Abstract
Astroviruses are known to cause enteric disease in several animal species, including turkeys. However, only human astroviruses have been well characterized at the nucleotide level. Herein we report the nucleotide sequence, genomic organization, and predicted amino acid sequence of a turkey astrovirus isolated from poults with an emerging enteric disease.
Astroviruses are small nonenveloped, positive-sense RNA viruses, which are distinct from other small round, positive-stranded RNA viruses such as caliciviruses and picornaviruses (5, 30, 39). Astroviruses have been reported to cause enteric disease in mammalian and avian species (2, 3, 9, 11, 14, 16, 18, 22, 26–28, 31, 35, 36, 38, 40, 41; R. E. Gough, M. S. Collins, E. Borland, and L. F. Keymer, Letter, Vet. Rec. 114:279, 1984), and is one of the major causes of diarrhea in infancy and childhood (17). Astroviruses also cause outbreaks of enteric disease in turkey poults. Astrovirus was first reported as a cause of gastroenteritis and mortality of turkeys 6 to 11 days of age in 1980 by McNulty et al. (28). Since then, there have been sporadic reports of astrovirus outbreaks in turkeys, mostly related to enteritis and growth depression (31). Sequence information has been reported from a previous turkey astrovirus (TAstV-1) isolate (21) (now referred to as TAstV-1 for clarity). However, this sequence is limited to 476 nucleotides (nt) from the 3′ end of the genome. Only the human astroviruses (HAstV) have been sequenced completely.
HAstV has been reported to contain approximately 6,800 nt and to be organized into three open reading frames (ORFs), ORF 1a, 1b, and 2. From the 5′ end of the genome, ORF 1a codes for the nonstructural proteins identified as a serine protease, transmembrane helices, and a nuclear localization signal, respectively (7). ORF 1b codes for the RNA-dependent RNA polymerase (RDRP). ORFs 1a and 1b overlap by approximately 70 nt. ORF 1b is brought into frame by a retrovirus-like frameshift sequence that produces a stem-loop (25). At the 3′ end of the genome is ORF 2, which codes for the capsid protein and is followed by an untranslated region and poly(A) tail. ORF 2 is transcribed into a subgenomic message of approximately 2,500 nt (7). In these studies we report the isolation and molecular characterization of a TAstV from poults with an emerging enteric disease of turkeys (33). Sequence analysis of this isolate has established that the TAstV genome contains 3 ORFs, each with predicted gene products, molecular characteristics, and organization consistent with previously reported astroviruses.
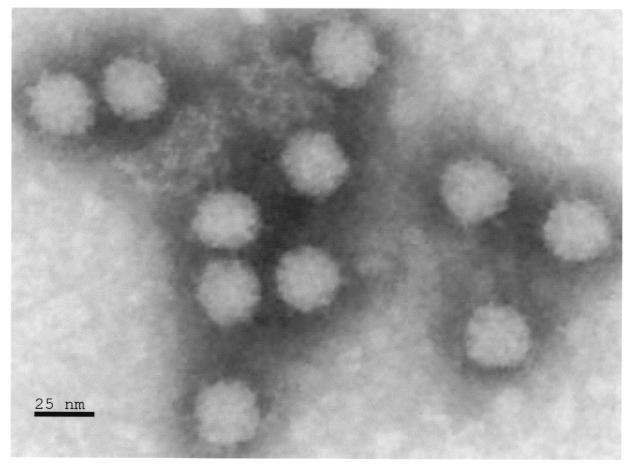
The putative astrovirus was isolated from the thymus and intestines of turkey poults affected with poult enteritis mortality syndrome (Fig. 1) (33). Briefly, the thymus and intestines from infected turkey poults (provided by H. John Barnes, North Carolina State College of Veterinary Medicine) were homogenized, filtered (0.2-μm-pore size; Fisher Scientific, Norcross, Calif.) and inoculated into the yolk sac of 20-day-old specific-pathogen-free (SPF) turkey embryos as described (33). At 5 days postinoculation intestines, intestinal fluid, and bursas were removed. After three passages the filtered tissue homogenate was centrifuged in a Sorvall fixed-angle rotor for 3 h at 23,425 × g, and the viral pellet was resuspended in 1 ml of 0.5 M Tris–0.25 M EDTA (TE) buffer, pH 7. The suspension was overlaid on a 27-37% CsCl gradient and centrifuged for 15 h at 130,000 × g in an SW28 rotor (L8-60M Ultracentrifuge; Beckman). A faint viral band, above the gradient layer, was removed with a syringe, brought to 15 ml with TE buffer, and centrifuged again at 23,425 × g for 3 h in a fixed-angle rotor. The purified viral pellet was resuspended in 200 μl of phosphate-buffered saline and a 50 μl aliquot of purified virus was negatively stained (15) and analyzed using a JEM-1210 transmission electron microscope (JEOL, Inc., Tokyo, Japan) at the College of Veterinary Medicine Electron Microscopy Laboratory, University of Georgia (Fig. 1).
FIG. 1.
Electron micrograph of 28-nm-diameter viral particles.
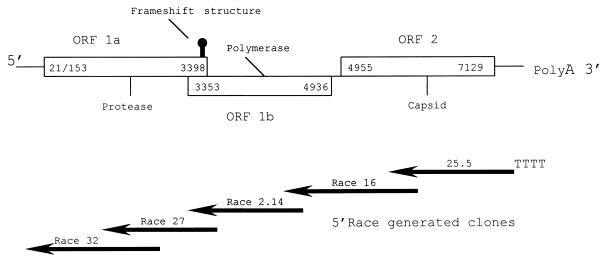
Total RNA was isolated from purified virus using TRIzol Total RNA Isolation Reagent (Life Technologies, Rockville, Md.) as previously described (6). An initial cDNA library was synthesized (12) using the SMART cDNA Library Construction Kit (Clonetech Laboratories, Inc., Palo Alto, Calif.) and then cloned using the TOPO-XL cloning system (29) according to manufacturer's directions (Invitrogen, San Diego, Calif.). Clones were screened using Luria-Bertani medium containing kanamycin (13). Approximately 25 clones, of varying length, were sequenced as previously described (34) and analyzed using DNASTAR (Madison, Wis.) and GeneWorks 2.3 (IntelliGenetics, Mountain View, Calif.) programs. These sequences were compared to reported sequences in the GenBank database using the basic local alignment search tool (BLAST) (1), and phylogenetic analysis was completed using parsimony (37). Two clones (p25.5 and p25.6) were identified as having putative amino acid similarities to TAstV and HAstV capsid protein (Fig. 2). These clones represented the last 1.5 kb of the 3′ end of the astrovirus genome and contained the poly(A) tail. Gene-specific primers were created using PRIMER2 software (Scientific and Educational Software, Stateline, Pa.) and were used to synthesize a series of cDNA libraries using the 5′ RACE System for rapid amplification of cDNA ends (8) (version 2.0; Life Technologies). Each cycle of subsequent cDNA synthesis was designed to overlap with its predecessor by at least 200 nt. Figure 2 illustrates the astrovirus genome and the sequencing strategy used. Each new clone, upstream of the previous sequence, was analyzed by BLAST individually. This was undertaken to reduce the chance of bias towards astrovirus when incorporated into the growing consensus data. Three or more clones from each 5′ RACE reaction were sequenced at least three times to generate one overall consensus sequence for TAstV. Finally, RNA was isolated from the intestines of experimentally infected turkey poults, and TAstV was resequenced directly from the reverse transcription-PCR (RT-PCR) product. Briefly, 1 μg of total RNA was incubated with 20 pmol of reverse primer, 20 pmol of each deoxynucleoside triphosphate, and 15 U of Superscript Reverse Transcriptase (Life Technologies) in a 20-μl reaction mixture for 60 min at 42°C. An aliquot (2 μl) of the first-strand product was amplified in a 50-μl reaction mixture containing 20 pmol of each primer, 20 pmol of each deoxynucleoside triphosphate 1.5 mM MgCl2, and 1.5 U of Taq polymerase (Life Technologies). Amplification was performed in a Perkin-Elmer 2400 DNA thermal cycler, and products were then purified using the Qiaquick PCR purification system (Qiagen, Valencia, Calif.) and sequenced. There was no significant difference between the sequence of the cloned cDNA and the RT-PCR products. The 5′ terminus of the virus was confirmed by synthesizing replicate 5′ RACE RT-PCR products using primers of different distances from the suspected end (all of the primers used were within 1 kb of the suspected end). These products were electrophoresed in a 1% agarose gel to confirm that each reaction yielded an amplicon of the expected size. These products were purified and sequenced as above.
FIG. 2.
Diagram of the astrovirus genomic organization. Numbers within the diagram represent the predicted start and stop sites. The two possible start sites given for ORF 1A are in the same reading frame; however, it is unclear which is the first codon. The overlapping clones generated by 5′ RACE, represented by black arrows, have amino acid similarities to the corresponding section of the astrovirus genome. The numbers above each arrow designates the clone used to generate that portion of the sequence data.
Analysis of the complete, 7,325-nt, TAstV sequence identified three ORFs. These ORFs were identified by BLAST analysis as coding for astrovirus-like proteins and being in a similar gene order. Each of these ORFs and their predicted gene products were compared to the corresponding reading frame of previously reported astroviruses.
The predicted amino acid sequence of ORF 2 was compared to the previously reported astrovirus capsid sequences (Table 1). TAstV was determined to be approximately 23.5% similar to the reported nucleotide sequence of TAstV-1. However, only the last 476 nt of TAstV-1 have been published; therefore, this comparison is limited to this extreme 3′ end of the virus. When the predicted amino acid sequence of this region was compared to the sequence obtained, the turkey isolates were found to be 21.9% similar. When the same region of the virus was compared with the mammalian isolates, TAstV was found to have a nucleotide similarity of ∼22% and a predicted amino acid similarity of ∼12% (Table 1). The same degree of similarity was observed between TAstV-1 and the mammalian isolates. In addition the mammalian isolates were determined to have nucleotide and amino acid similarities of 23 to 58% and 12 to 60%, respectively (data not shown).
TABLE 1.
Pairwise similarities of previously reported sequences to TAstVa
| Virus | % Similarity of:
|
||
|---|---|---|---|
| Last 476 nt | Last 114 a.a. | Predicted protein | |
| HAstV-1 | 22.9 | 11.4 | 17.4 |
| HAstV-2 | 23.3 | 12.3 | NDb |
| HAstV-3 | 26.5 | 12.3 | 18.1 |
| HAstV-4 | ND | 12.3 | 18.1 |
| HAstV-5 | 20.6 | 12.3 | 17.5 |
| HAstV-6 | 24.8 | 10.5 | 18.2 |
| HAstV-8 | 22.5 | 12.3 | 17.8 |
| FAstV | 22.9 | 16.7 | 18.1 |
| PAstV | 23.9 | 15.8 | NAc |
| SAstV | 20.4 | 12.3 | NA |
| TAstV-1 | 23.5 | 21.9 | NA |
Pairwise similarities of sequences of HAstV isolates, FAstV, porcine astrovirus (PAstV), sheep astrovirus (SAstV), and previously reported TAstV-1 to the TAstV sequence reported in the present work. Comparisons were done between the last 476 nt of each viral genome, the last 114 amino acids (a.a.) of the capsid protein from each virus, and the entire predicted amino acid sequence of the capsid protein.
ND, this comparison was not done.
NA, sequence information was not available for this comparison.
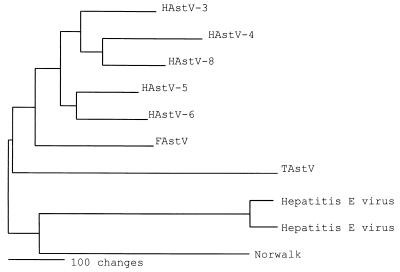
The predicted amino acid sequence of the entire TAstV capsid protein was also compared to published astrovirus sequences. However, only the feline astrovirus (FAstV) and HAstV capsid proteins have been completely sequenced. These sequences were used to determine amino acid similarities (Table 1) and phylogenetic relationships (Fig. 3). A heuristic search was completed with midpoint rooting. The phylogenetic tree in Fig. 3 shows that astroviruses clustered in one main branch, while the hepatitis E virus and Norwalk virus sequences clustered together as another branch.
FIG. 3.
Phylogenic relationship of predicted capsid proteins from HAstV-3, HAstV-4, HAstV-5, HAstV-6, HAstV-8, FAstV, TAstV, hepatitis E virus, and Norwalk virus. Analysis was done with heuristic search and midpoint rooting.
The amount of similarity between the sequence of this turkey isolate and the mammalian sequences was not surprising, as avian viruses can be quite different from their mammalian counterparts. However, we were surprised at the limited similarity observed between this isolate and TAstV-1. The previously reported sequence was used by Jonassen et al. to identify a region conserved among astroviruses within the 3′ noncoding sequence (21). This conserved region was not identified in our isolate, and furthermore, primers designed from the conserved region failed to produce RT-PCR product from our isolate. The reason for this lack of similarity is not understood. However, TAstV may represent a different serotype since antibodies against the TAstV-1 isolate failed to recognize our virus by Western blot analysis (data not shown).
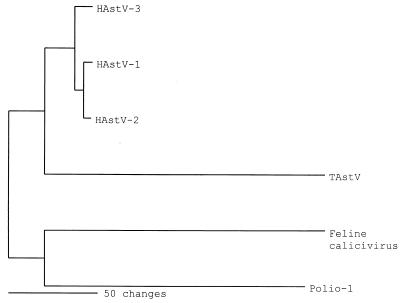
Predicted amino acid sequence of ORF 1B indicated the presence of the viral RNA polymerase active-site motif YGDD (data not shown) (19). The amino acid sequence of this putative polymerase was compared to the published astrovirus polymerase sequences. HAstVs are the only astroviruses completely sequenced; therefore, the TAstV polymerase sequence was aligned with the amino acid sequences of HAstV RDRPs. The similarity between TAstV and the HAstV isolates varied from 35 to 38%. Phylogenetic analysis of the entire amino acid sequence of the polymerases of TAstV, HAstV-1, HAstV-2, and HAstV-3 were completed using a heuristic search with midpoint rooting (Fig. 4). The polymerase proteins from a representative of the picornavirus family (poliovirus type 1) and a representative of the calicivirus family (feline calicivirus [FCV]) were used for comparison. The TAstV isolate reported here groups most closely with the HAstV isolates compared with the other positive-sense RNA virus-type polymerases analyzed. TAstV ORF 1B showed the greatest similarity to HAstV compared to the other ORFs. This was expected, since RDRPs are well-conserved proteins. The polymerase is the only highly conserved protein that is present in all positive-stranded RNA viruses; because of this they can be used to classify and separate related viruses (23).
FIG. 4.
Phylogenic relationship of the RDRP from astroviruses and representatives of other small, round RNA viruses. The amino acid sequence of FCV, human picornavirus, poliovirus type 1 (Polio-1) and HAstV-1, -2, and -3 were utilized to phylogenetically evaluate the putative TAstV polymerase. Analysis was done with heuristic search and midpoint rooting.
Analysis of ORF 1A identified a putative serine protease motif from the predicted amino acid sequence. This sequence was aligned with the serine protease sequences of HAstV types 1, 2, and 3. As presented in Fig. 5, there is sequence identity, particularly around the proposed catalytic amino acids (39). Position 40 is the putative location of the second catalytic amino acid (39). This position is occupied by the aspartic acid in the HAstV proteases. TAstV has the amide form of aspartic acid (asparagine) at position 40. Asparagine is less reactive than aspartic acid and is reported as a conserved substitution by BLAST analysis. TAstV does contain an aspartic acid residue in this region of the protein, at position 39 (Fig. 5). The reason for this mismatch is not understood; however, the putative catalytic histidine and serine residues of TAstV and HAstV are in alignment at position 7 and 104 (Fig. 5). Astrovirus is the only small, round, positive-sense RNA virus reported to code for a serine protease (39). Both caliciviruses and picornaviruses are reported to code for a cysteine protease (5, 10).
FIG. 5.
Comparison of the serine protease sequences of HAstV-1, -2, and -3 and the putative serine protease of TAstV. The suspected catalytic triad for each virus is underlined. Amino acid matches between TAstV and the HAstV isolates are shown in boldface type. ∧, position implicated in substrate binding (TAstV has glycine substituted for alanine); #, additional substrate binding site (histidines are present in all sequences compared).
Additional analysis of the TAstV genome revealed the presence of a possible frameshift sequence and secondary structure (data not shown) first described by Jiang et al. (20) in HAstV. The signal for this frameshift (AAAAAAU) is similar to that of the retrovirus mouse mammary tumor virus (4, 20). This sequence and structure are not found in other small, round RNA viruses and was one of the reasons for astrovirus classification in a family of its own (20, 30).
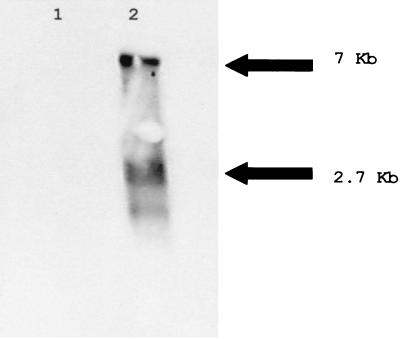
Finally, Northern blot analysis was used to detect two viral RNA species, one of approximately 7 kb and one of approximately 2.7 kb (Fig. 6). This was done using total RNA from intestines of control SPF embryos (Fig. 6, lane 1) and total RNA from intestinal fluid of virus-inoculated SPF turkey embryos (Fig. 6, lane 2). These RNAs were electrophoresed in a denaturing 1.2% agarose gel (24) and then transferred and cross-linked to nylon as described previously (32). The RNA was then probed using a 392-bp double-stranded DNA probe specific to base positions 588 to 979 of ORF 2. This probe was synthesized by PCR, using gene-specific primers, clones 25.5 (Fig. 2), and digoxigenin-11-dUTPs and then hybridized to the membrane, and detected by chemiluminescence following the manufacturer's instructions (Roche Molecular Biochemicals, Indianapolis, Ind.). The detected RNAs correspond to the reported size of the astrovirus genome and subgenomic RNA (20). There is also a faint third band visible beneath the subgenomic RNA. The identity of this band is not known.
FIG. 6.
Northern blot hybridization of total RNA from astrovirus-infected (lane 2) and astrovirus-uninfected (lane 1) turkey embryo intestines. The PCR probe synthesized from clone 25.5 (Fig. 1) hybridized to an ∼7-kb genomic RNA and an ∼2.7-kb subgenomic RNA (lane 2).
These data represent the first complete sequence and molecular characterization of a nonhuman astrovirus, to our knowledge. This is based on the identification of sequence similarities to astrovirus as well as the identification of several distinctive astrovirus properties. These comparisons showed TAstV to be 7,325 nt [excluding the poly(A) tail] in length, to contain three ORFs, and to produce one subgenomic RNA of approximately 2.7 kb. Each ORF coded for proteins with limited amino acid similarity to HAstV. The predicted proteins within this viral genome are designated as a serine protease, an RDRP, and a viral capsid protein. Also detected was a retroviruslike frameshift signal with potential secondary structure in the genome. Each of these elements is consistent with other sequenced astroviruses and establish this newly described virus as being a TAstV.
This TAstV was isolated from turkeys affected by an emerging disease, which is characterized by enteritis, high mortality, growth depression, lymphoid atrophy, and immunosuppression. It is because of this lymphoid atrophy and immunosuppression that we first examined the thymus as the source of disease agents, which led to the isolation and subsequent molecular characterization of this novel virus (33). This virus can readily be detected in the intestines, thymus, and bursa of infected poults, using both electron microscopy and RT-PCR (unpublished observation). More importantly, when naïve poults are given purified TAstV they exhibit many of the disease signs, including enteritis, mortality, lymphoid atrophy, and immunosuppression (unpublished data).
Nucleotide sequence accession numbers.
The nucleotide sequence of TAstV was submitted to GenBank and was given the accession number AF206663. The isolates used in this study for nucleotide and amino acid comparison to TAstV were HAstV-1 (L23513), HAstV-2 (L13745), HAstV-3 (AF141381), HAstV-4 (S45048), HAstV-5 (U15136), HAstV-6 (Z46658), HAstV-8 (Z66541), FAstV (AF056197), TAstV-1 (Y15936), porcine astrovirus (Y15938), sheep astrovirus (Y15937), hepatitis E virus (AF051830 and AF082843), Norwalk virus (AF093797), FCV (BAA143108), and poliovirus type 1 (3660269).
Acknowledgments
We acknowledge the invaluable work of Laura A. Kelley and Joyce Bennett. We also thank H. John Barnes at the Department of Food Animal and Equine Medicine, College of Veterinary Medicine, North Carolina State University, for providing tissue samples from infected turkey poults; Richard Remler at the National Animal Disease Center for SPF turkey eggs; and B. L. Steffans and Mary Ard of the College of Veterinary Medicine Electron Microscopy Laboratory, University of Georgia. Finally, we thank Valrie Simmons, Liz Turpin, Mark Jackwood, Steve Poet, David Suarez, Holly Sellers, and Darrell Kapczynski for many helpful discussions.
This work was supported by U.S. Egg and Poultry Association grant #311 and USDA CRIS project number 661232000020.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Aroonprasert D, Fagerland J A, Kelso N E, Zheng S, Woode G N. Cultivation and partial characterization of bovine astrovirus. Vet Microbiol. 1989;19:113–125. doi: 10.1016/0378-1135(89)90077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridger J C, Hall G A, Brown J F. Characterization of a calici-like virus (Newbury agent) found in association with astrovirus in bovine diarrhea. Infect Immun. 1984;43:133–138. doi: 10.1128/iai.43.1.133-138.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I. Ribosomal frameshifting viral RNAs. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 5.Carter M J. Genomic organization and expression of astroviruses and caliciviruses. Arch Virol Suppl. 1994;9:429–439. doi: 10.1007/978-3-7091-9326-6_42. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Cubitt W D. Historical background and classification of caliciviruses and astroviruses. Arch Virol Suppl. 1996;12:225–235. doi: 10.1007/978-3-7091-6553-9_24. [DOI] [PubMed] [Google Scholar]
- 8.Frohman M A. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 9.Geyer A, Steele A D, Peenze I, Lecatsas G. Astrovirus-like particles, adenoviruses and rotaviruses associated with diarrhoea in piglets. J S Afr Vet Assoc. 1994;65:164–166. [PubMed] [Google Scholar]
- 10.Gorbalenya A E, Donchenko A P, Blinov V M, Koonin E V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 1989;243:103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- 11.Gray E W, Angus K W, Snodgrass D R. Ultrastructure of the small intestine in astrovirus-infected lambs. J Gen Virol. 1980;49:71–82. doi: 10.1099/0022-1317-49-1-71. [DOI] [PubMed] [Google Scholar]
- 12.Gubler U, Hoffman B J. A simple and very efficient method for generating cDNA libraries. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D H. Techniques for transformation of E. coli. In: Glover D M, editor. DNA, cloning. I. A Practical Approach. Oxford, United Kingdom: IRL Press; 1985. pp. 366–369. [Google Scholar]
- 14.Harbour D A, Ashley C R, Williams P D, Gruffydd-Jones T J. Natural and experimental astrovirus infection of cats. Vet Rec. 1987;120:555–557. doi: 10.1136/vr.120.23.555. [DOI] [PubMed] [Google Scholar]
- 15.Hayat M A, Miller S E. Negative staining. New York, N.Y: McGraw-Hill Publishing Co.; 1990. [Google Scholar]
- 16.Herring A J, Gray E W, Snodgrass D R. Purification and characterization of ovine astrovirus. J Gen Virol. 1981;53:47–55. doi: 10.1099/0022-1317-53-1-47. [DOI] [PubMed] [Google Scholar]
- 17.Herrmann J E, Taylor D N, Echeverria P, Blacklow N R. Astroviruses as a cause of gastroenteritis in children. N Engl J Med. 1991;324:1757–1760. doi: 10.1056/NEJM199106203242501. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino Y, Zimmer J F, Moise N S, Scott F W. Detection of astroviruses in feces of a cat with diarrhea. Brief report. Arch Virol. 1981;70:373–376. doi: 10.1007/BF01320252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishihama A, Barbier P. Molecular anatomy of viral RNA-directed RNA polymerases. Arch Virol. 1994;134:235–258. doi: 10.1007/BF01310564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang B, Monroe S S, Koonin E V, Stine S E, Glass R I. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc Natl Acad Sci USA. 1993;90:10539–105343. doi: 10.1073/pnas.90.22.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonassen C M, Jonassen T O, Grinde B. A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J Gen Virol. 1998;79:715–718. doi: 10.1099/0022-1317-79-4-715. [DOI] [PubMed] [Google Scholar]
- 22.Kjeldsberg E, Hem A. Detection of astroviruses in gut contents of nude and normal mice. Brief report. Arch Virol. 1985;84:135–140. doi: 10.1007/BF01310560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. . (Erratum, 28:546.) [DOI] [PubMed] [Google Scholar]
- 24.Lehrach H, Diamond D, Wozney J M, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 25.Marczinke B, Bloys A J, Brown T D, Willcocks M M, Carter M J, Brierley I. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J Virol. 1994;68:5588–5595. doi: 10.1128/jvi.68.9.5588-5595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall J A, Healey D S, Studdert M J, Scott P C, Kennett M L, Ward B K, Gust I D. Viruses and virus-like particles in the faeces of dogs with and without diarrhoea. Aust Vet J. 1984;61:33–38. doi: 10.1111/j.1751-0813.1984.tb07186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsui S M, Greenberg H B. Astroviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields Virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 811–824. [Google Scholar]
- 28.McNulty M S, Curran W L, McFerran J B. Detection of astroviruses in turkey faeces by direct electron microscopy. Vet Rec. 1980;106:561. doi: 10.1136/vr.106.26.561. [DOI] [PubMed] [Google Scholar]
- 29.Mead D A, Pey N K, Herrnstadt C, Marcil R A, Smith L M. A universal method for the direct cloning of PCR amplified nucleic acid. Bio/Technology (New York) 1991;9:657–663. doi: 10.1038/nbt0791-657. [DOI] [PubMed] [Google Scholar]
- 30.Monroe S S, Jiang B, Stine S E, Koopmans M, Glass R I. Subgenomic RNA sequence of human astrovirus supports classification of Astroviridae as a new family of RNA viruses. J Virol. 1993;67:3611–3614. doi: 10.1128/jvi.67.6.3611-3614.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds D L, Saif Y M. Astrovirus: a cause of an enteric disease in turkey poults. Avian Dis. 1986;30:728–735. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual, 2 ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schultz-Cherry, S., D. R. Kapczynski, V. M. Simmons, M. D. Koci, C. Brown, and H. J. Barnes. Identifying agent(s) associated with poult enteritis mortality syndrome: importance of the thymus. Avian Dis., in press. [PubMed]
- 34.Seal B S, King D J, Locke D P, Senne D A, Jackwood M W. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J Clin Microbiol. 1998;36:1141–1145. doi: 10.1128/jcm.36.4.1141-1145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu M, Shirai J, Narita M, Yamane T. Cytopathic astrovirus isolated from porcine acute gastroenteritis in an established cell line derived from porcine embryonic kidney. J Clin Microbiol. 1990;28:201–206. doi: 10.1128/jcm.28.2.201-206.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snodgrass D R, Gray E W. Detection and transmission of 30 nm virus particles (astroviruses) in faeces of lambs with diarrhoea. Arch Virol. 1977;55:287–291. doi: 10.1007/BF01315050. [DOI] [PubMed] [Google Scholar]
- 37.Swofford D. PAUP: phylogenetic analysis using parsimony, 4.0 ed. Champaign: Illinois Natural History Survey; 1998. [Google Scholar]
- 38.Tzipori S, Menzies J D, Gray E W. Detection of astrovirus in the faeces of red deer. Vet Rec. 1981;108:286. doi: 10.1136/vr.108.13.286. [DOI] [PubMed] [Google Scholar]
- 39.Willcocks M M, Brown T D, Madeley C R, Carter M J. The complete sequence of a human astrovirus. J Gen Virol. 1994;75:1785–1788. doi: 10.1099/0022-1317-75-7-1785. [DOI] [PubMed] [Google Scholar]
- 40.Williams F P., Jr Astrovirus-like, coronavirus-like, and parvovirus-like particles detected in the diarrheal stools of beagle pups. Arch Virol. 1980;66:215–226. doi: 10.1007/BF01314735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woode G N, Bridger J C. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J Med Microbiol. 1978;11:441–452. doi: 10.1099/00222615-11-4-441. [DOI] [PubMed] [Google Scholar]