Abstract
Human immunodeficiency virus type 2 (HIV-2), like other lentiviruses, is capable of infecting nondividing T cells and macrophages. The present work shows that in HIV-2-infected cells, Vpx is necessary for efficient nuclear import of the preintegration complex. In agreement with this finding, the subcellular localization of a GFP-Vpx fusion protein was found to be predominantly nuclear. However, deletion of the proline-rich C-terminal 11 residues of Vpx resulted in a shift of the fusion protein to the cytoplasm. Furthermore, the same deletion in the context of the provirus resulted in a decrease in nuclear import of the preintegration complex and attenuated replication in macrophages.
A critical step in the process of retrovirus infection is the transfer of viral DNA into the nucleus of the infected cell (7). Once inside the nucleus, the linear proviral DNA integrates into the host genome, where it can be transcribed to form a full-length progeny RNA genome and mRNAs encoding viral proteins. Nuclear import of oncoretroviral DNA requires dissolution of the nuclear envelope with mitosis. In contrast, lentiviruses are also able to perform this task by exploiting cellular pathways for active nuclear import (23). Thus, lentiviruses are capable of infecting nondividing cells, such as terminally differentiated macrophages and memory T cells, which are important for viral dissemination and persistence (17).
The main cellular pathway for nuclear import is the importin pathway, wherein the import substrate binds via its nuclear localization signal (NLS) to the importin α/β heterodimer in the cytoplasm (14, 33). Importin α is responsible for binding to the NLS (14, 22, 46). The simplest NLS consists of a short stretch of 5 to 7 basic residues, whereas a bipartite NLS includes two basic domains separated by 10 to 11 residues. After importin α binds the NLS, importin β mediates the docking of the import substrate to the nuclear pore (15). Nuclear pore proteins, which often contain FXFG repeats, function as docking sites at the nuclear envelope (40, 41). Two other proteins, Ran and p10, mediate the translocation of the import complex across the nuclear pore (29, 30).
Upon entry of the human immunodeficiency virus (HIV) virion core into the newly infected cell, viral proteins required for nuclear import and integration remain associated with the viral nucleic acids (8). After reverse transcription, this high-molecular-weight nucleoprotein complex is referred to as the viral preintegration complex. Three proteins in HIV-1 have been implicated in the nuclear localization of this complex: matrix (MA), integrase (IN), and viral protein R (Vpr), although there is controversy about the role of MA in this process (10). Each of these proteins has been shown to bind importin α (12, 13, 38, 45). MA and IN bind in an NLS-dependent manner, and their binding is inhibited by a peptide including the simian virus 40 large T antigen NLS sequence. In contrast, Vpr binds importin α in an NLS-independent manner (12, 13). MA contains a putative basic-type NLS spanning residues 25 to 33. HIV-1 strains with mutations in this region of lysines to threonines (26KK→TT or 27K→T) are attenuated in their ability to replicate in nondividing cells when Vpr is also absent (MAΔNLSΔVPR) (3). The same mutation abrogates binding of MA to importin α (13). IN binds to importin α through an atypical bipartite NLS located within its C terminus. Mutation of this sequence in an MAΔNLSΔVPR virus results in a more complete block in nuclear import of the preintegration complex (12).
Vpr does not bind importin α at its NLS binding site (12, 13). This was demonstrated in experiments that showed that Vpr does not compete with MA for importin α binding. Instead, Vpr increases the affinity of importin α for the NLS of MA (38). In addition to binding importin α, Vpr also binds to FXFG repeat-containing nucleoporins (45) and has been postulated to stabilize docking of viral preintegration complexes to the nuclear pore (39).
When expressed in the absence of other viral proteins, Vpr localizes to the nucleus (5, 25, 27) and the nuclear envelope (45). Vpr does not contain a region which resembles an NLS. Mutations within an N-terminal α-helical region block nuclear localization (5, 26, 47). In addition, mutations in the leucine-isoleucine-rich domain and the arginine-rich C terminus of Vpr impair its nuclear targeting function (26, 49). Some of these mutations may alter subcellular localization due to global effects on Vpr conformation and/or stability, while others may specifically disrupt a domain critical for nuclear localization. Vpr may contain a novel nuclear targeting signal or perhaps a region important for protein-protein interactions with an NLS-containing protein (piggyback binding). Alternatively, the domain could be important for nuclear retention after passive diffusion of Vpr into the nucleus.
In addition to Vpr, HIV-2 and members of the simian immunodeficiency virus SIVsm/SIVmac lineage encode Vpx. Vpx and Vpr share considerable homology (42, 44). Like Vpr, Vpx is virion associated and recruited into virions through its interaction with the p6 portion of the Gag polyprotein (1, 34, 47). Also similar to Vpr, one study demonstrated that Vpx, when expressed in the absence of other viral components, is a nuclear protein (5). However, another group has described a perinuclear distribution for this accessory protein (47). Both Vpr and Vpx are found at high concentrations within the virion, in amounts comparable to that of Gag proteins. This suggests an important function early in infection. Despite the similarities of Vpr and Vpx, Fletcher and colleagues demonstrated that the two accessory proteins mediate distinct functions during SIV infection (9). In a highly pathogenic variant of SIV, SIVSM PBj1.9, the primary function of Vpr is induction of cell cycle arrest in G2, whereas SIVSM Vpx functions in the nuclear import of the preintegration complex. Cells expressing SIVSM Vpx, but not Vpr, were not arrested in G2, and viruses which retained Vpr but lacked Vpx were unable to efficiently infect nondividing cells. In contrast to the finding of redundant nuclear import signals present in the HIV-1 preintegration complex, this study showed that SIVSM PBj1.9 Vpx is both necessary and sufficient for the nuclear import of the preintegration complex.
The present study was designed to assess the role of HIV-2 Vpx in nuclear import processes. Similar to the observations with SIVSM PBj1.9 Vpx, HIV-2 Vpx is necessary for nuclear import of the viral preintegration complex. In addition, we have addressed the controversy concerning the cellular localization of Vpx. Finally, we sought to identify regions of the protein important for its subcellular localization. To accomplish this, we created green fluorescent protein (GFP) fusion proteins with wild-type HIV-2 Vpx and several mutant forms of the protein and assessed their distribution in transfected cells. Interestingly, the C-terminal proline-rich tail of Vpx appears to be important both for Vpx nuclear localization and nuclear import of the preintegration complex.
HIV-2 Vpx is necessary for nuclear import of viral cDNA.
In order to determine whether HIV-2 Vpx is sufficient for nuclear import in nondividing cells, we generated HIV-2ROD proviral clones with mutations in vpx, vpr, and the coding sequence for the NLS of MA. The functional HIV-2ROD proviral clone, pSE (20), was digested with SalI to remove flanking cellular sequences, which generated pES. Mutant pMX1+62, designated MX here, has been described previously (20) and includes mutations of both methionine codons of vpx. The pMAΔNLSMRMX clone has the MX1+62 vpx mutation, along with a G4700T change within vpr, which converts the seventh codon to a termination codon, and a 26K→T mutation in the NLS of MA. Mutagenesis of the MA NLS was carried out by a PCR-based overlap extension method, described previously (18). This MA mutation in HIV-1 blocks the ability of the virus to infect nondividing cells in the absence of functional Vpr (3). All mutations were confirmed by sequence analysis.
293T cells were transfected with 5 μg of the proviral DNAs by using Lipofectamine (Gibco). The supernatants of transfected cells were filtered 48 h posttransfection, and viral stocks were treated with 2 μg of DNase per ml in 10 mM MgCl2 for 30 min at 37°C. Viral stock concentrations were determined by p27 antigen-capture enzyme-linked immunosorbent assay (Coulter), and infectious titers were normalized by using CCR5-transfected Magi cells (21, 36).
U937 cells were maintained in RPMI1640 supplemented with 10% fetal bovine serum. To induce G1/S arrest, cells were synchronized by serum starvation for 48 h followed by release into complete medium containing 400 μM mimosine (31). Cells were infected with 50 ng of p27 of wild-type (ES) or mutant (MX, MAΔNLSMRMX, or ES-X101) DNase-treated HIV-2. Cells were collected at 6, 12, 24, and 48 h postinfection, and total DNA was isolated with the DNAzol reagent according to the manufacturer's protocol (Gibco).
Two-long terminal repeat (LTR) circle forms of HIV-2 DNA were amplified with 32P-end-labeled primers to the HIV-2 LTR U5 nucleotides 238 to 217 (5′-TTACTCAGGTGAACACCGAAT) and 278 to 299 (5′ACCGAGGCAGGAAAATCCCTA). Late reverse transcription products were amplified with end-labeled primers to LTR U5 nucleotides 278 to 299 and nucleotides 625 to 594 of gag (5′-GTCTTTCCCCCGGGCCGTAACCTCATTCTTTC). Standards were generated by conducting PCRs with serial dilutions of chronically infected CEM cells with the primers indicated.
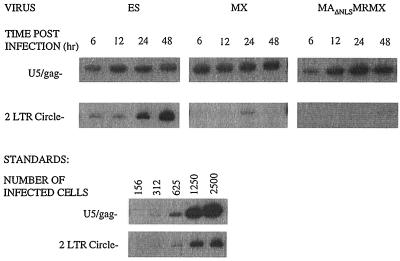
PCR analysis of cellular DNA obtained immediately after infection gave no visible product (data not shown). Linear reverse transcription products, which were synthesized in the cytoplasm of infected cells, were amplified with U5/gag primers and were detectable at equivalent levels in cells infected by all three viruses and within 6 h after infection (Fig. 1). Therefore, none of the mutations that we have introduced into the virus affect fusion, early postentry events, or reverse transcription. The two-LTR circular form of viral DNA, which is only found in the nuclei of infected cells and serves as a surrogate marker of nuclear import, was selectively amplified by using U5 primers. Markedly reduced levels of two-LTR circular DNA were found in both the MAΔNLSMRMX- and MX-infected cells, compared to ES-infected cells. Moreover, neither Vpr nor MA compensated for the loss of Vpx function in promoting nuclear import. Thus, HIV-2 Vpx is the predominant nuclear import factor of the viral preintegration complex, similar to what has been reported for SIVSM PBj1.9 Vpx (9).
FIG. 1.
Vpx is required for nuclear localization of viral reverse-transcribed DNA products. At the indicated times postinfection with ES, MX, or MAΔNLSMRMX, total cellular DNA was isolated from arrested U937 cells. PCR was performed with U5/gag primers and U5/U5 primers to detect viral DNA synthesis and nuclear localization, respectively. Standards were derived from reactions with the identical primers on serial dilutions of chronically infected cells. Representative results are shown from three different experiments which all yielded similar results.
Localization of GFP-Vpx fusion proteins.
In order to assess the subcellular localization of Vpx, wild-type and mutant forms of vpx were cloned into a plasmid which generates a fusion protein with GFP at the N terminus. The Vpx mutations that we studied included a deletion of residues 20 to 40 (AH), a region predicted to form an amphipathic helix, a truncation at residue 89 (X89), and a truncation at residue 101 (X101) (34). The last two mutations remove a highly conserved stretch of prolines found in C-terminal residues 102 to 112 of Vpx (11). Wild-type and mutant vpx constructs described previously (34) were amplified by PCR with primers which introduced a 5′ BspE1 site and a 3′ XhoI site. PCR products were then cloned into the pEGFP-C1 vector (Clontech), which had been digested with BspE1-XhoI. All constructs were confirmed by sequence analysis.
The DNAs were transfected into 293T or HeLa cells, and 48 h later, the distribution of GFP fusion proteins was determined by confocal microscopy. These cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, plated onto chamber tissue culture slides (Falcon), grown to 50 to 70% confluency, and transfected with the GFP DNAs and, in the indicated cases, with the HIV-2 gag-pol expression plasmid, pTM-GP2, described previously (19). After 19 h, cells transfected with pTM-GP2 were infected at a multiplicity of infection (MOI) of 10 with vaccinia virus vTF7-3, expressing T7 polymerase. After 24 h, the cells were fixed in 2% paraformaldehyde for 20 min and examined on a Zeiss axiovert microscope equipped with a Bio-Rad confocal scanning imaging system.
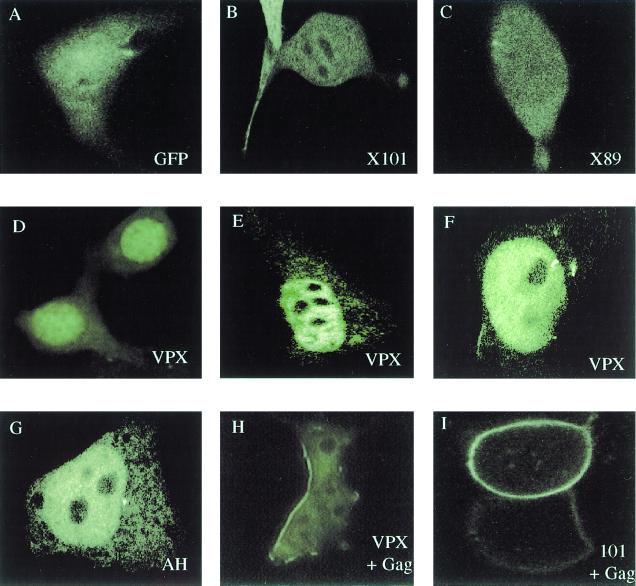
GFP alone is distributed diffusely throughout the cells (Fig. 2A). GFP-Vpx had a predominantly nuclear localization (Fig. 2D to F), although it was occasionally observed in a perinuclear distribution (data not shown). All of the fusion proteins showed a shift in localization to the plasma membrane when coexpressed with HIV-2 Gag (Fig. 2H and I, and data not shown), indicating that the GFP moiety did not interfere with the ability of Vpx to interact with Gag. GFP-AH had a distribution similar to that of GFP-Vpx (Fig. 2G). In contrast, both GFP-X89 and GFP-X101 had a diffuse cellular distribution, with both nuclear and cytoplasmic localization (Fig. 2B and C).
FIG. 2.
The C-terminal proline-rich tail of Vpx is important for nuclear localization. 293T cells were transfected with GFP constructs with or without HIV-2 Gag (pTM-GP2). The GFP fusion proteins included wild-type Vpx, X101, X89, and XAH. Cells transfected with pTM-GP2 were infected with vaccinia virus 19 h posttransfection and fixed in 2% paraformaldehyde 24 h posttransfection.
The size of the fusion proteins is relatively small (<47 kDa), allowing for their passive diffusion in and out of the nucleus. In the absence of an organelle targeting or retention signal, we would expect a diffuse cellular distribution. The distribution of GFP-X89 and GFP-X101 looks remarkably like that of GFP alone.
293T cells were transfected with GFP constructs and metabolically labeled overnight by using methionine- and cysteine-free medium supplemented with 50 μCi of Tran[35S]label (ICN) per ml. Cells were solubilized in radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.2 mM phenylmethylsulfonyl fluoride), clarified, and incubated with 5 μl of polyclonal anti-Vpx antiserum. Twenty microliters of protein A-Sepharose beads was added, and the incubations were continued for an additional hour. Precipitates were washed three times with RIPA buffer, boiled for 2 min in Laemmli sample buffer, and analyzed on an SDS–12% polyacrylamide gel. Labeled proteins were detected by autoradiography.
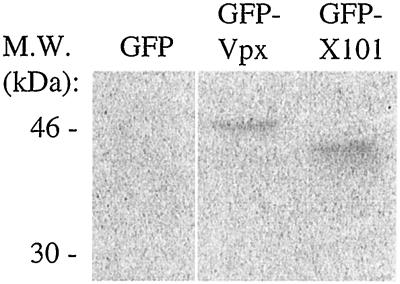
One potential explanation for these results is that GFP-X89 and GFP-X101 could be less stable than GFP-Vpx or GFP-XAH. However, the truncation of HIV-2 Vpx at either residue 89 or 101 did not affect protein stability (34). The GFP-X101 fusion protein was expressed in transfected 293T cells at levels comparable to those of GFP-Vpx (Fig. 3). Therefore, it appears that the C-terminal 11 residues of Vpx are important either for nuclear targeting or for nuclear retention of the protein.
FIG. 3.
GFP-Vpx101 and GFP-Vpx fusion proteins are expressed at equivalent levels. 293T cells transfected with GFP, GFP-Vpx, and GFP-Vpx101 were metabolically labeled overnight with Tran[35S] label. Protein expression was assessed by immunoprecipitation with anti-Vpx antiserum followed by SDS-polyacrylamide gel electrophoresis. M.W., molecular mass markers.
The C-terminal tail of Vpx is required for efficient nuclear targeting of viral cDNA.
In order to assess the functional importance of the C-terminal 11 residues of Vpx, we generated a provirus which contained Vpx 101 in place of the wild-type protein. The vpx mutant pES-X101 was generated by performing PCR-based overlap mutagenesis on pES to remove a SacI site at the 5′ end of vpr. Two flanking SacI sites were used for digestion and subcloning into pUC19 to generate pUC19-VpxMS. A StuI-XhoI fragment from vpx deletion mutant pTM-X101, described previously (34), was ligated into pUC19-VpxMS digested with StuI-XhoI. The entire SacI fragment was then religated back into pES.
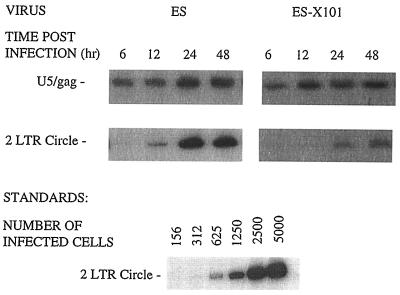
We used this virus in the PCR assay described above in order to determine its effects on nuclear import. A block in nuclear import is evident in cells infected with the ES-X101 virus, compared to cells infected with ES, as indicated by the decrease in accumulation of two-LTR circles (Fig. 4). Equivalent levels of cytoplasmic reverse transcription products were observed for ES and ES-X101, demonstrating that this mutation has not altered early infection events.
FIG. 4.
Deletion of the C-terminal tail of Vpx leads to a block in nuclear localization of viral DNA. Nondividing U937 cells were infected with ES or ES-X101, and total cellular DNA was isolated at the indicated times. Viral DNA was detected by using the same PCR primers as in Fig. 1, which differentiate between DNA synthesis and nuclear import events. Results from one of two experiments giving similar results are shown.
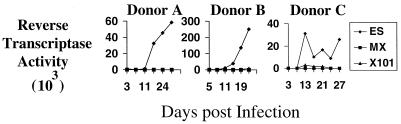
HIV-2 Vpx promotes productive infection of macrophages.
To ensure that the results obtained in the PCR assay are representative of events occurring during natural infection of nondividing cells, we examined the ability of viruses lacking Vpx or containing the truncated form of Vpx to elicit a spreading infection of monocyte-derived macrophages (MDMs). For infection of MDMs, blood monocytes were isolated from the peripheral blood of healthy blood donors by elutriation to >99% purity and allowed to differentiate in Iscove's medium containing 10% human serum and 500 U of macrophage colony-stimulating factor/ml. Infections were carried out by using 50 ng of p27 1 week after differentiation. Supernatants were collected every 3 to 4 days for 21 to 27 days and analyzed for exogenous reverse transcriptase (RT) activity (37).
Equivalent amounts of ES, MX, and ES-X101, based on titers in CCR5-expressing Magi cells, were used to infect macrophages (Fig. 5). In three independent experiments with macrophages from different donors, significantly higher levels of RT activity were generated from ES-infected macrophages, than from MX- or ES-X101-infected macrophages.
FIG. 5.
The C-terminal tail of Vpx is required for efficient HIV-2 replication in primary human macrophages. MDMs from three different donors were infected with equivalent amounts of infectious ES, MX, or ES-X101, as determined by assay on Magi-CCR5 cells. Virus expression was measured by RT activity measurements every 3 to 4 days.
Conclusions.
In this study, we demonstrated that HIV-2 Vpx is necessary for efficient nuclear import of viral DNA in nondividing cells. Our results indicate that the presence of intact MA and Vpr does not compensate for the lack of Vpx. This is in contrast to data reported for HIV-1, where MA, Vpr, and IN have redundant functions in terms of nuclear import of the preintegration complex (3, 12, 16). However, our findings are in agreement with the findings of Fletcher and colleagues using SIVSM PBj1.9 (9). One possible explanation for this discrepancy is that HIV-1 Vpr is the predominant mediator of nuclear import, and MA and IN are very weak karyophiles. Indeed, Fouchier and colleagues reported that the putative NLS within MA does not play a role in nuclear localization of the preintegration complex and that the 26KK→TT mutations introduced into this sequence affect posttranslational processing of Gag by the viral protease, rather than nuclear localization (10). They demonstrated that the replication kinetics of a 26KK→TT HIV-1 virus were equivalently decreased in both dividing and nondividing cells, in support of their hypothesis. However, these findings do not explain the ability of MA residues 25 to 33, when conjugated to bovine serum albumin (BSA), to direct its nuclear import (3). The 26KK→TT mutation within this sequence blocked nuclear import of BSA, further indicating that residues 25 to 33 comprise a functional NLS. Nor does it explain the observed interaction between MA and importin α, which is an NLS-dependent interaction (13). In contrast, Popov and colleagues suggest that there are multiple weak karyophilic components within the viral preintegration complex, including those in MA and IN (39). They suggest that the primary function of Vpr, or perhaps Vpx, is to stabilize the interaction between importin α and these relatively weak karyophiles. In support of this theory, Vpr binds to importin α at a different binding site than does MA, and the MA-importin α interaction is resistant to competition by the addition of exogenous NLS peptide in the presence of Vpr (39).
Some of the confusion surrounding the role of these various viral proteins in nuclear import may be attributable to subtle differences in experimental conditions. It has been reported that the role of integrase in nuclear import is apparent only when cells are infected at a relatively high MOI (12). At a high MOI, weaker karyophilic signals found in the preintegration complex, such as those found in MA and IN, could function in nuclear import to levels detectable by the PCR assay or macrophage infection experiments.
HIV-2 Vpx is primarily a nuclear protein, as indicated by GFP-Vpx fusion protein localization. However, a perinuclear distribution of the protein was observed in some cells, as reported by others (47). This appears to be in a discrete location outside of the nucleus, perhaps the endoplasmic reticulum or Golgi apparatus, rather than an association with the nuclear envelope as has been reported for HIV-1 Vpr (45). This suggests that Vpx may have another function in HIV-2-infected cells, separable from its role in nuclear localization. GFP-Vpx shifts to the plasma membrane when coexpressed with HIV-2 Gag, and it is efficiently packaged into HIV-2 virions when coexpressed with MX virus (data not shown). These findings support the notion that GFP-Vpx functions in a manner similar to that of Vpx alone. Furthermore, our localization results using GFP-Vpx are identical to those obtained by standard immunofluorescence techniques with Vpx-expressing cells (data not shown).
The current work sought to identify the domain of Vpx important for nuclear localization. Deletion of residues 20 to 40 within Vpx results in a predominantly nucleus-localized protein, similar to wild-type Vpx. This result differs from those obtained with HIV-1 Vpr where substitutions within the N-terminal amphipathic helix abrogated nuclear localization (5, 26, 48). It should not be surprising that the two accessory proteins do not use the same mechanism for nuclear import, since they also use distinct domains within Gag for virion incorporation (1, 24, 34, 47). Despite their homology, Vpr and Vpx have evolved distinct ways to interact with viral and cellular components.
Truncation of Vpx, at either residue 89 or 101, leads to a diffuse subcellular distribution, suggesting that a sequence important for nuclear localization has been perturbed. The C-terminal 10 residues of Vpx consist of a highly conserved motif of seven prolines followed by glycine, leucine, and valine (P7GLV). This motif is invariant in all of the Vpx proteins identified in HIV-2 and SIV isolates. Such strong sequence conservation suggests an important function for this domain, yet none has been identified. The proline-rich tail is dispensable for virion incorporation of Vpx, and it is dispensable for efficient SIV replication in dividing cells (35). Park and colleagues (35) suggested that this domain may be important for SIVmac Vpx protein stability, but we have previously demonstrated that this is not the case for HIV-2 Vpx (34). The data presented here suggest that this proline-rich domain may be important for the targeting of Vpx to the nucleus. However, it remains to be determined if residues 102 to 112 are sufficient to target a heterologous protein to the nucleus. A similar sequence is not found within Vpr, suggesting that it mediates nuclear import by a novel mechanism. At least two possibilities could explain how residues 102 to 112 function in nuclear localization. This portion of Vpx could contain a previously unidentified class of NLS, or the proline-rich domain could be important for protein-protein interactions with a second nucleus-targeted protein.
In terms of the first model, in which the C-terminal residues of Vpx contain a unique NLS, it is clear that the P7GLV motif does not resemble a canonical NLS. However, Shoya and colleagues identified two novel NLSs within the P phosphoprotein of Borna disease virus which are rich in prolines and lack basic amino acids (43). Mutational analyses of these sequences demonstrated that the prolines were required for nuclear targeting activity. It will be interesting to determine if mutation of the prolines of Vpx affects nuclear import.
With regard to the possibility that Vpx uses the C-terminal tail to interact with another protein and thereby piggyback into the nucleus, the proline-rich region of Vpx could be important for protein-protein interaction. SH3 domains bind proline-rich ligands through recognition of a PXXP motif (32). WW domains bind proline-rich ligands in WW-binding proteins by recognizing an expanding array of motifs. For example, the Yes-associated protein, YAP, has a WW domain that binds a PPXY motif (4), and the formin-binding protein, FBP11, recognizes a PPLP motif (2). It is interesting that the majority of WW-binding proteins identified to date are nuclear proteins (2). Proteins have been identified in yeast, Drosophila, mice, and humans, such as guanylate kinase MAGI-1, which contains both WW domains and NLSs (6, 28). It is tempting to speculate that proteins lacking a canonical NLS can use proline-rich domains for binding to WW domain proteins that can then escort them into the nucleus.
The current work has identified a primary function of HIV-2 Vpx and has mapped a domain critical for accomplishing this function. It will be interesting to determine if HIV-2 Vpx has a role in docking the viral preintegration complex to the nuclear pore, similar to what has been reported for Vpr in HIV-1. To address this, binding studies should determine if Vpx can interact with nucleoporins. It will also be interesting to elucidate the cellular pathway that Vpx exploits for its nuclear import. Binding studies between Vpx and importin α have not been done as of yet. Such studies may reveal that these two proteins can interact in an NLS-independent manner. Alternatively, Vpx may use a pathway distinct from the importin pathway, such as one involving the WW proteins or other proline-rich binding proteins. Such studies may prove useful in identifying alternative cellular pathways for nuclear import.
Acknowledgments
We thank Charles Rice for helpful discussions.
This work was supported by PHS grants AI36071 and AI34736 and training grant AI07172.
REFERENCES
- 1.Accola M A, Bukovsky A A, Jones M S, Göttlinger H G. A conserved dileucine-containing motif in p6gag governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIVmac and SIVagm. J Virol. 1999;73:9992–9999. doi: 10.1128/jvi.73.12.9992-9999.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford M T, Chan D C, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubei A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H I, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1993;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobrosotskaya I, Guy R K, James G L. MAGI-1, a membrane-associated guanylate kinase with a unique arrangement of protein-protein interaction domains. J Biol Chem. 1997;272:31589–31597. doi: 10.1074/jbc.272.50.31589. [DOI] [PubMed] [Google Scholar]
- 7.Emerman M. HIV-1, Vpr and the cell cycle. Curr Biol. 1996;6:1096–1103. doi: 10.1016/s0960-9822(02)00676-0. [DOI] [PubMed] [Google Scholar]
- 8.Farnet C N, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher T M, Brichacek B, Sharova N, Newman M A, Stivahtis G, Sharp P M, Emerman M, Hahn B H, Stevenson M. Nuclear import and cell cycle arrest of the HIV-1 vpr protein are encoded by two separate genes in HIV-2/SIVsm. EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier R A M, Meyer B E, Simon J H M, Fischer U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franchini G, Rusche J R, O'Keeffe T J, Wong-Staal F. The human immunodeficiency virus type 2 (HIV-2) contains a novel gene encoding a 16 kD protein associated with mature virions. AIDS Res Hum Retrovir. 1988;4:243–250. doi: 10.1089/aid.1988.4.243. [DOI] [PubMed] [Google Scholar]
- 12.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorlich D, Kostka H, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 15.Gorlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 16.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch V M, Sharkey M E, Brown C R, Brichacek B, Goldstein S, Wakefield J, Byrum R, Elkins W R, Hahn B H, Lifson J D, Stevenson M. Vpx is required for dissemination and pathogenesis of SIVsmPBj: evidence of macrophage-dependent viral amplification. Nat Med. 1998;4:1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Horton R, Spearman P, Ratner L. HIV-2 viral protein X association with the gag p27 capsid protein. Virology. 1994;199:453–457. doi: 10.1006/viro.1994.1144. [DOI] [PubMed] [Google Scholar]
- 20.Hu W, Vander Heyden N, Ratner L. Analysis of the function of viral protein X (VPX) of HIV-2. Virology. 1989;173:624–630. doi: 10.1016/0042-6822(89)90574-6. [DOI] [PubMed] [Google Scholar]
- 21.Hung C-S, Vander Heyden N, Ratner L. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J Virol. 1999;73:8216–8226. doi: 10.1128/jvi.73.10.8216-8226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamoto N, Tachibana T, Matsube M, Yoneda Y. A karyophilic protein forms a stable complex with cytoplasmic components prior to nuclear pore binding. J Biol Chem. 1995;270:8559–8565. doi: 10.1074/jbc.270.15.8559. [DOI] [PubMed] [Google Scholar]
- 23.Lewis P F, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y-L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triple repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y-L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahalingham S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner D B. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahalingham S, Khan S A, Jabbar M A, Monken C E, Collman R G, Srinivasan A. Identification of residues in the N-terminal acidic domain of HIV-1 vpr essential for virion incorporation. Virology. 1995;207:297–302. doi: 10.1006/viro.1995.1081. [DOI] [PubMed] [Google Scholar]
- 28.Maleszka R A, Lupas A, Hanes S D, Miklos G L G. The Dodo gene family encodes a novel protein involved in signal transduction and protein folding. Gene. 1997;203:89–93. doi: 10.1016/s0378-1119(97)00522-2. [DOI] [PubMed] [Google Scholar]
- 29.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 30.Moore M S, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosca P J, Dijkwel P A, Hamlin J L. The plant amino acid mimosine may inhibit initiation at origins of replication in Chinese hamster cells. Mol Cell Biol. 1992;12:4375–4383. doi: 10.1128/mcb.12.10.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musacchia A, Wilmanns M, Saraste M. Structure and function of the SH3 domain. Progress Biophys Mol Biol. 1994;61:283–297. doi: 10.1016/0079-6107(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 33.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–680. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 34.Pancio H A, Ratner L. Human immunodeficiency virus type 2 Vpx-Gag interaction. J Virol. 1998;72:5271–5275. doi: 10.1128/jvi.72.6.5271-5275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park I-W, Sodroski J. Amino acid sequence requirements for the incorporation of the vpx protein of simian immunodeficiency virus into virion particles. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10:506–510. [PubMed] [Google Scholar]
- 36.Pirounaki M, Vander Heyden N, Arens M, Ratner L. Rapid phenotypic drug susceptibility assay for HIV-1 with a CCR5 expressing indicator cell line. J Virol Methods. 2000;85:151–161. doi: 10.1016/s0166-0934(99)00163-9. [DOI] [PubMed] [Google Scholar]
- 37.Poiesz B J, Ruscetti F W, Gazdar A F, Burns P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popov S, Rexach M, Ratner L, Blobel G, Bukrinsky M. Viral protein R regulates docking of the HIV-1 preintegration complex to the nucleus pore complex. J Biol Chem. 1998;273:13347–13352. doi: 10.1074/jbc.273.21.13347. [DOI] [PubMed] [Google Scholar]
- 39.Popov S, Rexach M, Zybarth G, Reiling N, Lee M-A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 preintegration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radu A, Moore M S, Blobel G. The peptide repeat domain of Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 41.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 42.Sharp P M, Bailes E, Stevenson M, Emerman M, Hahn B. Gene acquisition in HIV and SIV. Nature. 1996;383:586–587. doi: 10.1038/383586a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoya Y, Kobayashi T, Koda T, Ikuta K, Kakinuma M, Kishi M. Two proline-rich nuclear localization signals in the amino- and carboxyl-terminal regions of the Borna disease virus phosphoprotein. J Virol. 1998;72:9755–9762. doi: 10.1128/jvi.72.12.9755-9762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tristem M, Marshall C, Karpas A, Hill F. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 1992;11:3405–3412. doi: 10.1002/j.1460-2075.1992.tb05419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vodicka M A, Koepp D M, Silver P A, Emerman M. HIV-1 vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 47.Wu X, Conway J A, Kim J, Kappes J C. Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein. J Virol. 1994;68:6161–6169. doi: 10.1128/jvi.68.10.6161-6169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao X-J, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y, Lu Y-L, Ratner L. Arginine residues in the C-terminus of HIV-1 vpr are important for nuclear localization and cell cycle arrest. Virology. 1998;242:414–424. doi: 10.1006/viro.1998.9028. [DOI] [PubMed] [Google Scholar]