluster with AMD Rome HDR IB at Expanse from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) program, which is supported by National Science Foundation grants BIO200057.97
Footnotes
Supporting Information. The following files are available free of charge. Detailed computational and experimental methods; folding free energy change upon mutation for 80 randomly generated single mutants of KE07-R7–2; constraints applied in the MD simulation; values of molecular dynamics-derived descriptors for the 15 variants; distribution of the snapshot electric field along the substrate breaking C–H bond for 14 KE07-R7–2 variants; primer sequences used in this study; Sanger sequencing chromatograms depicting site-directed mutagenesis in KE07-R7–2 variants; SDS-PAGE analysis of the purified KE07-R7–2 variants; scatter plots for the efficiency-enhancing free energy barrier changes upon mutation versus the substrate positioning index calculated using solvent-exclusive surface area; experimentally characterized kinetic parameters of the purified KE07-R7–2 variants; values of computed kinetic parameters for the 15 variants; decomposition of the change of transition state stabilization free energy upon mutation into kcat and KM components; correlation between the kcat and KM components of the effective activation free energy change upon mutation versus the substrate positioning index; solvent-accessible surface area decomposition of WT, N247W, and R154W mutants; the molecular mechanism underlying the impact of N247W and R154W mutations on enzyme kinetics; scatter plots for the correlation between the change of activation free energy versus the root-mean-square deviation from the idealized transition state. (PDF)
MD input files; workflow python script. (ZIP)
The authors declare no competing financial interest.
REFERENCES
- (1).Koeller KM; Wong C-H Enzymes for Chemical Synthesis. Nature 2001, 409, 232–240, DOI: 10.1038/35051706. [DOI] [PubMed] [Google Scholar]
- (2).Strohmeier GA; Pichler H; May O; Gruber-Khadjawi M Application of Designed Enzymes in Organic Synthesis. Chem. Rev 2011, 111, 4141–4164, DOI: 10.1021/cr100386u From NLM Medline. [DOI] [PubMed] [Google Scholar]
- (3).Petchey MR; Grogan G Enzyme-Catalysed Synthesis of Secondary and Tertiary Amides. Adv. Synth. Catal 2019, 361, 3895–3914, DOI: 10.1002/adsc.201900694. [DOI] [Google Scholar]
- (4).Austin HP; Allen MD; Donohoe BS; Rorrer NA; Kearns FL; Silveira RL; Pollard BC; Dominick G; Duman R; El Omari K; et al. Characterization and Engineering of a Plastic-Degrading Aromatic Polyesterase. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E4350–E4357, DOI: 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Knott BC; Erickson E; Allen MD; Gado JE; Graham R; Kearns FL; Pardo I; Topuzlu E; Anderson JJ; Austin HP; et al. Characterization and Engineering of a Two-Enzyme System for Plastics Depolymerization. Proc. Natl. Acad. Sci. U. S. A 2020, 117, 25476–25485, DOI: 10.1073/pnas.2006753117 From NLM Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tiso T; Narancic T; Wei R; Pollet E; Beagan N; Schroder K; Honak A; Jiang M; Kenny ST; Wierckx N; et al. Towards Bio-Upcycling of Polyethylene Terephthalate. Metab. Eng 2021, 66, 167–178, DOI: 10.1016/j.ymben.2021.03.011 From NLM Medline. [DOI] [PubMed] [Google Scholar]
- (7).Ellis LD; Rorrer NA; Sullivan KP; Otto M; McGeehan JE; Román-Leshkov Y; Wierckx N; Beckham GT Chemical and Biological Catalysis for Plastics Recycling and Upcycling. Nat. Catal 2021, 4, 539–556, DOI: 10.1038/s41929-02100648-4. [DOI] [Google Scholar]
- (8).Chundawat SP; Beckham GT; Himmel ME; Dale BE Deconstruction of Lignocellulosic Biomass to Fuels and Chemicals. Annu. Rev. Chem. Biomol. Eng 2011, 2, 121–145, DOI: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- (9).Yang B; Dai Z; Ding S-Y; Wyman CE Enzymatic Hydrolysis of Cellulosic Biomass. Biofuels 2011, 2, 421–449, DOI: 10.4155/bfs.11.116. [DOI] [Google Scholar]
- (10).Sweeney MD; Xu F Biomass Converting Enzymes as Industrial Biocatalysts for Fuels and Chemicals: Recent Developments. Catalysts 2012, 2, 244–263. [Google Scholar]
- (11).Horn SJ; Vaaje-Kolstad G; Westereng B; Eijsink VG Novel Enzymes for the Degradation of Cellulose. Biotechnol. Biofuels 2012, 5, 45, DOI: 10.1186/1754-6834-545 From NLM PubMed-not-MEDLINE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Gordon SR; Stanley EJ; Wolf S; Toland A; Wu SJ; Hadidi D; Mills JH; Baker D; Pultz IS; Siegel JB Computational Design of an Alpha-Gliadin Peptidase. J. Am. Chem. Soc 2012, 134, 20513–20520, DOI: 10.1021/ja3094795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Sun S; Jiang D; Fan M; Li H; Jin C; Liu W Selection of a Versatile Lactobacillus Plantarum for Wine Production and Identification and Preliminary Characterisation of a Novel Histamine-Degrading Enzyme. Int. J. Food Sci. Technol 2020, 55, 2608–2618, DOI: 10.1111/ijfs.14514. [DOI] [Google Scholar]
- (14).Samadi N; Heiden D; Klems M; Salzmann M; Rohrhofer J; Weidmann E; Koidl L; Jensen-Jarolim E; Untersmayr E Gastric Enzyme Supplementation Inhibits Food Allergy in a BALB/c Mouse Model. Nutrients 2021, 13, DOI: 10.3390/nu13030738 From NLM Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Saen-oon S; Quaytman-Machleder S; Schramm VL; Schwartz SD Atomic Detail of Chemical Transformation at the Transition State of an Enzymatic Reaction. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 16543–16548, DOI: 10.1073/pnas.0808413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Schwartz SD; Schramm VL Enzymatic Transition States and Dynamic Motion in Barrier Crossing. Nat. Chem. Biol 2009, 5, 552–559, DOI: 10.1038/nchembio.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Schwartz SD Protein Dynamics and Enzymatic Catalysis. J. Phys. Chem. B 2023, DOI: 10.1021/acs.jpcb.3c00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Henzler-Wildman KA; Lei M; Thai V; Kerns SJ; Karplus M; Kern D A Hierarchy of Timescales in Protein Dynamics Is Linked to Enzyme Catalysis. Nature 2007, 450, 913–U927, DOI: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- (19).Henzler-Wildman KA; Thai V; Lei M; Ott M; Wolf-Watz M; Fenn T; Pozharski E; Wilson MA; Petsko GA; Karplus M; et al. Intrinsic Motions Along an Enzymatic Reaction Trajectory. Nature 2007, 450, 838–U813, DOI: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- (20).Hanson JA; Duderstadt K; Watkins LP; Bhattacharyya S; Brokaw J; Chu JW; Yang H Illuminating the Mechanistic Roles of Enzyme Conformational Dynamics. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 18055–18060, DOI: 10.1073/pnas.0708600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Bhabha G; Lee J; Ekiert DC; Gam J; Wilson IA; Dyson HJ; Benkovic SJ; Wright PE A Dynamic Knockout Reveals That Conformational Fluctuations Influence the Chemical Step of Enzyme Catalysis. Science 2011, 332, 234–238, DOI: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Agarwal PK; Billeter SR; Rajagopalan PTR; Benkovic SJ; Hammes-Schiffer S Network of Coupled Promoting Motions in Enzyme Catalysis. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 2794–2799, DOI: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Cannon WR; Singleton SF; Benkovic SJ A Perspective on Biological Catalysis. Nat. Struct. Biol 1996, 3, 821–833, DOI: 10.1038/nsb1096-821. [DOI] [PubMed] [Google Scholar]
- (24).Epstein DM; Benkovic SJ; Wright PE Dynamics of the Dihydrofolate-Reductase Folate Complex - Catalytic Sites and Regions Known to Undergo Conformational Change Exhibit Diverse Dynamical Features. Biochemistry 1995, 34, 11037–11048, DOI: 10.1021/bi00035a009. [DOI] [PubMed] [Google Scholar]
- (25).Welborn VV Structural Dynamics and Computational Design of Synthetic Enzymes. Chem. Catalysis 2022, 2, 19–28, DOI: 10.1016/j.checat.2021.10.009. [DOI] [Google Scholar]
- (26).Petrovic D; Kamerlin SCL Molecular Modeling of Conformational Dynamics and Its Role in Enzyme Evolution. Curr. Opin. Struct. Biol 2018, 52, 50–57, DOI: 10.1016/j.sbi.2018.08.004. [DOI] [PubMed] [Google Scholar]
- (27).Kamerlin SCL; Warshel A At the Dawn of the 21st Century: Is Dynamics the Missing Link for Understanding Enzyme Catalysis? Proteins 2010, 78, 1339–1375, DOI: 10.1002/prot.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Radkiewicz JL; Brooks CL Protein Dynamics in Enzymatic Catalysis: Exploration of Dihydrofolate Reductase. J. Am. Chem. Soc 2000, 122, 225–231, DOI: 10.1021/ja9913838. [DOI] [Google Scholar]
- (29).Hur S; Bruice TC The near Attack Conformation Approach to the Study of the Chorismate to Prephenate Reaction. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 12015–12020, DOI: 10.1073/pnas.1534873100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Watney JB; Agarwal PK; Hammes-Schiffer S Effect of Mutation on Enzyme Motion in Dihydrofolate Reductase. J. Am. Chem. Soc 2003, 125, 3745–3750, DOI: 10.1021/ja028487u. [DOI] [PubMed] [Google Scholar]
- (31).Hammes-Schiffer S Impact of Enzyme Motion on Activity. Biochemistry 2002, 41, 13335–13343, DOI: 10.1021/bi0267137. [DOI] [PubMed] [Google Scholar]
- (32).Ramanathan A; Agarwal PK Evolutionarily Conserved Linkage between Enzyme Fold, Flexibility, and Catalysis. Plos. Biol. 2011, 9, DOI: 10.1371/journal.pbio.1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Norberg AL; Dybvik AI; Zakariassen H; Mormann M; Peter-Katalinić J; Eijsink VGH; Sørlie M Substrate Positioning in Chitinase a, a Processive Chito-Biohydrolase from Serratia Marcescens. FEBS Lett. 2011, 585, 2339–2344, DOI: 10.1016/j.febslet.2011.06.002. [DOI] [PubMed] [Google Scholar]
- (34).Bhowmick A; Sharma SC; Head-Gordon T The Importance of the Scaffold for de Novo Enzymes: A Case Study with Kemp Eliminase. J. Am. Chem. Soc 2017, 139, 5793–5800, DOI: 10.1021/jacs.6b12265. [DOI] [PubMed] [Google Scholar]
- (35).Bhowmick A; Sharma SC; Honma H; Head-Gordon T The Role of Side Chain Entropy and Mutual Information for Improving the de Novo Design of Kemp Eliminases KE07 and KE70. Phys. Chem. Chem. Phys 2016, 18, 19386–19396, DOI: 10.1039/c6cp03622h From NLM Medline. [DOI] [PubMed] [Google Scholar]
- (36).Ruscio JZ; Kohn JE; Ball KA; Head-Gordon T The Influence of Protein Dynamics on the Success of Computational Enzyme Design. J. Am. Chem. Soc 2009, 131, 14111–14115, DOI: 10.1021/ja905396s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Thielges MC; Chung JK; Fayer MD Protein Dynamics in Cytochrome P450 Molecular Recognition and Substrate Specificity Using 2D IR Vibrational Echo Spectroscopy. J. Am. Chem. Soc 2011, 133, 3995–4004, DOI: 10.1021/ja109168h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Liao QH; Kulkarni Y; Sengupta U; Petrovic D; Mulholland AJ; van der Kamp MW; Strodel B; Kamerlin SCL Loop Motion in Triosephosphate Isomerase Is Not a Simple Open and Shut Case. J. Am. Chem. Soc 2018, 140, 15889–15903, DOI: 10.1021/jacs.8b09378. [DOI] [PubMed] [Google Scholar]
- (39).Hong NS; Petrovic D; Lee R; Gryn’ova G; Purg M; Saunders J; Bauer P; Carr PD; Lin CY; Mabbitt PD; et al. The Evolution of Multiple Active Site Configurations in a Designed Enzyme. Nat. Commun 2018, 9, 3900, DOI: 10.1038/s41467-018-06305-y From NLM Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hamre AG; Jana S; Reppert NK; Payne CM; Sorlie M Processivity, Substrate Positioning, and Binding: The Role of Polar Residues in a Family 18 Glycoside Hydrolase. Biochemistry 2015, 54, 7292–7306, DOI: 10.1021/acs.biochem.5b00830. [DOI] [PubMed] [Google Scholar]
- (41).Patra N; Ioannidis EI; Kulik HJ Computational Investigation of the Interplay of Substrate Positioning and Reactivity in Catechol O-Methyltransferase. Plos One 2016, 11, DOI: 10.1371/journal.pone.0161868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Hu SS; Offenbacher AR; Thompson EM; Gee CL; Wilcoxen J; Carr CAM; Prigozhin DM; Yang V; Alber T; Britt RD; et al. Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The “Undoing” of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network. J. Am. Chem. Soc 2019, 141, 1555–1567, DOI: 10.1021/jacs.8b10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Mehmood R; Qi HW; Steeves AH; Kulik HJ The Protein’s Role in Substrate Positioning and Reactivity for Biosynthetic Enzyme Complexes: The Case of SyrB2/SyrB1. ACS Catal. 2019, 9, 4930–4943, DOI: 10.1021/acscatal.9b00865. [DOI] [Google Scholar]
- (44).Yabukarski F; Biel JT; Pinney MM; Doukov T; Powers AS; Fraser JS; Herschlag D Assessment of Enzyme Active Site Positioning and Tests of Catalytic Mechanisms through X-Ray-Derived Conformational Ensembles. Proc. Natl. Acad. Sci. U. S. A 2020, 117, 33204–33215, DOI: 10.1073/pnas.2011350117 From NLM Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Mehmood R; Vennelakanti V; Kulik HJ Spectroscopically Guided Simulations Reveal Distinct Strategies for Positioning Substrates to Achieve Selectivity in Nonheme Fe(II)/Alpha-Ketoglutarate-Dependent Halogenases. ACS Catal. 2021, 11, 12394–12408, DOI: 10.1021/acscatal.1c03169. [DOI] [Google Scholar]
- (46).Jiang Y; Yan B; Chen Y; Juarez RJ; Yang ZJ Molecular Dynamics-Derived Descriptor Informs the Impact of Mutation on the Catalytic Turnover Number in Lactonase across Substrates. J. Phys. Chem. B 2022, 126, 2486–2495, DOI: 10.1021/acs.jpcb.2c00142 From NLM Medline. [DOI] [PubMed] [Google Scholar]
- (47).Siegel JB; Zanghellini A; Lovick HM; Kiss G; Lambert AR; Clair JLS; Gallaher JL; Hilvert D; Gelb MH; Stoddard BL; et al. Computational Design of an Enzyme Catalyst for a Stereoselective Bimolecular Diels-Alder Reaction. Science 2010, 329, 309–313, DOI: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Blomberg R; Kries H; Pinkas DM; Mittl PRE; Grutter MG; Privett HK; Mayo SL; Hilvert D Precision Is Essential for Efficient Catalysis in an Evolved Kemp Eliminase. Nature 2013, 503, 418−+, DOI: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- (49).Broom A; Rakotoharisoa RV; Thompson MC; Zarifi N; Nguyen E; Mukhametzhanov N; Liu L; Fraser JS; Chica RA Ensemble-Based Enzyme Design Can Recapitulate the Effects of Laboratory Directed Evolution in Silico. Nat. Commun 2020, 11, DOI: 10.1038/s41467-020-18619-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Khersonsky O; Kiss G; Rothlisberger D; Dym O; Albeck S; Houk KN; Baker D; Tawfik DS Bridging the Gaps in Design Methodologies by Evolutionary Optimization of the Stability and Proficiency of Designed Kemp Eliminase KE59. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 10358–10363, DOI: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Haataja T; Gado JE; Nutt A; Anderson NT; Nilsson M; Momeni MH; Isaksson R; Valjamae P; Johansson G; Payne CM; et al. Enzyme Kinetics by GH7 Cellobiohydrolases on Chromogenic Substrates Is Dictated by Non-Productive Binding: Insights from Crystal Structures and MD Simulation. FEBS J. 2023, 290, 379–399, DOI: 10.1111/febs.16602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Offenbacher AR; Sharma A; Doan PE; Klinman JP; Hoffman BM The Soybean Lipoxygenase-Substrate Complex: Correlation between the Properties of Tunneling-Ready States and Endor-Detected Structures of Ground States. Biochemistry 2020, 59, 901–910, DOI: 10.1021/acs.biochem.9b00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Cameron CE; Benkovic SJ Evidence for a Functional Role of the Dynamics of Glycine-121 of Dihydrofolate Reductase Obtained from Kinetic Analysis of a Site-Directed Mutant. Biochemistry 1997, 36, 15792–15800, DOI: 10.1021/bi9716231. [DOI] [PubMed] [Google Scholar]
- (54).Rajagopalan PTR; Lutz S; Benkovic SJ Coupling Interactions of Distal Residues Enhance Dihydrofolate Reductase Catalysis: Mutational Effects on Hydride Transfer Rates. Biochemistry 2002, 41, 12618–12628, DOI: 10.1021/bi026369d. [DOI] [PubMed] [Google Scholar]
- (55).Wang L; Tharp S; Selzer T; Benkovic SJ; Kohen A Effects of a Distal Mutation on Active Site Chemistry. Biochemistry 2006, 45, 1383–1392, DOI: 10.1021/bi0518242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Roston D; Kohen A; Doron D; Major DT Simulations of Remote Mutants of Dihydrofolate Reductase Reveal the Nature of a Network of Residues Coupled to Hydride Transfer. J. Comput. Chem 2014, 35, 1411–1417, DOI: 10.1002/jcc.23629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Otten R; Padua RAP; Bunzel HA; Nguyen V; Pitsawong W; Patterson M; Sui S; Perry SL; Cohen AE; Hilvert D; et al. How Directed Evolution Reshapes the Energy Landscape in an Enzyme to Boost Catalysis. Science 2020, 370, 1442–1446, DOI: 10.1126/science.abd3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Hanoian P; Liu CT; Hammes-Schiffer S; Benkovic S Perspectives on Electrostatics and Conformational Motions in Enzyme Catalysis. Acc. Chem. Res 2015, 48, 482–489, DOI: 10.1021/ar500390e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Warshel A Electrostatic Origin of the Catalytic Power of Enzymes and the Role of Preorganized Active Sites. J. Biol. Chem 1998, 273, 27035–27038, DOI: 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- (60).Welborn VV; Head-Gordon T Computational Design of Synthetic Enzymes. Chem. Rev 2019, 119, 6613–6630, DOI: 10.1021/acs.chemrev.8b00399. [DOI] [PubMed] [Google Scholar]
- (61).Vaissier V; Sharma SC; Schaettle K; Zhang T; Head-Gordon T Computational Optimization of Electric Fields for Improving Catalysis of a Designed Kemp Eliminase. ACS Catal. 2018, 8, 219–227, DOI: 10.1021/acscatal.7b03151. [DOI] [Google Scholar]
- (62).Yang ZY; Liu F; Steeves AH; Kulik HJ Quantum Mechanical Description of Electrostatics Provides a Unified Picture of Catalytic Action across Methyltransferases. J. Phys. Chem. Lett 2019, 10, 3779–3787, DOI: 10.1021/acs.jpclett.9b01555. [DOI] [PubMed] [Google Scholar]
- (63).Wu YF; Fried SD; Boxer SG A Preorganized Electric Field Leads to Minimal Geometrical Reorientation in the Catalytic Reaction of Ketosteroid Isomerase. J. Am. Chem. Soc 2020, 142, 9993–9998, DOI: 10.1021/jacs.0c00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Fried SD; Boxer SG Electric Fields and Enzyme Catalysis. Annu. Rev. Biochem 2017, 86, 387–415, DOI: 10.1146/annurev-biochem-061516-044432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Fried SD; Boxer SG Measuring Electric Fields and Noncovalent Interactions Using the Vibrational Stark Effect. Acc. Chem. Res 2015, 48, 998–1006, DOI: 10.1021/ar500464j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Wolfenden R Transition State Analog Inhibitors and Enzyme Catalysis. Annu. Rev. Biophys. Bioeng 1976, 5, 271–306, DOI: 10.1146/annurev.bb.05.060176.001415. [DOI] [PubMed] [Google Scholar]
- (67).Bruice TC; Benkovic SJ Chemical Basis for Enzyme Catalysis. Biochemistry 2000, 39, 6267–6274, DOI: 10.1021/bi0003689. [DOI] [PubMed] [Google Scholar]
- (68).Schramm VL Enzymatic Transition States and Transition State Analogues. Curr. Opin. Struct. Biol 2005, 15, 604–613, DOI: 10.1016/j.sbi.2005.10.017. [DOI] [PubMed] [Google Scholar]
- (69).Simón L; Goodman JM Hydrogen-Bond Stabilization in Oxyanion Holes: Grand Jeté to Three Dimensions. Org. Biomol. Chem 2012, 10, 1905–1913, DOI: 10.1039/c2ob06717j. [DOI] [PubMed] [Google Scholar]
- (70).Burschowsky D; van Eerde A; Ökvist M; Kienhöfer A; Kast P; Hilvert D; Krengel U Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 17516–17521, DOI: 10.1073/pnas.1408512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Warshel A; Sharma PK; Kato M; Xiang Y; Liu HB; Olsson MHM Electrostatic Basis for Enzyme Catalysis. Chem. Rev 2006, 106, 3210–3235, DOI: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- (72).Lameira J; Bora RP; Chu ZT; Warshel A Methyltransferases Do Not Work by Compression, Cratic, or Desolvation Effects, but by Electrostatic Preorganization. Proteins 2015, 83, 318–330, DOI: 10.1002/prot.24717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Jindal G; Warshel A Misunderstanding the Preorganization Concept Can Lead to Confusions About the Origin of Enzyme Catalysis. Proteins 2017, 85, 2157–2161, DOI: 10.1002/prot.25381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Rothlisberger D; Khersonsky O; Wollacott AM; Jiang L; DeChancie J; Betker J; Gallaher JL; Althoff EA; Zanghellini A; Dym O; et al. Kemp Elimination Catalysts by Computational Enzyme Design. Nature 2008, 453, 190–195, DOI: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- (75).Shao Q; Jiang Y; Yang ZJ EnzyHTP: A High-Throughput Computational Platform for Enzyme Modeling. J. Chem. Inf. Model 2022, 62, 647–655, DOI: 10.1021/acs.jcim.1c01424. [DOI] [PubMed] [Google Scholar]
- (76).Alexandrova AN; Rothlisberger D; Baker D; Jorgensen WL Catalytic Mechanism and Performance of Computationally Designed Enzymes for Kemp Elimination. J. Am. Chem. Soc 2008, 130, 15907–15915, Article, DOI: 10.1021/ja804040s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Khersonsky O; Rothlisberger D; Dym O; Albeck S; Jackson CJ; Baker D; Tawfik DS Evolutionary Optimization of Computationally Designed Enzymes: Kemp Eliminases of the KE07 Series. J. Mol. Biol 2010, 396, 1025–1042, DOI: 10.1016/j.jmb.2009.12.031. [DOI] [PubMed] [Google Scholar]
- (78).Caselle EA; Yoon JH; Bhattacharya S; Rempillo JJL; Lengyel Z; D’Souza A; Moroz YS; Tolbert PL; Volkov AN; Forconi M; et al. Kemp Eliminases of the Alleycat Family Possess High Substrate Promiscuity. ChemCatChem 2019, 11, 1425–1430, DOI: 10.1002/cctc.201801994 From NLM PubMed-not-MEDLINE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Wang PY; Zhang J; Zhang SY; Lu DN; Zhu YS Using High-Throughput Molecular Dynamics Simulation to Enhance the Computational Design of Kemp Elimination Enzymes. J. Chem. Inf. Model 2023, 63, 1323–1337, DOI: 10.1021/acs.jcim.3c00002. [DOI] [PubMed] [Google Scholar]
- (80).Jiang Y; Stull SL; Shao Q; Yang ZJ Convergence in Determining Enzyme Functional Descriptors across Kemp Eliminase Variants. Electron. Struct 2022, 4, DOI: 10.1088/2516-1075/acad51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Gutierrez-Rus LI; Alcalde M; Risso VA; Sanchez-Ruiz JM Efficient Base-Catalyzed Kemp Elimination in an Engineered Ancestral Enzyme. Int. J. Mol. Sci 2022, 23, DOI: 10.3390/ijms23168934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Shao Q; Jiang Y; Yang ZJ EnzyHTP Computational Directed Evolution with Adaptive Resource Allocation. J. Chem. Inf. Model 2023, DOI: 10.1021/acs.jcim.3c00618 From NLM Publisher. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Park H; Bradley P; Greisen P; Liu Y; Mulligan VK; Kim DE; Baker D; DiMaio F Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. J. Chem. Theory Comput 2016, 12, 6201–6212, DOI: 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Frenz B; Lewis SM; King I; DiMaio F; Park H; Song YF Prediction of Protein Mutational Free Energy: Benchmark and Sampling Improvements Increase Classification Accuracy. Front. Bioeng. Biotech 2020, 8, DOI: 10.3389/fbioe.2020.558247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Park C Visual Interpretation of the Meaning of Kcat/Km in Enzyme Kinetics. J. Chem. Educ 2022, DOI: 10.1021/acs.jchemed.1c01268. [DOI] [Google Scholar]
- (86).Baath JA; Jensen K; Borch K; Westh P; Kari J Sabatier Principle for Rationalizing Enzymatic Hydrolysis of a Synthetic Polyester. JACS Au 2022, 2, 1223–1231, DOI: 10.1021/jacsau.2c00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Schaller KS; Molina GA; Kari J; Schiano-Di-Cola C; Sorensen TH; Borch K; Peters GHJ; Westh P Virtual Bioprospecting of Interfacial Enzymes: Relating Sequence and Kinetics. ACS Catal. 2022, 12, 7427–7435, DOI: 10.1021/acscatal.2c02305. [DOI] [Google Scholar]
- (88).Kari J; Schaller K; Molina GA; Borch K; Westh P The Sabatier Principle as a Tool for Discovery and Engineering of Industrial Enzymes. Curr. Opin. Biotechnol 2022, 78, DOI: 10.1016/j.copbio.2022.102843. [DOI] [PubMed] [Google Scholar]
- (89).Wodrich MD; Sawatlon B; Busch M; Corminboeuf C The Genesis of Molecular Volcano Plots. Acc. Chem. Res 2021, 54, 1107–1117, DOI: 10.1021/acs.accounts.0c00857. [DOI] [PubMed] [Google Scholar]
- (90).Xie WJ; Warshel A Natural Evolution Provides Strong Hints About Laboratory Evolution of Designer Enzymes. Proc. Natl. Acad. Sci. U. S. A 2022, 119, DOI: 10.1073/pnas.2207904119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Amber 2018; University of California, San Francisco, 2018. [Google Scholar]
- (92).Gaussian 16 Rev. C.01; Wallingford, CT, 2016. [Google Scholar]
- (93).Ufimtsev IS; Martinez TJ Quantum Chemistry on Graphical Processing Units. 3. Analytical Energy Gradients, Geometry Optimization, and First Principles Molecular Dynamics. J. Chem. Theory Comput 2009, 5, 2619–2628, DOI: 10.1021/ct9003004 From NLM PubMed-not-MEDLINE. [DOI] [PubMed] [Google Scholar]
- (94).Titov AV; Ufimtsev IS; Luehr N; Martinez TJ Generating Efficient Quantum Chemistry Codes for Novel Architectures. J. Chem. Theory Comput 2013, 9, 213–221, DOI: 10.1021/ct300321a From NLM PubMed-not-MEDLINE. [DOI] [PubMed] [Google Scholar]
- (95).The Pymol Molecular Graphics System , Version 2.4; 2015. [Google Scholar]
- (96).Graphpad Prism Version 8.3.1 for Macos; 2019.
- (97).Towns J; Cockerill T; Dahan M; Foster I; Gaither K; Grimshaw A; Hazlewood V; Lathrop S; Lifka D; Peterson GD; et al. XSEDE: Accelerating Scientific Discovery. Comput. Sci. Eng 2014, 16, 62–74, DOI: 10.1109/Mcse.2014.80. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


