Abstract
Mature human immunodeficiency virus type 1 (HIV-1) virions contain a typically cone-shaped core that encases the viral genome. In this study, we established conditions which allowed the efficient isolation of morphologically intact HIV-1 cores from virions. The isolated cores consisted mostly of cones which appeared uniformly capped at both ends but were heterogeneous with respect to the shape of the broad cap as well as the dimensions and angle of the cone. Vpr, a nonstructural virion component implicated in the nuclear import of the viral genome, was recovered in core preparations of HIV-1 and simian immunodeficiency viruses from African green monkeys. Unexpectedly, p6gag, a structural protein required for the incorporation of Vpr, was absent from HIV-1 core preparations. Taken together, our results indicate that the incorporation of Vpr into the virion core is a conserved feature of primate lentiviruses and that the interactions required for the uptake of Vpr into assembling particles differ from those which confine Vpr within the core.
Retroviruses undergo a process of maturation during which a distinct core structure is acquired (reviewed in reference 39). This morphological transformation depends on the presence of active viral protease, which cleaves the Gag polyprotein into its mature subunits, among which matrix (MA), capsid (CA), and nucleocapsid (NC) are found in all retroviruses. Human immunodeficiency virus type 1 (HIV-1) MA is myristylated at its N terminus and remains associated with the host cell-derived lipid envelope of the mature virion (reviewed in reference 13). CA forms the core structure, which in the case of HIV-1 is typically cone shaped, and NC is thought to cover the genomic viral RNA within the core (13).
The C terminus of the HIV-1 Gag polyprotein Pr55gag yields a peptide designated p6gag, which is found only in primate lentiviruses and whose location within the mature virion is unknown. The p6gag domain facilitates the release of assembled particles from the cell surface (15) and is essential for the virion association of the regulatory viral protein Vpr (21, 27, 29). Vpr is expressed by all primate lentiviruses, and one subgroup of primate lentiviruses in addition encodes a related virion-associated protein designated Vpx. Lentiviral Vpr proteins inhibit cell proliferation in the G2 phase of the cell cycle (8, 16, 19, 30, 31, 34, 36), perhaps because virus production is optimal in the G2 phase (14). HIV-1 Vpr also facilitates the nuclear import of the viral genome in nondividing host cells (17) and would thus be expected to be a component of the virion core. Indeed, Vpx, which shares the nuclear import function of HIV-1 Vpr (8), was found associated with mature HIV-2 cores (20). However, in the case of a very closely related simian immunodeficiency virus (SIV) from macaques, most or all of the Vpx protein present in virions was lost during the purification of mature core structures (25, 45).
To examine whether Vpr and its interaction partner p6gag are components of the mature HIV-1 core, we developed a method to isolate intact core structures from virions. The biochemical analysis of HIV-1 cores has been hampered by their marked tendency to dissociate in the presence of weak detergents. Therefore, we employed a previously described procedure in which virions are spun through a detergent layer to minimize the time of exposure (37). In initial experiments, a layer containing the detergent Igepal CO-630 in 15% sucrose was placed on top of a 20 to 70% sucrose gradient to separate cores by equilibrium centrifugation. As expected, CA peaked in fractions with densities of 1.14 and 1.16 g/ml in the absence of detergent. In contrast, when Igepal CO-630 was present in the 15% sucrose layer, most of CA was found near the top of the gradient and presumably represented material from completely disrupted virions. Even at 0.03% Igepal CO-630, the fractions with densities of between 1.14 and 1.21 g/ml contained very little CA, indicating that none of the input virus remained intact. However, at this low concentration of detergent, a distinct peak of CA was obtained at densities of between 1.24 and 1.28 g/ml, which is similar to the reported density of HIV-2 cores (20). Although these results indicated that HIV-1 cores can in principle be obtained with the “spin thru” technique, only a small fraction of CA banded at the expected density of cores (data not shown). Similarly poor yields of HIV-1 cores were recently reported by Kotov et al. (23), who also employed a “spin thru” procedure in combination with equilibrium density centrifugation.
In an effort to improve the yield of HIV-1 cores, we modified the “spin thru” method by replacing the sucrose gradient with a 30% sucrose cushion. This approach was chosen to avoid the exposure of isolated cores to higher concentrations of sucrose because of the possibility that these structures are osmotically sensitive. Furthermore, the results described above indicated that only cores remained sufficiently dense after centrifugation through detergent to pass through the sucrose cushion. 293T cells were transfected with pHXBH10ΔenvCAT/D116A (32), a noninfectious variant of the full-length proviral clone pHXBH10 with a deletion in env and an active-site mutation in integrase (IN), followed by metabolic labeling with [35S]cysteine. Virus-containing supernatant was split into two aliquots, and one half was loaded on top of a 20% sucrose layer containing 0.03% Igepal CO-630, which had been placed on top of a 30% sucrose cushion. The other half of the supernatant was loaded onto an identical step gradient, except that the detergent was omitted. After centrifugation at 4°C for 2 h in a Beckman SW41 rotor operated at 27,000 rpm, the pelleted material was resuspended and directly analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
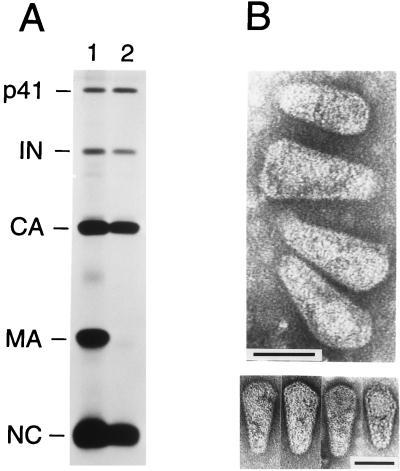
The mature Gag products were differentially affected by the presence of detergent in the 20% sucrose layer (Fig. 1A). PhosphorImager analysis indicated that about one-third of the CA present in the input virus was recovered in the particulate fraction after passage through the detergent layer. As in the case of intact virus, CA and NC were present in about equimolar quantities. In contrast, MA was almost entirely depleted from particulate material spun through a layer of Igepal CO-630. Also, it appeared that a trace amount that remained after passage through detergent migrated slightly faster than the bulk of virus-associated MA. The pol-encoded proteolytic cleavage product IN was recovered in similar molar amounts relative to CA or NC in both the presence and absence of detergent (Fig. 1A). These results suggested that brief exposure to detergent had effectively removed the viral envelope and the associated MA protein but had left a significant proportion of the viral cores intact.
FIG. 1.
Biochemical and electron microscopic analysis of HIV-1 core preparations. (A) Equal amounts of supernatant containing [35S]cysteine-labeled virus were loaded on top of 20% sucrose layers which either lacked detergent (lane 1) or contained 0.03% Igepal Co-630 (lane 2) and which were supported by 30% sucrose cushions. Following centrifugation, pelleted material was directly analyzed by SDS-PAGE and autoradiography. The product designated p41 is an intermediate Gag cleavage product (2). (B) Electron micrographs of isolated HIV-1 cores prepared by centrifugation through a detergent layer and a 30% sucrose cushion. The lower panel shows examples of cores with angular caps or with a spherical structure that is discernible at the broad end. Bars, 50 nm.
To confirm the isolation of HIV-1 cores, particulate material spun through a layer of 0.03% Igepal CO-630 and a 30% sucrose cushion was prepared for electron microscopy. The pelleted material was resuspended in a small volume of phosphate-buffered saline containing 2.5% glutaraldehyde as a fixative and was then allowed to sediment onto carbon grids, followed by negative staining with uranyl acetate. Transmission electron microscopy revealed the presence of numerous isolated cones which varied in length and diameter and which closely resembled the cone-shaped cores visible within mature HIV-1 virions (Fig. 1B). No intact viral particles could be detected; however, in addition to cones, tubes were occasionally seen. In some of the cones, a spherical density was discernible at or near the broad end, which may represent the condensed RNA-NC complex. The cones always appeared capped at both ends but were heterogeneous in the shape of the broad cap, which in some specimens had a distinctly angular profile (Fig. 1B, lower panel).
Because a model based on synthetic HIV-1 cores which predicts specific cone angles has recently been proposed (12), it was of particular interest to determine the cone angles of the core-like structures isolated from authentic viral particles. Our measurements revealed considerable variability among cone angles, from 10 to 22°, with a mean cone angle of about 17° (n = 53) and a standard deviation from the mean of 19.3% (data not shown). In the model of Ganser et al. (12), retroviral cores are composed of closed hexagonal lattices, and the conical shape of HIV-1 cores is dictated by the location of the 12 pentameric defects required to close the lattice. The narrowest cone angle allowed by this model is 19.2° (12). However, we cannot exclude that the small deviation of the average cone angle from the theoretical value observed in the present study was an artifact caused by fixation and negative staining.
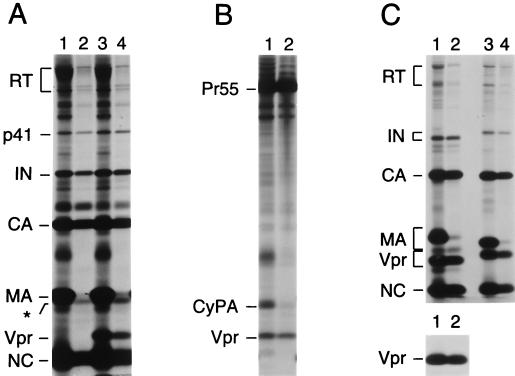
It has been shown that HIV-1 Vpr, a nonstructural component of the virion, remains associated with the viral preintegration complex and facilitates its nuclear import in nondividing cells (17). Vpr is thus expected to localize to the virion core during virus maturation. Having demonstrated that the modified “spin thru” method described above generates intact cores, we used this technique to determine whether Vpr associates with the virion core. [35S]cysteine-labeled HIV-1 virions were produced in 293T cells transfected with the infectious HXBH10 provirus, which is vpr negative, or with the vpr-positive variant HXBH10/R+ (7). As expected, the 14-kDa Vpr protein was observed only in virions produced by HXBH10/R+, confirming the identity of the product (Fig. 2A, lanes 1 and 3). Passage of virions through 0.03% Igepal CO-630 again removed most MA. However, the levels of residual MA were somewhat higher than in other experiments, and the existence of two closely spaced protein bands which comigrated with virion-associated MA became clearly evident (Fig. 2A, lanes 2 and 4). The more prominent of these two bands migrated slightly faster than the bulk of virion-associated MA. Additionally, a protein of unknown identity that migrated above CA was recovered in the core preparations shown in Fig. 2A. Apart from CA, NC, and IN, Vpr was clearly enriched relative to MA in particulate material spun through detergent, indicating that HIV-1 Vpr is a component of the virion core (Fig. 2A, lane 4). It is also noteworthy that the recovery of reverse transcriptase (RT) in the core preparations appeared to be less efficient than that of IN, raising the possibility that not all of the RT present in the virion is located within the core.
FIG. 2.
Association of Vpr with HIV-1 and SIVagm cores. [35S]cysteine-labeled wild-type HIV-1 virions (A), protease-defective immature HIV-1 virions (B), or wild-type SIVagm virions (C) were spun through step gradients which either lacked detergent (odd-numbered lanes) or contained detergent in the upper layer (even-numbered lanes) as described for Fig. 1. Pelleted material was analyzed directly by SDS-PAGE and autoradiography. Virus-containing supernatants were obtained by transfection of 293T cells with proviral plasmids and metabolic labeling. In panel A, the transfected HIV-1 proviruses were the infectious, vpr-negative molecular clone HXBH10 (lanes 1 and 2) and its vpr-positive variant HXBH10/R+ (lanes 3 and 4). The protease-defective HIV-1 provirus used in panel B was HXBH10/R+/PR− (4). The SIVagm proviruses in panel C were SIVagm 155-4 (lanes 1 and 2) and SIVagm gri-1 (lanes 3 and 4). In panel A, the p66 form of RT is partially obscured by a contaminant. The asterisk indicates the position of an unidentified core-associated protein that migrates slightly faster than the bulk of MA.
We considered the possibility that the minor [35S]cysteine-labeled species which comigrated with the front of MA from intact virions (indicated by an asterisk in Fig. 2A) was cyclophilin A (CyPA), a host protein that is specifically incorporated into HIV-1 virions via an interaction with CA (7, 9, 38). We therefore examined by Western blotting whether CyPA remained associated with viral particulate material spun through detergent. However, the amount of CyPA in the particulate fraction did not exceed the background levels obtained under identical conditions with a CA mutant defective for CyPA incorporation (5) or when CyPA incorporation was blocked by cyclosporin A (data not shown). Moreover, we observed that immature HIV-1 particles produced by a protease mutant were largely depleted of CyPA, but not of Vpr, after passage through a layer containing 0.03% Igepal CO-630 (Fig. 2B). In contrast to mature HIV-1 cores, immature capsids are relatively stable in the presence of detergent (40). The selective loss of CyPA both from mature and from immature capsids upon exposure to detergent suggests that virion-associated CyPA is only loosely bound to CA.
To examine whether the Vpr proteins of other primate lentiviruses behave similarly to that of HIV-1, we analyzed [35S]cysteine-labeled virions produced by full-length molecular clones of SIV strains from African green monkeys (SIVagm). Consistent with the high degree of sequence variation between SIVagm 155-4 and SIVagm gri-1 (18), the electrophoretic mobilities of the MA and Vpr proteins of these viruses differed considerably (Fig. 2C, lanes 1 and 3). Quantitation by PhosphorImager analysis and normalization for the number of cysteine residues present indicated that virions produced by both SIVagm strains contained about equimolar amounts of Gag and Vpr (Fig. 2C). In contrast, HIV-1 virions produced by different vpr-positive strains contained 5- to 10-fold-smaller amounts of Vpr (Fig. 2A and data not shown). PhosphorImager analysis also showed that centrifugation of SIVagm 155-4 virions through detergent removed more than 95% of MA. In contrast, about half of the CA, NC, and Vpr present in the input virus was recovered in the particulate fraction (Fig. 2C, lanes 1 and 2). Immunoprecipitation with a specific antiserum confirmed the efficient recovery of Vpr after passage through detergent (Fig. 2C, lower panel). Centrifugation of SIVagm gri-1 virions through detergent yielded similar results (Fig. 2C, lanes 3 and 4). Taken together, these observations indicate that the Vpr proteins of widely divergent primate lentiviruses associate with the core component of the virion.
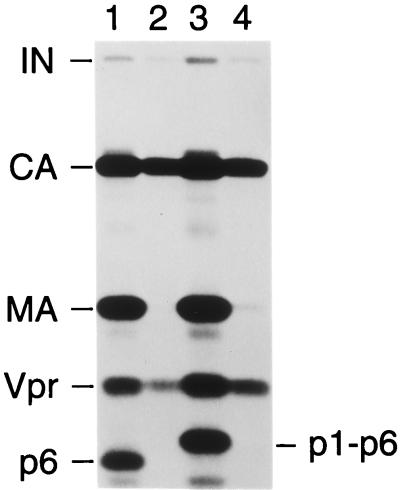
The incorporation of Vpr into assembling particles is mediated by p6gag, the C-terminal domain of the Gag polyprotein of primate lentiviruses (1, 21, 22, 27, 29, 35). Therefore, we examined whether p6gag is retained in HIV-1 core preparations together with Vpr. Because HIV-1 p6gag lacks methionine or cysteine residues, we used [3H]leucine for metabolic labeling. This approach also helped to distinguish p6gag from NC, which has a similar electrophoretic mobility in SDS-PAGE but lacks leucine residues. To unequivocally identify p6gag, we generated a mutant version of HXBH10/R+ that has the phenylalanine at the P1 position of the cleavage site between p1 and p6gag changed to isoleucine. Based on previous studies (2, 24), the presence of a β-branched amino acid at the P1 position was expected to block cleavage at the p1-p6 site and thus to induce a shift in the electrophoretic mobility of p6gag. Figure 3 shows the profile of [3H]leucine-labeled proteins in wild-type and mutant HXBH10/R+ virions pelleted through sucrose step gradients. If detergent was omitted from the upper layer of the step gradient, a prominently labeled product migrated at the predicted position of p6gag (lane 1) and, as expected, exhibited a reduced mobility if the p1-p6 cleavage site was mutated (lane 3). As before, passage through a layer containing 0.03% Igepal CO-630 resulted in the nearly complete loss of MA, whereas a significant portion of the CA protein present in intact virions remained particulate (lanes 2 and 4). Surprisingly, although Vpr was partially recovered, p6gag and the p1-p6gag fusion protein were completely removed from the particulate fraction by centrifugation of the wild-type or mutant virions through detergent (Fig. 3). Because our electron microscopy analysis demonstrated that the particulate fraction contained morphologically intact cores, it can be inferred that p6gag was absent from these isolated core structures.
FIG. 3.
Absence of p6gag from HIV-1 core preparations. [3H]leucine-labeled HIV-1 virions were spun through step gradients which either lacked detergent (odd-numbered lanes) or contained detergent in the upper layer (even-numbered lanes) as described for Fig. 1, and pelleted material was analyzed by SDS-PAGE. Virus was produced in 293T cells, and the transfected HIV-1 proviruses were the infectious, vpr-positive clone HXBH10/R+ (lanes 1 and 2) or a variant with a point mutation at the p1-p6 cleavage site (lanes 3 and 4).
MA was essentially absent from several of our HIV-1 core preparations (e.g., in Fig. 3), but appeared to be depleted to a lesser extent in HIV-1 core preparations recently obtained by Kotov et al. (23) and Welker et al. (42). In the latter study, HIV-1 virions were briefly exposed to detergent and cores were recovered by rapid centrifugation in a microcentrifuge. Electron microscopy analysis showed that the resulting core preparations were not completely pure (42), which may explain the presence of residual MA. However, Kotov et al. observed an enrichment of MA at the expected density of cores (23), suggesting that under certain isolation conditions some MA can remain associated with HIV-1 cores. A phosphorylated form of MA was previously detected in HIV-2 core preparations (11).
Because of the C-terminal location of p6gag within the Gag precursor, the absence of p6gag from our core preparations was unexpected. An early model of retroviral assembly (3), which is supported by recent cryoelectron microscopy studies (10, 44), proposes that the Gag polyprotein monomers are arranged radially in immature particles, with their N termini attached to the lipid envelope and their C termini oriented towards the center of the particle. Consistent with such an arrangement, the positions of the major Gag cleavage products within the precursor reflect their location within the mature virion (3), but it appears that p6gag does not conform to this rule. Interestingly, similar to HIV-1 p6gag, the p9gag protein of the nonprimate lentivirus equine infectious anemia virus, which occupies an analogous position in the Gag precursor, did not copurify with virion core components (33).
Despite the absence of p6gag, Vpr was recovered in core preparations, consistent with a function of Vpr early in the viral life cycle as a component of the reverse transcription complex (17). Our results are in good agreement with a very recent study by Welker et al. (42), who found Vpr enriched in HIV-1 core preparations obtained by a different method and also noted that p6gag was essentially absent. In apparent contrast, an earlier study localized Vpr immediately beneath the envelope of HIV-1 virions, suggesting that Vpr may not be a core component (41). However, the resolution of the immunelectron microscopy technique used in that study appears to be too low to unequivocally assign Vpr to a specific subviral compartment. In addition to Vpr, a small amount of the accessory viral protein Nef is detectable in HIV-1 virions (4, 28, 43), and a recent study indicates that Nef associates with the HIV-1 core (23). Furthermore, Vif, another accessory lentiviral protein, was found associated with HIV-1 core structures (26). However, whether Vif is a genuine virion component or a contaminant remains controversial, because others have reported that Vif is essentially absent from highly purified HIV-1 particles (6).
Because p6gag is essential for the incorporation of Vpr (21, 27, 29), our observations indicate that the interactions which concentrate Vpr in the core may differ from those which mediate its specific uptake into assembling particles. However, a role of p6gag as a vehicle which brings Vpr into the core cannot be entirely excluded, because our results with CyPA raise the possibility that p6gag was extracted from the core during the isolation procedure.
Acknowledgments
We thank Vanessa Hirsch for providing the SIVagm 155-4 and SIVagm gri-1 proviral clones and antiserum against SIVagm Vpr, and we thank Bettina Strack for helpful advice.
M.A.A. was supported by National Cancer Institute training grant T32 CA09141. This work was supported by National Institutes of Health grants AI29873 and AI28691 (Center for AIDS Research) and by a gift from the G. Harold and Leila Y. Mathers Charitable Foundation.
REFERENCES
- 1.Accola M A, Bukovsky A A, Jones M S, Göttlinger H G. A conserved dileucine-containing motif in p6gag governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIVmac and SIVagm. J Virol. 1999;73:9992–9999. doi: 10.1128/jvi.73.12.9992-9999.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola M A, Höglund S, Göttlinger H G. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolognesi D P, Montelaro R C, Frank H, Schäfer W. Assembly of type C oncornaviruses: a model. Science. 1978;199:183–186. doi: 10.1126/science.202022. [DOI] [PubMed] [Google Scholar]
- 4.Bukovsky A, Dorfman T, Weimann A, Göttlinger H G. Nef association with human immunodeficiency virus type 1 virions and cleavage by the viral protease. J Virol. 1997;71:1013–1018. doi: 10.1128/jvi.71.2.1013-1018.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukovsky A A, Weimann A, Accola M A, Göttlinger H G. Transfer of the HIV-1 cyclophilin-binding site to simian immunodeficiency virus from Macaca mulatta can confer both cyclosporin sensitivity and cyclosporin dependence. Proc Natl Acad Sci USA. 1997;94:10943–10948. doi: 10.1073/pnas.94.20.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dettenhofer M, Yu X F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorfman T, Göttlinger H G. The human immunodeficiency virus type 1 capsid p2 domain confers sensitivity to the cyclophilin-binding drug SDZ NIM 811. J Virol. 1996;70:5751–5757. doi: 10.1128/jvi.70.9.5751-5757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher T M, Brichacek B, Sharova N, Newman M A, Stivahtis G, Sharp P M, Emerman M, Hahn B H, Stevenson M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIVSM. EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 9.Franke E K, Yuan H E, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 10.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 11.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 12.Ganser B K, Li S, Klishko V Y, Finch J T, Sundquist W I. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 13.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 14.Goh W C, Rogel M E, Kinsey C M, Michael S F, Fultz P N, Nowak M A, Hahn B H, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 15.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson P R, Fomsgaard A, Allan J, Gravell M, London W T, Olmsted R A, Hirsch V M. Simian immunodeficiency viruses from African green monkeys display unusual genetic diversity. J Virol. 1990;64:1086–1092. doi: 10.1128/jvi.64.3.1086-1092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M-L, Chen I S Y. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kewalramani V N, Emerman M. Vpx association with mature core structures of HIV-2. Virology. 1996;218:159–168. doi: 10.1006/viro.1996.0176. [DOI] [PubMed] [Google Scholar]
- 21.Kondo E, Göttlinger H G. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J Virol. 1996;70:159–164. doi: 10.1128/jvi.70.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo E, Mammano F, Cohen E A, Göttlinger H G. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotov A, Zhou J, Flicker P, Aiken C. Association of Nef with the human immunodeficiency virus type 1 core. J Virol. 1999;73:8824–8830. doi: 10.1128/jvi.73.10.8824-8830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kräusslich H-G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liska V, Spehner D, Mehtali M, Schmitt D, Kirn A, Aubertin A-M. Localization of viral protein X in simian immunodeficiency virus macaque strain and analysis of its packaging requirements. J Gen Virol. 1994;75:2955–2962. doi: 10.1099/0022-1317-75-11-2955. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Wu X, Newman M, Shaw G M, Hahn B H, Kappes J C. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J Virol. 1995;69:7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y-L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandori M W, Fitch N J S, Craig H M, Richman D D, Spina C A, Guatelli J C. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J Virol. 1996;70:4283–4290. doi: 10.1128/jvi.70.7.4283-4290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Planelles V, Jowett J B M, Li Q-X, Xie Y, Hahn B, Chen I S Y. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J Virol. 1996;70:2516–2524. doi: 10.1128/jvi.70.4.2516-2524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Re F, Braaten D, Franke E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reil H, Bukovsky A A, Gelderblom H R, Göttlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts M M, Orozlan S. The preparation and biochemical characterization of intact capsids of equine infectious anemia virus. Biochem Biophys Res Commun. 1989;160:486–494. doi: 10.1016/0006-291x(89)92459-5. [DOI] [PubMed] [Google Scholar]
- 34.Rogel M E, Wu L I, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selig L, Pages J-C, Tanchou V, Preveral S, Berlioz-Torrent C, Liu L X, Erdtmann L, Darlix J-L, Benarous R, Benichou S. Interaction with the p6 domain of the Gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J Virol. 1999;73:592–600. doi: 10.1128/jvi.73.1.592-600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stivahtis G L, Soares M A, Vodicka M A, Hahn B H, Emerman M. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J Virol. 1997;71:4331–4338. doi: 10.1128/jvi.71.6.4331-4338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stromberg K, Hurley N E, Davis N L, Rueckert R R, Fleissner E. Structural studies of avian myeloblastosis virus: comparison of polypeptides in virion and core component by dodecyl sulfate-polyacrylamide gel electrophoresis. J Virol. 1972;13:513–528. doi: 10.1128/jvi.13.2.513-528.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Göttlinger H G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 39.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–69. [PubMed] [Google Scholar]
- 40.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J J, Lu Y-L, Ratner L. Particle assembly and Vpr expression in human immunodeficiency virus type 1-infected cells demonstrated by immunoelectron microscopy. J Gen Virol. 1994;75:2607–2614. doi: 10.1099/0022-1317-75-10-2607. [DOI] [PubMed] [Google Scholar]
- 42.Welker R, Hohenberg H, Tessmer U, Huckhagel C, Kräusslich H-G. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J Virol. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welker R, Kottler H, Kalbitzer H R, Kräusslich H-G. Human immunodeficiency virus type 1 Nef protein is incorporated into virus particles and specifically cleaved by the viral proteinase. Virology. 1996;219:228–236. doi: 10.1006/viro.1996.0240. [DOI] [PubMed] [Google Scholar]
- 44.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X, Matsuda Z, Yu Q-C, Lee T-H, Essex M. Vpx of simian immunodeficiency virus is localized primarily outside the virus core in mature virions. J Virol. 1993;67:4386–4390. doi: 10.1128/jvi.67.7.4386-4390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]