Abstract
The cytoplasmic tail (R peptide) sequence is able to regulate the fusion activity of the murine leukemia virus (MuLV) envelope (Env) protein. We have previously shown that this sequence exerts a profound inhibitory effect on the fusion activity of simian immunodeficiency virus (SIV)-MuLV chimeric Env proteins which contain the extracellular and transmembrane domains of the SIV Env protein. Recent studies have shown that SIV can utilize several alternative cellular coreceptors for its fusion and entry into the cell. We have investigated the fusion activity of SIV and SIV-MuLV chimeric Env proteins using cells that express different coreceptors. HeLa cells were transfected with plasmid constructs that carry the SIV or SIV-MuLV chimeric Env protein genes and were overlaid with either CEMx174 cells or Ghost Gpr15 cells, which express the Gpr15 coreceptor for SIV, or Ghost CCR5 cells, which express CCR5, an alternate coreceptor for SIV. The R-peptide sequence in the SIV-MuLV chimeric proteins was found to inhibit the fusion with CEMx174 cells or Ghost Gpr15 cells. However, a significant level of fusion was still observed when HeLa cells expressing the chimeric Env proteins were cocultivated with Ghost CCR5 cells. These results show that the R-peptide sequence exerts differential effects on the fusion activity of SIV Env proteins using target cells that express alternative coreceptors.
The envelope (Env) proteins of retroviruses play an important role in virus infection and replication, as they mediate virus entry into the cells through interacting with virus receptors and then induce fusion between viral and cellular membranes. Retrovirus Env proteins are type I transmembrane proteins. They are synthesized as precursor proteins which are subsequently processed into two subunits, the surface protein (SU) and the transmembrane protein (TM), by a cellular protease (21, 24, 40). The Env proteins of human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) are unusual in that they contain very long cytoplasmic domains (with more than 150 amino acids) in comparison with most other viral Env proteins, which contain cytoplasmic domains with less than 50 amino acids. It has been reported that mutations in the cytoplasmic domain of the HIV and SIV Env proteins affect their incorporation into virus particles and their membrane fusion activities (19, 45). For SIV, it has been shown that passage in human T-cell lines, such as CEMx174 cells, results in the truncation of its long cytoplasmic domain to 18 amino acids, and such a truncation of the cytoplasmic domain increases the membrane fusion activity of the SIV Env protein and enhances SIV replication in these cell lines (7, 23, 35, 45). However, due to an unknown mechanism, the full-length Env protein of SIV confers an advantage for virus replication in rhesus peripheral blood mononuclear cells (23, 26). Previous studies have shown that the truncation of the SIVmac239 Env protein cytoplasmic domain results in a conformational change in its extracellular domain (38). However, the mechanism by which the cytoplasmic domain affects the replication of SIV is still not clearly understood.
Regulation of the fusion activity of viral Env proteins by changes in the cytoplasmic domain has also been observed with other viruses. In murine leukemia virus (MuLV), the processing of the Env protein removes a C-terminal fragment of 16 amino acids, which has been designated the R peptide (20, 22). It has been shown that removal of the R peptide is important for activating the protein's membrane fusion activity (32, 33). Cells expressing the truncated MuLV Env protein, but not the full-length form, induce extensive syncytium formation when they are cocultivated with cells expressing receptors for MuLV. Similar observations have also been reported for the Env protein of type D retroviruses (3, 37). The regulation of the membrane fusion activity of the Env proteins by its processing in virions may be an advantage for the virus, since it may prevent the cytopathic effect of the Env protein that otherwise might be detrimental to virus production. We have shown that the MuLV R peptide also exerts a profound inhibitory effect on cell fusion activity of SIV-MuLV chimeric Env proteins, in which the cytoplasmic domain of the SIV Env protein is replaced by that of the MuLV Env protein (42).
In recent studies, it has been found that in addition to CD4, membrane fusion induced by HIV and SIV Env proteins requires the presence of specific coreceptors, which are seven-transmembrane proteins also serving as receptors for chemokines (1, 10, 13, 14, 17). It has been shown that the SIV Env protein can utilize CCR5, Gpr1 (Bonzo), and Gpr15 (BOB) as coreceptors (2, 4, 9, 11, 15, 29, 36). The discovery of the coreceptors for the HIV and SIV Env proteins has shed new light on the mechanism of their membrane fusion activities. Several studies have reported that soluble CD4 enhances the interaction of the HIV and SIV Env proteins with their coreceptors (28, 30). Furthermore, it has been shown that interaction of the SU protein with CD4 results in a conformational change in the HIV Env protein (25, 27, 41). By analogy with influenza virus hemagglutinin (HA), it was postulated that the interaction with CD4 induces a conformational change in the ectodomain of the HIV and SIV Env proteins which enhances their interaction with coreceptors and, in turn, results in the extension of the fusion peptide at the N terminus of the TM protein to insert it into the target membrane to initiate the fusion process (12, 31).
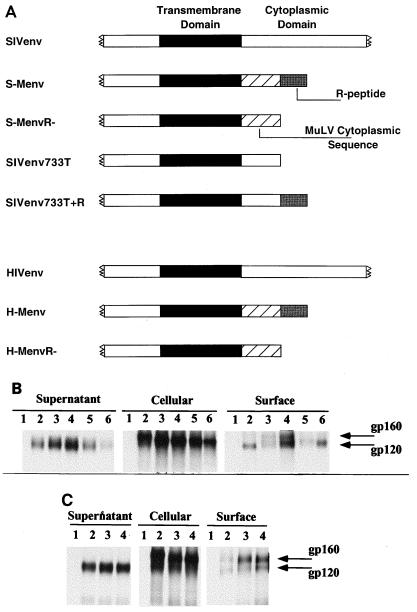
In this study, to further investigate the mechanism of fusion inhibition by the MuLV R peptide, we examined the cell fusion activities of SIV-MuLV chimeric Env proteins in relation to their coreceptor usage. We also constructed HIV-MuLV chimeric Env proteins and compared their abilities to induce cell fusion. Shown in Fig. 1A is a schematic diagram of the chimeric Env proteins used for this study. As shown in Fig. 1B and C, the expression levels of the chimeric proteins were comparable with each other as well as with those of their wild-type Env protein counterparts. Surface expression and secretion of gp120 by each chimeric Env protein were also found to be similar to those of their wild-type counterparts. Therefore, the replacement of the HIV or SIV Env protein cytoplasmic domain with the cytoplasmic domain of the MuLV Env protein does not significantly affect the expression or transport of the chimeric proteins.
FIG. 1.
(A) Schematic diagram of the transmembrane region of Env protein constructs used in this study. Black boxes represent the transmembrane domains of the SIV or HIV Env protein, hatched boxes represent cytoplasmic sequences of MuLV origin, gray boxes represent the MuLV R-peptide sequence, and white boxes represent the SU or cytoplasmic domain sequences of SIV or HIV origin (the SU and full-length cytoplasmic domains of SIV and HIV Env proteins are not drawn to scale). Construction of genes encoding the chimeric Env proteins was carried out by following standard cloning procedures described previously (42). (B) Expression of SIV and SIV-MuLV chimeric Env proteins. Lanes 1, mock transfection; lanes 2, full-length SIV Env; lanes 3, S-Menv; lanes 4, S-MenvR-; lanes 5, SIVenv733T+R; lanes 6, SIVenv733T. (C) Expression of HIV and HIV-MuLV chimeric Env proteins. Lanes 1, mock transfection; lanes 2, full-length HIV Env; lanes 3, H-Menv; lanes 4, H-MenvR-. Proteins were expressed in HeLa cells using the vaccinia virus T7 expression system (18). Surface biotinylation and immunoprecipitation were carried out as described previously (42).
Fusion activity of SIV-MuLV chimeric Env proteins in the presence of coreceptor CCR5.
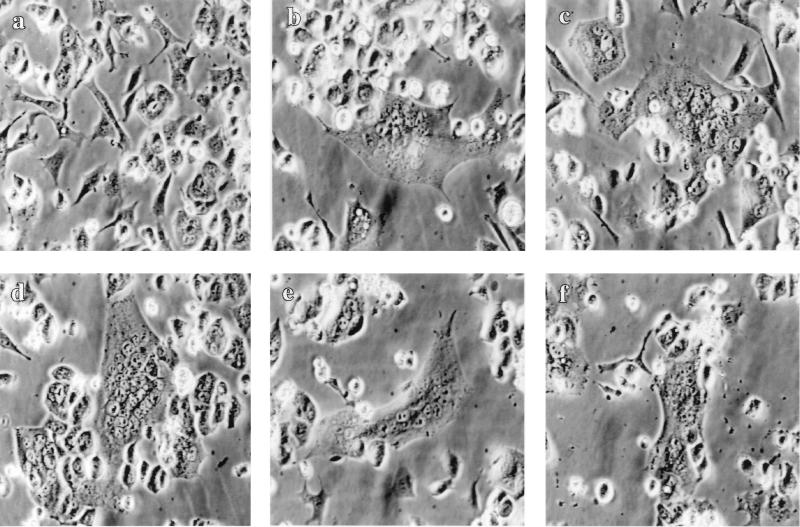
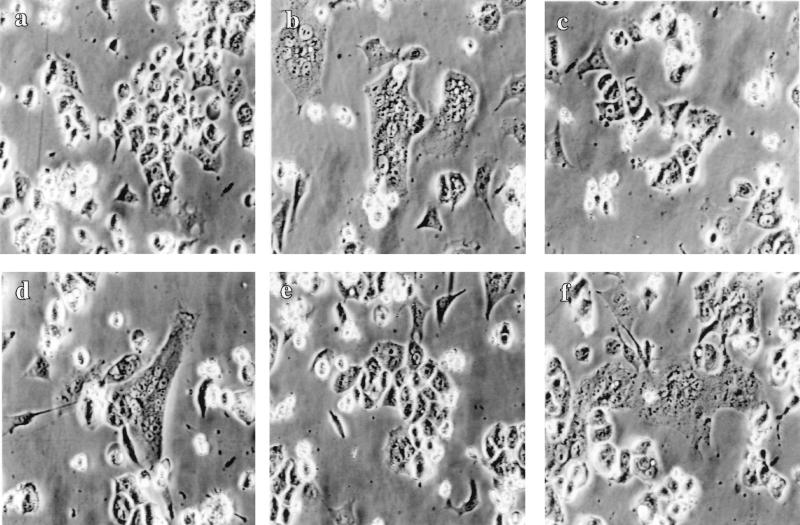
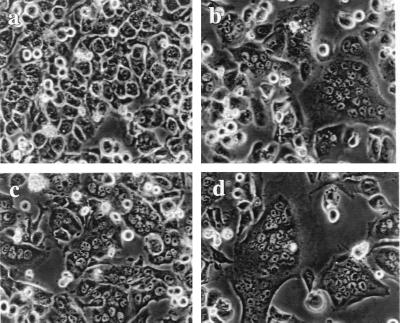
It has been reported that SIVmac239, which we used for construction of SIV-MuLV chimeric proteins, can use CCR5 as its coreceptor (9, 29). We therefore tested the cell fusion activity of SIV-MuLV chimeric Env proteins in Ghost cells expressing CD4 and CCR5 (Ghost CCR5 cells, Ghost Gpr15 cells, and Ghost CXCR4 cells were obtained from the National Institutes of Health [NIH] AIDS Research and Reference Reagent Program, and Ghost [3] parental cells were obtained from Vineet N. KewalRamani and Dan R. Littman). To our surprise, as shown in Fig. 2, all SIV-MuLV chimeric Env proteins, with or without the MuLV R peptide, induced extensive cell fusion in Ghost CCR5 cells, at a level similar to that induced by the wild-type SIV Env protein. In contrast, when we tested the same constructs in CEMx174 cells, the results reconfirmed our previous finding that the presence of the MuLV R peptide greatly inhibited the fusion activity of the SIV-MuLV chimeric Env proteins in these cells (data not shown) (42). Since the same cells (HeLa) were used for Env protein expression in both experiments, the possibility of differences in expression of chimeric proteins can be excluded. These results indicated that the presence of the MuLV R-peptide sequence in the cytoplasmic domain of the SIV-MuLV chimeric Env proteins does not inhibit their cell fusion activity in the presence of the coreceptor CCR5, indicating that there are factors in CEMx174 cells important for the fusion inhibition activity of the MuLV R peptide. It should be noted that CEMx174 cells and the Ghost CCR5 cells are of different origin and that CEMx174 cells are suspension cells, while the Ghost CCR5 cells are attached cells which form a monolayer in culture.
FIG. 2.
Fusion activity of SIV and SIV-MuLV chimeric Env proteins in cells expressing the coreceptor CCR5. Env proteins were expressed in HeLa cells using the vaccinia virus T7 expression system. HeLa cells were infected by recombinant virus VTF7-3 (a recombinant vaccinia virus expressing the T7 polymerase, provided by B. Moss) for 1 h and then transfected with DNA constructs encoding the Env proteins. At 12 h posttransfection, HeLa cells were overlaid with Ghost cells (obtained from the NIH AIDS Research and Reference Reagent Program) which express CD4 and CCR5. Cell fusion pictures were taken at 8 h after overlay of Ghost CCR5 cells under a Nikon microscope. (a) Mock transfection; (b) full-length SIV Env; (c) S-Menv; (d) S-MenvR-; (e) SIVenv733T+R; (f) SIVenv733T.
Fusion activity of SIV-MuLV chimeric Env proteins in the presence of coreceptor Gpr15.
It was discovered that CEMx174 cells do not express the coreceptor CCR5. Rather, they express an orphan chemokine receptor, Gpr15, which can serve as the coreceptor for SIV infection (11, 15, 36). We therefore obtained Ghost cells which express CD4 and Gpr15 and used them for the study of the cell fusion activities of the SIV-MuLV chimeric Env proteins. As shown in Fig. 3, both the full-length and truncated (SIVenv733T) SIV Env proteins induced extensive cell fusion, while the control, in which HeLa cells were transfected with vector plasmid, did not show any cell fusion (compare Fig. 3b and f with a). Regarding the SIV-MuLV chimeric Env proteins, S-Menv did not induce observable syncytium formation above the control level (Fig. 3c), while S-MenvR-, in which the R peptide in the MuLV cytoplasmic domain of the chimeric protein is truncated, induced syncytium formation at a level similar to that induced by the SIV Env proteins (Fig. 3d). Moreover, SIVenv733T+R, in which the MuLV R-peptide sequence is attached to the C terminus of SIVenv733T, also did not induce any significant level of cell fusion (Fig. 3e). The difference in cell fusion activities was not the result of the differences in the length of the cytoplasmic domain, based on our results in previous studies showing that extensive cell fusion was induced by a truncated SIV Env protein with a 33-amino-acid cytoplasmic domain, similar in length to that of the MuLV Env protein (42). These results are in agreement with our observation that the presence of the MuLV R peptide in the cytoplasmic domain of the SIV-MuLV chimeric Env proteins can profoundly inhibit their cell fusion activity in CEMx174 cells. Therefore, we conclude from these results that the fusion activity of the SIV-MuLV chimeric Env proteins is inhibited by the MuLV R peptide when Gpr15 is used as the coreceptor, but not when CCR5 is used as the coreceptor in the same cell type.
FIG. 3.
Fusion activity of SIV and SIV-MuLV chimeric Env proteins in cells expressing the coreceptor Gpr15. Env proteins were expressed in HeLa cells using the vaccinia virus T7 expression system. HeLa cells were infected by recombinant virus VTF7-3 for 1 h and then transfected with DNA constructs encoding the Env proteins. At 12 h posttransfection, HeLa cells were overlaid with Ghost cells (obtained from the NIH AIDS Research and Reference Reagent Program) which express CD4 and Gpr15. Cell fusion pictures were taken at 8 h after overlay of Ghost Gpr15 cells under a Nikon microscope. (a) Mock transfection; (b) SIVenv; (c) S-Menv; (d) S-MenvR-; (e) SIVenv733T+R; (f) SIVenv733T.
Fusion activity of HIV-MuLV chimeric Env proteins.
We have shown that the presence of the MuLV R peptide can profoundly affect the cell fusion activity of the SIV-MuLV chimeric Env proteins with cells expressing the coreceptor Gpr15, such as Ghost Gpr15 and CEMx174 cells. To further understand the mechanism of fusion inhibition by the MuLV R peptide, we also studied its effect on the cell fusion activity of HIV-MuLV chimeric Env proteins in comparison with that of the wild-type HIV Env protein. The HIV Env protein backbone we used to generate HIV-MuLV chimeric Env proteins is from HIV-IIIB, which utilizes CXCR4 as a coreceptor (17). We expressed the HIV-MuLV chimeric Env proteins in HeLa cells and then overlaid them with HeLa T4 cells. As shown in Fig. 4, the HIV-MuLV chimeric Env proteins induced extensive cell fusion, similar to that observed with the wild-type HIV Env protein. We also tested the fusion activity of the HIV-MuLV chimeric Env proteins with Ghost CXCR4 cells, and a similar result was observed (data not shown). These results indicate that unlike SIV-MuLV chimeric Env proteins, the presence of the MuLV R peptide in the HIV-MuLV chimeric Env protein does not suppress its cell fusion activity.
FIG. 4.
Fusion activity of HIV and HIV-MuLV chimeric Env proteins. Env proteins were expressed in HeLa cells using the vaccinia virus T7 expression system. HeLa cells were infected by recombinant virus VTF7-3 for 1 h and then transfected with DNA constructs encoding the Env proteins. At 12 h posttransfection, HeLa cells were overlaid with HeLa T4 cells (obtained from the NIH AIDS Research and Reference Reagents Program) which express CD4 and fusin. Cell fusion pictures were taken at 8 h after overlay of HeLa cells under a Nikon microscope. (a) Mock transfection; (b) full-length HIV Env; (c) H-Menv; (d) H-MenvR-.
We have investigated the cell fusion activities of the SIV-MuLV and HIV-MuLV chimeric Env proteins in relation to their coreceptor usage. Our results show that the presence of the MuLV R peptide in the cytoplasmic domain of the HIV-MuLV chimeric proteins does not affect their cell fusion activity. More interestingly, we found that the R peptide in the SIV-MuLV chimeric Env proteins showed no fusion inhibition activity with Ghost cells which express CD4 and the coreceptor CCR5. However, when Ghost cells which express CD4 and the coreceptor Gpr15 were used for fusion studies, the fusion activity of R peptide-containing constructs was greatly reduced in comparison with that of the wild-type SIV Env protein or other chimeric proteins which lacked the R peptide. Our results thus demonstrate that the inhibitory effect of the MuLV R peptide on cell fusion activities of the SIV Env proteins depends on the coreceptor expressed by the target cells. The observed differences in fusion inhibition activity of the MuLV R peptide also indicate that the SIV Env protein may utilize different domains to interact with alternate coreceptor molecules and that the presence of the R peptide may selectively affect one of these domains. We also observed that the chimeric Env protein S-MenvR- showed a higher relative surface expression level than did other Env proteins (Fig. 1B, lane 4). However, this would not account for the differences seen in syncytium formation, particularly the finding that the presence of the R peptide in the chimeric Env proteins affects syncytium formation only when Gpr15 is used as the coreceptor.
One possible mechanism for the fusion inhibition activity of the MuLV R peptide is that its presence in the cytoplasmic domain may affect the folding and conformation of the viral Env proteins. Studies with influenza virus HA have shown that it is activated to induce membrane fusion through sequential conformational changes induced by a low pH (5, 6). It has been found that there are two heptad repeat regions in HA and that they play a critical role in the conformational changes and membrane fusion activity of HA (6). Such heptad repeat regions have also been found in the TM subunit of the HIV, SIV, and MuLV Env proteins and have been shown to form a triple-stranded coiled-coil structure (6, 8, 16, 39). The discovery of coreceptors for infection by HIV and SIV has greatly increased our understanding of the mechanism of membrane fusion induced by the HIV and SIV Env proteins. By analogy with membrane fusion induced by the influenza virus HA protein, it was proposed that the HIV and SIV Env proteins first bind to CD4 on cells, which induces a conformational change in the Env protein, allowing it to interact with a coreceptor and resulting in another conformational change and membrane fusion (12, 31). Our results suggest that the presence of the MuLV R peptide profoundly affects the interaction between the external domain of the SIV Env proteins and the coreceptor Gpr15. Apparently, binding of the Env protein to CD4 is not affected by the presence of the MuLV R peptide, as the R-peptide-containing SIV-MuLV chimeric Env proteins can still induce cell fusion in the presence of CCR5. However, the presence of the R peptide has an impact on the conformation of the chimeric Env proteins, and as a result, their functional interaction with the coreceptor Gpr15 is impaired. Alternatively, it is also possible that the presence of the R peptide may affect the affinity of the Env proteins for both coreceptors CCR5 and Gpr15. However, the affinity of the Env proteins for the coreceptor CCR5 is much stronger, or less affected by the presence of the R peptide, than is the affinity of the Env proteins for the coreceptor Gpr15. It will be interesting for future studies to determine whether the presence of the R peptide reduces the affinity between the chimeric Env protein and Gpr15 or if it prevents the chimeric Env protein from assuming a fusogenic conformation upon interaction with Gpr15.
Functional studies of the MuLV Env proteins have shown that Env proteins of different tropism can form heterooligomers and complement each other in inducing membrane fusion (34, 43, 44). In the studies by Rein et al. (34), the Env protein of ecotropic MuLV with a truncated R peptide was found to form heterooligomers with the full-length Env protein of amphotropic MuLV. Such Env protein heterooligomers were found to induce fusion in cells expressing only the amphotropic MuLV receptor, while each Env protein by itself failed to induce any cell fusion. These studies suggested that truncation of the MuLV R peptide may render the MuLV Env protein into a fusion-competent conformation, while binding to the receptor is required to bring the target membrane into its vicinity. Alternatively, it is possible that fusion by MuLV Env proteins may also involve a secondary coreceptor which is yet to be identified. Our study suggests that the interaction between the MuLV Env protein and its receptor is similar to the interaction between the SIV Env protein and its coreceptors, as indicated by the similar sensitivities of these Env proteins to fusion inhibition by the MuLV R peptide. Future studies on the mechanism of fusion inhibition by the MuLV R peptide should further dissect the process of membrane fusion induced by viral Env proteins.
Acknowledgments
We thank Tanya Cassingham for help with preparation of the manuscript.
This study was supported by NIH grants CA 18611 and AI 47018.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Liao F, Berger E A, Farber J M, Peden K W. A new SIV co-receptor, STRL33. Nature. 1997;388:238. doi: 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- 3.Brody B A, Rhee S S, Hunter E. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J Virol. 1994;68:4620–4627. doi: 10.1128/jvi.68.7.4620-4627.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bron R, Klasse P J, Wilkinson D, Clapham P R, Pelchen-Matthews A, Power C, Wells T N C, Kim J, Peiper S C, Hoxie J A, Marsh M. Promiscuous use of CC and CXC chemokine receptors in cell-to-cell fusion mediated by a human immunodeficiency virus type 2 envelope protein. J Virol. 1997;71:8405–8415. doi: 10.1128/jvi.71.11.8405-8415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti L, Emerman M, Tiollais P, Sonigo P. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J Virol. 1989;63:4395–4403. doi: 10.1128/jvi.63.10.4395-4403.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio R, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 11.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 12.Doms R W, Peiper S C. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 13.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 14.Dragic T, Litwin V, Allaway G P, Martin S R, Hunag Y, Nagashima K A, Cayanan C, Maddon P A, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 15.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–874. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 18.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk H-D, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 22.Henderson L E, Sowder R, Copeland T D, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52:492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch V M, Edmondson P, Murphey-Corb M, Arbeille B, Johnson P R, Mullins J L. SIV adaption to human cells. Nature. 1989;341:573–574. doi: 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]
- 24.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 25.Kang C-Y, Hariharan K, Posner M R, Nara P. Identification of a new neutralizing epitope conformationally affected by the attachment of CD4 to gp120. J Immunol. 1993;151:449–457. [PubMed] [Google Scholar]
- 26.Kodama T, Wooley D P, Naidu Y M, Kestler III H W, Daniel M D, Li Y, Desrosiers R C. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong P, Wyatt R, Robinson J, Sweet R, Sodroski J, Hendrickson W. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 29.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard N, Gerard C, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin K, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 31.Montefiori D, Moore J P. HIV vaccines. Magic or the occult? Science. 1999;283:336–337. doi: 10.1126/science.283.5400.336. [DOI] [PubMed] [Google Scholar]
- 32.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rein A, Yang C, Haynes J A, Mirro J, Compans R W. Evidence for cooperation between murine leukemia virus Env molecules in mixed oligomers. J Virol. 1998;72:3432–3435. doi: 10.1128/jvi.72.4.3432-3435.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritter G D, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 36.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommerfelt M A, Petteway S R, Jr, Dreyer G B, Hunter E. Effect of retroviral proteinase inhibitors on Mason-Pfizer monkey virus maturation and transmembrane glycoprotein cleavage. J Virol. 1992;66:4220–4227. doi: 10.1128/jvi.66.7.4220-4227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spies C P, Ritter G D, Jr, Mulligan M J, Compans R W. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters conformation of the external domain. J Virol. 1994;68:585–591. doi: 10.1128/jvi.68.2.585-591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 40.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Biosynthesis, cleavage and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 42.Yang C, Compans R W. Analysis of the cell fusion activities of chimeric simian immunodeficiency virus-murine leukemia virus envelope proteins: inhibitory effects of the R peptide. J Virol. 1996;70:248–254. doi: 10.1128/jvi.70.1.248-254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y, Lee S, Anderson W F. Functional interactions between monomers of the retroviral envelope protein complex. J Virol. 1997;71:6967–6972. doi: 10.1128/jvi.71.9.6967-6972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, Zhu L, Benedict C A, Chen D, Anderson W F, Cannon P M. Functional domains in the retroviral transmembrane protein. J Virol. 1998;72:5392–5398. doi: 10.1128/jvi.72.7.5392-5398.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zingler K, Littman D R. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J Virol. 1993;67:2824–2831. doi: 10.1128/jvi.67.5.2824-2831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]