Abstract
Motivation
Electronic health records (EHRs) represent a comprehensive resource of a patient’s medical history. EHRs are essential for utilizing advanced technologies such as deep learning (DL), enabling healthcare providers to analyze extensive data, extract valuable insights, and make precise and data-driven clinical decisions. DL methods such as recurrent neural networks (RNN) have been utilized to analyze EHR to model disease progression and predict diagnosis. However, these methods do not address some inherent irregularities in EHR data such as irregular time intervals between clinical visits. Furthermore, most DL models are not interpretable. In this study, we propose two interpretable DL architectures based on RNN, namely time-aware RNN (TA-RNN) and TA-RNN-autoencoder (TA-RNN-AE) to predict patient’s clinical outcome in EHR at the next visit and multiple visits ahead, respectively. To mitigate the impact of irregular time intervals, we propose incorporating time embedding of the elapsed times between visits. For interpretability, we propose employing a dual-level attention mechanism that operates between visits and features within each visit.
Results
The results of the experiments conducted on Alzheimer’s Disease Neuroimaging Initiative (ADNI) and National Alzheimer’s Coordinating Center (NACC) datasets indicated the superior performance of proposed models for predicting Alzheimer’s Disease (AD) compared to state-of-the-art and baseline approaches based on F2 and sensitivity. Additionally, TA-RNN showed superior performance on the Medical Information Mart for Intensive Care (MIMIC-III) dataset for mortality prediction. In our ablation study, we observed enhanced predictive performance by incorporating time embedding and attention mechanisms. Finally, investigating attention weights helped identify influential visits and features in predictions.
Availability and implementation
1 Introduction
Electronic health records (EHRs) represent a comprehensive resource that provide a historical collection of a patient’s medical record and health-related data. This holistic resource includes structured data, such as patient medical conditions, medications, clinical measurements, and demographics, and unstructured data, exemplified by clinical notes (Yadav et al. 2018, Lyu et al. 2022, Hossain et al. 2023). EHRs have become the cornerstone in modeling the classification of patients and the progression and sub-typing of diseases through advanced technologies (e.g. machine and deep learning [DL]), enabling healthcare providers to analyze vast volumes of data, extract valuable insights, and make more precise and data-driven clinical decisions (Xu et al. 2022, Yang et al. 2022). Structured EHRs are longitudinal in nature, providing a dynamic and chronological representation of a patient’s medical history. The longitudinal nature of EHRs reflects their capacity to capture and document a patient’s health information over an extended time. Unlike static data snapshots, the temporal patterns embedded in EHR provide valuable insights into the progression of a patient’s health. This, in turn, enables AI-based techniques to make precise clinical decisions. However, machine learning methods such as Random Forest (RF) (Breiman 2001), Support Vector Machine (SVM) (Cortes and Vapnik 1995), and neural networks (McCulloch and Pitts 1943) lack the capacity to handle temporal relations in EHR. These methods usually rely on a specific time point within the EHR, such as the baseline or the most recent clinical visit. Alternatively, decisions can be made by aggregating data across all time points. Furthermore, EHRs present data analysis challenges such as varying numbers of visits for patients, irregular time intervals between visits, and the presence of missing values. Consequently, AI-based methods for modeling EHR need to address these inherent issues.
To preserve the temporal nature of EHR and address varying number of visits per patient, recurrent neural networks (RNN) models such as Long Short-Term Memory (LSTM) (Hochreiter and Schmidhuber 1997) and Gated Recurrent Unit (GRU) (Cho et al. 2014), along with Transformer (Vaswani et al. 2017) have been employed (Yadav et al. 2018, Xu et al. 2022, Yang et al. 2022, 2023, Herp et al. 2023, Hossain et al. 2023).
In Lee et al. (2019), a multimodal GRU-based approach was employed on EHR data related to Alzheimer’s Disease (AD) to predict conversion from Mild Cognitive Impairment (MCI) to AD. In Li and Fan (2019), an LSTM-based DL model was proposed for early prediction of AD using EHR data related to AD. In Nguyen et al. (2020), an RNN-based model was applied to AD EHR data to predict the diagnosis of patients up to six years. In Venugopalan et al. (2021), an integrative classification approach was proposed for the early detection of AD stage. In Fouladvand et al. (2021), a multi-stream Transformer-based approach was applied on EHR to predict opioid use disorder. Although RNN- and Transformer-based methods handle longitudinal EHR data, they do not consider irregular time intervals between clinical visits. RNN (e.g. LSTM and GRU) assume that temporal gaps between time points are equal while absolute position encoding is used in the original Transformer architecture (Vaswani et al. 2017).
Recent studies have proposed to extend LSTM to handle irregular time intervals. Time-aware LSTM (T-LSTM) was developed with the purpose of learning a subspace decomposition of the cell memory, enabling time decay to discount the memory content according to the elapsed time (Baytas et al. 2017). In a subsequent study (Luong and Chandola 2019), T-LSTM’s effectiveness was assessed on synthetic data and real EHR data from kidney patients. Despite its success in handling synthetic data, the results showed challenges in effectively sub-typing chronic kidney disease using real EHR data. In Liu et al. (2022), KIT-LSTM expands the LSTM model by incorporating two time-aware gates: one for the time gap between two consecutive visits and another for the time gap between consecutive measurements for each clinical feature. In Al Olaimat et al. (2023), an RNN-based DL architecture called Predicting Progression of AD (PPAD) was proposed. PPAD addresses irregular time intervals by using patients’ age in each visit as a feature to indicate time changes between successive visits. Although most recent RNN-based studies were able to mitigate the irregular time intervals, they exhibit limited interpretability, making the explanation of their prediction results challenging.
To improve model’s interpretability while considering irregular time intervals, RETAIN (Choi et al. 2016) was proposed. This model aims to identify the significant time points and features influencing the prediction of heart disease using EHR. The methodology incorporates two attention mechanisms, facilitated by two distinct GRUs, to capture interactions between time points and features. Additionally, to address irregular time intervals, time information, such as duration between consecutive visits or cumulative number of days since the initial visit, are optionally introduced as an additional feature at each time point. Although the tool is available, its application is limited to MIMIC-III data, requiring algorithm reimplementation to use with different data formats. Furthermore, the effectiveness of incorporating time information as an additional feature remains unassessed. In Zhang et al. (2019), another RNN-based architecture called ATTAIN was proposed to handle irregular time intervals. This approach incorporates both time intervals and an attention mechanism. The current prediction is formulated by aggregating information from all or some of the preceding memory cells, with regularization of the most recent memory cell guided by weights generated through the attention mechanism and time intervals, employing a time decay function. DATA-GRU (Tan et al. 2020) was proposed to predict patients’ mortality risk. This model incorporates a time-aware mechanism to handle irregular time intervals through internally incorporating time intervals into DATA-GRU to adjust the hidden status in the previous memory cell. Additionally, a dual-attention mechanism is employed to address missing values by considering both data quality and medical knowledge perspectives. However, a notable limitation of this model lies in the excessive measures taken to address missing values and time intervals problems, leading to an increase in the model’s complexity. This includes aspects such as the number of RNN cells employed, the number of learnable parameters, and the preprocessing steps conducted before training to generate the unreliability scores of the data. The Multi-Integration Attention Module (MIAM) (Lee et al. 2022) was proposed for different downstream tasks to capture complex missing patterns in EHR. This involves combining missing indicators and time intervals, followed by integrating observations and missing patterns within the representation space using a self-attention mechanism.
Nevertheless, these approaches do not embed the time intervals directly into the visits data. Instead, they introduce them as additional features to handle irregular time intervals or internally modify the hidden states within RNN by updating the memory cells with the time intervals. Furthermore, most of these approaches lack a mechanism to interpret their results.
In this study, we present two interpretable DL architectures based on RNN, namely attention-based time-aware RNN (TA-RNN) and TA-RNN-autoencoder (TA-RNN-AE) for early prediction of patient clinical outcomes at the next visit and multiple visits ahead, respectively. To mitigate the effect of irregular time intervals, we propose incorporating elapsed times between consecutive visits into visit data through a time embedding layer. Additionally, to enhance the interpretability of the model's results, we propose utilizing a dual-level attention mechanism that operates between visits and features within each visit to identify the significant visits and features influencing the model’s prediction. To demonstrate the robustness of our proposed models, we evaluated them on three real-world datasets: (i) early prediction of conversion to AD by training and testing the models on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset; (ii) early prediction of conversion to AD by training the models on the ADNI dataset and testing them on the National Alzheimer’s Coordinating Center (NACC) dataset; (iii) prediction of mortality by training and testing the models on the Medical Information Mart for Intensive Care (MIMIC-III) dataset. In the task of predicting AD conversion, our experiments demonstrated that our proposed models outperformed all baseline models across most prediction scenarios, particularly in terms of F2 and sensitivity. Regarding the mortality prediction task, the results indicated that TA-RNN outperformed RETAIN in AUC. We also illustrated that integrating time embedding into the input data contributed to improved model performance by effectively handling irregular time intervals between consecutive visits. Furthermore, the results demonstrated the remarkable ability of the dual-level attention mechanism to interpret the results of our proposed models.
The main contributions of this study can be summarized as follows:
We propose two interpretable DL architectures based on RNN, namely attention-based TA-RNN and TA-RNN-autoencoder (TA-RNN-AE) for early prediction of patient clinical outcomes at the next visit and multiple visits ahead, respectively.
We propose incorporating time embedding of the elapsed times between consecutive visits into visits’ data through the utilization of a time embedding layer. This approach aims to mitigate the impact of the irregular time intervals.
We propose employing a dual-level attention mechanism that operates between visits and features within each visit to identify notable visits and features influencing the model’s predictions. This approach is intended to improve the interpretability of the model’s results.
We demonstrate that the proposed methods exhibit superior performance compared to the state of the art (SOTA) and baseline approaches in downstream tasks. This was achieved by leveraging longitudinal multimodal data and cross-sectional demographic data from two large AD databases, namely ADNI and NACC, for early prediction of conversion to AD. Additionally, we validated the effectiveness of our methods on a real-world EHR dataset obtained from MIMIC-III for predicting mortality.
2 Materials and methods
2.1 Datasets
In this study, three datasets were used to evaluate the proposed models. The first dataset consisted of longitudinal and cross-sectional data from the ADNI database (https://adni.loni.usc.edu/). The ADNI was launched in 2003 as a public–private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Since it has been launched, the public–private cooperation has contributed to significant achievements in AD research by sharing data to researchers from all around the world (Jack et al. 2010, Jagust et al. 2010, Risacher et al. 2010, Saykin et al. 2010, Trojanowski et al. 2010, Weiner et al. 2010, 2013). The second dataset consisted of longitudinal and cross-sectional data from the NACC database (Besser et al. 2018). The NACC database is a centralized asset for AD research, designed specifically to facilitate and expedite investigations into the causes, diagnosis, and treatment of AD. It comprises information from various study sites throughout the United States, encompassing a range of data such as demographics, cognitive assessments, genetic details, and MRI data. The third dataset was from the MIMIC-III database (Johnson et al. 2016a,b, Goldberger et al. 2000). The MIMIC-III database serves as a comprehensive resource of EHR designed for researchers interested in gaining insights into critical care practices and patient clinical outcomes, with a specific focus on patients of the intensive care unit (ICU). The MIMIC-III databases consist of multiple set of clinical data, including vital signs, laboratory results, conditions, medical procedures, medications, and clinical notes.
We employed three distinct experimental setups to train and evaluate our proposed models utilizing these three datasets (Table 1). In the first experimental setup, we split the ADNI dataset into 70% training and 30% testing in a random stratified manner for model training and testing to predict conversion from MCI to AD, respectively. The ADNImerge R package (available at https://adni.bitbucket.io/) was employed to gather longitudinal and cross-sectional data from all ADNI studies, including ADNI1, ADNI2, and ADNI-GO (Jiang et al. 2020). We have conducted a preprocessing using the same steps as implemented in the PPAD (Al Olaimat et al. 2023). Briefly, in the preprocessing phase, we performed the removal of irrelevant features and visits, handled missing values through data imputation using K-nearest neighbors (KNN), and normalized the features. Following the preprocessing, the final dataset comprised 20 longitudinal and five cross-sectional demographic features for a total of 1169 patients and 5759 visits (Supplementary Tables S1 and S2). For generalization, this entire process was iterated across three random splits.
Table 1.
Experimental setups that were employed for the proposed models.a
| Experimental setup | Training data | Validation data | Test data | Downstream task |
|---|---|---|---|---|
| First | 70% of ADNI | 5-fold cross-validation | 30% of ADNI | Predicting conversion of MCI to AD |
| Second | Entire ADNI | 5-fold cross-validation | Entire NACC | Predicting conversion of MCI to AD |
| Third | 70% of MIMIC-III | 10% of MIMIC-III | 20% of MIMIC-III | Predicting mortality |
In the first and second experimental setups, 5-fold cross-validation was employed on the training data to tune the hyperparameters.
In the second experimental setup, the entire ADNI dataset served as the training data, while the NACC dataset served as an external dataset for model testing to predict conversion from MCI to AD. We conducted a data harmonization step to collect common features from the ADNI and the NACC datasets. Subsequently, the harmonized datasets were preprocessed, following the same steps as outlined in the PPAD (Al Olaimat et al. 2023). Ultimately, we successfully harmonized nine features between the ADNI and NACC datasets. The final ADNI dataset consisted of 1205 patients and 6066 visits (Supplementary Tables S3 and S4). The difference in patient and visit numbers between the first and second experimental setups in the ADNI data arises due to the preprocessing step. Since the second experimental setup used fewer features, the missing rate was lower, hence more visits were kept. The final NACC dataset encompassed 6118 patients and 35 423 visits.
For the third experimental setup, the MIMIC-III dataset was utilized to evaluate the proposed models for mortality prediction task. We extracted patients’ visits, mortality labels, and time information from the MIMIC-III dataset using the same procedures that have been employed in RETAIN (Choi et al. 2016). Patients’ visits include the International Classification of Diseases, Ninth Revision (ICD-9) codes, with dataset includes 942 unique ICD-9 codes for 7537 patients. To train the proposed models, the dataset underwent a stratified random split, allocating 70% for training, 20% for testing, and 10% for validation (Supplementary Table S9).
In all experimental setups, datasets have been preprocessed such that each sample represents a unique patient with at least two visits. Consequently, the input data is structured in three dimensions, delineated as (samples, visits, and features).
2.1.1 Dataset notations
Let denote the longitudinal EHR data with samples (ie, patients). where each sample represents measurements of features collected over time points (i.e. visits): . For each visit, , represents a vector of features of sample at visit . For each feature , represents the feature value of sample over visits, and represents the feature value of sample at visit . In , each sample has a corresponding elapsed time data , where each represents the elapsed times for sample collected over time points (visits): . For each visit , represents the elapsed time at visit , such that , where represents the visit date at visit . For ADNI and NACC data, the unit of was in years, whereas for MIMIC-III data, the unit was in days. Finally, in each sample has a corresponding diagnosis or clinical outcome for each time point: . In this study, for each visit , where 0 denotes MCI and 1 denotes AD in the MCI to AD conversion prediction task, and 0 denotes absence of mortality and 1 denotes mortality in the mortality prediction task.
2.2 The proposed method
In this study, we developed two interpretable RNN-based DL architectures to predict the clinical outcome at the next visit and the multiple visits ahead. To demonstrate the robustness of the proposed models, we evaluated them using two downstream tasks, namely predicting conversion from MCI to AD and patient mortality prediction. In both architectures, we employed a time embedding layer that incorporates elapsed time between consecutive visits in EHR into input data to address lack of consideration of irregular time intervals between consecutive inputs by RNN models. Furthermore, we employed a dual-level attention mechanism that operates between visits and features within each visit to identify significant visits and features influencing predictions. Implementing the dual-level attention mechanism, in turn, improves the interpretability of the model. Finally, the RNN cell type was considered as a hyperparameter, offering options such as LSTM, GRU, bidirectional LSTM (Bi-LSTM), and bidirectional GRU (Bi-GRU).
2.2.1 Time embedding layer
To alleviate the limitation of RNN models with irregular time intervals, we employed a time embedding layer. The layer takes the longitudinal () and elapsed time () data as inputs to generate a new representation in a manner where the time information is integrated with the original data. This layer is a variation of positional encoding designed for continuous time values as input (Lee et al. 2022). It transforms these values into an encoded vector representation () (Equation (1)), which is an adapted version of the positional encoding formula proposed in Vaswani et al. (2017).
| (1) |
In Equation (1), represents the elapsed time since the previous visit, represents the index in the embedding space, represents the model dimension or size, and represents the maximum elapsed time of time data. Then, the longitudinal input data is added to the time embedding to generate a new embedding or representation () (Equation (2)) of that has time information based on .
| (2) |
In Equation (2), represents the new embedding for the input data that incorporates the temporal representation. In the following sections, will be the input to the proposed models.
2.2.2 A predictive model for clinical outcome at the next visit
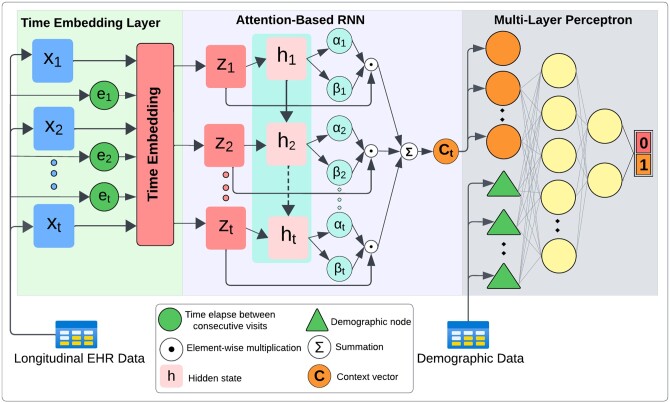
TA-RNN comprises three components, namely, a time embedding layer, attention-based RNN, and a prediction layer based on multi-layer perceptron (MLP) (Fig. 1). In this model, to address the irregular time intervals, first, the time embedding layer is used to generate the new input embedding based on the original input data and time embedding (Equation (2), Section 2.2.1). Then, is fed to an RNN cell (Equation (3)) that employs a dual sets of attention weights (for visits and features) and generates the hidden state for each time point .
| (3) |
Figure 1.
TA-RNN architecture for predicting clinical outcomes in EHR at the next visit.
To control the impact of each visit’s embedding , the scalars () that represent visit-level attention weights are calculated using hidden states of the RNN cell (Equation (4)).
| (4) |
In Equation (4), and are the trainable parameters. On the other hand, to control the impact of features in each visit’s embedding , the vectors () that represent feature-level attention weights are calculated using hidden states of the RNN cell (Equation (5)) where and are the trainable linear transformation matrix and the bias vector, respectively.
| (5) |
Then, the obtained visits and features attention weights are utilized to generate the context vector (Equation (6)) of a sample in the EHR up to the visit.
| (6) |
In Equation (6), represents the element-wise multiplication operation. Finally, if the demographic data () is available, the is concatenated with it to train an MLP for predicting the clinical outcome of the next visit (Equation (7)).
| (7) |
In Equation (7), represents the predicted clinical outcome, and are the trainable linear transformation matrices, and are the trainable bias vectors, represents the sigmoid function, and represents the concatenation operation.
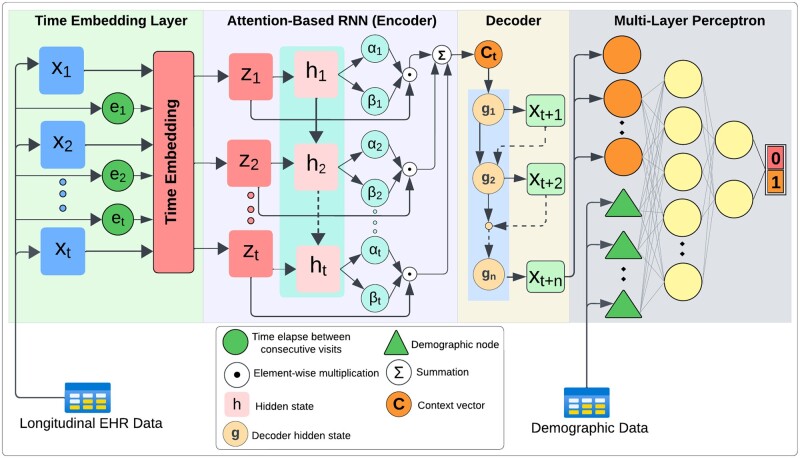
2.2.3 A predictive model for clinical outcome at multiple visits ahead
To predict clinical outcomes in EHR at multiple visits ahead, we developed another predictive model called TA-RNN-AE. TA-RNN-AE comprises time embedding, attention-based RNN autoencoder, and MLP (Fig. 2). In this model, the encoder component learns of the longitudinal data as described in the previous section (Equation (6)). Subsequently, the decoder component generates multiple upcoming visits’ representations using up to visits in an autoregressive manner. Initially, both the input and the initial hidden state to the decoder are set to . Subsequently, for each subsequent time step, the input to the decoder is derived from the output of the previous time step (Equation (8)).
| (8) |
Figure 2.
TA-RNN-AE architecture for predicting clinical outcome in EHR at multiple visits ahead.
Finally, if demographic data is available, the MLP model is then trained by concatenating it with the last visit’s representation generated by the decoder . This training is designed to predict the clinical outcome in the EHR at the visit, as outlined in (Equation (9)).
| (9) |
2.2.4 Parameter learning and evaluation metrics
In both proposed architectures, trainable parameters of all components are jointly learned using a customized binary cross-entropy loss function (Equation (10)). This customized loss function assigns greater weight to predicting positive cases to minimize predicting false negative cases which in turn increases the sensitivity of the predictive model.
| (10) |
In Equation (10), is a hyperparameter real number between 0 and 1 to define the relative weight of positive prediction and is the true diagnosis. In this study, we set to 0.7 and 0.65 for predicting AD conversion and mortality tasks, respectively. For the optimization process, all models underwent training with the adaptive moment estimation (Adam) optimizer (Kingma and Ba 2014), utilizing the default learning rate 0.001. Hyperparameters such as the RNN cell type, number of epochs, batch size, dropout rate, L2 regularization, hidden size, embedding size, and were tuned using 5-fold cross-validation and 10% validation data for predicting AD conversion and mortality tasks, respectively. Model evaluation was performed using the F2 score (Equation (11)) and sensitivity.
| (11) |
In Equation (11), recall is given times more emphasis than precision. We set to 2 in this study.
3 Results and discussion
This study proposes two RNN-based frameworks: TA-RNN and TA-RNN-AE to predict the clinical outcome in EHR at the next visit and at multiple visits ahead, respectively. The proposed frameworks were evaluated based on three experimental setups (see Section 2.1 for details). We also conducted an ablation study to assess the importance of the time embedding and dual-attention components. Finally, we have assessed the interpretability of the model by examining the visit- and feature-based attention weights for the entire test set and for an individual patient. The details of these analyses are described below.
3.1 Predicting clinical outcome at next visits
To evaluate the predictive performance of TA-RNN, in the first and second experimental setups, we trained four models using two, three, five, and six preceding visits to predict the conversion to AD at the subsequent visit. For hyperparameter tuning, a grid search with 5-fold cross-validation was performed. The optimal hyperparameters of the models in the first and second experimental setup are shown in Supplementary Tables S5 and S6, respectively.
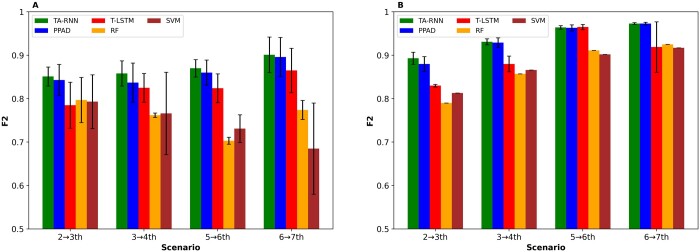
We compared TA-RNN to RF, SVM, T-LSTM, and PPAD. PPAD was trained using the longitudinal features including the age while RF and SVM, which are not designed for longitudinal data, were trained on the aggregated longitudinal data after averaging each feature’s values across all visits. On the other hand, T-LSTM and TA-RNN were trained without utilizing the age feature to evaluate the effectiveness of their mechanisms of handling irregular time intervals. The entire process was repeated 15 times in the second experimental setup and for each of the three random data splits in the first experimental setup to ensure generalizability. The outcomes in the first and second experimental setups demonstrated the superiority of TA-RNN upon all baseline models in terms of F2 (Fig. 3A and B) and sensitivity (Supplementary Fig. S1A and B) except one case in the second experimental setup (Fig. 3B) and (Supplementary Fig. S1B). Moreover, this superior performance highlights TA-RNN’s capability to address the irregular time intervals issue better than T-LSTM and PPAD. We observed that the performance differences between TA-RNN and RF, SVM, and T-LSTM were statistically significant (t-test P-value ≤ .05) for all cases except for one. Although the performance difference between TA-RNN and PPAD was not statistically significant in most cases (Supplementary Tables S11 and S12), TA-RNN’s interpretability feature through its dual-level attention mechanism is a clear advantage over PPAD.
Figure 3.
F2 scores for TA-RNN models predicting conversion to AD at the next visit. (A) Models evaluated on held-out samples in ADNI after training with two, three, five, and six preceding visits in ADNI. (B) Models evaluated on NACC after training with two, three, five, and six preceding visits in ADNI. Error bars represent standard deviations. Statistical significance between TA-RNN and other tools was assessed using t-test (see Supplementary Tables S11 and S12 for P-values).
For the third experimental setup, our evaluation focused only on TA-RNN and excluded TA-RNN-AE as 70% and 18% of patients in the MIMIC-III dataset have only two and three visits, respectively. TA-RNN was trained on MIMIC-III data and compared only with RETAIN, which was designed to enhance model’s interpretation, for mortality prediction at the last hospital stay. For hyperparameter tuning, 10% validation data was utilized. The optimal hyperparameters are shown in Supplementary Table S10. The entire process was repeated 15 times to ensure generalizability. We observed that TA-RNN outperformed the RETAIN model in AUC slightly (Table 2). Although the difference between their performances was marginally significant (P value = .05), TA-RNN exhibits lower complexity compared to RETAIN as TA-RNN utilizes only one RNN cell. On the other hand, RETAIN employs two separate RNN cells—one for handling visit attention weights and the other for handling feature attention weights. Reduced complexity makes TA-RNN faster to train with less trained data compared to RETAIN.
Table 2.
AUC score for the TA-RNN model for mortality prediction at the next visit.
| Model | AUC |
|---|---|
| RETAIN | 0.731 ± 0.002 |
| TA-RNN | 0.733 ± 0.003 |
3.2 Predicting clinical outcome at multiple visits ahead
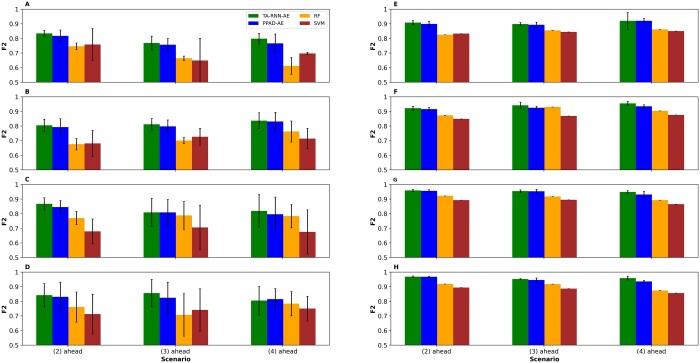
To evaluate the predictive performance of TA-RNN-AE, we conducted the training for various scenarios, including models trained on two, three, five, and six visits to predict the conversion to AD at the subsequent second, third, and fourth visits. This was accomplished using both the first and second experimental setup. For instance, a trained model on datasets from two clinical visits was evaluated for its ability to predict the conversion to AD at the fourth, fifth, and sixth visits. For hyperparameter tuning, grid search with 5-fold cross-validation was performed. The optimal hyperparameters of the models in the first and second experimental setup are shown in Supplementary Tables S7 and S8, respectively. We also repeated the entire process 15 times in the second experimental setup and for each of the three random data splits in the first experimental setup to ensure generalizability. Because T-LSTM and RETAIN do not have the ability to predict the clinical outcome in EHR at multiple visits ahead, TA-RNN-AE comparison was limited to RF, SVM, and PPAD-AE. The results demonstrated the superiority of TA-RNN-AE upon all baseline and SOTA models in terms of F2 (Fig. 4) and sensitivity (Supplementary Fig. S2) except for one case where PPAD-AE outperformed TA-RNN-AE slightly in terms of F2 (Fig. 4D). The performance difference between TA-RNN-AE and RF and SVM was statistically significant (t-test P value ≤ .05) for 45 out of 48 cases in the first and second experimental setup. TA-RNN-AE outperformed PPAD-AE significantly for 3 and 7 out of 12 scenarios in the first and second experimental setup, respectively (Supplementary Tables S13 and S14). Furthermore, unlike PPAD-AE, TA-RNN-AE provides higher interpretability through its dual-level attention mechanism.
Figure 4.
F2 scores for TA-RNN-AE models predicting conversion to AD at the next second, third, and fourth visits ahead. (A–D) Models were tested on held-out samples in ADNI after training using two, three, five, and six visits in ADNI, respectively. (E–H) Models were tested on NACC after training using two, three, five, and six visits in ADNI, respectively. Error bars represent standard deviations. Statistical significance between TA-RNN-AE and other tools was assessed using t-test (see Supplementary Tables S13 and S14 for P-values).
3.3 Ablation study
We conducted an ablation study for TA-RNN and TA-RNN-AE based on the first and second experimental setups to investigate the contribution of the time embedding layer that addresses the irregular time interval issue and the dual-level attention mechanism that improves the proposed models' interpretability. Here, we evaluated the predictive performance of the proposed models with two variations (i) without using the time embedding nor elapsed time, but using dual-level attention (i.e. A-RNN and A-RNN-AE); (ii) without using the dual-level attention mechanism but using time embedding (i.e. T-RNN and T-RNN-AE). We observed that the performance of A-RNN was lower compared to TA-RNN in all cases for the first (Table 3) and the second experimental setup (Supplementary Table S15). The results also showed that the performance of T-RNN was slightly lower compared to TA-RNN in two out of four cases for the first experimental setup (Table 3) and three out of four cases for the second experimental setup (Supplementary Table S15). In addition, we observed that A-RNN-AE performed lower than TA-RNN-AE in 7 out of 12 cases and remained unchanged in one case for the first experimental setup (Supplementary Table S16), and eleven out of twelve cases for the second experimental setup (Supplementary Table S17). Finally, the performance of T-RNN-AE was lower compared to TA-RNN-AE in all cases except for one case for the first experimental setup (Supplementary Table S16) and all cases for the second experimental setup (Supplementary Table S17). The ablation study showed a decrease in the performance of the proposed models across most scenarios, indicating the significance of the time embedding layer and dual-level attention mechanism in enhancing both the performance and interpretability of the models.
Table 3.
TA-RNN F2 scores for the ablation of dual-level attention and time embedding.a
| Model\Scenario | 2→1 | 3→1 | 5→1 | 6→1 |
|---|---|---|---|---|
| A-RNN | 0.844 ± 0.040 | 0.850 ± 0.035 | 0.855 ± 0.029 | 0.887 ± 0.035 |
| T-RNN | 0.852 ± 0.031 | 0.855 ± 0.019 | 0.869 ± 0.020 | 0.905 ± 0.036 |
| TA-RNN (proposed) | 0.851 ± 0.022 | 0.858 ± 0.029 | 0.870 ± 0.020 | 0.901 ± 0.041 |
A-RNN, TA-RNN without time embedding. T-RNN, TA-RNN without dual attention. TA-RNN was evaluated on held-out samples in ADNI after training using two, three, five, and six preceding visits in ADNI. Best F2 score in each case (column) is shown in bold.
3.4 Interpretations of the models’ predictions
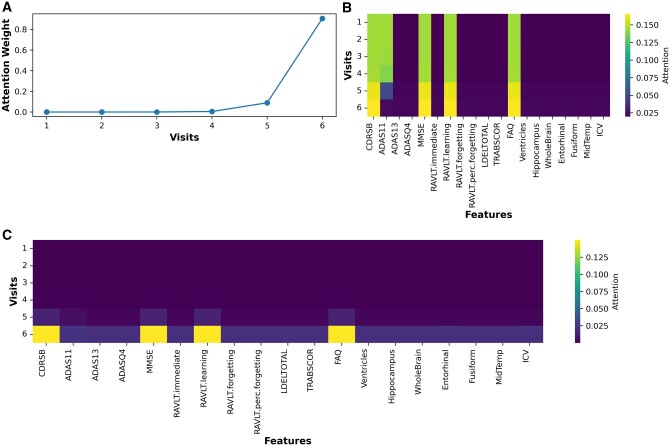
To demonstrate the interpretability of the models, we visualized their behavior during the prediction process using the generated attention weights for visits and features. In the first experimental setup, for example, TA-RNN model that was trained on six visits to predict the clinical outcome in the next visit generated six visit-level attention weights (i.e. one for each visit). The attention weight values for each visit range from 0 to 1, and the sum of all six weights equals to 1. Additionally, the model generated 19 attention weights, one for each feature (except for the age feature, which was not utilized in our models) at each visit, representing the feature-level attention. These weights provide insights into the influence of each visit and feature during the model's prediction of the clinical outcome. Figure 5, visualizing the attention weights at the visit level, demonstrates that for most samples the model prioritizes on the final visits—specifically the fifth and sixth visits—during predictions. Additionally, Fig. 6 shows the average attention weight at the feature level across all visits for the same example. We observed that CDRSB, MMSE, RAVLT.learning, and FAQ features exhibit the most substantial influence on the model’s predictions, aligning with observations in the literature (Nguyen et al. 2020, Velazquez and Lee 2021, Al Olaimat et al. 2023, Perera and Ganegoda 2023, Zhang et al. 2024). In addition, we evaluated TA-RNN’s interpretability in predicting MCI to AD conversion using a patient from the test dataset who converted from MCI to AD in the subsequent visit (seventh), where the model accurately predicted the conversion. We made the same observation that fifth and sixth visits are the most influential visit for this patient (Fig. 7A).
Figure 5.
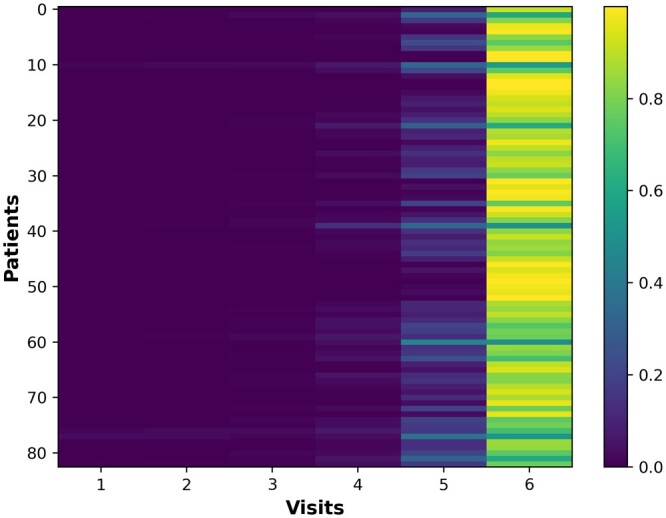
Attention weights at the visit level for TA-RNN. TA-RNN was evaluated on held-out samples in ADNI after training using six preceding visits in ADNI.
Figure 6.
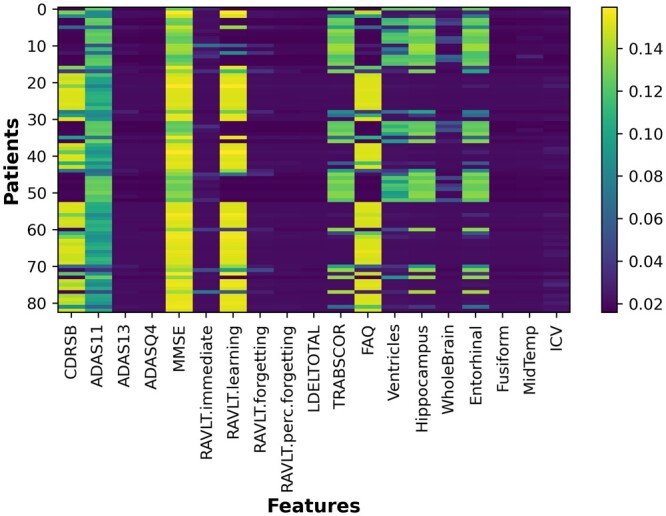
Average attention weights at the feature level for TA-RNN. TA-RNN was evaluated on held-out samples in ADNI after training using six preceding visits in ADNI.
Figure 7.
Visualization of TA-RNN’s behavior during correctly predicting the conversion from MCI to AD for a patient. A–C represent visits attention weights, features attention weights, and element-wise multiplication of attention weights, respectively. TA-RNN was evaluated on held-out samples in ADNI after training using six preceding visits to ADNI.
Furthermore, several cognitive test features were important regardless all visits (Fig. 7B). After combining feature and visit attention values, the model primarily focuses on CSRSB, MMSE, RAVLT.learning, and FAQ features in the model’s prediction (Fig. 7C). In light of the preceding observations, the dual-attention mechanism of our model holds significant promise for enhancing the interpretability and utility of our model.
4 Conclusion
In this study, we propose two RNN-based architectures, namely TA-RNN and TA-RNN-AE for the prediction of clinical outcomes in EHR at the next visit and multiple visits ahead, respectively. The proposed models were able to address the irregular time interval issue in longitudinal EHR through a time embedding layer that incorporates time intervals with original data. Additionally, model predictions were made interpretable by utilizing a dual-level attention mechanism that identifies the significant visits and features influencing each prediction. To evaluate the effectiveness of our proposed models, we employed three experimental setups using two extensive EHR datasets obtained from ADNI and NACC for predicting MCI to AD conversion, as well as data from MIMIC-III for mortality prediction. In all experimental setups, the results showed that TA-RNN and TA-RNN-AE outperformed baseline and SOTA methods in almost all cases. TA-RNN and TA-RNN-AE source code and documentation are publicly available at https://github.com/bozdaglab/TA-RNN/.
Supplementary Material
Contributor Information
Mohammad Al Olaimat, Department of Computer Science and Engineering, University of North Texas, Denton, TX 76203, United States; BioDiscovery Institute, University of North Texas, Denton, TX 76203, United States.
Serdar Bozdag, Department of Computer Science and Engineering, University of North Texas, Denton, TX 76203, United States; BioDiscovery Institute, University of North Texas, Denton, TX 76203, United States; Department of Mathematics, University of North Texas, Denton, TX 76203, United States.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Acknowledgement
Data collection and sharing for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health [U01 AG024904]) and DOD ADNI (Department of Defense award number W81XWH-12–2-0012). ADNI was funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. The NACC database was funded by NIA/NIH [U24 AG072122]. NACC data are contributed by the NIA-funded ADRCs: P30 AG062429 (PI James Brewer, MD, PhD), P30 AG066468 (PI Oscar Lopez, MD), P30 AG062421 (PI Bradley Hyman, MD, PhD), P30 AG066509 (PI Thomas Grabowski, MD), P30 AG066514 (PI Mary Sano, PhD), P30 AG066530 (PI Helena Chui, MD), P30 AG066507 (PI Marilyn Albert, PhD), P30 AG066444 (PI John Morris, MD), P30 AG066518 (PI Jeffrey Kaye, MD), P30 AG066512 (PI Thomas Wisniewski, MD), P30 AG066462 (PI Scott Small, MD), P30 AG072979 (PI David Wolk, MD), P30 AG072972 (PI Charles DeCarli, MD), P30 AG072976 (PI Andrew Saykin, PsyD), P30 AG072975 (PI David Bennett, MD), P30 AG072978 (PI Neil Kowall, MD), P30 AG072977 (PI Robert Vassar, PhD), P30 AG066519 (PI Frank aFerla, PhD), P30 AG062677 (PI Ronald Petersen, MD, PhD), P30 AG079280 (PI Eric Reiman, MD), P30 AG062422 (PI Gil Rabinovici, MD), P30 AG066511 (PI Allan Levey, MD, PhD), P30 AG072946 (PI Linda Van Eldik, PhD), P30 AG062715 (PI Sanjay Asthana, MD, FRCP), P30 AG072973 (PI Russell Swerdlow, MD), P30 AG066506 (PI Todd Golde, MD, PhD), P30 AG066508 (PI Stephen Strittmatter, MD, PhD), P30 AG066515 (PI Victor Henderson, MD, MS), P30 AG072947 (PI Suzanne Craft, PhD), P30 AG072931 (PI Henry Paulson, MD, PhD), P30 AG066546 (PI Sudha Seshadri, MD), P20 AG068024 (PI Erik Roberson, MD, PhD), P20 AG068053 (PI Justin Miller, PhD), P20 AG068077 (PI Gary Rosenberg, MD), P20 AG068082 (PI Angela Jefferson, PhD), P30 AG072958 (PI Heather Whitson, MD), P30 AG072959 (PI James Leverenz, MD).
Funding
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM133657 and the startup funds from the University of North Texas.
Data availability
The datasets were derived from sources in the public domain: ADNI: https://adni.loni.usc.edu/. NACC: https://naccdata.org/MIMIC-III: https://physionet.org/.
References
- Al Olaimat M, Martinez J, Saeed F. et al. ; Alzheimer’s Disease Neuroimaging Initiative. PPAD: a deep learning architecture to predict progression of Alzheimer’s disease. Bioinformatics 2023;39:i149–57. 10.1093/bioinformatics/btad249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baytas IM, Xiao C, Zhang X. et al. Patient subtyping via time-aware LSTM networks. In: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, pp. 65–74. Association for Computing Machinery (ACM), 2017. 10.1145/3097983.3097997. [DOI]
- Besser L, Kukull W, Knopman DS. et al. ; Neuropsychology Work Group, Directors, and Clinical Core leaders of the National Institute on Aging-funded US Alzheimer’s Disease Centers. Version 3 of the National Alzheimer’s Coordinating Center’s uniform data set. Alzheimer Disease Assoc Disord 2018;32:351–8. 10.1097/WAD.0000000000000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach Learn 2001;45:5–32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Cho K, van Merrienboer B, Bahdanau D. et al. On the Properties of Neural Machine Translation: Encoder-Decoder Approaches, 2014. arXiv preprint arXiv:1409.1259. [Google Scholar]
- Choi E, Bahadori MT, Sun J. et al. RETAIN: an interpretable predictive model for healthcare using reverse time attention mechanism. In: Lee D, Sugiyama M, Luxburg U, Guyon I, Garnett R (eds.), Advances in Neural Information Processing Systems, Barcelona, Spain, Vol. 29. Curran Associates, Inc., 2016, pp. 3504–12. https://proceedings.neurips.cc/paper_files/paper/2016/file/231141b34c82aa95e48810a9d1b33a79-Paper.pdf. [Google Scholar]
- Cortes C, Vapnik V.. Support-vector networks. Mach Learn 1995;20:273–97. 10.1007/BF00994018. [DOI] [Google Scholar]
- Fouladvand S, Talbert J, Dwoskin LP. et al. Predicting Opioid Use Disorder from Longitudinal Healthcare Data using Multi-Stream Transformer. 2021. arXiv preprint arXiv:2103.08800. [PMC free article] [PubMed]
- Goldberger AL, Amaral LAN, Glass L. et al. PhysioBank, PhysioToolkit, and PhysioNet. Circulation 2000;101:e215–20. 10.1161/01.CIR.101.23.e215. [DOI] [PubMed] [Google Scholar]
- Herp J, Braun J-M, Cantuaria ML. et al. Modeling of electronic health records for time-variant event learning beyond bio-markers—a case study in prostate cancer. IEEE Access 2023;11:50295–309. 10.1109/ACCESS.2023.3272745. [DOI] [Google Scholar]
- Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997;9:1735–80. 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Hossain E, Rana R, Higgins N. et al. Natural language processing in electronic health records in relation to healthcare decision-making: a systematic review. Comput Biol Med 2023;155:106649. 10.1016/j.compbiomed.2023.106649. [DOI] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Borowski BJ. et al. ; Alzheimer’s Disease Neuroimaging Initiative. Update on the magnetic resonance imaging core of the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement 2010;6:212–20. 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Bandy D, Chen K. et al. ; Alzheimer’s Disease Neuroimaging Initiative. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimers Dement 2010;6:221–9. 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Lin H, Chen Y. et al. Sex difference in the association of APOE4 with cerebral glucose metabolism in older adults reporting significant memory concern. Neurosci Lett 2020;722:134824. 10.1016/j.neulet.2020.134824. [DOI] [PubMed] [Google Scholar]
- Johnson A, Pollard T, Mark R.. MIMIC-III clinical database (version 1.4). PhysioNet 2016a;10:2. [Google Scholar]
- Johnson AEW, Pollard TJ, Shen L. et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016b;3:160035. 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma DP, Ba J. Adam: a method for stochastic optimization, 2014. https://arxiv.org/pdf/1412.6980.
- Lee G, Nho K, Kang B, for Alzheimer’s Disease Neuroimaging Initiative et al. Predicting Alzheimer’s disease progression using multi-modal deep learning approach. Sci Rep 2019;9:1952. 10.1038/s41598-018-37769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jun E, Choi J. et al. Multi-view integrative attention-based deep representation learning for irregular clinical time-series data. IEEE J Biomed Health Inform 2022;26:4270–80. 10.1109/JBHI.2022.3172549. [DOI] [PubMed] [Google Scholar]
- Li H, Fan Y. Early prediction of Alzheimer’s disease dementia based on baseline hippocampal MRI and 1-year follow-up cognitive measures using deep recurrent neural networks. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, pp. 368–71, 2019. 10.1109/ISBI.2019.8759397. [DOI] [PMC free article] [PubMed]
- Liu LJ, Ortiz-Soriano V, Neyra JA. et al. KIT-LSTM: knowledge-guided time-aware LSTM for continuous clinical risk prediction. In: Proceedings – 2022 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2022, Las Vegas, NV, USA, pp. 1086–91, 2022. 10.1109/BIBM55620.2022.9994931. [DOI] [PMC free article] [PubMed]
- Luong DTA, Chandola V. Learning deep representations from clinical data for chronic kidney disease. In: 2019 IEEE International Conference on Healthcare Informatics (ICHI), Xi'an, China, pp. 1–10. 2019. 10.1109/ICHI.2019.8904646. [DOI]
- Lyu W, Dong X, Wong R. et al. A multimodal transformer: fusing clinical notes with structured EHR data for interpretable in-hospital mortality prediction. AMIA Annu Symp Proc 2022;2022:719–28. [PMC free article] [PubMed] [Google Scholar]
- McCulloch WS, Pitts W.. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophy 1943;5:115–33. 10.1007/BF02478259. [DOI] [PubMed] [Google Scholar]
- Nguyen M, He T, An L. et al. ; Alzheimer’s Disease Neuroimaging Initiative. Predicting Alzheimer’s disease progression using deep recurrent neural networks. NeuroImage 2020;222:117203. 10.1016/j.neuroimage.2020.117203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera LRD, Ganegoda GU. Alzheimer’s disease prediction using clinical data approach. In: 2023 8th International Conference on Information Technology Research (ICITR), Colombo, Sri Lanka, pp. 1–6, 2023. 10.1109/ICITR61062.2023.10382737. [DOI]
- Risacher SL, Shen L, West JD. et al. ; Alzheimer’s Disease Neuroimaging Initiative (ADNI). Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiol Aging 2010;31:1401–18. 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud TM. et al. ; Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement 2010;6:265–73. 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Ye M, Yang B. et al. DATA-GRU: dual-attention time-aware gated recurrent unit for irregular multivariate time series. AAAI 2020;34:930–7. 10.1609/aaai.v34i01.5440. [DOI] [Google Scholar]
- Trojanowski JQ, Vandeerstichele H, Korecka M. et al. ; Alzheimer’s Disease Neuroimaging Initiative. Update on the biomarker core of the Alzheimer’s Disease Neuroimaging Initiative subjects. Alzheimers Dement 2010;6:230–8. 10.1016/j.jalz.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaswani A, Shazeer N, Parmar N. et al. Attention is all you need. In: Guyon I, Von Luxburg U, Bengio S, Wallach H, Fergus R, Vishwanathan S, Garnett R (eds.), Advances in Neural Information Processing Systems, Long Beach, USA, Vol. 30. La Jolla, USA: Neural Information Processing Systems (NIPS), 2017. https://proceedings.neurips.cc/paper_files/paper/2017/file/3f5ee243547dee91fbd053c1c4a845aa-Paper.pdf. [Google Scholar]
- Velazquez M, Lee Y; Alzheimer’s Disease Neuroimaging Initiative. Random Forest model for feature-based Alzheimer’s disease conversion prediction from early mild cognitive impairment subjects. PLoS One 2021;16:e0244773. 10.1371/journal.pone.0244773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopalan J, Tong L, Hassanzadeh HR. et al. Multimodal deep learning models for early detection of Alzheimer’s disease stage. Sci Rep 2021;11:3254. 10.1038/s41598-020-74399-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Aisen PS, Jack CR. et al. ; Alzheimer’s Disease Neuroimaging Initiative. The Alzheimer’s disease Neuroimaging Initiative: progress report and future plans. Alzheimers Dement 2010;6:202–11.e7. 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS. et al. The Alzheimer’s disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer’s Dement 2013;9:e111–94. 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xi X, Chen J. et al. A survey of deep learning for electronic health records. Appl Sci 2022;12:11709. 10.3390/app122211709. [DOI] [Google Scholar]
- Yadav P, Steinbach M, Kumar V. et al. Mining electronic health records (EHRs). ACM Comput Surv 2018;50:1–40. 10.1145/3127881. [DOI] [Google Scholar]
- Yang S, Varghese P, Stephenson E. et al. Machine learning approaches for electronic health records phenotyping: a methodical review. J Am Med Inform Assoc 2023;30:367–81. 10.1093/jamia/ocac216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chen A, PourNejatian N. et al. A large language model for electronic health records. NPJ Digit Med 2022;5:194. 10.1038/s41746-022-00742-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shen S, Li X. et al. A multiclass extreme gradient boosting model for evaluation of transcriptomic biomarkers in Alzheimer’s disease prediction. Neurosci Lett 2024;821:137609. 10.1016/j.neulet.2023.137609. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang X, Ivy J. et al. ATTAIN: attention-based time-aware LSTM networks for disease progression modeling. In: Proceedings of the 28th International Joint Conference on Artificial Intelligence, 2019, pp. 4369–75.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets were derived from sources in the public domain: ADNI: https://adni.loni.usc.edu/. NACC: https://naccdata.org/MIMIC-III: https://physionet.org/.