Abstract
The complete genome sequences of a number of diverse members of the Baculoviridae including both nucleopolyhedroviruses (NPVs) and granuloviruses (GVs) revealed that they lack a homolog of GP64, the envelope fusion protein of the budded form of Autographa californica multinucleocapsid NPV (AcMNPV) and its close relatives. Computer-assisted analyses of the genome of one of these viruses, Lymantria dispar MNPV (LdMNPV), revealed a single open reading frame (ld130) whose product had the predicted properties of a membrane protein. Characterization of the localization of the products of the full-length ld130 gene and of an ld130-enhanced green fluorescent protein gene (egfp) fusion using both immunofluorescence and fluorescence microscopy revealed that LD130 accumulates at the plasma membranes of cells infected with LdMNPV or transfected with ld130-egfp. In addition, cells transfected with either ld130 or ld130-egfp or infected with wild-type virus undergo membrane fusion at pH 5. Western blot analyses indicate that LD130 is present in infected cells as an 83-kDa protein and is also present in budded virions as a protein doublet containing bands of 81 and 83 kDa. Tunicamycin treatment of infected cells resulted in an immunoreactive band of about 72 kDa, indicating that LD130 is N-glycosylated. Whereas the distribution of gp64 appears to be confined to a relatively closely related group of NPVs, homologs of ld130 are present in a diverse number of both NPVs and GVs. This suggests that LD130 may be the primordial baculovirus envelope fusion protein.
The Baculoviridae are a large family of occluded, rod-shaped viruses with circular, supercoiled, double-stranded DNA genomes of 100 to 180 kb depending on the virus strain. Two baculovirus genera have been described; the nucleopolyhedroviruses (NPVs) (34) have multiple virions present in large polyhedron-shaped occlusion bodies, whereas the granuloviruses (GVs) (42) normally display a single nucleocapsid embedded in a small granular occlusion body. NPVs are characterized by the production of two virion forms, or phenotypes (40). Occlusion-derived virions are found in occlusion bodies and initiate infection in midgut cells upon ingestion by susceptible hosts. In contrast, the budded virus (BV) form is not occluded, is produced early in infection, and spreads the infection within the infected insect prior to occlusion body production. Although the nucleocapsids for the two virion forms appear to have similar polypeptide contents, the compositions and structures of their envelopes appear to be distinct (7). BV infection by both Autographa californica multinucleocapsid NPV (AcMNPV) and Orgyia pseudotsugata MNPV (OpMNPV) has been investigated, and a glycoprotein called GP64 has been found to be associated with the BV form of both these viruses (3, 39, 41). GP64 is expressed both early and late in infection and is transported to and incorporated into the cell membrane. As nucleocapsids bud through the cell membrane and exit the cell, they become enveloped in the GP64-modified cell membrane. GP64 appears to be required for the spread of the infection to other cells and for the virus to exit from an infected cell (23, 26, 28). BV enters cells via an endocytic pathway, and, upon acidification of the endocytic vesicle, the structure of GP64 is altered and fusion of the viral and endosomal membranes occurs (17, 20, 22). This results in the release of the nucleocapsid into the cytoplasm. Surprisingly, GP64 is closely related to the envelope fusion protein of the Thogoto virus genus of the Orthomyxoviridae, which are minus strand RNA viruses (24, 25).
Recently the sequences of the 161-kb Lymantria dispar MNPV (LdMNPV) (18), 136-kb Spodoptera exigua MNPV (15), and 178-kb Xestia c-Nigrum GV (XcGV) (11) genomes have been reported. A striking feature of these genomes is the lack of an identifiable homolog to gp64. The fact that the life cycle of LdMNPV is similar to those of AcMNPV and OpMNPV suggested that it must express a different envelope fusion protein. Computer-assisted analyses of the LdMNPV genome identified a single 676-amino-acid product of LdMNPV open reading frame (ORF) 130 (ld130), which was predicted to have both a signal sequence and a transmembrane domain (18). In this report we demonstrate that LD130 has the properties of a low-pH-dependent envelope fusion protein. In addition, we describe its location in infected and transfected cells and its association with BV.
MATERIALS AND METHODS
Virus and cell lines.
The LdMNPV strains 56-1 (36) and A21 (2, 37) were used for infections and were propagated in L. dispar cell line Ld-652Y grown in TNM-FH media (38) supplemented with 10% fetal bovine serum, penicillin G (50 U/ml), streptomycin (50 μg/ml; Whittaker Bioproducts), and amphotericin B (Fungizone; 375 ng/ml; Flow Laboratories). Spodoptera frugiperda (Sf-9) cells were cultured in SF900II media (Gibco-BRL) as previously described (9).
Transfection.
Log phase insect cells were seeded into 35-mm-diameter plates at a density of 0.8 × 106 cells/well. After incubation overnight to allow attachment, the cells were transfected at 27°C with 7 μg of plasmid DNA using Cellfectin liposomes (Gibco-BRL). After 5 h the transfection mixture was replaced with TNM-FH medium, and the cells were further incubated at 27°C.
PCR, cloning, DNA sequence analysis, and DNA purification.
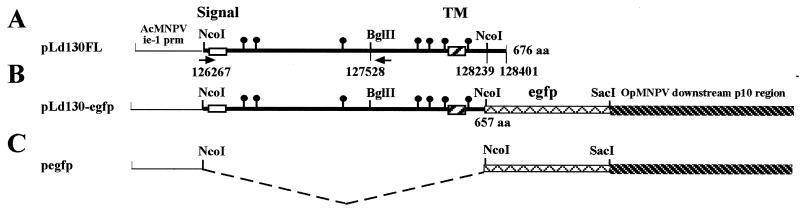
Clones of ld130 were constructed as follows. Two primers to the ld130 region were synthesized. In one (5′ ATT CGT CGC CAT GGC GCC G) three nucleotides (nt) (underlined) were altered, creating an NcoI site containing the ATG initiation codon and changing the second codon from specifying S to A. The other primer (5′ CAG CCA GTT GTT GTA GTC GG) started at nt 127546 just downstream of a BglII site at nt 127528 (18) (Fig. 1A). PCR, using LdMNPV cosmid F (P288) (29, 36) as the template, was performed according to the Perkin-Elmer, Inc., GeneAmp protocol using Amplitaq DNA polymerase in 2 mM MgCl2. Template DNA (10 ng) and primers (20 μM each) were subjected to the following: 1 cycle of 2 min at 95°C, 35 cycles of 60 s at 95°C and 60 s at 60°C, and a final cycle of 7 min at 72°C. The resulting product was digested with NcoI and BglII and gel purified. Expression plasmid pBSie-1pro was constructed by cloning an AcMNPV SphI-EcoRV fragment (nt 126726 to 127352) (1) containing the promoter and the first 155 nt of ie-1, with bases surrounding the ATG modified to produce an NcoI site (30), into SphI-HincII-cut pBS. The purified ld130 PCR product was cloned into NcoI-BglII-cut pBSie-1pro. A BglII-BamHI fragment (nt 127528 to 128401) from cosmid F containing the 3′ region of ld130 was then inserted into the BglII site, and a clone containing this fragment in the correct orientation was isolated. The resulting clone, called pLd130FL (Fig. 1A), contained the full-length ld130 gene under the control of the AcMNPV ie-1 promoter as well as 104 nt of the 3′ ld130 untranslated sequence. Sequence analysis showed that the sequence was correct. An enhanced green fluorescent protein (EGFP) expression plasmid was constructed by first cloning a 717-bp BamHI-NotI fragment containing the egfp gene from pEGFP-N1 (Clontech, Inc.) into BamHI-NotI-cut pBKS (Stratagene, Inc.) in order to obtain a SacI site on the 3′ end. This plasmid was then cut with NcoI and SacI, and the egfp gene fragment was cloned into an NcoI-SacI-cut plasmid containing the AcMNPV ie-1 promoter (nt 126600 to 127198) upstream of the NcoI site and the OpMNPV p10 downstream flanking region (nt 112503 to 114068) downstream of the SacI site (30) (Fig. 1C). This plasmid (pegfp) was cut with NcoI and ligated with a 1,976-bp fragment purified from NcoI-cut Ld130FL. The resulting construct, pLd130-egfp (Fig. 1B), encodes a protein which lacks the 17 C-terminal amino acids located downstream of the predicted transmembrane domain.
FIG. 1.
Maps of plasmids used to characterize LD130. The numbers for the ld130 region are from Kuzio et al. (18). Dashed line, sequence not present; solid circles, locations of N-glycosylation consensus sequences. The locations of the predicted signal and transmembrane (TM) domains are shown. Arrows (A) show locations of the 5′ and 3′ PCR primers. The numbers of amino acids (aa) in the full-length LD130 protein (A) and EGFP fusion (B) are shown. AcMNPV ie-1 prm shows the position of the ie-1 promoter used for expressing all three constructs.
All plasmid DNAs used for transfections and sequencing were purified on Qiagen columns. Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs and were used according to the manufacturer's instructions.
Envelope fusion assay.
To test the ability of genes to express proteins that cause membrane fusion at reduced pH, a protocol similar to that described by Blissard and Wenz (4) was employed. At 24 h after transfection the TNM-FH medium, pH 6.0, was replaced with fusion medium (Graces insect medium [Life Technologies] containing 10% fetal bovine serum and antibiotics at pH 5 or 6). The cells were observed 24 h after the pH shift.
Construction of a pMalcR1-ld130 gene fusion and antibody production.
A maltose binding protein (MBP)-LD130 fusion was produced by cloning a portion of the ld130 ORF into the bacterial expression plasmid pMalcR1 (New England Biolabs). To accomplish this, pMalcR1 was digested with BamHI, filled in, cut with PstI, and gel purified. The pLd130FL clone was digested with NcoI, filled in, digested with PstI (at nt 127997), and inserted into pMalcR1. This resulted in an in-frame fusion beginning at amino acid 1 and extending through amino acid 437. Sequence analysis confirmed that the construct was in frame. The fusion protein was expressed, isolated, and used for inducing rabbit polyclonal antiserum as previously described (35). The immune serum used in this report was collected 8 days after the fifth boost.
BV purification, protein isolation, Western blot analysis, and tunicamycin treatment.
BV from 56 ml of medium at 7 days postinfection (p.i.) was purified as previously described (3) and resuspended in 25 μl of phosphate-buffered saline (PBS), and 12 μl was used in Western blot analysis. For protein isolation, L. dispar cells were infected with LdMNPV at a multiplicity of infection (MOI) of 10. Approximately 9 × 106 cells were harvested at various times p.i. and mixed with TRIzol reagent (Life Technologies), and protein was isolated by following the manufacturer's protocol.
Samples were electrophoresed through sodium dodecyl sulfate (SDS)–10% polyacrylamide gels (19) and electroblotted onto nitrocellulose (Micron Separations Inc.) for 1.5 h at 185 mA, and Western blot analysis was performed using a chemiluminescent method according to the manufacturer's instructions (Boehringer Mannheim). The Ld130 antiserum was used at 1:1,000, and the second antibody, goat anti-rabbit horseradish peroxidase (Promega), was used at 1:2,500.
L. dispar cells were infected at an MOI of 5 in the presence or absence of tunicamycin (Sigma) (10 μg/ml). Three days p.i., cells were harvested (32) and used for Western blot analysis.
Immunofluorescence microscopy.
Monolayers of L. dispar cells were grown on coverslips and infected with LdMNPV strain 56-1 at an MOI of 5. At various times p.i., the coverslips were washed twice in PBS (Sigma) and then fixed in 3.7% formaldehyde for 10 min, followed by 2 min in −20°C methanol. The samples were air dried and stored at −20°C until used. Immunofluorescence staining was carried out as previously described (3) except that PBS–1% bovine serum albumin was used as the buffer. The MBP-LD130 antiserum was diluted 1:800, and the Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G antibody (Molecular Probes) was diluted 1:200. Phase and fluorescence microscopy was performed using a Zeiss Axiovert 35 microscope with ×5 and ×32 phase 1 Achrostigmat lenses and a ×40 Neofluar lens. A standard fluorescein isothiocyanate filter was used. Pictures were taken with an Hamamatsu C2400 SIT camera. Images were generated using National Institutes of Health Image, version 1.60, and Adobe Photoshop, version 4.0, software.
RESULTS
LdMNPV expresses an envelope fusion protein.
Analysis of the complete LdMNPV genome sequence led to the observation that it does not encode a homolog of GP64, the envelope fusion protein of AcMNPV and several closely related viruses (18). Computer-assisted analyses of the LdMNPV ORFs identified a single ORF, ld130, whose product was predicted to contain features of a membrane protein, including a signal sequence and a transmembrane domain (18). To determine if LdMNPV encodes a low-pH-dependent envelope fusion protein, L. dispar cells were infected with LdMNPV and 48 h later the pH of the medium was reduced from 6.0 to 5.0. Under these conditions fused cells were observed (not shown) suggesting that LdMNPV encodes a membrane fusion protein. Therefore, we investigated whether the product of ld130 had the properties of an envelope fusion protein.
Cell membrane fusion induced by LD130.
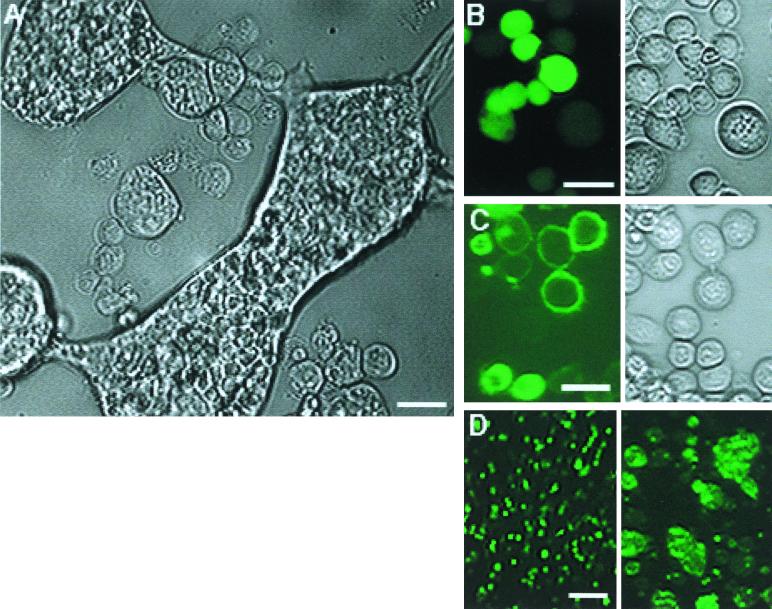
In order to determine if ld130 encodes an envelope fusion protein, three plasmids in which reading frames were expressed from the AcMNPV ie-1 promoter were constructed (Fig. 1). These included pLd130FL, containing the full-length ld130 (Fig. 1A), pLd130-egfp, in which the EGFP gene (egfp) is fused to ld130 downstream of the predicted transmembrane domain (Fig. 1B), and pegfp, which contains egfp alone (Fig. 1C). These plasmids were transfected into either L. dispar or Sf-9 cells, and their effects were characterized microscopically. Similar results were observed for both cell types, but only those for the transfected Sf-9 cells are shown in this report. When cells were transfected with pLd130FL and the pH of the medium was reduced from 6 to 5, extensive arrays of multinucleate fused cells were observed (Fig. 2A; compare cell size to sizes of unfused cells in Fig. 2B and C). These results are similar to those reported for cells transfected with OpMNPV gp64 (4) and suggest that ld130 encodes a membrane fusion protein. To facilitate the localization of LD130, two egfp expression plasmids were used. Cells transfected with the control plasmid, pegfp, exhibited fluorescence that was distributed throughout the cell at both pH 6 (not shown) and pH 5, and no cell fusion was observed (Fig. 2B). In contrast, expression of ld130-egfp resulted in localization of fluorescence to the cell membrane at pH 6.0 in many of the cells (Fig. 2C). However, in a few cells overall fluorescence was observed (Fig. 2C, lower left). Similar to cells transfected with pLd130FL (Fig. 2A), cells transfected with pLd130-egfp formed numerous large multinucleate fused cells upon reduction of the pH from 6.0 to 5.0 (Fig. 2D).
FIG. 2.
Expression of LD130 in insect cells. (A) Phase-contrast image of LD130 expressed in Sf-9 cells and treated at pH 5. Cells were transfected with pLd130FL, and 24 h later the pH was reduced from 6 to 5. Photographs were taken after an additional 24 h of incubation. (B) Fluorescence and phase-contrast images of Sf-9 cells expressing pegfp at pH 5. (C) Fluorescence and phase-contrast images of Sf-9 cells transfected with pLd130-egfp at pH 6. (D) Fluorescence images of Sf-9 cells expressing pLd130-egfp at pH 6 (left) and 5 (right). Bars, 20 μm (A to C) and 100 μm (D). In panels B to D, treatments were the same as in panel A.
Western blot analysis of LD130.
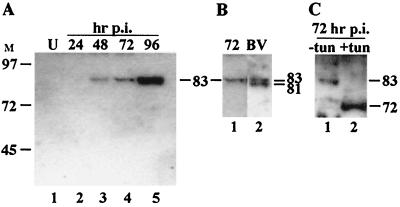
In order to study the expression of the ld130 gene product, an antibody to MBP-LD130 fusion protein was produced. Time courses of LdMNPV-infected L. dispar cells were analyzed by Western blotting using this antiserum. A major immunoreactive band of approximately 83 kDa was first observed at 48 h (Fig. 3A, lane 3). The band stained more intensely as the infection progressed, suggesting that it accumulated in association with cells at late time points (Fig. 3A, lane 5).
FIG. 3.
Western blot analysis of LD130 in extracts of infected cells and LdMNPV BV. (A) Time course of LD130 expression. Extracts from approximately 3.6 × 105 cells were loaded in each lane. (B) Analyses of LdMNPV BV. Lanes: 1, LdMNPV-infected L. dispar cells at 72 h p.i.; 2, BV. (C) Glycosylation of LD130. Lanes: 1, LdMNPV-infected L. dispar cells at 72 h p.i.; 2, Same as lane 1 except virus was grown in the presence of 10 μg tunicamycin (tun)/ml. The sizes (kilodaltons) of selected prestained markers are shown on the left of panel A. The estimated sizes of the immunoreactive bands are indicated.
To determine if the protein recognized by the LD130 antiserum was associated with BV, Western blot analysis was performed on LdMNPV BV and these results were compared to those for an infected cell extract from 72 h p.i. In the BV preparation the antiserum reacted with a doublet of 81 and 83 kDa (Fig. 3B, lane 2). The size of the 83-kDa band was similar to that observed in infected-cell extracts (Fig. 3B, compare lanes 1 and 2).
To determine if LD130 is glycosylated, cells were infected and grown in the presence of tunicamycin, which blocks N-linked glycosylation. This treatment resulted in a reduction in the size of the immunoreactive band from 83 kDa to about 72 kDa (Fig. 3C; compare lanes 1 and 2). This is less than the size expected from the sequence (76.7 kDa) (18) and may reflect in part the removal of the predicted N-terminal signal peptide of about 20 amino acids from this population of cell-associated molecules as well as experimental error inherent to determining protein size using the SDS-polyacrylamide gel electrophoresis technique. These results suggest that LD130 is modified by the addition of about 10 kDa of sugar residues.
Immunofluorescence localization of Ld130 in LdMNPV-infected cells.
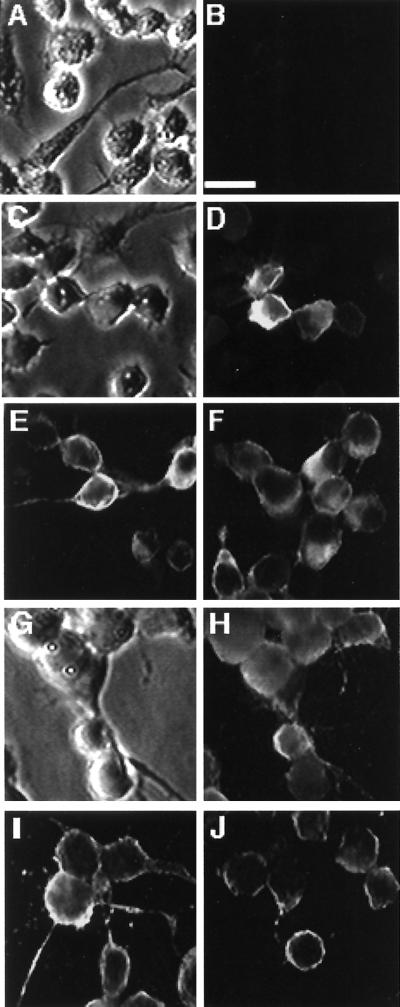
In order to characterize the localization of LD130 in infected cells, immunofluorescence microscopy of LdMNPV-infected L. dispar cells sampled at different times p.i. was carried out. Mock-infected cells showed little fluorescence when immune serum against LD130 was used (Fig. 4B). However by 24 h p.i. some of the infected cells showed specific fluorescence that was mostly associated with the cell membrane (Fig. 4D). At 48 h p.i., more cells showed fluorescence and it was concentrated at the membranes and long pseudopodia-like structures (Fig. 4E and F). By 72 h p.i. most of the fluorescence was associated with the cell membrane (Fig. 4H to J) and some of the cells contained polyhedra (Fig. 4G).
FIG. 4.
Immunofluorescence staining with LD130 antiserum of L. dispar cells infected with LdMNPV. (A and B) Phase-contrast and fluorescence images of mock-infected cells; (C and D) phase-contrast and fluorescence images of cells at 24 h p.i.; (E and F) fluorescence images of cells at 48 h p.i.; (G and H) phase-contrast and fluorescence images of cells at 72 h p.i.; (I and J) fluorescence images of cells at 72 h p.i. Bar, 20 μm. Cells were infected at an MOI of 5 and fixed at the time points shown. For details see Materials and Methods.
DISCUSSION
In this report we demonstrate that LD130 is a glycosylated low-pH-activated fusion protein that is associated with BV and that localizes to the plasma membranes of cells infected with LdMNPV or transfected with a pLd130-egfp expression plasmid. Plasmids expressing both the full-length ld130 and ld130 linked to egfp were capable of causing low-pH-induced cell fusion. To our knowledge, this is the first example of a low-pH-activated membrane fusion protein that is capable of inducing membrane fusion when linked to EGFP. This suggests that the barrel-shaped EGFP structure (21) does not significantly compromise the localization of the LD130 to the membrane or the conformational change of the protein, which likely occurs under low-pH conditions and which is involved in membrane fusion.
LD130 was first observed at 48 h p.i. on Western blots, which is consistent with the finding that the rate of LdMNPV infection is slower than the rate of AcMNPV infection (33). Although it appeared as an 83-kDa protein in extracts of infected cells, it was present as a doublet of 81 and 83 kDa associated with BV. This could reflect a mixture of the protein with various patterns of glycosylation as well as anomalous migration of glycoproteins seen by SDS-polyacrylamide gel electrophoresis (16).
Recently completed genome sequences from a number of diverse members of the Baculoviridae, including representatives of both NPVs and GVs, revealed that they lacked homologs of gp64, which encodes an envelope fusion protein demonstrated to be essential for cell-to-cell transmission of AcMNPV (23). In addition to their presence in AcMNPV, homologs of gp64 have been found in a number of relatively closely related baculoviruses including OpMNPV (3), Choristoneura fumiferana MNPV (12), Anagrapha falcipera MNPV (6), Epiphyas postvittana NPV (14), Bombyx mori NPV (8), and Anticarsia gemmatalis MNPV (27). In all these viruses the predicted GP64 proteins are over 74% identical at the amino acid level. In addition, GP64 has remarkable levels of amino acid sequence identity (up to 30%) with the envelope glycoproteins from two members of the Orthomyxoviridae that are vectored by ticks (24, 31).
In contrast to the high level of relatedness and limited distribution among baculoviruses of gp64, homologs of ld130 are found in all baculovirus genomes that have been completely sequenced. These include not only viruses that lack homologs of gp64, including LdMNPV (18), XcGV (11), Plutella xylostella GV (10), S. exigua MNPV (15), Cydia pomonella GV (T. Luque, R. Finch, D. Winstanley, and D. O'Reilly, Program Abstr. XXXI Meet. Soc. Invert. Pathol., p. 55, 1999), and Heliocoverpa zea SNPV (A. Lu, personal communication) but also AcMNPV, OpMNPV, and B. mori NPV, which contain gp64. Although the amino acid identity of the predicted sequences is low (e.g., 20 to 40%), all the LD130 homologs have predicted signal sequences and transmembrane domains, are of similar size (540 to 690 amino acids), and have 11 invariant cysteine residues. Furthermore, when the genome of XcGV was analyzed for ORFs with properties predicted for a membrane protein, only the homolog of ld130 was identified (11). Because of their presence in all baculovirus genomes that have been sequenced, it is possible that the homologs of ld130 encode the envelope fusion proteins in viruses which lack gp64. This may include all of the GVs and the majority of the NPVs. The high degree of relatedness of gp64 among a small group of baculoviruses suggests that relatively recently a gp64 homolog became incorporated into an NPV and subsumed the envelope fusion function of the ld130 homolog (see below). This could have resulted in the evolution of a branch of the Baculoviridae consisting of viruses that are dependent on GP64 for cell-to-cell spread.
The presence of gp64 is likely a defining characteristic of a subgroup of NPVs called group I that have been classified based on molecular evolutionary analyses of polyhedrin and other predicted protein sequences (5, 13, 14, 43; for a further discussion see reference 27). Although the genomes of many of the viruses that fall into group I have not been completely sequenced, many are known to be variants of, or closely related to, those of AcMNPV and OpMNPV (see, e.g., reference 14). All the NPVs that have been completely sequenced, analyzed phylogenetically, and classified as group II viruses lack gp64 (5, 13, 14, 43). However, although GVs have not been placed in group II, based on their lack of gp64, they also appear to be members of this category.
The fact that LD130 is an envelope fusion protein in LdMNPV suggests that its homologs may play a similar role for most baculoviruses lacking gp64, but it is unclear what role LD130 plays in viruses that express GP64. In initial investigations of the function of the OpMNPV ld130 homolog (orf21), we found no evidence that it functions as a low-pH envelope fusion protein in cells transfected with either the wild-type ORF or an orf21-egfp fusion (unpublished data). Therefore, in viruses lacking gp64, ld130 may have multiple roles in the infection cycle. When the LD130 envelope fusion function was displaced by GP64, the gene may have been retained because it performed other essential functions.
These investigations also suggest that gp64 has invaded two viral families including the Baculoviridae and the Orthomyxoviridae (24, 31). Influenza virus infections are initiated by the binding of the viral hemagglutinin to sialic acid-containing oligosaccharides. It has been suggested that incorporation of the gp64 homolog into certain members of the Orthomyxoviridae (Thogoto and Dhori viruses) permitted vectoring by ticks which lack sialic acid in their cell membrane proteins (25). The fact that the orthomyxoviruses encoding GP64 are vectored by ticks and the gp64-containing baculoviruses replicate in Lepidoptera may indicate that an invertebrate homolog of gp64 recombined into the genomes of these viral groups on two independent occasions.
ACKNOWLEDGMENTS
We thank James Slavicek for the LdMNPV viruses. We also thank Douglas Leisy for helpful suggestions and criticisms of the manuscript.
This research was supported by grants from the NSF (9982536) and USDA (95 37302-1920 and 97 02081) and contributions from Lawrence and Margaret Noall.
Footnotes
Technical report 11650 from the Oregon State University Agricultural Experiment Station.
REFERENCES
- 1.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff D S, Slavicek J M. Characterization of the Lymantria dispar nucleopolyhedrovirus 25K FP gene. J Gen Virol. 1996;77:1913–1923. doi: 10.1099/0022-1317-77-8-1913. [DOI] [PubMed] [Google Scholar]
- 3.Blissard G W, Rohrmann G F. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology. 1989;170:537–555. doi: 10.1016/0042-6822(89)90445-5. [DOI] [PubMed] [Google Scholar]
- 4.Blissard G W, Wenz J R. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J Virol. 1992;66:6829–6835. doi: 10.1128/jvi.66.11.6829-6835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulach D M, Kumar C A, Zaia A, Liang B, Tribe D E. Group II nucleopolyhedrovirus subgroups revealed by phylogenetic analysis of polyhedrin and DNA polymerase gene sequences. J Invertebr Pathol. 1999;73:59–73. doi: 10.1006/jipa.1998.4797. [DOI] [PubMed] [Google Scholar]
- 6.Federici B A, Hice R H. Organization and molecular characterization of genes in the polyhedrin region of the Anagrapha falcifera multinucleocapsid NPV. Arch Virol. 1996;142:333–348. doi: 10.1007/s007050050080. [DOI] [PubMed] [Google Scholar]
- 7.Funk C J, Braunagel S, Rohrmann G F. Baculovirus structure. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. [Google Scholar]
- 8.Gomi S, Majima K, Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol. 1999;80:1323–1337. doi: 10.1099/0022-1317-80-5-1323. [DOI] [PubMed] [Google Scholar]
- 9.Harwood S H, Li L, Ho P S, Preston A K, Rohrmann G F. AcMNPV late expression factor-5 interacts with itself and contains a zinc ribbon domain that is required for maximal late transcription activity and is homologous to elongation factor TFIIS. Virology. 1998;250:118–134. doi: 10.1006/viro.1998.9334. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto Y, Fujita T, Ueno Y. DNA sequence analysis of Plutella xylostella granulovirus. RIKEN Rev. 1999;22:30–33. doi: 10.1006/viro.2000.0530. [DOI] [PubMed] [Google Scholar]
- 11.Hayakawa T, Ko R, Okano K, Seong S, Goto C, Maeda S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology. 1999;262:277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- 12.Hill J E, Faulkner P. Identification of the gp67 gene of a baculovirus pathogenic to the spruce budworm, Choristoneura fumiferana, multinucleocapsid nuclear polyhedrosis virus. J Gen Virol. 1994;75:1811–1813. doi: 10.1099/0022-1317-75-7-1811. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z H, Arif B M, Jin F, Martens J W, Chen X W, Sun J S, Zuidema D, Goldbach R W, Vlak J M. Distinct gene arrangement in the Buzura suppressaria single-nucleocapsid nucleopolyhedrovirus genome. J Gen Virol. 1998;79:2841–2851. doi: 10.1099/0022-1317-79-11-2841. [DOI] [PubMed] [Google Scholar]
- 14.Hyink O, Graves S, Fairbairn F M, Ward V K. Mapping and polyhedrin gene analysis of the Epiphyas postvittana nucleopolyhedrovirus genome. J Gen Virol. 1998;79:2853–2862. doi: 10.1099/0022-1317-79-11-2853. [DOI] [PubMed] [Google Scholar]
- 15.Ijkel W F J, van Strien E A, Jeldens J G M, Broer R, Zuidema D, Goldbach R W, Vlak J M. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J Gen Virol. 1999;80:3289–3304. doi: 10.1099/0022-1317-80-12-3289. [DOI] [PubMed] [Google Scholar]
- 16.Jarvis D L, Wills L, Burow G, Bohlmeyer D W. Mutational analysis of the N-linked glycans on Autographa californica nucleopolyhedrovirus gp64. J Virol. 1998;72:9459–9469. doi: 10.1128/jvi.72.12.9459-9469.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kingsley D H, Behbahani A, Rashtian A, Blissard G W, Zimmerberg J. A discrete stage of baculovirus GP64-mediated membrane fusion. Mol Biol Cell. 1999;10:4191–4200. doi: 10.1091/mbc.10.12.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzio J, Pearson M N, Harwood S H, Funk C J, Evans J T, Slavicek J, Rohrmann G F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Markovic I, Pulyaeva H, Sokoloff A, Chernomordik L V. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J Cell Biol. 1998;143:1155–1166. doi: 10.1083/jcb.143.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misteli T, Spector D L. Applications of the green fluorescent protein in cell biology and biotechnology. Nat Biotechnol. 1997;15:961–964. doi: 10.1038/nbt1097-961. [DOI] [PubMed] [Google Scholar]
- 22.Monsma S A, Blissard G W. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus GP64 envelope fusion protein. J Virol. 1995;69:2583–2595. doi: 10.1128/jvi.69.4.2583-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monsma S A, Oomens A G P, Blissard G W. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J Virol. 1996;70:4607–4616. doi: 10.1128/jvi.70.7.4607-4616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morse M A, Marriott A C, Nuttall P A. The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein gp64. Virology. 1992;186:640–646. doi: 10.1016/0042-6822(92)90030-s. [DOI] [PubMed] [Google Scholar]
- 25.Nuttall P A, Morse M A, Jones L D, Portela A. Adaptation of members of the Orthomyxoviridae family to transmission by ticks. In: Gibbs A J, Calisher C H, García-Arenal F, editors. Molecular basis of virus evolution. New York, N.Y: Cambridge University Press; 1995. pp. 416–425. [Google Scholar]
- 26.Oomens A G, Blissard G W. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology. 1999;254:297–314. doi: 10.1006/viro.1998.9523. [DOI] [PubMed] [Google Scholar]
- 27.Oomens A G P. Functional and structural analysis of GP64, the major envelope glycoprotein of the budded virus phenotype of Autographa californica and Orgyia pseudotsugata multicapsid nucleopolyhedroviruses. Ph.D. dissertation. Wageningen, The Netherlands: University of Wageningen; 1999. [Google Scholar]
- 28.Oomens A G P, Monsma S A, Blissard G W. The baculovirus GP64 envelope fusion protein: synthesis, oligomerization, and processing. Virology. 1995;209:592–603. doi: 10.1006/viro.1995.1291. [DOI] [PubMed] [Google Scholar]
- 29.Pearson M N, Rohrmann G F. Characterization of a baculovirus-encoded ATP-dependent DNA ligase. J Virol. 1998;72:9142–9149. doi: 10.1128/jvi.72.11.9142-9149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson M N, Rohrmann G F. Splicing is required for transactivation by the immediate early gene 1 of the Lymantria dispar multicapsid nuclear polyhedrosis virus. Virology. 1997;235:153–165. doi: 10.1006/viro.1997.8687. [DOI] [PubMed] [Google Scholar]
- 31.Portela A, Jones L D, Nuttall P. Identification of viral structural polypeptides of Thogoto virus (a tick-borne orthomyxo-like virus) and functions associated with the glycoprotein. J Gen Virol. 1992;73:2823–2830. doi: 10.1099/0022-1317-73-11-2823. [DOI] [PubMed] [Google Scholar]
- 32.Quant-Russell R L, Pearson M N, Rohrmann G F, Beaudreau G S. Characterization of baculovirus p10 synthesis using monoclonal antibodies. Virology. 1987;160:9–19. doi: 10.1016/0042-6822(87)90038-9. [DOI] [PubMed] [Google Scholar]
- 33.Riegel C I, Slavicek J M. Characterization of the replication cycle of the Lymantria dispar nuclear polyhedrosis virus. Virus Res. 1997;51:9–17. doi: 10.1016/s0168-1702(97)00075-0. [DOI] [PubMed] [Google Scholar]
- 34.Rohrmann G F. Nuclear polyhedrosis viruses. In: Webster R G, Granoff A, editors. Encyclopedia of virology. 2nd ed. London, United Kingdom: Academic Press; 1999. pp. 146–152. [Google Scholar]
- 35.Russell R L Q, Funk C J, Rohrmann G F. Association of a baculovirus encoded protein with the capsid basal region. Virology. 1997;227:142–152. doi: 10.1006/viro.1996.8304. [DOI] [PubMed] [Google Scholar]
- 36.Slavicek J M. Temporal analysis and spatial mapping of Lymantria dispar nuclear polyhedrosis virus transcripts and in vitro translation products. Virus Res. 1991;20:223–236. doi: 10.1016/0168-1702(91)90077-9. [DOI] [PubMed] [Google Scholar]
- 37.Slavicek J M, Mercer M J, Kelly M E, Hayes-Plazolles N. Isolation of a baculovirus variant that exhibits enhanced polyhedra production stability during serial passage in cell culture. J Invertebr Pathol. 1996;67:153–160. [Google Scholar]
- 38.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agricultural Experiment Station Bulletin 1555. College Station, Tex: Texas A&M University; 1987. [Google Scholar]
- 39.Volkman L E, Goldsmith P A, Hess R T, Faulkner P. Neutralization of budded Autographa californica NPV by a monoclonal antibody: identification of the target antigen. Virology. 1984;133:354–362. doi: 10.1016/0042-6822(84)90401-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkman L E, Summers M D, Hsieh C H. Occluded and nonoccluded nuclear polyhedrosis virus grown in Trichoplusia ni: comparative neutralization, comparative infectivity, and in vitro growth studies. J Virol. 1976;19:820–832. doi: 10.1128/jvi.19.3.820-832.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitford M, Stewart S, Kuzio J, Faulkner P. Identification and sequence analysis of a gene encoding gp67, an abundant envelope glycoprotein of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol. 1989;63:1393–1399. doi: 10.1128/jvi.63.3.1393-1399.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winstanley D, O'Reilly D. Granuloviruses. 2nd ed. London, United Kingdom: Academic Press; 1999. [Google Scholar]
- 43.Zanotto P M D A, Sampaio M J A, Johnson D W, Rocha T L, Maruniak J E. The Anticarsia gemmatalis nuclear polyhedrosis virus polyhedrin gene region: sequence analysis, gene product and structural comparisons. J Gen Virol. 1992;73:1049–1056. doi: 10.1099/0022-1317-73-5-1049. [DOI] [PubMed] [Google Scholar]