Sobemoviruses are plant RNA viruses named after their type species, Southern bean mosaic virus (SBMV). In 1969, Walters proposed combining single-component-RNA beetle-transmitted viruses into a southern bean mosaic virus group (107). In 1977, Hull recommended establishment of this plant virus group on the basis of similarities in protein subunit molecular weight, capsid stabilization, sedimentation coefficient, and distribution of particles within the cell (44). The group was accepted by the International Committee on Taxonomy of Viruses (ICTV) under the name sobemovirus (63). In 1995 the ICTV recognized the group as an unassigned genus, Sobemovirus (47). Presently, the genus contains 11 definitive species (Table 1) (20). The most recent change in the list of sobemovirus species was the distinguishing of SBMV and Southern cowpea mosaic virus (SCPMV) as separate species (20). These viruses were previously described as the bean and cowpea strains of SBMV, respectively. Due to differences in host range and antigenicity and substantial sequence differences, they are now accepted as independent Sobemovirus species (M. C. Fauquet, personal communication).
TABLE 1.
Viruses of the genus Sobemovirus and their biological properties
| Virus | Natural host | Vector | Seed transmission | Reference(s) |
|---|---|---|---|---|
| Definitive species | ||||
| BSSV | Vaccinium corymbosum, Vaccinium angustifolium | Aphids | No | 77 |
| CfMV | Dactylis glomerata, Triticum aestivum | Beetles | No | 65, 91 |
| LTSV | Medicago sativa | NDa | No | 7, 22 |
| RYMV | Oryza sativa, Oryza longistaminata | Beetles | No | 5 |
| SNMoV | Solanum nodiflorum, Solanum nitidibaccatum, Solanum nigrum | Beetles | No | 32, 51 |
| SBMV | Phaseolus vulgaris | Beetles | Yes | 100 |
| SCPMV | Vigna unguiculata | Beetles | Yes | 100 |
| SoMV | Chenopodium spp., Chenopodium murale, Vitis sp., Prunus domestica, Atriplex suberecta | Leafminers, leafhoppers | Yes | 52, 53 |
| SCMoV | Trifolium subterraneum | ND | Yes | 23 |
| TRoV | Brassica campestris subsp. napus, Brassica campestris subsp. rapa | Beetles | ND | 42 |
| VTMoV | Nicotiana velutina | Mirid | No | 78 |
| Tentative species | ||||
| CfMMV | Phleum pratense, Dactylis glomerata, Agrostis stolonifera, Bromus mollis, Festuca pratensis, Poa trivialis | Aphids, beetles | No | 48 |
| CnMoV | Cynosurus cristatus, Lolium perenne, Agrostis tenuis, Agrostis stolonifera | Aphids | ND | 65 |
| GCFV | Zingiber officinale | ND | ND | 97 |
ND, not determined.
Tentative species of the genus Sobemovirus are Cocksfoot mild mosaic virus (CfMMV), Cynosurus mottle virus (CnMoV), and Ginger chlorotic fleck virus (GCFV) (20).
(The official abbreviation for cocksfoot mild mosaic virus in the seventh ICTV report is CMMV. In the same report, cocksfoot mottle virus is abbreviated as CoMV. Fauquet and Mayo point out that cocksfoot is one of the plant names whose abbreviations cause discrepancies in virus taxonomy (20). Co is also used as an abbreviation for coffee and cole, whereas C alone is used for many different plant species. Since CfMV is a widely used abbreviation for cocksfoot mottle virus in the sobemovirus literature, we propose that Cf be used as an abbreviation for cocksfoot in the future. The two viruses will then be abbreviated as CfMMV and CfMV, respectively. These abbreviations will be used throughout this article.)
It should be noted that three other viruses recently regarded as tentative species of the genus Sobemovirus (Maize chlorotic mottle virus [genus Machlomovirus], Olive latent virus 1 [genus Necrovirus], and Panicum mosaic virus [genus Panicovirus]) have now been assigned to the family Tombusviridae. However, several viruses presently not recognized by the ICTV have been proposed to be closely related to sobemoviruses. These viruses include, for instance, Rottboellia yellow mottle virus (VIDE database, http://biology.anu.edu.au/Groups/MES/videl/), Ryegrass mottle virus (VIDE database, http://biology.anu.edu.au/Groups/MES/vide/), and Sesbania mosaic virus (6).
Viruses in the genus Sobemovirus are icosahedral particles of about 30 nm in diameter (47). The virions contain a single coat protein (approximately 30 kDa in size), a genomic RNA, and one subgenomic RNA (sgRNA) molecule. The capsid is constructed of 180 subunits according to T=3 symmetry. The genomic RNA is a single-stranded messenger-sense molecule, approximately 4 to 4.5 kb in size. The 5′ terminus of the RNA has a genome-linked protein (VPg), and the 3′ end lacks a poly(A) tail.
In addition to their genomic RNA, some sobemoviruses encapsidate a viroid-like satellite RNA (satRNA) that is dependent on a helper virus for replication. The presence of viroid-like satRNAs has been reported for Lucerne transient streak virus (LTSV), Rice yellow mottle virus (RYMV), Subterranean clover mottle virus (SCMoV), Solanum nodiflorum mottle virus (SNMoV), and Velvet tobacco mottle virus (VTMoV) (13, 23, 25, 31, 50, 51, 78, 90, 99). The sizes of these circular satRNAs range from 220 to 390 nucleotides (nt) (2, 13, 14, 39, 54). At 220 nt, the RYMV satRNA is the smallest naturally occurring viroid-like RNA known today (13).
Several interesting interactions between the satRNA, helper virus, and host plant have been described among sobemoviruses. For example, LTSV supports the replication of satRNA of SNMoV (51). On the contrary, SNMoV does not replicate LTSV satRNA (51). The replication of satRNA of LTSV is also supported by sobemoviruses that are normally devoid of satRNA, including CfMV, SBMV, Sowbane mosaic virus (SoMV), and Turnip rosette virus (TRoV) (2, 72, 90). The replication of satRNA of LTSV is dependent not only on the helper virus but also on the host plant. For instance, TRoV supports the replication of the LTSV satRNA in Brassica rapa, Raphanus raphanistrum, and Sinapsis arvensis, but not in Thlaspi arvense or Nicotiana bigelovii (90). Furthermore, satRNA of LTSV replicates effectively and is encapsidated in the presence of CfMV in two monocotyledonous species, Triticum aestivum and Dactylis glomerata (90).
Sobemoviruses are transmitted by vectors or by seeds and are readily transmitted mechanically (Table 1). CfMV, RYMV, SNMoV, SBMV, SCPMV, and TRoV are transmitted by beetles. Blueberry shoestring virus (BSSV) is transmitted by aphids, and VTMoV is transmitted by mirids. SoMV is transmitted by leafminers and leafhoppers. The natural host range of each virus species is relatively narrow. However, sobemoviruses in general infect plant species from not less than 15 different families, including dicotyledonous and monocotyledonous species. The first sobemovirus to be isolated was SBMV, in Louisiana and California (115). Later it was demonstrated that sobemoviruses are spread all over the world, colonizing plants in countries from Scandinavia to New Zealand and throughout tropical Africa.
GENOME ORGANIZATION
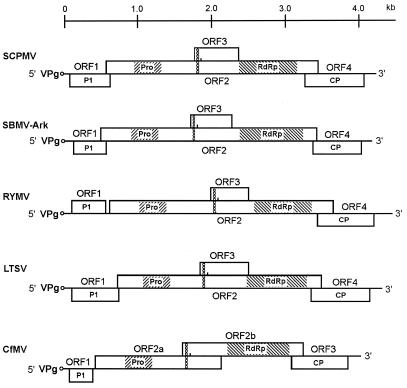
The complete nucleotide sequences of several sobemoviruses have been determined (58, 61, 114, 70, 86, 112; A. C. Jeffries, J. P. Rathjen, and R. H. Symons, GenBank accession no. U31286; Y. M. Pinto and D. C. Baulcombe, GenBank accession no. U23142). The sobemovirus genome is compact, and most of the predicted open reading frames (ORFs) overlap. The coding region regularly contains four ORFs (Fig. 1).
FIG. 1.
Genomic organization of sobemoviruses. ORFs are illustrated as boxes. VPg, the peptide covalently attached to the 5′ terminus of the RNA genome, is shown as a small circle. The approximate locations of the putative protease and putative RdRp domains are labeled Pro and RdRp, respectively. The sites of −1 ribosomal frameshift consensus signals are indicated by vertical chains. The positions of the first AUGs in ORF3 of SCPMV, SBMV-Ark, RYMV, and LTSV and in ORF2b of CfMV are indicated by short vertical lines. P1, ORF1-encoded protein; CP, coat protein.
All sequenced sobemoviruses contain a small ORF, ORF1, at the 5′ end of the genome and a 3′-proximal ORF which encodes the viral coat protein. Exceptionally, the LTSV genome nucleotide sequence available from the GenBank database (Jeffries et al., GenBank accession no. U31286) indicates the presence of two small ORFs at the beginning of the genome, ORF1a and ORF1b. Later, however, this was shown to be a sequencing error (J. P. Rathjen, personal communication). Thus, LTSV, like other sobemoviruses, contains a single ORF1.
Based on the organizational differences in the central part of the genome (encoding the virus polyprotein), the sobemoviruses can be subdivided into SCPMV-like and CfMV-like viruses. The polyprotein of SCPMV is encoded by the large continuous ORF2 (112). The genome of SCPMV also contains an internal coding region, ORF3, situated in the −1 reading frame within ORF2. Similar genomic organizations have been reported for the Arkansas isolate of SBMV (SBMV-Ark), the Ivory Coast isolate of RYMV (RYMV-CI), and LTSV (58, 114; Jeffries et al., GenBank accession no. U31286).
In contrast, the Norwegian (CfMV-NO) and Russian (CfMV-RU) isolates of CfMV lack the continuous ORF2 and a nested coding region similar to ORF3 of SCPMV (61, 86). Instead, CfMV has two overlapping ORFs, ORF2a and ORF2b, and the polyprotein is expressed through a −1 ribosomal frameshift mechanism. A similar genomic organization is characteristic of the Nigerian isolate of RYMV (RYMV-NG) (Y. M. Pinto, personal communication).
The only exception with regard to these subdivisions of sobemoviruses is the genome of SBMV (70). SBMV lacks the small ORF3 characteristic of SCPMV and SBMV-Ark, and the three remaining ORFs do not overlap. Unlike SBMV, SBMV-Ark contains four putative overlapping ORFs, making it more similar in genomic organization (but not in amino acid sequences of individual proteins [see below]) to SCPMV (58). It has been assumed that the considerable differences in the genomic organizations of SBMV and SBMV-Ark result from mutations or sequencing errors in SBMV that resulted in misidentification of the ORFs (58).
The CfMV-type genome arrangement resembles that reported for Mushroom bacilliform virus (MBV; genus Barnavirus, family Barnaviridae) (79). This similarity extends to the presence of both an ORF1 and a 3′-proximal coat protein ORF, as well as two overlapping ORFs for polyprotein expression. In addition, the arrangement of the CfMV ORFs in the 5′ half of the genome is similar to that of members of the genus Polerovirus (formerly known as subgroup II luteoviruses—Potato leafroll virus [PLRV] [101], Beet western yellows virus [BWYV] [104], Beet mild yellowing virus [BMYV] [34], Cereal yellow dwarf virus-RPV [CYDV-RPV, formerly BYDV-RPV] [105], and Cucurbit aphid-borne yellows virus [CABYV] [33]) and the genus Enamovirus (Pea enation mosaic virus 1 [PEMV-1; formerly RNA1 of PEMV] [15]) in the family Luteoviridae. The 5′ gene cluster of these viruses contains a small ORF0 and overlapping ORFs 1 and 2, and the polyprotein is expressed as a translational frameshift fusion of the ORF1 and -2 products.
GENE PRODUCTS AND THEIR FUNCTIONS
The first data on proteins encoded by sobemoviruses (besides the structural coat protein) were obtained from in vitro translation experiments. The RNAs of several sobemoviruses have been translated in rabbit reticulocyte lysate and wheat germ extract (WGE) systems. The RNAs of SBMV and SCPMV induce the translation of four major proteins in cell-free systems: 105-, 75-, 29-, and 14-kDa proteins and 100-, 70-, 30-, and 20-kDa proteins, respectively (62, 87). Four polypeptides can also be translated from CfMV-NO (60, 62), LTSV (67), SNMoV (55), and TRoV (66) RNAs, with only slight differences in their molecular weights being displayed (Table 2).
TABLE 2.
In vitro translation products of some sobemoviruses
Previous studies have demonstrated that the 100- and 70-kDa proteins of SCPMV are related and are translated from the full-length RNA, and the 20-kDa protein is presumably encoded by ORF1 (29, 62, 112). It has also been proposed that the polyprotein encoded by ORF2 of SCPMV is processed by proteolytic cleavage to give the 70-kDa translation product (112). The 30-kDa protein (viral coat protein) was shown to be translated from a smaller, sgRNA. An sgRNA has indeed been detected in sobemovirus-infected tissues as well as in virus particles. For instance, both SBMV and SCPMV encapsidate the subgenomic component into viral particles (85, 108). The sgRNA in the molecular weight range of 0.3 × 106 to 0.4 × 106 of SCPMV and SBMV has a VPg linked to its 5′ end, as does the genomic RNA (29, 62). Sobemovirus RNAs of less than genomic length have also been reported for RYMV-CI (about 1 kb), CfMV-NO (1.2 kb), and CfMV-RU (about 1 kb) (8, 61, 86). Like those of SBMV and SCPMV, the sgRNA molecules of CfMV are encapsidated (86, 95).
Recently, the individual genes of CfMV-NO from which the in vitro translation products are synthesized were identified by immunoprecipitation with specific antibodies (95). The 12-, 71-, and 100-kDa CfMV proteins are synthesized from the bicistronic genomic RNA of the virus. The CfMV 12-kDa protein is produced from ORF1, its 71-kDa protein is produced from ORF2a, and the 100-kDa protein is a polyprotein encoded by ORF2a and -2b. The CfMV 34-kDa in vitro translation product is the coat protein synthesized from the sgRNA. Based on these findings, it was postulated that the 70-kDa in vitro translation product of SCPMV might represent the ORF2-ORF3 transframe fusion protein and that it is not the product of proteolytic cleavage, as was hypothesized earlier.
Knowing which polypeptides are synthesized from which genes, we characterize below the individual gene products and their expression mechanisms and possible functions in more detail.
P1.
All of the sobemoviruses characterized encode a small P1 protein from the 5′-terminal ORF1. However, the ORF1 nucleotide sequences as well as the P1 primary sequences of the different members of the genus Sobemovirus are not similar. They are also unrelated to any other known proteins. The theoretical or experimental molecular masses of different sobemovirus P1 proteins range between 11.7 and 24.3 kDa. It was hypothesized that ORF1 of RYMV-CI, unlike the corresponding proteins of other Sobemovirus species, encodes two polypeptides, with molecular masses of 18 and 19 kDa, due to readthrough of the UGA stop codon at nt 553 (114). However, in vitro translation of constructs containing the ORF1 coding region with two stop codons (at nt 553 and 599) or with one stop codon only (at nt 553) both produced two products, of 18 and 19 kDa (8). Therefore, it is unlikely that the 19-kDa product of RYMV-CI is translated by a readthrough of the stop codon at nt 533. It was assumed that the proteins detected in vitro and in vivo likely resulted from degradation, posttranslational modification(s), or structural or other characteristics of RYMV-CI P1 (8).
Recently, full-length cDNA clones of RYMV-CI and SCPMV were constructed and used to study the functions of P1 in the viral life cycle (8, 93). Analysis of mutants incapable of producing P1 or producing only truncated versions of that protein indicated that the RYMV-CI or SCPMV P1 is not needed for virus replication (8, 93). At the same time, the absence of full-length P1 abolished cell-to-cell and systemic movement of the virus in rice and cowpea plants, respectively. The RYMV-CI mutant that did not express P1, due to a mutation at the initiation codon, replicated efficiently in rice protoplasts, but at a level lower than the wild-type cDNA transcript. Transgenic rice plants expressing wild-type P1 were able to complement this initiation codon mutant and exhibited systemic infections. These data demonstrate that one or more of the P1 functions act in trans and are essential during infection of plants.
Most plant viruses produce movement proteins which presumably facilitate cell-to-cell movement of the virus through the plasmodesmata. Virus-like particles of RYMV have been reported in plasmodesmata (69). Because movement protein functions are not attached to any sobemovirus gene products, and the functions of P1 have remained largely unknown, it is tempting to propose that P1 is a sobemovirus movement protein. Our recent study has focused on the RNA binding activity of the recombinant CfMV-NO P1 protein. His-tagged P1 was able to interact with single-stranded RNA (ssRNA) transcripts in a non-sequence-specific manner (96). The biological significance of RNA binding and the domain(s) essential for binding remain to be elucidated, but movement proteins, in general, are indeed nucleic acid-binding proteins.
It is interesting that RYMV symptoms appeared more rapidly in P1-overexpressing plants inoculated with the full-length RYMV-CI transcript than in nontransformed plants (8). The observation that expression of P1 in trans enhances the infection process in planta suggests that P1 can act as an enhancing factor for genome amplification. Recently, it was reported that P1 of RYMV-NG functions as a suppressor of posttranscriptional gene silencing (PTGS) (106). PTGS of a green fluorescent protein transgene in Nicotiana benthamiana plants was reversed by RYMV-NG P1 expressed from the potato virus X vector. It was concluded that P1 protein of RYMV-NG is the suppressor of maintenance of PTGS in N. benthamiana, although it is encoded in the genome of a virus that is not infectious on Nicotiana species.
Polyprotein.
All of the sobemoviruses characterized encode a relatively large protein with a molecular mass of around 100 kDa. The genomic structures of SBMV, SBMV-Ark, SCPMV, RYMV-CI, and LTSV allow the synthesis of this protein from a continuous single ORF. The C-terminal half of the CfMV and RYMV-NG polyprotein is encoded by a separate ORF2b and is translated as part of the polyprotein by a −1 ribosomal frameshifting mechanism (60; Y. M. Pinto, personal communication). The consensus signals for a −1 ribosomal frameshift event can be found at the beginning of the region of overlap between CfMV ORF2a and ORF2b. These signals are the heptanucleotide (slippery) sequence 5′ UUUAAAC (nt 1634 to 1640) and a predicted stem-loop structure located 7 nt downstream from the heptamer. Mäkinen et al. (60) examined CfMV-NO frameshifting in vitro by inserting the cDNA fragment representing nt 1621 to 2521 of CfMV-NO RNA into the middle of the GUS sequence. This CfMV-NO region contains the overlapping region of ORF2a and ORF2b, including the consensus sequences for frameshifting. When translated in a WGE, this sequence directed frameshifting at a level of 26 to 29%. This experiment showed also that no CfMV-NO-specific sequences upstream of the consensus signals are required for an efficient −1 ribosomal frameshifting event. We demonstrated that the polyprotein of CfMV-NO was produced with an efficiency of 10.6% ± 1.4% (mean ± standard deviation) when the construct containing the entire ORF2a-ORF2b region was transcribed and translated in a WGE (95).
Translation of the sobemovirus polyprotein is not initiated from a corresponding sgRNA, since no such RNA has been found. Experimental evidence supports the idea that the genomic RNA of sobemoviruses functions as a bicistronic mRNA; the translation of ORF1 and ORF2 from the genomic RNA occurs by initiation at their respective AUG codons. Products of the expected size from both ORFs were observed after translation of the genomic RNAs of SCPMV (94) and CfMV-NO (95). A comparison of the sequence surrounding the initiation codon for ORF1 of sobemoviruses with the consensus sequences established for plant mRNAs shows that the sequence surrounding the first AUG codon is in a poor context for translation by plant ribosomes. It is characterized by the presence of a pyrimidine at position −3 and by the absence of a G at position +4 (11, 59). In contrast, the ORF2 initiation codon is present in a context more favorable for translation, with a purine at position −3. The 5′-terminal half of the sobemovirus genome resembles the genome of poleroviruses, enamoviruses, and barnaviruses. In all of these cases, the start codon of the first ORF is flanked by suboptimal bases compared to those flanking the second ORF start site (15, 64, 80).
The mechanism of SCPMV ORF2 translation initiation was further investigated by Sivakumaran and Hacker (94). In vitro and in vivo studies showed that the addition of one or two AUG codons in a sequence context favorable for translation initiation reduced ORF2 expression and that the elimination of the ORF1 initiation codon resulted in an increase in ORF2 expression. In vivo studies demonstrated that addition of 19 AUG codons in the 5′ untranslated region abolished ORF2 expression and that placement of the ORF1 initiation codon within a sequence context optimal for translation initiation reduced ORF2 expression. These results indicate that ORF2 of SCPMV is translated by leaky scanning rather than by internal ribosome binding or coupled termination-reinitiation.
The cotranslational disassembly of destabilized SBMV was suggested as a mechanism for uncoating of viral nucleic acid (109). In contrast to some other isometric viruses, which appear to release their RNA rapidly prior to translation, the particles of the Ghana strain of SBMV can disassemble only after their RNA has initiated translation (92). According to this model, the ribosome has to “find” the 5′ end of the RNA, and translation of the ORFs starts. Further removal of coat protein subunits occurs as ribosome translocation proceeds. The proposed model supports the finding that leaky scanning is the mechanism by which ORF2 of sobemoviruses is translated, since the ribosomes must start from the 5′ end of the RNA.
Based on experimental data as well as on analysis in silico, one can distinguish at least the following functional domains in the polyproteins of sobemoviruses: a serine protease-like domain, VPg, and an RNA-dependent RNA polymerase (RdRp)-like domain (30, 58, 61, 70, 86, 103, 112, 114; K. Mäkinen, K. Mäkeläinen, N. Arshava, T. Tamm, E. Truve, S. Zavriev, and M. Saarma, unpublished data). Presently it is not clear whether these are the only functions of the polyprotein.
Putative serine protease.
Gorbalenya et al. (30) identified a putative serine protease motif in the N-terminal part of the polyprotein of SCPMV by sequence comparisons with cellular and viral proteases. This putative protease is similar to the 3CPro cysteine proteases of picornaviruses and is characteristic of all sobemoviruses, poleroviruses, PEMV-1, and MBV. The serine protease motif is located in the N-terminal third of the polyprotein encoded by ORF2 in the case of SBMV, SBMV-Ark, SCPMV, RYMV, and LTSV, and it is encoded by ORF2a in the case of CfMV. The proposed protease sequence is unique among plant virus proteases in that it resembles a cellular serine protease in possessing a serine residue instead of cysteine in the catalytic triad (18, 30). The consensus amino acid sequence of the catalytic triad is H(X32–35)[D/E](X61–62)TXXGXSG (56). The glycine and histidine residues downstream from the putative catalytic residues are suggested to be the site of substrate binding. However, a biochemical demonstration of protease activity has not been made.
VPg.
A 12-kDa protein has been reported to be covalently linked to the 5′ end of the SBMV genome (29), and a 10-kDa VPg is attached to the SCPMV genome (62). Recently, the N-terminal sequences of the VPgs of two sobemoviruses, CfMV-RU and SBMV, have been determined (103; Mäkinen et al., unpublished data). The amino acid sequences obtained started at position 320 of the ORF2a product of CfMV-RU and at position 327 of the ORF2 product of SBMV, respectively. A comparison of the N-terminal sequence of SBMV VPg and residues 326 to 345 of the SCPMV ORF2 product revealed 63% identity (103). The N-terminal sequence of the CfMV-RU VPg was 100% identical to the corresponding amino acid sequence of the CfMV-NO polyprotein (Mäkinen et al., unpublished data).
The position of the VPg in the sobemovirus genome differs from the genome arrangement characteristic of many RNA virus families, including the Picornaviridae and Comoviridae, i.e., VPg-protease-RdRp (57). The N-terminal sequence of VPg places it between the serine protease and the RdRp motifs in the sobemovirus polyprotein. A similar polyprotein arrangement, protease-VPg-RdRp, has also been shown for PLRV, PEMV-1, and MBV (80, 102, 110). As indicated, these viruses exhibit several common features of sobemoviruses.
In all sobemoviruses, a conserved WAD or WGD amino acid sequence followed by a D- and E-rich region is present in proved and putative VPg proteins (61). A similar motif can also be found in PLRV, BWYV, PEMV-1, MBV, and Human astrovirus 2 (genus Astrovirus, family Astroviridae) upstream of the −1 frameshift signals. The sizes of the VPg proteins determined for CfMV, MBV, PLRV, and SBMV indicate that the WAD or WGD plus D- and E-rich region is present in the VPgs of these viruses (80, 102, 103; Mäkinen et al., unpublished data). This motif is the only sequence element conserved among the VPg proteins of these viruses. It is possible that this conserved motif is characteristic of the VPg proteins of viruses with a sobemovirus- or polerovirus-like genome arrangement.
The identification of the proteolytic cleavage sites at N termini of the VPg proteins of two sobemoviruses gave the first indication of how the sobemovirus polyprotein is processed in vivo. Gorbalenya et al. (30) proposed that the sobemovirus serine protease could cleave at E/T or E/S sites. In the SBMV and CfMV-RU polyprotein sequences, the N-terminal residue of VPg (T and N, respectively) is preceded by a glutamic acid residue, indicating that the N-terminal proteolytic processing site consists of the residues E/T or E/N. This finding is not in full accordance with the previously predicted cleavage sites. Although there are several putative cleavage sites conserved among the polyproteins of sobemoviruses (61), there are not sufficient experimental data to allow the proposal of a polyprotein processing model for these viruses.
Putative RdRp.
The C-terminal region of the sobemovirus polyprotein is predicted to encode a putative RdRp based on the presence of the GDD motif and surrounding conserved motifs characteristic of RdRps (56, 57). The putative sobemovirus RdRps show extensive similarities to RdRps of a number of positive-strand ssRNA viruses, which include again poleroviruses (PLRV, BWYV, BMYV, CYDV-RPV, and CABYV), an enamovirus (PEMV-1), and a barnavirus (MBV). Such similarities have been used to evaluate the taxonomic position of SCPMV in relation to other positive-strand RNA viruses (56, 57). SCPMV, BWYV, PLRV, and PEMV-1 have been grouped into the Sobemo lineage of polymerase supergroup 1 (57), indicating that the RdRps of poleroviruses are more similar to those of the sobemoviruses than they are to those of luteoviruses (formerly known as subgroup I luteoviruses).
Relatively little is known about the replication signals needed for initiation of plus- and minus-strand synthesis in sobemoviruses. Primer extension experiments have been used to identify the precise 5′ ends of the genomic RNAs as a start point for plus-strand synthesis. The 5′-terminal nucleotides of the SCPMV genomic RNA were identified as ACAAAA (36). The 5′ ends of the genomic RNAs of two other sobemoviruses, LTSV and RYMV-CI, have been reported to be ACAAA and ACAA, respectively (114; Jeffries et al., GenBank accession no. U31286). The 5′ ends of the SBMV and SBMV-Ark genomic RNAs start with a similar motif, CACAAAA (58, 70). At the same time, the 5′ ends of the SCPMV and SBMV sgRNAs map to bases 3241 and 3163, respectively (36), and, based on the sequences of the SCPMV and SBMV genomes (70, 112), also possess the sequence ACAAAA. These results demonstrate that the genomic RNAs and sgRNAs of sobemoviruses start with very similar primary sequences and that these are potentially important for virus replication. In contrast, this sequence motif is not present at the 5′ end of the CfMV genome (61, 86). All sobemoviruses have a polypurine tract, including the sequence aAGgAAA (lowercase indicates less conservation of that base) just at the beginning of the genomic RNA (36).
First of all, using the similarity between the 5′ ends of the genomic and subgenomic RNAs, it is possible to predict the 5′ termini of as-yet-uncharacterized sobemovirus sgRNAs. The sequence ACAAAA (nt 3222 to 3227) is present upstream from the SBMV-Ark coat protein initiation codon. For RYMV-CI, the sequence ACAAA (nt 3441 to 3445) is located 6 nt upstream of the ORF4 AUG codon. The putative transcription start site for the LTSV sgRNA also has the ACAAAA sequence motif (nt 3285 to 3290). Unfortunately, it is not possible to predict the 5′ end of the sgRNA of CfMV because this sequence motif is not present upstream from the coat protein coding region.
The ACAAAa sequence is present at the 5′ termini of genomic RNAs and sgRNAs of poleroviruses (64). The same sequence motif is found at the 5′ end of Red clover necrotic mosaic virus (genus Dianthovirus, family Tombusviridae) RNA1 and at the 5′ end of its sgRNA (113, 116). The 5′ termini of the MBV genomic RNAs and sgRNAs also begin with this sequence (79, 81). The conservation of this sequence at the 5′ ends of genomic RNAs and sgRNAs of several viruses belonging to different groups suggests that it or its complementary sequence in the minus-strand RNA may function in viral RNA synthesis. It has been proposed that the minus-strand sequence complementary to the ACAAA domain may act as a promoter or enhancer for viral replicase binding and initiation of RNA synthesis (64). So far the role(s) of the above sequences in sgRNA and genomic RNA synthesis in sobemo- and poleroviruses has not been tested. Even less is known about the replication signals needed to initiate minus-strand synthesis at the 3′ ends of sobemovirus genomic RNAs. A potential tRNA-like structure has been attributed to the 3′ end of some sobemovirus genomic RNAs (86, 114). However, for SBMV and SCPMV, it has been impossible to find an RNA sequence at the 3′ end that has the potential to fold into a tRNA-resembling secondary structure (70, 112). It should be emphasized that these reports are based on computer modeling. No experimental data are available on RNA secondary structures characteristic of the 5′ or 3′ ends of sobemovirus RNAs.
P3.
The genomes of SCPMV, SBMV-Ark, RYMV-CI, and LTSV have a small ORF3 nested in the middle of ORF2 in the −1 reading frame. The N-terminal half of each of these proteins is similar to the N-terminal part of the ORF2b-encoded protein of CfMV (60) as well as to the N-terminal part of the PLRV and BWYV ORF2-encoded proteins and to MBV and PEMV-1 ORF3-encoded products (86). At the moment, the function(s) and the translational mechanism of this ORF are unknown. The genome-length cDNA clone of SCPMV has been used to characterize the phenotypes of ORF3 mutants (93). A mutant expressing a truncated P3 was not infectious in cowpeas, indicating that P3 is needed for SCPMV infectivity in plants. Protoplast experiments demonstrated also that various mutations in ORF3 had no effect on viral RNA synthesis or SCPMV assembly.
The consensus signals for −1 ribosomal frameshifting, similar to those characterized for CfMV, can be found in the genomes of all sobemoviruses. The slippery sequence, UUUAAAC followed by a putative stem-loop structure, is located upstream from the potential translational initiation codon of ORF3 of SCPMV, SBMV-Ark, RYMV-CI, and LTSV. There are no stop codons present between the slippery sequence and the initiation codon of ORF3 in either reading frame. It has been assumed that ORF3 is likely to be translated by a −1 ribosomal frameshift mechanism similar to that shown for ORF2b in CfMV, but this has not been verified experimentally (60, 95). Five facts support this hypothesis: (i) no in vitro translation product has been attributed to this ORF; (ii) no sgRNA corresponding to this region has been found; (iii) an SCPMV mutant which had an in-frame stop codon between the predicted frameshift site and the potential initiation codon in the ORF3 reading frame was unable to infect cowpea, suggesting that the ORF3 protein was not expressed (93); (iv) the 70-kDa in vitro translation product of SCPMV may represent an ORF2-ORF3 transframe fusion protein, since the calculated molecular mass for the ORF2-ORF3 fusion of SCPMV is 65.8 kDa (95); and (v) the similarity of P3 to the N-terminal part of the ORF2b-encoded protein of CfMV as well as to the N-terminal part of the PLRV and BWYV ORF2-encoded proteins and the MBV and PEMV-1 ORF3-encoded products starts upstream from the putative ORF3 translation initiation signal (60, 86).
Coat protein.
The coat proteins of sobemoviruses are encoded by their 3′-proximally located ORFs (ORF4 in the case of the SCPMV-type genome and ORF3 for the CfMV type). The amino acid sequence of the SCPMV coat protein reported by Hermodson et al. (40) showed that translation initiation occurs at the second AUG (nt 3271 to 3273) of ORF4 and is followed by the hydrolysis of the N-terminal methionine and acetylation of the subsequent alanine. Direct sequencing of the N terminus of the RYMV-CI and CfMV-NO coat proteins showed that they commence at the first AUG codon of the last ORF, at nt 3447 and 3093, respectively (61, 114).
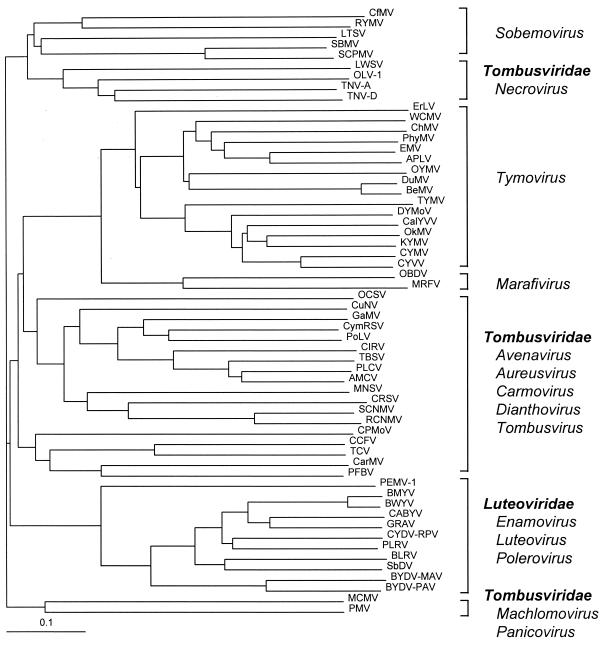
A tentative phylogenetic tree generated by aligning the coat protein sequence of SCPMV with those of several other viruses consisted of three distinct subdivisions (17). Interestingly, the coat protein of SCPMV grouped together with the Tobacco necrosis virus (TNV; genus Necrovirus, family Tombusviridae) coat protein rather than with the coat proteins of poleroviruses. The coat protein of RYMV-CI is also more closely related to that of TNV (114), indicating that coat proteins of sobemoviruses and necroviruses are phylogenetically related (Fig. 2) (17). This is different from sobemovirus proteases, VPgs, and RdRps, which are most closely related to those of poleroviruses, as indicated above.
FIG. 2.
Phylogenic tree of coat protein sequences of plus-strand RNA plant viruses with icosahedral particles. The tree was generated by using the PHYLIP program (version 3.573c) (21) CONSENSE data with TreeView (71). The families and genera into which individual virus species are classified are indicated on the right. GenBank accession numbers are as follows: CfMV, Z48630; RYMV, L20893; LTSV, U31286; SCPMV, M23021; SBMV, AF055887; LWSV (Leek white stripe virus), X94560; OLV-1 (Olive latent virus 1), X85989; TNV-A, M33002; TNV-D, U62546; ErLV (Erysimum latent virus), AF098523; WCMV (Wild cucumber mosaic virus), AF035633; ChMV (Chayote mosaic virus), AF195000; PhyMV (Physalis mottle virus), Y16104; EMV (Eggplant mosaic virus), J04374; APLV (Andean potato latent virus), AF035402; OYMV (Ononis yellow mosaic virus), J04375; DuMV (Dulcamara mottle virus), AF035634; BeMV (Belladonna mottle virus), X54529; TYMV (Turnip yellow mosaic virus), J04373; DYMoV (Desmodium yellow mottle virus), AF035201; CalYVV (Calopogonium yellow vein virus), U91413; OkMV (Okra mosaic virus), AF035202; KYMV (Kennedya yellow mosaic virus), AF035198; CYMV (Cacao yellow mosaic virus), X54354; CYVV (Clitoria yellow vein virus), AF035200; OBDV (Oat blue dwarf virus), U87832; MRFV (Maize rayado fino virus), U97725; OCSV (Oat chlorotic stunt virus), X83964; CuNV (Cucumber necrosis virus), M25270; GaMV (Galinsoga mosaic virus), Y13463; CymRSV (Cymbidium ringspot virus), X15511; PoLV (Pothos latent virus), AJ243370; CIRV (Carnation Italian ringspot virus), X85215; TBSV (Tomato bushy stunt virus), M21958; PLCV (Pelargonium leaf curl virus), S58174; AMCV (Artichoke mottled crinkle virus), X62493; MNSV (Melon necrotic spot virus), D00562; CRSV (Carnation ringspot virus), L18870; SCNMV (Sweet clover necrotic mosaic virus), L07884; RCNMV (Red clover necrotic mosaic virus), J04357; CPMoV (Cowpea mottle virus), U20976; CCFV (Cardamine chlorotic fleck virus), L16015; TCV (Turnip crinkle virus), M22445; CarMV (Carnation mottle virus), AF173979; PFBV (Pelargonium flower break virus), AJ003153; PEMV-1, L04573; BMYV (Beet mild yellowing virus), X83110; BWYV, X13063; CABYV, X76931; GRAV (Groundnut rosette assistor virus), Z68894; CYDV-RPV, L25299; PLRV, Y07496; BLRV (Bean leafroll virus), X53865; SbDV (Soybean dwarf virus), L24049; BYDV-MAV, D11028; BYDV-PAV, D11032; MCMV (Maize chlorotic mottle virus), X14736; and PMV (Panicum mosaic virus), U55002.
The coat protein is the single protein required to build sobemovirus isometric particles. The three-dimensional structure of SCPMV has been determined to a resolution of 2.8 Å (1), and that of SeMV has been determined to 3-Å resolution (6). Each icosahedral unit of the particle comprises three quasiequivalent subunits, A, B, and C, whose individual conformations may differ slightly. The A subunits cluster about the fivefold axes, whereas sets of three B and three C subunits cluster about quasi-sixfold vertices.
According to the X-ray structure, the coat protein is composed of two functional domains, the R (random) domain and the S (shell or surface) domain, connected by an arm (1, 40, 83). The S domain is composed of eight antiparallel β-sheets (termed a β-barrel) and five α-helices. The S domain is responsible for subunit-subunit interactions in the virus particle (40, 83). The R domain is formed by the N-terminal part of the polypeptide chain and is rich in arginine, lysine, proline, and glutamine. The basic residues located on the R domain (together with some similar residues on the inner surface of the S domain) are responsible for coat protein contacts with the RNA (40, 83). The pattern of the basic residues on the coat protein surface facing the RNA is able to dock a 9-bp double-helical A-RNA structure with surprising accuracy. The basic residues are each associated with a different phosphate, and the protein can interact with five bases in the minor groove. The total number of positive charges associated with the RNA is around 2,340, sufficient to cancel about half the negative charges of the nucleic acid. A recombinant R domain of the coat protein of SCPMV (amino acid residues 1 to 54) expressed in Escherichia coli was shown to have nonspecific RNA binding activity in vitro (S. K. Lee and D. Hacker, Abstr. 18th Annu. Meet. Am. Soc. Virol. 1999, abstr. W-23, p. 100, 1999). Studies of alanine substitution mutants revealed that the N-terminal arginine-rich motif was required for RNA binding. The interactions between the CfMV-NO coat protein and RNA have also been studied (96). The recombinant coat protein expressed in E. coli and the native coat protein purified from virus particles both bound ssRNA in a non-sequence-specific manner in vitro and were selective for ssRNA over double-stranded DNA molecules.
Hull (45) studied the particle stabilization of TRoV and other sobemoviruses and concluded that the particles of these viruses are stabilized by three types of bonds: pH-dependent interactions between subunits, protein-RNA interactions, and divalent-cation protein-protein bonds. The important cations forming protein-protein bonds in the particle are calcium and magnesium ions. The major Ca2+-binding site is on the quasi-threefold axis between the A, B, and C subunits (40). The sequence shows that Glu194 is the ligand associated with this position. Interestingly, in SBMV this residue is lysine (62), suggesting a different mode of subunit association. The second Ca2+-binding site is also between the quasi-threefold-related subunits, with interactions between Asp138 and Asp141 on one subunit and the main-chain carbonyls 199 and 259 on the other. The proposed Mg2+-binding sites contain residues His132, Glu229, and Glu77 (84). The virus swells upon removal of cations at alkaline pH values (45).
The importance of RNA-coat protein interactions in the assembly of SCPMV was demonstrated by Savithri and Erickson (88). The assembly of T=3 particles from the SCPMV protein and viral RNA requires Ca2+ at a neutral or alkaline pH. A low-molecular-weight RNA component and coat protein formed T=1 particles at acidic and neutral pHs. It is possible to convert swollen SBMV into T=1 particles by digestion with trypsin, which removes the basic N-terminal segment of the coat protein (89). Erickson and Rossmann (19) also showed that T=1 particle formation did not require RNA when purified and partially digested coat protein, lacking the basic N-terminal portion, was used. Native coat protein failed to assemble in either the T=1 or T=3 mode in the absence of RNA, indicating that initial RNA-protein interactions are needed and that the formation of the T=3 particles requires interaction between viral RNA and the basic arm of the coat protein. These studies did not demonstrate the requirement for specific coat protein-RNA interactions in SCPMV assembly but indicated that the N-terminal R domain is important for RNA binding.
However, dissociation of SBMV in a high-salt solution (0.4 M KCl) at a neutral pH yields a ribonucleoprotein complex (RNPC) composed of the viral RNA and about six coat protein subunits (43). Hacker demonstrated that coat protein subunits present in the RNPC, resulting from SCPMV dissociation, bind to a specific region of the viral RNA which potentially folds into a hairpin (35). The specific coat protein binding site is located within the protease coding region in SCPMV ORF2. This region is the most highly conserved region of ORF2 among sobemoviruses. Unfortunately, these results do not directly demonstrate that coat protein binding to this region serves to nucleate SCPMV assembly.
Full-length cDNA clones of RYMV-CI and SCPMV have been used to study the function(s) of the coat protein (9, 93). The mutants tested (C-terminal deletion and frameshift mutants of RYMV-CI; initiation codon and insertion mutants of SCPMV) were not infectious in plants. No virus accumulation was detected in the inoculated or systemic leaves, indicating that coat protein is essential for cell-to-cell and systemic virus movement. At the same time, it was found that the coat protein was not required for RYMV-CI or SCPMV RNA synthesis in rice and cowpea protoplasts, respectively. In rice plants, RNA replication for both RYMV-CI mutants was detected in leaves 4 weeks after inoculation, indicating the importance of coat protein particularly in long-distance virus movement.
Direct evidence that coat protein determines the range of systemic hosts of sobemoviruses was provided by characterizing the resistance-breaking mutant of SBMV-Ark, SBMV-S (58). SBMV-S is able to move systemically in bean cultivars Pinto and Great Northern, although the wild-type SBMV-Ark is restricted to the inoculated leaves of this host. Sequence analysis of the genomes of SBMV-Ark and SBMV-S revealed 7 nt differences but only four deduced amino acid changes. Three amino acid changes were identified in the R domain of the virus coat protein. Changes in this R domain may have an effect on specific coat protein-RNA interactions needed for correct capsid assembly and disassembly and thereby determine the long-distance movement of the virus.
Mixed infections of bean plants with SCPMV, which cannot infect beans systemically, and Sunn-hemp mosaic virus (SHMV; genus Tobamovirus) were limited to short-distance movement of SCPMV in the primary inoculated leaves, and only SHMV spread systemically (26). This SCPMV movement limitation was not due to the inability of the bean cells to support virus replication, since SCPMV replicated efficiently in bean protoplasts. It was proposed that the failure of SCPMV to move systemically in bean plants was due to a lack of normal SCPMV virion formation (27). Examination of thin sections of primary bean leaves doubly infected with SHMV and SCPMV revealed the presence of SCPMV virions having a T=1 structure and coat protein clumps in the vacuoles of mesophyll cells. These results show that short-distance (cell-to-cell) and long-distance (vascular) movements of SCPMV are distinct and separate processes in bean plants and that the formation of normal T=3 virions is a prerequisite for long-distance movement.
Recently, experiments were carried out to determine whether the host range restriction of SCPMV in bean plants could be complemented in trans by a related virus such as SBMV (37). It was demonstrated that SCPMV accumulates in the inoculated and systemically infected leaves of bean plants following coinoculation with SBMV. SCPMV recovered from coinfected bean plants was encapsidated, suggesting that heterologous encapsidation might play a role in the movement of SCPMV in this host. However, while the RNA synthesis and assembly of SBMV were not restricted in cowpea protoplasts, SCPMV did not complement the lack of systemic movement of SBMV in that host.
Supportive evidence for the requirement of encapsidation for long-distance transport comes from studies in which RYMV-CI virions were found in systemically infected leaves (69). RYMV-CI virions accumulated in large numbers in xylem parenchyma cells and vessels. The predominant location of RYMV virions within xylem implies that the upward flow pattern through xylem may facilitate the systemic spread of infection. Long-distance movement through xylem is rare for plant viruses. In addition to the accumulation of virions in mature xylem cells, association of RYMV-CI with intervascular pit membranes was observed (69). According to the proposed model for the translocation of RYMV in xylem, the partial digestion of pit membranes that occurs during programmed cell death may permit virus migration through these membranes. During the process of hydrolysis of pit membranes, the displacement of Ca2+ from the membranes to virus particles may contribute to the further disruption of the membranes and enable systemic virus transport. Presently it is not known whether this transport route is characteristic only of RYMV or is a feature common to all sobemoviruses. So far it is not known if the encapsidation is also needed for short-distance movement of RYMV, although virus-like particles have been identified within the plasmodesmata connecting mesophyll cells of RYMV-CI-infected leaves (69).
PATHOLOGY
Sobemovirus infections can cause a variety of disease symptoms in plants: mild or severe chlorosis and mottling, stunting, necrotic lesions, vein clearing, and sterility (12, 23, 42, 46, 78). Outcomes range from symptomless infections to severe diseases and death of plants.
Most sobemoviruses occur at relatively high concentrations in infected plants (46). They have been detected predominantly in mesophyll and vascular tissues but have been reported also, for instance, in epidermal, guard, and bundle sheath cells (38, 69, 76; VIDE database, http://biology.anu.edu.au/Groups/MES/vide/). In vascular tissues, the localization of virus particles in xylem parenchyma cells and xylem vessels seems to be characteristic, since only the occasional presence of particles has been reported in phloem parenchyma cells and sieve elements (38, 69; VIDE database, http://biology.anu.edu.au/Groups/MES/vide/). However, CfMV has been reported to localize in phloem of vascular tissues (12, 76).
Subcellularly, particles of sobemoviruses have been detected in the cytoplasm and vacuoles of infected cells (24, 44, 46). These viruses also form crystalline arrays in the cytoplasm (44, 69). Cells infected with several sobemoviruses contain cytoplasmic fibrils, some of which are enveloped in endoplasmic reticulum-derived vesicles (24). Characteristic tubules, often aggregated into bundles, are found in cells of plants infected, for example, with RYMV, BSSV, or SNMoV. The nature of these structures is unknown. No particles have been detected in either chloroplasts or mitochondria of cells infected with any of these viruses (46).
Particles are also found in the cell nuclei of plants infected with sobemoviruses (24). Yassi et al. (114) noted that the N-terminal part of the RYMV-CI coat protein (amino acid residues 3 to 22) contains a sequence which is identical to the bipartite nuclear targeting motif (16). A similar bipartite nuclear targeting motif can be found at the N termini of all sobemoviral coat proteins (61, 114). This finding may explain the observation that during sobemovirus infection, virus particles have been found in the nuclei of infected cells. Except for this observation, no molecular determinants have been attributed to the subcellular or tissue-specific localization of sobemovirus particles. Nearly nothing is known about the subcellular localization of the nonstructural sobemovirus proteins.
In susceptible hosts, several sobemoviruses can cause severe diseases with concurrent economic losses. This is true, for example, for CfMV in Norwegian and United Kingdom cocksfoot varieties, for RYMV in African rice, and for SCMoV in Australian subterranean clover. Studying varieties of hosts displaying total or partial resistance to the corresponding sobemoviruses has been of great help. Natural resistance to sobemoviruses has been detected for CfMV in cocksfoot (10, 82), for SCMoV in subterranean clover (111), for CnMoV in Cynosurus cristatus (10), for RYMV in Orza sativa (28, 68, 98), and in Oryza glaberrima (4, 68, 73, 98), and for SBMV in beans (115). For SCPMV, it has been demonstrated that resistance in cowpeas is controlled by a single gene according to a classical gene-to-gene interaction model (41). Resistance to RYMV in several O. glaberrima cultivars and in O. sativa indica cultivar Gigante is determined by a single recessive gene (4, 68, 73, 98). Partial RYMV resistance in many japonica rice cultivars is a polygenic trait (3, 28, 75). Nothing is known about the molecular features of genes encoding resistance to sobemoviruses. Few natural resistance sources are available for RYMV, making it the most rapidly spreading disease of rice in Africa. Here, recent advances in constructing transgenic rice plants expressing the putative RdRp sequence of RYMV and displaying resistance to several different RYMV strains have been reported (74). The protection achieved was based on RNA homology-dependent resistance. It is the only reported case of transgenic resistance to sobemoviruses.
CONCLUSIONS
In recent years there has been a substantial accumulation of knowledge on sobemoviruses. Some progress has been made in terms of understanding sobemovirus gene function and expression by (i) comparison with genes of known function, (ii) use of reporter gene assays in vitro and in vivo, (iii) construction of full-length infectious cDNA constructs and analysis of mutants, (iv) N-terminal sequencing of viral VPg proteins, and (v) mapping of the transcriptional start site of sgRNA. However, a better understanding of the mechanisms of transmission, replication, and movement would provide new insights into important aspects of the sobemovirus life cycle.
Both genomic organization and the primary structures of several sobemovirus nonstructural proteins (except P1) indicate that these viruses are related to the genera Polerovirus and Enamovirus in the family Luteoviridae. This similarity has been noticed several times and has allowed the authors to define a so-called Sobemo lineage in the evolution of viral RdRps (57). Besides substantial differences in biological properties (noncirculative transmission by beetles, in contrast with circulative transmission by aphids for luteoviruses), the main difference between sobemovirus proteins and those of poleroviruses and enamoviruses lies in their coat protein. It is more similar to the coat proteins of another genus of small isometric plant viruses, the necroviruses. The latter are fungus transmitted but have coat proteins similar to those of sobemoviruses. Members of the family Luteoviridae encode a specific protein needed for aphid transmission. Such a protein has so far not been identified for sobemoviruses.
The similarity between sobemoviruses and MBV is intriguing. This similarity extends both to the genome arrangement and to gene function and expression. The genome organization of MBV resembles that of CfMV. The similarities of expression strategies include (i) translation initiation for ORF2 by a leaky scanning mechanism, (ii) posttranslational proteolysis of the polyprotein, (iii) coat protein translation from an sgRNA, and (iv) expression of the putative RdRp as a fusion protein with the ORF2-encoded protein by −1 ribosomal frameshifting. Another virus taxon with molecular properties similar to those of sobemoviruses are the animal viruses of the family Astroviridae. Schematically, we can characterize astroviruses as animal sobemoviruses without the small 5′ ORF1. Indeed, the putative astrovirus protease and RdRp exhibit similarity to the corresponding proteins of sobemoviruses (49). Moreover, the putative RdRp of astroviruses is expressed as a fusion with the protease via a −1 ribosomal frameshift mechanism. Based solely on polyprotein sequences, we propose that the astrovirus VPg is also encoded downstream from the protease domain in the polyprotein, similarly to sobemo-, polero-, and enamoviruses. Taken together, the genomic organization and the primary structures of some nonstructural proteins characteristic of sobemoviruses have been conserved among viruses infecting fungi, plants, and animals.
The genus Sobemovirus contains viruses which have similar biological properties (transmission, subcellular localization, and symptomatology) and some similar molecular properties (particle structure, putative protease motif, putative RdRp motif, and coat protein sequence). The analysis of the amino acid sequences of coat proteins clearly shows that sobemoviruses cluster separately from other plant RNA viruses with similar particle morphologies (Fig. 2). However, the genomic organizations of these viruses, and therefore polyprotein expression, differ within the genus. We consider it possible that in the future a new taxonomic unit, the family Sobemoviridae, will be acknowledged. This would help in the recognition of the differences in genome organization (and in vectors for transmission) of sobemoviruses at the genus level. Whether it is relevant remains indistinct until the genomic structures of viruses transmitted by vectors other than beetles (BSSV, SoMV, and VTMoV) have been determined.
ACKNOWLEDGMENTS
We thank C. Fauquet, D. Hacker, K. Mäkinen, Y. Pinto, and J. Rathjen for sharing unpublished data. M.-A. Laane, K. Mäkinen, and M. Piirsoo are acknowledged for critical readings of the manuscript.
Research on sobemoviruses in our group has been supported by the Estonian Science Foundation (grant 2308) and by the EC INCO-Copernicus Programme (ERBIC15CT-96-0907).
REFERENCES
- 1.Abad-Zapatero C, Abdel-Meguid S S, Johnson J E, Leslie A G W, Rayment I, Rossmann M G, Suck D, Tsukihara T. Structure of southern bean mosaic virus at 2.8 Å resolution. Nature. 1980;286:33–39. doi: 10.1038/286033a0. [DOI] [PubMed] [Google Scholar]
- 2.AbouHaidar M G, Paliwal Y C. Comparison of the nucleotide sequences of the viroid-like satellite RNA of the Canadian and Australasian strains of lucerne transient streak virus. J Gen Virol. 1988;69:2369–2373. [Google Scholar]
- 3.Albar L, Lorieux M, Ahmadi N, Rimbault I, Pinel A, Sy A A, Fargette D, Ghesquière A. Genetic basis and mapping of the resistance to rice yellow mottle virus. I. QTLs identification and relationship between resistance and plant morphology. Theor Appl Genet. 1998;97:1145–1154. [Google Scholar]
- 4.Attere A F, Fatokun C A. Reaction of Oryza glaberrima accessions to rice yellow mottle virus. Plant Dis. 1983;67:420–421. [Google Scholar]
- 5.Bakker W. Characterisation and ecological aspects of rice yellow mottle virus in Kenya. Agricultural Research Report no. 829. Wageningen, The Netherlands: Agricultural University; 1974. [Google Scholar]
- 6.Bhuvaneshwari M, Subramanya H S, Gopinath K, Savithri H S, Nayudu M V, Murthy M R. Structure of sesbania mosaic virus at 3 Å resolution. Structure. 1995;3:1021–1030. doi: 10.1016/s0969-2126(01)00238-6. [DOI] [PubMed] [Google Scholar]
- 7.Blackstock J M. Lucerne transient streak and lucerne latent, two new viruses of lucerne. Aust J Agric Res. 1978;29:291–304. [Google Scholar]
- 8.Bonneau C, Brugidou C, Chen L, Beachy R N, Fauquet C. Expression of the rice yellow mottle virus P1 protein in vitro and in vivo and its involvement in virus spread. Virology. 1998;244:79–86. doi: 10.1006/viro.1998.9100. [DOI] [PubMed] [Google Scholar]
- 9.Brugidou C, Holt C, Yassi M N, Zhang S, Beachy R, Fauquet C. Synthesis of an infectious full-length cDNA clone of rice yellow mottle virus and mutagenesis of the coat protein. Virology. 1995;206:108–115. doi: 10.1016/s0042-6822(95)80025-5. [DOI] [PubMed] [Google Scholar]
- 10.Catherall P L. Resistances of grasses to two sobemoviruses, cocksfoot mottle and cynosurus mottle. Grass Forage Sci. 1985;40:311–316. [Google Scholar]
- 11.Cavener D R, Ray S C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chamberlain J A, Catherall P L. Electron microscopy of some grasses and cereals infected with cocksfoot mottle, Phleum mottle and cocksfoot mild mosaic viruses. J Gen Virol. 1976;30:41–50. [Google Scholar]
- 13.Collins R F, Gellatly D L, Sehgal O P, Abouhaidar M G. Self-cleaving circular RNA associated with rice yellow mottle virus is the smallest viroid-like RNA. Virology. 1998;241:269–275. doi: 10.1006/viro.1997.8962. [DOI] [PubMed] [Google Scholar]
- 14.Davies C, Haseloff J, Symons R H. Structure, self-cleavage, and replication of two viroid-like satellite RNAs (virusoids) of subterranean clover mottle virus. Virology. 1990;177:216–224. doi: 10.1016/0042-6822(90)90475-7. [DOI] [PubMed] [Google Scholar]
- 15.Demler S A, de Zoeten G A. The nucleotide sequence and luteovirus-like nature of RNA 1 of an aphid non-transmissible strain of pea enation mosaic virus. J Gen Virol. 1991;72:1819–1834. doi: 10.1099/0022-1317-72-8-1819. [DOI] [PubMed] [Google Scholar]
- 16.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 17.Dolja V V, Koonin E V. Phylogeny of capsid proteins of small icosahedral RNA plant viruses. J Gen Virol. 1991;72:1481–1486. doi: 10.1099/0022-1317-72-7-1481. [DOI] [PubMed] [Google Scholar]
- 18.Dougherty W G, Semler B L. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erickson J W, Rossmann M G. Assembly and crystallization of a T = 1 icosahedral particle from trypsinized southern bean mosaic virus coat protein. Virology. 1982;116:128–136. doi: 10.1016/0042-6822(82)90408-1. [DOI] [PubMed] [Google Scholar]
- 20.Fauquet M C, Mayo M A. Abbreviations for plant virus names—1999. Arch Virol. 1999;144:1249–1273. doi: 10.1007/s007050050584. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J. PHYLIP version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 22.Forster R L S, Jones A T. Properties of lucerne transient streak virus, and evidence of its affinity to southern bean mosaic virus. Ann Appl Biol. 1979;93:181–189. [Google Scholar]
- 23.Francki R I B, Randles J W, Hatta T, Davies C, Chu P W G, McLean G D. Subterranean clover mottle virus: another virus from Australia with encapsidated viroid-like RNA. Plant Pathol. 1983;32:47–59. [Google Scholar]
- 24.Francki R I B, Milne R G, Hatta T. Sobemovirus group. In: Francki R I B, Milne R G, Hatta T, editors. Atlas of plant viruses. Vol. 1. Boca Raton, Fla: CRC Press; 1985. pp. 153–169. [Google Scholar]
- 25.Francki R I B, Grivell C J, Gibb K S. Isolation of velvet mottle virus capable of replication with and without a viroid-like RNA. Virology. 1986;148:381–384. doi: 10.1016/0042-6822(86)90335-1. [DOI] [PubMed] [Google Scholar]
- 26.Fuentes A L, Hamilton R I. Sunn-hemp mosaic virus facilitates cell-to-cell spread of southern bean mosaic virus in a nonpermissive host. Phytopathology. 1991;81:1302–1305. [Google Scholar]
- 27.Fuentes A L, Hamilton R I. Failure of long-distance movement of southern bean mosaic virus in a resistant host is correlated with lack of normal virion formation. J Gen Virol. 1993;74:1903–1910. doi: 10.1099/0022-1317-74-9-1903. [DOI] [PubMed] [Google Scholar]
- 28.Ghesquière A, Albar L, Lorieux M, Ahmadi N, Fargette D, Huang N, McCouch S R, Notteghem J L. A major quantitative trait locus for rice yellow mottle virus resistance maps to a cluster of blast resistance on chromosome 12. Phytopathology. 1997;87:1243–1249. doi: 10.1094/PHYTO.1997.87.12.1243. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh A, Rutgers T, Ke-Qiang M, Kaesberg P. Characterization of the coat protein mRNA of southern bean mosaic virus and its relationship to the genomic RNA. J Virol. 1981;39:87–92. doi: 10.1128/jvi.39.1.87-92.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorbalenya A E, Koonin E V, Blinov V M, Donchenko A P. Sobemovirus genome appears to encode a serine protease related to cysteine proteases of picornaviruses. FEBS Lett. 1988;236:287–290. doi: 10.1016/0014-5793(88)80039-5. [DOI] [PubMed] [Google Scholar]
- 31.Gould A R, Hatta T. Studies on encapsidated viroid-like RNA. III. Comparative studies on RNAs isolated from velvet tobacco mottle virus and Solanum nodiflorum mottle virus. Virology. 1981;109:137–147. doi: 10.1016/0042-6822(81)90478-5. [DOI] [PubMed] [Google Scholar]
- 32.Greber R S. Some characteristics of solanum nodiflorum virus—a beetle-transmitted isometric virus from Australia. Aust J Biol Sci. 1981;34:369–378. [Google Scholar]
- 33.Guilley H, Wipf-Scheibel C, Richards K, Lecoq H, Jonard G. Nucleotide sequence of cucurbit aphid-borne yellows luteovirus. Virology. 1994;202:1012–1017. doi: 10.1006/viro.1994.1429. [DOI] [PubMed] [Google Scholar]
- 34.Guilley H, Richards K E, Jonard G. Nucleotide sequence of beet mild yellowing virus RNA. Arch Virol. 1995;140:1109–1118. doi: 10.1007/BF01315419. [DOI] [PubMed] [Google Scholar]
- 35.Hacker D L. Identification of a coat protein binding site on southern bean mosaic virus RNA. Virology. 1995;207:562–565. doi: 10.1006/viro.1995.1117. [DOI] [PubMed] [Google Scholar]
- 36.Hacker D L, Sivakumaran K. Mapping and expression of southern bean mosaic virus genomic and subgenomic RNAs. Virology. 1997;234:317–327. doi: 10.1006/viro.1997.8667. [DOI] [PubMed] [Google Scholar]
- 37.Hacker D L, Fowler B C. Complementation of the host range restriction of southern cowpea mosaic virus in bean by southern bean mosaic virus. Virology. 2000;266:140–149. doi: 10.1006/viro.1999.0072. [DOI] [PubMed] [Google Scholar]
- 38.Hartmann J X, Bath J E, Hooper G R. Electron microscopy of virus like particles from shoestring-diseased high bush blueberry, Vaccinium corymbosum L. Phytopathology. 1973;63:432–436. [Google Scholar]
- 39.Haseloff J, Symons R H. Comparative sequence and structure of viroid-like RNAs of two plant viruses. Nucleic Acids Res. 1982;10:3681–3691. doi: 10.1093/nar/10.12.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermodson M A, Abad-Zapatero C, Abdel-Meguid S S, Pundak S, Rossmann M G, Tremaine J H. Amino acid sequence of southern bean mosaic virus coat protein and its relation to the three-dimensional structure of the virus. Virology. 1982;119:133–149. doi: 10.1016/0042-6822(82)90071-x. [DOI] [PubMed] [Google Scholar]
- 41.Hobbs H A, Kuhn C W, Papa K E, Brantley B B. Inheritance of nonnecrotic resistance to southern bean mosaic virus in cowpea. Phytopathology. 1987;77:1624–1629. [Google Scholar]
- 42.Hollings M, Stone O M. Turnip rosette virus. CMI/AAB description of plant viruses no. 125. Kew, Surrey: Commonwealth Mycological Institute; 1973. , and Association of Applied Biologists, Wellesbourne, Warwick, England. [Google Scholar]
- 43.Hsu C H, White J A, Sehgal O P. Assembly of southern bean mosaic virus from its two subviral intermediates. Virology. 1977;81:471–475. doi: 10.1016/0042-6822(77)90162-3. [DOI] [PubMed] [Google Scholar]
- 44.Hull R. The grouping of small spherical plant viruses with single RNA components. J Gen Virol. 1977;36:289–295. [Google Scholar]
- 45.Hull R. The stabilization of the particles of turnip rosette virus and of other members of the southern bean mosaic virus group. Virology. 1977;79:58–66. doi: 10.1016/0042-6822(77)90334-8. [DOI] [PubMed] [Google Scholar]
- 46.Hull R. The sobemovirus group. In: Koenig R, editor. The plant viruses. Vol. 3. New York, N.Y: Plenum Press; 1988. pp. 113–146. [Google Scholar]
- 47.Hull R. Sobemovirus. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 376–378. [Google Scholar]
- 48.Huth W, Paul H L. Cocksfoot mosaic virus. CMI/AAB description of plant viruses no. 107. Kew, Surrey: Commonwealth Mycological Institute; 1972. , and Association of Applied Biologists, Wellesbourne, Warwick, England. [Google Scholar]
- 49.Jiang B, Monroe S S, Koonin E V, Stine S E, Glass R I. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc Natl Acad Sci USA. 1993;90:10539–10543. doi: 10.1073/pnas.90.22.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones A T, Mayo M A. Interaction of lucerne transient streak virus and the viroid-like RNA-2 of Solanum nodiflorum mottle virus. J Gen Virol. 1983;64:1771–1774. [Google Scholar]
- 51.Jones A T, Mayo M A. Satellite nature of the viroid-like RNA-2 of Solanum nodiflorum mottle virus and the ability of other plant viruses to support the replication of viroid-like RNA molecules. J Gen Virol. 1984;65:1713–1721. [Google Scholar]
- 52.Kado C I. Biological and biochemical characterisation of sowbane mosaic virus. Virology. 1967;31:217–229. doi: 10.1016/0042-6822(67)90165-1. [DOI] [PubMed] [Google Scholar]
- 53.Kado C I. Sowbane mosaic virus. CMI/AAB description of plant viruses no. 64. Kew, Surrey: Commonwealth Mycological Institute; 1971. , and Association of Applied Biologists, Wellesbourne, Warwick, England. [Google Scholar]
- 54.Keese P, Bruening G, Symons R H. Comparative sequence and structure of circular RNAs from two isolates of lucerne transient streak virus. FEBS Lett. 1983;159:185–190. [Google Scholar]
- 55.Kiberstis P A, Zimmern D. Translational strategy of Solanum nodiflorum mottle virus RNA: synthesis of a coat protein precursor in vitro and in vivo. Nucleic Acids Res. 1984;12:933–943. doi: 10.1093/nar/12.2.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 57.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 58.Lee L, Anderson E J. Nucleotide sequence of a resistance breaking mutant of southern bean mosaic virus. Arch Virol. 1998;143:2189–2201. doi: 10.1007/s007050050451. [DOI] [PubMed] [Google Scholar]
- 59.Lütcke H A, Chow K C, Mickel F S, Moss K A, Kern H F, Scheele G A. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mäkinen K, Næss V, Tamm T, Truve E, Aaspõllu A, Saarma M. The putative replicase of the cocksfoot mottle sobemovirus is translated as a part of the polyprotein by −1 ribosomal frameshift. Virology. 1995;207:566–571. doi: 10.1006/viro.1995.1118. [DOI] [PubMed] [Google Scholar]
- 61.Mäkinen K, Tamm T, Næss V, Truve E, Puurand U, Munthe T, Saarma M. Characterization of cocksfoot mottle sobemovirus genomic RNA and sequence comparison with related viruses. J Gen Virol. 1995;76:2817–2825. doi: 10.1099/0022-1317-76-11-2817. [DOI] [PubMed] [Google Scholar]
- 62.Mang K-Q, Ghosh A, Kaesberg P. A comparative study of the cowpea and bean strains of southern bean mosaic virus. Virology. 1982;116:264–274. doi: 10.1016/0042-6822(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 63.Matthews R E F. Classification and nomenclature of viruses. Fourth report of the International Committee on Taxonomy of Viruses. Intervirology. 1982;17:1–199. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- 64.Miller W A, Dinesh-Kumar S P, Paul C P. Luteovirus gene expression. Crit Rev Plant Sci. 1995;14:179–211. [Google Scholar]
- 65.Mohamed N A, Mossop D W. Cynosurus and cocksfoot mottle viruses: a comparison. J Gen Virol. 1981;55:63–74. [Google Scholar]
- 66.Morris-Krsinich B A M, Hull R. Translation of turnip rosette virus RNA in rabbit reticulocyte lysates. Virology. 1981;114:98–112. doi: 10.1016/0042-6822(81)90256-7. [DOI] [PubMed] [Google Scholar]
- 67.Morris-Krsinich B A M, Forster R L S. Lucerne transient streak virus RNA and its translation in rabbit reticulocyte lysate and wheat germ extract. Virology. 1983;128:176–185. doi: 10.1016/0042-6822(83)90328-8. [DOI] [PubMed] [Google Scholar]
- 68.Ndjiondjop M N, Albar L, Fargette D, Fauquet C, Ghesquière A. The genetic basis of high resistance to rice yellow mottle virus (RYMV) in cultivars of two cultivated rice species. Plant Dis. 1999;83:931–935. doi: 10.1094/PDIS.1999.83.10.931. [DOI] [PubMed] [Google Scholar]
- 69.Opalka N, Brugidou C, Bonneau C, Nicole M, Beachy R N, Yeager M, Fauquet C. Movement of rice yellow mottle virus between xylem cells through pit membranes. Proc Natl Acad Sci USA. 1998;95:3323–3328. doi: 10.1073/pnas.95.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Othman Y, Hull R. Nucleotide sequence of the bean strain of southern bean mosaic virus. Virology. 1995;206:287–297. doi: 10.1016/S0042-6822(95)80044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Page R D M. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 72.Paliwal Y C. Interaction of the viroid-like RNA 2 of lucerne transient streak virus with southern bean mosaic virus. Can J Plant Pathol. 1984;6:93–97. [Google Scholar]
- 73.Paul C N, Ng N Q, Ladeinde T. Diallel analysis of resistance to rice yellow mottle virus in African rice Oryza glaberrima Steud. J Genet Breed. 1995;49:217–222. [Google Scholar]
- 74.Pinto Y M, Kok R A, Baulcombe D C. Resistance to rice yellow mottle virus (RYMV) in cultivated African rice varieties containing RYMV transgenes. Nat Biotechnol. 1999;17:702–707. doi: 10.1038/10917. [DOI] [PubMed] [Google Scholar]
- 75.Pressoir G, Albar L, Ahmadi N, Rimbault I, Lorieux M, Fargette D, Ghesquière A. Genetic basis and mapping of the resistance to rice yellow mottle virus. II. Evidence of a complementary epistasis between two QTLs. Theor Appl Genet. 1998;97:1155–1161. [Google Scholar]
- 76.Rabenstein F, Stanarius A. Untersuchungen zum Knaulgrasscheckungs-Virus (cocksfoot mottle virus) Arch Phytopathol Pflanzenschutz. 1984;20:15–31. [Google Scholar]
- 77.Ramsdell D C. Blueberry shoestring virus. CMI/AAB description of plant viruses no. 204. Kew, Surrey: Commonwealth Mycological Institute; 1979. , and Association of Applied Biologists, Wellesbourne, Warwick, England. [Google Scholar]
- 78.Randles J W, Davies C, Hatta T, Gould A R, Francki R I B. Studies on encapsidated viroid-like RNA. I. Characterisation of velvet tobacco mottle virus. Virology. 1981;108:111–122. doi: 10.1016/0042-6822(81)90531-6. [DOI] [PubMed] [Google Scholar]
- 79.Revill P A, Davidson A D, Wright P J. The nucleotide sequence and genome organization of mushroom bacilliform virus: a single-stranded RNA virus of Agaricus bisporus (Lange) Imbach. Virology. 1994;202:904–911. doi: 10.1006/viro.1994.1412. [DOI] [PubMed] [Google Scholar]
- 80.Revill P A, Davidson A D, Wright P J. Mushroom bacilliform virus RNA: the initiation of translation at the 5′ end of the genome and identification of the VPg. Virology. 1998;249:231–237. doi: 10.1006/viro.1998.9345. [DOI] [PubMed] [Google Scholar]
- 81.Revill P A, Davidson A D, Wright P J. Identification of a subgenomic mRNA encoding the capsid protein of mushroom bacilliform virus, a single-stranded RNA mycovirus. Virology. 1999;260:273–276. doi: 10.1006/viro.1999.9839. [DOI] [PubMed] [Google Scholar]
- 82.Rognli O A, Aastveit K, Munthe T. Genetic variation in cocksfoot (Dactylis glomerata L.) populations for mottle virus resistance. Euphytica. 1995;83:109–116. [Google Scholar]
- 83.Rossmann M G, Abad-Zapatero C, Hermodson M A, Erickson J W. Subunit interactions in southern bean mosaic virus. J Mol Biol. 1983;166:37–73. doi: 10.1016/s0022-2836(83)80049-7. [DOI] [PubMed] [Google Scholar]
- 84.Rossmann M G. Constraints on the assembly of spherical virus particles. Virology. 1984;134:1–11. doi: 10.1016/0042-6822(84)90267-8. [DOI] [PubMed] [Google Scholar]
- 85.Rutgers T, Salerno-Rife T, Kaesberg P. Messenger RNA for the coat protein of southern bean mosaic virus. Virology. 1980;104:506–509. doi: 10.1016/0042-6822(80)90355-4. [DOI] [PubMed] [Google Scholar]
- 86.Ryabov E V, Krutov A A, Novikov V K, Zheleznikova O V, Morozov S Y, Zavriev S K. Nucleotide sequence of RNA from the sobemovirus found in infected cocksfoot shows a luteovirus-like arrangement of the putative replicase and protease genes. Phytopathology. 1996;86:391–397. [Google Scholar]
- 87.Salerno-Rife T, Rutgers T, Kaesberg P. Translation of southern bean mosaic virus RNA in wheat embryo and rabbit reticulocyte extracts. J Virol. 1980;34:51–58. doi: 10.1128/jvi.34.1.51-58.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Savithri H S, Erickson J W. The self-assembly of the cowpea strain of southern bean mosaic virus: formation of T = 1 and T = 3 nucleoprotein particles. Virology. 1983;126:328–335. doi: 10.1016/0042-6822(83)90482-8. [DOI] [PubMed] [Google Scholar]
- 89.Sehgal O P, Hsu C H, White J A, Van M. Enzymic sensitivity of conformationally altered virions of southern bean mosaic virus. Phytopathol Z. 1979;95:167–177. [Google Scholar]
- 90.Sehgal O P, Sinha R C, Gellatly D L, Ivanov I, AbouHaidar M G. Replication and encapsidation of the viroid-like satellite RNA of lucerne transient streak virus are supported in divergent hosts by cocksfoot mottle virus and turnip rosette virus. J Gen Virol. 1993;74:785–788. doi: 10.1099/0022-1317-74-4-785. [DOI] [PubMed] [Google Scholar]
- 91.Serjeant E P. Some properties of cocksfoot mottle virus. Ann Appl Biol. 1967;59:31–38. [Google Scholar]
- 92.Shields S A, Brisco M J, Wilson T M, Hull R. Southern bean mosaic virus RNA remains associated with swollen virions during translation in wheat germ cell-free extracts. Virology. 1989;171:602–606. doi: 10.1016/0042-6822(89)90630-2. [DOI] [PubMed] [Google Scholar]
- 93.Sivakumaran K, Fowler B C, Hacker D L. Identification of viral genes required for cell-to-cell movement of southern bean mosaic virus. Virology. 1998;252:376–386. doi: 10.1006/viro.1998.9489. [DOI] [PubMed] [Google Scholar]
- 94.Sivakumaran K, Hacker D L. The 105-kDa polyprotein of southern bean mosaic virus is translated by scanning ribosomes. Virology. 1998;246:34–44. doi: 10.1006/viro.1998.9183. [DOI] [PubMed] [Google Scholar]
- 95.Tamm T, Mäkinen K, Truve E. Identification of genes encoding for the cocksfoot mottle virus proteins. Arch Virol. 1999;144:1557–1567. doi: 10.1007/s007050050610. [DOI] [PubMed] [Google Scholar]
- 96.Tamm T, Truve E. RNA-binding activities of cocksfoot mottle sobemovirus proteins. Virus Res. 2000;66:197–207. doi: 10.1016/s0168-1702(00)00125-8. [DOI] [PubMed] [Google Scholar]
- 97.Thomas J E. Gringer chlorotic fleck virus. CMI/AAB description of plant viruses no. 328. Kew, Surrey: Commonwealth Mycological Institute; 1988. , and Association of Applied Biologists, Wellesbourne, Warwick, England. [Google Scholar]
- 98.Thottapilly G, Rossel H W. Evaluation of resistance to rice yellow mottle virus in Oryza species. Indian J Virol. 1993;9:65–73. [Google Scholar]
- 99.Tien-Po, Davies C, Hatta T, Francki R I B. Viroid-like RNA encapsidated in lucerne transient streak virus. FEBS Lett. 1981;132:353–356. [Google Scholar]
- 100.Tremaine J G, Hamilton R I. Southern bean mosaic virus. CMI/AAB descriptions of plant viruses no. 274. Kew, Surrey: Commonwealth Mycological Institute; 1983. , and Association of Applied Biologists, Wellesbourne, Warwick, England. [Google Scholar]
- 101.van der Wilk F, Huisman M J, Cornelissen B J, Huttinga H, Goldbach R. Nucleotide sequence and organization of potato leafroll virus genomic RNA. FEBS Lett. 1989;245:51–56. doi: 10.1016/0014-5793(89)80190-5. [DOI] [PubMed] [Google Scholar]
- 102.van der Wilk F, Verbeek M, Dullemans A M, van den Heuvel J F J M. The genome-linked protein of potato leafroll virus is located downstream of the putative protease domain of the ORF1 product. Virology. 1997;234:300–303. doi: 10.1006/viro.1997.8654. [DOI] [PubMed] [Google Scholar]
- 103.van der Wilk F, Verbeek M, Dullemans A, van den Heuvel J F J M. The genome-linked protein (VPg) of southern bean mosaic virus is encoded by the ORF2. Virus Genes. 1998;17:21–24. doi: 10.1023/a:1008044715899. [DOI] [PubMed] [Google Scholar]
- 104.Veidt I, Lot H, Leiser M, Scheidecker D, Guilley H, Richards K, Jonard G. Nucleotide sequence of beet western yellows virus RNA. Nucleic Acids Res. 1988;16:9917–9932. doi: 10.1093/nar/16.21.9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vincent J R, Lister R M, Larkins B A. Nucleotide sequence analysis and genomic organization of the NY-RPV isolate of barley yellow dwarf virus. J Gen Virol. 1991;72:2347–2355. doi: 10.1099/0022-1317-72-10-2347. [DOI] [PubMed] [Google Scholar]
- 106.Voinnet O, Pinto Y M, Baulcombe D C. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Walters H J. Beetle transmission of plant viruses. Adv Virus Res. 1969;15:339–363. doi: 10.1016/s0065-3527(08)60879-9. [DOI] [PubMed] [Google Scholar]
- 108.Weber K A, Sehgal O P. Subgenomic RNAs in virions of southern bean mosaic virus. Phytopathology. 1982;72:909–913. [Google Scholar]
- 109.Wilson T M. Nucleocapsid disassembly and early gene expression by positive-strand RNA viruses. J Gen Virol. 1985;66:1201–1207. doi: 10.1099/0022-1317-66-6-1201. [DOI] [PubMed] [Google Scholar]
- 110.Wobus C E, Skaf J S, Schultz M H, de Zoeten G A. Sequencing, genomic localization and initial characterization of the VPg of pea enation mosaic enamovirus. J Gen Virol. 1998;79:2023–2025. doi: 10.1099/0022-1317-79-8-2023. [DOI] [PubMed] [Google Scholar]
- 111.Wroth J M, Jones R A C. Subterranean clover mottle sobemovirus: its host range, resistance in subterranean clover and transmission through seed and by grazing animals. Ann Appl Biol. 1992;121:329–343. [Google Scholar]
- 112.Wu S, Rinehart C A, Kaesberg P. Sequence and organization of southern bean mosaic virus genomic RNA. Virology. 1987;161:73–80. doi: 10.1016/0042-6822(87)90172-3. [DOI] [PubMed] [Google Scholar]
- 113.Xiong Z, Lommel S A. The complete nucleotide sequence and genome organization of red clover necrotic mosaic virus RNA-1. Virology. 1989;171:543–554. doi: 10.1016/0042-6822(89)90624-7. [DOI] [PubMed] [Google Scholar]
- 114.Yassi M N, Ritzenthaler C, Brugidou C, Fauquet C, Beachy R N. Nucleotide sequence and genome characterization of rice yellow mottle virus RNA. J Gen Virol. 1994;75:249–257. doi: 10.1099/0022-1317-75-2-249. [DOI] [PubMed] [Google Scholar]
- 115.Zaumeyer W J, Harter L L. Inheritance of symptom expression of bean mosaic virus 4. J Agric Res. 1943;67:295–300. [Google Scholar]
- 116.Zavriev S K, Hickey C M, Lommel S A. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology. 1996;216:407–410. doi: 10.1006/viro.1996.0076. [DOI] [PubMed] [Google Scholar]