Abstract
Summary
Artificial intelligence (AI) is increasingly used in genomics research and practice, and generative AI has garnered significant recent attention. In clinical applications of generative AI, aspects of the underlying datasets can impact results, and confounders should be studied and mitigated. One example involves the facial expressions of people with genetic conditions. Stereotypically, Williams (WS) and Angelman (AS) syndromes are associated with a “happy” demeanor, including a smiling expression. Clinical geneticists may be more likely to identify these conditions in images of smiling individuals. To study the impact of facial expression, we analyzed publicly available facial images of approximately 3500 individuals with genetic conditions. Using a deep learning (DL) image classifier, we found that WS and AS images with non-smiling expressions had significantly lower prediction probabilities for the correct syndrome labels than those with smiling expressions. This was not seen for 22q11.2 deletion and Noonan syndromes, which are not associated with a smiling expression. To further explore the effect of facial expressions, we computationally altered the facial expressions for these images. We trained HyperStyle, a GAN-inversion technique compatible with StyleGAN2, to determine the vector representations of our images. Then, following the concept of InterfaceGAN, we edited these vectors to recreate the original images in a phenotypically accurate way but with a different facial expression. Through online surveys and an eye-tracking experiment, we examined how altered facial expressions affect the performance of human experts. We overall found that facial expression is associated with diagnostic accuracy variably in different genetic conditions.
1 Introduction
Artificial intelligence (AI) applications have made important inroads in many areas of biomedicine, including within the field of medical genetics (Schaefer et al., 2020; Hanchard et al., 2022). One important example in medical genetics involves the use of deep learning (DL) to examine photos of people with suspected genetic conditions to help determine what condition they may have (Hsieh et al., 2022). Despite the individual rarity of most genetic conditions and the fact that datasets are relatively small, leveraging pre-trained models has allowed robust performance (and rapid adoption) of DL-based tools in this context (Duong et al., 2022b). This technological approach may help ameliorate issues like long wait times to see clinical genetics specialists, which arises primarily from a lack of expert clinicians relative to the prevalence of people with genetic conditions (Tekendo-Ngongang et al., 2020; DeGrave et al., 2021; Porras et al., 2021). While these types of DL-based tools have become popular in medical genetics, important questions arise about their performance, especially in the context of rare diseases. As one of many questions, it remains unclear how patient characteristics (e.g. patient age or ancestry, as well as other confounders and potential artifacts) may affect DL model results (Duong et al., 2022a). For example traditional textbooks may depict certain genetic conditions in stereotypical ways (which may not accurately reflect the real-life range of manifestations across affected individuals), and these depictions can bias computational models as well as human clinicians. These types of challenges can lead to delays in the ability to provide precise and accurate genetic diagnoses, which in turn are often necessary for optimal clinical care (Solomon et al., 2013; Incerti et al., 2022).
Human facial expressions represent a complex type of nonverbal communication that is essential for the display of emotions and social cues. Computationally assessing the subtleties of facial expressions has been an area of active research in DL, especially since the introduction of datasets and models such as the Facial Action Coding System (FACS), which has allowed for in-depth and wide-ranging studies (Ekman et al., 1976; Tian et al., 2001; Skiendziel et al., 2019; Rosenberg and Ekman, 2020; Correia-Caeiro et al., 2022). Although many investigations have been performed on facial expressions in general (Li and Deng, 2022; Peña et al., 2021; Zhi et al., 2021; Wehrli et al., 2022), there is still a significant gap in the medical domain, such as how facial expressions may be associated with specific genetic conditions. For example, people with Williams syndrome (WS) and Angelman syndrome (AS) are often portrayed in the medical literature as smiling and/or as having “happy” demeanors (Jones et al., 2021). While these observations may be based on astute clinical observation, this may also mean that diagnoses may be missed when a patient with one of these conditions is not smiling when assessed, or otherwise exhibits atypical characteristics (Tanzi and Bertram, 2001; Richmond et al., 2018). Our previous work (Duong et al., 2022a) suggested that clinician performance changes significantly with respect to the facial expressions of images of people with WS and 22q11.2 deletion syndrome (22q). For example, with WS, we found that clinicians were more likely to correctly suspect this condition for those patients with a smiling expression. We did not observe a similar phenomenon with 22q (see details in Supplementary Table 1). This is expected based on textbook explanations of these conditions, but as described above, can be a barrier to efficient diagnosis and appropriate medical care. As individuals are often diagnosed in medical situations, such as through a visit to a clinician or an inpatient admission, clinicians in our group (and those with whom we have discussed this question) have anecdotally reported that patients may be less prone to smiling in such scenarios, which may affect clinicians’ diagnostic assessments in light of what is expected based on textbook descriptions.
Following our preliminary observations, we wanted to expand this investigation by including other conditions besides WS and 22q. Moreover, since the above-mentioned cohorts, which were divided based on “smile versus no smile” expressions, were made up of different individuals, it is possible that one group of individuals was simply easier to identify via other syndromic factors besides the face expressions. Hence, in this article, we further conducted the experiments in a manner that allows us to better control for these potential confounders.
Specifically, we used Generative Adversarial Network inversion (GAN-inversion) technique to systematically modify the facial expressions of real individuals. Next, we assessed how the altered facial expressions affect the performance of a DL classifier and human clinicians. For human clinician assessment, we employed web-based surveys to evaluate how well clinicians identify individuals with genetic conditions, and eye-tracking experiments, which allow us to observe human visual attention with respect to specific facial regions in images of people with genetic conditions. We anticipate that these results can demonstrate how a specific confounder can influence a DL classifier and human clinicians in the assessment medical conditions. We hope this work can contribute to improved diagnosis and management of people and families affected by these disorders.
2 Methods
2.1 Data collection
Following our previous work (Duong et al., 2022a, 2022b) we identified and used publicly available images depicting individuals with four conditions: 22q (OMIM #611867), AS (OMIM #105830), Noonan syndrome (NS) (OMIM #163950, 605275, 609942, 610733, 611553, 613706, 615355, 616559, 616564, 619087), and WS (OMIM #194050). We selected these genetic conditions because they have recognizable facial features and are rare but still occur relatively frequently compared to many other genetic conditions. Additionally, we chose AS and WS because of the “textbook definitions” of the facial expressions of affected individuals. For example, individuals with AS or WS are often described as having a happy demeanor or a smiling expression, whereas individuals with 22q and NS are not typically depicted this way (Jones et al., 2021).
For each image, we documented reported age and gender when this information was provided by the source. We sought images of individuals with diverse ancestry, intentionally aiming for diverse representation. However, as the genomic ancestry details were not always available for all individuals, we did not want to assume a person’s specific genomic ancestry if not clearly provided.
We also used images of individuals with other genetic conditions to train the ResNet-50 image classifier and GAN image generator (see condition names in Table 1). These other conditions may exhibit similar syndromic features to our diseases of interest. Hence, having images of these other conditions may help us better classify and generate images of 22q, AS, NS, and WS.
Table 1.
Fivefold cross-validation performance of ResNet-50 based on the 3484 training images (excluding the 32 test images in the clinician survey and eye-tracking experiment).
| Precision | Recall | F1-score | Sample size | |
|---|---|---|---|---|
| 22q | 0.7198 | 0.8256 | 0.7691 | 585 |
| AS | 0.7847 | 0.7969 | 0.7908 | 453 |
| BWS | 0.6974 | 0.6156 | 0.6539 | 307 |
| CdLS | 0.7500 | 0.7804 | 0.7649 | 123 |
| Down | 0.8591 | 0.8615 | 0.8603 | 354 |
| KS | 0.8302 | 0.7364 | 0.7804 | 239 |
| NS | 0.7454 | 0.7620 | 0.7536 | 269 |
| PWS | 0.6039 | 0.5700 | 0.5865 | 107 |
| RSTS1 | 0.8152 | 0.6944 | 0.7500 | 108 |
| Unaffected | 0.6619 | 0.5840 | 0.6205 | 238 |
| WHS | 0.7446 | 0.7865 | 0.7650 | 178 |
| WS | 0.8790 | 0.8757 | 0.8773 | 523 |
| Average | 0.7576 | 0.7408 | 0.7477 | 3484 |
Accuracy metrics are averaged across the five folds. The average top-1 accuracy over all the diseases is 0.7715 (not shown table).
Abbreviations: 22q, 22q11.2 deletion syndrome; AS, Angelman syndrome; BWS, Beckwith–Wiedemann syndrome; CdLS, Cornelia de Lange syndrome; Down, Down syndrome; KS, Kabuki syndrome; NS, Noonan syndrome; PWS, Prader–Willi syndrome; RSTS1, Rubinstein–Taybi syndrome type 1; WHS, Wolf–Hirschhorn syndrome; WS, Williams syndrome.
In total, we collected 3528 facial images of individuals affected with 11 genetic conditions, as well as images of unaffected individuals, including 590 22q images, 456 AS images, 327 NS images, and 529 WS images. We set aside 32 images (eight for each of these four genetic conditions) so that we could evaluate these images in the human assessment experiments. These 32 test images were chosen showing individuals from 2 to 20 years of age since optimal medical management depends on early clinical diagnosis, and as these conditions are all recognizable in childhood.
Twelve images were excluded due to lack of recognizable facial landmarks. This results in a dataset of 3484 (3528 minus 32 human test images and 12 images with unrecognizable landmarks) for training ResNet-50.
Our GAN image generator was trained with 3516 images (3528 minus 12 images with unrecognizable landmarks). We note that the 3516 includes the 32 test images. Our key objective was to train the image generator on these test images. This allows us to generate high-quality (i.e. realistic and clinically accurate) versions of these test images with different facial expressions.
To summarize, images in this study were either (1) the real images, (2) the reconstructed versions of these real images done via GAN-inversion, which we will refer to as “reconstructed images” (see Section GAN-inversion), and (3) the reconstructed images with modified facial expressions which we will refer to as “expression-manipulated images”. Reconstructed images primarily depicted individuals with 22q, AS, NS, or WS having the typical facial expressions of these syndromes (i.e. AS and WS with a smiling expression and 22q and NS with a non-smiling expression). Expression-manipulated images were computationally altered to have the opposite facial expressions (i.e. AS and WS with a non-smiling expression and 22q and NS with a smiling expression).
2.2 Syndromic image classifier
To classify the images with respect to their genetic conditions, we based our ResNet-50 classifier on the approach of (Sümer et al., 2023). This ResNet-50 classifier was pre-trained on the VGG Face-2 dataset (Cao et al., 2018b; Krawitz et al., 2021; Sümer et al., 2023). Our training dataset size for the fivefold cross-validation is 3484 images (as explained in the Data Collection section of the Methods).
For data preprocessing, all images were rescaled to 224 × 224 pixel resolution. Next, we followed the StyleGAN2 approach to standardize image sizes and facial landmark alignment (Abdal et al., 2019). We selected StyleGAN2 data preprocessing because it was also used by the GAN-inversion approach HyperStyle (see next section).
For the fivefold cross-validation, we trained and evaluated ResNet-50 five times, each time using a different fold as the validation set and the remaining four folds as the training set. We followed the hyperparameter setting in Sümer et al. (2023), and used 32 batch size and SGD optimizer with learning rate 0.001, momentum 0.9, and Cosine annealing warm start. We trained for 35 epochs (about 5 h on one p100 GPU) and selected the saved model with the best validation loss out of these epochs.
For a test image, the predicted probability is computed by averaging the outcome across all folds. The training and inference script for this ResNet-50 is available at our GitHub (https://github.com/pateltanvi2992/genetic-conditions-image-classifier.).
2.3 GAN-inversion and facial expression manipulation
A typical GAN image generator maps an input vector (usually randomly drawn from a normal distribution) into a new output image that looks realistic with respect to the training dataset. To reverse the process of the image generator, given an image of interest, one would need to solve for the vector that can recreate an output appearing very similar to this image of interest. We note that this image of interest can be either a fake or real image. This reverse process is known as GAN-inversion and its output is called the “reconstructed image” of the original image of interest (Alaluf et al., 2022). By solving for the vector that can be used to reconstruct the input image, GAN-inversion allows us to edit this image by manipulating its corresponding vector representation.
StyleGAN is one of several GAN architectures for creating high-quality synthetic images, including the ability to generate realistic and diverse human faces. Although StyleGAN can generate fake images, it cannot reliably change the face expressions of real images. We opted for the GAN-inversion approach HyperStyle, which has two main advantages over StyleGAN: (1) HyperStyle uses a hypernetwork to efficiently update StyleGAN weights according to our unique datasets, and (2) HyperStyle can edit the facial expressions of real images (Abdal et al., 2019; Von Oswald et al., 2019; Alaluf et al., 2022).
For human faces, almost all GAN-like methods are trained with datasets like Flickr-Faces-HQ (FFHQ) or CelebFaces Attribute (CelebA) (Guo et al., 2016; Cao et al., 2018a; Karras et al., 2019; Zhao et al., 2020). The original authors of HyperStyle trained their model on FFHQ, which does not contain faces of individuals affected with the genetic conditions as in our analysis. Hence, we could not reliably apply their pre-trained HyperStyle to reconstruct the images in our dataset. Initially, we finetuned HyperStyle on just our small dataset of 3516 images (see Data Collection section), but as this sample size is too small, finetuning with various hyperparameter options did not provide high-quality outputs.
For this reason, we combined our dataset with 70 000 FFHQ images. Specifically, we first randomly partitioned the 70 000 FFHQ images into 59 600 and 10 400 for training and validation, respectively. Next, we added our 3516 syndromic images into the training set. This yields a final training dataset of size 63 116 (from 59 600 and 3516) which enabled us to reliably train HyperStyle. Next, we followed the exact hyperparameter settings as described by the original authors of HyperStyle and trained their model from-scratch on the combined dataset. Our GitHub (https://github.com/pateltanvi2992/Analyzing-Facial-Expressions-with-Generative-AI-in-Clinical-Genetics/.). provides the training and inference script to reproduce the results in this article. Here, we briefly describe a few key hyperparameters. Following the setting in Alaluf et al. (2022) we used batch size 8 and the ranger optimizer (Wright, 2019) with learning rate 0.0001. We trained until the validation loss converged, which took 109 000 epochs and 10 days on one v100x GPU.
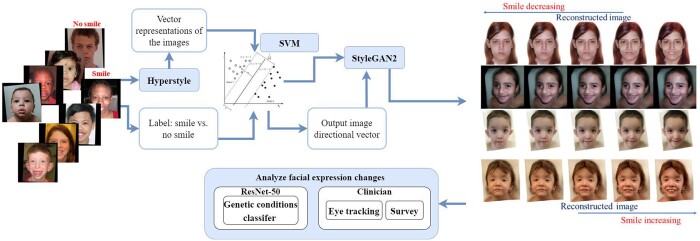
During the inference phase, we applied HyperStyle to obtain the vector representations of the original images, and then manipulated these vectors to create new face expressions for the original images (see Figure 1 and Supplementary Figure 1). That is, in the first step, we solve for the vector representations that can generate outputs (i.e. the reconstructed images) that look like the original images. In the second step, we manipulated the facial expressions of the reconstructed images to produce the expression-manipulated outputs.
Figure 1.
Manipulating facial expressions in images of people with genetic conditions via HyperStyle and StyleGAN2. We started with our real images, and manually classified these facial expressions into smile versus no smile. We finetuned HyperStyle, where the goal is to find an image embedding that can reconstruct the original input image (via StyleGAN2). Next, we manipulated the embedding of the reconstructed image by using SVM as shown in InterfaceGAN (Pisner and Schnyer, 2020). Finally, we passed the newly edited embedding into StyleGAN2 to create a new image with a new facial expression that still closely resembles the original input. On the far right, the examples illustrate the conditions we analyzed. All appropriate permissions have been obtained to reuse images and the citations for each are provided in Supplementary Table 5.
To alter the face expressions, we followed the concept of InterfaceGAN (Shen et al., 2020). After obtaining the vector representations of the reconstructed images, we applied Support Vector Machine (SVM) to approximate the “direction vector” that separates the “boundaries” between images with and without smiling expressions. This direction vector allowed us to change the facial expressions by modifying the vector representations of the reconstructed images; this step produces the expression-manipulated images. For each of the four genetic conditions, we found their own unique direction vector via their own unique SVM (Figure 1).
Subjectively, our own clinicians deemed the reconstructed images and their corresponding expression-manipulated outputs to be realistic representations of the given conditions. We further verified these reconstructed images and their expression-manipulated versions via an independent classifier built to analyze genetic conditions (Hsieh et al., 2022). This is an important validation as the presence of a genetic condition can influence the appearance of facial expressions beyond whether a person is smiling or not. However, reconstruction may still introduce undetected changes that may affect the ability of clinicians to recognize the conditions.
2.4 Clinician surveys and eye-tracking experiments
After generating expression-manipulated syndromic images, we compared the accuracy of human experts (clinical geneticists) on the reconstructions of the original images versus the expression-manipulated images via electronic surveys sent using Qualtrics (Provo, Utah, United States). Using the reconstructions of the original images would include GAN artifacts, which would also be found in the expression-manipulated images. Hence, this would allow for a fair comparison between smile and no smile images. We used two versions of the survey, allowing for a reconstructed image to be shown to one group and the corresponding expression-manipulated image to be shown to the other group. Each survey included 32 total images, with an equal number of reconstructed and expression-manipulated images. Participants were asked to classify each image as showing a person with either 22q, AS, NS, WS, or who was unaffected by a genetic condition. While there were no unaffected images included in the test set, we included this option in our surveys to investigate whether expression affected the participants’ perceptions of whether the image showed a person affected by a genetic condition or not. Example surveys are available at our GitHub (https://github.com/pateltanvi2992/Analyzing-Facial-Expressions-with-Generative-AI-in-Clinical-Genetics/tree/main/survey/.).
Participants were recruited via email. To identify survey respondents, we obtained email addresses through professional networks, departmental websites, journal publications, and other publicly available lists.
We also conducted a visual attention experiment involving images of individuals whose facial images were first reconstructed and then expression-manipulated to study how clinicians observe facial expressions. Our objective was to investigate whether clinicians’ visual attention was affected by changes in facial expression for individuals with genetic conditions. This experiment also extends prior research, highlighting distinctions in the assessment of genetic conditions between human observers and DL models (Bours et al., 2018).
For this part of our study, the formatted images were embedded in a screen-based eye-tracking system (Tobii Pro X3-120, Stockholm, Sweden). To examine the effects of expression changes on general clinicians, we recruited 10 clinicians at the National Institutes of Health (NIH) in Bethesda, Maryland, United States to take part in the eye-tracking experiments. Participants were instructed to view the images as if assessing patients in a clinic.
After calibration, each participant viewed the 32 images for 7 s per image. We chose 7 s for the viewing time after extensive initial testing and per our previous work using eye-tracking in a related way, as subjective feedback and preliminary assessments showed that this amount of time was sufficient to assess an image but minimized participants visually revisiting areas of the image in a way that might not inform the assessment (Duong et al., 2023).
At the end of the experiment, we analyzed the amount of time the participants looked at the custom area-of-interest (AOI) drawn around the mouth. This would approximate the effect of the facial expressions on human visual attention. To minimize bias, we used the same AOI for each image (see examples in Figure 2).
Figure 2.
Example outputs of the eye-tracking experiment. Each image illustrates the visual heatmap averaged over 10 clinicians. Each set of images includes the reconstructed image (left) and its corresponding expression-manipulated version (right). All appropriate permissions have been obtained to reuse images and the citations for each are provided in Supplementary Table 5.
The study was formally approved as IRB exempt by the NIH IRB [IRB# 001686 (clinical geneticist survey), 001684 (clinician eye-tracking)].
3 Results
3.1 Model performance on validation set
Table 1 shows the overall performance of ResNet-50 at identifying each of the genetic conditions on the validation set; the metrics are averaged over five folds. This result indicates that our trained ResNet-50 can adequately recognize our disease of interests: 22q, AS, NS, and WS.
Next, we assessed ResNet-50 performance with respect to the facial expressions that were manually annotated by our clinicians (Table 2).
Table 2.
Measuring the effects of facial expression on classifier accuracy.
| Expression | No. of images | Prediction probability | P-value | Top-1 accuracy | P-value | |
|---|---|---|---|---|---|---|
| 22q (not associated with smile) | No smile | 425 | 0.8247 | 0.1457 | 0.8400 | 0.1362 |
| Smile | 160 | 0.7778 | 0.7875 | |||
| AS (associated with smile) | No smile | 111 | 0.6481 | 0.6937 | ||
| Smile | 346 | 0.8128 | 0.8324 | |||
| NS (not associated with smile) | No smile | 226 | 0.7847 | 0.0709 | 0.7965 | 0.0322 |
| Smile | 39 | 0.6670 | 0.6410 | |||
| WS (associated with smile) | No smile | 276 | 0.8227 | 0.8333 | ||
| Smile | 247 | 0.9156 | 0.9231 |
Our facial image dataset was manually labeled as “smile” or “no smile”. We assessed the classifier performance when assessing expression-labeled images, which we refer to as the “average prediction probability”. Additionally, we assess the overall performance of the model in accurately classifying the primary category, which we refer to as “average top-1 accuracy”. WS and AS are classically associated with smile, while 22q and NS are not.
Besides using the manually labels for facial expressions, we also automatically measured the smile intensity using the FACS (Rosenberg and Ekman, 2020). Since FACS is a systematic approach, it may capture more subtle changes (e.g. continuous or incremental changes) in facial expression than a human labeler may report (e.g. binary label of smile versus not smiling). Regardless of using human labeling or FACS, we still expect that there should be a correlation between diagnostic accuracy and facial expressions for most of the four chosen conditions.
We first selected images that our ResNet-50 classifier correctly identified via top-1 accuracy. Then, we measured smile intensities via FACS, focusing on AU6 (Action Unit: Cheek Raiser) and AU12 (Action Unit: Lip Corner Puller), which together indicate a genuine and spontaneous smile (Tian et al., 2001). Overall, in the context of FACS measurement, we observed significant changes in disease classification accuracy with respect to the facial expression in WS, NS, and 22q (Figure 3).
Figure 3.
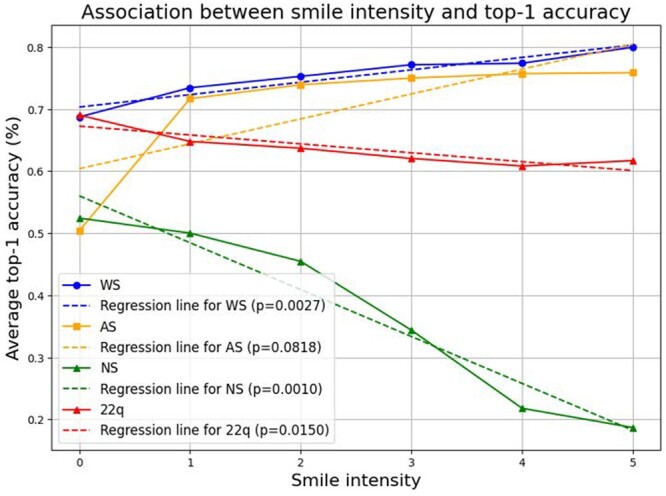
Top-1 accuracy with respect to smile intensity as determined by Facial Action Coding System (FACS). Smile intensity was calculated for each facial image by adding facial action units 6 (cheek raiser) to 12 (lip corner puller). Top-1 accuracy refers to the ability of the classifier to predict the correct genetic condition (highest prediction probability). With higher smile intensity, there is a considerable decrease in correct prediction of top-1 accuracy in NS and a more modest decrease in 22q. Conversely, there is an increase in top-1 accuracy in AS and WS with greater smile intensity.
Figure 3 shows significant changes in top-1 accuracy for WS () and NS () with respect to smile intensity. This result agrees with Table 2, which shows significant changes for WS and NS () and (), respectively. Interestingly, for AS, there is a significant difference observed in Table 2 () but not in Figure 3, as the P-value for the association between accuracy and smile intensity is 0.0818. However, in AS the accuracy increases slightly after the first smile intensity level, with the most significant improvement happening during the transition between smile intensity 0 and 1. For 22q, Table 2 does not show a significant difference () while Figure 3 does (). Overall, there are no major contradictory outcomes between FACS and our human labeling, except for slight differences in AS and 22q.
3.2 Model performance on reconstructed and expression-manipulated test images
Our test dataset consisted of 32 images, with eight images from each of the following categories: WS, NS, AS, and 22q. These images were also used to evaluate the performance of human clinicians (see below). We evaluated the classifier performance on the reconstructed images and expression-manipulated images of these 32 real images.
All reconstructed images of WS, 22q, and NS were correctly identified by the classifier. In the case of AS, two incorrect predictions were made, with prediction probabilities of 33.59% and 41.34% for the correct disease label, respectively. Table 3 illustrates the comparison between the average accuracies of reconstructed and expression-manipulated images. We observed significant changes in accuracies when altering facial expression for different genetic conditions. For 22q and NS, classifier accuracy dropped when changing the expression to a smile. Conversely, accuracy for AS decreased when transitioning from smile to no smile. However, the accuracy in WS remained consistently high, changing only slightly with expression-manipulated ().
Table 3.
Model performance on reconstructed and expression-manipulated test images.
| Expression | No. of images | Average ground truth prediction probability | P-value | |
|---|---|---|---|---|
| 22q (not associated with smile) | Reconstructed | 8 | 0.9420 | 0.0098 |
| Smile manipulated | 8 | 0.8452 | ||
| AS (associated with smile) | Reconstructed | 8 | 0.7254 | 0.0236 |
| No smile manipulated | 8 | 0.5969 | ||
| NS (not associated with smile) | Reconstructed | 8 | 0.8788 | 0.0353 |
| Smile manipulated | 8 | |||
| WS (associated with smile) | Reconstructed | 8 | 0.9836 | 0.3442 |
| No smile manipulated | 8 | 0.9878 |
A total of 32 images were included in the experiments conducted with human clinicians, with eight images from each of the 22q, AS, NS, and WS categories. The first column indicates the classically expected expression (or lack thereof) for each condition. The second column indicates that the expression was manipulated to the opposite expression. For example, AS is associated with smile, so the expression manipulated images were changed to no smile. The expression-manipulated images showed a decrease in accuracy for the 22q, AS, and NS image groups.
3.3 Clinician performance on reconstructed and expression-manipulated test images
A total of 314 clinical geneticists were contacted and asked to complete the surveys. In response, 56 surveys were completed (17.8% total response rate), with 23 and 33 surveys completed for the two survey versions, respectively. A survey was considered complete if all survey questions were answered and the survey was submitted. See Supplemental Table 2 for a description of survey respondents.
The average accuracies for the two surveys were 53.3% and 52.0%, which were not statistically different (P = 0.575) (see Supplementary Table 3). Both surveys were combined to determine the diagnostic accuracy for each of the four conditions (i.e. the accuracies for the reconstructed images and their expression-manipulated versions). For all conditions, clinician performance decreased for the images manipulated to have the opposite facial expression with respect to their classically expected representation (Table 4). The largest differences were observed in 22q and WS. When non-smiling 22q images were manipulated to show a smiling facial expression, accuracy decreased from 60.7% to 50% (). When smiling WS images were manipulated to a more neutral facial expression, accuracy decreased from 65.6% to 52.7% () (Table 4). Unlike the model, the average clinician performance did not statistically decrease in AS and NS. Figure 4 shows the confusion matrices averaged over all the participants who saw the reconstructed images and their expression-manipulated versions, respectively. See Supplemental Table 4 for the correlations between the performance of the classifier and clinical geneticists.
Table 4.
Clinical geneticist performance at classifying reconstructed and expression-manipulated images.
| Overall accuracy (%) | Average accuracy for reconstructed images (%) | Average accuracy for expression-manipulated images (%) | P-value | |
|---|---|---|---|---|
| 22q | 55.40 | 60.70 | 50 | 0.0225 |
| AS | 38.50 | 41.30 | 35.70 | 0.2295 |
| NS | 56.90 | 60.30 | 53.60 | 0.1531 |
| WS | 59.15 | 65.60 | 52.70 | 0.005 |
A total of 56 clinical geneticists participated in the survey. See also Supplemental Table 2.
Figure 4.
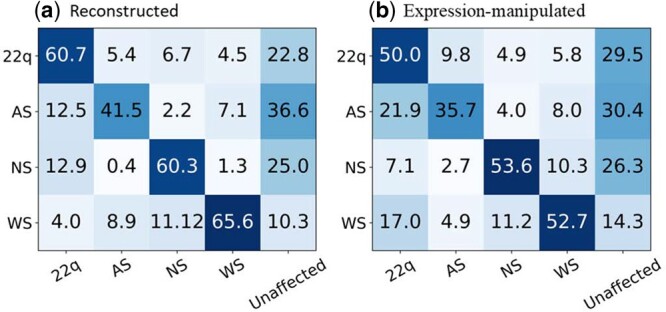
Confusion matrices for the reconstructed (a) and expression-manipulated images (b) based on the performances of the clinical geneticists. In total, 56 clinical geneticists classified 32 images with one of five labels (classification was done via a Qualtrics survey).
For the eye-tracking experiment, over all the images, there was not a significant difference for the visual attention to the AOI between smile and no smile groups (). There was also not a significant difference for each individual condition, though we observed a small, almost significant trend for WS (), which aligns with the largest diagnostic accuracy drop in Table 4 (65.60% versus 52.70%, , between reconstructed versus expression-manipulated images). For each condition, results of smile versus no smile were as follows: 22q: ; AS: ; NS: ; WS: .
4 Discussion
Our analyses approximate how changing specific variables like facial expression affects the identification of individuals with 22q, AS, NS, and WS (Oliver et al., 2007). For the ResNet-50 classifier, we observed a significantly higher prediction probability for the ground-truth labels in AS and WS with smiling versus non-smiling expression, but did not observe a significant difference for NS or 22q.
Since the groups of smiling and non-smiling images are not of the same individuals, there can be bias if syndromic features of one group can be more easily recognized. Thus, we next employed HyperStyle, which enabled us to alter facial expressions of the real images while maintaining high fidelity with respect to the genetic conditions (i.e. ensuring the edited image still retained features specific to a particular genetic condition). Although there are other recent tools to manipulate images (Ramesh et al., 2021; Rombach et al., 2022), which can be amusing and impressive for creative undertakings, they do not reliably produce medically accurate images.
For the expression-manipulated images, ResNet-50 accuracy drops for AS when the expression changed from smile to no smile. Similarly, the accuracy dropped when NS and 22q image expressions were changed from no smile to smile. In addition to the limitations of our study (see below), there may be intrinsic differences in the ability to identify WS. That is, people with WS may have additional features that make this condition easier to identify regardless of facial expression.
Besides evaluating the DL image classifier, we also investigated how facial expression affects the performance of human clinicians. Unlike the classifier, we found that expression changes had a significant effect on the clinical geneticists when assessing WS but not AS. We did not observe any major differences in clinician visual attention to the facial regions encompassing the smile (i.e. the areas around the mouth). Overall, participants spend approximately the same amount of time viewing the mouth area with or without a smiling expression. However, the change in accuracy that we observed implies that other features besides the mouth contribute to diagnostic accuracy. These results suggest that there may be key syndromic features associated with smiling rather than simply the mouth expression alone. For example smiling may affect the appearance of the eyes or other parts of the face, and this may occur in a different way for different genetic conditions. This conclusion stems from the fact that we used the same set of patient images with and without the smiling expression (i.e. the same patient with an edited face expression). This inference would not be possible if the two sets of tested images were of different patients. We note that this is an approximation, since the survey-based participants and the eye-tracking participants are not the same individuals and were intentionally chosen to represent different types of clinicians with differing familiarity with genetic conditions (clinical geneticists for the surveys and general clinicians for the eye-tracking experiments).
Due to our small sample size, we could not further assess differences besides the mouth expression in the eye-tracking experiments, but future work could be done to further determine which other regions of the face contribute to classifier and human accuracy with different facial expressions. For example, future experiments to further test the effects of expression changes via eye-tracking could involve repeating the eye-tracking experiments with the area of the mouth masked.
Related to these points, our study has multiple limitations. Due to the small number of images in this current study (in both the training and testing set) our future work would focus on collecting more images of different racial, ethnic, age, and gender groups, and of many other genetic conditions (Kruszka et al., 2017; Tekendo-Ngongang et al., 2020; Duong et al., 2022b; Kruszka and Tekendo-Ngongang, 2023; Solomon et al., 2023). Our current dataset was also not perfectly balanced with respect to different types of facial expressions. For example, there was a noticeable disparity in expression in the NS images, with fewer smiling versus non-smiling faces. Thus, while we find our results intriguing, they require additional inquiry using larger datasets. A larger and more diverse dataset could also ensure that generative AI methods work equitably across different populations.
Additionally, our choice of HyperStyle may fail for images that were not seen during training. This was our rationale for including the images we used for human testing into the training set for HyperStyle. Future studies will focus on how to efficiently build generative models to edit new, unseen images. We note that there are other image editing approaches (e.g. diffusion-based methods), and planned work would include evaluating these other techniques (Rombach et al., 2022).
Another limitation was that we could only recruit a limited number of clinicians. In future studies, it may be interesting to further assess how different facial expressions affect non-expert geneticist clinicians, who may be less familiar with the overall range of presentation of genetic conditions, but who may more frequently encounter these patients due to the dearth of clinical genetics experts.
Despite these limitations, one overall interesting conclusion, which echoes some of our previous work, is that a DL model and humans simply perform differently under different circumstances (Duong et al., 2023). As AI tools are adopted in clinical scenarios, better understanding of these differences will be important to ensure that they are useful in helping to diagnose and care for people affected by genetic conditions.
We predict that generative AI tools will become increasingly used in medical settings (Waikel et al., 2023). We hope that the use of these tools can be helpful to clinicians and patients in many situations. In genetics, these approaches may help diagnose patients more quickly and cost-effectively, perhaps especially in underserved communities and parts of the world where there is less access to subspecialists who are very familiar with rare genetic conditions (Solomon et al., 2023). However, we emphasize that it is critical to carefully study DL tools to better understand their strengths and limitations. Even before embarking on clinical implementation studies, it will be important to understand ways to maximize benefits and minimize risks.
Supplementary Material
Contributor Information
Tanviben Patel, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Amna A Othman, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Ömer Sümer, Institute of Computer Science, Augsburg University, Augsburg, Bavaria 86159, Germany.
Fabio Hellman, Institute of Computer Science, Augsburg University, Augsburg, Bavaria 86159, Germany.
Peter Krawitz, Institute for Genomic Statistics and Bioinformatics, University of Bonn, Bonn, North Rhine-Westphalia 53113, Germany.
Elisabeth André, Institute of Computer Science, Augsburg University, Augsburg, Bavaria 86159, Germany.
Molly E Ripper, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Chris Fortney, Social and Behavioral Research Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Susan Persky, Social and Behavioral Research Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Ping Hu, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Cedrik Tekendo-Ngongang, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Suzanna Ledgister Hanchard, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Kendall A Flaharty, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Rebekah L Waikel, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Dat Duong, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Benjamin D Solomon, Medical Genomics Unit, Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MA 20892, United States.
Author contributions
T.P., R.L.W., D.D., and B.D.S. were responsible for conceiving and conducting experiments, analyzing the results, and writing and reviewing the manuscript. A.O., P.H., S.L.H., and C.F. were responsible for conducting experiments, analyzing the results, and reviewing the manuscript. O.S., F.H., P.K., E.A., M.E.R., P.H., C.T.-N., S.P., and K.A.F. were responsible for conceiving experiments and reviewing the manuscript.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
No competing interest is declared.
Funding
This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. This work utilized the computational resources of the National Institute of Health (NIH) Biowulf cluster.
Data availability
The data and code underlying this article are available in the article and supplementary materials, with links to code and additional data sources provided in online supplementary material.
References
- Abdal R, Qin Y, Wonka P. Image2StyleGAN: how to embed images into the StyleGAN latent space? In: Proceedings of the IEEE/CVF International Conference on Computer Vision, p. 4432–41. 2019.
- Alaluf Y, Tov O, Mokady R. et al. Hyperstyle: StyleGAN inversion with hypernetworks for real image editing. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, p. 18511–21. 2022.
- Bours CCAH, Bakker-Huvenaars MJ, Tramper J. et al. Emotional face recognition in male adolescents with autism spectrum disorder or disruptive behavior disorder: an eye-tracking study. Eur Child Adolesc Psychiatry 2018;27:1143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Li Y, Zhang Z. Celeb-500k: a large training dataset for face recognition. In: 2018 25th IEEE International Conference on Image Processing (ICIP) p. 2406–10. IEEE, 2018a.
- Cao Q, Shen L, Xie W. et al. VGGFace2: a dataset for recognising faces across pose and age. In: 2018 13th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2018), p. 67–74. IEEE, 2018b.
- Correia-Caeiro C, Burrows A, Wilson DA. et al. Callifacs: the common marmoset facial action coding system. PLoS One 2022;17:e0266442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrave AJ, Janizek JD, Lee S-I.. AI for radiographic covid-19 detection selects shortcuts over signal. Nat Mach Intell 2021;3:610–9. [Google Scholar]
- Duong D, Hu P, Tekendo-Ngongang C. et al. Neural networks for classification and image generation of aging in genetic syndromes. Front Genet 2022a;13:864092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong D, Johny AR, Hanchard SL. et al. Human and computer attention in assessing genetic conditions. medRxiv. 2023.
- Duong D, Waikel RL, Hu P. et al. Neural network classifiers for images of genetic conditions with cutaneous manifestations. Hum Genet Genom Adv 2022b;3:100053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman P, Friesen WV, Hager J.. Environmental psychology & nonverbal behavior. J Nonverbal Behav 1976;1:56–75. [Google Scholar]
- Guo Y, Zhang L, Hu Y. et al. MS-Celeb-1M: a dataset and benchmark for large-scale face recognition. In: Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11–14, 2016, Proceedings, Part III 14, p. 87–102. Springer, 2016.
- Hanchard SEL, Dwyer MC, Liu S. et al. Scoping review and classification of deep learning in medical genetics. Genet Med 2022;24:1593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T-C, Bar-Haim A, Moosa S. et al. Gestaltmatcher facilitates rare disease matching using facial phenotype descriptors. Nat Genet 2022;54:349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incerti D, Xu X-M, Chou JW. et al. Cost-effectiveness of genome sequencing for diagnosing patients with undiagnosed rare genetic diseases. Genet Med 2022;24:109–18. [DOI] [PubMed] [Google Scholar]
- Jones KL, Jones MC, Del Campo M.. Smith’s Recognizable Patterns of Human Malformation – E-book. Elsevier Health Sciences, 2021. [Google Scholar]
- Karras T, Laine S, Aila T. A style-based generator architecture for generative adversarial networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, p. 4401–10. 2019. [DOI] [PubMed]
- Krawitz P, Hsieh T-C, Bar-Haim A. et al. Gestaltmatcher: overcoming the limits of rare disease matching using facial phenotypic descriptors. 2021.
- Kruszka P, Porras AR, Addissie YA. et al. Noonan syndrome in diverse populations. Am J Med Genet A 2017;173:2323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka P, Tekendo-Ngongang C.. Application of facial analysis technology in clinical genetics: considerations for diverse populations. Am J Med Genet C Semin Med Genet 2023;193:e32059. [DOI] [PubMed] [Google Scholar]
- Li S, Deng W.. A deeper look at facial expression dataset bias. IEEE Trans Affective Comput 2022;13:881–93. [Google Scholar]
- Oliver C, Horsler K, Berg K. et al. Genomic imprinting and the expression of affect in Angelman syndrome: what’s in the smile? J Child Psychol Psychiatry 2007;48:571–9. [DOI] [PubMed] [Google Scholar]
- Peña A, Morales A, Serna I. et al. Facial expressions as a vulnerability in face recognition. In: 2021 IEEE International Conference on Image Processing (ICIP), p. 2988–92. IEEE, 2021.
- Pisner DA, Schnyer DM.. Support vector machine. In: Machine Learning, p. 101–121. Amsterdam, The Netherlands: Elsevier, 2020. [Google Scholar]
- Porras AR, Rosenbaum K, Tor-Diez C. et al. Development and evaluation of a machine learning-based point-of-care screening tool for genetic syndromes in children: a multinational retrospective study. Lancet Digit Health 2021;3:e635–43. [DOI] [PubMed] [Google Scholar]
- Ramesh A, Pavlov M, Goh G. et al. Zero-shot text-to-image generation. In; International Conference on Machine Learning, p. 8821–31. PMLR, 2021.
- Richmond S, Howe LJ, Lewis S. et al. Facial genetics: a brief overview. Front Genet 2018;9:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombach R, Blattmann A, Lorenz D. et al. High-resolution image synthesis with latent diffusion models. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, p. 10684–95. 2022.
- Rosenberg EL, Ekman P.. 2020. What the Face Reveals: Basic and Applied Studies of Spontaneous Expression using the Facial Action Coding System (FACS). Oxford: Oxford University Press. [Google Scholar]
- Schaefer J, Lehne M, Schepers J. et al. The use of machine learning in rare diseases: a scoping review. Orphanet J Rare Dis 2020;15:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yang C, Tang X. et al. Interfacegan: interpreting the disentangled face representation learned by GANS. IEEE Trans Pattern Anal Mach Intell 2020;44:2004–18. [DOI] [PubMed] [Google Scholar]
- Skiendziel T, Rösch AG, Schultheiss OC.. Assessing the convergent validity between the automated emotion recognition software noldus FaceReader 7 and facial action coding system scoring. PLoS One 2019;14:e0223905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Adam MP, Fong C-T. et al. Perspectives on the future of dysmorphology. Am J Med Genet A 2023;191:659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Nguyen A-D, Bear KA. et al. Clinical genomic database. Proc Natl Acad Sci U S A 2013;110:9851–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sümer Ö, Waikel RL, Hanchard SEL. et al. Region-based saliency explanations on the recognition of facial genetic syndromes. In: Machine Learning for Healthcare Conference, p. 712–36. PMLR, 2023.
- Tanzi RE, Bertram L.. New frontiers in Alzheimer’s disease genetics. Neuron 2001;32:181–4. [DOI] [PubMed] [Google Scholar]
- Tekendo-Ngongang C, Owosela B, Fleischer N. et al. Rubinstein–Taybi syndrome in diverse populations. Am J Med Genet A 2020;182:2939–50. [DOI] [PubMed] [Google Scholar]
- Tian Y-I, Kanade T, Cohn JF.. Recognizing action units for facial expression analysis. IEEE Trans Pattern Anal Mach Intell 2001;23:97–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Oswald J, Henning C, Grewe BF. et al. Continual learning with hypernetworks. arXiv preprint arXiv:1906.00695. 2019.
- Waikel RL, Othman AA, Patel T. et al. Generative artificial intelligence methods for pediatric genetics education. medRxiv, p. 2023–8. 2023.
- Wehrli S, Hertweck C, Amirian M. et al. Bias, awareness, and ignorance in deep-learning-based face recognition. AI Ethics 2022;2:509–22. [Google Scholar]
- Wright L. 2019. Ranger – a synergistic optimizer. https://github.com/lessw2020/Ranger-Deep-Learning-Optimizer.
- Zhao S, Liu Z, Lin J. et al. Differentiable augmentation for data-efficient GAN training. Adv Neural Inform Process Syst 2020;33:7559–70. [Google Scholar]
- Zhi R, Zhou C, Li T. et al. Action unit analysis enhanced facial expression recognition by deep neural network evolution. Neurocomputing 2021;425:135–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and code underlying this article are available in the article and supplementary materials, with links to code and additional data sources provided in online supplementary material.