Abstract
Carotenoids in plant foods provide health benefits by functioning as provitamin A. One of the vital provitamin A carotenoids, β-cryptoxanthin, is typically plentiful in citrus fruit. However, little is known about the genetic basis of β-cryptoxanthin accumulation in citrus. Here, we performed a widely targeted metabolomic analysis of 65 major carotenoids and carotenoid derivatives to characterize carotenoid accumulation in Citrus and determine the taxonomic profile of β-cryptoxanthin. We used data from 81 newly sequenced representative accessions and 69 previously sequenced Citrus cultivars to reveal the genetic basis of β-cryptoxanthin accumulation through a genome-wide association study. We identified a causal gene, CitCYP97B, which encodes a cytochrome P450 protein whose substrate and metabolic pathways in land plants were undetermined. We subsequently demonstrated that CitCYP97B functions as a novel monooxygenase that specifically hydroxylates the β-ring of β-cryptoxanthin in a heterologous expression system. In planta experiments provided further evidence that CitCYP97B negatively regulates β-cryptoxanthin content. Using the sequenced Citrus accessions, we found that two critical structural cis-element variations contribute to increased expression of CitCYP97B, thereby altering β-cryptoxanthin accumulation in fruit. Hybridization/introgression appear to have contributed to the prevalence of two cis-element variations in different Citrus types during citrus evolution. Overall, these findings extend our understanding of the regulation and diversity of carotenoid metabolism in fruit crops and provide a genetic target for production of β-cryptoxanthin-biofortified products.
Key words: carotenoids, β-cryptoxanthin, cytochrome P450, CYP97B, hydroxylation
This study reports the identification of the causal gene (CitCYP97B) responsible for differences in β-cryptoxanthin accumulation in citrus pulp. CitCYP97B is a novel β-cryptoxanthin monohydroxylase that converts β-cryptoxanthin to zeaxanthin. Introgression/hybridization during citrus evolution appear to have induced allelic variations in the CitCYP97B promoter, and two critical structural cis-element variations contribute to increased expression of CitCYP97B.
Introduction
Carotenoids, a group of lipophilic isoprenoids, are synthesized by photosynthetic organisms and several non-photosynthetic microorganisms (Zheng et al., 2020; Sun et al., 2022). In plants, carotenoids play an essential role in photoprotection and contribute to pigmentation (Stanley et al., 2020). Carotenoids contribute to human health as dietary antioxidants and provitamin A (Krinsky and Johnson, 2005). Insufficient quantities of provitamin A carotenoids in plant foods have stimulated interest in developing bioengineering technologies that will be useful for generation of biofortified crops.
There are three major types of provitamin A carotenoids: α-carotene, β-carotene, and β-cryptoxanthin. β-cryptoxanthin, an intermediate in the conversion of β-carotene to zeaxanthin, is associated with a reduced risk of particular cancers and many chronic diseases (Tanaka et al., 2000; Kohno et al., 2001; Sugiura et al., 2011; Lim and Wang, 2020). β-cryptoxanthin is rare in nature but rich in a few horticultural crops, such as citrus, papaya, chili pepper, and persimmon (Burri et al., 2016; Ma et al., 2020). Citrus is one of the most important fruit trees worldwide, producing 158.5 million metric tons of fruit in 2021 (FAO, https://www.fao.org/faostat/en/#data/). Citrus fruits are a major dietary source of provitamin A because they contain abundant β-cryptoxanthin (O’Connell et al., 2007; Burri et al., 2011; Turner et al., 2013; Burri, 2015). Citrus includes seven common taxa: three basic species (mandarin, pummelo, and citron) and additional interspecific hybrids (sour orange, sweet orange, grapefruit, and lemon) (Wu et al., 2018). Most mandarin fruits are rich in carotenoids, particularly in β-cryptoxanthin. Pummelo and sweet orange accumulate much lower levels of β-cryptoxanthin (Matsumoto et al., 2007). The different levels of β-cryptoxanthin in different types of citrus fruit provide an ideal system for investigating β-cryptoxanthin metabolism and crop biofortification. However, the genetic basis for differences in β-cryptoxanthin content among different citrus taxa remains unclear.
Dissection of the carotenoid biosynthetic pathway provides the molecular basis for metabolic engineering of biofortified crops. Previous studies have reported two types of hydroxylases that catalyze the conversion of carotenes to lutein, β-cryptoxanthin, and zeaxanthin in plants, including non-heme di-iron enzymes (BCH1 and BCH2) and heme-containing cytochrome P450 (CYP) enzymes (CYP97A3, CYP97B, and CYP97C1) (Ma et al., 2016; Zheng et al., 2020). BCH1, BCH2, and CYP97A hydroxylate the β-rings of α-carotene and β-carotene, and CYP97C hydroxylates the ε-ring of α-carotene (Kim et al., 2009; Niu et al., 2020). Citrus carotene hydroxylases, including CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C, have been characterized by Ma et al. (2016). However, the role of CYP97B in xanthophyll biosynthesis has not been confirmed. Overexpression of AtCYP97B in Arabidopsis thaliana led to regulation of carotenoid accumulation. However, no accumulation of hydroxylated carotenes was detected in a quadruple Arabidopsis mutant (bch1bch2cyp97c1cyp97a3), and the function of CYP97B remains controversial, with an undetermined substrate and metabolic pathway in plants (Kim et al., 2009, 2010; Fiore et al., 2012; Ma et al., 2016). In recent decades, metabolic engineering of carotenoid biosynthesis has been used to produce carotenoid-biofortified crops to combat vitamin A deficiency. Overexpression of one or more biosynthetic enzymes enhanced carotenoid flux and content of provitamin A carotenoids in crops such as cassava (Welsch et al., 2010), wheat (Wang et al., 2014), maize (Zhu et al., 2008), and the famous example of Golden Rice (Paine et al., 2005). Silencing of BCH genes increased β-carotene content in potato tubers and biofortified orange fruit (Diretto et al., 2007; Pons et al., 2014). Manipulation of BCH led to an increase in β-carotene content and a decrease in β-cryptoxanthin content in kiwifruit (Xia et al., 2022), and provitamin A equivalents were dramatically increased in β-cryptoxanthin-biofortified maize (Liu et al., 2012; Heying et al., 2014).
Although great efforts have been made to investigate the mechanism of carotenoid metabolism and improve fruit quality (Sun et al., 2018; Wurtzel, 2019), gaps in our understanding of the carotenoid biosynthetic pathway and the mechanisms that regulate carotenoid accumulation have limited our ability to use genetic and metabolic engineering approaches to biofortify crops. We need more information on the genetic basis of β-cryptoxanthin accumulation to successfully biofortify crops with provitamin A. In the present study, we aimed to clarify the evolutionary mechanism and genetic basis of β-cryptoxanthin accumulation in citrus fruit and provide necessary information for enhancement of fruit quality and biofortification of crops with increased levels of provitamin A carotenoids. We demonstrated the characteristics of β-cryptoxanthin accumulation with a widely targeted metabolomic analysis of carotenoids in citrus. Using a genome-wide association study (GWAS), we identified CitCYP97B as the carotenogenic gene responsible for differences in β-cryptoxanthin levels among different types of citrus fruit. These findings refine our understanding of β-cryptoxanthin biosynthesis and provide a potential genetic target for improving the nutritional value of citrus fruit and perhaps other crops.
Results
Genetic diversity and carotenoid accumulation diversity of 150 citrus germplasms
To determine the genetic mechanisms that control carotenoid accumulation in citrus fruit, we constructed a variant map using 150 citrus accessions (Supplemental Figure 1A) that included mandarin, pummelo, citron, and interspecific hybrids (sweet orange, sour orange, grapefruit, and lemon). Sequences from 69 accessions were obtained from previous studies, and 81 accessions were newly sequenced for this study using whole-genome re-sequencing (36× genome coverage) (Supplemental Table 1). We identified 5 157 365 high-quality biallelic single-nucleotide polymorphisms (SNPs) relative to the reference genome (Citrus grandis (L.) Osbeck cv. ‘Wanbaiyou’ v.1.0), with ∼15 SNPs per kilobase, 760 916 insertions/deletions (indels), and 34 795 structural variants (SVs). Maximum-likelihood phylogenetic analysis showed that the 150 accessions could be clustered into seven groups (Figure 1A). This interpretation was supported by principal component analysis (PCA) and population structure analysis (Supplemental Figure 1).
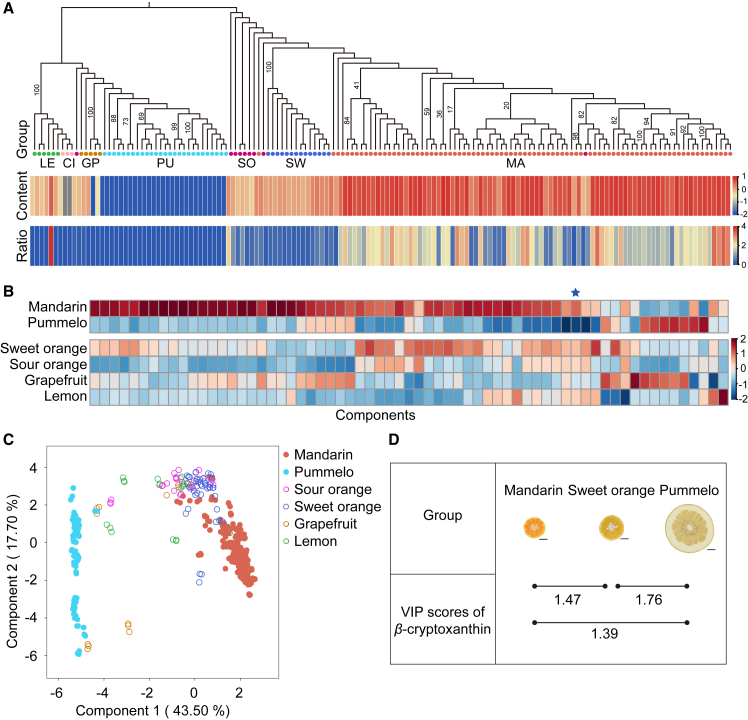
Figure 1.
Carotenoid profiles of pulp from different varieties of citrus.
(A) Maximum-likelihood phylogenetic tree based on the SNP dataset (top) and heatmaps of normalized β-cryptoxanthin abundance and β-cryptoxanthin-to-violaxanthin ratios (bottom). The colored circles represent citrus categories defined in previous studies (Wang et al., 2018; Wu et al., 2018), including mandarin (MA), pummelo (PU), citron (CI), sweet orange (SW), sour orange (SO), grapefruit (GP), and lemon (LE).
(B) Heatmap of carotenoid abundance in citrus pulp. Each column indicates one component. The star indicates β-cryptoxanthin.
(C) Sparse PLS-DA of 148 accessions based on carotenoid abundance.
(D) VIP scores of β-cryptoxanthin from mandarin, pummelo, and sweet orange based on an orthogonal PLS-DA. Scale bar, 3 cm.
We characterized carotenoid variation in citrus fruit by performing a widely targeted metabolomic analysis of 65 major carotenoids and carotenoid derivatives (Supplemental Table 2) using ultra-high-performance liquid chromatography–high-resolution tandem mass spectrometry and liquid chromatography–triple-quadrupole mass spectrometry. A heatmap analysis revealed a rich diversity of 64 carotenoids in mandarin and a reduced diversity of 52 carotenoids in pummelo. We found intermediate levels of carotenoid diversity in the citrus hybrids sweet orange, sour orange, lemon, and grapefruit (Figure 1B). Sparse partial-least-squares–discriminant analysis (PLS-DA) of carotenoid concentrations in 148 accessions (excluding two citron accessions that do not develop pulp) distinguished mandarin and pummelo and placed the interspecific hybrids in intermediate positions (Figure 1C). Similar data were obtained from PCA and PLS-DA (Supplemental Figure 2). These findings provide evidence for taxon-specific accumulation of carotenoids in citrus fruit.
Variable importance in projection (VIP) score is an indicator for evaluating the intensity and explanatory power of differentially accumulated metabolites. We assessed the differentially accumulated carotenoids (VIP ≥ 1) in mandarin, pummelo, and sweet orange using orthogonal PLS-DA. β-cryptoxanthin, one of the important provitamin A carotenoids, exhibited a taxon-specific accumulation pattern with high VIP scores (Figure 1A; Supplemental Figure 3). Correlations between β-cryptoxanthin and other carotenoids revealed a strong correlation between β-cryptoxanthin and violaxanthin (Pearson correlation coefficient r = 0.87). Mandarin and sweet orange are reported to accumulate predominantly β-cryptoxanthin and violaxanthin, respectively, and these two carotenoids are critical determinants for citrus classification (Fanciullino et al., 2006). We therefore quantified the β-cryptoxanthin-to-violaxanthin ratio in citrus pulp and used the proportion of β-cryptoxanthin as a measure of carotenoid characteristics in different citrus varieties. Interestingly, although most mandarin varieties accumulated high proportions of β-cryptoxanthin, we found a low proportion of β-cryptoxanthin in a subset of mandarin varieties (Figure 1A; Supplemental Table 3). These results demonstrate the diversity of carotenoid accumulation in different citrus varieties and indicate that β-cryptoxanthin is a candidate carotenoid biomarker that is useful for distinguishing different citrus varieties.
Identification of the causal gene for diversity in β-cryptoxanthin content of citrus pulp
To assess the performance of a genome-wide association study (GWAS) in this population, we performed a GWAS for the content of β-citraurinene, a C30 apocarotenoid that is significantly associated with the red color of citrus peel and natural variation in the CCD4b promoter (Zheng et al., 2019, 2021). An association signal peak was detected on chromosome 8. The lead SNP was located in the promoter of the CCD4b gene (Supplemental Figure 4). Subsequently, we performed GWASs for the proportion and abundance of β-cryptoxanthin using the SNP and SV datasets. Suggestive thresholds were set at 9.69 × 10−9 for SNPs and 1.44 × 10−6 for SVs using a Bonferroni correction. We detected an association signal peak on chromosome 6 in both the SNP-GWAS and the SV-GWAS for β-cryptoxanthin accumulation, suggesting that a gene encoding a master regulator of β-cryptoxanthin level is located in this region (Figure 2).
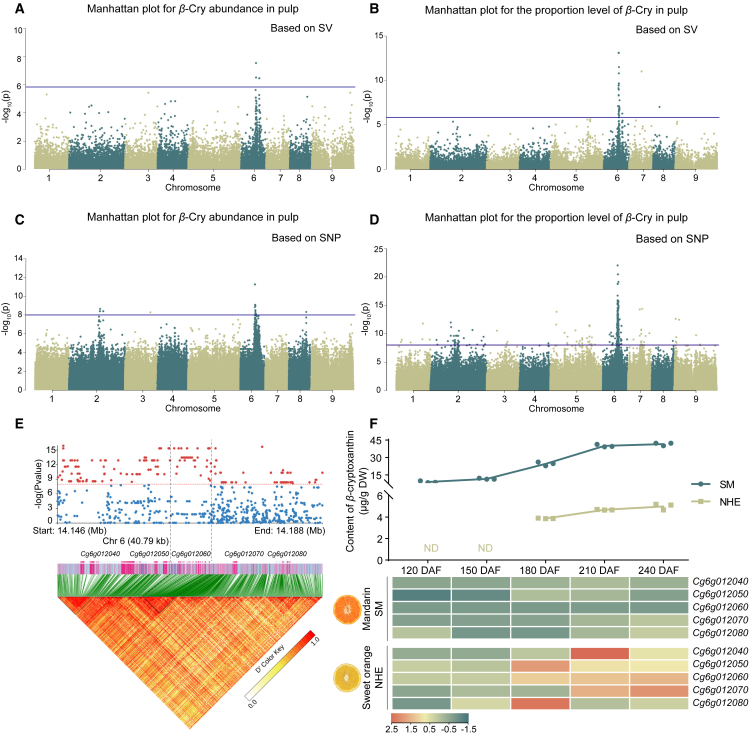
Figure 2.
Identification of the causal gene Cg6g012060.
(A–D) SV-GWASs for (A) the abundance of β-cryptoxanthin and (B) the proportion of β-cryptoxanthin. SNP-GWASs for (C) the abundance of β-cryptoxanthin and (D) the proportion of β-cryptoxanthin. β-Cry, β-cryptoxanthin.
(E) Enlarged SNP-GWAS and LD block of CRC6.
(F) β-cryptoxanthin content and expression levels of candidate genes at five stages of fruit development. DAF, days after flowering; SM, Satsuma mandarin; NHE, Newhall navel orange.
The estimated linkage disequilibrium (LD) decay rate was approximately 50 kb for the whole population (Supplemental Figure 5). We analyzed 100-kb intervals upstream and downstream of associated loci that exceeded the suggestive threshold value. We focused on a 40.69-kb intersection interval that contained five annotated protein-coding genes and named this locus of β-cryptoxanthin on chromosome 6 (CRC6) (Figure 2E; Supplemental Table 4). The lead SNP (chr6:14146788, P = 1.2408 × 10−16) was located 2.49 kb from Cg6g012040. Several SNPs with the second-lowest P value (P = 4.3156 × 10−16) were located within the gene body and promoter region of Cg6g012060. We compared the expression patterns of these five genes in β-cryptoxanthin-rich Satsuma mandarin and β-cryptoxanthin-poor Newhall navel orange. Cg6g012060, annotated as encoding a CYP97B3 protein, was expressed at higher levels and had a broader expression pattern during fruit ripening in Newhall navel orange than in Satsuma mandarin (Figure 2F) and was thus considered the candidate gene for CRC6.
CitCYP97B catalyzes the conversion of β-cryptoxanthin to zeaxanthin
A phylogenetic analysis of amino acid sequences confirmed that Cg6g012060 encodes a CYP protein from the CYP97B subfamily (Figure 3A) that was named CitCYP97B by Ma et al. (2016). Subcellular localization experiments showed that CitCYP97B co-localized with a plastid marker protein in Nicotiana benthamiana (Figure 3B), and carotenoids are known to be biosynthesized and stored in plastids (Sun et al., 2018). The subcellular location of plastid-localized CitCYP97B thus differed from that of other endoplasmic reticulum membrane-localized CYP proteins (Mizutani and Ohta, 2010), consistent with participation of CitCYP97B in carotenoid biosynthesis.
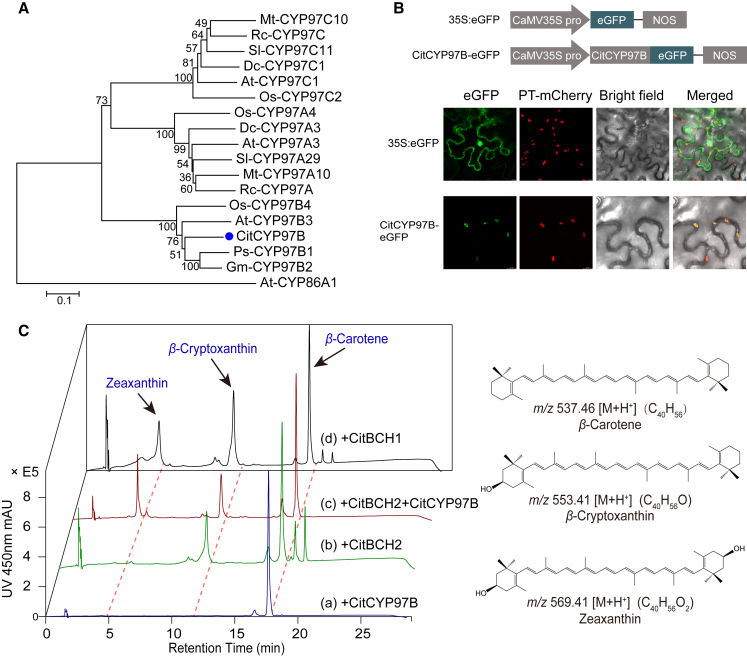
Figure 3.
Subcellular localization and functional complementation assays with CitCYP97B.
(A) Phylogeny of CYP97-family amino acid sequences. Abbreviations and amino acid sequence accession numbers used to construct the neighbor-joining tree are listed in Supplemental Table 5.
(B) Subcellular localization of a CitCYP97B-eGFP fusion protein in N. benthamiana leaves. PT-mCherry is a plastid marker protein.
(C) Analysis of carotenoids in β-carotene-accumulating E. coli strains. E. coli BL21 (DE3) cells harboring pACCARΔ16crtX were co-transformed with (a) pRSFDuet-CitCYP97B, (b) pRSFDuet-CitBCH2, (c) pRSFDuet-CitBCH2-CitCYP97B, or (d) pRSFDuet-CitBCH1 (left). Mass spectrometry data and structures for the carotenoids are shown at right.
Given the undetermined substrate and metabolic pathway of CYP97B in land plants, we performed functional complementation assays to investigate the substrate specificity of CitCYP97B. We used Escherichia coli as a heterologous host, as it is reported to be an effective platform for research on the functions of carotenoid hydroxylases (Quinlan et al., 2007; Ma et al., 2016). No detectable hydroxylated product was observed when we expressed CitCYP97B in β-carotene-accumulating E. coli BL21 (DE3) cells (Figure 3C). However, co-expression of CitCYP97B with CitBCH2, a hydroxylase that converts β-carotene into β-cryptoxanthin (Zhang et al., 2023), led to production of zeaxanthin in β-carotene-accumulating E. coli BL21 (DE3) cells (Figure 3C). These results provide evidence that CitCYP97B participates in the β branch of carotenoid biosynthesis by serving as a novel monohydroxylase that hydroxylates β-cryptoxanthin to yield zeaxanthin, distinct from CYP97A and CYP97C.
CitCYP97B negatively regulates β-cryptoxanthin accumulation in citrus fruit
To determine whether CitCYP97B contributes to carotenoid metabolism in vivo, we knocked down CitCYP97B in orange peel using RNA interference. Compared with control regions, regions in which CitCYP97B expression was transiently suppressed had a higher color contribution index (CCI) value (Figure 4A). Relative expression levels of PSY1, BCH2, CYP97A3, CYP97C1, and CCD4b were significantly upregulated in the CitCYP97B-suppressed regions, and relative expression levels of BCH1, LCYB1, LCYB2, and LCYE were significantly downregulated (Figure 4B and 4C). We found a slight increase in β-cryptoxanthin content (14.11 ± 0.20 μg/g DW) of 11.3% compared with the control. We also detected increases in total carotenoid content and in phytoene, phytofluene, and β-citraurinene levels (Figure 4D and 4E). These findings indicated that a moderate reduction in CitCYP97B expression slightly influenced the carotenoid content of citrus fruit.
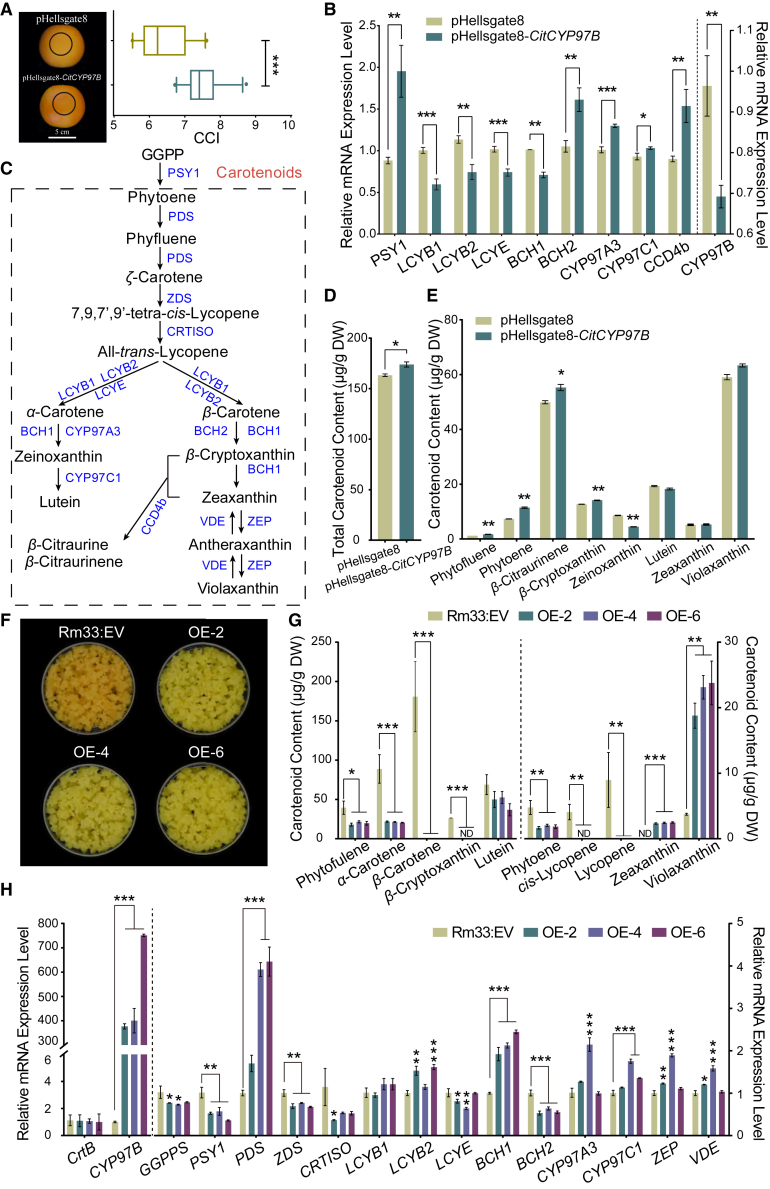
Figure 4.
Knockdown and overexpression of CitCYP97B in citrus.
(A) Phenotypes and CCI values of fruit with RNAi-suppressed CitCYP97B. Different sides of fruits were transiently transfected with an RNAi empty vector (pHellsgate8) and pHellsgate8-CitCYP97B.
(B) Expression levels of carotenogenic genes in fruit transiently transfected with pHellsgate8-CitCYP97B and the interference vector pHellsgate8.
(C) Carotenoid biosynthetic pathway.
(D and E) Total carotenoid content (D) and carotenoid content of pHellsgate8 and pHellsgate8-CitCYP97B infiltration sites (E).
(F) Phenotypes of transgenic lines of Rm33 callus.
(G and H) Carotenoid content (G) and relative expression levels of carotenogenic genes (H) in transgenic lines of Rm33 callus. Data are presented as mean values ± standard error in triplicate. Asterisks indicate statistically significant differences relative to the control line determined using Student’s t-test; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. The data on the left side of the dashed line correspond to the left y axis, and the data on the right side correspond to the right y axis.
Citrus callus is an effective in planta system for investigating the activities of enzymes related to the carotenoid/apocarotenoid pathway in perennial woody trees with prolonged juvenile periods (Cao et al., 2012; Zheng et al., 2021). We performed a transformation experiment to overexpress CitCYP97B in carotene-rich citrus callus Rm33, a cell-engineering model derived from callus Rm that overexpresses a bacterial phytoene synthase gene (CrtB) (Cao et al., 2012). We selected three transgenic lines (OE-2, OE-4, and OE-6) with stable phenotypes for carotenoid analysis. Compared with orange callus containing the empty vector (Rm33:EV), the CitCYP97B-overexpressing transgenic lines developed a bright-yellow phenotype (Figure 4F). Levels of carotenoids were significantly lower in the transgenic lines, with the exceptions of zeaxanthin and violaxanthin (Figure 4G). β-cryptoxanthin accumulated to a concentration of 26.33 ± 0.23 μg/g DW in Rm33 but was not detectable in the CitCYP97B-overexpressing lines. These data provide more evidence that CitCYP97B catalyzes the conversion of β-cryptoxanthin to zeaxanthin. To further explore the molecular mechanism of carotenoid regulation, we analyzed expression levels of endogenous carotenogenic genes using RT–qPCR. We found that expression levels of carotenogenic genes that act upstream of lycopene biosynthesis were downregulated, except for PDS (Figure 4C and 4H). LCYB2, BCH1, ZEP, and VDE, which participate in the β-branch biosynthetic pathway, were upregulated in the transgenic lines. However, expression of LCYE was downregulated, indicating that overexpression of CitCYP97B could regulate the flux of carotenoids into the β branch. These results demonstrate that carotenoid accumulation in citrus is regulated by the collective action of carotenogenic genes, with a pivotal role for CitCYP97B in the hydroxylation of β-cryptoxanthin.
Variations in the CitCYP97B promoter influence transcription
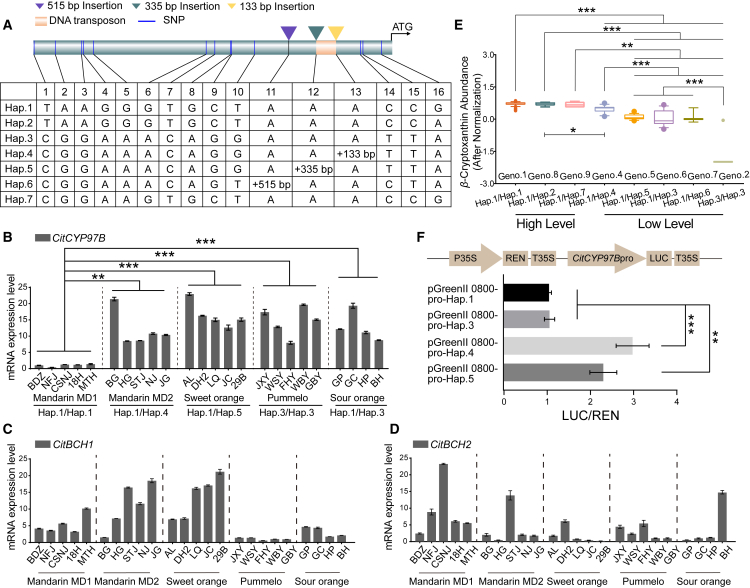
The genome-wide association results showed that significant variants were enriched in the promoter region of the differentially expressed gene CitCYP97B (Figure 2E and 2F). To investigate the contribution of natural variations to CitCYP97B transcription, we analyzed the promoter haplotypes of different Citrus types. We found seven major haplotypes of CitCYP97B promoters (≥3 accessions per haplotype), containing 13 SNPs and 3 SVs (Figure 5A; Supplemental Table 6). Satsuma mandarin showed low CitCYP97B expression (Figure 2F) and was homozygous for Hap.1. By contrast, Newhall navel orange showed high CitCYP97B expression and was heterozygous for Hap.1 and Hap.5. PCR amplification and molecular markers confirmed the taxon-specific distribution of SVs (Supplemental Figure 6). Cultivated mandarin MD2 from the south Nanling Mountains harbored Hap.1 and Hap.4, characterized by a 133-bp miniature inverted-repeat transposable element. Sweet orange harbored Hap.1 and Hap.5, characterized by a 335-bp insertion (Supplemental Figures 6 and 7). Citron was homozygous for Hap.6, characterized by a 515-bp insertion.
Figure 5.
Variations in the CitCYP97B promoter.
(A) Polymorphisms in the CitCYP97B promoter in different haplotypes (Hap.1–Hap.7).
(B–D) Expression levels of CitCYP97B(B), CitBCH1(C), and CitBCH2(D) in mature citrus varieties with different haplotypes. Relative expression was quantified by RT–qPCR. Data are presented as mean values ± standard error in triplicate.
(E) Abundance of β-cryptoxanthin in mature citrus pulp from different genotypes.
(F) Activities of the CitCYP97B promoter from different haplotypes are represented as LUC/REN ratios. Asterisks indicate statistically significant differences determined with Student’s t-test. ∗∗P < 0.01; ∗∗∗P < 0.001.
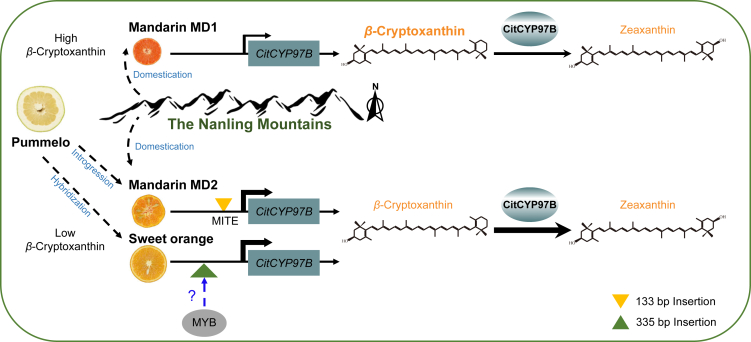
Figure 7.
Proposed model for diversity in β-cryptoxanthin accumulation influenced by CitCYP97B during citrus evolution.
Mandarin MD1 is the cultivated mandarin group from the north Nanling Mountains, and mandarin MD2 is the cultivated mandarin group from the south Nanling Mountains. Low expression of CitCYP97B in mandarin MD1 suppresses hydroxylation of β-cryptoxanthin, thus promoting β-cryptoxanthin accumulation in citrus pulp. The introgression and hybridization of mandarin and pummelo induced a 133-bp insertion and a 335-bp insertion in the promoter region of CitCYP97B in mandarin MD2 and sweet orange, respectively, which enhance expression of CitCYP97B and promote hydroxylation of β-cryptoxanthin. The blue dashed arrow indicates potential regulation by transcription factors in sweet orange.
We randomly selected citrus varieties harboring different haplotypes and quantified relative expression levels of CitCYP97B in mature fruits. We found that β-cryptoxanthin-rich mandarin was homozygous for Hap.1 and had the lowest CitCYP97B expression of the studied species and hybrids (Figure 5B). There was a negative correlation between CitCYP97B expression level and β-cryptoxanthin level in citrus, and the expression pattern of CitCYP97B differed from those of the other two carotene hydroxylase genes (Figure 5B–5E; Supplemental Figure 8). Promoter activity analyses in N. benthamiana provided further evidence that promoters containing SVs (Hap.4 and Hap.5) had higher activities than Hap.1, which was consistent with the higher rates of transcription on alleles of CitCYP97B from Hap.4 and Hap.5 (Figure 5B and 5F). These findings suggest that SVs in the CitCYP97B promoter lead to increases in promoter activity and thus enhance CitCYP97B transcription.
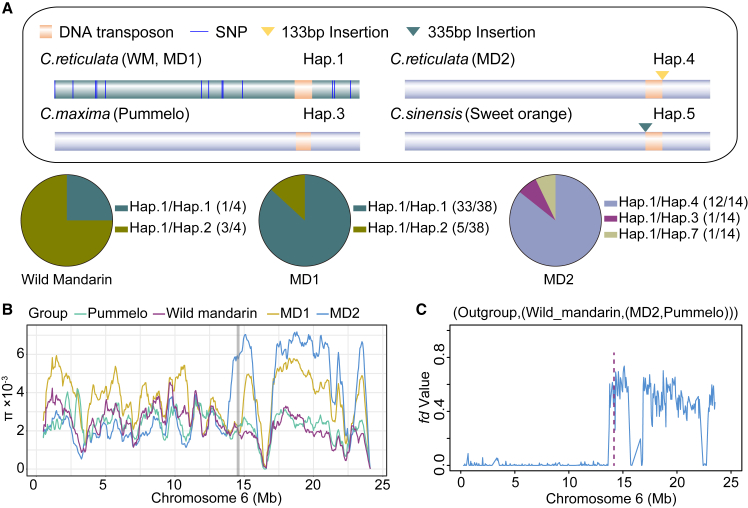
Hybridization has influenced polymorphisms in the CitCYP97B promoter
To investigate allelic variations in the CitCYP97B promoter and their effects on citrus, we analyzed the distribution of distinct promoter alleles among citrus varieties. Wild mandarin ‘Mangshan’ underwent two independent domestication events that yielded two groups of cultivated mandarin—MD1 and MD2—around the Nanling Mountains (Wang et al., 2018). We noticed a taxon-specific distribution of allelic variations of the CitCYP97B promoter. Most MD1 varieties were homozygous for Hap.1 (86.84%), whereas most MD2 varieties were heterozygous for Hap.1 and Hap.4 (85.71%). All pummelo varieties were homozygous for Hap.3. Sweet oranges were heterozygous for Hap.1 and Hap.5 (Figure 6A). We estimated the genetic diversity of citrus groups by calculating the nucleotide diversity and fixation index. The nucleotide diversity (π) of the CitCYP97B locus was significantly higher in cultivated mandarin MD2 than in other groups (Figure 6B). Population differentiation (FST) at the CitCYP97B locus (Supplemental Figure 9) was high between wild mandarin and pummelo (FST = 0.6598) and between wild mandarin and cultivated mandarin MD2 (FST = 0.3242). These data indicate that divergence may contribute to shaping β-cryptoxanthin diversity.
Figure 6.
Genetic population analysis of citrus populations.
(A) Schematic of CitCYP97B promoter polymorphisms.
(B) Nucleotide diversity (π) of citrus populations. The gray line indicates the window containing CitCYP97B.
(C)fd statistics and topology for chromosome 6 from wild mandarin, cultivated mandarin MD2, and pummelo. The pink line indicates the window containing CitCYP97B.
We calculated fd statistics in sliding windows to explore the origin of genetic variation in CitCYP97B, and we found a clear introgression signal between 13.7 and 15.5 Mb on chromosome 6 (Figure 6C). This interval contains the CitCYP97B locus (fd 0.64). The introgression signal in MD2 displayed a similar pattern in the divergence signal from nucleotide diversity among wild mandarin, pummelo, and cultivated mandarin MD2 (Figure 6B and 6C). Topology weighting confirmed the introgression from pummelo to cultivated mandarin MD2 (Supplemental Figure 9). These data indicate that CitCYP97B was introgressed and associated with the domestication of mandarin. Introgression and interspecific hybridization of citrus have thus contributed to the polymorphisms of CitCYP97B, accompanied by two independent structural-variation events that induced novel SVs specific to cultivated mandarin MD2 (Hap.4) and sweet orange (Hap.5).
Discussion
Carotenoids are secondary metabolites important for plant and human health (Krinsky and Johnson, 2005). Elucidating the carotenoid biosynthetic pathway is a prerequisite for metabolic engineering of biofortified crops. The diverse accumulation of carotenoids in citrus fruit provides an ideal system for dissecting the genetic basis of carotenoid metabolism. In this study, we combined GWASs and functional analyses to identify a causal gene, CitCYP97B, responsible for differences in β-cryptoxanthin accumulation among citrus fruits. Our results provide evidence that CitCYP97B encodes a monohydroxylase that catalyzes the conversion of β-cryptoxanthin to zeaxanthin and negatively regulates β-cryptoxanthin content in citrus. Allelic variations in the CitCYP97B promoter contribute to increased CitCYP97B expression and thus modulate β-cryptoxanthin accumulation in different types of citrus fruit. These findings extend our current understanding of plant carotenoid biosynthetic pathways and provide a potential genetic target for creation of carotenoid-biofortified crops.
Functional differentiation of the monooxygenase CYP97B
Previous studies have demonstrated that CYP97B, the oldest subfamily of the CYP97 family with β-ring-specific activity, is conserved from green algae to vascular plants and probably originated before the formation of extant algal groups (Nelson et al., 2008; Cui et al., 2013). Although a red algae homolog of CYP97B has dihydroxylation activity and catalyzes the conversion of β-carotene to zeaxanthin (Yang et al., 2014), the role of CYP97B is controversial and requires clarification in land plants (Kim et al., 2009, 2010; Fiore et al., 2012; Ma et al., 2016; Niu et al., 2020). We identified CitCYP97B by performing a GWAS on β-cryptoxanthin accumulation and demonstrated that CitCYP97B hydroxylates β-cryptoxanthin to zeaxanthin using a heterologous expression system in E. coli (Figure 3C). Functional complementation assays in E. coli have been reported to be a suboptimal system for determining the substrate specificity of CYP enzymes. Our results did illustrate the β-ring hydroxylation activity of CitCYP97B in E. coli. However, the enzyme activity may have been limited by culture conditions, insufficient expression level of endogenous reductase, and codon usage preference, consistent with the interpretation of Quinlan et al. (2007), and our assays could therefore be improved in the future. Moreover, CYP97B is encoded by a single-copy gene in green algae and higher plants but encoded by two genes with different functions in diatoms. The hydroxylated product of β-cryptoxanthin is zeaxanthin, an oxygenated carotenoid that participates in the xanthophyll cycle (Esteban et al., 2009). Functional differentiation of CYP97B may have occurred during algal differentiation, possibly owing to different photosynthetic requirements and the need for photoprotection. Our data provide evidence of how CYP97B catalyzes carotenoid biosynthesis in land plants and suggest that the duplication and functional divergence of CYP97B have contributed to evolution (Cui et al., 2019).
Cooperation of hydroxylases in carotenoid β-branch metabolism
Carotenoid hydroxylation is an essential branch point for the biosynthesis of oxygenated carotenoids. Compared with the characterized carotene hydroxylases BCH1, BCH2, CYP97A, and CYP97C (Ma et al., 2016; Zhang et al., 2023), CitCYP97B appears to be a special monohydroxylase in carotenoid metabolism, as supported by the following lines of evidence. First, CitCYP97B participates in the biosynthesis of β-branch carotenoids. By contrast, CYP97A and CYP97C synergistically catalyze the conversion of α-carotene to lutein. Second, the substrate of CitCYP97B is β-cryptoxanthin. By contrast, the non-heme di-iron carotene hydroxylases BCH1 and BCH2 use β-carotene as a substrate. In addition, the activity of heme-containing CitCYP97B requires molecular oxygen and redox protein partners, a feature of CYP proteins (Mizutani and Ohta, 2010).
Transient suppression of CitCYP97B yielded a slight increase in the content of β-cryptoxanthin in orange but failed to reduce the content of downstream β,β-xanthophylls. The slight change in carotenoids might be attributed to the moderate suppression of CitCYP97B and the differential regulation of carotenogenic gene expression, such as the increased expression of PSY1, which could increase carotenoid flux (Figure 4). Previous research demonstrated that silencing of Csβ-CHX (BCH1) in sweet orange led to an increase in β-carotene content and a decrease in total carotenoid content. However, β, β-xanthophylls remained the primary carotenoid in the pulp of transgenic fruit (Pons et al., 2014). Our results showed that CitBCH1 expression is lower in mandarin than in sweet orange but is still relatively high (Supplemental Figure 8), consistent with the results of previous studies (Kato et al., 2016; Zhang et al., 2023). We suggest that sufficient carotenoid flux and the expression of CitBCH1 could sustain the synthesis of β-cryptoxanthin, as supported by the results of Kato et al. (2016), who proposed that high expression of upstream carotenogenic genes (e.g., PSY1, PDS, ZDS, and LCYb) supports sufficient carotenoid flux and that BCH1 predominantly catalyzes the conversion of β-carotene to β-cryptoxanthin. We found that the proportion of β-cryptoxanthin was higher in mandarin MD1 than in mandarin MD2 and sweet orange, but the expression level of CitCYP97B was higher in mandarin MD2 and sweet orange (Figure 5). Thus, we suggest that both CitBCH1 and CitCYP97B contribute to greater β-cryptoxanthin accumulation in mandarin relative to other citrus taxa. High levels of β-cryptoxanthin accumulation should require sufficient upstream carotenoid flux and limited downstream hydroxylation, consistent with the model proposed by Kato et al. (2016) in which β-cryptoxanthin accumulation is attributable to an imbalance in the expression of upstream and downstream carotenogenic genes. Low expression of CitCYP97B also hinders the hydroxylation of β-cryptoxanthin and could cooperate with CitBCH1 to promote β-cryptoxanthin accumulation in mandarin MD1. The delicate regulation of CitCYP97B expression has great potential for manipulation of β-cryptoxanthin accumulation, expanding the synthetic biology toolkit for carotenoid engineering and providing a genetic target for creation of β-cryptoxanthin-biofortified crops.
Evolution of diversity in Citrus carotenoid accumulation
Introgression has been a vital feature in the evolution of perennial crops, possibly owing to incomplete reproductive isolation or longer generation times compared with annual crops (Gaut et al., 2015). In our study, ABBA–BABA statistics and topology analysis confirmed that introgression occurred from pummelo to the cultivated mandarin MD2 (Figure 6C) and aligned with the introgressed region that is specific to MD2 mandarin, which is widespread in the south Nanling Mountains (Wang et al., 2018). Sweet orange is regarded as a hybrid variety of citrus that was derived from mandarin and pummelo (Wu et al., 2018), and it accumulates intermediate levels of carotenoids (Figure 1). Thus, interspecific hybridization appears to have played an essential role in shaping the diversity of carotenoid content in citrus. Given that the CitCYP97B gene is located within the region of the introgression signal (Figure 6), we suggest that taxon-specific variations in the CitCYP97B promoter gave rise two independent structural variations, enhancing the transcription of CitCYP97B and consequently the capacity to metabolize β-cryptoxanthin in fruits. The abundance of β-cryptoxanthin in wild mandarin and the cultivated mandarin MD1 may contribute to their high antioxidant content (Bunea et al., 2014) and their capacity to tolerate stress.
On the basis of our study, we propose a specific model to illustrate the genetic mechanism by which CitCYP97B mediates β-cryptoxanthin diversity in citrus fruits (Figure 7). During domestication of the cultivated mandarin MD2 population in the Nanling Mountains, introgression of pummelo induced the polymorphisms of CitCYP97B, accompanied by a novel SV in the CitCYP97B promoter. In addition, hybridization between mandarin and pummelo gave rise to another novel SV in the CitCYP97B promoter of sweet orange. Both SVs promoted the increased expression of CitCYP97B and enhanced the hydroxylation of β-cryptoxanthin, thus regulating the differential accumulation of β-cryptoxanthin and shaping the diversity of carotenoid accumulation in different types of citrus.
Natural variations in promoters play critical roles in shaping agronomic traits by regulating gene expression (Springer et al., 2019). The taxon-specific SV in the CitCYP97B promoter of sweet orange (Hap.5) was predicted to contain light-responsive elements and an MYB-binding site (Supplemental Figure 7). Previous studies have reported that CYP97B expression responds to light intensity and light quality and that high-intensity light induces increases in zeaxanthin content of red algae (Cui et al., 2019; Xie et al., 2020). However, citrus pulp is not a light-exposed tissue, and there is insufficient evidence to demonstrate that light signals directly regulate the expression of carotenogenic genes in citrus pulp. We speculate that transcription factors, like MYBs, may play critical roles in upregulating CitCYP97B in sweet orange, a possibility that requires further investigation. The promoter activity of CitCYP97B that originated from pummelo (i.e., Hap.3) was not significantly higher than that from wild mandarin (Hap.1) in N. benthamiana leaves. However, the expression levels of CitCYP97B were higher in pummelo and sour orange than in cultivated mandarin harboring homozygous Hap.1 (Figure 5B and 5F), a result that may possibly be explained by different regulatory systems in N. benthamiana leaves and citrus fruit. Natural variations in the CitCYP97B promoter (Hap.1 and Hap.3) include variations in cis-elements containing the W-box motif (Supplemental Figure 7), which may influence the binding affinity of WRKY transcription factors that have been reported to regulate carotenoid metabolism (Yuan et al., 2022).
In conclusion, our work provides insight into the diversity of carotenoid accumulation in citrus pulp. A novel monohydroxylase, CitCYP97B, which catalyzes the conversion of β-cryptoxanthin to zeaxanthin, was identified in citrus using GWAS. SVs in the promoter of CitCYP97B may contribute to its increased expression and thus enhance the metabolism of β-cryptoxanthin. This work refines our current understanding of plant carotenoid biosynthetic pathways and provides new insights for research on the carotenoid biofortification of citrus and perhaps other crops.
Methods
Plant materials
The 150 citrus accessions used in this study were collected from the National Citrus Breeding Center at Huazhong Agricultural University, the National Citrus Germplasm Repository (Chongqing), Hunan Province, Guangdong Province, Guangxi Province, and Jiangxi Province in 2019 and 2020. Information about the citrus accessions, including common names and classifications, is listed in Supplemental Table 1. The fruits used for metabolite analysis were of average size, healthy, and ripe. Fruits from each variety were randomly separated into three replicates. Fruits from Satsuma mandarin (Citrus unshiu Marc.) and Newhall navel orange (Citrus sinensis Osbeck) were collected at five developmental stages from the National Citrus Breeding Center at Huazhong Agricultural University. As previously described, citrus callus Rm33 was derived from Marsh grapefruit (Citrus paradisi Macf.) and harbors a highly expressed bacterial phytoene synthase gene (CrtB). Citrus callus Rm33 was subcultured at 25°C on solid MT medium (Cao et al., 2012). All samples used for metabolite analysis were separated, frozen in liquid nitrogen, and stored at −80°C.
Carotenoid identification and analysis
Citrus fruit and callus were lyophilized and crushed into a powder before extraction. To quantify carotenoid levels in citrus fruit, 0.5 g of dry citrus pulp powder was extracted and analyzed using a high-throughput identification and quantification procedure with a liquid chromatography–tandem mass spectrometry system (Thermo Fisher QExactive Plus and TSQ Quantis triple-quadrupole mass spectrometer) as described previously (Zhu et al., 2022). TraceFinder 4.0 (Thermo Fisher Scientific, San Jose, CA, USA) was used for relative quantification of carotenoids and carotenoid derivatives based on integrated peak areas. Heatmap construction and PCA of metabolomic data with log10 transformation and autoscaling were performed with MetaboAnalyst 5.0 (Pang et al., 2022). Quantification of carotenoids from citrus fruit and callus was performed using a high-performance liquid chromatography system (Waters, Framingham, MA, USA). Detection and quantitative analysis were performed as described in a previous report (Zheng et al., 2019). Each sample was analyzed in triplicate. Statistically significant differences were determined using GraphPad Prism v.8.0 (Student’s t-test).
Color measurement
A color analyzer (KONICA MINOLTA CM-5, Japan) was used to measure color parameters (L∗, a∗, and b∗) at five points around the injection site. Twenty biological replicates were analyzed. The CCI value (= 1000 × a/(L × b)) was calculated as described previously (Luo et al., 2015).
Resequencing and variant calling
In this study, 81 citrus accessions were newly sequenced. We also used data from 69 other citrus accessions that were sequenced in previous studies (Wu et al., 2014, 2018; Wang et al., 2018, 2021; Liang et al., 2020), as indicated in Supplemental Table 1. Genomic DNA from each accession was used to construct a paired-end sequencing library with a 150-bp read length and sequenced on the Illumina platform. FastQC (v.0.11.5) was used to check sequence quality after removal of adapter and low-quality sequences from the raw reads (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped to the high-quality reference genome (C. grandis (L.) Osbeck cv. ‘Wanbaiyou’ v.1.0 from NCBI) using the Burrows–Wheeler Aligner (BWA-MEM, v.0.7.17) and sorted using SAMtools (v.1.7) (Li and Durbin, 2009; Li et al., 2009). Duplicate reads were marked using GATK v.4.2.3 (McKenna et al., 2010). For SNPs and indels, the GATK HaplotypeCaller was used to call variants and produce GVCF files, followed by genotyping and merging with the GATK Best-Practices pipeline (https://software.broadinstitute.org/gatk/best-practices/). Parameters for variant filtration were QD > 2.0, FS < 60.0, MQ > 20.0, MQRankSum > 12.5, and ReadPosRankSum > 8.0. A total of 5 157 365 biallelic SNPs were retained with a minor allele frequency >0.05 and a missing rate <0.1. SNPs and indels were annotated using SnpEff software (Cingolani et al., 2012). Four software packages were used to call SVs: Delly (Rausch et al., 2012), Manta (Chen et al., 2016), Smoove (https://hpc.nih.gov/apps/smoove.html), and GRIDSS (Cameron et al., 2021). The SV results were merged using Jasmine (Kirsche et al., 2023) and genotyped with Paragraph (Chen et al., 2019).
Population structure and genome-wide association study
Four-fold synonymous third-codon transversion sites were extracted from the SNP data with Reseqtools (https://github.com/BGI-shenzhen/Reseqtools) and used to construct a maximum-likelihood phylogenetic tree in RAxML (v.8.2.12) with 1000 bootstraps (Kozlov et al., 2019). The population genetic structure was inferred using ADMIXTURE v.1.3.0 (K values from 2 to 10) with 5 157 365 biallelic SNPs (Alexander et al., 2009). PCA with the top 20 principal components was estimated using PLINK v.1.90 (Purcell et al., 2007) and GCTA v.1.93.2 (Yang et al., 2011).
We performed a GWAS for carotenoid abundance using the efficient mixed-model association eXpedited (EMMAX) algorithm with default parameters (Kang et al., 2010). Manhattan and quantile–quantile plots were created using the R package qqman. LD was calculated using PopLDdecay (Zhang et al., 2019), and LD and haplotype blocks were analyzed using LDBlockShow (Dong et al., 2021).
RNA extraction and quantitative RT–PCR analysis
To investigate the expression patterns of candidate genes during fruit development, we collected fruits of Satsuma mandarin and Newhall navel orange at five developmental stages. Total RNA was extracted from citrus pulp of Satsuma mandarin, Newhall navel orange, and other mature varieties as described previously (Zhang et al., 2020). cDNA synthesis was performed using HiScript II RT SuperMix for qPCR (+gDNA wiper, Vazyme). RT–qPCR primers used in this study are listed in Supplemental Table 7. The relative expression levels of candidate genes were quantified using qPCR SYBR Green Master Mix (YEASEN, Shanghai) and a Roche LightCycler 480 system (Roche, https://www.roche.com). The procedures and calculations used for RT–qPCR analysis have been reported previously (Zhang et al., 2020). Each sample was analyzed independently with three replicates.
Gene cloning and sequence alignment
The CitCYP97B (Cg6g012060) reference sequence was obtained from the Citrus Pan-genome to Breeding Database (http://citrus.hzau.edu.cn/). The coding sequence of CitCYP97B was amplified from cDNA of ‘Nanfeng’ mandarin pulp (Citrus reticulata Blanco) using gene-specific primers (Supplemental Table 7). Sequences of CYP97B homologs from other plants were obtained from NCBI (https://www.ncbi.nlm.nih.gov/protein/) and are listed in Supplemental Table 5. Multiple sequence alignments and neighbor-joining phylogenetic trees were constructed using MEGA 6 with At-CYP86A1 (NP_200694.1).
Subcellular localization in N. benthamiana
To determine the subcellular localization of the CitCYP97B protein, we fused the coding sequence of CitCYP97B in frame with the coding sequence of an enhanced green fluorescent protein (eGFP) in pRI121 and transformed the resulting plasmid into Agrobacterium strain GV3101. The plastid marker PT-mCherry was obtained from a previous study in which a plasmid was constructed by amplifying the small subunit of tobacco rubisco and ligating it into an expression vector encoding mCherry (Dabney-Smith et al., 1999; Nelson et al., 2007). Strains containing pRI121-CitCYP97B and PT-mCherry were co-infiltrated into N. benthamiana leaves. We detected fluorescence from the CitCYP97B-eGFP fusion protein and the plastid marker protein using a confocal laser scanning microscope (Leica Microsystems, Germany) 48 h after infiltration.
Heterologous expression of CitCYP97B in E. coli
The coding sequence of CitCYP97B without the transit peptide (residues 46–581) was cloned into the E. coli expression vector pRSFDuet-1 (Novagen). We co-transformed pACCARΔ16crtX with pRSFDuet-CitCYP97B and pRSFDuet-CitBCH2-CitCYP97B into E. coli BL21 (DE3). E. coli BL21 (DE3) transformed with pACCARΔ16crtX and pRSFDuet-CitBCH1 and E. coli BL21 (DE3) transformed with pACCARΔ16crtX and pRSFDuet-CitBCH2 were used as positive and negative controls, respectively. Colonies were cultured in lysogeny broth medium at 37°C in the dark until the optical density at 600 nm (OD600) reached 0.6. Protein synthesis was induced by addition of isopropyl β-D-thiogalactoside at a final concentration of 0.5 mM, and cultures were maintained at 28°C with shaking at 160 rpm for 3 d. Cells were collected and stored at −80°C for further analysis. All experiments were conducted with three replicates.
RNA interference in citrus fruit
Gateway recombination (Invitrogen, Thermo Fisher Scientific, USA) was used to insert a gene-specific region of CitCYP97B into pHellsgate8-HG to yield plasmids for RNA interference (RNAi) assays. Transient expression was conducted in ‘Lane Late’ navel orange after the color transition. Agrobacterium strain GV3101-pSoup-p19 transformed with pHellsgate8-CitCYP97B was grown overnight at 28°C in liquid lysogeny broth medium until OD600 reached 0.6. As described previously, cells were collected and resuspended in an infiltration medium (Gong et al., 2021). One site on the fruit was infiltrated with a strain harboring the pHellsgate8-HG empty vector control. Another site was injected with a strain harboring pHellsgate8-CitCYP97B. Tissue at the infiltration site was collected after 7 d.
Citrus callus transformation
The coding sequence of CitCYP97B was cloned into the MT-GFP overexpression vector (Wang et al., 2022) containing a CaMV35S promoter and eGFP. Citrus callus Rm33 was transformed with this plasmid using Agrobacterium tumefaciens strain EHA105 as described previously (Cao et al., 2012). Citrus callus was subcultured on solid MT medium with antibiotics (cefotaxime sodium and herbicides) every 20 d at 25°C in the dark. Transgenic lines were identified using GFP fluorescence, expression levels of CrtB and CitCYP97B, and antibiotic selection. Callus used for subsequent studies was collected and frozen in liquid nitrogen.
Promoter sequence analysis and dual-luciferase transactivation assays
cis-acting regulatory elements were predicted using the online PlantCARE tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Different haplotypes of the CitCYP97B promoter (2 kb upstream of the translational start codon) were amplified and cloned into the pGreenII 0800-LUC vector containing the LUC and Renilla luciferase (REN) reporter genes. All vectors were introduced into Agrobacterium strain GV3101 containing plasmid pSoup-p19. N. benthamiana leaves were infiltrated with these Agrobacterium strains. After 3 d, the LUC and REN activities were quantified using the NIGHTSHADE imaging system (LC 985, Berthold) and a Dual-Luciferase Reporter Assay Kit (Vazyme). LUC activity was normalized to REN activity. The primers used for vector construction are listed in Supplemental Table 7.
Genetic diversity and introgression analysis
The nucleotide diversity (π) for each group (wild mandarin, pummelo, and cultivated mandarin MD1 and MD2) and the fixation index (FST) between populations were calculated using VCFtools (0.1.16) with a 500-kb sliding window size and a 50-kb step size (Danecek et al., 2011). To investigate introgression events and detect differences in allele sharing between citrus populations, we performed the ABBA–BABA test (D statistic) and calculated fd statistics according to the regular pipeline (Martin et al., 2015). We added three varieties of Atalantia buxifolia (Amo, ARO, HKC) that had been sequenced in a previous study (Wang et al., 2017) as outgroups. SNP variant calling was performed as described above. Introgressed genomic fragments were evaluated using fd statistics in a sliding window of 50 kb using the ABBABABAwindows.py script (https://github.com/simonhmartin/genomics_general). Wild mandarin was defined as Population A, cultivated mandarin MD2 as Population B, and pummelo as Population C. A. buxifolia was set as the outgroup to estimate the ancestral state of alleles. Topology weighting was computed using the SNP dataset in a sliding window of 20 kb using Twisst to support introgression analysis (Martin and Van Belleghem, 2017).
Data availability and statistical analysis
Data supporting the findings of this work are available in the paper and its supplemental information. Whole-genome resequencing data are accessible through NCBI. The accession numbers are listed in Supplemental Table 1. The accession numbers for CYP97 family homologs from other plants are listed in Supplemental Table 5. The sequence data used in this study can be found at NCBI GenBank (https://www.ncbi.nlm.nih.gov/) with the following accession numbers: CitCYP97B, LC143647.1; CitBCH1, AF296158.2; and CitBCH2, KY612512.1. Unless otherwise noted, data are expressed as mean values ± standard error based on triplicate measurements. Statistically significant differences were calculated using GraphPad Prism v.8.0.
Funding
This research was supported by the National Key Research and Development Program of China (2022YFF1003100), the National Natural Science Foundation of China (31930095), and Modern Agro-industry Technology Research System (CARS-26).
Author contributions
X.D. conceived the project and supervised the experiments. Y.Z. and X.D. designed the experiments. Y.Z. performed the experiments and analyzed the data with contributions from J.J., D.F., S.Z., Z.W., S.L., and Z.X.; they helped with sample collection and metabolite analysis. Y.Z. and X.D. wrote the manuscript with contributions from N.W., Q.S., J.Y., and L.C. All authors read and approved the manuscript.
Acknowledgments
We thank Prof. Norihiko Misawa for providing the pACCARΔ16crtX plasmid and Prof. Pengwei Wang for providing the pRI121, PT-mCherry, pHellsgate-HG, and MT-GFP plasmids. We thank Prof. Mei Liang, Dr. Xiongjie Zheng, Dr. Yin Zhang, and Dr. Kaijie Zhu for critically reading the manuscript and providing suggestions. We thank Prof. Robert M. Larkin for suggestions on writing and language embellishment. No conflict of interest is declared.
Published: February 19, 2024
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunea A., Socaciu C., Pintea A. Xanthophyll esters in fruits and vegetables. Not Bot Horti Agrobo. 2014;42:310–324. [Google Scholar]
- Burri B.J. Beta-cryptoxanthin as a source of vitamin A. J. Sci. Food Agric. 2015;95:1786–1794. doi: 10.1002/jsfa.6942. [DOI] [PubMed] [Google Scholar]
- Burri B.J., Chang J.S.T., Neidlinger T.R. β-Cryptoxanthin- and α-carotene-rich foods have greater apparent bioavailability than β-carotene-rich foods in Western diets. Br. J. Nutr. 2011;105:212–219. doi: 10.1017/S0007114510003260. [DOI] [PubMed] [Google Scholar]
- Burri B.J., La Frano M.R., Zhu C. Absorption, metabolism, and functions of β-cryptoxanthin. Nutr. Rev. 2016;74:69–82. doi: 10.1093/nutrit/nuv064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron D.L., Baber J., Shale C., Valle-Inclan J.E., Besselink N., van Hoeck A., Janssen R., Cuppen E., Priestley P., Papenfuss A.T. GRIDSS2: comprehensive characterisation of somatic structural variation using single breakend variants and structural variant phasing. Genome Biol. 2021;22:202. doi: 10.1186/s13059-021-02423-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Zhang J., Xu J., Ye J., Yun Z., Xu Q., Xu J., Deng X. Comprehending crystalline β-carotene accumulation by comparing engineered cell models and the natural carotenoid-rich system of citrus. J. Exp. Bot. 2012;63:4403–4417. doi: 10.1093/jxb/ers115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- Chen S., Krusche P., Dolzhenko E., Sherman R.M., Petrovski R., Schlesinger F., Kirsche M., Bentley D.R., Schatz M.C., Sedlazeck F.J., Eberle M.A. Paragraph: a graph-based structural variant genotyper for short-read sequence data. Genome Biol. 2019;20:291. doi: 10.1186/s13059-019-1909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Yu X., Wang Y., Cui Y., Li X., Liu Z., Qin S. Evolutionary origins, molecular cloning and expression of carotenoid hydroxylases in eukaryotic photosynthetic algae. BMC Genom. 2013;14:457. doi: 10.1186/1471-2164-14-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Ma H., Cui Y., Zhu X., Qin S., Li R. Cloning, identification and functional characterization of two cytochrome P450 carotenoids hydroxylases from the diatom Phaeodactylum tricornutum. J. Biosci. Bioeng. 2019;128:755–765. doi: 10.1016/j.jbiosc.2019.06.008. [DOI] [PubMed] [Google Scholar]
- Dabney-Smith C., van Den Wijngaard P.W., Treece Y., Vredenberg W.J., Bruce B.D. The C Terminus of a Chloroplast Precursor Modulates Its Interaction with the Translocation Apparatus and PIRAC∗. J. Biol. Chem. 1999;274:32351–32359. doi: 10.1074/jbc.274.45.32351. [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diretto G., Welsch R., Tavazza R., Mourgues F., Pizzichini D., Beyer P., Giuliano G. Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol. 2007;7:11. doi: 10.1186/1471-2229-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S.-S., He W.-M., Ji J.-J., Zhang C., Guo Y., Yang T.-L. LDBlockShow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 2021;22:bbaa227. doi: 10.1093/bib/bbaa227. [DOI] [PubMed] [Google Scholar]
- Esteban R., Martínez B., Fernández-Marín B., María Becerril J., García-Plazaola J.I. Carotenoid composition in Rhodophyta: insights into xanthophyll regulation in Corallina elongata. Eur. J. Phycol. 2009;44:221–230. [Google Scholar]
- Fanciullino A.-L., Dhuique-Mayer C., Luro F., Casanova J., Morillon R., Ollitrault P. Carotenoid diversity in cultivated citrus is highly influenced by genetic factors. J. Agric. Food Chem. 2006;54:4397–4406. doi: 10.1021/jf0526644. [DOI] [PubMed] [Google Scholar]
- Fiore A., Dall’Osto L., Cazzaniga S., Diretto G., Giuliano G., Bassi R. A quadruple mutant of Arabidopsis reveals a β-carotene hydroxylation activity for LUT1/CYP97C1 and a regulatory role of xanthophylls on determination of the PSI/PSII ratio. BMC Plant Biol. 2012;12:50. doi: 10.1186/1471-2229-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B.S., Díez C.M., Morrell P.L. Genomics and the contrasting dynamics of annual and perennial domestication. Trends Genet. 2015;31:709–719. doi: 10.1016/j.tig.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Gong J., Zeng Y., Meng Q., Guan Y., Li C., Yang H., Zhang Y., Ampomah-Dwamena C., Liu P., Chen C., et al. Red light-induced kumquat fruit coloration is attributable to increased carotenoid metabolism regulated by FcrNAC22. J. Exp. Bot. 2021;72:6274–6290. doi: 10.1093/jxb/erab283. [DOI] [PubMed] [Google Scholar]
- Heying E.K., Tanumihardjo J.P., Vasic V., Cook M., Palacios-Rojas N., Tanumihardjo S.A. Biofortified orange maize enhances β-cryptoxanthin concentrations in egg yolks of laying hens better than tangerine peel fortificant. J. Agric. Food Chem. 2014;62:11892–11900. doi: 10.1021/jf5037195. [DOI] [PubMed] [Google Scholar]
- Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.-Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M. Mechanism of ß-cryptoxanthin accumulation in citrus fruits. Acta Hortic. 2016;1:1–10. doi: 10.17660/ActaHortic.2016.1135.1. [DOI] [Google Scholar]
- Kim J.-E., Cheng K.M., Craft N.E., Hamberger B., Douglas C.J. Over-expression of Arabidopsis thaliana carotenoid hydroxylases individually and in combination with a beta-carotene ketolase provides insight into in vivo functions. Phytochemistry. 2010;71:168–178. doi: 10.1016/j.phytochem.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Kim J., Smith J.J., Tian L., Dellapenna D. The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol. 2009;50:463–479. doi: 10.1093/pcp/pcp005. [DOI] [PubMed] [Google Scholar]
- Kirsche M., Prabhu G., Sherman R., Ni B., Battle A., Aganezov S., Schatz M.C. Jasmine and Iris: population-scale structural variant comparison and analysis. Nat. Methods. 2023;20:408–417. doi: 10.1038/s41592-022-01753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H., Taima M., Sumida T., Azuma Y., Ogawa H., Tanaka T. Inhibitory effect of mandarin juice rich in beta-cryptoxanthin and hesperidin on 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced pulmonary tumorigenesis in mice. Cancer Lett. 2001;174:141–150. doi: 10.1016/s0304-3835(01)00713-3. [DOI] [PubMed] [Google Scholar]
- Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinsky N.I., Johnson E.J. Carotenoid actions and their relation to health and disease. Mol. Aspect. Med. 2005;26:459–516. doi: 10.1016/j.mam.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Cao Z., Zhu A., Liu Y., Tao M., Yang H., Xu Q., Wang S., Liu J., Li Y., et al. Evolution of self-compatibility by a mutant Sm-RNase in citrus. Nat. Plants. 2020;6:131–142. doi: 10.1038/s41477-020-0597-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.Y., Wang X.-D. Mechanistic understanding of β-cryptoxanthin and lycopene in cancer prevention in animal models. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865 doi: 10.1016/j.bbalip.2020.158652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-Q., Davis C.R., Schmaelzle S.T., Rocheford T., Cook M.E., Tanumihardjo S.A. β-Cryptoxanthin biofortified maize (Zea mays) increases β-cryptoxanthin concentration and enhances the color of chicken egg yolk. Poultry Sci. 2012;91:432–438. doi: 10.3382/ps.2011-01719. [DOI] [PubMed] [Google Scholar]
- Luo T., Xu K., Luo Y., Chen J., Sheng L., Wang J., Han J., Zeng Y., Xu J., Chen J., et al. Distinct carotenoid and flavonoid accumulation in a spontaneous mutant of Ponkan (Citrus reticulata Blanco) results in yellowish fruit and enhanced postharvest resistance. J. Agric. Food Chem. 2015;63:8601–8614. doi: 10.1021/acs.jafc.5b02807. [DOI] [PubMed] [Google Scholar]
- Ma G., Zhang L., Yungyuen W., Tsukamoto I., Iijima N., Oikawa M., Yamawaki K., Yahata M., Kato M. Expression and functional analysis of citrus carotene hydroxylases: unravelling the xanthophyll biosynthesis in citrus fruits. BMC Plant Biol. 2016;16:148. doi: 10.1186/s12870-016-0840-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Zhang L., Sugiura M., Kato M. The Genus Citrus. 2020. Citrus and health; pp. 495–511. [Google Scholar]
- Martin S.H., Van Belleghem S.M. Exploring evolutionary relationships across the genome using topology weighting. Genetics. 2017;206:429–438. doi: 10.1534/genetics.116.194720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.H., Davey J.W., Jiggins C.D. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 2015;32:244–257. doi: 10.1093/molbev/msu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H., Ikoma Y., Kato M., Kuniga T., Nakajima N., Yoshida T. Quantification of carotenoids in citrus fruit by LC-MS and comparison of patterns of seasonal changes for carotenoids among citrus varieties. J. Agric. Food Chem. 2007;55:2356–2368. doi: 10.1021/jf062629c. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Ohta D. Diversification of P450 genes during land plant Evolution. Annu. Rev. Plant Biol. 2010;61:291–315. doi: 10.1146/annurev-arplant-042809-112305. [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Nelson D.R., Ming R., Alam M., Schuler M.A. Comparison of cytochrome P450 genes from six plant genomes. Trop. Plant Biol. 2008;1:216–235. [Google Scholar]
- Niu G., Guo Q., Wang J., Zhao S., He Y., Liu L. Structural basis for plant lutein biosynthesis from α-carotene. Proc. Natl. Acad. Sci. USA. 2020;117:14150–14157. doi: 10.1073/pnas.2001806117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell O.F., Ryan L., O’Brien N.M. Xanthophyll carotenoids are more bioaccessible from fruits than dark green vegetables. Nutr. Res. 2007;27:258–264. [Google Scholar]
- Paine J.A., Shipton C.A., Chaggar S., Howells R.M., Kennedy M.J., Vernon G., Wright S.Y., Hinchliffe E., Adams J.L., Silverstone A.L., Drake R. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- Pang Z., Zhou G., Ewald J., Chang L., Hacariz O., Basu N., Xia J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022;17:1735–1761. doi: 10.1038/s41596-022-00710-w. [DOI] [PubMed] [Google Scholar]
- Pons E., Alquézar B., Rodríguez A., Martorell P., Genovés S., Ramón D., Rodrigo M.J., Zacarías L., Peña L. Metabolic engineering of β-carotene in orange fruit increases its in vivo antioxidant properties. Plant Biotechnol. J. 2014;12:17–27. doi: 10.1111/pbi.12112. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan R.F., Jaradat T.T., Wurtzel E.T. Escherichia coli as a platform for functional expression of plant P450 carotene hydroxylases. Arch. Biochem. Biophys. 2007;458:146–157. doi: 10.1016/j.abb.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T., Zichner T., Schlattl A., Stütz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N., de León N., Grotewold E. Challenges of translating gene regulatory information into agronomic improvements. Trends Plant Sci. 2019;24:1075–1082. doi: 10.1016/j.tplants.2019.07.004. [DOI] [PubMed] [Google Scholar]
- Stanley L.E., Ding B., Sun W., Mou F., Hill C., Chen S., Yuan Y.-W. A tetratricopeptide repeat protein regulates carotenoid biosynthesis and chromoplast development in monkeyflowers (Mimulus) Plant Cell. 2020;32:1536–1555. doi: 10.1105/tpc.19.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M., Nakamura M., Ogawa K., Ikoma Y., Ando F., Shimokata H., Yano M. Dietary patterns of antioxidant vitamin and carotenoid intake associated with bone mineral density: findings from post-menopausal Japanese female subjects. Osteoporos. Int. 2011;22:143–152. doi: 10.1007/s00198-010-1239-9. [DOI] [PubMed] [Google Scholar]
- Sun T., Yuan H., Cao H., Yazdani M., Tadmor Y., Li L. Carotenoid metabolism in plants: The role of plastids. Mol. Plant. 2018;11:58–74. doi: 10.1016/j.molp.2017.09.010. [DOI] [PubMed] [Google Scholar]
- Sun T., Rao S., Zhou X., Li L. Plant carotenoids: recent advances and future perspectives. Mol. Hortic. 2022;2:3. doi: 10.1186/s43897-022-00023-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kohno H., Murakami M., Shimada R., Kagami S., Sumida T., Azuma Y., Ogawa H. Suppression of azoxymethane-induced colon carcinogenesis in male F344 rats by mandarin juices rich in beta-cryptoxanthin and hesperidin. Int. J. Cancer. 2000;88:146–150. [PubMed] [Google Scholar]
- Turner T., Burri B.J., Jamil K.M., Jamil M. The effects of daily consumption of β-cryptoxanthin-rich tangerines and β-carotene-rich sweet potatoes on vitamin A and carotenoid concentrations in plasma and breast milk of Bangladeshi women with low vitamin A status in a randomized controlled trial. Am. J. Clin. Nutr. 2013;98:1200–1208. doi: 10.3945/ajcn.113.058180. [DOI] [PubMed] [Google Scholar]
- Wang C., Zeng J., Li Y., Hu W., Chen L., Miao Y., Deng P., Yuan C., Ma C., Chen X., et al. Enrichment of provitamin A content in wheat (Triticum aestivum L.) by introduction of the bacterial carotenoid biosynthetic genes CrtB and CrtI. J. Exp. Bot. 2014;65:2545–2556. doi: 10.1093/jxb/eru138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu Y., Zhang S., Cao L., Huang Y., Cheng J., Wu G., Tian S., Chen C., Liu Y., et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 2017;49:765–772. doi: 10.1038/ng.3839. [DOI] [PubMed] [Google Scholar]
- Wang L., He F., Huang Y., He J., Yang S., Zeng J., Deng C., Jiang X., Fang Y., Wen S., et al. Genome of wild mandarin and domestication history of mandarin. Mol. Plant. 2018;11:1024–1037. doi: 10.1016/j.molp.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Wang L., Huang Y., Liu Z., He J., Jiang X., He F., Lu Z., Yang S., Chen P., Yu H., et al. Somatic variations led to the selection of acidic and acidless orange cultivars. Nat. Plants. 2021;7:954–965. doi: 10.1038/s41477-021-00941-x. [DOI] [PubMed] [Google Scholar]
- Wang K., Gao E., Liu D., Wu X., Wang P. The ER network, peroxisomes and actin cytoskeleton exhibit dramatic alterations during somatic embryogenesis of cultured citrus cells. Plant Cell Tissue Organ Cult. 2022;148:259–270. [Google Scholar]
- Welsch R., Arango J., Bär C., Salazar B., Al-Babili S., Beltrán J., Chavarriaga P., Ceballos H., Tohme J., Beyer P. Provitamin A accumulation in Cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell. 2010;22:3348–3356. doi: 10.1105/tpc.110.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.A., Prochnik S., Jenkins J., Salse J., Hellsten U., Murat F., Perrier X., Ruiz M., Scalabrin S., Terol J., et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014;32:656–662. doi: 10.1038/nbt.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.A., Terol J., Ibanez V., López-García A., Pérez-Román E., Borredá C., Domingo C., Tadeo F.R., Carbonell-Caballero J., Alonso R., et al. Genomics of the origin and evolution of Citrus. Nature. 2018;554:311–316. doi: 10.1038/nature25447. [DOI] [PubMed] [Google Scholar]
- Wurtzel E.T. Changing form and function through carotenoids and synthetic biology. Plant Physiol. 2019;179:830–843. doi: 10.1104/pp.18.01122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Zhou Y., Lin Z., Guo Y., Liu X., Wang T., Wang J., Deng H., Lin L., Deng Q., et al. Characterization and functional validation of β-carotene hydroxylase AcBCH genes in Actinidia chinensis. Hortic. Res. 2022;9 doi: 10.1093/hr/uhac063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu X., Wang L., He L., Wang G. High light intensity increases the concentrations of β-carotene and zeaxanthin in marine red macroalgae. Algal Res. 2020;47 [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.-E., Huang X.-Q., Hang Y., Deng Y.-Y., Lu Q.-Q., Lu S. The P450-type carotene hydroxylase PuCHY1 from Porphyra suggests the evolution of carotenoid metabolism in red algae: P450-type carotenoid hydroxylase from red algae. J. Integr. Plant Biol. 2014;56:902–915. doi: 10.1111/jipb.12229. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Ren S., Liu X., Su L., Wu Y., Zhang W., Li Y., Jiang Y., Wang H., Fu R., et al. SlWRKY35 positively regulates carotenoid biosynthesis by activating the MEP pathway in tomato fruit. New Phytol. 2022;234:164–178. doi: 10.1111/nph.17977. [DOI] [PubMed] [Google Scholar]
- Zhang C., Dong S.-S., Xu J.-Y., He W.-M., Yang T.-L. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ye J., Liu C., Xu Q., Long L., Deng X. Citrus PH4-Noemi regulatory complex is involved in proanthocyanidin biosynthesis via a positive feedback loop. J. Exp. Bot. 2020;71:1306–1321. doi: 10.1093/jxb/erz506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jin J., Zhu S., Sun Q., Zhang Y., Xie Z., Ye J., Deng X. Citrus β-carotene hydroxylase 2 (BCH2) participates in xanthophyll synthesis by catalyzing the hydroxylation of β-carotene and compensates for BCH1 in citrus carotenoid metabolism. Hortic. Res. 2023;10 doi: 10.1093/hr/uhac290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Zhu K., Sun Q., Zhang W., Wang X., Cao H., Tan M., Xie Z., Zeng Y., Ye J., et al. Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in citrus peel. Mol. Plant. 2019;12:1294–1307. doi: 10.1016/j.molp.2019.04.014. [DOI] [PubMed] [Google Scholar]
- Zheng X., Giuliano G., Al-Babili S. Carotenoid biofortification in crop plants: citius, altius, fortius. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865 doi: 10.1016/j.bbalip.2020.158664. [DOI] [PubMed] [Google Scholar]
- Zheng X., Yang Y., Al-Babili S. Exploring the diversity and regulation of apocarotenoid metabolic pathways in plants. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.787049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Naqvi S., Breitenbach J., Sandmann G., Christou P., Capell T. Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc. Natl. Acad. Sci. USA. 2008;105:18232–18237. doi: 10.1073/pnas.0809737105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Chen H., Zhang Y., Liu Y., Zheng X., Xu J., Ye J., Deng X. In: Methods in Enzymology. Wurtzel E.T., editor. Academic Press; 2022. Chapter Six - Carotenoid extraction, detection, and analysis in citrus; pp. 179–212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this work are available in the paper and its supplemental information. Whole-genome resequencing data are accessible through NCBI. The accession numbers are listed in Supplemental Table 1. The accession numbers for CYP97 family homologs from other plants are listed in Supplemental Table 5. The sequence data used in this study can be found at NCBI GenBank (https://www.ncbi.nlm.nih.gov/) with the following accession numbers: CitCYP97B, LC143647.1; CitBCH1, AF296158.2; and CitBCH2, KY612512.1. Unless otherwise noted, data are expressed as mean values ± standard error based on triplicate measurements. Statistically significant differences were calculated using GraphPad Prism v.8.0.