Abstract
Global warming has a severe impact on the flowering time and yield of crops. Histone modifications have been well-documented for their roles in enabling plant plasticity in ambient temperature. However, the factor modulating histone modifications and their involvement in habitat adaptation have remained elusive. In this study, through genome-wide pattern analysis and quantitative-trait-locus (QTL) mapping, we reveal that BrJMJ18 is a candidate gene for a QTL regulating thermotolerance in thermotolerant B. rapa subsp. chinensis var. parachinensis (or Caixin, abbreviated to Par). BrJMJ18 encodes an H3K36me2/3 Jumonji demethylase that remodels H3K36 methylation across the genome. We demonstrate that the BrJMJ18 allele from Par (BrJMJ18Par) influences flowering time and plant growth in a temperature-dependent manner via characterizing overexpression and CRISPR/Cas9 mutant plants. We further show that overexpression of BrJMJ18Par can modulate the expression of BrFLC3, one of the five BrFLC orthologs. Furthermore, ChIP-seq and transcriptome data reveal that BrJMJ18Par can regulate chlorophyll biosynthesis under high temperatures. We also demonstrate that three amino acid mutations may account for function differences in BrJMJ18 between subspecies. Based on these findings, we propose a working model in which an H3K36me2/3 demethylase, while not affecting agronomic traits under normal conditions, can enhance resilience under heat stress in Brassica rapa.
Subject terms: Agricultural genetics, Plant breeding, Abiotic
The study reveals that the BrJMJ18 gene, encoding an H3K36me2/3 Jumonji demethylase, is a candidate gene for a QTL regulating thermotolerance in a thermotolerant Brassica rapa subspecies, and its allele (BrJMJ18Par) can modulate flowering time, plant growth, and chlorophyll biosynthesis in a temperature-dependent manner.
Introduction
Temperature plays a crucial role in determining the pace of plant development. Given the current scenario of climate change, rising temperatures can accelerate flowering and shorten developmental phases in crops. This may lead to significant reductions in agricultural yields, posing a widespread risk of food insecurity. Primary production needs to increase, and crops that use resources more efficiently and display increased resilience to unpredictable climatic events need to be developed urgently. The use of genetically modified crops are largely limited; therefore, the use of natural genetic variations in plant breeding continues to underpin the improvement of all major crops. It’s vital to uncover the mechanisms underlying the selection of genetic variants—both derived and preexisting—that enable crops to thrive in new and challenging environments. By comprehending these mechanisms, we can gather knowledge and germplasms essential for crafting resilient crop varieties capable of confronting the ongoing threats posed by global warming.
Brassica rapa L. (B. rapa) is an important oilseed and vegetable crop and is the subgenome donor of two other important Brassica crops, Brassica napus and Brassica juncea. B. rapa demonstrates remarkable adaptability, thriving across diverse habitats spanning from sea level to elevations exceeding 4500 meters and also from cold to tropical areas. All these subspecies or variants occur over a wide range of ecological conditions, and some individual species have adapted to particular microclimates to meet human needs or their survival1,2. Plant flowering time is an important life history trait underlying reproductive fitness and is sensitive to local growing conditions. Wide variation in flowering time exists between and within B. rapa morphotypes; therefore, the widely cultivated B. rapa crops are thus ideal objects to illustrate the diversity that can be created by domestication or/and breeding3,4. Among these morphotypes, B. rapa subsp. chinensis var. parachinensis (abbreviated to Par) is an understudied member within the realm of B. rapa. Par is a vegetable primarily valued for its edible components, encompassing the stems, stem leaves, and terminal inflorescences, which are commonly consumed in Asia. The Par variety displays two distinctive domestication traits not found in other leafy B. rapa crops. Firstly, it exhibits vernalization-independent flowering, in alignment with its suitability as a year-round vegetable. Secondly, building upon the first characteristic, Par was further domesticated to maintain stable flowering time under the warm conditions in southern China. This stability is crucial for preserving the quality and yield of the commercial organ of Par since flowering at inappropriate times can impact them. In other words, the recent domestication of Par primarily focuses on its flowering time adaptation to high temperatures, however, the procedure and genetic basis of this stepwise domestication is unknown.
High temperatures can impact the flowering time of B. rapa in various ways. The timing of the heat stress is crucial since its impact varies during the vegetative growth period and the reproductive growth period5. Here, we would like to discuss the effect of high temperatures on the flowering time of B. rapa plants during their vegetative growth. Del Olmo et al. 6 studied R-o-18, an oilseed rapa variety, and found that plants grown at 28 °C flowered later than those at 21 °C6. While some studies on vegetative B. rapa, like summer-planting Chinese cabbage and non-heading Chinese cabbage, have shown that high-temperature exposure during the seedling stage leads to premature bolting, affecting yield and quality7,8. Some Par varieties, as reported by Lu (2022), can start flowering prematurely when temperatures exceed 30 °C, leading to stunted growth and slow development, significantly affecting their production9. Rameeh (2012) investigated the effects of high temperature on the flowering time of Indian mustard (B. juncea)10, revealing that late-planted Indian mustard experienced early flowering due to heat stress.
The definitions of ′thermotolerance′ and ′heat stress′ are influenced by the plant’s optimal range and the temperature conditions in its cultivation region. As reported by Morrison and Stewart11, during the flowering stage, the critical threshold temperature causing seed yield losses for all Brassica species were found to be 29.5 °C and mean maximum temperatures of more than 29 °C during vegetative development led to a decline in flower numbers across all Brassica species. We also obtained meteorological data from the China Meteorological Administration regarding the average summer temperatures in Guangdong province, the region where Par domestication predominantly occurs. These temperatures have exhibited an average of 29.4 °C over the past half-century. By integrating this information, we propose that the approximate threshold for Par domestication can be established at around 29 °C.
The present study used var. parachinensis, which has adapted to gain plasticity of flowering time and growth duration under hot conditions, to explore the genetic variations recorded in its genome by sequencing a collection of different B. rapa subspecies and determined the genomic signatures that underlie B. rapa’s improved high-temperature responses. We demonstrated a stepwise model for the speciation of subsp. chinensis var. parachinensis during domestication, and further identified BrJMJ18Par (the BrJMJ18 allele from Par) as a candidate gene for a QTL regulating thermotolerance in var. parachinensis. Through investigations involving overexpression and CRISPR/Cas9 mutant plants, we demonstrated that BrJMJ18Par plays a pivotal role in mediating flowering time and plant growth in response to temperature fluctuations. Overexpression of BrJMJ18Par can adjust the expression of BrFLC3 and flowering time, while the outcome of BrFLC3 is less than direct in the WT Par plants. These findings not only advance our understanding of speciation during domestication but also offer insights and germplasm resources for developing resilient B. rapa crop varieties in response to the challenges of global warming.
Results
A stepwise domestication model of Brassica rapa subsp. chinensis var. parachinensis (Par)
The germplasm collection used in this study included a new set of 169 accessions and a previous set of 41 subsp. rapifera germplasms12 from all over the world (Supplementary Fig. 1a and Supplementary Table 1). We sequenced the genomes of the plants in the new set, resulting in a sequencing dataset from a total of 210 varieties of six Brassica rapa morphotypes, including 15 subsp. oleifera (Ole), 51 subsp. rapifera (Raf), 9 subsp. chinensis var. narinosa (Nar), 50 subsp. chinensis (Pak-Choi, PC), 41 subsp. chinensis DG (Dark Green, DG), and 44 subsp. chinensis var. parachinensis (Par). The dataset consisted of 2.9 Tb of 125 bp paired-end reads, with more than 10 × coverage on average for each sample. Using the B. rapa genome v3.0 as reference genome (downloaded from BRAD http://brassicadb.cn/), we obtained a total of 2,690,680 and 210,664 high-quality single nucleotide polymorphisms (SNPs) and insertions/deletions (InDels; Table 1), respectively.
Table 1.
General information on genetic variation in the 210 B. rapa lines
| all | Raf | Ole | PC+Nar | DG | Par | ||
|---|---|---|---|---|---|---|---|
| Sample Size | 210 | 51 | 15 | 59 | 41 | 15 | |
| Variants | SNP | 2,690,680 | 2,237,437 | 2,026,926 | 1,808,392 | 1,750,704 | 1,371,766 |
| Unique SNPa | NA | 142,903 | 11,606 | 25,649 | 8,434 | 7,737 | |
| InDel | 210,664 | 169,012 | 149,831 | 1,921,857 | 135,267 | 104,795 | |
| Unique InDela | NA | 12366 | 880 | 2504 | 833 | 778 | |
| PIb | 0.002507 | 0.002329 | 0.001788 | 0.002044 | 0.001896 | 0.001524 | |
| taijima D | 3.172689 | 2.120287 | 0.783791 | 1.623865 | 1.533449 | 1.201749 | |
| LD | LDc 0.2 | 1.72 | 0.63 | 120.4 | 6.85 | 11.8 | 37.8 |
| LD half | 0.11 | 0.05 | 0.19 | 0.5 | 0.98 | 2.84 | |
aUnique single nucleotide polymorphisms (SNPs) and insertion/deletions (InDels) are variations specific to each group.
bPI, nucleotide diversity within each group. PI was calculated based on the genotypes of each line at the SNP positions using BioPerl.
cLD (linkage disequilibrium) blocks were calculated based on SNPs with minor allele frequency (MAF) greater than 0.05 using Haploview software. LD decay was calculated based on the squared correlation coefficient (r2 = 0.2 or 0.5) by pairwise physical distance between SNPs.
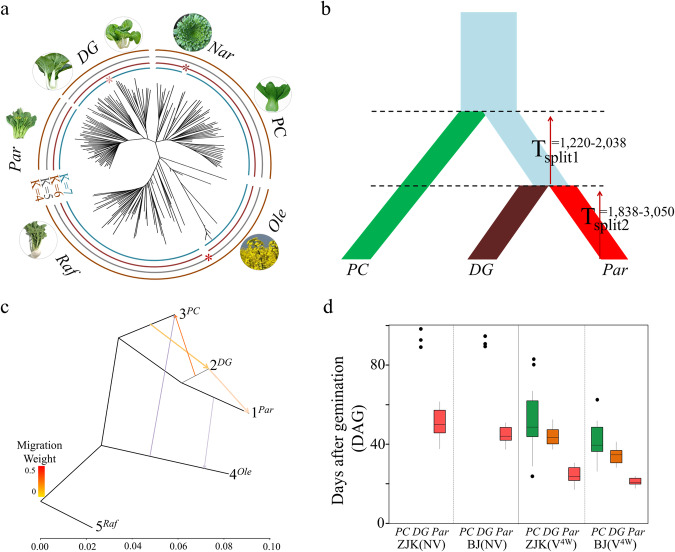
We then determined the phylogenetic relationships of the 210 lines. Analyses of the genetic structure recovered seven clusters using STRUCTURE (Fig. 1a and Supplementary Fig. 1b). The results showed that Par was positioned at the most distant point from the ancient Raf group, while PC was closest to the root and DG was found at an intermediate position between Par and PC (Fig. 1a and Supplementary Fig. 1c, d). In the PCA plot, the transitional feature of DG between Par and PC was more evident (Supplementary Fig. 1c). To further understand the evolutionary history of Par, we used ∂a∂i analysis to estimate the demographic modeling of PC, DG, and Par (Fig. 1b). The best model supported an initial common ancestor split from PC followed by a subsequent divergence into DG and Par. The time span from the initial divergence of PC to the split of PC was approximately 1220–2038 years, and the divergence of DG and Par occurred approximately 1838–3050 years ago (Fig. 1b and Supplementary Table 2). Besides, the data of gene flow evaluation and nucleotide diversity divergence of the three groups further revealed a transitional feature of DG during Par evolution (Fig. 1c and Supplementary Fig. 2).
Fig. 1. A stepwise model for speciation of subsp. chinensis var. parachinensis (Par).
a Neighbor-joining (N-J) tree of the 210 lines. An unrooted phylogenetic tree of the 210 lines was constructed using the N-J method under the Kimura 2-parameter model implemented in MEGA-CC (v 7.0). Bootstrap support > 50% calculated from 1000 replicates is shown. b The best demographic model evaluated using ∂a∂i. At time Tsplit. The time of the split was estimated in the generation time, which was converted to years, assuming one generation per year. We used STRUCTURE K = 6 as the likelihood ratio, which can also satisfy the requirements of both STRUCTURE and empirical classification. c Inferred B. rapa tree showing the directions of gene flow in each group. Arrows indicate the direction of gene flow, while the line colors represent the migration weight based on the sample number. The horizontal branch length is proportional to the amount of genetic drift that has occurred on the branch. Scale bars represent 100-fold average standard error (SE) for the entries in the sample covariance matrix. d Population phenotypic differentiation for the bolting time of PC, DG, and Par in Zhangjiakou (ZJK) and Beijing (BJ), respectively. Flowering time of 50 PC, 41 DG and 44 Par accessions from our germplasm collection were investigated. The flowering time was evaluated with (denoted as V4W) or without (denoted as NV) 4 weeks vernalization in ZJK and BJ, respectively. Bolting time, days after germination (DAG), was defined as the number of days from sowing to the appearance of the visible bud. The box encompasses two middle quartiles, with a central line showing the median. Whiskers extend to the furthest data point within 1.5 times the interquartile range. Par represents B. rapa subsp. chinensis var. parachinensis, DG represents B. rapa subsp. chinensis var. Dark-green, PC represents B. rapa subsp. chinensis (pak choi).
In terms of phenotypic analyses, flowering time varied greatly among the three groups (Fig. 1d). Par was the only subspecies that could bloom without vernalization; however, three accessions of DG could still flower without cold, suggesting that some early-flowering genetic components have started to deposit in DG during selection (Fig. 1d, the left panel). A 4-week vernalization was then imposed on the three subspecies to determine flowering-time variations. We found that PC exhibited the longest bolting duration, Par bloomed earliest, while DG fell in between (Fig. 1d, the right panel). With regard to other morphological features, the three subspecies resemble each other at the seedling stage (Supplementary Fig. 3, upper panel); while at the adult juvenile stage, PC and DG share similar plant architecture, and DG and Par have similar the leaf shapes and color (Supplementary Fig. 3, bottom panel). All these molecular and phenotypic findings suggest a stepwise selection for Par’s evolution: DG originated from a particular PC population, and Par subsequently diverged from DG through the enrichment of early-flowering mutations and other adapted traits for local conditions.
Genetic basis of subsp. chinensis var. parachinensis domestication identified via Coupling genome-wide pattern analysis and quantitative-trait-locus (QTL) mapping
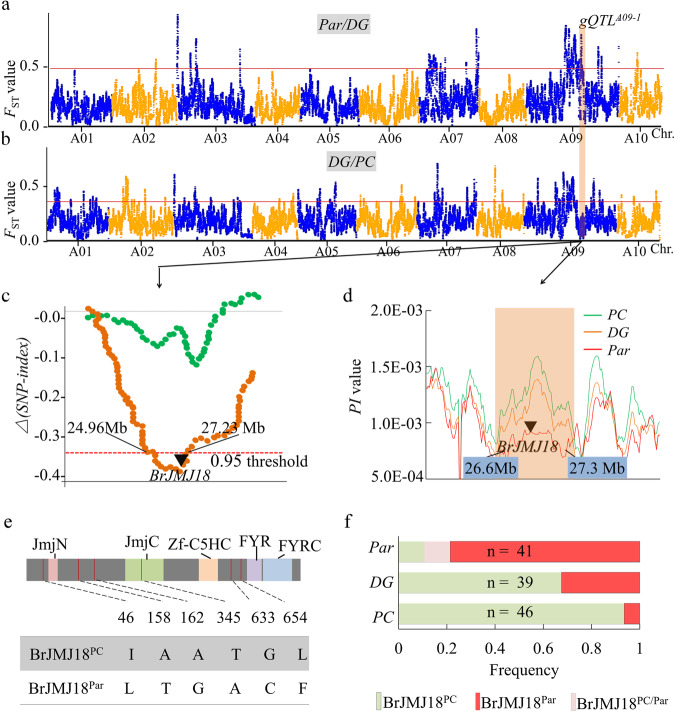
We divided Par’s speciation into two stages: DG/PC (from PC to DG) and Par/DG (from DG to Par). The close level of nucleotide diversity (PI) of the three groups (Supplementary Fig. 2b) indicated very weak bottlenecks during selection. However, for the second step, due to significant flowering time variation yet low PI value between DG and Par, we speculated a mild but precise selection in Par’s speciation. We identified candidate genomic regions selected during each step. Calculation of the FST (5% threshold) detected 85 and 51 selected regions of the first and second steps, respectively. The selected regions in DG/PC and Par/DG span 21.0 Mb (4.4% of the genome) and 25.8 Mb (5.4%), harboring 4632 and 3089 genes, respectively (Fig. 2a, b and Supplementary Data 1). Among these selective genes, 1964 (42.4%) and 1184 (38.2%) showed differential expression during floral transition according to the RNA seq data (Supplementary Data 1). Gene Ontology (GO) analyses13 revealed 171 specific (p < 0.05) enriched GO terms in DG/PC comparison, whereas only 45 terms were enriched in Par/DG comparison (Supplementary Data 2), confirming a precise selection during the second step.
Fig. 2. Genetic basis of subsp. chinensis var. parachinensis domestication.
a–b Genetic basis of subsp. chinensis var. parachinensis selection. (a) and (b) show highly divergent regions between Par and DG, and DG and PC, respectively. The FST values are plotted against the position on each chromosome. The horizontal pink lines indicate the genome-wide thresholds (5%) of differentiation signals. The peach bar on Chr. A09 intersecting (a) and (b) indicates the candidate selection sweep (A09: 26,630,001…27,300,000) studied. c A temperature-responsive QTL(HT) co-locates with the region of gQTLA09-1. An F2 population established from a cross between Par and PC, was used for the analysis. Half of the 4-week-old seedlings continued to grow under normal conditions (NC), while the other half of the seedlings were moved to high temperature (HT) conditions until flowering. Flowering-related quantitative trait loci (ftQTLs) responding to different temperatures were then identified. The green and brown dot lines indicate QTLs responding to NC and HT, respectively. The horizontal red line indicates the 95% confidence interval of Δ(SNP-index). gQTL represents QTLs (quantitative trait locus) associating with Par domestication generated from (a). Black triangle, the genome position of BrJMJ18. d The local π value features for the selection sweep on chromosome A09 (26,630,001…27,300,000) of PC, DG, and Par around gQTLA09-1. Black triangle, the genome position of BrJMJ18. e, f Sequence polymorphisms and haplotype analysis of BrJMJ18 in PC, DG, and Par. Red lines in (e) indicate different nucleotide alterations and corresponding amino acid substitutions are shown. JmjN: Jumonji N-terminal domain; JmjC: Jumonji C-terminal domain; Zf-C5HC2: C5HC2 zinc finger domain; FYRN: FY-rich domain, N-terminal region; FYRC: FY-rich domain, C-terminal region. The alteration of allelic frequencies of BrJMJ18 in PC, DG, and Par are shown in (f). BrJMJ18PC and BrJMJ18Par represent the BrJMJ18 allele coming from PC and Par, respectively; n denotes the number of lines of PC, DG, and Par used for the analysis.
Given Par’s domestication focus on stable flowering time under warm temperatures, we expected the enrichment of genes related to flowering time and temperature response in its selection. Compared with PC and DG, the higher thermotolerance of Par was preliminarily phenotypically confirmed (Supplementary Fig. 4). Besides, the flowering time of Par was also less sensitive to cultivation environment change (Fig. 1d and Supplementary Fig. 5). We then recalled all the GO terms associated with reproduction and abiotic stress, and confirmed that these terms were specifically (p < 0.05) enriched in the Par/DG comparison but not, or less frequently, in the DG/PC comparison (Supplementary Fig. 6a). To define Par-speciation-related genes, we concentrated on loci solely distinguishing Par from DG, rather than DG from PC. This yielded 24 loci and 964 candidate genes (Supplementary Fig. 6b and Supplementary Data 3), including 373 differentially expressed genes (DEGs). To further narrow down target genes, we phenotyped Par × PC F2 population for flowering time under normal (22/22 °C, 16/8 h; hereafter shortened as NC) and high temperature (29/29 °C, 16/8 h; hereafter shortened as HT) chamber conditions and used bulked segregant analysis sequencing (BSA-seq) approach for mapping heat-responsive QTLs. A total of 13 HT-associated quantitative trait loci (QTL(HT)s), solely responding to HT, were then identified (Supplementary Data 4).
BrJMJ18 is a candidate gene for a QTL regulating thermotolerance in Par
We concentrated in a selection sweep on chromosome A09 (26,630,001…27,300,000, designated as gQTLA09-1, marked in Fig. 2a–d), because 1) the flanking region of gQTLA09-1 did not co-locate within any of the reported flowering time (ft) ftQTLs in B. rapa, and 2) it overlapped with the region of one of the QTL(HT)s (Fig. 2d). A total of 77 genes, the expression of 18 of which, including 8 DEGs, were detectable during flowering, were mapped to the gQTLA09-1 region (Supplementary Data 3). Most of the 77 genes were independent of floral transition, except for three Jumonji C (JMJC) domain-containing proteins (Supplementary Data 3). In Arabidopsis, ten of the 21 JMJ genes are experimentally confirmed to affect flowering time, and fourteen of them respond to short-term heat stress (Supplementary Fig. 7). We then found that BrJMJ18 (BraA09g034190.3 C) was the most highly expressed BrJMJ, while the other two were undetectable; and BrJMJ18 was differently expressed during floral transition in DG and Par, but not PC, under NC conditions (Supplementary Data 5). Therefore, we proposed BrJMJ18 as the best candidate gene contributing to the speciation of Par in gQTLA09-1.
We then used the sequenced genomes of 58 PC, 41 DG, and 44 Par accessions (Supplementary Table 1) to scan for selection signatures surrounding BrJMJ18. A sharp reduction of nucleotide diversity was found between Par and DG at the BrJMJ18 locus, while by contrast, no significant reduction was found between DG and PC (Fig. 2d). In addition, BrJMJ18 was found to be located in a linkage disequilibrium (LD) block of Par/DG (Supplementary Fig. 8). Furthermore, we conducted a haplotype analysis of BrJMJ18 in PC, DG, and Par. A total of 71 DNA polymorphisms, comprising 13 in the promoter and 26 in the exons, were found in the gene body of BrJMJ18. Since no significant differences of the expression pattern of BrJMJ18 were found during flowering (Supplementary Data 5) and in the response to high temperature (Supplementary Fig. 9) between DG and Par, therefore, we only used the six nonsynonymous SNPs, resulting in six amino acid substitutions of BrJMJ18, for haplotype classification (Fig. 2e). A total of 126 of the 135 genotypes were then classified into two major haplotype groups: BrJMJ18PC and BrJMJ18Par (BrJMJ18PC is the prominent haplotype in PC and DG, while BrJMJ18Par mainly existed in Par) (Fig. 2f). The frequency of BrJMJ18Par was very low (6.5%, n = 46) in PC, and increased to 23.1% (n = 39) in DG, and finally expanded to 90.2% (n = 41) in Par (Fig. 2f). We further noticed that BrJMJ18PC and BrJMJ18Par were evenly represented in the ancient subsp. rapifera and subsp. oleifera groups, respectively (Supplementary Fig. 10). These observations suggested that the variations at BrJMJ18Par were not important for early B. rapa speciation, but conferred advantages to subsequent speciation or/and local adaptation.
Overexpression of BrJMJ18Par delays flowering under both greenhouse and field high temperature conditions
The effect of BrJMJ18PC and BrJMJ18Par on thermotolerance was then preliminary confirmed in the natural DG and Par group and the “Par × PC” F2 population mentioned above under NC and HT conditions, respectively, expressing as high temperature exerted a stronger effect of flowering on BrJMJ18PC-carrying lines than on BrJMJ18Par-carrying lines (Supplementary Fig. 11).
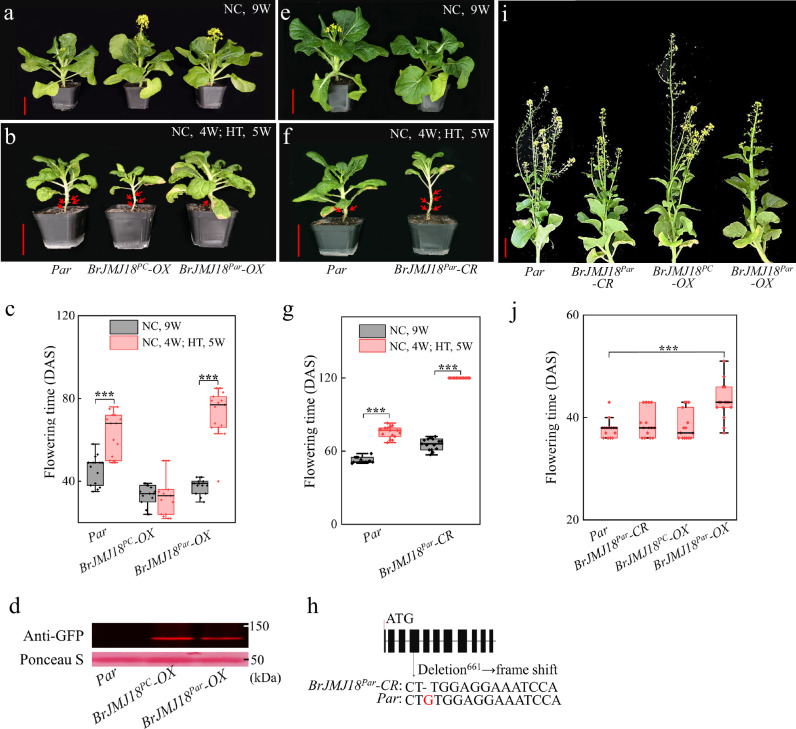
To characterize BrJMJ18PC and BrJMJ18Par, BrJMJ18PC- and BrJMJ18Par-OX plants of Par grown in a greenhouse were used for analysis (Fig. 3). The open reading frames (ORFs) of BrJMJ18PC and BrJMJ18Par driven by the cauliflower mosaic virus 35 S promoter (35 S), respectively, were transformed into Par plants, Transgenic T1 lines with similar protein expressions were used for study (Fig. 3d). Under NC, both BrJMJ18PC- and BrJMJ18Par-OX plants bolted earlier than the Par controls (Fig. 3a, c). While under HT, high temperature significantly delayed bolting in BrJMJ18Par-OX, but did not affect bolting in BrJMJ18PC-OX plants (Fig. 3b, c). To further characterize BrJMJ18Par, the BrJMJ18 knockout lines of Par plants, denoted as BrJMJ18Par-CR, generated by Clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), were used for analysis (Fig. 3e–h). Loss of BrJMJ18Par induced late flowering under both NC and HT conditions, and the flowering time delay induced by high temperature was also significantly broadened in BrJMJ18Par-CR plants, which is surprisingly similar to the changing trend of BrJMJ18Par-OX plants (BrJMJ18Par-CR did not bolt in 120 days under HT, and DAS was then set as 120). We also analyzed the flowering time of BrJMJ18 transgenic lines (AtJMJ18::BrJMJ18orf-GFP) in Arabidopsis (Supplementary Fig. 12), and found that under HT, the AtJMJ18::BrJMJ18Par-GFP plants flowered much later than the AtJMJ18::BrJMJ18PC-GFP plants.
Fig. 3. Flowering characterizations of the BrJMJ18 transgenic Par plants under greenhouse and field conditions, respectively.
a–h Phenotypes of the BrJMJ18 transgenic Par plants grown in the greenhouse under different temperature conditions. a BrJMJ18 transgenic Par plants grown under normal conditions (NC) for 9 weeks. Scale bar, 5 cm. b Phenotypes of the BrJMJ18 transgenic Par plants grown under NC conditions for 4 weeks, following another 5 weeks under high temperature (HT) conditions. Scale bar, 5 cm. Red arrows, withered leaves. c Flowering time of plants is shown in (a) and (b). DAS, days after sowing. Data are means ± SD, n = 15. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t-test (***p < 0.001) Par, p = 2.53 × 10−5; BrJMJ18Par-OX, p = 9.28 × 10−9. d Confirmation of BrJMJ18 protein expression in the transgenic plants by immunoblotting analysis. Ponceau S staining was used to assess equal loading. e, f Flowering characterizations of BrJMJ18 knockout Par line, BrJMJ18Par-CR, in greenhouse under different temperatures. g Flowering time of plants shown in (e) and (f). Red arrows, withered leaves. Data are means ± SD, n = 15. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t-test (***p < 0.001) Par, p = 2.32 × 10−10; BrJMJ18Par-CR, p = 1.47 × 10−16. The experiments in (a–f) were repeated three times with similar results. h Confirmation of BrJMJ18Par-CR by Sanger sequencing. A single base pair deletion (G → /) at the + 661 position after the ATG leads to a frame-shift mutation of the BrJMJ18 protein. i, j Par, BrJMJ18PC- and BrJMJ18Par-OX plants were planted in a tunnel greenhouse under natural field conditions in the summer (15th, Jul. … 29th, Sept.) of 2021 to simulate extremely high-temperature conditions for flowering time characterization. Data are means ± SD, n = 20. Asterisks indicate significant differences between Par and BrJMJ18Par-OX, two-tailed Student’s t-test (***p < 0.001), p = 8.19 × 10−5. For box plots (c, g, and j), the box encompasses two middle quartiles, with a central line showing the median. Whiskers extend to the furthest data point within 1.5 times the interquartile range. Source data are provided as a Source Data file.
We then planted Par, BrJMJ18PC- and BrJMJ18Par-OX plants in a tunnel greenhouse in the summer (15th, Jul. … 29th, Sept. 2021) to simulate extreme field high temperature conditions for further flowering time characterization. In the tunnel, daytime temperatures reached up to 52 °C and dropped to a low of 31 °C, while nighttime temperatures ranged from 33 °C to 24 °C. We found that BrJMJ18Par-OX plants flowered later than controls, in line with the changing direction in the greenhouse under HT (Fig. 3i, j). Besides, the BrJMJ18Par-OX plants demonstrated superior commercial quality compared with other plants (Fig. 3i).
All the above results established that both alleles of BrJMJ18 promote flowering under NC, but BrJMJ18Par evolved to obtain the function of delaying flowering under HT conditions. Moreover, because of the consistent flowering phenotype of BrJMJ18Par-OX and BrJMJ18Par-CR Par plants under high-temperature conditions, we hypothesized that JMJ18Par could be deactivated by high temperature, which indicated that BrJMJ18Par functions in flowering time control in a temperature-dependent manner.
BrJMJ18 is a histone H3 lysine 36 demethylase
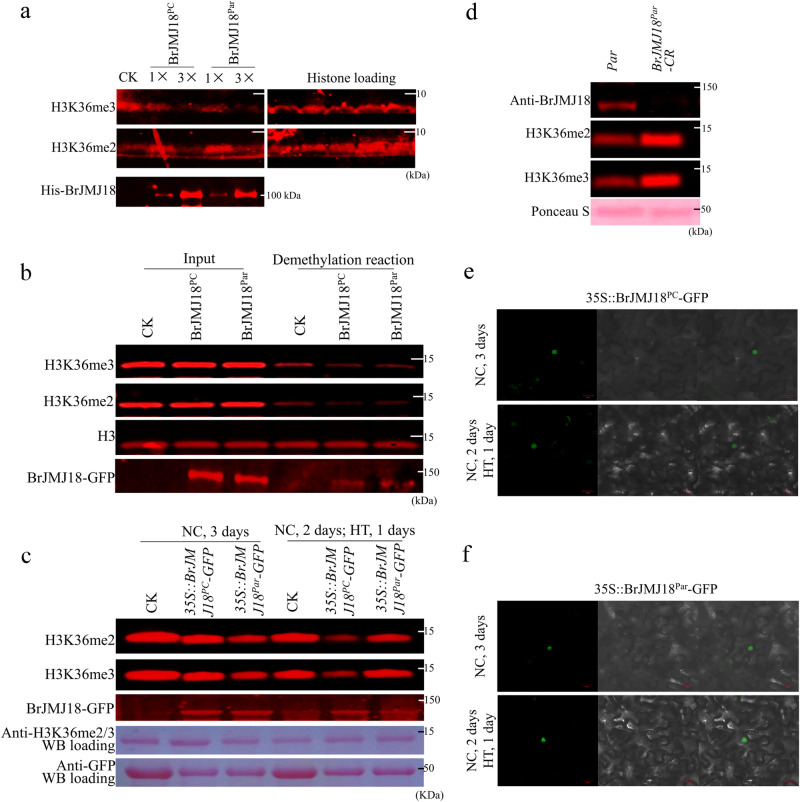
BrJMJ18 was classified in the clade of the H3K4me3/2 demethylase family, with the highest homology (87%) at the amino acid level to Arabidopsis AtJMJ18 (Supplementary Fig. 13). AtJMJ18 displayed demethylase activity toward H3K4me3/214. We then employed affinity-purified His-BrJMJ18PC/Par proteins and synthesized histone H3 peptides containing specific modifications, including H3K4me3, H3K9me3, H3K27me3, and H3K36me2/3, to perform demethylase activity assays. The His-BrJMJ18 protein demonstrated efficient demethylation of H3K36me2/3 peptides while keeping the methylation levels of H3K4me3, H3K9me3, and H3K27me3 peptides unchanged; and there was no difference in demethylation activity between the BrJMJ18PC and BrJMJ18Par proteins at H3K36me3/2 (Fig. 4a and Supplementary Fig. 14a). We next immunoaffinity-purified the two allelic BrJMJ18-GFP proteins from BrJMJ18 transgenic Arabidopsis lines (AtJMJ18::BrJMJ18orf-GFP) and conducted in vitro demethylase analysis. Both allelic BrJMJ18-GFP proteins demethylated H3K36me3 and H3K36me2, but not at H3K4me3/2, H3K9me3/2, and H3K27me3/2 (Fig. 4b and Supplementary Fig. 14b). Finally, to confirm whether BrJMJ18PC and BrJMJ18Par can exert this demethylase activity in vivo, we transiently expressed BrJMJ18PC and BrJMJ18Par proteins (35 S::BrJMJ18orf-GFP) in tobacco leaves. We showed that H3K36me3/2 underwent a dramatic global reduction in both BrJMJ18PC and BrJMJ18Par overexpression leaves under NC (Fig. 4c); meanwhile, we noticed that high temperature led to stronger H3K36me3/2 reduction in BrJMJ18PC-OX than in BrJMJ18Par-OX leaves (Fig. 4c). We used BrJMJ18Par-CR plants for further analysis and observed that signals at H3K36me3/2 were markedly increased in the BrJMJ18Par-CR plants (Fig. 4d).
Fig. 4. BrJMJ18 is an H3K36me2/3 demethylase.
a–d both BrJMJ18PC and BrJMJ18Par demethylate H3K36me3 and H3K36me2 in vitro and in vivo. a E. coli expressed His-BrJMJ18PC and His-BrJMJ18Par proteins demethylate H3K36me3 and H3K36me2 in vitro. Synthesized Histone H3 peptide with H3K4me3, H3K9me3, H3K27me3, H3K36me2, and H3K36me3 modifications were used as substrate. Purified His was used as a negative control. b Both BrJMJ18PC-GFP and BrJMJ18Par-GFP demethylate H3K36me3 and H3K36me2 in vitro. The two allelic BrJMJ18-GFP proteins were immunoaffinity-purified from BrJMJ18 transgenic Arabidopsis lines (AtJMJ18::BrJMJ18orf-GFP) and subjected to in vitro demethylase analysis using histone from calf thymus as a substrate. Col-0 was used as a negative control. Immunoaffinity-purified BrJMJ18-GFP proteins were detected with anti-GFP antibodies to confirm equal loading of BrJMJ18PC-GFP and BrJMJ18Par-GFP. H3, detected by blotting with anti-H3 antibodies, served as a loading control. c H3K36me3 and H3K36me2 status in 35 S::BrJMJ18PC/Par -GFP expressing tobacco leaves under normal conditions (NC) for 3 days and NC for 2 days following 1-day of HT, respectively 35 S::GFP expressing tobacco leaves were used as a negative control. BrJMJ18-GFP proteins were detected with anti-GFP antibodies to confirm equal expression of exogenous genes. Ponceau S staining was used to assess equal loading. d H3K36me3 and H3K36me2 status in BrJMJ18 loss-of-function mutants of Par under NC. Western blotting analysis of BrJMJ18 and H3K36me2/3 were conducted using anti-BrJMJ18 and anti-H3K36me2/3 antibodies in the plants. Five-week-old plants of Par grown under NC conditions were used for analysis. Ponceau S staining was used to assess equal loading. All the western blot experiments were repeated at least three times with similar results. e, f BrJMJ18PC/Par-GFP subcellular localization in transiently expressing tobacco plants under normal conditions (NC) for 3 days (e) and NC for 2 days following 1-day of HT (f), respectively. BrJMJ18-GFP protein was viewed using the confocal laser scanning system, Zeiss LSM510. Images of at least 30 epidermis cells expressing each allele of BrJMJ18-GFP protein under NC or HT conditions were acquired using the confocal laser scanning system Zeiss LSM510, respectively. Source data are provided as a Source Data file.
Nuclear localization is a necessary context to enable a Jumonji protein to conduct its function. The two allelic BrJMJ18 proteins localized to the nucleus in transiently expressed tobacco leaves (Fig. 4e, f). Moreover, we revealed that the nuclear localization of BrJMJ18PC or BrJMJ18Par were not affected by HT (Fig. 4e, f). Taken together, our results showed that BrJMJ18 is a nuclear H3K36me3/2 demethylase under different temperatures; however, the activity amplitude between different temperatures is weaker in BrJMJ18Par-expressing plants than that of BrJMJ18PC-expressing plants.
Overexpression of BrJMJ18Par alters the expression of BrFLC3 and modulates flowering
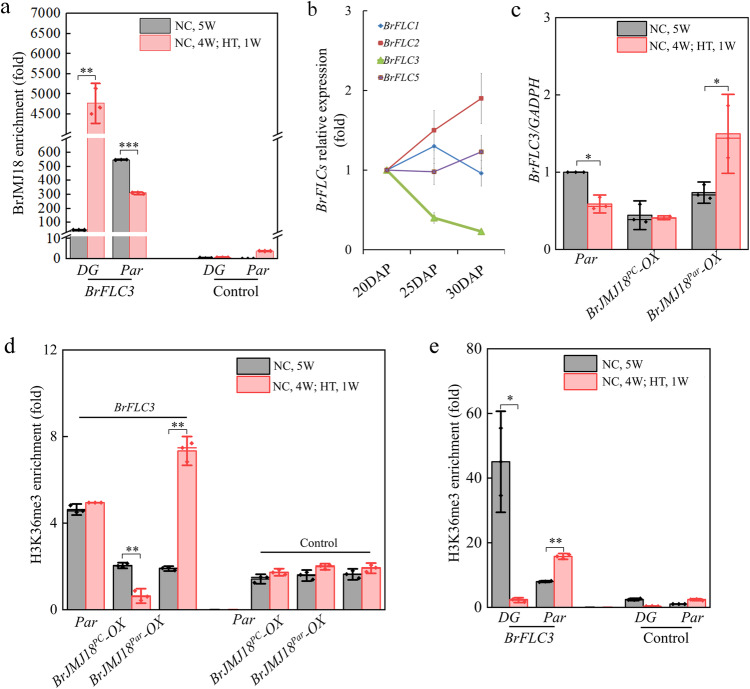
To assess the downstream genes of BrJMJ18, we conducted a chromatin immunoprecipitation (ChIP)-seq experiment using a polyclonal antibody recognizing BrJMJ18 in Par plants under NC (the specificity of the antibody was shown in Supplementary Fig. 15). We identified 6456 genes (12,547 loci) as candidate targets of BrJMJ18 (Supplementary Data 6). FLOWERING LOCUS C (BrFLC) family genes, including BrFLC1-3, were among the list of the flowering genes pulled down by the BrJMJ18 protein. We then validated the ChIP-seq results by carrying out quantitative PCR to quantify the gene bodies of BrFLCs in DG (BrJMJ18PC-carrying) and Par (BrJMJ18Par-carrying) plants, respectively. We found that BrJMJ18PC bound strongly to BrFLC1-3, and high temperature aggravated this binding markedly in DG. While in Par, BrJMJ18Par only bound strongly to BrFLC3, and opposite of that in DG, high temperature thoroughly disassociated the binding of BrJMJ18Par and BrFLC3 (Fig. 5a and Supplementary Fig. 16a). Similar results were obtained in the anti-GFP ChIP-qPCR using BrJMJ18PC/Par-OX Par plants grown under NC and HT, respectively (Supplementary Fig. 16b).
Fig. 5. Overexpression of BrJMJ18Par moderates flowering by altering the expression of BrFLC3.
a Chromatin immunoprecipitation (ChIP) analysis of the BrJMJ18 level across BrFLC3 in BrJMJ18Par-carrying Par and BrJMJ18PC-carrying DG plants. The 5th−7th young and healthy rosette leaves of five-week-old plants were used for the analysis. Rabbit IgG was used as a control. GADPH was used as a BrJMJ18-independent control. Control is a locus gene desert region where BrJMJ18 does not bind. The values are the mean ± standard deviation from three biological replicates. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t-test (**0.01 < p < 0.05, ***p < 0.001). DG, p = 0.0016; Par, p = 0.00027. b Expression patterns of BrFLC1, 2, 3 and 5 in Par before and after bolting under natural field conditions. The 5th−7th young and healthy rosette leaves were used for qPCR test. The values are the mean ± standard deviation from three biological replicates. The Par bolted at 25DAP. DAP, days after planting (14-day Par seedlings grown in cultivation pots in greenhouse were transplanted into natural field). c BrFLC3 expression patterns in BrJMJ18PC/Par- overexpressing plants. The 5th−7th young and healthy rosette leaves of Par plants were used for qPCR test. The values are the mean ± standard deviation from three biological replicates. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t test (*0.01 < p < 0.05). BrJMJ18PC-OX, p = 0.0032; BrJMJ18Par-OX, p = 0.0023. d ChIP analysis of H3K36me3 enrichment on the BrFLC3 locus in BrJMJ18 overexpression Par lines. The values are the mean ± standard deviation from three biological replicates. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t test (** 0.001 < p < 0.01). Par, p = 0.012; BrJMJ18Par-OX, p = 0.034. e The H3K36me3 level at BrFLC3 in DG (BrJMJ18PC-carrying) and Par (BrJMJ18Par-carrying). The values are the mean ± standard deviation from three biological replicates. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t-test (*p < 0.05, **0.001 <p < 0.01,). DG, p = 0.021; Par, p = 0.0016. Source data are provided as a Source Data file.
Furthermore, we noticed that BrFLC3 was the only downregulated BrFLC during floral transition in Par (Fig. 5b) and was the dominant expressed BrFLC during flowering in Par (Supplementary Fig. 17). Besides, we noticed that BrFLC3 is an effective FLC regulating flowering in yellow sarson, another type of B. rapa that can flower without vernalization15. Thus we hypothesized that BrFLC3 is an indispensable regulator in adjusting flowering time in Par. The linking of BrJMJ18 and BrFLC3 was further supported by testing BrFLC3 expression in transgenic Par plants. Under NC, BrFLC3 expression decreased markedly in both BrJMJ18PC- and BrJMJ18Par-OX plants, while under HT, it was decreased in BrJMJ18PC-OX but increased in BrJMJ18Par-OX plants (Fig. 5c), which is in line with their flowering time variation under HT (Fig. 3b, c). In Arabidopsis, similar results for AtFLC were observed in transgenic plants (Supplementary Fig. 18). Therefore, all these findings implicated that in BrJMJ18Par-OX plants, BrFLC3 levels increase to delay flowering at high temperatures.
As described in Fig. 5c, we speculated that BrJMJ18Par dissociates from BrFLC3 via a mechanism by which the expression of BrFLC3 is consequently activated upon heat in BrJMJ18Par-OX plants. We were then interested in verifying whether the BrFLC3 expression pattern is reflected in its H3K36me3 context. ChIP-PCR was carried out and we found that the H3K36me3 level at the BrFLC3 locus was consistent with the expression pattern of BrFLC3 in BrJMJ18PC- and BrJMJ18Par-overexpressing plants under both temperature conditions (Fig. 5d). We also observed consistent results of the expression pattern and H3K36me3 methylation status at the AtFLC locus in transgenic AtJMJ18::BrJMJ18PC/Par-GFP Arabidopsis plants (Supplementary Fig. 19). To further confirm if this was the case in non-transgenic DG and Par plants, we explored the H3K36me3 level at the BrFLC3 locus and found that the H3K36me3 level was downregulated in DG (BrJMJ18PC-carrying) but upregulated in Par (BrJMJ18Par-carrying) under HT, which agreed with the enrichment of H3K36me3 at BrFLC3 demonstrated in BrJMJ18PC- and BrJMJ18Par-OX plants in Fig. 5e. These results proposed that in BrJMJ18Par-OX plants the late flowering primarily results from the dissociation of BrJMJ18Par from BrFLC3, leading to the inability to repress its expression, while the effect of BrFLC3 is less than straightforward in the WT Par plants. (Fig. 3g, j).
Overexpression of BrJMJ18Par mediates plant growth under both greenhouse and field high- temperatures conditions
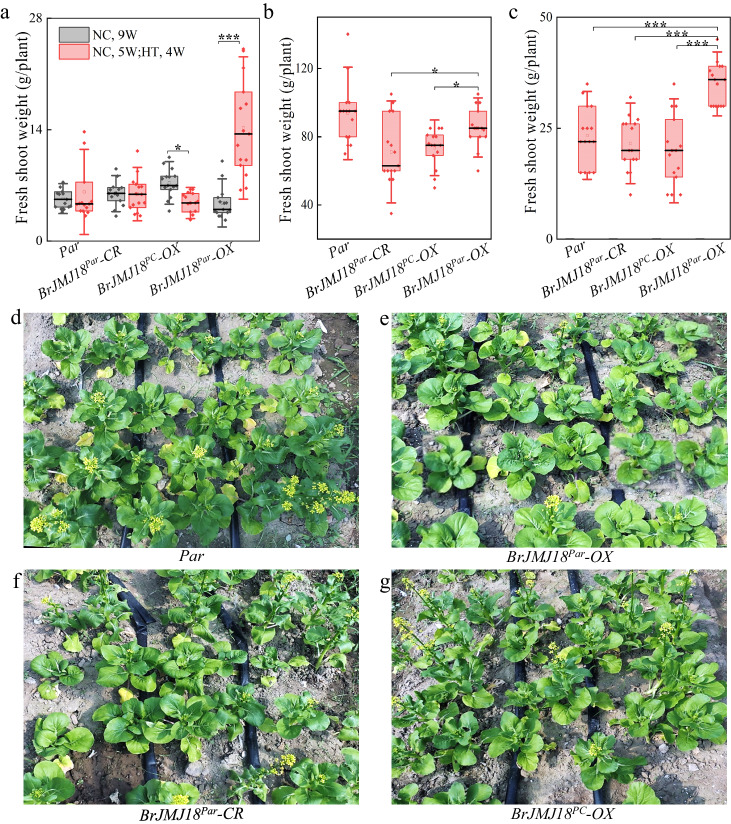
We evaluated the effects of the overexpression of the two allelic BrJMJ18s on major commercial qualities of different transgenic Par plants under both greenhouse and field conditions. For the plants grown in greenhouse under NC, BrJMJ18Par- and BrJMJ18PC-OX and BrJMJ18Par-CR plants displayed no or slight decrease in the aboveground biomass (fresh shoot weight) and leaf number compared with Par controls (Fig. 6a and Supplementary Fig. 20). However, under HT, increased biomass and leaf number was found for BrJMJ18Par-OX but not the other three plants (Fig. 6a and Supplementary Fig. 20). Besides, BrJMJ18Par-OX had fewer aging leaves than the other plants under HT (Fig. 3b). In addition, we observed that BrJMJ18Par-OX plants exhibited little or no changes in morphology under HT comparing with the other plants (Fig. 3a, b).
Fig. 6. Overexpression of BrJMJ18Par mediates plant growth under both greenhouse and field high temperature conditions.
a The aboveground biomass evaluation of the BrJMJ18 transgenic Par plants grown in the greenhouse under different temperature conditions. BrJMJ18 transgenic Par plants grown under NC conditions for 9 weeks, and under NC conditions for 4 weeks, following another 5 weeks under high temperature (HT) conditions, respectively, were used for yield test. Data are means ± SD, n = 15. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t-test (*p < 0.05, **p < 0.01, ***p < 0.001). BrJMJ18PC-OX, p = 0.0011; BrJMJ18Par-OX, p = 8.51×10−5. b, c The aboveground biomass evaluation of the BrJMJ18 transgenic Par plants grown in tunnel greenhouse under field conditions. The day maximum and minimum temperatures in the tunnel greenhouse were 52 °C and 31 °C, respectively, while the night maximum and minimum temperatures were 33 °C and 24 °C, respectively. b Plants were harvested at 55 days after sowing. c Plants were harvested at their proper picking stages. Data are means ± SD, n = 15. Asterisks indicate significant differences between BrJMJ18Par-OX and the other three plants, two-tailed Student’s t test (*p < 0.05). In (b), BrJMJ18Par-OX and BrJMJ18PC-OX, p = 0.029; BrJMJ18Par-OX and BrJMJ18Par-CR, p = 0.03. In (c), BrJMJ18Par-OX and BrJMJ18PC-OX, p = 4.87E×10−6; BrJMJ18Par-OX and BrJMJ18Par-CR, p = 2.5 × 10−6; BrJMJ18Par-OX and Par, p = 7.56 × 10−5. d–g Photos of the BrJMJ18 transgenic Par plants grown in tunnel greenhouse under field conditions. d, Par controls; e, BrJMJ18Par-OX plants; f, BrJMJ18Par-CR plants; g, BrJMJ18PC-OX plants. For box plots (a–c), the box encompasses two middle quartiles, with a central line showing the median. Whiskers extend to the furthest data point within 1.5 times the interquartile range. Source data are provided as a Source Data file.
We then evaluated the yield of BrJMJ18 transgenic plants grown in the tunnel greenhouse under natural field conditions. We harvested the edible parts of the plants in two different ways. Firstly, we picked all the plants at the same time, and the production of BrJMJ18Par-OX plants was comparable with that of control Par, but higher than BrJMJ18Par-CR and BrJMJ18PC-OX plants (Fig. 6b). However, at this point only BrJMJ18Par-OX plants exhibited appropriate exterior quality (Fig. 3i). We alternatively harvested the plants at their proper picking stages, and the BrJMJ18Par-OX line displayed much higher yield than the other plants (Fig. 6c). Interestingly, despite exhibiting a similar delayed flowering phenotype to BrJMJ18Par-OX plants, BrJMJ18Par-CR plants did not display increased aboveground biomass and leaf numbers under HT (Fig. 6 and Supplementary Fig. 20). This suggests that the enhanced vegetable growth of BrJMJ18Par-OX under HT is not simply due to the deactivation of BrJMJ18Par. We hypothesized that high temperatures enable BrJMJ18Par to acquire new functions to mediate plant growth.
BrJMJ18Par modulates chlorophyll biosynthesis under high temperatures
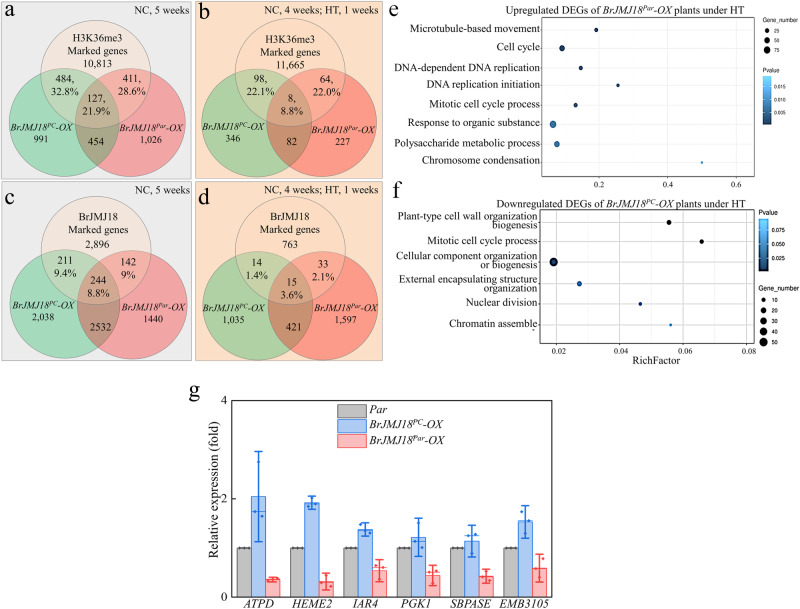
Given the phenotypic differences between BrJMJ18Par- and BrJMJ18PC -OX plants, we conducted a ChIP-seq assay with anti-GFP antibody using BrJMJ18PC-OX, BrJMJ18Par-OX, and Par plants grown under NC and HT, respectively, to explore their functional differences comprehensively. A total of 2056 and 2018 genes were enriched by BrJMJ18PC and BrJMJ18Par under NC, respectively, while the corresponding numbers under HT were 535 and 382 (Supplementary Data 7). The decreased number of enriched genes in both allelic BrJMJ18s under HT revealed that high temperature impeded their binding activities; while compared with BrJMJ18PC, BrJMJ18Par was affected more. Previous studies identified that at least 11,835 B. rapa genes are targeted by H3K36me316. We found that under NC, 32.8% of enriched genes in BrJMJ18PC-OX and 28.6% in BrJMJ18Par-OX overlapped with H3K36me3-targeted genes. Under HT, these percentages dropped to 22.1% and 20.0% (Fig. 7a, b and Supplementary Data 7). We further found that 71.7% and 71.2% of the enrichment genes under NC were specific to BrJMJ18PC-OX and BrJMJ18Par-OX plants, respectively, while the corresponding frequencies under HT increased to 82.9% and 76.2% (Fig. 7a, b). Notably, GO analysis revealed that under NC, the enriched terms for BrJMJ18PC and BrJMJ18Par showed remarkable consistency (Supplementary Fig. 21a, b). In contrast, under HT conditions, the enriched GO terms diverged. Specifically, BrJMJ18PC’s targets were more enriched in entries related to cellular microscopic structure, while BrJMJ18Par was more centralized in items associated with macromolecule metabolism (Supplementary Fig. 21c, d). We then conducted a RNA-seq assay using the same samples above. A total of 5025 and 4358 DEGs were identified in BrJMJ18PC-OX and BrJMJ18Par-OX plants under NC, respectively, while the corresponding DEGs under HT were 1457 and 2019, respectively (Supplementary Data 8). Over one-third of DEGs in BrJMJ18PC-OX and BrJMJ18Par-OX plants under HT overlapped with heat-responsive genes from Yue et al. 17 (Supplementary Data 8). Under NC, 40.5% of the DEGs in BrJMJ18PC-OX plants and 45.6% in BrJMJ18Par-OX plants overlapped with reported H3K36me3-targeted genes, while under HT the corresponding frequencies decreased to 31.2% and 27.9% (Supplementary Data 8). We further found that 18.2% (455 genes) of the DEGs in BrJMJ18PC-OX plants and 17.8% (386 genes) of the DEGs in BrJMJ18Par-OX plants are BrJMJ18-target genes identified by ChIP-seq under NC, while the corresponding frequencies under HT decreased to 5.0% (29 genes) and 5.7% (38 genes) (Supplementary Data 8). 44.8% and 36.3% of the DEGs under NC were specific to BrJMJ18PC-OX and BrJMJ18Par-OX plants, respectively, while the corresponding frequencies under HT increased to 71.0% and 79.1% (Fig. 7c, d and Supplementary Data 8), further indicating a function divergence of BrJMJ18PC and BrJMJ18Par under HT. We then performed GO analysis of the DEGs in BrJMJ18PC- and BrJMJ18Par-OX plants under NC and HT (Fig. 7e, f and Supplementary Fig. 22), respectively, to characterize BrJMJ18PC and BrJMJ18Par. Under NC, both the upregulated and downregulated DEGs in BrJMJ18PC-OX and BrJMJ18Par-OX plants showed enrichment in similar GO categories (Supplementary Fig. 22a). However, there’s a notable difference under HT conditions. In BrJMJ18Par-OX plants, the upregulated DEGs were predominantly associated with cell division. Surprisingly, the downregulated DEGs in the BrJMJ18PC-OX line now showed enrichment in cell division-related GO terms (Fig. 7e, f and Supplementary Fig. 22b). We also observed that the downregulated DEGs in BrJMJ18Par-OX were notably enriched in processes such as chlorophyll and tetrapyrrole biosynthesis, cofactor metabolic processes, and chlorophyll biosynthetic processes (Supplementary Fig. 22b). Interestingly, this enrichment pattern closely mirrors the GO items enriched among the downregulated genes in the heat-resistant Chinese cabbage under heat treatment reported by Zhang et al. 18. We performed qPCR analysis on six heat-responsive genes associated with chlorophyll biosynthesis and carbon metabolism to assess BrJMJ18Par’s role in thermotolerance. These genes, identified by Zhang et al. 18, were also found to be DEGs in BrJMJ18Par-OX plants under HT. Compared with control Par, all six genes showed slight induction in BrJMJ18PC-OX plants under HT, while they were significantly downregulated in BrJMJ18Par-OX plants. These findings suggest significant functional differences between BrJMJ18PC and BrJMJ18Par under high temperatures, with BrJMJ18Par demonstrating greater effectiveness in regulating chlorophyll biosynthesis and plant growth.
Fig. 7. Integrated analysis of RNA-seq and ChIP-seq data reveled BrJMJ18Par modulates chlorophyll biosynthesis r under high temperature.
a Diagram showing the overlap of genes targeted by BrJMJ18PC and BrJMJ18Par, respectively, and reported H3K36me3-regulated genes under NC. b Diagram showing the overlap of genes targeted by BrJMJ18PC and BrJMJ18Par, respectively, and reported H3K36me3-regulated genes under HT. c Diagram showing the overlap of the DEGs of BrJMJ18PC-OX and BrJMJ18Par-OX plants, respectively, and the identified BrJMJ18-targeted genes under NC. Genes showing a 2-fold change within a 95% confidence interval were considered to be differentially expressed. d Diagram showing the overlap of the DEGs of BrJMJ18PC-OX and BrJMJ18Par-OX plants, respectively, and the identified BrJMJ18-targeted genes under HT. Genes showing a 2-fold change within a 95% confidence interval were considered to be differentially expressed. e, f Functional categorization by Gene Ontology of the DEGs of BrJMJ18PC-OX and BrJMJ18Par-OX plants under HT. Only significantly enriched entries are shown. e, GO analysis demonstrated that the upregulated DEGs in BrJMJ18Par-OX plants under HT were mainly enriched in cell division. f, the cell division-related GO entries were enriched in the downregulated DEGs in the BrJMJ18PC-OX line. g Expression of heat stress-related genes reported by Zhang et al. in Par and BrJMJ18-OX plants. 5-week-old plants grown under NC, and 4-week-old plants grown under NC followed by 1 week under HT were used for Q-PCR. Relative to Par plants, all six genes exhibited slight induction in BrJMJ18PC-OX plants under high temperatures, while they were significantly downregulated in BrJMJ18Par-OX plants. Photosynthesis-related gene BraA9g029800.3 C (ATP synthase DELTA-subunit gene, ATPD), porphyrin and chlorophyll metabolism-related gene BraA04g028660.3 C (Uroprophyrinogen decarboxylase, HEME2) and carbon metabolism-related genes BraA07g034950.3 C (Pyruvate dehydrogenase E1a-like subunit, IAR4), BraA03g035490.3 C (Phosphoglycerate kinase 1, PGK1), BraA07g0022160.3 C (Sedoheptulose- bisphosphatase, SBPASE), BraA08g004460.3 C (Embryo defective 3105, EMB3105). GADPH was used as the internal control. The values are the mean ± standard deviation from three biological replicates. Source data are provided as a Source Data file.
Amino acid mutations of T345A, Y633C and L654F contribute to functional divergence of BrJMJ18PC and BrJMJ18Par
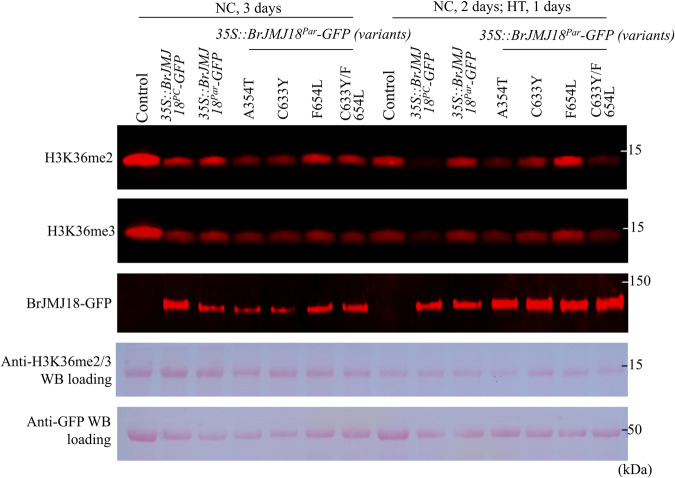
Compared with BrJMJ18PC, BrJMJ18Par displayed decreased binding and catalytic activities under HT (Figs. 4–7). BrJMJ18 contains five distinct functional domains, including the JmjN, JmjC, C5HC2 zinc finger (Zf-C5HC2), F/Y-rich N terminus (FYRN), and F/Y-rich C-terminal (FYRC) domains (Fig. 2f). Among all the six amino acid mutations, T345A occurred in the known catalytic JmjC domain, while both Y633C and L654F localized in the interspace between the DNA-binding Zf-C5HC2 domain and the FYRN domain, commonly found in chromatin-associated proteins19,20. Therefore, we reasoned that the functional diversity of BrJMJ18PC and BrJMJ18Par was embodied in the mutations of T345A, Y633C, and L654F. We then transiently expressed BrJMJ18PC, BrJMJ18Par, and variant proteins of BrJMJ18Par(A345T), BrJMJ18Par(C633Y), BrJMJ18Par(F654L), and BrJMJ18Par(C633Y)/(F654L) in tobacco leaves to explore their catalytic functions. These variants were generated by introducing specific substitutions (A345T, C633Y, and F654L) into BrJMJ18Par to match the BrJMJ18PC sequence. Under NC, the demethylase activities of BrJMJ18Par(A345T) and BrJMJ18Par(C633Y) were found to be higher than that of BrJMJ18PC, whereas BrJMJ18Par(F654L) and BrJMJ18Par(C633Y)/(F654L) exhibited lower activities (Fig. 8). However, under HT, BrJMJ18Par(A345T) and BrJMJ18Par(C633Y633)/(F654L) displayed similar H3K36me3/2 demethylase activity to BrJMJ18PC, but higher than that of BrJMJ18Par (Fig. 8). These findings indicate that mutations T345A, Y633C, and L654F contribute to the catalytic differences between BrJMJ18PC and BrJMJ18Par, which was also supported by our phenotypic assessments of transgenic Arabidopsis plants (Supplementary Fig. 23).
Fig. 8. Amino acid mutations of T345A, Y633C and L654F contribute to functional divergence of BrJMJ18PC and BrJMJ18Par.
(BrJMJ18Par(A345T) is identical to the sequence of BrJMJ18Par except with a substitution at position 345 at which A was replaced with T, which is specific to BrJMJ18PC; the same goes for BrJMJ18Par(C633Y), BrJMJ18Par(F654L), and BrJMJ18Par(C633Y)/(F654L)). BrJMJ18PC-GFP, BrJMJ18Par-GFP, and its variant proteins were transiently overexpressed in tobacco leaves. H3K36me3 and H3K36me2 were detected using anti-H3K36me3 and anti-H3K36me2 antibodies in 35S::BrJMJ18-GFP expressing leaves under NC for 3 days and NC for 2 days following 1-day of HT, respectively. BrJMJ18-GFP proteins were detected with anti-GFP antibodies to confirm equal expression of exogenous genes. Ponceau S staining was used to assess equal loading of total protein extracts. All the western blot experiments were repeated at least three times with similar results. Source data are provided as a Source Data file.
Discussion
It is reported that East Asian leafy B. rapa crops likely evolved from turnips2,21,22. We cross-referenced the estimated demographic modeling with historical records of leafy B. rapa domestication events in China. This comparison confirmed that the genomic inferences of domestication events of PC, DG and Par are corroborated by documented historical records, as depicted in Supplementary Fig. 24. Par has acquired the capacity to maintain its flowering time and nutritional growth with minimal disruption in response to temperature fluctuations during its domestication in the warm regions of southern China. However, this crop has received limited attention. Heat tolerance seems to be polygenic, which might explain why the genetic basis of heat stress tolerance in plants is poorly understood23–25. Although different mapping methodologies and populations have been used to define QTLs and genes involved in thermotolerance in different plants and crops (e.g., in Arabidopsis, azuki bean, barley, brassica, cowpea, maize, potato, rice, sorghum, tomato, and wheat)26–29, only a few causal genes have been identified. Here, we employed various genetic and biochemical approaches to demonstrate the thermotolerant characteristics of an epigenetic remodeler, BrJMJ18, during the domestication of B. rapa crops. We show a model that summarizes our hypothesis (Supplementary Fig. 25). Briefly, BrJMJ18PC functions as a “throttle” for flowering, regardless of whether it is under NC conditions or HT. On the other hand, BrJMJ18Par exhibits functional divergence at different temperatures. Under NC conditions, BrJMJ18Par promotes flowering, but under high temperatures, it acts as a “brake” for flowering. This working mechanism can explain why under HT conditions, BrJMJ18PC-OX or BrJMJ18 knockout Par plants appear stressed, whereas BrJMJ18Par-OX plants did not (Figs. 3 and 6).
BrJMJ18 is an H3K36me2/3 demethylase
Tri-methylation at H3K36me3/2 is widely distributed in the whole genome in various species30–33. In Arabidopsis, more than four-fifths of genes are enriched with H3K36me334. These facts suggest that the establishment and maintenance of H3K36me3 is indispensable during organic evolution. However, compared with the well-documented establishment of H3K36me3, the H3K36me3 erasure is barely described. To the best of our knowledge, only two Jumonji C-domain containing proteins, Arabidopsis JMJ30 and INCURVATA11 (ICU11), were reported as possible H3K36me3 demethylases in plants34–36; however, it is also interesting to note that AtJMJ30 and its homolog AtJMJ32 are primarily associated with inhibiting early flowering at higher temperatures by eliminating the H3K27me3 on the AtFLC locus37. Here, we showed that BrJMJ18 is an H3K36me3/2 demethylase using in vivo and in vitro demethylation activity analysis, and the increased and decreased levels at H3K36me3/2 in BrJMJ18 overexpressing and knockout lines of Par, respectively. The H3K36me3/2 demethylation activity of BrJMJ18 was also confirmed by the fact that: 1) BrJMJ18-OX and sdg8, loss-of-function mutation of the H3K36me3 methylase SDG8, Arabidopsis plants show similar flowering phenotype34, and 2) ~ 30% the immunoprecipitated genes by BrJMJ18PC or BrJMJ18Par are reported H3K36me3-targeted genes (Fig. 7 a, b). H3K27me3 has an opposing relationship with H3K36me3 accumulation. According to Mehraj et al.16, there are at least 10,445 B. rapa genes marked by H3K27me3. We further found that under NC, only 15% of enriched genes immunoprecipitated by BrJMJ18PC or BrJMJ18Par overlap with the reported H3K27me3-marked genes, and these proportions decrease to 13.0% and 10.5% under HT (Supplementary Data 7). These data indicate that the genes precipitated by BrJMJ18 are specifically targeted by H3K36me2/3.
Epigenetic mechanisms regulate diverse signaling pathways in response to environmental stresses. The flowering integrator gene, AtFLC, offers a classic example demonstrating the interaction between epigenetic modifications and environmental adaptation38. Under standard growth conditions, FLC chromatin is enriched in a number of histone modifications associated with actively transcribed genes39, including H3K4me3/2 and H3K36me3/2. Upon cold, these markers are dynamically removed from the locus and are replaced with H3K27me3/240,41. We noticed that H3K27me3 and H3K36me3 show opposing profiles under cold42, and this antagonism is thought to be functionally important in the establishment of mutually exclusive chromatin states. This model also predicted a physical association between antagonistic histone modifiers, specifically the H3K36me3 methyltransferase SDG8 and the H3K27me3 demethylase ELF6; such a prediction was recently confirmed experimentally42,43. Recently, it was reported that H3K27me3 is also important for flowering time control under ambient temperature conditions, in which Jumonji proteins (JMJ30/JMJ32/JMJ13) mediate H3K27me3 demethylation at FLC, constituting one of the balancing mechanisms37. In addition, studies in A. thaliana revealed that heat memory is also partially mediated by the sustained demethylation of H3K27me3 on small HSPs44,45. Compared with H3K27me3, the establishment and maintenance of H3K36me3 and its biological significance are relatively unreported. It is interesting to find that Pajoro et al. report that H3K36me3 methyltransferase mutants (sdg8-2 and sdg26-1) exhibited comparable flowering times under varying temperature conditions34, while H3K36me3 demethylase mutant jmj30-1 showed heightened sensitivity to high temperatures, indicating the necessity of H3K36me3 modification for temperature-responsive flowering. We speculated that BrJMJ18 and the H3K27me3 demethylases might form a balanced or feedback loop in histone methylation, contributing to a resilient mechanism for plant growth.
Functional divergence of BrFLCs
In Arabidopsis, FLC is a common regulator in vernalization-mediated flowering. Recent reports suggested that FLC moderates flowering at elevated temperatures37,46–48. Beyond flowering control, pleiotropic functions of FLC were assigned to the development of flowers49, leaf shape, trichome number50, seed dormancy/germination51,52, water usage regulation53, and some fitness-related traits54. Brassicaceae genomes have undergone three rounds of whole genome duplication (WGD) after speciation from Arabidopsis55,56. We believe that during the evolution of B. rapa, BrFLC paralogs with specific neofuncations could have diverged from the ancestor FLC. Multiple lines of evidence show BrFLCs (BrFLC1–3 and BrFLC5) functionally diverging. First, all four functional BrFLCs have been genetically mapped to flowering time loci in B. rapa, varying based on G × E (population × growth conditions) (Supplementary Table 3). Secondly, all four BrFLCs can produce different splice variants57–59. Thirdly, COOLAIR is a cold-induced antisense RNA transcribed from the FLC locus which has been proposed to facilitate FLC silencing. In B. rapa, however, only COOLAIRs of BrFLC2 have been detected, suggesting that BrFLC2 might have a more important function than the other BrFLCs in vernalization-mediated flowering58. Whether the remaining BrFLC genes have distinct COOLAIRs and their respective functions remain unexplored. Lastly, different BrFLCs were genetically linked with traits other than flowering. For instance, a key QTL that regulates turnip development was assigned to the BrFLC2 gene locus60, while BrFLC1 was proposed as a candidate influencing reproductive fitness traits51,61. The above information strongly suggests BrFLCs’ functional divergence after gene duplication in B. rapa evolution.
Our data, together with other pieces of evidence, implied that BrFLC3 is an active player in flowering time control: (1) BrFLC3 is a dominant expressed BrFLC in Par (Supplementary Fig. 17); (2) BrFLC3 expression decreased rapidly upon cold in different B. rapa morphotypes58,62; (3) BrFLC3 overexpression caused a comparable flowering delay to BrFLC1 and BrFLC2 overexpressing Arabidopsis plants; and 4) BrFLC3-overexpressing Chinese cabbage plants showed a dramatically delayed flowering time63, and (5) in yellow sarson, a type of B. rapa that can flowering without vernalization, ELF6 controls flowering by regulating BrFLC3 expression15. However, the genetic correlation of BrFLC3 with flowering time in B. rapa was only reported by two studies57,64. BrFLC3’s distinct genomic dissimilarity to AtFLC among the four BrFLCs (Supplementary Fig. 26) raises the possibility of neo-functionalization during evolution. Beyond, we noticed that BrFLC3 is the only downregulated BrFLC in Par. We speculated that Par may have developed a pathway involving BrFLC3 for sensing specific environmental cues. Based on our findings, under HT conditions, BrFLC3 levels increase to delay flowering, as evidenced by the comparison of H3K36me2/3 enrichment and BrFLC3 expression in BrJMJ18Par-OX plants. This correlation is specifically observed in BrJMJ18Par-OX lines. However, in non-transgenic Par plants, the scenario differs: the H3K36me3 level at BrFLC3 remains largely unchanged (Fig. 5d), and there is no significant increase in BrFLC3 levels at high temperatures (Fig. 5c). We hypothesize that these disparities in response likely stem from the multi-level regulation of FLC, including chromatin remodeling, transcriptional control, and co-transcriptional RNA metabolism. Specifically, in BrJMJ18Par-OX plants, an overabundance of BrJMJ18Par proteins may disrupt the balanced regulation of BrFLC3 seen in wild-type Par plants. This disruption thereby exacerbates BrJMJ18Par’s influence and leading to pronounced increases in H3K36me2/3 levels and BrFLC3 expression under HT conditions. In contrast, in WT Par plants, the regulation of BrFLC3 expression involves a complex interplay of mechanisms, resulting in less pronounced changes compared to the BrJMJ18-overexpressing lines. Moreover, both WT Par and BrJMJ18Par-OX plants exhibit consistent flowering time variations under HT conditions, but the latter experience more significant changes. All these findings suggested that in BrJMJ18Par-OX plants BrJMJ18Par can adjust the expression of BrFLC3 and flowering time, while the effect of BrFLC3 is less than straightforward in the WT Par plants. Additionally, specific BrFLC3 haplotypes for a B. rapa crop were not identified (Supplementary Fig. 27). This could elucidate the limited genetic mapping of BrFLC3, suggesting its expression plasticity as a potential determinant for thermotolerance.
Breeding high-yielding and resilient cultivars without significant fitness costs poses challenges. Ideally, resistance responses would remain inactive in the absence of stimuli, and stress-related gene expression would be tightly activated only during stress. Natural variants aligning with the operational model of BrJMJ18, as outlined here, can offer advantages for stress-resistant plant breeding. Furthermore, considering the reversible nature of H3K36me3/2 on chromatin, epigenetic modifications can introduce adaptable, short- or long-term gene expression changes in response to varying environmental stresses, potentially enhancing adaptive capabilities in the face of environmental shifts65,66.
Methods
Plant materials and growth condition
Genotype selection, planting, and phenotyping of natural B. rapa collection
A collection of 210 varieties of different B. rapa morphotypes, including 15 subsp. oleifera (Ole), 51 subsp. rapifera (Raf), 9 subsp. chinensis var. narinosa (Nar), 50 subsp. chinensis (Par-Choi, PC), 41 subsp. chinensis DG (DarkGreen, DG), and 44 subsp. chinensis var. parachinensis (Par) were used for genetic structure and selection analyses. All of these lines were self-pollinated for at least six generations. A separate F2 population generated from PC and Par was used to investigate the impact of allelic effects on flowering time.
The flowering time of the varieties collection was evaluated under NV and V4W conditions in Zhangjiakou (ZJK, 114:55E/40:51 N) and Beijing (BJ,116:28E/39:54 N), China, respectively, from 20th June to 20th September, 2018. NV, 14-day seedlings grown under a long-day (LD) regime (16/8 h day/night) and 22/22 °C at a photon flux density of 100 mol/m2/s (denoted as NC (Normal Conditions) hereafter), were moved to natural field conditions in ZJK and BJ, respectively, until bolting; V4W, 14-day seedlings grown under NC conditions were moved to a long-day regime (16/8 h day/night) and 4/4 °C conditions for 4-weeks (denoted as V4W (Vernalization for 4 weeks) hereafter), and then transplanted to natural field conditions in ZJK and BJ respectively, until bolting. Flowering time, as days after germination (DAG), was defined as the number of days from sowing to the appearance of the visible bud.
The flowering time of the F2 population derived from Par × PC was investigated in the Beijing Vegetable Research Center (BVRC) greenhouse. The germinated seeds were planted in soil and cultivated under NC conditions at 22 /22 °C with a 16-h light and 8-h dark period for 4 weeks. Following this, half of the seedlings were kept under NC conditions until their flowering time data were recorded, while the other half were transferred to HT conditions at 29 /29 °C with a 16-h light and 8-h dark period until they flowered. Flowering-related quantitative trait loci (ftQTLs) responding to different temperatures were then identified by using BSA analysis.
Planting of transgenic B. rapa and Arabidopsis plants
Seeds of inbred lines or transgenic plants of different B. rapa morphotypes, including PC, DG, and Par, were germinated under NC for 36 h, and then transferred to soil and grown under NC until the collection of phenotypic data. For high-temperature treatment, 4-week-old seedlings grown under NC were transferred to HT conditions until the collection of phenotypic data.
Arabidopsis thaliana Col-0 and transgenic plant seeds were surface sterilized, treated in the dark for 4 days at 4 °C, and sown on 0.5 × Murashige and Skoog (MS) medium. Seven-day-old seedlings were transferred from the plates to the soil and grown under NC. For HT treatment, 2-week-old seedlings grown under NC were transferred to a 29/29 °C growth room with an LD photoperiod at a photon flux density of 100 mol/m2/s.
Phenotyping of transgenic plants
Flowering time: The flowering time of the transgenic Arabidopsis and Col-0 plants was determined by the number of rosette leaves at the time of bolting. At least 15 individual plants of each line were measured. The flowering time of transgenic Par plants was determined by the days from germination to bolting. At least 15 individual plants of each line were measured.
Fresh weight: Shoots of transgenic Arabidopsis or Par plants were excised and weighed as the fresh weight.
Leaf number: The number of Arabidopsis or Par plants true leaves was counted as leaf number.
Uncropped photos of plants phenotype are provided in the Source Data file.
Data processing and SNP calling
We conducted data processing and SNP calling as described by Su et al.67, except that we used the B. rapa genome v3.0 (http://brassicadb.cn/#/Download/) as the reference.
Population genetic analyses and gene flow estimates
Population genetic analyses and gene flow estimates were conducted as described by Su et al.67.
Demographic modeling
Models of the demographic history of PC, DG, and Par were evaluated using diffusion approximations to the allele frequency spectrum (AFS) with ∂a∂i68. We implemented a model testing hierarchy to accommodate ∂a∂i’s limit of analyzing three populations at a time. We first evaluated one- and two-population models for each morphotype group to estimate the timing of divergence among the derived groups. The results of these initial analyses informed our subsequent demographic modeling of three different three-population model groups. In our model, we tested whether the DG and Par groups were independently derived from PC or whether a group ancestral to both DG and Par split from PC and then split into DG and Par.
Analyses of putative selective sweeps
To detect genomic regions that have potentially differentiated during domestication or improvement of Par, the FST scores were calculated for DG/PC and Par/DG groups using PopGenome69 to estimate population differentiation. The FST scores were estimated for 200 kb sliding windows with a step size of 5 kb. The average FST of all sliding windows was considered as the value at the whole-genome level across different groups. Sliding windows with FST values greater than the 95th percentile of the genome-wide FST values were selected and regarded as significantly different windows. Overlapping significance windows were then merged into one fragment. These fragments were regarded as highly diverged regions across the groups. Nucleotide diversity, п, is often used as a measurement of the degree of genotype variability within a population or species. To improve the prediction accuracy, we then evaluated the nucleotide diversity of the DG/PC and Par/DG comparisons, respectively. Values of п were calculated for 200 kb overlapping windows (5 kb steps) across the genome using the BioPerl module PopGen (Stajich and Hahn, 2005). We then calculated the п ratios for each chromosome and identified potential candidate selection regions following the method described in70.
RNA and DNA extraction
Total RNA was isolated from the 5th–7th healthy rosette leaves of transgenic Arabidopsis and PC, DG, and Par plants and corresponding controls using an RNAprep pure plant kit (DP441, Tiangen, Beijing, China).
Total DNA of Arabidopsis, and PC, DG, and Par plants, was isolated from rosette leaves using the cetyltrimethylammonium bromide (CTAB) method71.
RNA sequencing
For Arabidopsis, the 5th–7th healthy rosette leaves were collected from Col-0 and overexpression plants (AtJMJ18::BrJMJ 18PC-GFP and AtJMJ18::BrJMJ18Par-GFP) grown under NC for 3 weeks or 2 weeks under NC following 1-week of HT. For Brassica, PC (PC261, ZYC), DG (HN129, ZJHYQ), and Par (CX268, SNLBLY70TCX) plants were germinated and planted in the BVRC under natural field conditions from March to June, 2019. PC, DG, and Par bolted at the 10th week, 6th week, and 4th week, respectively. BrJMJ18PCT/Par-OX and wildtype Par plants were germinated and planted in the BVRC greenhouse under NC for 5 weeks or 4 weeks following 1 weeks under HT. The 5th–7th healthy rosette leaves were collected from the third week after planting at 3 to 4 pm every Monday weekly. The leaf samples were collected at the end of the day. RNA sequencing analyses were performed by Igenecode Company (Beijing, China).
RNA-seq transcriptome library was prepared following TruSeq TM RNA sample preparation Kit from Illumina (San Diego, CA) using 1 μg of total RNA. After quantified by TBS380, the paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq xten/NovaSeq 6000 sequencer (2 × 150 bp read length). The raw paired-end reads were trimmed and quality-controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. Then clean reads were separately aligned to the reference genome with orientation mode using HISAT2 (http://ccb.jhu.edu/software/hisat2/index.shtml software. The mapped reads of each sample were assembled by StringTie (https://ccb.jhu.edu/software/stringtie/index.shtml? t = example) in a reference-based approach. To identify DEGs (differential expression genes) between two different samples, the expression level of each transcript was calculated according to the transcripts per million reads (TPM) method. RSEM was used to quantify gene abundances. Essentially, differential expression analysis was performed using the DESeq2[4]/DEGseq[5]/EdgeR[6]with Q value ≤ 0.05, DEGs with |log2FC | >1 and Q value < = 0.05(DESeq2 or EdgeR) /Q value < = 0.001(DEGseq) were considered to be significantly different expressed genes). In addition, functional-enrichment analysis including GO and KEGG were performed to identify which DEGs were significantly enriched in GO terms and metabolic pathways at Bonferroni-corrected P-value ≤ 0.05 compared with the whole-transcriptome background. GO functional enrichment and KEGG pathway analysis were carried out by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS.
Bulked segregation analyses (BSA)
Bulked segregation analyses were performed by BoYunHuaKang Company (Beijing, China). Sequencing libraries were generated using NEB Next® Ultra DNA Library Prep Kit for Illumina®(NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using HiSeq 4000 PE Cluster Kit (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 4000 platform and 150 bp paired-end reads were generated. Two parameters, SNP-index and Δ (SNP-index)72 were calculated to identify candidate regions. An SNP-index is the proportion of reads harboring the SNP that are different from the reference sequence. Δ (SNP-index) was obtained by subtraction of the SNP-index of two pools. For each read depth, 95 % confidence intervals of Δ (SNP-index) were obtained.
Quantitative real-time reverse transcription PCR (qRT-PCR)
First-strand cDNA was synthesized using a Prime Script™ reagent Kit with gDNA Eraser (RR047A, Takara, Dalian, China). qPCR was performed using the SYBR Green PCR master mix (04887352001, Roche, Basel, Switzerland), and a LightCycler 480 Real-Time PCR system (Roche, Basel, Switzerland). Actin2 (Arabidopsis) and GAPDH (B. rapa) were used as internal controls. qPCR primers are listed in Supplemental Supplementary Table 4.
Vector and transgenic plants constructions
The 2 kb upstream region of AtJMJ18 and the coding region of BrJMJ18PC and BrJMJ18Par were obtained by PCR and confirmed by sequencing. The 2 kb upstream region of AtJMJ18 was used as a promoter and inserted into vector pCMBIA130073 to substitute for the CaMV35S promoter. The SpeI/KpnI fragments containing the open reading frames of BrJMJ18PC and BrJMJ18Par were inserted into a plasmid with a green fluorescent protein (GFP) tag at the 3′ end. For the overexpression vectors, XbaI/SalI fragments of BrJMJ18PC and BrJMJ18Par coding regions were inserted into vector pCMBIA2300 vector with CaMV35S promoter and a GFP tag at the 3′ ends. The resultant constructs were transformed into Agrobacterium tumefaciens strain GV3101. pCMBIA1300-AtJMJ18pro-BrJMJ18PC/Par vectors were used to generate of Arabidopsis BrJMJ18PC/Par overexpression transgenic plants, denoted as AtJMJ18::BrJMJ18PC-GFP and AtJMJ18::BrJMJ18Par-GFP, respectively; while pCMBIA2300-35S-BrJMJ18PC/Par vectors were used to generate BrJMJ18PC/Par overexpression plants of Par, denoted as 35S::BrJMJ18PC-GFP (BrJMJ18PC-OX) and 35S::BrJMJ18Par-GFP (BrJMJ18Par-OX), respectively. For prokaryotic expression vectors, the EcoRI/SalI fragments of BrJMJ18PC/Par coding region were inserted into the pET28a (+) vector with His Tag at the 3’ end. The resultant constructs were transformed into Escherichia coli strain BL21. To construct CRISPR/Cas9 vectors, single guide RNA (sgRNA) sequences were designed using the web server CRISPR-P3. Using pCBC-DT1T2 as the template, two AtU6 promoter-sgRNA-AtU6 terminator cassettes were amplified using PCR. The PCR fragments were inserted into pKSE40174 and confirmed using Sanger sequencing. The primers used are listed in Supplementary Table 4.
Arabidopsis transgenic plants were generated using the flower-dipping method75
The T1 BrJMJ18 transgenic plants were screened using 15 mg/L hygromycin. Transgene expression was detected by immunoblotting using an anti-GFP antibody (HT801-01, TransGene, Beijing, China). Progeny of AtJMJ18::BrJMJ18PC-GFP lines 4#, 6#, and N1#; and AtJMJ18::BrJMJ18Par-GFP lines 2#, 8#, and N3# revealed a 3:1 separation ratio against hygromycin. T2 seeds of these lines were used for further experiments.
B. rapa transgenic plants were constructed as following
Agrobacterium Preparation: Transform Agrobacterium strain GV3101 via electroporation with the constructs of interest. Inoculate single colonies into 10 mL LB cultures (50 mg/L kanamycin and 50 mg/L rifampicin) and incubate at 28 °C, 220 rpm for 48 h. Centrifuge the culture at 3000 × g for 15 min, and re-suspend the pellet in liquid MS at OD650 = 0.05.
Seed Sterilization and Germination: Sterilize Par inbred line 16A-1 seeds with 75% ethanol (1 min) and 5% sodium hypochlorite (15 min). Germinate seeds on GM plates at 25 °C for 4 days.
Explant Isolation, Inoculation, and Co-cultivation: Cut 1–2 mm cotyledonary petioles of 4-day-old seedlings and place them on CIM-C plates. Inoculate by dipping petiole ends in Agrobacterium suspension. Return explants to the same CIM-C plates and Incubate in a growth chamber at 25 °C in dim darkness for 72 h.
Selection and Transfer of Plants to Greenhouse. After 72 h on CIM-C plates, the explants were transferred to the SIM plates and moved to a growth chamber at 25/22 °C with a 16/8-h (light/dark) photoperiod (photon flux density 100 mol/m2/s) for 2 weeks. Then the explants were moved to fresh SIM plates for a further 1-3 weeks until the emergence of green shoots. Green shoots were transferred to SEM plates for a further 1-2 weeks in the same growth chamber. The well-developed green shoots were transferred to the RM plates in the same growth chamber for 1-3 weeks. Rooted green plants were acclimatized in a growth chamber under 25 °C with 70–80% humidity for 2 days and then transferred into the soil.
Transgenic Plant Verification: For overexpression lines, transgene expression was confirmed by immunoblotting with an anti-GFP antibody (HT801-01, TransGene, Beijing, China) for BrJMJ18PC/Par-OX plants. T0 plants of both lines were self-crossed to generate seeds. In the case of CRISPR/Cas9 plants, the target gene BrJMJ18 from T0 plants was amplified, purified, and cloned into a vector using the pMD™18-T Vector Cloning Kit (Takara) for sequencing. Progeny T1 seedlings were grown on 0.5 × MS medium without kanamycin. Loss-of-function BrJMJ18 seedlings, confirmed through PCR and Sanger sequencing, were used for subsequent analysis.
Plant culture Media are listed as below:
Liquid MS: 4.3 g/L Murashige and Skoog (MS) basal salts (M519, Phytotech, USA), 30 g/L Sucrose, pH 5.7.
Germination media (GM): 4.3 g/L Murashige and Skoog (MS) basal salts, 30 g/L Sucrose, pH 5.7, 8 g/L agar.
Callus induction media for cocultivation (CIM-C): GM plus 500 mg/L 2-(N-morpholino) ethanesulfonic acid (MES), 4 mg/L 6-BA, 0.667 mg/L IAA.
Shoot induction media (SIM): CIM-C medium plus 160 mg/L Timentin, 2 mg/L AgNO3 and 25 mg/L kanamycin.
Shoot elongation media (SEM): GM plus 500 mg/L MES, 160 mg/L Timentin, 50 μg/L 6-BA, 2 mg/L AgNO3 and 25 mg/L kanamycin.
Rooting medium (RM): 3.05 g/L Gamborg’s B5 salts (G398, PhytoTech, USA), 10 g/L sucrose, pH5.7, 8 g/L agar, 160 mg/L Timentin, 1 mg/L IBA, 2 mg/L AgNO3 and 25 mg/L kanamycin.
Transient transformation
Tobacco (Nicotiana benthamiana) leaf transient transformation was performed as described previously76. Infiltrated tobacco plants grown under NC for three days and under NC for two days following 1 day of HT, respectively, were used for further analysis. BrJMJ18-GFP protein expression was viewed using the confocal laser scanning system Zeiss LSM510 (Carl Zeiss, Jena, Germany). BrJMJ18-GFP protein and Histone H3K36 methylation levels were determined using immunoblotting analysis using anti-GFP (HT801-01, TransGene, Beijing, China) and anti-Histone H3 di/tri methyl K36 antibodies (ab176921/ab195489, Abcam, Cambridge, MA, USA).
Protein extraction and immunoblotting analysis
For BrJMJ18 protein detection, total proteins were extracted from Arabidopsis and Par young rosette leaves or transiently transformed tobacco leaves using extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1% Triton X-100 and 1 mM Phenylmethanesulfonyl fluoride). For histone methylation detection, total proteins were extracted using histone extraction buffer (50 mM Tris-HCl, pH 7.5, 0.5 M sucrose, 1 mM MgCl2, 1 mM EDTA, 5 mM DTT, 1 mM Phenylmethanesulfonyl fluoride and 1 × protease inhibitor cocktail). After centrifuged at 14,000 × g for 30 min at 4 °C, the proteins in the supernatant were separated by SDS-PAGE and transferred to a 0.4 μm nitrocellulose blotting membrane (10600002, GE Healthcare, Chicago, IL, USA) using a Bio-Rad Tran-Blot Turbo transfer system (Bio-Rad, Hercules, CA, USA).
Antibodies against the following proteins were used: GFP (HT801-01, TransGen Biotech, Beijing, China), β-Tubulin (HC101, TransGen Biotech), Histone H3 (ab1791, Abcam, Cambridge, MA, USA), Histone H3 mono/di/tri methyl K4 (ab8895/ab32356/ab213224, Abcam, Cambridge, MA, USA), Histone H3 mono/di/tri methyl K9 (ab176880/ab32521/ab176916, Abcam), Histone H3 Histone H3 mono/di/tri methyl K27 (ab194688/ab24684/ab195477, Abcam), and Histone H3 mono/di/tri methyl K36 (ab176920/ab176921/ab195489, Abcam). β-tubulin and Ponceau S staining were used as loading controls. IRDye 680RD Goat anti-mouse (926-68070, LI-COR, Lincoln, NE, USA) or anti-rabbit (926-68071, LI-COR) antibodies were used as secondary antibodies. Anti-BrJMJ18 antibodies were produced by immunizing rabbits with a recombinantly expressed BrJMJ18 protein fragment (amino acids 1–133) expressed in Escherichia coli and purified using an BrJMJ18 antigen column (HuaBio, Hangzhou, China). Uncropped scans of immunoblotting blots are provided in the Source Data file.
Preparation of recombinant protein
Escherichia coli cells transformed with pET28a (+)-BrJMJ18PC/Par constructs were cultured, and isopropyl-D-thiogalactopyranoside was added to induce the recombinant protein expression. His-BrJMJ18PC/Par proteins were purified using a Ni2 + -NTA agarose column (40724, QIAGEN, Germany).
In vitro histone demethylase assay using recombinant BrJMJ18-GFP protein
The histone demethylase assay was performed as previously reported37 with some modifications. Total proteins of young rosette leaves of BrJMJ18 transgenic plants were extracted using extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1% Triton X-100 and 1 mM Phenylmethanesulfonyl fluoride) and centrifuged at 14,000 × g for 30 min at 4 °C. The supernatant was incubated with anti-GFP monoclonal antibody (mAb) agarose (D153-8, MBL, Woburn, MA, USA) at 4 °C for 3 h. The agarose was collected by centrifugation and washed with wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2) three times. Then, the agarose was added to demethylation buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 50 μM (NH4)2Fe(SO4)2, 1 mM α-ketoglutarate, 2 mM ascorbic acid, and 0.1 mg/mL Histone from calf thymus (1223565001, Roche, Basel, Switzerland) and incubated at 37 °C for 4 h.
In vitro histone demethylase assay using recombinant His-BrJMJ18 protein
The histone demethylase assay was performed as previously reported77 with some modifications. 2.5 μg synthesized Histone H3 peptide with H3K4me3, H3K9me3, H3K27me3, H3K36me2, H3K36me3 modification (R-1023, R-1028, R-1034, R-1038, R-1039, Epigentek, USA) were incubated with affinity-purified recombinant His-BrJMJ18PC/Par protein (approximately 0.8 μg-2.4 μg) in 30 μL reaction buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 50 μM (NH4)2Fe(SO4)2, 1 mM α-ketoglutarate, 2 mM ascorbic acid) for 4 h at 37 °C. Purified His was used as a negative control.
The proteins in the reaction solution were separated by SDS-PAGE and transferred to 0.4 μm nitrocellulose membrane (10600002, GE Healthcare, Chicago, IL, USA) to detect histone H3 methylation using immunoblotting analysis with anti-histone H3 mono/di/tri methyl K4, mono/di/tri methyl K9, mono/di/tri methyl K27, and mono/di/tri methyl K36 antibodies. The amount of H3 in each sample (detected with anti-histone H3 antibody (ab1791, Abcam, Cambridge, MA, USA)) served as a loading control. His-BrJMJ18PC/Par proteins were detected with anti-BrJMJ18 antibody. The agarose was boiled in 1 × SDS buffer for 5 mins and the proteins in the supernatant were separated by SDS-PAGE and transferred to 0.4 μm nitrocellulose membranes (10600002, GE Healthcare, Chicago, IL, USA) to detect BrJMJ18-GFP protein using immunoblotting analysis with anti-GFP antibodies.
Chromatin immunoprecipitation (ChIP)
For ChIP-Sequencing using anti-BrJMJ18 antibodies, the 5th–7th healthy rosette leaves of 5-week-old Par plants grown under NC were used. And the leaf samples were collected at the end of the day. For ChIP-seq using anti-GFP antibodies (ab290, Abcam, Cambridge, MA, USA), the 55th–7th healthy rosette leaves of BrJMJ18PC/Par-OX and wild-type Par plants grown under NC for 5 weeks or 4 weeks following 1 weeks under HT were used. The chIP-seq experiment was carried out by IGENEBOOK Biotechnology Company (Wuhan, China) using rabbit polyclonal anti-BrJMJ18 antibodies and according to a previously described method78. Two biological replicates were performed for each sample. Briefly, 1.5 g samples were washed twice in cold phosphate-buffered saline (PBS), cross-linked with 1% formaldehyde for 10 min at room temperature, and then quenched by the addition of glycine (125 mmol/L final concentration). Afterwards, samples were lysed and chromatin was obtained on ice. Chromatin was sonicated to get soluble sheared chromatin (average DNA length of 200–500 bp). Then, 20 μL of chromatin was reserved at –20 °C as input DNA, and 100 μL of chromatin was used for immunoprecipitation using 10 μg of anti-BrJMJ18 at 4 °C overnight. The next day, 30 μL of protein beads were added and the samples were further incubated for 3 h. The beads were then washed once with 20 mM Tris/HCL (pH 8.1), 50 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS; twice with 10 mM Tris/HCL (pH 8.1), 250 mM LiCl, 1 mM EDTA, 1% NP-40, 1% deoxycholic acid; and twice with TE buffer 1 × (10 mM Tris-Cl at pH 7.5. 1 mM EDTA). Bound material was then eluted from the beads in 300 μL of elution buffer (100 mM NaHCO3, 1% SDS), treated first with RNase A (final concentration 8 μg/mL) for 6 h at 65 °C and then with proteinase K (final concentration 345 μg/mL) overnight at 45 °C. Immunoprecipitated DNA was used to construct sequencing libraries following the protocol provided by the I NEXTFLEX® ChIP-Seq Library Prep Kit for Illumina® Sequencing (NOVA-5143-02, Bioo Scientific, Austin, TX, USA) and sequenced on Illumina Xten with the PE 150 method (Illumina Inc. San Diego, CA, USA). Trimmomatic (version 0.38) was used to filter out low-quality reads (Bolger et al., 2014). Clean reads were mapped to the B. rapa genome v3.0 using Bwa (version 0.7.15) (Li and Durbin, 2009). Samtools (version 1.3.1) was used to remove potential PCR duplicates79. MACS2 software (version 2.1.1.20160309) was used to call peaks using default parameters (bandwidth, 300 bp; model fold, 5, 50; q value, 0.05) using input as control. If the summit of a peak is located closest to the transcription start site (TSS) of one gene, the peak was assigned to that gene80. Gene Ontology (GO) enrichment analysis was performed using the EasyGO gene ontology enrichment analysis tool (http://bioinformatics.cau.edu.cn/easygo/)81. And BrJMJ18 target genes under NC/HT were obtained using wild-type Par plants under NC/HT as control. The GO term enrichment was calculated using hypergeometric distribution, with a P value cutoff of 0.01. P values obtained by Fisher’s exact test were adjusted by the false discovery rate (FDR) for multiple comparisons to detect overrepresented GO terms.
For ChIP-QPCR, experiments were performed using an Imprint Chromatin Immunoprecipitation Kit (CHP1, Sigma-Aldrich, St. Louis, MO, USA). For Brassica crops, plants grown under NC for 5 weeks or 4 weeks under NC following 1-week of HT, respectively, were used as materials. For Arabidopsis, Col-0 and AtJMJ18::BrJMJ18PC-GFP, and AtJMJ18::BrJMJ18Par-GFP plants grown under NC for 3 weeks or 2 weeks under NC following 1-week of HT, respectively, were used for analysis. Young and healthy rosette leaves (40 mg) of the above mentioned plants were used for ChIP qPCR experiments. Rabbit polyclonal anti-BrJMJ18 antibodies and rabbit polyclonal anti-Histone H3 di/tri methyl K36 antibodies (ab176921/ab195489, Abcam) were used in the immunoprecipitations and normal rabbit IgG (12-370, Sigma-Aldrich) was use as the negative control. Actin2 (Arabidopsis) was used as the internal control. The level of the protein in the negative control was set as 1 and enrichment represented the relative fold-change fold in relation to the negative control. Primers used are listed in Supplementary Table 4.
Statistical analysis
For bar graphs, error bars represent the SD, data are presented as means ± SD from n = 3. For box-and-whisker plots, boxes show the interquartile range (i.e., the 1st and 3rd quartile) framing the median. The whiskers show the minimal and maximal values except if these values are higher than the interquartile range multiplied by 1.5. In this case, the whiskers show the interquartile range by 1.5 and the dots are outliers. The significance of the difference in the columns/boxplots were calculated with a two-tailed Student’s t test for the pairwise comparisons (*p < 0.05; **p < 0.01; ***p < 0.001).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Source data
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2022YFF1003003, T.S.), the National Natural Science Foundation of China (32172557, T.S.), the Innovation and Capacity-Building Project of BAAFS (KJCX20240408, S.Y., and KJCX20230431, X.X.), the Outstanding Scientists Training program of BAAFS (JKZX202406, T.S.), and the China Agriculture Research System of MOF and MARA (CARS-A03, T.S.).
Author contributions
T.S., X.X., S.Y., and F.Z. designed the experiments. T.S., X.X., and P.L. carried out the analyses. W.W., X.Z., G.J., J.W., L.S., Y.Y., and D.Z. collected the phenotypes. T.S. and X.X. wrote and revised the paper. All authors discussed the results and commented on the manuscript.
Peer review
Peer review information
Nature Communications thanks Sarah-Veronica Schiessl and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
ChIP-seq data have been deposited on NCBI with accession number GSE223969. All the raw sequencing data are archived at the Genome Sequence Archive of China National Center for Bioinformation with the accession number PRJCA025632. Source data are provided with this paper.
Competing interests
The authors declared no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fenglan Zhang, Email: zhangfenglan@nercv.org.
Shuancang Yu, Email: yushuancang@nercv.org.
Tongbing Su, Email: sutongbing@nercv.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-49721-z.
References
- 1.Elvis, K., Quezada, D., Ihien, E., Vasquez-Teuber, P. & Mason, A. Interspecific hybridization for Brassica crop improvement. Crop Breed. Genet. Genom.10.20900/cbgg20190007 (2020).
- 2.Qi X, et al. Genomic inferences of domestication events are corroborated by written records in Brassica rapa. Mol. Ecol. 2017;26:3373–3388. doi: 10.1111/mec.14131. [DOI] [PubMed] [Google Scholar]
- 3.Prakash, S., Wu, X.i. & Bhat, S. R. History, Evolution, and Domestication of Brassica Crops, Plant Breed. Rev.35, 19–84 (2011).
- 4.Purugganan M, Boyles AL, Suddith JI. Variation and selection at the CAULIFLOWER floral homeotic gene accompanying the evolution of domesticated Brassica oleracea. Genetics. 2000;155:855–862. doi: 10.1093/genetics/155.2.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClung CR, Lou P, Hermand V, Kim JA. The importance of ambient temperature to growth and the induction of flowering. Front. plant sci. 2016;7:1266. doi: 10.3389/fpls.2016.01266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Olmo I, Poza-Viejo L, Piñeiro M, Jarillo JA, Crevillén P. High ambient temperature leads to reduced FT expression and delayed flowering in Brassica rapa via a mechanism associated with H2A.Z dynamics. Plant j. 2019;100:343–356. doi: 10.1111/tpj.14446. [DOI] [PubMed] [Google Scholar]
- 7.Yui S, Yoshikawa H. Breeding of bolting resistance in Chinese cabbage. critical day length for flower induction of late bolting material with no chilling requirement. Engei Gakkai Zasshi. 1992;61:565–568. doi: 10.2503/jjshs.61.565. [DOI] [Google Scholar]
- 8.Wei, Q., et al. The New Variation in the Promoter Region of FLOWERING LOCUS T Is Involved in Flowering in Brassica rapa. Genes13, 1162 (2022) [DOI] [PMC free article] [PubMed]
- 9.Lu Y. Evaluation of heat tolerance and analysis of allele variation of heat tolerance candidate gene in flowering Chinese cabbage. Doctoral dissertation, Guangzhou University, (2020).
- 10.Rameeh, V. Correlation analysis in different planting dates of rapeseed varieties. J. Agric. Sci.7, 76–84 (2012)
- 11.Morrison MJ, Stewart DW. Heat stress during flowering in summer Brassica. Crop Sci. 2002;42:797–803. doi: 10.2135/cropsci2002.7970. [DOI] [Google Scholar]
- 12.Cheng F, et al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016;48:1218–1224. doi: 10.1038/ng.3634. [DOI] [PubMed] [Google Scholar]
- 13.Goto, S. et al. Organizing and computing metabolic pathway data in terms of binary relations. Pac. Symp. Biocomput.1997, 175–186 (1997). [PubMed]
- 14.Yang H, et al. A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet. 2012;8:e1002664. doi: 10.1371/journal.pgen.1002664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poza-Viejo L, et al. Conserved and distinct roles of H3K27me3 demethylases regulating flowering time in Brassica rapa. Plant Cell Environ. 2022;45:1428–1441. doi: 10.1111/pce.14258. [DOI] [PubMed] [Google Scholar]
- 16.Mehraj H, et al. Characterization of histone H3 Lysine 4 and 36 Tri-methylation in Brassica rapa L. Front. Plant Sci. 2021;12:659634. doi: 10.3389/fpls.2021.659634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue L, et al. Gene co-expression network analysis of the heat-responsive core transcriptome identifies hub genes in Brassica rapa. Planta. 2021;253:111. doi: 10.1007/s00425-021-03630-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, et al. Heat stress response in Chinese cabbage (Brassica rapa L.) revealed by transcriptome and physiological analysis. PeerJ. 2022;10:e13427. doi: 10.7717/peerj.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balciunas D, Ronne H. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem. Sci. 2000;25:274–276. doi: 10.1016/S0968-0004(00)01593-0. [DOI] [PubMed] [Google Scholar]
- 20.Clissold PM, Ponting CP. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem. Sci. 2001;26:7–9. doi: 10.1016/S0968-0004(00)01700-X. [DOI] [PubMed] [Google Scholar]
- 21.McAlvay AC, et al. Brassica rapa domestication: untangling wild and feral forms and convergence of crop morphotypes. Mol. biol. Evol. 2021;38:3358–3372. doi: 10.1093/molbev/msab108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo G. A analysis of historical materials about the time and place of the origin of the peking cabbage. Stud. Hist. Nat. Sci. 1992;2:171–176. [Google Scholar]
- 23.Ainsworth EA, Ort DR. How do we improve crop production in a warming world? Plant Physiol. 2010;154:526–530. doi: 10.1104/pp.110.161349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.As W, Galani S, Ashraf M, Foolad M. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- 25.Collins NC, Tardieu F, Tuberosa R. Quantitative trait loci and crop performance under abiotic stress: where do we stand? Plant Physiol. 2008;147:469–486. doi: 10.1104/pp.108.118117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jha, U. & Singh, N. Heat stress in crop plants: Its nature, impacts and integrated breeding strategies to improve heat tolerance. Plant Breed.133, 679–701 (2014).
- 27.Li XM, et al. Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 2015;47:827–833. doi: 10.1038/ng.3305. [DOI] [PubMed] [Google Scholar]
- 28.Cappetta E, et al. Tomato genomic prediction for good performance under high-temperature and identification of loci involved in thermotolerance response. Hortic. Res. 2021;8:212. doi: 10.1038/s41438-021-00647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, et al. Overcoming reproductive compromise under heat stress in wheat: physiological and genetic regulation, and breeding strategy. Front. plant sci. 2022;13:881813. doi: 10.3389/fpls.2022.881813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Martelot G, et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10:e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Smolle M, et al. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pajoro A, Severing E, Angenent GC, Immink RGH. Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol. 2017;18:102. doi: 10.1186/s13059-017-1235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones MA, Morohashi K, Grotewold E, Harmer SL. Arabidopsis JMJD5/JMJ30 acts independently of LUX ARRHYTHMO within the plant circadian clock to enable temperature compensation. Front. Plant Sci. 2019;10:57. doi: 10.3389/fpls.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bloomer RH, et al. The Arabidopsis epigenetic regulator ICU11 as an accessory protein of polycomb repressive complex 2. Proc. Natl. Acad. Sci. USA. 2020;117:16660–16666. doi: 10.1073/pnas.1920621117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan ES, et al. Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun. 2014;5:5098. doi: 10.1038/ncomms6098. [DOI] [PubMed] [Google Scholar]
- 38.Whittaker C, Dean C. The FLC Locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 2017;33:555–575. doi: 10.1146/annurev-cellbio-100616-060546. [DOI] [PubMed] [Google Scholar]
- 39.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 40.Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young MD, et al. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 2011;39:7415–7427. doi: 10.1093/nar/gkr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H, Howard M, Dean C. Physical coupling of activation and derepression activities to maintain an active transcriptional state at FLC. Proc. Natl. Acad. Sci. USA. 2016;113:9369–9374. doi: 10.1073/pnas.1605733113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crevillén P, et al. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature. 2014;515:587–590. doi: 10.1038/nature13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019;29:379–390. doi: 10.1038/s41422-019-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi N, et al. H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat. Commun. 2021;12:3480. doi: 10.1038/s41467-021-23766-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bac-Molenaar JA, et al. Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant cell. 2015;27:1857–1874. doi: 10.1105/tpc.15.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blázquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- 48.Mateos JL, et al. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015;16:31. doi: 10.1186/s13059-015-0597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuchiya T, Eulgem T. The Arabidopsis defense component EDM2 affects the floral transition in an FLC-dependent manner. Plant J. Cell Mol. Biol. 2010;62:518–528. doi: 10.1111/j.1365-313X.2010.04169.x. [DOI] [PubMed] [Google Scholar]
- 50.Willmann MR, Poethig RS. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development. 2011;138:677–685. doi: 10.1242/dev.057448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen M, Penfield S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science. 2018;360:1014–1017. doi: 10.1126/science.aar7361. [DOI] [PubMed] [Google Scholar]
- 52.Yuan W, et al. mapping of germination speed in Arabidopsis thaliana. Mol. Ecol. 2016;25:4177–4196. doi: 10.1111/mec.13768. [DOI] [PubMed] [Google Scholar]
- 53.Schmalenbach I, Zhang L, Reymond M, Jiménez-Gómez JM. The relationship between flowering time and growth responses to drought in the Arabidopsis Landsberg erecta x Antwerp-1 population. Front. Plant Sci. 2014;5:609. doi: 10.3389/fpls.2014.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ågrena J, Oakley CG, McKay JK, Lovell JT, Schemske DW. Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2013;110:21077–21082. doi: 10.1073/pnas.1316773110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 56.Cai X, et al. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021;22:166. doi: 10.1186/s13059-021-02383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitamoto N, Yui S, Nishikawa K, Takahata Y, Yokoi S. A naturally occurring long insertion in the first intron in the Brassica rapaFLC2 gene causes delayed bolting. Euphytica. 2014;196:213–223. doi: 10.1007/s10681-013-1025-9. [DOI] [Google Scholar]
- 58.Li X, Zhang S, Bai J, He Y. Tuning growth cycles of Brassica crops via natural antisense transcripts of BrFLC. Plant Biotechnol. J. 2016;14:905–914. doi: 10.1111/pbi.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schranz ME, et al. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics. 2002;162:1457–1468. doi: 10.1093/genetics/162.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lou P, et al. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 2007;58:4005–4016. doi: 10.1093/jxb/erm255. [DOI] [PubMed] [Google Scholar]
- 61.Dechaine JM, Brock MT, Weinig C. QTL architecture of reproductive fitness characters in Brassica rapa. BMC Plant Biol. 2014;14:66. doi: 10.1186/1471-2229-14-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao D, et al. Genetic dissection of flowering time in Brassica rapa responses to temperature and photoperiod. Plant Sci. Int. J. Experiment. Plant Biol. 2019;280:110–119. doi: 10.1111/jipb.12768. [DOI] [PubMed] [Google Scholar]
- 63.Kim SY, et al. Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa L.: ssp. pekinensis) Plant Cell Rep. 2007;26:327–336. doi: 10.1007/s00299-006-0243-1. [DOI] [PubMed] [Google Scholar]
- 64.Xiao D, et al. The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. J. Experiment. Bot. 2013;64:4503–4516. doi: 10.1093/jxb/ert264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burggren, W. Epigenetic Inheritance and Its Role in Evolutionary Biology: Re-Evaluation and New Perspectives. Biology5, 24 (2016) [DOI] [PMC free article] [PubMed]
- 66.McCaw BA, Stevenson TJ, Lancaster LT. Epigenetic responses to temperature and climate. Integr. Comp. Biol. 2020;60:1469–1480. doi: 10.1093/icb/icaa049. [DOI] [PubMed] [Google Scholar]
- 67.Su T, et al. A genomic variation map provides insights into the genetic basis of spring chinese cabbage (Brassica rapa ssp. pekinensis) selection. Mol. Plant. 2018;11:1360–1376. doi: 10.1016/j.molp.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duan N, et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 2017;8:249. doi: 10.1038/s41467-017-00336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Teng D, Tian F, Guan Q, Wang J. Comparison of three DNA extraction methods for feed products and four amplification methods for the 5’-junction fragment of Roundup Ready soybean. J. Agric. Food Chem. 2012;60:4586–4595. doi: 10.1021/jf300827q. [DOI] [PubMed] [Google Scholar]
- 72.Abe A, et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012;30:174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- 73.Cao X, et al. Characterization of DUF724 gene family in Arabidopsis thaliana. Plant Mol. Biol. 2010;72:61–73. doi: 10.1007/s11103-009-9551-5. [DOI] [PubMed] [Google Scholar]
- 74.Xing HL, et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 76.Xu J, et al. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008;283:26996–27006. doi: 10.1074/jbc.M801392200. [DOI] [PubMed] [Google Scholar]
- 77.Yang W, Jiang D, Jiang J, He Y. A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. Cell Mol. Biol. 2010;62:663–673. doi: 10.1111/j.1365-313X.2010.04182.x. [DOI] [PubMed] [Google Scholar]
- 78.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics. 2010;11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou X, Su Z. EasyGO: Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species. BMC Genomics. 2007;8:246. doi: 10.1186/1471-2164-8-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
ChIP-seq data have been deposited on NCBI with accession number GSE223969. All the raw sequencing data are archived at the Genome Sequence Archive of China National Center for Bioinformation with the accession number PRJCA025632. Source data are provided with this paper.