Abstract
The complete surface glycoprotein (SU) nucleotide sequences of three French isolates of caprine arthritis-encephalitis virus (CAEV) were determined and compared with those of previously described isolates: three American isolates and one French isolate. Phylogenetic analyses revealed the existence of four distinct and roughly equidistant evolutionary CAEV subtypes. Four conserved and five variable domains were identified in the SU. The fine specificities of antibodies produced against these domains during natural infection were examined using a pepscan analysis. Nine immunogenic segments were delineated throughout the conserved and variable domains of SU, two of them corresponding to conserved immunodominant epitopes. Antigenic determinants which may be involved in the immunopathogenic process induced by CAEV were identified. These results also provide sensitive and specific antigen peptides for the serological detection and differentiation of CAEV and visna/maedi virus infections.
The surface glycoprotein (SU) of lentiviruses contains determinants important for cellular host range, infectivity, cytopathogenicity, and disease progression. The region of the envelope gene encoding the SU displays a particularly high level of sequence variation, resulting in hypervariable domains interspersed with less variable domains throughout the protein. Both variable and conserved domains are major targets for the host immune response, including virus-neutralizing antibodies and cell-mediated cytotoxicity. Therefore, SU has been an obvious candidate in vaccine trials and diagnostic assays of infection by lentiviruses, such as human, simian, and feline immunodeficiency viruses (HIV, SIV, and FIV, respectively) (for reviews, see references 10, 19, 33, 34, and 38).
Caprine arthritis-encephalitis virus (CAEV) is a lentivirus causing slow and persistent inflammatory diseases in goats, primarily arthritis and mastitis (9, 42). These inflammatory diseases are the result of viral infection of cells of monocyte/macrophage lineage, which are the main target cells in vivo (13, 43, 44). The results of a recent experiment using live attenuated CAEV vaccine in goats have demonstrated the development of some protection against challenge with the pathogenic homologous virus (17), indicating the effectiveness of an immunological control of virus replication. However, this protective immunity did not prevent the development of clinical signs of disease, although the lesions were not as severe as those found in wild-type CAEV-infected goats. Previous investigations have indicated that the presence and severity of arthritic lesions are specifically correlated with the predominant humoral immune response directed against the SU and transmembrane (TM) CAEV envelope glycoproteins (3, 22, 30, 35). Collateral experiments have demonstrated that infected goats having early dominant anti-SU antibody responses (48) as well as goats challenged with CAEV during persistent CAEV infection or after vaccination with inactivated virus (37) developed more rapidly progressing and severe arthritis. Conversely, long-term infected nonprogressor goats are characterized by a lack of clinical pathology and by low anti-CAEV antibody titers, compared to arthritic goats (30, 48). These observations suggest that antigenic determinants of envelope glycoproteins of CAEV may be involved in the immunopathogenic process leading to inflammatory diseases.
Precise knowledge of the immunogenic domains of CAEV glycoproteins would provide useful information on the antigenic structures to be included in candidate vaccines. Four immunodominant epitopes have been identified in the TM ectodomain of CAEV (3). Three of them have been shown to be associated with clinical arthritis. In contrast, the immunogenic epitopes of the SU are still unknown. Our objective is to provide the basic framework for understanding the CAEV-induced pathogenic process and for vaccine development. In this study, we have defined the variability profile of the SU, and we have precisely mapped epitopes within conserved and variable domains which elicit humoral immune responses during natural CAEV infection.
Variability of CAEV SU.
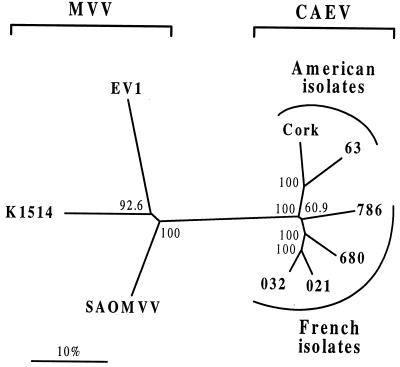
At present, the complete SU nucleotide sequences (29, 50, 61, 62) of only one French (strain 680) and three American (strains Cork, 63, and 1244) CAEV isolates have been analyzed. Expanded surveys of CAEV isolates are required to explore the extent and nature of SU diversity. In the present study, the complete SU nucleotide sequences of three new French CAEV isolates (named 021, 032, and 786) selected for their relative great divergence with the prototype Cork and 680 strains using heteroduplex mobility assay (61; unpublished data) were determined. Genomic DNAs were purified (Isoquick; Microprobe) from explanted goat synovial membrane cells (strain 786) or cocultures of milk mononuclear cells with goat synovial membrane cells (strains 021 and 032) harvested at maximum cytopathic effects. One microgram of DNA was subjected to 35 cycles of PCR amplification using oligonucleotide primers 5084 and 5087 as previously described (61), and the resulting 2.2-kb PCR products containing the entire SU sequences were cloned into pGEM-1 vector. For each strain, three independent rounds of PCR and cloning were done, and at least three clones were sequenced and aligned to determine a consensus sequence and rule out PCR artifacts or intrastrain variability. To provide information about the evolutionary relationships of these newly identified French CAEV isolates with previously reported prototype CAEV isolates, a phylogenetic tree was constructed from the full-length SU coding sequence (1.6 kb). In addition, three previously published prototype SU sequences (strains K1514, EV-1, and SAOMVV) (49, 52, 58) of visna/maedi virus (MVV), an ovine lentivirus antigenically and genetically closely related to CAEV (14, 65), were also included. Phylogenetic relationships were determined by using the neighbor-joining algorithm with the Kimura two-parameter distance matrix (PHYLIP) (11). Sites at which there was a gap in any of the aligned sequences were excluded from all comparisons. To evaluate the consistency of the phylogenetic groupings, branching order reliability was evaluated by 1,000 replications of bootstrap resampling analysis. As shown in Fig. 1, all CAEV isolates form a related group on a clearly separate branch from the MVV group. Since previous phylogenetic studies using short nucleotide sequences have reported the existence of French ovine lentiviruses that were more closely related to CAEV than to the prototype MVV strains (32), phylogenetic trees were also constructed using different small regions of the SU coding sequence. These analyses yielded virtually identical trees, confirming the existence of a distinct CAEV cluster and ruling out possible recombination events. Genetic distances between CAEV and MVV SU sequences were estimated by means of the CLUSTAL W program (Table 1) (60). The intergroup Hamming distances of amino acid sequences ranged from 29.2 to 36.7%, indicating that a large distance exists between CAEV isolates and prototype MVV isolates. Phylogenetic relationships among CAEV isolates revealed the existence of a cluster of four distinct subtypes; one of these harbored exclusively American isolates, while the three others represented French subtypes with nearly 12% genetic diversity between each pair of subtypes. The intragroup distances of amino acid sequences ranged from 7.8 to 20.1% in the CAEV group. The amino acid distances were 14.9 to 20.1% between French and American CAEV isolates and 7.8 to 15.7% among the four French isolates, confirming that French isolates distinctly differ from the American isolates and have evolved in different but closely related subtypes.
FIG. 1.
Phylogenetic relationships of French CAEV isolates to prototype CAEV and MVV strains. The SU multiple sequence alignment was resampled by the bootstrap method (1,000 data sets); an unrooted tree generated by neighbor-joining analysis is shown. The horizontal and vertical orientations of branches are noninformative and for clarity only; branching patterns and branch lengths reflect phylogenetic distance relationships. The numbers on the nodes represent the percentage of bootstrap samples.
TABLE 1.
Pairwise distances between SU sequences of CAEV and MVV lentivirusesa
| Type | Isolate | Hamming distance with:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cork | 63 | 1244 | 680 | 021 | 032 | 786 | K1514 | EV-1 | SAOb | ||
| CAEV | Cork | 11.5 | 16.9 | 16.2 | 14.9 | 15.7 | 17.3 | 33.0 | 36.2 | 32.1 | |
| 63 | 10.8 | 18.5 | 17.8 | 17.8 | 18.5 | 18.5 | 32.9 | 36.7 | 32.3 | ||
| 1244 | —c | — | 20.1 | 18.8 | 19.9 | 20.1 | 32.4 | 36.7 | 31.2 | ||
| 680 | 14.9 | 15.4 | — | 11.3 | 11.9 | 15.7 | 31.7 | 35.3 | 31.0 | ||
| 021 | 14.5 | 16.2 | — | 10.1 | 7.8 | 14.4 | 32.3 | 36.2 | 31.0 | ||
| 032 | 14.9 | 16.4 | — | 10.0 | 6.3 | 14.6 | 31.2 | 35.6 | 29.2 | ||
| 786 | 15.7 | 15.8 | — | 13.3 | 13.0 | 13.0 | 32.3 | 35.6 | 32.7 | ||
| MVV | K1514 | 32.1 | 31.0 | — | 30.1 | 30.9 | 30.5 | 29.8 | 22.6 | 16.0 | |
| EV-1 | 32.2 | 32.1 | — | 31.7 | 33.4 | 33.0 | 31.8 | 22.8 | 22.2 | ||
| SAOb | 30.2 | 28.4 | — | 29.0 | 29.0 | 27.9 | 27.9 | 20.4 | 22.5 | ||
Pairwise Hamming distances (expressed as percentage of total SU sequence length) are presented in a triangular matrix. Nucleotide distances are presented in the lower half of each matrix, and amino acid distances are presented in the upper half of each matrix.
SAO, SAOMVV strain.
—, not determined.
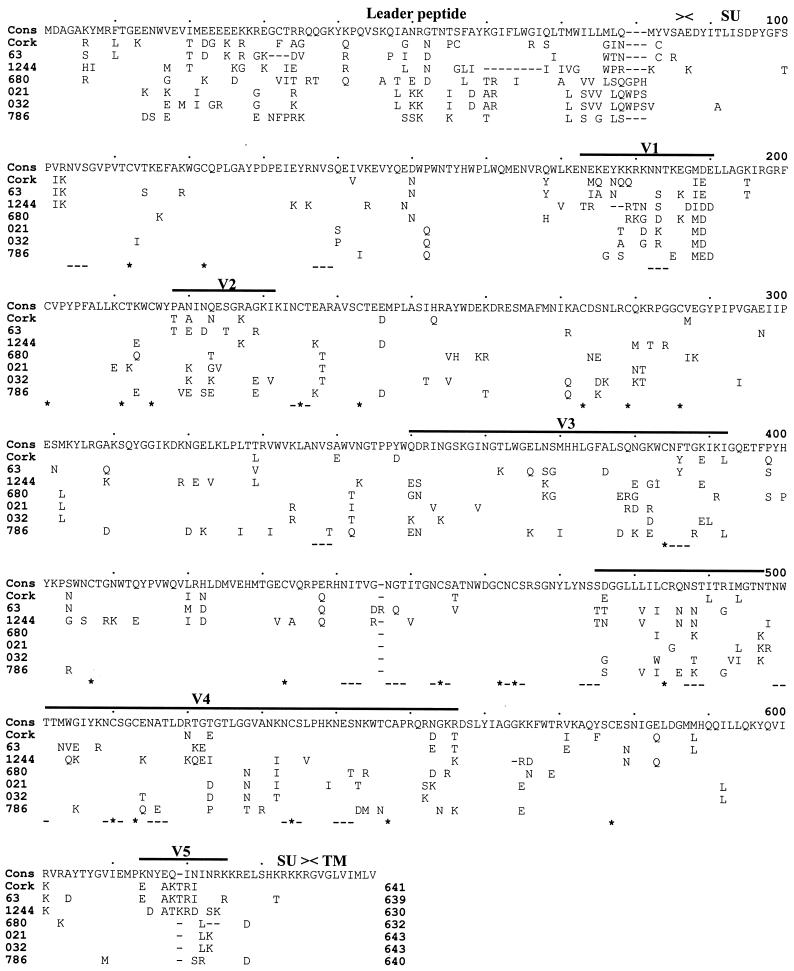
The amino acid sequence alignment of all CAEV SU precursor sequences (four French and three American isolates) is shown in Fig. 2. A hypervariability was observed for the large leader peptide, including insertions and deletions. The three French isolates (021, 032, and 786) characterized in this study exhibited an intact open reading frame without unusual insertions or deletions, resulting in a length of 548 ± 2 amino acids for the mature SU. The 2-amino-acid length variation among French SU sequences corresponded to a single deletion located at the carboxy terminus of SU of the first reported French CAEV isolate (strain 680). This deletion allowed the loss of one of the two potential cleavage sites between the SU and TM of the strain 680, while these two sites were well conserved in the three newly described French isolates. A perfect conservation of the 22 cysteine residues in the SU was observed among all isolates. Of the 24 potential N-linked glycosylation sites, 17 (71%) were conserved and, curiously, 30% of the cysteine residues were located within or just beside these conserved N-linked glycosylation sites. Five variable regions (V1 to V5) and four conserved regions (C1 to C4) were identified. The relative distribution of these variable and conserved domains of the mature SU is presented in Fig. 3, which illustrates the variability profile of SU based on the frequency of amino acid substitutions relative to the consensus sequence and smoothed using a 10-amino-acid window size. The first conserved region (C1) spanning 86 amino acids corresponded to the amino terminus of SU. This region was followed by two contiguous short hypervariable regions (V1 and V2). The V1 region was consistently the most variable but contained a perfectly conserved potential N-linked glycosylation site (position 182). The V2 region was flanked by two stretches of conserved amino acids, particularly cysteine residues. The C3 region spanning 81 residues (positions 393 to 474) in the central part of the mature SU was highly conserved, with 80% conservation among all isolates. This C3 region, together with the neighboring V4 region, contains the majority of the conserved N-linked glycosylation sites (11 of 17) and cysteine residues (10 of 22), suggesting that they form a highly constrained and surface-exposed domain. It had been proposed that the V4 region of CAEV and MVV may be analogous to the V3 principal neutralizing domain of HIV-type 1 (HIV-1) (29, 57). The V5 region located at the carboxy terminus overlaps the first of the two putative SU-TM cleavage sites.
FIG. 2.
Alignment of predicted SU precursor amino acid sequences of CAEV isolates. The sequences were aligned with a consensus sequence (Cons) consisting of the most frequent residue at a given position. Dashes in the sequence alignment represent deletions. Variable domains (V1 to V5) are delineated by overlines. ∗, conserved cysteine residue; –––, conserved potential N-linked glycosylation site.
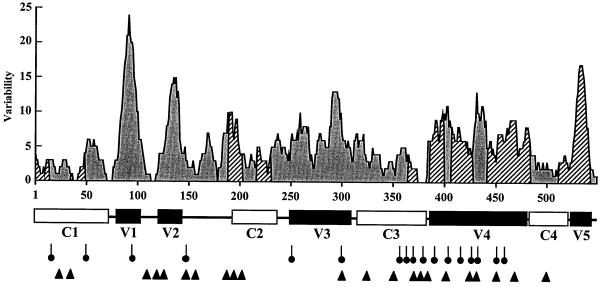
FIG. 3.
Relationships between immunogenicity and variability profiles of CAEV SU. Variable and conserved domains of the SU were established from all available sequences (those for three American and four French isolates). The variability profile is based on the frequency of amino acid substitutions relative to the consensus sequence and smoothed using a 10-amino-acid window size. The amino acid numbers on the x axis start with the mature SU amino-terminal Glu as residue 1 (29). The immunogenic sites identified in this study are delineated by hatched areas. Below the variability profile, the conserved regions are shown as white boxes and the variable regions are shown as black boxes. Stick with circle, conserved potential N-linked glycosylation site; arrowhead, conserved cysteine residue.
Immunogenicity of CAEV SU.
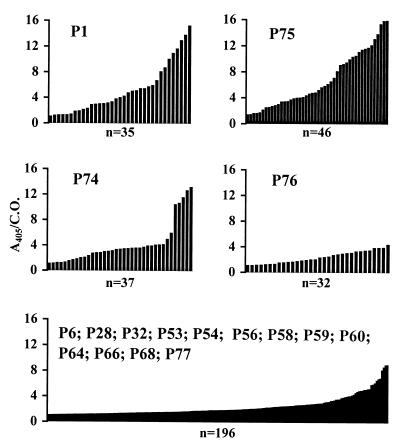
To precisely map linear epitopes in the CAEV SU glycoprotein which elicit humoral immune responses during natural infection, a pepscan analysis was performed using 77 synthetic peptides. Overlapping peptides were constructed to cover the entire amino acid sequence of the mature SU of the French CAEV strain 680 (Table 2). The peptides were designed to be 14 amino acids long and to overlap each other by 7 amino acids. The residues for each peptide were numbered relatively to the known amino terminal Glu1 of the mature SU, corresponding to the Glu87 of the envelope precursor (29). The peptides were synthetized by automated 9-fluorenylmethyloxycarbonyl chemistry (Chiron Mimotopes Peptide Systems, Clayton, Australia) (4). An amino-terminal biotinylated tetrapeptide (Ser-Gly-Ser-Gly) was added to all peptides to facilitate epitope accessibility and absorption to streptavidin-coated wells. Fifty-five immune sera were collected from naturally infected goats originating from different flocks in France. All infected animals were PCR positive and seropositive when tested by commercially available enzyme-linked immunosorbent assays (ELISAs) employing whole-virus preparations as antigens (Chekit CAEV/MVV; Behring). Serum samples were tested at a dilution of 1:100 against each peptide in a standard ELISA according to the manufacturer's instructions, and optical densities were recorded on an automatic ELISA plate reader (Labsystem) at 405 nm. The cutoff level for positivity for each peptide was defined as the mean absorbance value of a panel of 32 negative control sera plus 3 standard deviations. As shown in Table 2, a wide range of peptide immunoreactivities (0 to 84%) was observed. Seventeen immunoreactive peptides (P1, P6, P28, P32, P53, P54, P56, P58, P59, P60, P64, P66, P68, P74, P75, P76, and P77), defined by ≥20% positive reactivity with goat immune sera, were identified. By correlating the relative reactivities of immunoreactive peptides with adjacent or overlapping peptides, nine major immunogenic segments were identified in the SU (Table 2; Fig. 3). These have been tentatively assigned to the regions of amino acids Glu1-Ser14, Cys36-Tyr49, Asn190-Gly203, Lys218-Lys231, Gly365-Ser378, Asn386-Gln399, Met407-Glu427, Val442-Arg483, and Val512-Leu539. The most immunoreactive peptides (P1, P74, P75, and P76), i.e., the immunodominant peptides, recognized by at least 58% of immune sera, were located exclusively at the amino (residues 1 to 14) and carboxy (residues 512 to 539) termini of the SU. The amino-terminal peptide (P1, residues 1 to 14) of SU reacted with 67% of the goat immune sera tested. Immune serum reactivity fell to 2% with peptide P2 (residues 8 to 21), delineating the immunodominant amino-terminal epitope to the first 14 amino acids of mature SU. The immunodominant peptide P75 (residues 519 to 532) located at the carboxy terminus of SU displayed the strongest relative reactivity of all peptides tested, exhibiting 84% serological reactivity with the goat immune sera. The comparison between the reactivity of peptide P75 with those of the two overlapping immunoreactive peptides P74 and P76 suggests that there may be at least two major linear epitopes in the region encompassing residues 519 to 532. In addition, the level of immunoreactivities (absorbance/cutoff ratio) of positive immune sera to immunoreactive peptides (Fig. 4) revealed that immunodominant peptides P1 and P75 were strongly recognized by the majority of sera tested. Using a combination of only five immunoreactive peptides (P28, P64, P74, P75, and P76) the sensitivity increased to 96.4%, indicating that the development of an ELISA based on such peptides would give a highly sensitive and specific test for the detection of CAEV infection. The relationships between localization of the immunogenic peptide determinants and the variability profile of CAEV SU are illustrated in Fig. 3. As expected, all conserved regions (C1 to C4) contained immunoreactive linear B epitopes. No group-specific epitopes were identified within the V1 and V2 hypervariable regions and the less-variable V3 region. The V4 region showed clearly different immunogenic patterns: 7 out of 17 (40%) immunoreactive peptides described above were distributed throughout the V4 region. Remarkably, a high concentration of conserved cysteine residues and N-linked glycosylation sites was also found within this region, suggesting that these immunogenic determinants are well-exposed in the native protein structure. Despite a high degree of variability, the short V5 region located at the carboxy terminus of SU contained one of the two immunodominant epitopes included in peptide P75, suggesting that the sequences of field isolates from which immune sera were randomly collected were closely related to that of the prototype CAEV 680 used as a basis for peptide synthesis. Indeed, examination of amino acid sequences in the V5 region (Fig. 2) revealed a high sequence homology among French isolates, compared to the great divergence observed between French and American isolates.
TABLE 2.
Reactivity of sera from naturally CAEV-infected goats to SU-derived synthetic peptides
| Peptide code | Amino acidsa | Sequenceb (one-letter code) | No. (%) of reactive serac | Peptide code | Amino acidsa | Sequenceb (one-letter code) | No. (%) of reactive serac | |
|---|---|---|---|---|---|---|---|---|
| P1 | 1–14 | EDYITLISDPYGFS | 35 (67) | |||||
| P2 | 8–21 | SDPYGFSPVRNVSG | 1 (2) | |||||
| P3 | 15–28 | PVRNVSGVPVTCVT | 1 (2) | |||||
| P4 | 22–35 | VPVTCVTKKFAKWG | 4 (7) | |||||
| P5 | 29–42 | KKFAKWGCQPLGAY | 9 (16) | |||||
| P6 | 36–49 | CQPLGAYPDPEIEY | 15 (27) | |||||
| P7 | 43–56 | PDPEIEYRNVSQEI | 3 (5) | |||||
| P8 | 50–63 | RNVSQEIVKEVYQE | 2 (4) | |||||
| P9 | 57–70 | VKEVYQENWPWNTY | 0 (0) | |||||
| P10 | 64–77 | NWPWNTYHWPLWQM | 2 (4) | |||||
| P11 | 71–84 | HWPLWQMENVRHWL | 1 (2) | |||||
| P12 | 78–91 | ENVRHWLKENEKEY | 4 (7) | |||||
| P13 | 85–98 | KENEKEYKRKGNDT | 9 (16) | |||||
| P14 | 92–105 | KRKGNDTKKGMDEL | 5 (9) | |||||
| P15 | 99–112 | KKGMDELLAGKIRG | 9 (16) | |||||
| P16 | 106–119 | LAGKIRGRFCVPYP | 8 (15) | |||||
| P17 | 113–126 | RFCVPYPFALLKCT | 3 (5) | |||||
| P18 | 120–133 | FALLKCTQWCWYPA | 5 (9) | |||||
| P19 | 127–140 | WCWYPANINTESG | 7 (13) | |||||
| P20 | 134–147 | NINTESGRAGKIKI | 2 (4) | |||||
| P21 | 141–154 | RAGKIKINCTETRA | 7 (13) | |||||
| P22 | 148–161 | NCTETRAVSCTEEM | 9 (16) | |||||
| P23 | 155–168 | VSCTEEMPLASIHR | 5 (9) | |||||
| P24 | 162–175 | PLASIHRVHWDKRD | 6 (11) | |||||
| P25 | 169–182 | VHWDKRDRESMAFM | 2 (4) | |||||
| P26 | 176–189 | RESMAFMNIKACNE | 4 (7) | |||||
| P27 | 183–196 | NIKACNENLRCQKR | 5 (9) | |||||
| P28 | 190–203 | NLRCQKRPGGCIKG | 15 (27) | |||||
| P29 | 197–210 | PGGCIKGYPIPVGA | 3 (5) | |||||
| P30 | 204–217 | YPIPVGAEIIPESL | 4 (7) | |||||
| P31 | 211–224 | EIIPESLKYLRGAK | 0 (0) | |||||
| P32 | 218–231 | KYLRGAKSQYGGIK | 19 (35) | |||||
| P33 | 225–238 | SQYGGIKDKNGELK | 6 (11) | |||||
| P34 | 232–245 | DKNGELKLPLTTRV | 1 (2) | |||||
| P35 | 239–252 | LPLTTRVWVKLANV | 2 (4) | |||||
| P36 | 246–259 | WVKLANVSAWTNGT | 1 (2) | |||||
| P37 | 253–266 | SAWTNGTPPYWGNR | 2 (4) | |||||
| P38 | 260–273 | PPYWGNRINGSKGI | 7 (13) | |||||
| P39 | 267–280 | INGSKGINGTLWGE | 5 (9) |
| P40 | 274–287 | NGTLWGELKGMHHL | 3 (5) |
| P41 | 281–294 | LKGMHHLGFALERG | 1 (2) |
| P42 | 288–301 | GFALERGGKWCNFT | 3 (5) |
| P43 | 295–308 | GKWCNFTGKIRIGQ | 4 (7) |
| P44 | 302–315 | GKIRIGQETFSYPY | 2 (4) |
| P45 | 309–322 | ETFSYPYKPSWNCT | 6 (11) |
| P46 | 316–329 | KPSWNCTGNWTQYP | 4 (7) |
| P47 | 323–336 | GNWTQYPVWQVLRH | 1 (2) |
| P48 | 330–343 | VWQVLRHLDMVEHM | 2 (4) |
| P49 | 337–350 | LDMVEHMTGECVQR | 6 (11) |
| P50 | 344–357 | TGECVQRPERHNIT | 10 (18) |
| P51 | 351–364 | PERHNITVGNGTIT | 5 (9) |
| P52 | 358–371 | VGNGTITGNCSATN | 10 (18) |
| P53 | 365–378 | GNCSATNWDGCNCS | 12 (22) |
| P54 | 372–385 | WDGCNCSRSGNYLY | 11 (20) |
| P55 | 379–392 | RSGNYLYNSSDGGL | 5 (9) |
| P56 | 386–399 | NSSDGGLLLIICRQ | 15 (27) |
| P57 | 393–406 | LLIICRQNKTITRI | 7 (13) |
| P58 | 400–413 | NKTITRIMGTKTNW | 11 (20) |
| P59 | 407–420 | MGTKTNWTTMWGIY | 19 (35) |
| P60 | 414–427 | TTMWGIYKNCSGCE | 19 (35) |
| P61 | 421–434 | KNCSGCENATLDRT | 8 (16) |
| P62 | 428–441 | NATLDRTGTGTLGN | 7 (13) |
| P63 | 435–448 | GTGTLGNVANINCS | 10 (18) |
| P64 | 442–455 | VANINCSLPHKNET | 16 (29) |
| P65 | 449–462 | LPHKNETNRWTCAP | 10 (18) |
| P66 | 456–469 | NRWTCAPRQRDGRR | 12 (22) |
| P67 | 463–476 | RQRDGRRDSLYIAG | 9 (16) |
| P68 | 470–483 | DSLYIAGGKNFWER | 11 (20) |
| P69 | 477–490 | GKNFWERVKAQYSC | 7 (13) |
| P70 | 484–497 | VKAQYSCESNIGEL | 1 (2) |
| P71 | 491–504 | ESNIGELDGMMHQQ | 2 (4) |
| P72 | 498–511 | DGMMHQQILLQKYQ | 1 (2) |
| P73 | 505–518 | ILLQKYQVIRVKAY | 9 (16) |
| P74 | 512–525 | VIRVKAYTYGVIEM | 37 (67) |
| P75 | 519–532 | TYGVIEMPKNYEQI | 46 (84) |
| P76 | 526–539 | PKNYEQINLKKRDL | 32 (58) |
| P77 | 533–546 | NLKKRDLSHKRKKR | 21 (38) |
The amino acid numbers start with the mature SU amino-terminal Glu as residue 1 (29).
Linear immunogenic sites have tentatively been assigned to the underlined amino acids.
Serological reactivity of synthetic peptides against a panel of 55 goat immune sera.
FIG. 4.
Reactivity profiles of ELISA-positive immune sera to immunogenic CAEV SU synthetic peptides. Each graph represents ELISA reactivities to one peptide or to different peptides as indicated. Each bar in the different serum reactivity profiles represents the ELISA reactivity (absorbance value/cutoff [A405/C.O.]) of one positive immune serum to an individual peptide. The number of positive immune sera to an individual immunodominant peptide or to combined immunoreactive peptides is indicated below each panel.
In summary, results from combined analyses of variability and immunogenicity of CAEV SU have revealed significant structural similarities between the SU of CAEV and other lentiviruses, despite the lack of substantial sequence homology. Comparison of the SU variation profile of CAEV with that of HIV-1 (40, 59) revealed that analogous conserved and variable domains exist between the respective SU glycoproteins of the two lentiviruses. Five variable regions and four conserved regions were identified in the CAEV SU. We found that conserved C1 (peptide 1) and C4 (peptide 74) regions located at the amino and carboxy termini, respectively, of CAEV SU contain immunodominant determinants displaying 67% reactivity with immune sera from natural infections. Examination of caprine humoral immune reactivities allowed the identification of a distinct immunodominant epitope (peptide 75, residues 526 to 532) in the carboxy terminus of SU, located in the V5 region just downstream from the immunodominant epitope (residues 519 to 525) of the C4 region. This epitope reacted with 84% of caprine immune sera tested. Comparison of amino acid sequences in the V5 region revealed a high sequence homology among French isolates whereas sequences greatly differed between American and French isolates, suggesting that this immunogenic determinant is conserved between geographically linked CAEV isolates, a situation reinforced by extensive commercial exchanges of infected animals among neighboring regions. This is further supported by the inability of immune sera from goats experimentally infected by the American Cork strain to react with peptides P75 and P76 (data not shown).
As for the HIV-1 SU, the first conserved region of CAEV SU was followed by two contiguous hypervariable regions, V1 and V2. These variable domains and the V3 region do not contain group-specific epitopes, as is also the case for the analogous regions of HIV-1 SU (41, 45), which are the targets of type-specific neutralizing antibodies (12, 15, 39, 45). The large V4 variable region of CAEV SU shows several striking features. Firstly, most of the antigenic determinants identified in this study are located within this region. Secondly, the glycosylation pattern of CAEV SU is not uniformly distributed but mapped essentially in the C3-V4 region, which also contains a high concentration of conserved cysteine residues. Finally, the V4 region of CAEV SU is in an analogous position to the V4-V5 region of SIV and FIV (47), which are the major targets of neutralizing antibodies (21, 55). In the case of these two lentiviruses, amino acid mutations in these regions of envelope protein allow the virus to escape from neutralization (27, 56). Recently, a type-specific discontinuous neutralization epitope has been identified in the V4 region of MVV SU (57). The neutralization phenotype was found to map within 39 amino acids corresponding to the immunogenic region encompassing peptides 64 to 68 (residues 442 to 483) in the V4 region of CAEV SU. Taken together, these observations suggest that the V4 region of CAEV SU is a highly conformational and well-exposed immunogenic domain which could be involved in the emergence of neutralization escape variants. Furthermore, it has been shown that the extensive glycosylation of CAEV SU, which accounts for nearly 50% of the total molecular weight of the protein, contributes to the inaccessibility of the SU epitopes to neutralizing antibodies (20). This may explain, at least in part, the apparent low neutralizing antibody titers developed by some CAEV-infected goats (7, 8, 36). In contrast, sheep infected by MVV develop relatively high neutralizing antibody titers against SU epitopes. Interestingly, one of two perfectly conserved N-linked glycosylation sites in CAEV sequences (amino acids 533 and 540 in Fig. 2) is missing in the equivalent MVV region containing the major neutralization epitope (57), suggesting that conservation of a relatively high number of N-linked glycosylation sites in the V4 region of CAEV SU would allow the virus to escape the host neutralizing immune response.
Using a pepscan analysis, several studies have demonstrated that the terminal ends of SU glycoproteins of HIV-1 (16, 46) and equine infectious anemia virus (1) also contain highly conserved, immunodominant linear B epitopes. It appears that extremities of these lentiviral SU glycoproteins are exposed and immunogenic in their native state. We have found that peptide 1 and particularly peptide 75 exhibited a much higher level of serological reactivity than did all of the other CAEV SU peptides tested. In the CAEV infection model, the results of several studies have demonstrated a direct correlation between the level of anti-Env antibodies and the development of arthritic lesions (3, 22, 30, 35), together with high virus loads in synovial tissue of CAEV-infected goats (8, 25, 28). Particularly, increased anti-CAEV SU antibody titers and a dominant population of SU-reactive T-helper 2-like lymphocytes are associated with disease progression (6, 48, 64). In vaccine experiments, immunization with inactivated CAEV resulted in exacerbation of arthritic lesions after in vivo challenge (37). This concern has been described after immunization with viral envelope glycoprotein subunit vaccines in other lentiviral vaccine models such as SIV, FIV, and equine infectious anemia virus (53, 54, 63). Finally, an enhancement of viral binding and entry into macrophages, the principal target cells in vivo, have been observed in vitro by using nonneutralizing antibodies to CAEV (23). These observations, together with our results, suggest that conserved immunodominant epitopes located at both amino and carboxy termini of CAEV SU may contribute to the enhancement of virus replication and disease. The localization of one of the main immunodominant domains (P75), about 10 amino acids upstream from the SU-TM cleavage site, seems very intriguing. A parallel can be established between this situation and the role of a glycosylation site located about 10 amino acids upstream from the influenza virus hemagglutinin cleavage site; it has been reported that hemagglutinin cleavability and influenza virus virulence are directly related to the nature of the sequence at this site (18, 26). It could be proposed that in CAEV-infected goats, the level of recognition of the immunodominant site by antibodies would have a direct effect on the SU-TM cleavability and consequently on CAEV virulence and pathogenicity. Further analyses would be necessary to evaluate the role of the immune response against this immunodominant site during the course of natural CAEV infection.
As previously mentioned, a dominant humoral immune response to viral envelope glycoproteins is a general feature of CAEV infection. In addition, experimental infection studies have revealed a characteristic evolution of antibody responses towards CAEV antigens during the course of viral infection (3). Sera of infected goats recognized both SU and Gag proteins during the first weeks postinfection, and then anti-Gag antibody titers decreased slowly whereas anti-TM antibodies appeared much later. In contrast, anti-SU antibodies were maintained over time. Similar kinetics of antibody responses have been observed in experimentally MVV-infected sheep (24). Thus, SU appears to be an obvious candidate for diagnostic assays of CAEV infection. Our results revealed the existence of conserved immunodominant epitopes in the SU which could be used as potential diagnostic peptide antigens. No individual peptide was reactive with all of the CAEV-infected goat sera tested, with the highest sensitivity being 84%. However, the combination of only five peptides improved the sensitivity of CAEV antibody detection, raising it to 96.4%. In addition, the overall divergence between CAEV isolates of distant geographical regions did not exceed 20%, which is approximately 2.5 times lower than that observed for HIV-1 isolates (31), and would not drastically impede serological diagnostic tests. Finally, CAEV and MVV are closely related viruses having >60% amino acid homology and antigenically cross-reactive structural proteins (14, 50). Earlier experimental evidence, demonstrating that sheep can be infected with CAEV and that goats can be infected with MVV (2), has supported the possibility of cross-species transmission. More recently, phylogenetic analyses from partial nucleotide sequences of pol and/or env genes of European and American ovine lentiviruses have led to the suggestion that these ovine isolates may have originated from a CAEV-like virus (5, 32, 66). Therefore, the inability of current serological methods to differentiate between the two small ruminant lentiviruses (SRLVs) has markedly limited the interpretation of results obtained in seroepidemiological studies (51). Our results on diversity and immunogenicity of CAEV SU provide potential peptide substrates for the design of not only group-specific serological tests but also group-cross-reactive assays. The immunodominant epitope located at the amino terminus of CAEV SU (amino acids 1 to 14) that differs by 50% in amino acid sequence from MVV SU, would be used as a group-specific antigen. In contrast, the immunodominant epitope located at the carboxy terminus of the protein (amino acids 512 to 525) that differs by only one amino acid substitution between the two viruses could be used for the detection of all SRLVs. Thus, it would be interesting to conduct a seroepidemiological survey using such immunodominant synthetic peptides to obtain a comprehensive view on the relationships between the two SRLV clusters, i.e., CAEV and MVV.
Nucleotide sequence accession numbers.
EMBL accession no. AJ400718 to AJ400721 have been assigned to the SU nucleotide sequences of CAEV isolates 680, 021, 032, and 786, respectively.
Acknowledgments
We thank Thierry Vidard for excellent technical assistance and Bernadette Trentin and Kathryn Mayo for critically reviewing the English usage of the manuscript.
This work was supported in part by grants from the ANRS and the Establissements Publics Régionaux d'Aquitaine et de Poitou-Charentes.
REFERENCES
- 1.Ball J M, Rushlow K E, Issel C J, Montalero R C. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J Virol. 1992;66:732–742. doi: 10.1128/jvi.66.2.732-742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banks K L, Adams D S, McGuire T C, Carlson J. Experimental infection of sheep by caprine arthritis-encephalitis virus and goats by progressive pneumonia virus. Am J Vet Res. 1983;44:2307–2311. [PubMed] [Google Scholar]
- 3.Bertoni G, Zahno M L, Zanoni R, Vogt H R, Peterhans E, Ruff G, Cheevers W P, Sonigo P, Pancino G. Antibody reactivity to the immunodominant epitopes of the caprine arthritis-encephalitis virus gp38 transmembrane protein associates with the development of arthritis. J Virol. 1994;68:7139–7147. doi: 10.1128/jvi.68.11.7139-7147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpino L A, Han G H. The 9-fluoronylmethoxycarbonyl function, a new base sensitive amino protecting group. J Org Chem. 1970;37:5748–5749. [Google Scholar]
- 5.Chebloune Y, Karr B, Sheffer D, Leung K, Narayan O. Variations in lentiviral gene expression in monocyte-derived macrophages from naturally infected sheep. J Gen Virol. 1996;77:2037–2051. doi: 10.1099/0022-1317-77-9-2037. [DOI] [PubMed] [Google Scholar]
- 6.Cheevers W P, Beyer J C, Knowles D P. Type 1 and type 2 cytokine expression by viral gp135 surface protein-activated T lymphocytes in caprine arthritis-encephalitis lentivirus infection. J Virol. 1997;71:6259–6263. doi: 10.1128/jvi.71.8.6259-6263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheevers W P, McGuire T C, Norton L K, Cordery-Cotter R, Knowles D P. Failure of neutralizing antibody to regulate CAE lentivirus expression in vivo. Virology. 1993;196:835–839. doi: 10.1006/viro.1993.1542. [DOI] [PubMed] [Google Scholar]
- 8.Cheevers W P, Knowles D P, Norton L K. Neutralization-resistant antigenic variants of caprine arthritis-encephalitis lentivirus associated with progressive arthritis. J Infect Dis. 1991;164:679–685. doi: 10.1093/infdis/164.4.679. [DOI] [PubMed] [Google Scholar]
- 9.Cheevers W P, McGuire T C. The lentiviruses: maedi/visna, caprine arthritis-encephalitis, and equine infectious anemia. Adv Virus Res. 1988;34:189–215. doi: 10.1016/s0065-3527(08)60518-7. [DOI] [PubMed] [Google Scholar]
- 10.Emini E A, Putney S D. Human immunodeficiency virus. Bio/Technology. 1992;20:309–326. doi: 10.1016/b978-0-7506-9265-6.50019-1. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. PHYLIP (phylogeny interference package) version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 12.Fung M S C, Sun C R Y, Gordon W L, Liou R-S, Chang T W, Sun W N C, Daar E S, Ho S D. Identification and characterization of a neutralization site within the second variable region of human immunodeficiency virus type 1 gp120. J Virol. 1992;66:848–856. doi: 10.1128/jvi.66.2.848-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gendelman H E, Narayan O, Molineau S, Clements J E, Ghotbi Z. Slow persistent replication of lentiviruses: role of tissue macrophages and macrophage precursors in bone marrow. Proc Natl Acad Sci USA. 1985;82:7086–7090. doi: 10.1073/pnas.82.20.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gogolewski R P, Adams D S, McGuire T C, Banks K L, Cheevers W P. Antigenic cross-reactivity between caprine arthritis-encephalitis, visna and progressive pneumonia viruses involves all virion-associated proteins and glycoproteins. J Gen Virol. 1985;66:1233–1240. doi: 10.1099/0022-1317-66-6-1233. [DOI] [PubMed] [Google Scholar]
- 15.Goudsmit J, Debouck C, Meloen R H, Smith L, Bakker M, Asher D M, Wolff A V, Gibbs C J, Gajdusek D C. Human immunodeficiency virus type 1 neutralization epitope with conserved architecture elicits early type-specific antibodies in experimentally infected chimpanzees. Proc Natl Acad Sci USA. 1988;85:4478–4482. doi: 10.1073/pnas.85.12.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goudsmit J, Boucher C A B, Meloen R H, Epstein L G, Smit L, Van der Hoek L, Bakker M. Human antibody response to a strain-specific HIV-1 gp120 epitope associated with cell fusion inhibition. AIDS. 1988;2:157–164. [PubMed] [Google Scholar]
- 17.Harmache A, Vitu C, Guiguen F, Russo P, Bertoni G, Pepin M, Vigne R, Suzan M. Priming with tat-deleted caprine arthritis-encephalitis virus (CAEV) proviral DNA or live virus protects goats from challenge with pathogenic CAEV. J Virol. 1998;72:6796–6804. doi: 10.1128/jvi.72.8.6796-6804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horimoto T, Kawaoka Y. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J Virol. 1994;68:3120–3128. doi: 10.1128/jvi.68.5.3120-3128.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S L. Recombinant subunit vaccines against primate lentiviruses. AIDS Res Hum Retrovir. 1996;12:451–453. doi: 10.1089/aid.1996.12.451. [DOI] [PubMed] [Google Scholar]
- 20.Huso D L, Narayan O, Hart G W. Sialic acids on the surface of caprine arthritis encephalitis virus define the biological properties of the virus. J Virol. 1988;62:1974–1980. doi: 10.1128/jvi.62.6.1974-1980.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Javaherian K, Langlois A J, Schmidt S, Kaufmann M, Cates N, Langedijk J P M, Meloen R H, Desrosiers R C, Burns D P W, Bolognesi D P, LaRosa G J, Putney S D. The principal neutralization determinant of simian immunodeficiency virus differs from that of human immunodeficiency virus. Proc Natl Acad Sci USA. 1992;89:1418–1422. doi: 10.1073/pnas.89.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson G C, Barbet A F, Klevjer-Anderson P, McGuire T C. Preferential immune response to virion surface glycoproteins by caprine arthritis-encephalitis virus-infected goats. Infect Immun. 1983;41:657–665. doi: 10.1128/iai.41.2.657-665.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolly P E, Huso D, Hart G, Narayan O. Modulation of lentivirus replication by antibodies. Non-neutralizing antibodies to caprine arthritis-encephalitis virus enhance early stages of infection in macrophages, but do not cause increased production of virions. J Gen Virol. 1989;70:2221–2226. doi: 10.1099/0022-1317-70-8-2221. [DOI] [PubMed] [Google Scholar]
- 24.Juste R A, Kwang J, de la Concha-Bermejillo A. Dynamics of cell-associated viremia and antibody response during the early phase of lentivirus infection in sheep. Am J Vet Res. 1998;59:563–568. [PubMed] [Google Scholar]
- 25.Jutila M A, Banks K L. Increased macrophage division in the synovial fluid of goats infected with caprine arthritis-encephalitis virus. J Infect Dis. 1988;157:1193–1202. doi: 10.1093/infdis/157.6.1193. [DOI] [PubMed] [Google Scholar]
- 26.Kawaoka Y, Webster R G. Interplay between carbohydrate in the stalk and the length of the connecting peptide determines the cleavability of influenza virus hemagglutinin. J Virol. 1989;63:3296–3300. doi: 10.1128/jvi.63.8.3296-3300.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinsey N E, Anderson M G, Unangst T J, Joag S V, Narayan O, Zink M C, Clements J E. Antigenic variation of SIV: mutations in V4 alter the neutralization profile. Virology. 1996;221:14–21. doi: 10.1006/viro.1996.0348. [DOI] [PubMed] [Google Scholar]
- 28.Klevjer-Anderson P, Adams D S, Anderson L W, Banks K L, McGuire T C. A sequential study of virus expression in retrovirus-induced arthritis of goats. J Gen Virol. 1984;65:1519–1525. doi: 10.1099/0022-1317-65-9-1519. [DOI] [PubMed] [Google Scholar]
- 29.Knowles D, Cheevers W, McGuire T, Brassfield A, Hardwood W, Stem T. Structure and genetic variability of envelope glycoproteins of two antigenic variants of caprine arthritis-encephalitis lentivirus. J Virol. 1991;65:5744–5750. doi: 10.1128/jvi.65.11.5744-5750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knowles D J, Cheevers W, McGuire T, Stem T, Gorham J. Severity of arthritis is predicted by antibody response to gp135 in chronic infection with caprine arthritis encephalitis virus. J Virol. 1990;64:2396–2398. doi: 10.1128/jvi.64.5.2396-2398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korber B T, Allen E E, Farmer A D, Myers G L. Heterogeneity of HIV-1 and HIV-2. AIDS. 1995;9(Suppl. A):S5–S18. [PubMed] [Google Scholar]
- 32.Leroux C, Chastang J, Greenland T, Mornex J F. Genomic heterogeneity of small ruminant lentiviruses: existence of heterogeneous population in sheep and of the same lentiviral genotypes in sheep and goats. Arch Virol. 1997;142:1125–1137. doi: 10.1007/s007050050147. [DOI] [PubMed] [Google Scholar]
- 33.Letvin N L. Animal models for the study of human immunodeficiency virus infections. Curr Opin Immunol. 1992;4:481–485. doi: 10.1016/s0952-7915(06)80043-9. [DOI] [PubMed] [Google Scholar]
- 34.Lutz H, Hofmann-Lehmann R, Leutenegger C, Allenspach K, Cuisinier A M, Cronier J, Duquesne V, Aubert A. Vaccination of cats with recombinant envelope glycoprotein of feline immunodeficiency virus: decreased viral load after challenge infection. AIDS Res Hum Retrovir. 1996;12:431–433. doi: 10.1089/aid.1996.12.431. [DOI] [PubMed] [Google Scholar]
- 35.McGuire T C, Knowles D, David W, Brassfield A, Stem T, Cheevers W. Transmembrane protein oligomers of caprine arthritis-encephalitis lentivirus are immunodominant in goats with progressive arthritis. J Virol. 1992;66:3247–3250. doi: 10.1128/jvi.66.5.3247-3250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGuire T C, Norton L K, O'Rourke K I, Cheevers W P. Antigenic variation of neutralization sensitive epitopes of caprine arthritis-encephalitis lentivirus during persistent infection. J Virol. 1988;62:3488–3492. doi: 10.1128/jvi.62.9.3488-3492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGuire T C, Adams D S, Johnson G C, Klevjer-Anderson P, Barbee D D, Gorham J R. Acute arthritis in caprine arthritis-encephalitis virus challenge exposure of vaccinated or persistently infected goats. Am J Vet Res. 1986;47:537–540. [PubMed] [Google Scholar]
- 38.McKeating J A, Balfe P. The role of the viral glycoprotein in HIV-1 persistence. Immunol Lett. 1999;65:63–70. doi: 10.1016/s0165-2478(98)00126-6. [DOI] [PubMed] [Google Scholar]
- 39.McKeating J A, Shotten C, Cordell J, Graham S, Balfe P, Sullivan N, Charles M, Page M, Bolmstedt A, Olofsson S, Kayman S C, Wu Z, Pinter A, Dean C, Sodroski J, Weiss R A. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J Virol. 1993;67:4932–4944. doi: 10.1128/jvi.67.8.4932-4944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modrow S, Hahn B H, Shaw G M, Gallo R C, Wong-Staal F, Wolf H. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J Virol. 1987;61:570–578. doi: 10.1128/jvi.61.2.570-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore J P, Sattentau Q, Yoshiyama H, Thali M, Charles M, Sullivan N, Poon S-W, Fung M S, Traincard F, Robinson J E, Ho D D, Sodroski J. Probing the structure of the V2 domain of the human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: the human immune response to the V1 and V2 domains. J Virol. 1993;67:6136–6151. doi: 10.1128/jvi.67.10.6136-6151.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narayan O, Cork L C. Lentiviral diseases of sheep and goats: chronic pneumonia, leukoencephalomyelitis and arthritis. Rev Infect Dis. 1985;7:89–98. doi: 10.1093/clinids/7.1.89. [DOI] [PubMed] [Google Scholar]
- 43.Narayan O, Kennedy-Stoskopf S, Sheffer D, Griffin D E, Clements J E. Activation of caprine arthritis-encephalitis virus expression during maturation of monocytes to macrophages. Infect Immun. 1983;41:67–73. doi: 10.1128/iai.41.1.67-73.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narayan O, Wolinsky J, Clements J, Strandberg J, Griffin D, Cork L. Slow virus replication: the role of macrophages in the persistence and expression of visna viruses of sheep and goats. J Gen Virol. 1982;59:345–356. doi: 10.1099/0022-1317-59-2-345. [DOI] [PubMed] [Google Scholar]
- 45.Palker T J, Clark M E, Langlois A J, Matthews T J, Weinhold K J, Randall R R, Bolognesi D P, Haynes B F. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci USA. 1988;85:1932–1936. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palker T J, Matthews T J, Clark M E, Cianciolo G C, Randall R R, Langlois A J, White G C, Safai B, Snyderman R, Bolognesi D P, Haynes B F. A conserved region at the COOH terminus of human immunodeficiency virus gp120 envelope protein contains an immunodominant epitope. Proc Natl Acad Sci USA. 1987;84:2479–2483. doi: 10.1073/pnas.84.8.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pancino G, Ellerbrok H, Sitbon M, Sonigo P. Conserved framework of envelope glycoproteins among lentiviruses. Curr Top Microbiol Immun. 1993;188:77–100. doi: 10.1007/978-3-642-78536-8_5. [DOI] [PubMed] [Google Scholar]
- 48.Perry L L, Wilkerson M J, Hullinger G A, Cheevers W P. Depressed CD4+ T lymphocyte proliferative response and enhanced antibody response to viral antigen in chronic lentivirus-induced arthritis. J Infect Dis. 1995;171:328–334. doi: 10.1093/infdis/171.2.328. [DOI] [PubMed] [Google Scholar]
- 49.Quérat G, Audoly G, Sonigo P, Vigne R. Nucleotide sequence analysis of SA-OMVV, a visna-related ovine lentivirus: phylogenetic history of lentiviruses. Virology. 1990;175:434–447. doi: 10.1016/0042-6822(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 50.Saltarelli M, Quérat G, Konings D A, Vigne R, Clements J E. Nucleotide sequence and transcriptional analysis of molecular clones of CAEV which generate infectious virus. Virology. 1990;179:347–364. doi: 10.1016/0042-6822(90)90303-9. [DOI] [PubMed] [Google Scholar]
- 51.Saman E, Eynde G V, Lujan L, Extramiana B, Harkiss G, Tolari F, Gonzalez L, Amorena B, Watt N, Badiola J. A new serological assay for detection of lentivirus infections in small ruminants. Clin Diagn Lab Immun. 1999;6:734–740. doi: 10.1128/cdli.6.5.734-740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sargan D R, Bennet I D, Cousens C, Roy D J, Blacklaws B A, Dalziel R G, Watt N J, McConnell I. Nucleotide sequence of EV1, a British isolate of maedi-visna virus. J Gen Virol. 1991;72:1893–1903. doi: 10.1099/0022-1317-72-8-1893. [DOI] [PubMed] [Google Scholar]
- 53.Schlienger K, Montefiori D C, Mancini M, Riviere Y, Tiollais P, Michel M L. Vaccine-induced neutralizing antibodies directed in part to the simian immunodeficiency virus (SIV) V2 domain were unable to protect rhesus monkeys from SIV experimental challenge. J Virol. 1994;68:6578–6588. doi: 10.1128/jvi.68.10.6578-6588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siebelink K H, Tijhaar E, Huisman R C, de Ronde A, Darby I H, Francis M J, Rimmelzwaan G F, Osterhaus A D M E. Enhancement of feline immunodeficiency virus infection after immunization with envelope glycoprotein subunit vaccines. J Virol. 1995;69:3704–3711. doi: 10.1128/jvi.69.6.3704-3711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siebelink K H J, Huisman W, Karlas J A, Rimmelzwaan G F, Bosch M L, Osterhaus A D M E. Neutralization of feline immunodeficiency virus by polyclonal feline antibody: simultaneous involvement of hypervariable regions 4 and 5 of the surface glycoprotein. J Virol. 1995;69:5124–5127. doi: 10.1128/jvi.69.8.5124-5127.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siebelink K H J, Rimmelzwaan G F, Bosch M L, Meloen R H, Osterhaus A D M E. A single amino acid substitution in hypervariable region 5 of the envelope protein of feline immunodeficiency virus allows escape from virus neutralization. J Virol. 1993;67:2202–2208. doi: 10.1128/jvi.67.4.2202-2208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skraban R, Matthiasdottir S, Torsteinsdottir S, Agnarsdottir G, Gudmundsson B, Georgsson G, Meloen R H, Andresson O S, Staskus K A, Thormar H, Andresdottir V. Naturally ocurring mutations within 39 amino acids in the envelope glycoprotein of maedi-visna virus alter the neutralization phenotype. J Virol. 1999;73:8064–8072. doi: 10.1128/jvi.73.10.8064-8072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonigo P, Alizon M, Staskus K, Klatzmann D, Cole S, Danos O, Retzel E, Tiollais P, Haase A, Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985;42:369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- 59.Starcich B R, Hahn B H, Shaw G M, McNeely P D, Modrow S, Wolf H, Parks W P, Josephs S F, Gallo R C, Wong-Staal F. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 60.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valas S, Benoit C, Guionaud C, Perrin G, Mamoun R Z. North American and French caprine arthritis-encephalitis viruses emerge from ovine maedi-visna viruses. Virology. 1997;237:307–318. doi: 10.1006/viro.1997.8800. [DOI] [PubMed] [Google Scholar]
- 62.Wain-Hobson S, Sonigo P, Guyader M, Gazit A, Henry M. Erratic G to A hypermutation within complete caprine arthritis-encephalitis virus (CAEV) provirus. Virology. 1995;209:297–303. doi: 10.1006/viro.1995.1261. [DOI] [PubMed] [Google Scholar]
- 63.Wang S Z-S, Rushlow K E, Issel C J, Cook R F, Cook S J, Raabe M L, Chong Y-H, Costa L, Montelaro R C. Enhancement of EIAV replication and disease by immunization with a baculovirus-expressed recombinant envelope surface glycoprotein. Virology. 1994;199:247–251. doi: 10.1006/viro.1994.1120. [DOI] [PubMed] [Google Scholar]
- 64.Wilkerson M J, Davis W C, Baszler T V, Cheevers W P. Immunopathology of chronic lentivirus-induced arthritis. Am J Pathol. 1995;146:1433–1443. [PMC free article] [PubMed] [Google Scholar]
- 65.Zanoni R G. Phylogenetic analysis of small ruminant lentiviruses. J Gen Virol. 1998;79:1951–1961. doi: 10.1099/0022-1317-79-8-1951. [DOI] [PubMed] [Google Scholar]
- 66.Zanoni R G, Nauta I M, Kuhnert P, Pauki U, Pohl B, Peterhans E. Genomic heterogeneity of small ruminant lentiviruses detected by PCR. Vet Microbiol. 1992;33:341–351. doi: 10.1016/0378-1135(92)90061-w. [DOI] [PubMed] [Google Scholar]