Abstract
We have constructed two novel adenovirus (Ad) replication-competent vectors, named KD1 and KD3, that may have use in anticancer therapy. The vectors have two key features. First, they markedly overexpress the Ad death protein (ADP), an Ad nuclear membrane glycoprotein required at late stages of infection for efficient cell lysis and release of Ad from cells. Overexpression of ADP was achieved by deleting the E3 region and reinserting the adp gene. Because ADP is overexpressed, KD1 and KD3 are expected to spread more rapidly and effectively through tumors. Second, KD1 and KD3 have two E1A mutations (from the mutant dl1101/1107) that prevent efficient replication in nondividing cells but allow replication in dividing cancer cells. These E1A mutations preclude binding of E1A proteins to p300 and pRB. As a result, the virus should not be able to drive cells from G0 to S phase and therefore should not be able to replicate in normal tissues. We show that KD1 and KD3 do not replicate well in quiescent HEL-299 cells or in primary human bronchial epithelial cells, small airway epithelial cells, or endothelial cells; however, they replicate well in proliferating HEL-299 cells and human A549 lung carcinoma cells. In cultured A549 cells, KD1 and KD3 lyse cells and spread from cell to cell more rapidly than their control virus, dl1101/1107, or wild-type Ad. They are also more efficient than dl1101/1107 or wild-type Ad in complementing the spread from cell to cell of an E1− E3− replication-defective vector expressing β-galactosidase. A549 cells form rapidly growing solid tumors when injected into the hind flanks of immunodeficient nude mice; however, when A549 cells were infected with 10−4 PFU of KD3/cell prior to injection into mice, tumor formation was nearly completely suppressed. When established A549 tumors in nude mice were examined, tumors injected with buffer grew 13.3-fold over 5 weeks, tumors injected with dl1101/1107 grew 8-fold, and tumors injected with KD1 or KD3 grew 2.6-fold. Hep 3B tumors injected with buffer grew 12-fold over 3.5 weeks, whereas tumors injected with KD1 or KD3 grew 4-fold. We conclude that KD1 and KD3 show promise as anticancer therapeutics.
Human adenoviruses (Ad) in subgroup C (e.g., Ad5) are ubiquitous DNA viruses that cause relatively mild respiratory infections in young children and induce lifelong immunity (21). Ad are being widely considered as gene therapy vectors to treat cancer (3, 8, 33). Before discussing such vectors, we will briefly outline the Ad replication cycle. In Ad5, the immediate-early E1A proteins induce expression of the delayed-early proteins encoded by the E1B, E2, E3, and E4 transcription units. Viral DNA begins to replicate at about 7 h postinfection (p.i.), and then late proteins derived from the major late transcription unit are synthesized. The major late mRNAs are formed by alternative splicing and polyadenylation of a large pre-mRNA initiated at the single major late promoter and extending to the right end of the genome. All late mRNAs have a tripartite leader (leaders 1, 2, and 3) at their 5′ termini that facilitates translation. Virions begin to assemble in the cell nucleus at 20 to 24 h p.i., and then after 2 to 3 days the cells begin to lyse and release virions, with lysis complete by about 5 to 6 days (37, 38).
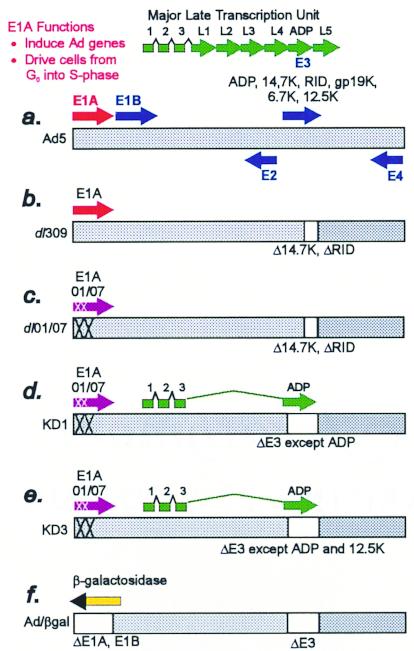
There are two basic types of Ad anticancer vectors, replication defective and replication competent (Fig. 1). The former vectors have the genes in the E1A, E1B, and usually the E3 transcription units deleted (Fig. 1f). The E1A proteins have two main functions: they force quiescent cells (e.g., lung epithelia) into S phase so that Ad DNA can replicate efficiently, and they induce transcription of other Ad genes (5). The E1A and E1B regions (collectively known as E1) are replaced by a therapeutic gene expressed from a promoter or enhancer. Numerous therapeutic proteins have been expressed, including the herpes simplex virus thymidine kinase (HSV TK), cytokines, and proteins such as p53 that induce apoptosis (33). Some therapeutic proteins only affect the vector-infected tumor cell; others, e.g., HSV TK, which generates gancyclovir monophosphate, exhibit a bystander effect on neighboring cells. However, in general, replication-defective vectors are limited because they only infect a small fraction of the cells in the tumor.
FIG. 1.
Schematic of the adenovirus genome and vectors. (a) Schematic of Ad5. The horizontal bar indicates the duplex DNA genome of 36 kbp encoding ca. 34 genes. The arrows depict transcription units. The E3 proteins are indicated. The adp gene is located in the E3 transcription unit, and ADP is expressed at low levels in early stages of infection (39). However, at late stages, splicing of the major late pre-mRNA occurs such that ADP is expressed at high levels (39). (b) Schematic of dl309. dl309 is identical to Ad5 except it has a deletion in the E3 transcription unit that removes the genes for the RID and 14.7K proteins (2). (c) Schematic of dl01/07. dl01/07 is identical to dl309 except it has two small deletions (XX) in E1A. (d and e) Schematics of the replication-competent vectors named KD1 (d) and KD3 (e). Both KD1 and KD3 contain two small E1A deletions derived from dl01/07, which is otherwise identical to dl309 (22). KD1 has adp but lacks all other E3 genes. KD3 has the adp and 12.5K genes but lacks all other E3 genes. The adp gene is reinserted into the E3 deletion such that the ADP major late mRNA will be formed abundantly, presumably because there is less competition for splicing for the ADP mRNA. (f) E1− E3− replication-defective vector expressing β-galactosidase from the Rous sarcoma virus promoter.
The second type of vector is competent for replication and therefore capable of infecting more cells in the tumor. Such vectors kill cells as part of the natural Ad life cycle. In theory, these vectors should be designed such that their replication is restricted to tumor cells and such that normal tissues are not damaged. Only a few vectors of this type have been described. One such vector, ONYX-015 (the Ad mutant dl1520 [1]), lacks the gene for E1B-55K (4, 19, 20). E1B-55K is required for efficient cytoplasmic accumulation and translation of Ad mRNAs (15). It also binds the tumor suppressor p53 and represses p53-responsive promoters (27), inhibiting p53-induced apoptosis and enabling cells to enter S phase. ONYX-015 was originally reported to replicate only in cells lacking wild-type p53 and thus to be selective for tumors lacking functional p53 (4, 19). However, it is now clear that its replication can be independent of p53 (11, 12, 15, 18, 31, 40). Phase I and II clinical trials of ONYX-015 in head and neck cancer have been encouraging (24, 25) and support the use of Ad replication-competent vectors in cancer treatment. Other workers have also constructed E1B-55K mutant vectors; one vector expresses HSV TK (42, 43), another expresses a TK-cytosine deaminase chimeric protein (10), and another infects gliomas more efficiently (34). Two other types of vectors have been described that are restricted to tumors or at least tissues by having E1A gene expression dependent on tumor-specific promoters or enhancers, namely, prostate-specific antigen for prostate cancer (30, 48) and α-fetoprotein for liver cancer (13). There are no clinical trial results published for replication-competent vectors other than ONYX-015.
Here we describe two replication-competent Ad vectors, named KD1 and KD3 (Fig. 1d and e), that have a combination of unique features not found in other Ad vectors. First, they are more cytolytic than Ad5 or dl309, resulting in faster spread of vector from cell to cell in culture. dl309 is an Ad5 E3 deletion mutant that is commonly used as wild-type Ad and is so used in our studies. KD1 and KD3 are more cytolytic than dl309 because they overexpress the Ad death protein (ADP), an integral membrane palmitoylated glycoprotein (17, 32). dl309 expresses the levels of ADP observed with Ad5. In Ad5 and Ad2, ADP is synthesized at very late stages of infection when virus has assembled in the cell nucleus (39). ADP mediates efficient cell lysis and release of Ad from cells, as indicated by studies with adp mutants (37, 38).
The second feature of KD1 and KD3 is that they contain mutations in the E1A gene that affect some but not all aspects of E1A function and that are predicted to allow vector growth in cancer cells but not normal cells. There are two E1A proteins, of 289 and 243 amino acids, named 289R and 243R, respectively, that are translated from alternatively spliced 13S and 12S mRNAs (5, 9, 41, 44). The 5′-terminal exon of the 13S mRNA for 289R encodes three protein domains named conserved region 1, conserved region 2, and conserved region 3 (CR1, CR2, and CR3, respectively). The 243R protein has CR1 and CR2 but not CR3. The E1A proteins exert their functions by interacting with various cellular proteins that affect transcription. For example, CR3 of E1A binds many transcription factors and facilitates their assembly onto promoters. CR3 is required for efficient transcription from the Ad delayed-early promoters. The 243R protein alters the cell to become an efficient host for virus replication, e.g., to contain the metabolites and enzymes required for DNA synthesis. Viewed simply, the expression of S-phase genes is controlled in noncycling differentiated cells by the binding of the tumor suppressor pRB protein to the transcription factor E2F. The pRB-E2F complex is tethered to E2F sites in S-phase-specific promoters and represses transcription. In Ad-infected cells, the 243R protein, acting through its CR2 and CR1 domains, binds pRB and liberates E2F. Free E2F bound to E2F sites activates transcription of S-phase-specific genes. Another cellular protein, the p300 (highly related to CBP) transcriptional coadaptor, activates transcription of many cellular genes, presumably including genes that maintain cell differentiation and inhibit the cell cycle (5, 9, 41, 44). The E1A proteins, acting via the CR1 domain and a region near the NH2 terminus of E1A, form a complex with p300/CBP, inhibit its intrinsic histone acetyltransferase activity, and repress its ability to activate transcription (6, 14, 29). KD1 and KD3 contain two small deletions in E1A (22), one near the NH2 terminus of the E1A proteins and the other in CR2, that abolish the binding of E1A to pRB and p300/CBP but leave intact CR3 and the ability of E1A to transactivate viral genes. In theory, since the mutated E1A cannot drive cells from G0 or G1 into S phase, our vectors should not replicate well in quiescent or primary cells. Since the mutated E1A can transactivate Ad genes, our vectors should replicate in cancer cells, which must cycle at some rate through S phase. Our presumption is that cancer cells, which have mutations that deregulate the cell cycle, will resemble the state induced in differentiated cells by the binding of E1A to pRB and p300.
The third feature of KD1 and KD3 is that they lack all or nearly all of the E3 genes. There are seven known E3 genes situated in the following order in the Ad5 genome: 12.5K, 6.7K, gp19K, adp, RIDα, RIDβ, and 14.7K. KD1 lacks all the E3 genes other than adp, and KD3 lacks all but 12.5K and adp (Fig. 1b; Table 1). Deletion of the E3 genes has no effect on Ad replication in cultured cells. Some of the E3 proteins (gp19K, RIDα, RIDβ, and 14.7K) function to prevent apoptosis induced by cytotoxic T lymphocytes and other cells that kill through the tumor necrosis factor receptor 1 and Fas pathways (26, 45–47). Because KD1 and KD3 lack E3 proteins, infected tumor cells might be killed not only by vector replication, but also by immune killer cells. In addition, cytotoxic T lymphocytes specific to tumor antigens might be more easily generated. Also, the vectors should be more safe than Ad5 because, should they spread in the body, they will be unable to prevent immune killer cells from destroying vector-infected cells. Thus, the features of KD1 and KD3 are overexpression of ADP, two small deletions in E1A that knock out binding of E1A to pRB and p300, and lack of E3 genes.
TABLE 1.
Genotype of viruses
| Virus | E1A | ADP | E3 genes deleted |
|---|---|---|---|
| dl309 | Wild-type | Wild-type | RID, 14.7K |
| dl327 | Wild-type | Deleted | All but 12.5K |
| pm734.1 | Wild-type | Mutant | None |
| dl01/07 | Mutant | Wild-type | RID, 14.7K |
| KD1 | Mutant | Overexpressed | All |
| KD2 | Mutant | Wild-type | gp19K, RID, 14.7K |
| KD3 | Mutant | Overexpressed | All but 12.5K |
MATERIALS AND METHODS
Cell lines.
Human cancer cell lines A549 (alveolar carcinoma), Hep 3B (hepatocellular carcinoma), HeLa (cervical epithelioid carcinoma), C-33A (cervix carcinoma), DU 145 (prostate carcinoma), and PC-3 (prostate adenocarcinoma) were obtained from the American Type Culture Collection (ATCC) and grown in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). Human diploid lung fibroblasts HEL-299 were obtained from the ATCC and grown in DMEM (10% FBS). Human primary small airway epithelial cells and human primary normal bronchial epithelial cells were obtained and grown in defined medium from Clonetics Corporation (San Diego, Calif.). Human primary pulmonary artery endothelial cells were obtained from Cascade Biologics (Portland, Oreg.) and grown in Medium 200 supplemented with Low Serum Growth Supplement. Human embryonic kidney 293 cells were obtained from Microbix (Toronto, Ontario, Canada) and propagated in DMEM with 10% FBS.
Viruses.
Ad5, dl309 (23), dl327 (35), and pm734.1 (38) were described previously. Mutant dl1101/1107 (dl01/07) (obtained from Stanley Bayley, McMaster University) is similar to dl01/07/520, an Ad5 E1A mutant that is defective in inducing DNA synthesis in primary baby rat kidney cells (22). dl01/07 has two E1A deletions, named 01 (residues 4 to 25 near the NH2 terminus of E1A) and 07 (residues 111 to 123 in CR2 of E1A) (22). The 01 and 07 deletions abolish binding of E1A to p300 and pRB, respectively. dl01/07 expresses both 243R and 289R proteins. It also has a deletion in the E3 region that is identical to the E3 deletion in dl309 (2, 22).
The viruses named KD1, KD2, and KD3 were constructed as follows. Shuttle plasmid pLKH, containing sequences of the Ad5 genome from 60 to 100 map units with the E3 region deleted, was used to introduce mutations into the E3 region. Using a PCR-based protocol, three different versions of the plasmid were constructed. pKD1 has a deletion of Ad5 bp 27858 to 27860 (7) and an insert of TAA; deletion of Ad5 bp 27982 to 28134; deletion of Ad5 bp 28395 to 29397 and insert of CCTTAATTAAA; and deletion of Ad5 bp 29783 to 30883 and insert of TTAATTAAGG. These mutations remove the 12.5K open reading frame (ORF), leave the L4 poly(A) site, remove all E3 sequences between the L4 poly(A) site and a site (bp 30883) downstream of the E3B poly(A) sites and upstream of the fiber ORF, and create a PacI site (TTAATTAA [sequence underlined above]). A PCR fragment, comprising bp 29397 to 29783 (the ADP ORF is at bp 29491 to 29772) and flanking PacI sites, is cloned into the PacI site. pKD2 has a dl309 E3 background, with a deletion of Ad5 bp 28788 to 28789 and an insert of TTAATTAA. The precursor plasmid to pKD3 has a deletion of Ad5 bp 28598 to 30469, with an XbaI site at the deletion. In pKD3, a PCR fragment comprising bp 29397 and 29783 (containing the ADP ORF) and flanking XbaI sites is cloned into the XbaI site; thus, pKD3 has deletions of Ad5 bp 28598 to 29397 and 29783 to 30469. The plasmids were cotransfected into HEK 293 cells along with dl01/07 virion DNA digested with EcoRI. The resulting plaques for virus vectors KD1, KD2, and KD3 were screened for the expected genome structure and were plaque purified three times on A549 cells. E1− E3− replication-deficient Ad/βgal virus expressing β-galactosidase under the control of Rous sarcoma virus promoter was constructed using pN30 and pBHG11 (Microbix). All replication-competent viruses were grown in suspension cultures of KB cells and banded in CsCl, and titers were determined by plaque assay on A549 cells (36). Ad/βgal was grown in suspension cultures of 293 cells and titered on 293 cell monolayers. Suspension cultures were grown in Joklik's medium with 5% horse serum.
Viruses used in this study are shown in Fig. 1 and described in Table 1. Mutant dl01/07 (22) is the control for KD1 and KD3 because it has the 01/07 mutation in E1A, it expresses “normal” levels of ADP in cells, and some E3 genes are lacking (Fig. 1c). dl309 (2, 23) is identical to dl01/07 except it has wild-type E1A (Fig. 1b).
Immunoblots.
A549 cells were infected with 50 PFU of different viruses per cell and then were harvested at various times p.i. The protein concentration was determined with the BIO-RAD DC protein assay kit (Bio-Rad Laboratories, Hercules, Calif.) Ten micrograms of protein per sample were electrophoresed on sodium dodecyl sulfate-polyacrylamide (10 or 15%) gels. The proteins were electroblotted onto Immobilon polyvinylidene difluoride membranes (Millipore, Bedford, Mass.), using a BIO-RAD Trans-BLOT SD semidry electrophoretic transfer cell. The membranes were incubated overnight at 4°C in TBST (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.2% Tween 20) containing 10% dry milk (Carnation) and then probed with a rabbit polyclonal antibody raised against residues 63-77 of ADP (39) (Fig. 2a), the mouse M73 monoclonal antibody against E1A (16) (Fig. 2b), or a rabbit anti-Ad5 polyclonal antibody (ATCC) (Fig. 2b). The secondary antibodies were goat anti-rabbit horseradish peroxidase and goat anti-mouse horseradish peroxidase (Cappel, Durham, N.C.). The bands were visualized via the ECL protocol (Amersham Pharmacia, Arlington Heights, Ill.).
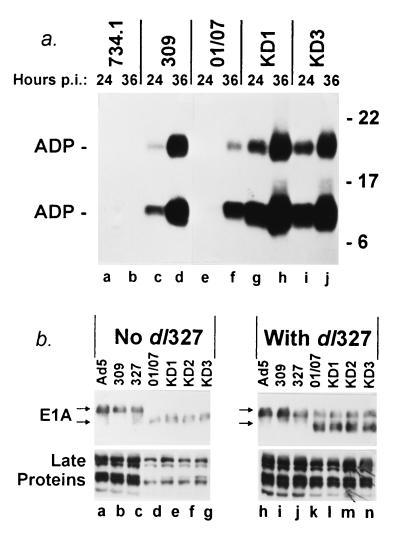
FIG. 2.
KD1 and KD3 overexpress ADP and express the expected smaller E1A proteins. (a) Overexpression of ADP. A549 cells were infected with 50 PFU of the indicated viruses per cell. At 24 or 36 h p.i., proteins were extracted and ADP was detected by immunoblotting. The upper ADP band is glycosylated, and the lower band is a proteolytic cleavage product that is probably functional (39). (b) Expression of E1A and Ad late proteins. In the left panel, parallel cultures of A549 cells were infected with 50 PFU of the indicated viruses per cell. The E1A proteins (top) were extracted at 15 h p.i., and late proteins (bottom) were extracted at 24 h p.i., and then proteins were detected by immunoblotting. The conditions in the right panel were identical, except the cells were coinfected with 25 PFU of dl327 per cell.
Virus replication in growing and growth-arrested HEL-299 cells.
HEL-299 cells were planted at semiconfluency into 60-mm-diameter dishes and grown in DMEM with 10% FBS. For growth-arrested (quiescent) studies (Fig. 3a), cells were grown to confluent monolayers and starved for three days in growth medium containing 0.2% FBS. Growing and growth-arrested cells were infected with 100 PFU of virus/cell. Cells combined with medium were frozen at the indicated time points after infection. Cells were frozen and thawed three times, and the titers of viruses were determined by plaque assay on A549 cells.
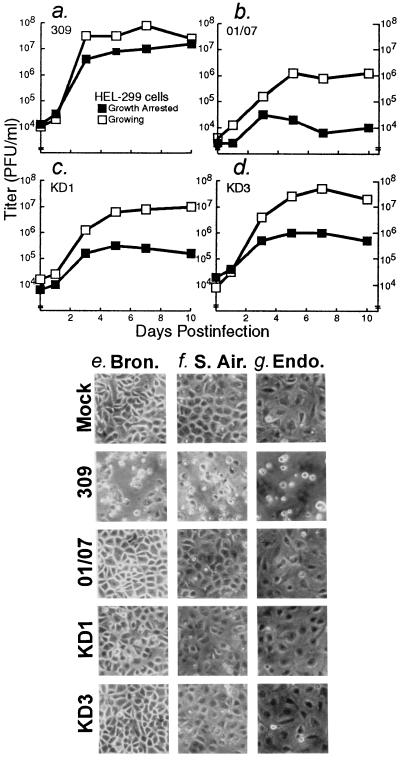
FIG. 3.
KD1 and KD3 do not replicate well in nongrowing cells and primary human cells. (a to d) HEL-299 cells growing in 10% FBS or growth arrested in 0.2% FBS were infected with 100 PFU/cell of the indicated viruses. At the indicated days p.i., virus was extracted from cells, and titers were determined on A549 cells. (e) Human primary bronchial epithelial cells, infected at 30% confluency with 10 PFU of the viruses indicated at left per cell and photographed at 5 days p.i. (f) Human primary small airway epithelial cells infected at 50% confluency with the indicated virus (10 PFU/cell), 5 days p.i. (g) Human primary endothelial cells infected at confluency with the indicated virus (10 PFU/cell), 8 days p.i.
Virus-induced cell killing, virus spread, and infectious-center assays.
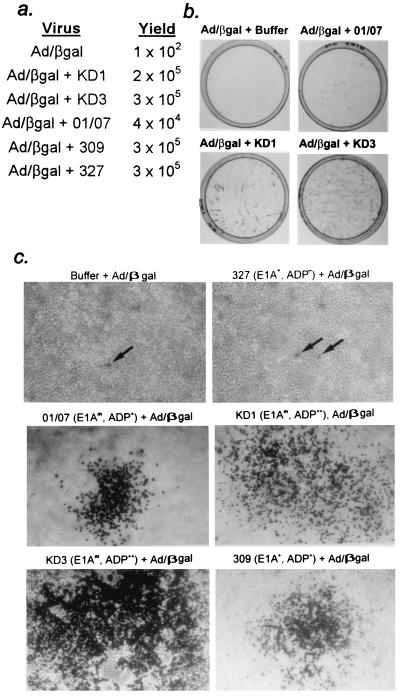
Trypan blue exclusion and lactate dehydrogenase release assays on A549 cells (Fig. 4a and b) were conducted as described previously (38). For each data point in the trypan blue experiment, about 400 cells were examined and the percentage of blue cells was calculated. This experiment was conducted five times, with similar results. For the lactate dehydrogenase release experiment, each data point is the mean of triplicate values. In the virus spread assay (Fig. 4e), A549 cells were seeded onto 48-well plates and infected with serial dilutions of viruses. At 7 days p.i., monolayers were fixed and stained with crystal violet. In the infectious-center assay (Fig. 5b and c), A549 cells were coinfected as described in the text. After 2 h, cells were trypsinized, collected, and diluted in DMEM, and 103 infected cells were seeded onto uninfected monolayers of A549 cells. At 4 days p.i., monolayers were fixed, stained for β-galactosidase with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and photographed.
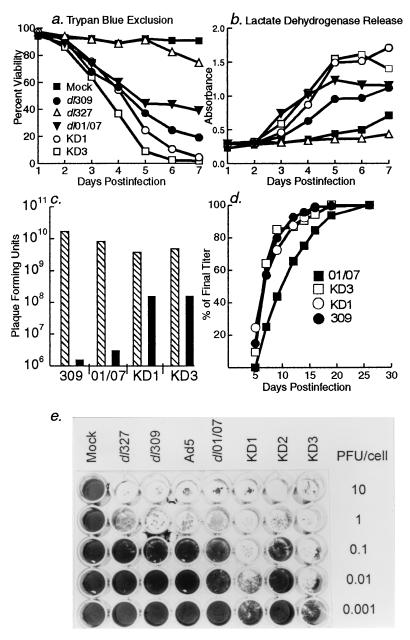
FIG. 4.
KD1 and KD3 lyse cells and spread from cell to cell efficiently. (a and b) A549 cells were infected with 20 PFU of the indicated viruses per cell, and then at the indicated days p.i. they were assayed for cell lysis by trypan blue exclusion (a) or release of lactate dehydrogenase into the medium (b). (c) A549 cells were infected with 20 PFU of virus per cell, and then at 2 days p.i. the amount of virus within the cells (hatched bars) and in the culture supernatant (black bars) was determined by plaque assay on A549 cells. (d) Plaque development assay on A549 cells. The number of plaques seen on a given day of the assay (x axis) is plotted as a percentage of plaques seen on the final day of the assay (y axis). (e) Virus spread assay. A549 cells were infected with different viruses, as indicated above the columns, and with different amounts of virus, as indicated beside the rows. At 7 days p.i., cells remaining on the plate were fixed and stained with crystal violet. Note the comet-shaped plaques of missing cells formed by KD1 and KD3 at 0.001 PFU/cell; each plaque arose from a single virus.
FIG. 5.
KD1 and KD3 efficiently complement the replication and cell-to-cell spread of Ad/βgal, an E1− E3− adp− replication-defective Ad vector expressing β-galactosidase. (a) A549 cells were infected with Ad/βgal alone (10 PFU/cell) or with Ad/βgal (10 PFU/cell) plus KD1, KD3, dl01/07, dl309, or dl327 (10 PFU/cell). At 2 days p.i., virus was extracted and Ad/βgal titers were determined by β-galactosidase expression (blue cells) on A549 cells. (b) Infectious-center assay. (c) Typical blue cells or blue foci (comets) seen in panel b and plates for dl309 and dl327 from the same experiment.
Tumor xenograft experiments.
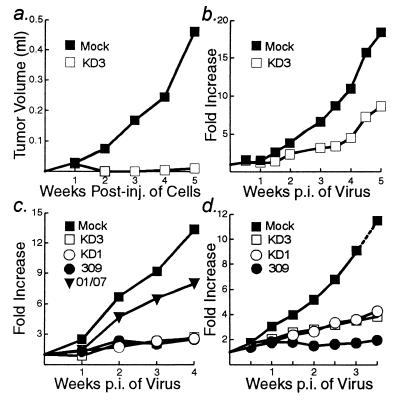
Female athymic nu/nu mice at 4 to 5 weeks of age were obtained from Harlan Sprague-Dawley Corporation, and quarantined for 1 week before entering the study. Mice were housed three per cage in filter-top cages and fed with Purina rodent chow (Ralston-Purina, St. Louis, Mo.) and tap water ad libitum. Institutional and federal guidelines for animal care were strictly followed. For Fig. 6a, 103 A549 cells either pre-infected with 10 PFU of KD3/cell or mock-infected were mixed with 107 uninfected A549 cells, injected into flanks of nude mice (n = 3), and measured for growth at weekly intervals. Tumors were measured with a Mitutoyo digital caliper, and the volumes were calculated according to the formula length × width2/2. For Fig. 6b, 107 A549 cells were injected into the flanks of mice (n = 12). Tumors of about 75 μl were injected with a single dose of 5 × 108 PFU of KD3 in 50 μl of DMEM lacking serum (the KD3 stock was diluted 1:50 in DMEM). For Fig. 6c, 107 A549 cells in 200 μl of DMEM were injected into flanks of mice and allowed to grow to about 50 to 70 μl. For Fig. 6d, 107 Hep 3B cells in 200 μl of DMEM plus 10% Matrigel (Becton Dickinson Labware, Bedford, Mass.) were similarly injected and allowed to grow to about 100 μl. Preestablished tumors (n = 6 for A549; n = 10 for Hep 3B) were injected with 50 μl of DMEM or 5 × 107 PFU of the indicated viruses in DMEM. Injections of viruses were repeated weekly for A549 tumors to a total dose of 2 × 108 PFU per tumor, or twice weekly for Hep 3B tumors to a total dose of 3.5 × 108 PFU per tumor. Tumor size measurements were taken just prior to injection. For Fig. 6b to d, data are represented as means of increase in tumor size relative to the size at the initial injection.
FIG. 6.
KD1 and KD3 reduce the growth of human tumors in nude mice. In all experiments, 107 cells were injected into each hind flank of nude mice. (a) A549 cells were mock infected or infected with KD3 (10 PFU/cell). After 3 h, the cells were trypsinized and added to trypsinized uninfected A549 cells such that 1 of 10,000 cells was infected. Cells were injected, and tumor growth was measured over 5 weeks. P = 0.007. (b) A549 cells were injected. When the tumors reached about 50 to 70 μl, they were injected with DMEM (mock) or KD3 (5 × 108 PFU). The fold increase in tumor size was measured over 5 weeks. P = 0.048. (c) Same as in panel b except that tumors were measured and then injected with 5 × 107 PFU of the indicated viruses on day 0 and at weeks 1, 2, and 3. For the following comparisons P was as indicated: KD3 and mock, 0.015; KD1 and mock, 0.015; 309 and mock, 0.014; dl01/07 and mock, 0.107; KD3 and dl01/07, 0.010; KD3 and KD1, 0.473; KD3 and 309, 0.461. (d) Hep 3B cells were injected. When tumors reached about 100 μl, they were injected with 5 × 107 PFU of the indicated viruses on day 0 and twice per week until 3 weeks. For the following comparisons, P was as indicated: KD3 and mock, 0.030; KD1 and mock, 0.017; 309 and mock, 0.002; KD3 and KD1, 0.282; KD3 and 309, 0.003; KD1 and 309, 0.029.
RESULTS
KD1 and KD3 overexpress ADP.
The E3 transcription unit is expressed during early stages of infection, when the multiple overlapping E3 mRNAs are synthesized by alternative splicing and polyadenylation of pre-mRNAs derived from the E3 promoter (39) (Fig. 1a). The E3 region is of further interest because it is embedded within the major late transcription unit (39). As a consequence, some E3 mRNAs are synthesized at late stages of infection by alternative splicing and polyadenylation of major late pre-mRNAs initiated at the major late promoter (Fig. 1a). The mRNA for ADP is relatively scarce at early times, and ADP is only made in low amounts. However, at late stages, ADP is synthesized in large amounts from mRNAs derived from the major late promoter, typical of major late proteins (39). In designing KD1 and KD3, we sought to increase ADP expression further by removing the ORFs in the E3 region except the one for ADP, and therefore to increase the probability that a major late pre-mRNA would become spliced such that ADP-specific mRNA is synthesized in abundance. KD1 has the adp gene and no other E3 genes, and KD3 has only the adp and 12.5K genes. The function of the E3-12.5K protein is unknown.
To confirm that KD1 and KD3 overexpress ADP, human A549 lung carcinoma cells were infected and then analyzed for ADP by immunoblot. As shown in Fig. 2a, KD1 and KD3 greatly overexpressed ADP at 24 h p.i. as compared to dl309 and dl01/07 (compare lanes g and i with lanes c and e). KD1 and KD3 also made more ADP than dl01/07 at 36 h p.i. (compare lanes h and j with lane f) and perhaps a little more than dl309 (lane d). The two ADP bands shown represent posttranslationally modified forms of ADP (32, 39). The upper band is full-length ADP that contains N-linked high-mannose oligosaccharides. The lower band is a proteolytically processed form of ADP.
The second key feature of KD1 and KD3 is that they contain the version of E1A with the 01/07 deletion. As expected, dl01/07, KD1, KD2, and KD3 all expressed the expected smaller version of the E1A proteins (Fig. 2b, lanes d to g). (KD2 is similar to KD1 and KD3 except that it has a smaller E3 deletion and only slightly overexpresses ADP; it is included as a negative control. In addition to expressing ADP, KD2 is expected to express the E3 12.5K and 6.7K proteins. It lacks the genes for the E3 gp19K, RIDα, RIDβ, and 14.7K proteins.)
The next question addressed was how the 01/07 mutation and ADP overexpression would affect the time course of the infection. We found that the kinetics of Ad late gene expression were somewhat delayed (by about 10 h) in the viruses with the 01/07 mutation, as indicated by reduced late protein synthesis at 24 h p.i. (Fig. 2b, compare lanes d to g with lanes a to c). This delay is due to the 01/07 mutation inasmuch as when cells were coinfected with E1A mutants and dl327 (35), which has wild-type E1A and lacks ADP, late proteins were made at levels comparable to Ad5, dl309, and dl327 alone (Fig. 2b, compare lanes k to n with lanes h to j). That is, the E1A expressed by dl327 compensated for the E1A mutation in the viruses with the 01/07 mutation. After 2 days, Ad protein accumulation and virus yield is equivalent in all the viruses. It is noteworthy that overexpression of ADP by KD1 and KD3 did not noticeably alter late protein synthesis as compared to dl01/07.
KD1 and KD3 replicate poorly in primary and quiescent cells.
The 01/07 mutation in KD1 and KD3 should limit their replication in quiescent cells (22, 28). Human diploid HEL-299 fibroblasts were cultured in 10% FBS or rendered quiescent in 0.2% FBS. Cells were infected with 100 PFU of KD1, KD3, dl01/07, or dl309 per cell, and then at different days p.i., virus was extracted and titers were determined by plaque assay on A549 cells. This relatively high multiplicity was chosen in order to rigorously test whether replication is blocked in quiescent cells. In 10% serum, KD1, KD3, and dl01/07 replicated well, reaching titers of 106 to 107 PFU/ml, only slightly less than dl309 levels (Fig. 3a to d). However, in quiescent cells, KD1, KD3, and dl01/07 levels were 1.5 to 2 logs lower, ranging from 104 to 2 × 105 PFU/ml. dl309 reached 107 PFU/ml, nearly the level achieved in growing cells. Similar results were obtained with WI-38 human fibroblasts (not shown). When examined morphologically, quiescent HEL-299 and WI-38 cells infected with KD1, KD3, or dl01/07 did not show cytopathic effects (CPE) until about 17 days p.i., whereas those infected with dl309 showed CPE at about 8 days p.i. (not shown).
The vectors were also limited in inducing CPE in primary human cells. In primary bronchial epithelial cells, primary small airway epithelial cells, and primary pulmonary artery endothelial cells, dl309 induced CPE at 3 days p.i., whereas KD1, KD3, and dl01/07 did not induce CPE until about 10 days p.i. Fig. 3e to g show typical results after 5 days for the epithelial cells and after 8 days for the endothelial cells. The endothelial cells were confluent when they were initially infected. The epithelial cell monolayers were 30 to 50% confluent when initially infected and then grew to confluency. This result implies that both proliferating and nonproliferating primary cells are refractory to KD1 and KD3.
KD1 and KD3 replicate well and are cytolytic in cancer cells.
We next tested whether the vectors would grow and be cytolytic in cancer cells. When the viability of A549 cells was examined by trypan blue exclusion (38), only 25% of the KD1-infected cells and 9% of the KD3-infected cells were alive at 5 days p.i., compared to 44 and 38% of cells infected with dl01/07 or dl309, respectively (Fig. 4a). With dl327 (ADP− E1A+), 94% of the cells were alive. When cell lysis was estimated by release of lactate dehydrogenase (38), KD1 and KD3 once again lysed cells faster than dl01/07 or dl309, and dl327 caused little lysis (Fig. 4b). Thus, ADP is required for efficient cell lysis, and overexpression of ADP increases the rate of cell lysis.
If ADP mediates cell lysis and virus release, as shown earlier using adp mutants (37, 38), then increased virus release should be observed with KD1 and KD3. A549 cells were infected with KD1, KD3, dl01/07, or dl309 and the titers of intracellular and extracellular virus were determined after 2 days. The total virus observed with each group was similar, about 1010 total PFU. However, 2 logs more KD1 and KD3 than dl01/07 or dl309 was released (Fig. 4c).
Replication and spread from cell to cell of Ad can be semiquantitated in a plaque development assay (38). In this assay, the number of plaques seen on a given day of the plaque assay are calculated as a percentage of the number of plaques seen at the end of the assay. That is, this assay measures the size and rate of development of plaques. We have shown that adp mutants have small plaques that are slow to develop (38). As expected from the overexpression of ADP, KD1 and KD3 formed plaques more rapidly than dl01/07 (Fig. 4d). The rate of plaque formation was about the same or a little faster than that seen with dl309 (Fig. 4d); the probable reason that KD1 and KD3 did not form plaques much faster than dl309 is that the 01/07 E1A mutation in KD1 and KD3 is slightly attenuating in A549 cells (Fig. 2b). That is, plaque formation is a balance between the rate of intracellular replication and the lysis of cells mediated by ADP.
The ability of KD1 and KD3 to spread from cell to cell was measured in a virus spread assay. A549 cells were infected with 10−3, 10−2, 10−1, 1.0, or 10 PFU of dl327, dl309, Ad5, dl01/07, KD1, KD2, or KD3 per cell. At low PFU/cell concentrations, the viruses must go through two or more rounds of replication, cell lysis, reinfection, and cell lysis in order to infect and destroy every cell in the monolayer. At 1.0 and 10 PFU/cell, the monolayer should be destroyed by the virus that initially infected the cells. Remarkably, at 7 days p.i., monolayers were virtually eliminated by KD1 and KD3 at 0.01 PFU/cell, whereas a concentration of 1.0 PFU/cell was required for dl01/07, KD2, dl309, and Ad5 (Fig. 4e). Much of the monolayer was destroyed by KD1 or KD3 at 0.001 PFU/cell. This result attests to the potency of ADP in mediating cell lysis and virus spread. KD1 and KD3 were also more effective than dl01/07 in killing other types of human cancer cell lines. At 7 to 10 days p.i., they killed HeLa (cervix), DU 145 (prostate), and PC-3 (prostate) cells at 10−2 PFU/cell and ME-180 (cervix) and Hep 3B (liver) cells at 10−1 PFU/cell (data not shown). From 10- to 100-fold more dl01/07 was required to kill these cells. These results indicate that the function of ADP is manifested in many cancer cell lines.
As another means to demonstrate the enhanced cytolytic and cell spreading ability of KD1 and KD3, we asked whether they could complement the growth and cell-to-cell spread of an E1− E3− ADP− vector expressing β-galactosidase (Ad/βgal) (Fig. 1f). As discussed, replication-defective vectors such as Ad/βgal can infect cells efficiently and express their transgene, but they cannot replicate in cells or spread from cell to cell. However, if the same cell is infected with Ad/βgal plus KD1 or KD3, then Ad/βgal should replicate and spread together with KD1 or KD3. Because KD1 and KD3 overexpress ADP, they should be more efficient in complementing the spread of Ad/βgal than the other viruses used in this study. Initially, we examined complementation of intracellular growth. A549 cells were infected with Ad/βgal alone, or with Ad/βgal plus KD1, KD3, dl01/07, dl309, or dl327. At 2 days p.i., Ad/βgal titers were determined by β-galactosidase expression (blue cells) in A549 cells. As expected, 3 logs more Ad/βgal was obtained from the Ad/βgal plus KD1 or KD3 coinfections than infections with Ad/βgal alone (Fig. 5a). The other viruses also complemented to about the same extent because all cells were infected with each virus, and ADP does not affect virus growth within cells (38) (Fig. 4c).
The ability of KD1 and KD3 to complement the spread of Ad/βgal from cell to cell was determined in an infectious-center assay. A549 cells were infected with 10 PFU of Ad/βgal alone per cell or with 10 PFU of Ad/βgal per cell plus 10 PFU of KD1, KD3, dl01/07, dl309, or dl327 per cell. After 2 h, cells were trypsinized, collected, diluted, and seeded onto monolayers of fresh A549 cells. Two hours is sufficient time for infection but not for most viral gene expression. After 4 days, the cells were stained with X-Gal. Four days is sufficient time for one or more rounds of replication, cell lysis, and reinfection. With Ad/βgal alone, only individual blue cells were obtained, no doubt the original infected cell (not visible in the print in Fig. 5b but indicated by the arrow in Fig. 5c). With Ad/βgal plus KD1 or KD3, numerous comet-shaped foci of blue cells were seen resulting from the spread of Ad/βgal from one originally infected cell (Fig. 5b). Most of the cells within a focus were stained with X-Gal (Fig. 5c). dl01/07 and dl309 formed fewer and smaller foci than KD1 and KD3 (Fig. 5c). With dl327 (ADP−), there was little spread from the original infected cell (Fig. 5c). In summary, KD1 and KD3 complement the replication of Ad/βgal to a similar extent as do dl01/07 and wild-type Ad (dl309), but the former are considerably more efficient than dl01/07 and dl309 in complementing the spread of Ad/βgal.
KD1 and KD3 reduce the growth of human cancer cells as tumors in nude mice.
KD1 and KD3 were examined for their ability to suppress tumor growth in a human cancer cell xenotransplant model in immunodeficient mice. Mouse or rat tumors could not be used because these species do not support replication of human Ads. A549 cells were mock-infected or infected with KD3, diluted into uninfected cells such that 1 in 10,000 cells was infected, and then implanted subcutaneously into each hind flank of nude mice. Tumor growth was measured over 5 weeks. Mock-infected cells grew into tumors averaging 461 μl, whereas KD3-infected cells barely grew (Fig. 6a). This result is consistent with the virus spread data (Fig. 4e) and supports the notion that KD3 can spread from cell to cell in tumors.
KD3 was effective when a single dose of 5 × 108 PFU in DMEM was injected directly into preformed tumors (Fig. 6b). The vectors also reduced A549 tumor growth at lower doses (5 × 107 PFU) given at weekly intervals (total of 2 × 108 PFU) (Fig. 6c). Tumors that received DMEM alone (mock infected) grew 13.3-fold over 4 weeks (Fig. 6c). Tumors injected with KD3 or KD1 grew only 2.6-fold, similar to dl309. With dl01/07, the control for KD3 and KD1, tumors grew eightfold.
KD3 and KD1 also reduced the growth in nude mice of human Hep 3B liver cancer cells. Tumors of about 100 μl were injected twice per week for 3 weeks with 5 × 107 PFU of KD1, KD3, or dl309. Tumors that received buffer alone grew 9-fold over 3 weeks and were projected to grow about 12-fold over 3.5 weeks (Fig. 6d) (after 3 weeks the mice had to be sacrificed because the tumors were becoming too large). Tumors injected with KD1 or KD3 grew fourfold, and those injected with dl309 grew twofold (Fig. 6d).
DISCUSSION
We have described two similar vectors, KD1 and KD3, that show promise as anticancer vectors. KD1 and KD3 were engineered to have three unique features. First, they overexpress ADP, an Ad protein that functions after virion assembly is complete to mediate cell lysis, the release of Ad from cells, and the spread of Ad to other cells. We reasoned that vectors that overexpress ADP should spread more effectively through tumors than other types of Ad vectors. Second, KD1 and KD3 contain two small deletions in E1A, transferred from the mutant dl01/07 (22); we reasoned that these mutations would allow vector replication in cancer cell lines but severely restrict their replication in primary or quiescent cells (22, 28). Thus, in a cancer treatment protocol where KD1 or KD3 is injected directly into the tumor, the vector should spread throughout the tumor while at the same time sparing noncancerous tissue. Third, the vectors lack the E3 proteins that counteract immunosurveillance; we reasoned that the vectors' inability to counter immunosurveillance might result in increased immune attack on the tumor and at the same time increase the safety of the vectors. We could not examine this feature because we used immunodeficient mice as our model.
Our results are consistent with most of our expectations. KD1 and KD3 markedly overexpress ADP as compared to their control, dl01/07. Interestingly, they also express ADP earlier in the infection cycle than dl01/07 and dl309, viruses that express wild-type levels of ADP. As expected from overexpression of ADP, KD1 and KD3 lysed cells and spread from cell to cell more efficiently than dl01/07. This superiority over dl01/07 was seen in nearly all cancer cell lines examined, indicating that ADP exerts its function in multiple cell types (see also reference 37). In A549 cells, KD1 and KD3 were also more efficient than dl309. These results are in accord with our earlier proposal, based on the phenotype of adp mutants, that ADP mediates efficient cell lysis and Ad spread (37, 38). KD3 was somewhat more efficient than KD1, conceivably because it expresses the E3-12.5K protein whereas KD1 does not. Both KD1 and KD3 were more effective than dl01/07 and equally as effective as dl309 in suppressing the growth of A549 tumors in nude mice. They were about twofold less effective than dl309 in suppressing Hep 3B tumors.
Although KD1 and KD3 express ADP earlier in infection than dl01/07, the time of synthesis of other late proteins was similar. This result is consistent with studies with adp mutants which indicated that the presence or absence of ADP does not affect the appearance of other late proteins (38). Although KD1 and KD3 lysed cells faster than dl01/07, the titers of CsCl-banded stocks were similar when infected KB suspension cultures were harvested after 2 or 3 days. This indicates that ADP overexpression does not cause cells to lyse before virus has assembled. This observation is important to understanding the role of ADP in Ad biology, and it is highly relevant to the use of KD1 and KD3 in cancer gene therapy.
The inclusion of the 01/07 mutation in KD1 and KD3 had the intended effect: the vectors grew in cancer cell lines, grew poorly in quiescent HEL-299 and WI-38 cells, and were much delayed in inducing CPE in primary cells. ONYX-015 also induced CPE in primary cells (15, 19). The 01/07 mutation causes a varied growth attenuation in cancer cells, as determined by the virus spread assay. The attenuation is mild in A549, DU 145, and PC-3 cells and is more restrictive in Hep 3B and ME-180 cells.
The regions in E1A deleted in 01/07 must induce numerous changes in the cell: the E1A-p300/CBP interaction will cause repression of genes; the E1A-pRB interaction will cause relief of pRB/E2F repression and the induction of E2F-mediated gene expression (5, 29, 41, 44). These alterations are required for efficient Ad replication in normal cells. Apparently, cancer cell lines exhibit different aspects of the E1A-induced cellular environment, and accordingly they differ in their ability to complement the 01/07 mutation. In fact, we do not know whether the ability of these cancer cells to complement KD1 and KD3 is directly related to whether they are dividing; it could be due to other changes in gene expression that are not directly related to cell division.
An interesting aspect of KD1 and KD3 is their ability to complement the replication, spread, and transgene expression of replication-defective Ad vectors. KD1 and KD3 complemented Ad/βgal in cell culture, and in preliminary experiments they complemented a replication-defective Ad vector expressing luciferase in Hep 3B tumors growing in nude mice. Coinfecting a tumor with KD3 and a vector that expresses an anticancer therapeutic protein should result in much more therapeutic protein in many more tumor cells than could be achieved by the replication-defective vector alone. The replication-defective vector should not grow in noncancerous cells because of the 01/07 mutation in KD3 (the vector requires KD3 for growth). Dissemination of the vector should be minimized because the vector and KD3 lack the E3 proteins that counter immunosurveillance. Tumors should be destroyed by both KD3 replication and the action of the therapeutic protein. Recombination may occur between KD3 and the replication-defective vector, but the end product will be a virus similar to dl01/07 or a replication-defective vector containing the gene for ADP.
An important question is whether the vectors will destroy noncancerous proliferating cells in the body. The vectors replicated in HEL-299 and WI-38 cells growing in 10% serum, but it is unlikely that these cells are truly normal. Importantly, the vectors were severely restricted in proliferating primary epithelial cells (Fig. 3e and f). It appears that, as with at least some cancer cell lines, the ability of cells to divide is not sufficient to support efficient replication of 01/07 mutants, and other characteristics must be present. If so, then proliferating normal cells in the body may not be a problem. We did not directly examine whether the vectors replicated in the normal tissues of the nude mice, because mice are well known to be nonpermissive for human Ad. We can say, however, that the tumor-bearing mice infected with KD1 or KD3 did not exhibit increased morbidity as compared to mock-, dl01/07-, or dl309-infected mice. In further consideration of safety, it should be noted that, in nature, subgroup C Ad are benign in adults except in cases of severe immunodeficiency (21). Further, the 01/07 mutation in E1A should preclude the remote possibility of E1A-mediated oncogenic cell transformation (22).
ACKNOWLEDGMENTS
K.D., K.T. and M.K. contributed equally to this work.
We thank Liqian Zhang and Jeffrey A. Whitsett for participating in the early phase of the research, Stanley Bayley and Thomas Shenk for viruses, Lynda Hawkins for pLKH, Nripendranath Mandel for pN30, Pat Farrar and Meribeth Broadway for advice, Chris Wells and Shari O'Brien for technical assistance, Joyce Weber for photography, and Jayma Mikes and Sue Sulkey for preparation of the figures and manuscript.
This work was supported by grant CA71704 from the National Institutes of Health.
REFERENCES
- 1.Barker D D, Berk A J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156:107–121. doi: 10.1016/0042-6822(87)90441-7. [DOI] [PubMed] [Google Scholar]
- 2.Bett A J, Krougliak V, Graham F L. DNA sequence of the deletion/insertion in early region 3 of Ad5 dl309. Virus Res. 1995;39:75–82. [PubMed] [Google Scholar]
- 3.Bilbao G, Contreras J L, Gomez-Navarro J, Curiel D T. Improving adenoviral vectors for cancer gene therapy. Tumor Targeting. 1998;3:59–79. [Google Scholar]
- 4.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 5.Branton P E. Early gene expression. In: Seth P, editor. Adenoviruses: basic biology to gene therapy. R. G. Austin, Tex: Landes Co.; 1999. pp. 39–58. [Google Scholar]
- 6.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 7.Chroboczek J, Bieber F, Jacrot B. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology. 1992;186:280–285. doi: 10.1016/0042-6822(92)90082-z. [DOI] [PubMed] [Google Scholar]
- 8.Crystal R G. In vivo and ex vivo gene therapy strategies to treat tumors using adenovirus gene transfer vectors. Cancer Chemother Pharm. 1999;43(Suppl.):S90–S99. doi: 10.1007/s002800051105. [DOI] [PubMed] [Google Scholar]
- 9.Flint J, Shenk T. Viral transactivating proteins. Annu Rev Genet. 1997;31:177–212. doi: 10.1146/annurev.genet.31.1.177. [DOI] [PubMed] [Google Scholar]
- 10.Freytag S O, Rogulski K R, Paielli D L, Gilbert J D, Kim J H. A novel three-pronged approach to kill cancer cells selectively: concomitant viral, double suicide gene, and radiotherapy. Hum Gene Ther. 1998;9:1323–1333. doi: 10.1089/hum.1998.9.9-1323. [DOI] [PubMed] [Google Scholar]
- 11.Goodrum F D, Ornelles D A. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J Virol. 1998;72:9479–9490. doi: 10.1128/jvi.72.12.9479-9490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall A R, Dix B R, O'Carroll S J, Braithwaite A W. p53-dependent cell death/apoptosis is required for a productive adenovirus infection. Nat Med. 1998;4:1068–1072. doi: 10.1038/2057. [DOI] [PubMed] [Google Scholar]
- 13.Hallenbeck P L, Chang Y-N, Hay C, Golightly D, Stewart D, Lin J, Phipps S, Chiang Y L. A novel tumor-specific replication-restricted adenoviral vector for gene therapy of hepatocellular carcinoma. Hum Gene Ther. 1999;10:1721–1733. doi: 10.1089/10430349950017725. [DOI] [PubMed] [Google Scholar]
- 14.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 15.Harada J N, Berk A J. p53-independent and -dependent requirements for E1B-55K in adenovirus type 5 replication. J Virol. 1999;73:5333–5344. doi: 10.1128/jvi.73.7.5333-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow E, Franza B R J, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haussman J, Ortmann D, Witt M, Veit M, Seidel W. Adenovirus death protein, a transmembrane protein encoded in the E3 region, is palmitoylated at the cytoplasmic tail. Virology. 1998;244:343–351. doi: 10.1006/viro.1998.9135. [DOI] [PubMed] [Google Scholar]
- 18.Hay J G, Shapiro N, Sauthoff H, Heitner S, Phupakdi W, Rom W N. Targeting the replication of adenoviral gene therapy vectors to lung cancer cells: the importance of the adenoviral E1b-55kD gene. Hum Gene Ther. 1999;10:579–590. doi: 10.1089/10430349950018652. [DOI] [PubMed] [Google Scholar]
- 19.Heise C, Sampson-Johannes A, Williams A, McCormick F, Von Hoff D D, Kirn D H. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3:639–645. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 20.Heise C C, Williams A M, Xue S, Propst M, Kirn D H. Intravenous administration of ONYX-015, a selectively replicating adenovirus, induces antitumoral efficacy. Cancer Res. 1999;59:2623–2628. [PubMed] [Google Scholar]
- 21.Horwitz M S. Adenoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2149–2171. [Google Scholar]
- 22.Howe J A, Mymryk J S, Egan C, Branton P E, Bayley S T. Retinoblastoma growth suppressor and a 300-kDa protein appear to regulate cellular DNA synthesis. Proc Natl Acad Sci USA. 1990;87:5883–5887. doi: 10.1073/pnas.87.15.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones N, Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979;17:683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 24.Kirn D, Hermiston T W, McCormick F. ONYX-015: clinical data are encouraging. Nat Med. 1998;4:1341–1342. doi: 10.1038/3902. [DOI] [PubMed] [Google Scholar]
- 25.Kirn D H. Development of an E1B, 55 kDa gene-deleted, selectively replicating adenovirus for the treatment of cancer: ONYX-015. In: Seth P, editor. Adenoviruses: basic biology to gene therapy. R. G. Austin, Tex: Landes Co.; 1999. pp. 201–206. [Google Scholar]
- 26.Mahr J A, Gooding L R. Immune evasion by adenoviruses. Immunol Rev. 1999;168:121–130. doi: 10.1111/j.1600-065x.1999.tb01287.x. [DOI] [PubMed] [Google Scholar]
- 27.Martin M E, Berk A J. Corepressor required for adenovirus E1B 55,000-molecular-weight protein repression of basal transcription. Mol Cell Biol. 1999;19:3403–3414. doi: 10.1128/mcb.19.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montell C, Courtois G, Eng C, Berk A. Complete transformation by adenovirus 2 requires both E1A proteins. Cell. 1984;36:951–961. doi: 10.1016/0092-8674(84)90045-x. [DOI] [PubMed] [Google Scholar]
- 29.Prives C. Signaling to p53: breaking the MDM2-p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez R, Schuur E R, Lim H Y, Henderson G A, Simons J W, Henderson D R. Prostate attenuated replication competent adenovirus (ARCA) CN706: a selective cytotoxic for prostate-specific antigen-positive prostate cancer cells. Cancer Res. 1997;57:2559–2563. [PubMed] [Google Scholar]
- 31.Rothmann T, Hengstermann A, Whitaker N J, Scheffner M, zur Hausen H. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J Virol. 1998;72:9470–9478. doi: 10.1128/jvi.72.12.9470-9478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scaria A, Tollefson A E, Saha S K, Wold W S M. The E3-11.6K protein of adenovirus is an Asn-glycosylated integral membrane protein that localizes to the nuclear membrane. Virology. 1992;191:743–753. doi: 10.1016/0042-6822(92)90250-s. [DOI] [PubMed] [Google Scholar]
- 33.Seth P, Katayose Y, Rakkar A N S. Adenoviral vectors for cancer gene therapy. In: Seth P, editor. Adenoviruses: basic biology to gene therapy. R. G. Austin, Tex: Landes Co.; 1999. pp. 103–120. [Google Scholar]
- 34.Shinoura N, Yoshida Y, Tsunoda R, Ohashi M, Zhang W, Asai A, Kirino T, Hamada H. Highly augmented cytopathic effect of a fiber-mutant E1B-defective adenovirus for gene therapy of gliomas. Cancer Res. 1999;59:3411–3416. [PubMed] [Google Scholar]
- 35.Thimmappaya B, Weinberger C, Schneider R J, Shenk T. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell. 1982;31:543–551. doi: 10.1016/0092-8674(82)90310-5. [DOI] [PubMed] [Google Scholar]
- 36.Tollefson A E, Hermiston T W, Wold W S M. Preparation and titration of CsCl-banded adenovirus stocks. In: Wold W S M, editor. Adenovirus methods and protocols. Totowa, N.J: Humana Press, Inc.; 1998. pp. 1–9. [Google Scholar]
- 37.Tollefson A E, Ryerse J S, Scaria A, Hermiston T W, Wold W S M. The E3-11.6kDa adenovirus death protein (ADP) is required for efficient cell death: characterization of cells infected with adp mutants. Virology. 1996;220:152–162. doi: 10.1006/viro.1996.0295. [DOI] [PubMed] [Google Scholar]
- 38.Tollefson A E, Scaria A, Hermiston T W, Ryerse J S, Wold L J, Wold W S M. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tollefson A E, Scaria A, Saha S K, Wold W S M. The 11,600-MW protein encoded by region E3 of adenovirus is expressed early but is greatly amplified at late stages of infection. J Virol. 1992;66:3633–3642. doi: 10.1128/jvi.66.6.3633-3642.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnell A S, Grand R J, Gallimore P H. The replicative capacities of large E1B-null group A and group C adenoviruses are independent of host cell p53 status. J Virol. 1999;73:2074–2083. doi: 10.1128/jvi.73.3.2074-2083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White E. Regulation of apoptosis by adenovirus E1A and E1B oncogenes. Semin Virol. 1998;8:505–513. [Google Scholar]
- 42.Wildner O, Blaese R M, Morris J C. Therapy of colon cancer with oncolytic adenovirus is enhanced by the addition of herpes simplex virus-thymidine kinase. Cancer Res. 1999;59:410–413. [PubMed] [Google Scholar]
- 43.Wildner O, Morris J C, Vahanian N N, Ford H J, Ramsey W J, Blaese R M. Adenoviral vectors capable of replication improve the efficacy of HSVtk/GCV suicide gene therapy of cancer. Gene Ther. 1999;6:57–62. doi: 10.1038/sj.gt.3300810. [DOI] [PubMed] [Google Scholar]
- 44.Wold W S M, Chinnadurai G. Adenovirus proteins that regulate apoptosis. In: Cann A J, editor. DNA virus replication. Oxford, United Kingdom: Oxford University Press; 2000. pp. 200–232. [Google Scholar]
- 45.Wold W S M, Tollefson A E. Adenovirus-host interactions to subvert the host immune system. In: Seth P, editor. Adenoviruses: basic biology to gene therapy. R. G. Austin, Tex: Landes Co.; 1999. pp. 243–250. [Google Scholar]
- 46.Wold W S M, Doronin K, Toth K, Kuppuswamy M, Lichtenstein D L, Tollefson A E. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr Opin Immunol. 1999;11:380–386. doi: 10.1016/S0952-7915(99)80064-8. [DOI] [PubMed] [Google Scholar]
- 47.Wold W S M, Tollefson A E. Adenovirus E3 proteins: 14.7K, RID, and gp19K inhibit immune-induced cell death; adenovirus death protein promotes cell death. Semin Virol. 1998;8:515–523. [Google Scholar]
- 48.Yu D-C, Chen Y, Seng M, Dilley J, Henderson D R. The addition of adenovirus type 5 region E3 enables Calydon virus 787 to eliminate distant prostate tumor xenografts. Cancer Res. 1999;59:4200–4203. [PubMed] [Google Scholar]