Abstract
Background:
Uveal coloboma, a developmental eye defect, is caused by failed development of the optic fissure, a ventral structure in the optic stalk and cup where axons exit the eye and vasculature enters. The Hedgehog (Hh) signaling pathway regulates optic fissure development: loss-of-function mutations in the Hh receptor ptch2 produce overactive Hh signaling and can result in coloboma. We previously proposed a model where overactive Hh signaling disrupts optic fissure formation by upregulating transcriptional targets acting both cell- and non-cell-autonomously. Here, we examine the Netrin family of secreted ligands as candidate Hh target genes.
Results:
We find multiple Netrin ligands upregulated in the zebrafish ptch2 mutant during optic fissure development. Using a gain-of-function approach to overexpress Netrin in a spatiotemporally specific manner, we find that netrin1a or netrin1b overexpression is sufficient to cause coloboma and disrupt wild-type optic fissure formation. We used loss-of-function alleles, CRISPR/Cas9 mutagenesis, and morpholino knockdown to test if loss of Netrin can rescue coloboma in the ptch2 mutant: loss of netrin genes does not rescue the ptch2 mutant phenotype.
Conclusion:
These results suggest that Netrin is sufficient but not required to disrupt optic fissure formation downstream of overactive Hh signaling in the ptch2 mutant.
Keywords: netrin1a, coloboma, eye development, morphogenesis, ptch2
Introduction
The proper three-dimensional structure of the eye is critical for vision, as structural defects commonly account for visual impairment in newborns. One such defect, uveal coloboma, is caused by failed development of the optic fissure, a transient seam along the ventral surface of the optic stalk and optic cup that forms a conduit during development for retinal ganglion cell axons to exit the eye and vasculature to enter. Uveal coloboma is a significant cause of pediatric blindness worldwide, yet we lack a basic understanding of the cellular and molecular mechanisms disrupted 1–5. Through human genetic studies and findings using animal models, we know that the genetic underpinnings of coloboma are heterogeneous and include mutations in multiple signaling pathways 4,6.
One pathway central to optic fissure development is the Hedgehog (Hh) signaling pathway: mutations upstream, within, and downstream of this pathway can result in coloboma 4. Human mutations in the Hh receptor PTCH cause Gorlin syndrome 7,8, in which affected individuals are typically diagnosed with medulloblastoma or basal cell carcinoma, along with numerous additional phenotypes including coloboma 9.
In zebrafish, the ptch2 loss-of-function mutant displays coloboma: given the function of Ptch2 as a negative regulator of Hh signaling, these mutations lead to overactive Hh signaling. The ptch2 mutant coloboma phenotype has been described in detail 10, however, molecular mechanisms directly driving disruption of optic fissure morphogenesis are still unclear. We previously determined the cellular mechanisms by which the ptch2 mutant phenotype initially arises 11. Using multidimensional timelapse microscopy and cell tracking, we identified the cells that give rise to the optic fissure in wild-type embryos. In the ptch2 mutant, these cells do not move to their correct position; as a result, the optic fissure fails to form. Additional analyses of cells in the optic stalk revealed morphological defects at the single cell level: cells are less elongated compared to the wild-type optic stalk. Downstream transcriptional targets are upregulated in the ptch2 mutant, and Gli activity is required for the mutant phenotype. In addition, the ptch2 mutant phenotype is regulated by both cell autonomous and non-cell autonomous mechanisms 11. Taken together, these data suggest that overactive Hh signaling in the ptch2 mutant disrupts cell movements and morphology via misregulation of downstream transcriptional targets acting intra- and inter-cellularly, resulting in aberrant optic fissure and stalk formation. We hypothesize that this disruption of optic fissure and stalk formation underlies the ptch2 mutant coloboma phenotype.
Therefore, downstream transcriptional targets of Hh signaling are likely the key factors that directly disrupt optic fissure and stalk cell movements and cause coloboma. To identify these downstream factors, we have taken a candidate approach, focusing initially on intercellular signaling molecules that are known transcriptional targets of Hh signaling and are expressed at the appropriate time and place to influence optic fissure and stalk morphogenesis. Using these criteria, we have identified an initial candidate: Netrin, a family of laminin-related secreted molecules, largely studied in the context of axon guidance. In this study we examine the genetic interaction between netrin and the Hh signaling pathway to determine if upregulation of Netrin is, in part, responsible for the ptch2 mutant coloboma phenotype.
Netrin family proteins are diffusible molecules that can regulate diverse developmental processes, including but not limited to axonal guidance, cell survival, and cell-cell adhesion 12–15. Zebrafish contain five netrin genes: netrin 1a (ntn1a), netrin 1b (ntn1b), netrin 2 (ntn2), netrin 4 (ntn4), and netrin 5 (ntn5) 16–19. Roles for Netrin1 in eye development have previously been described in zebrafish, chick and mouse. For example, Ntn1a acts as a retinal ganglion cell axon guidance molecule expressed along the optic fissure 20. However, ntn1a is also expressed at an earlier stage in the nasal optic vesicle and optic vesicle junction with the forebrain 21. This suggests that ntn1a is expressed at the right time and in the right location to act downstream of Hh signaling in optic fissure and stalk formation. Further, both ntn1a and ntn1b expression have been established as being responsive to Hh signaling: in sonic hedgehog a (shha) and smoothened (smo) mutants, both of which have decreased Hh signaling, ntn1a and ntn1b mRNA levels are decreased 22,23. In response to increased Hh signaling, for example, sonic Hh (Shh) and a dominant negative form of protein kinase A (dnPKA) overexpression, ntn1a expression is ectopically expanded in all regions, including the head and eyes 24,25. Although ntn1a expression has been assayed at timepoints relevant to optic fissure and stalk formation, other Netrins have only been analyzed at later stages 16,18,26. Of interest, two recent studies utilized optic fissure transcriptomic approaches to identify novel coloboma causing genes and found Netrin1 mediates optic fissure closure later in development 27,28.
Here, we characterize a novel role for Netrin during early eye development: we asked whether netrin might be a key downstream target of Hedgehog signaling in the ptch2 mutant, in which overactive signaling leads to defective optic fissure formation and coloboma. We find that in wild-type embryos, upregulation of netrin in Hh-responding cells is sufficient to disrupt optic fissure formation and can lead to coloboma. Despite finding that spatiotemporally specific overexpression of netrin is sufficient to lead to coloboma, these ligands are not required for the ptch2 mutant phenotype; removal of multiple netrin ligands from the ptch2 mutant is unable to rescue coloboma. This suggests that although Netrin may be involved in both formation and fusion of the optic fissure, there is likely functional redundancy in this complex morphological defect, particularly in the formation step. Together, this work uncovers molecular mechanisms regulating optic fissure formation and, in turn, coloboma.
Results
netrin genes are expressed in the head during optic cup morphogenesis and are upregulated in response to overactive Hh signaling
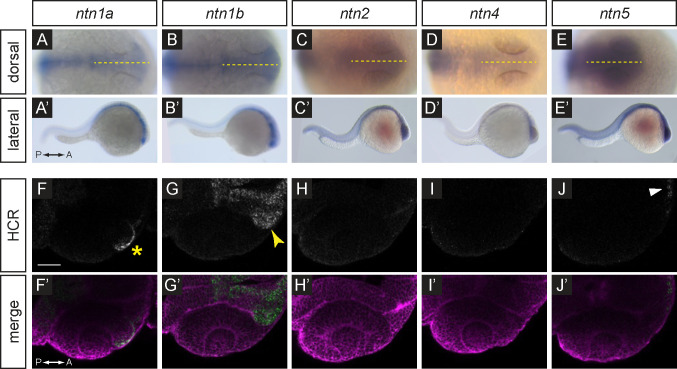
Research in different animal models has demonstrated that netrin genes are expressed widely during development and in a variety of organs 12,14,29–34. In zebrafish, it has been previously shown that both ntn1a and ntn1b are expressed in the optic vesicle 21,22, however it remains unknown if ntn2, ntn4, or ntn5 are expressed during early eye development in zebrafish. Thus, we first examined which ligands are expressed in the early embryo to determine which might regulate optic fissure morphogenesis. We performed whole mount in situ hybridization for all 5 genes in 24 hours post fertilization (hpf) wild-type embryos (Fig. 1A–E’). We found that ntn1a and ntn1b are expressed in a distinct pattern restricted to the embryonic midline, and potentially within the optic stalk (Fig. 1A–B’). ntn2, ntn4, and ntn5 appear lowly expressed and not spatially restricted at this stage (Fig. 1C–E’).
Figure 1. Expression patterns of netrin ligands.
(A-E’) Whole-mount in situ hybridization for ntn1a, ntn1b, ntn2, ntn4, and ntn5 in wild-type embryos at 24 hpf, (A-E) dorsal orientation, (A’-E’) lateral orientation. Yellow dotted lines (A-E) indicate embryo midline.
(F-J’) HCR RNA-FISH for ntn1a, ntn1b, ntn2, ntn4, and ntn5 in wild-type embryos harboring the Tg(bactin2:EGFP-CAAX) transgene at 24 hpf, (F-J) HCR signal alone. (F’-J’) Merge of HCR (green) and Tg(bactin2:EGFP-CAAX) transgene (magenta) to visualize tissue morphology. Yellow asterisk (F) indicates ntn1a signal in the nasal margin of the optic fissure; yellow arrowhead (G) indicates ntn1b signal in the optic stalk; white arrowhead (J) indicates faint ntn5 in the telencephalon. Scale bar, 50 μm.
In order to visualize gene expression with greater spatial resolution, we utilized hybridization chain reaction RNA fluorescence in situ (HCR RNA-FISH) technology 35. Probe target sites were selected to be specific to each netrin gene, and HCR was performed on embryos fixed at 24 hpf from a ptch2+/−; Tg(bactin2:EGFP-CAAX) incross. In wild-type embryos, we observe a similar expression pattern for ntn1a and ntn1b at this stage as we did with colorimetric in situ hybridization. ntn1a is expressed in the midline, in addition to the anterior rim of the optic cup and nasal margin of the optic fissure (Fig. 1F–F’; yellow asterisk marks nasal margin of the fissure). ntn1b is expressed in a broader pattern than ntn1a and includes expression within the optic stalk, but not the optic cup (Fig. 1G–G’; yellow arrowhead indicates stalk). Expression of ntn2 and ntn4 is not detected within the head at this stage (Fig. 1H–I’). ntn5 expression appears restricted to only the few most anterior, ventral cells within the forebrain (Fig. 1J–J’; white arrowhead indicates forebrain).
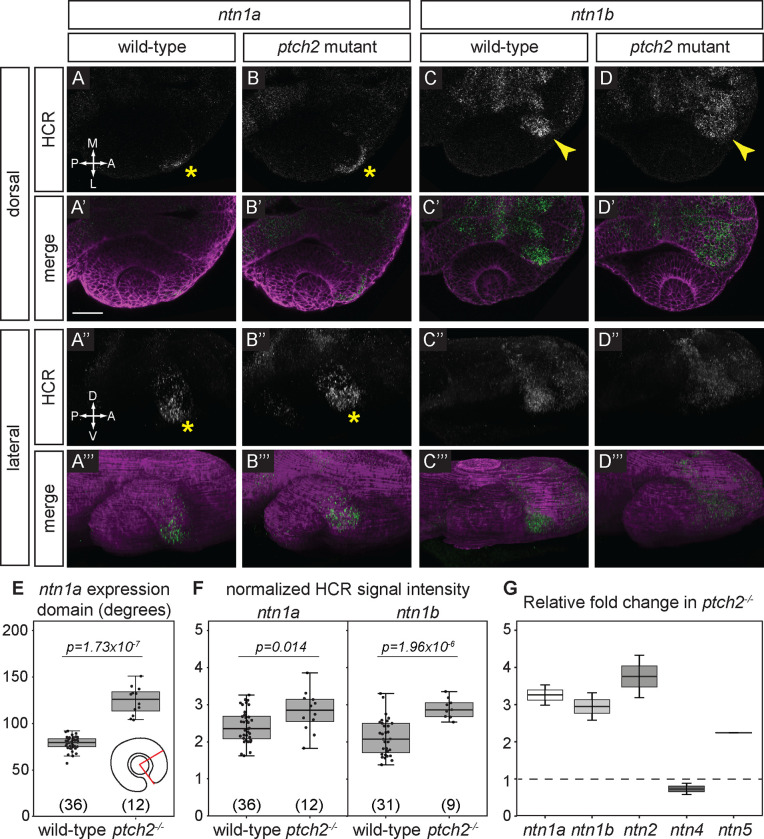
To determine if Netrin ligands are responsive to Hh signaling we assayed gene expression in ptch2 mutants and siblings. We observe that ntn1a expression is apparent in the optic fissure in ptch2−/− embryos similar to wild-type, however expression appears expanded (Fig. 2A–A’”, B-B’”; asterisks indicate expression in the nasal optic fissure). We quantified expression by measuring the domain angle in degrees (schematized in Fig. 2E) and found that indeed the domain of expression is expanded in ptch2−/− compared to wild-type siblings (Fig. 2E; sibling 78.69 ± 7.51°; ptch2−/− 124.5 ±15.08°). Similar to wild-type, ntn1b expression in ptch2−/− is observed in the optic stalk, but expression appears increased (Fig. 2C–C’”, D-D’”; arrowheads indicate stalk). We further measured ntn1a and ntn1b expression in wild-type and ptch2−/− embryos by quantifying and normalizing fluorescence intensity in the nasal margin of the optic fissure (ntn1a), or the optic stalk (ntn1b); see Methods for details. We observe a statistically significant increase in ntn1a and ntn1b expression in ptch2−/− embryos compared to wild-type siblings (Fig. 2F). ntn2 and ntn4 expression remain absent in the ptch2−/− head at 24 hpf, unchanged from wild-type (data not shown). ntn5 expression appears stronger and slightly expanded in ptch2−/−, although this is still restricted to a narrow region in the forebrain, distant from the optic fissure and stalk (data not shown). These data suggest that Ntn2, Ntn4, and Ntn5 may be less likely to regulate optic fissure formation.
Figure 2. Responsiveness of netrin ligand expression to Hedgehog signaling.
(A-D’”) HCR RNA-FISH for ntn1a and ntn1b in wild-type embryos (A-A’”, C-C’”) and ptch2−/− embryos (B-B’”, D-D’”) at 24 hpf. Dorsal view (A-D, A’-D’) and lateral views of 3D renderings (A”-D”, A’”-D’”). The merged images (A’-D’, A’”-D’”) are HCR signal (green) and Tg(bactin2:EGFP-CAAX) transgene (magenta) to visualize tissue morphology. Yellow asterisks (A, A”, B, B”) indicate ntn1a signal in the nasal margin of the optic fissure, and yellow arrowheads (C, D) indicate ntn1b signal in the optic stalk.
(E, F) Quantification of HCR data, for (E) the extent of the ntn1a expression domain; and (F) normalized fluorescence intensity for ntn1a in the optic fissure and ntn1b in the optic stalk.
(G) RT-qPCR quantification showing relative fold change of netrin genes in ptch2−/− embryos compared to wild-type embryos at 24 hpf using the ΔΔCt method, where the relative quantity of each gene was normalized to the reference gene eef1a1l1. Scale bar, 50 μm.
As further evidence for increased netrin gene expression in ptch2−/− embryos, we performed reverse transcription-quantitative PCR (RT-qPCR) in whole embryos at 24 hpf. We calculated the fold change in gene expression for each netrin gene normalized to a conventional housekeeping gene, eef1a1l1, in ptch2−/− embryos compared to wild-type using the ΔΔCt method 36. We find that expression of ntn1a, ntn1b, ntn2, and ntn5 genes are each at least 2-fold upregulated in ptch2−/−embryos at 24 hpf (Fig. 2G). Together with our findings from in situ hybridization, we conclude that ntn1a and ntn1b are expressed in the optic fissure and stalk respectively and are upregulated in response to overactive Hh signaling. Therefore, moving forward, we examined potential roles for ntn1a and ntn1b in optic fissure formation.
Overexpression of Netrin in a spatiotemporally specific manner is sufficient to disrupt optic fissure formation and cause coloboma
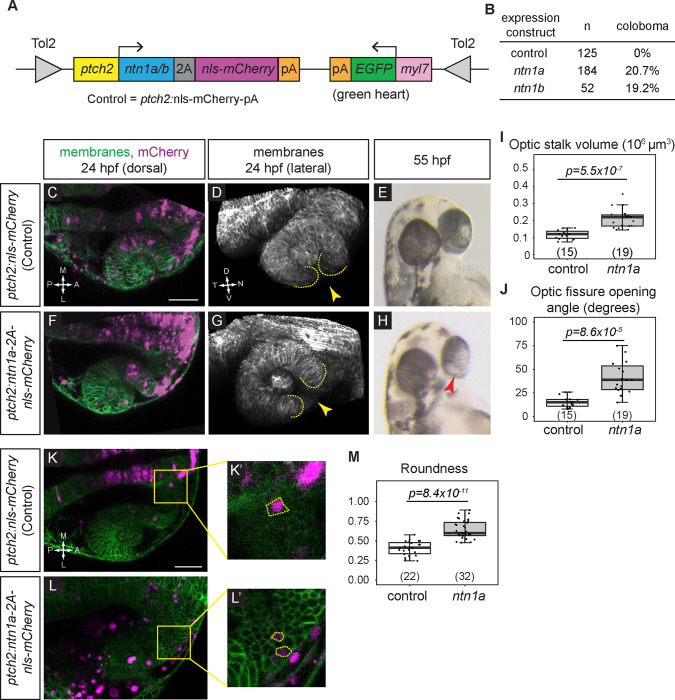
We next asked whether Netrin overexpression might be sufficient to disrupt optic fissure formation and lead to coloboma, thereby phenocopying the ptch2 mutant. This would suggest that Netrin might be a key downstream target of overactive Hh signaling to cause coloboma. In order to test whether overexpression of Netrin is sufficient to disrupt optic fissure and stalk formation and cause coloboma, we used the Tol2kit system 37, which permits transient transgenesis in injected embryos via Tol2 transposon-mediated insertion. We used the Hh-responsive promoter GBS-ptch2 38 to overexpress Ntn1a or Ntn1b. The GBS-ptch2 promoter restricts overexpression to cells responding to Hh ligand, the cell population that is defective in the ptch2 mutant 11. This construct additionally contains a viral 2A peptide followed by a fluorescent marker, nuclear-localized mCherry (nls-mCherry), to allow identification of cells expressing Netrin (schematized in Fig. 3A; ptch2:ntn1a-2A-nlsmCherry or ptch2:ntn1b-2A-nlsmCherry). As a control, we injected a construct using the same promoter driving only nls-mCherry (ptch2:nls-mCherry). Each DNA construct was injected into wild-type embryos with Tol2 transposase RNA to catalyze genomic insertion.
Figure 3. netrin overexpression is sufficient to disrupt cause coloboma, disrupt optic fissure formation, and perturb optic stalk cell morphology.
(A) Schematic illustrating transient transgenesis expression construct (GBS-ptch2:ntn1a(or ntn1b)-2A-nls-mCherry). The control construct drives expression of only nls-mCherry.
(B) Quantification of coloboma in wild-type embryos injected with the control construct, the ntn1a, or ntn1b overexpression construct. Transient transgenic overexpression of ntn1a or ntn1b is sufficient to cause coloboma in 20.7% or 19.2% of wild-type embryos, respectively.
(C-M) Analysis of optic fissure formation and optic stalk cell morphology. (C-E) Wild-type embryo injected with control (nls-mCherry) transgene expression construct. (F-H) Wild-type embryo injected with the ntn1a overexpression transgene expression construct. (C, F) Single optical sections, dorsal view, 24 hpf. Green, membranes (Tg(bactin2:EGFPCAAX)); magenta, nuclei (nls-mCherry from the transgene expression construct). (D, G) Lateral views of 3D renderings show optic fissure margins (dotted lines) and opening (yellow arrowhead), 24 hpf. Grayscale, membranes only. (E, H) Optic fissure phenotypes at 55 hpf. The control embryo optic fissure is largely fused (E); (H) shows a representative ntn1a-overexpressing embryo with coloboma (red arrowhead indicates open, unfused fissure).
(I-J) Quantification of optic stalk volume (I) and optic fissure opening angle (J), both of which are significantly increased in ntn1a-overexpressing embryos.
(K-M) Analysis of optic stalk cell morphology, 24 hpf, dorsal view, single optical section. (K, K’) Wild-type embryo injected with control (nls-mCherry) transgene expression construct. (L, L’) Wild-type embryo injected with experimental (ntn1a overexpression) transgene expression construct. Green, membranes (Tg(bactin2:EGFPCAAX)); magenta, nls-mCherry from the transgene expression construct. (K’, L’) Zoomed views of individual transgene-expressing cells in the optic stalk, as marked by nls-mCherry fluorescence. Dotted lines show cell morphology, as visualized with (Tg(bactin2:EGFPCAAX)). (M) Quantification of cell elongation using the Roundness metric; netrin1a-overexpressing optic stalk cells are significantly less elongated than their control counterparts.
Numbers in parentheses at base of graphs indicate n. Scale bar, 50 μm.
We first examined embryos for coloboma at 55 hpf, when the eye is pigmented and the optic fissure is nearly fused in wild-type embryos. When we assay coloboma following injection of the control construct, ptch2:mCherry, we observe no instances of coloboma in wild-type embryos (0/125). When ntn1a is overexpressed in Hh-responsive cells using the ptch2:ntn1a DNA construct, we observe coloboma in 20.7% (38/184) of injected embryos; injection of the ptch2:ntn1b DNA construct results in 19.2% (10/52) of injected embryos with coloboma (Fig. 3B). Although lower than the ptch2 mutant (60–100% coloboma), this penetrance is nonetheless striking: since this experimental strategy utilizes transient overexpression, the number of transgenic cells and level of overexpression can vary widely between individual embryos. This suggests that even mosaic overexpression of ntn1a or ntn1b can cause coloboma. Since the phenotypes caused by ntn1a or ntn1b overexpression appeared similar, for the sake of simplicity, we continued our experiments using the ptch2:ntn1a DNA construct only.
Despite the appearance of coloboma, such a phenotype could be caused by disruption of many processes linked to optic fissure development. To examine the phenotype more closely, we asked whether netrin overexpression can disrupt optic fissure and stalk formation, similar to the ptch2 mutant. Therefore, we assayed embryos at 24 hpf for optic fissure and stalk formation. Using a transgenic line to label cell membranes and provide tissue morphology, Tg(bactin2:EGFP-CAAX)z200 11, we imaged the optic cup at 24 hpf in live embryos injected with the control (ptch2:nls-mCherry) or experimental (ptch2:ntn1a-2A-nls-mCherry) constructs (Fig. 3C, D, F, G). In both control and experimental embryos, nls-mCherry is largely restricted to Hh-responding cells in the brain and eye 11, although ectopic expression is also seen, as expected in transient transgenic experiments (Fig. 3C, F). Lateral views of three-dimensional renderings reveal optic fissure morphology: at the ventral side of the control (ptch2:nls-mCherry) eye, the optic fissure is visible as a narrow cleft with closely apposed tissue margins (Fig. 3D, yellow arrowhead), whereas the ntn1a-overexpressing optic fissure appears wide and open (Fig. 2G, yellow arrowhead). We quantified optic stalk and optic fissure formation, as, at optic cup stage, overactive Hh signaling results in a larger optic stalk volume and wider optic fissure opening angle than wild-type 11. Optic stalk volume is significantly increased in embryos overexpressing ntn1a compared to control embryos overexpressing mCherry (Fig. 3I; control 0.117 ± 0.007×106 μm3; ntn1a overexpression 0.213 ± 0.013×106 μm3), and optic fissure opening angle is significantly larger in embryos overexpressing ntn1a compared to control embryos (Fig. 3J; control 15.53 ± 1.585°; ntn1a overexpression 43.74 ± 5.590°). Both of these phenotypes are reminiscent of the ptch2 mutant 11 (optic stalk volume 0.51 ± 0.08×106 μm3; and optic fissure opening angle 59.6 ± 6.2°), although both phenotypes are quantitatively less severe in this ntn1a-overexpression condition, as might be expected for transient transgenesis. To determine whether embryos with increased optic stalk volume or optic fissure opening angle go on to exhibit coloboma, we raised the imaged embryos to screen for coloboma at 55 hpf. Embryos injected with the ntn1a construct that display a 24 hpf optic stalk and fissure phenotype go on to exhibit coloboma by 55 hpf with incomplete penetrance, again similar to the ptch2 mutant (Fig. 3E, H), a phenotype that is not observed in the control injections.
Taken together, these data suggest that spatiotemporally regulated overexpression of netrin is sufficient to cause coloboma, and the phenotype is similar to overactive Hh signaling in the ptch2 mutant, with disrupted optic stalk and fissure formation.
netrin1a overexpression disrupts single cell morphology in the optic stalk
We took these observations of aberrant tissue morphology a step further: we previously found that in the ptch2 mutant, cell morphology in the optic stalk is disrupted 11. Optic stalk cells typically exhibit an elongated morphology, while in the ptch2 mutant, optic stalk cells are significantly rounder and less elongated. We therefore asked whether the morphology of ntn1a-overexpressing optic stalk cells is affected similarly. We quantified optic stalk cell elongation using the metric “roundness”, a measure of aspect ratio, in Fiji 39, in our transient transgenic embryos (Fig. 3K–L’, yellow dashed outline indicates cell’s perimeter). We find that compared to control, mCherry-positive optic stalk cells are less elongated when overexpressing ntn1a (Fig. 3M; 0 represents infinitely elongated and 1 represents a perfect circle; control 0.40 ± 0.09; ntn1a overexpression 0.64 ± 0.11). Our observations suggest that overexpressing ntn1a in a subset of cells in wild-type embryos is sufficient to disrupt the optic stalk in a manner similar to loss of ptch2.
Netrin ligands are not required for the ptch2 mutant coloboma phenotype
Having established that overexpression of netrin in wild-type embryos is sufficient to reproduce the ptch2 mutant phenotype, we sought to determine whether Netrin is necessary for the coloboma phenotype in ptch2 mutants. We tested if Netrin is required for the ptch2 mutant phenotype using a genetic epistasis approach: we acquired a stable loss-of-function mutant allele for ntn1b 40, and we additionally targeted ntn1a, ntn2, and ntn5 using the Alt-R CRISPR-Cas-9 system, which enables efficient editing in F0 injected embryos 41,42. Because we did not find ntn4 upregulated in ptch2−/− embryos, we did not test the requirement for this gene. All injected embryos were quantitatively analyzed for mutagenesis using capillary gel electrophoresis 43; only embryos with >70% alleles mutated for ntn1a, ntn2, and ntn5 were included in phenotypic scoring.
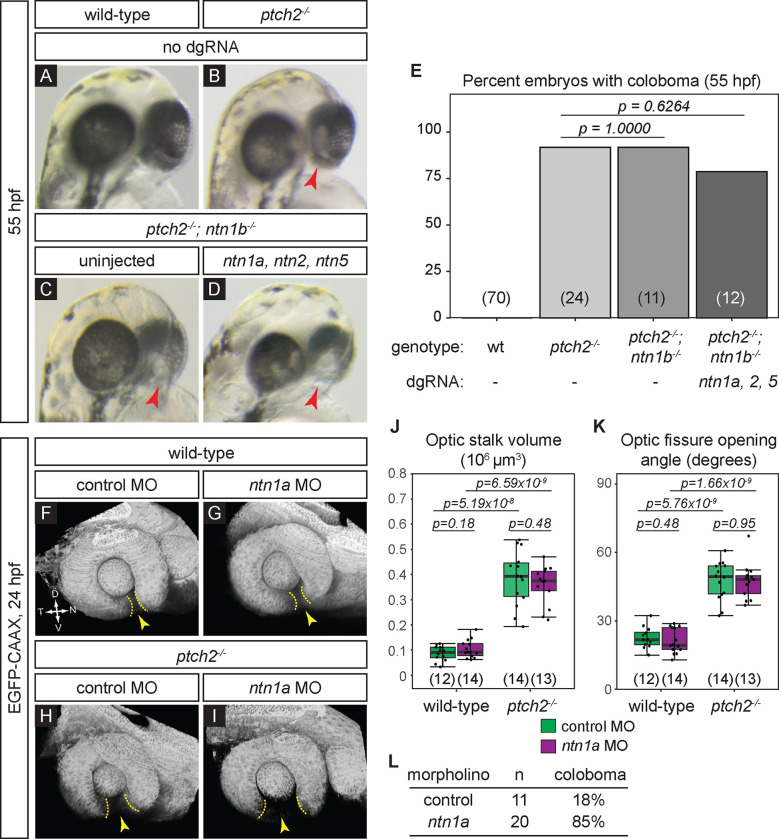
To evaluate effects on eye development, we scored embryos for coloboma at 55 hpf (Fig. 4A–E). As expected, loss of ptch2 results in coloboma with incomplete penetrance (Fig. 4B, E; 91.67±8.33%). Removal of ntn1b in the ptch2 mutant background resulted in coloboma, with no change in penetrance of the phenotype (Fig. 4C, E; 91.67±8.33%). Finally, to remove the complete suite of upregulated Netrins (Fig. 2G), we injected guides against ntn1a, ntn2, and ntn5 in the ptch2; ntn1b compound mutant background. We observed a similar penetrance of coloboma among uninjected and injected ptch2; ntn1b mutants, despite mutation of ntn1a, ntn2, and ntn5 (Fig. 4D, E; 78.57±21.43%). This result suggests that Netrin is not required for the coloboma phenotype resulting from overactive Hh signaling via the ptch2 mutant. Because these are CRISPR-injected embryos with >70% alleles mutated for all three genes, there is the possibility that some unmutated cells could provide sufficient residual Netrin activity. Additionally, it has been reported that loss of Netrins can itself result in coloboma at 2 dpf 27,28; this could confound a potential rescue of the ptch2 mutant phenotype at earlier (optic cup) stages. Therefore, we sought to determine whether loss of Netrin might rescue ptch2 mutant optic stalk and fissure phenotypes at optic cup stage (24 hpf).
Figure 4. Netrin ligands are not required for the ptch2−/− coloboma phenotype.
(A-E) Loss of netrin genes using CRISPR mutagenesis does not rescue the ptch2−/− coloboma phenotype. All embryos were evaluated for coloboma, genotyped, and gRNA-injected embryos were then individually quantified for mutagenesis efficiency. For gRNA-injected embryos, only ptch2 −/−;ntn1b−/− embryos with >70% alleles mutated for the remaining 3 genes were analyzed. (A) wild-type (wt), (B) ptch2−/−, (C) ptch2−/− ; ntn1b−/−, (D) ptch2−/− ; ntn1b−/− injected with ntn1a, ntn2, and ntn5 dgRNA, 55 hpf. Red arrowheads, coloboma. (E) Percentage of embryos with coloboma. Numbers in parentheses at base of graph indicate n.
(F-L) Morpholino-mediated knockdown of ntn1a does not rescue the ptch2−/− optic fissure and stalk phenotypes. (F) wild-type injected with control MO; (G) wild-type injected with ntn1a MO; (H) ptch2−/− injected with control MO; (I) ptch2−/− injected with ntn1a MO. Tissue morphology visualized with Tg(bactin2:EGFP-CAAX) transgene (grayscale), 24 hpf. Yellow dotted lines, optic fissure margins; yellow arrowheads, optic fissure opening.
(J, K) Quantification of optic stalk volume (J) and optic fissure opening angle (K) at 24 hpf, neither of which is affected by ntn1a MO injection.
(L) Quantification of coloboma in embryos from ptch2+/−; Tg(bactin2:EGFP-CAAX) incross injected with the control MO, or the ntn1a MO. Knockdown of ntn1a in the ptch2−/− results in a coloboma phenotype in 85% of embryos. n = total number of embryos screened.
We sought a second alternative method to impair netrin, as a complement to the CRISPR strategy, therefore, we turned to a morpholino oligonucleotide (MO) knockdown approach. We knocked down the netrin gene expressed in the optic fissure, ntn1a (Fig. 1, 2), using a previously validated translation-blocking ntn1a morpholino 27,28. ntn1a MO or standard control MO (0.5 pmol) was injected into embryos from a ptch2 heterozygous incross carrying the Tg(bactin2:EGFP-CAAX)z200 transgene to label cell membranes, allowing evaluation of eye morphology. Morpholino oligonucleotide efficacy was evaluated by scoring some embryos for coloboma at 52–55 hpf: as expected from prior reports 27,28, 17/20 (85%) ntn1a MO-injected embryos displayed coloboma, whereas 2/11 (18%) control MO-injected embryos displayed coloboma, within the range expected for a ptch2 heterozygous incross (Fig. 4L). Embryos were imaged at 24 hpf: optic stalk volume and optic fissure opening angle were measured, and embryos subsequently genotyped for ptch2. We find that in wild-type embryos, control or ntn1a MO injection has no effect on optic stalk volume (Fig. 4F, G, J; control MO 0.09 ± 0.03×106 μm3; ntn1a MO 0.1 ± 0.036×106 μm3). Optic fissure opening angle is also unaffected (Fig. 4F, G, K; control MO 22.5 ± 4.7°; ntn1a MO 21.1 ± 5.5°). ptch2 mutant embryos have a large optic stalk volume (Fig. 4H, J; control MO 0.385 ± 0.11×106 μm3); this is unaffected by injection of ntn1a MO (Fig. 4I, J; ntn1a MO 0.359 ± 0.078×106 μm3). Similarly, injection of ntn1a MO did not affect the larger optic fissure opening angle (Fig. 4H, I, K; ptch2−/−+control MO 47.62 ± 8.5°; ptch2−/−+ntn1a MO 47.42 ± 7.86°).
Taken together, these data suggest that reduced netrin expression, either via genome editing or morpholino knockdown, does not rescue the ptch2 mutant phenotype. Therefore, the most parsimonious interpretation of these data is that Netrin, while sufficient, is not solely required for the ptch2 mutant eye phenotype.
Discussion
Based on our prior work 11, our model is that overactive Hh signaling in the ptch2 mutant acts through both cell- and non-cell-autonomous mechanisms to cause coloboma. We interpret this to mean that a combination of cell-intrinsic and intercellular signaling factors are responsible for disrupting cell movements to give rise to coloboma. While we demonstrate that ntn1a overexpression is sufficient to disrupt optic fissure and stalk formation, the factors required to act together to produce the ptch2 mutant coloboma phenotype are still unknown.
Our model that Netrins may be sufficient but not necessary for the ptch2 mutant phenotype is not entirely unexpected. In the context of axon guidance, these molecules are often observed to be sufficient but not necessary 40,44,45. Functional redundancy and compensatory mechanisms between signaling pathways and gene regulatory networks in in vivo systems ensure phenotypic robustness of crucial biological processes. Our results highlight the complexity and robustness of optic fissure morphogenesis, a process that likely has similar mechanisms in place to prevent perturbations.
Understanding how Netrin genes function in normal development within these tissues, and if canonical receptors, such as DCC, Unc5, or Neogenin, are involved remains an open question. Zebrafish contain numerous Netrin receptors 46–49, yet there is also evidence that Netrins directly bind Integrins and Laminin to modulate cell adhesion 12,50. These will be important pathways to examine in the future.
In this work, we focused on Netrins as one candidate downstream target of Hh signaling in eye development, but our working model implicates additional effectors that are altered in the context of overactive Hh signaling to disrupt optic fissure formation. The strategies described in this study can be used to evaluate many other potential Hh downstream targets, and in turn, uncover new molecular mechanisms controlling optic fissure morphogenesis and impacting coloboma.
Experimental Procedures
Zebrafish husbandry and mutant/transgenic lines
All zebrafish (Danio rerio) husbandry was performed under standard conditions in accordance with University of Utah Institutional Animal Care and Use Committee (IACUC) Protocol approval (Protocol #1647). Embryos (Tu or TL strains) were raised at 28.5–30 °C and staged according to time post fertilization and morphology 51. Mutant lines used include: ptch2/blowouttc294z 10,11,52,53; ntn1bp210 40. Transgenic alleles used were: Tg(bactin2:EGFP-CAAX)z200 11.
For genotyping, genomic DNA was extracted from single embryos or adult fins, incubated at 95 °C in 0.05 M NaOH for 30 m, then neutralized with 1 M Tris pH 8.0. The ptch2 locus was genotyped using an HRMA protocol 11,54 with the following primers: ptch2HRMA_F: 5′- CTGCACCTTCCTGGTGTGTG-3′, ptch2HRMA_R: 5′- GGTAGAAATGGATTAGAGTGAGAGGAA-3′. The ntn1b locus was genotyped using a CAPS assay 55 with the following primers: ntn1b_F: 5’- ATGATAAGGATTTTGGTAACGTGCG-3’, ntn1b_R: 5’- CTTCCCGAAAGCGGAGTTCAC-3’. Full length PCR product was run on a 3% gel to distinguish bands.
RNA synthesis and nucleic acid injections
pCS2 template (pCS2-Transposase) was linearized with NotI-HF (R3189L, New England Biolabs) and capped RNA was synthesized using the mMessage mMachine SP6 kit (AM1340, Invitrogen). RNA was purified using the RNeasy Mini Kit (74104, Qiagen) and ethanol precipitated. For Tol2 injections, 25 pg Transposase RNA and 25 pg assembled DNA construct were co-injected into one-cell embryos. For CRISPR-Cas9 injections, 5 μM dgRNA and 5 μM Cas9 protein (1074181, Integrated DNA Technologies) were co-injected into one-cell stage embryos.
Morpholino oligonucleotide injection
Ntn1a translation blocking morpholino (5’ - CATCAGAGACTCTCAACATCCTCGC - 3’) was used, as in 27,28. The standard GeneTools control morpholino was used as our control. For our experiments, 0.5 pmol (4.17 ng) was injected into one-cell stage embryos.
In situ hybridization
Embryos were fixed at 24 hpf in 4% paraformaldehyde overnight at 4 °C and dehydrated in 100% methanol. Colorimetric in situ hybridization was performed as described previously 56. In situ riboprobes were synthesized from linearized templates: pBluescript II SK− (netrin1a), pBluescript II KS+ (netrin1b), pGEM-Teasy (netrin2 and netrin4; 18,19). Netrin5 was synthesized from a PCR fragment amplified from cDNA using primers described previously with T7 RNA polymerase 16.
HCR RNA-FISH
HCR was performed using an adapted version of the publicly available protocol, “HCR RNA-FISH protocol for whole-mount zebrafish embryos and larvae” (Molecular Instruments; 35). For each netrin gene, one HCR RNA-FISH bundle per target RNA was ordered and contained the HCR split-initiator probe set and the HCR amplifier, and the reaction was performed with HCR RNA-FISH buffers. For all target mRNAs, amplifier B1–647 was used.
Reverse transcription-quantitative PCR
Embryos were pooled at 24 hpf (n=30) and immediately homogenized using the QIAshredder (79654, Qiagen). Total RNA was then extracted using the RNeasy Mini Kit (74104, Qiagen) and stored at −80°C until use. cDNA was synthesized using the iScript cDNA Synthesis kit (1708890, Bio-Rad) following the manufacturer’s recommendations, such that 1 μg of RNA was loaded into each reaction. Three biological replicates were collected for each condition.
RT-qPCR primers were designed to span exon-exon junctions and produce amplicons of ~100 bp in length. Primer sequences used are: netrin1a_F1: 5’- GCTGTGTTTCAGCACAGGAG-3’, netrin1a_R1: 5’- CCTTTGAGCACACAGCAGAG-3’, netrin1b_F1: 5’- GGCAAGATGAAGGTCACCA-3’, netrin1b_R1: 5’- CACCGATATGATGTTGATGG-3’, netrin2_F1: 5’- AAGAGGCCAACGAGTGCTTA-3’, netrin2_R1: 5’- CACACTCCTCCACTCTTTCG-3’, netrin4_F1: 5’- TGAGCACTATGGAGCTGACG-3’, netrin4_R1: ATTTCCCATGCGTGGATTAC-3’, netrin5_F1: 5’- GCTCCGCCTGAATATCTGTC-3’, netrin5_R1: 5’- GGAGAGGGTCTGGAAAGGAG-3’, eef1a1l1_F: 5’- CCTCTTTCTGTTACCTGGCAAA-3’, eef1a1l1_R: 5’- CTTTTCCTTTCCCATGATTGA-3’. During optimization, products were both gel analyzed and sequenced to ensure product specificity. All reactions utilized the PowerUp SYBR Green Master Mix (A25741, Applied BioSystems) and were performed on an Applied BioSystems 7900HT instrument. Cycling parameters were: 50°C (2 min) followed by 40 cycles of 95°C (2 min), 58°C (15 s), 72°C (1 min), then followed by a dissociation curve. Applied BioSystems software SDSv2.4 was used to determine cycle threshold (Ct) values and melting curves. All reactions were performed in triplicate with a ‘no-template’ control.
RT-qPCR analysis was performed in Microsoft Excel using the ΔΔCt method 36,57. The relative quantity (RQ) of each gene was normalized to the reference gene eef1a1l1 58, and the normalized relative quantity (NRQ) was determined by normalizing 24 hpf ptch2−/− expression to normalized 24 hpf wild-type expression.
Generation of transient transgenesis expression constructs
For the GBS-ptch2:nls-mCherry control construct, the GBS-ptch2 promoter (p5E-GBSptch2; 11,38) was recombined with a middle entry clone of nuclear localized mCherry (pME-nlsmCherry) and a 3’ clone of the SV40 late polyA signal sequence (p3E-polyA) into the Tol2 transposon-flanked destination vector, pDestTol2CG2 37.
For the GBS-ptch2:ntn1a-2A-nls-mCherry and GBS-ptch2:ntn1b-2A-nls-mCherry experimental constructs, the GBS-ptch2 promoter (p5E-GBSptch2) was recombined with a middle entry clone of the open reading frames of netrin1a (pME-ntn1a) or netrin1b (pME-ntn1b) lacking a stop codon, and a 3’ clone of the PTV-2A peptide, nuclear localized mCherry and the SV40 late polyA signal sequence (p3E-2A-NLSmCherry-pA) into the Tol2 transposon-flanked destination vector, pDestTol2CG2 37.
Coloboma scoring
Embryos were individually screened and scored for coloboma at 52–55 hpf using an Olympus SZX16 stereomicroscope. The phenotype was scored by viewing the back of the eye and focusing at the depth of the RPE; embryos that were scored as positive for coloboma had eyes that displayed an expanded region lacking pigmentation in the area of the optic nerve head either unilaterally or bilaterally. This area was distinctly wider and more open than the rest of the optic fissure that was undergoing fusion at the ventral side of the optic cup. All genetic experiments were scored blindly. Embryos were subsequently genotyped as described above.
CRISPR-Cas9 mutagenesis
gRNA target sites were identified using the web programs CHOpCHOP (http://chopchop.cbu.uib.no). Genomic DNA sequences from Ensembl GRCz11 (http://useast.ensembl.org/Danio_rerio/Info/Index) were used for target site searches. Mutagenesis was performed using the Alt-R CRISPR-Cas9 system (Integrated DNA Technologies). CRISPR RNAs (crRNAs) were designed to target exon 1 of ntn1a (5′- CAUCCCCGUCUUCGUAAACGCGG-3′), exon 1 of ntn2 (5′- CCAACCGCAUAAUAGUACGUCGG-3′), and exon 1 of ntn5 (5′- UGGACUUUGAUAGUUCCCCUAGG-3′). The crRNA and trans-activating crRNA (tracrRNA) were annealed into a dual guide dgRNA complex at a 1:1 ratio and stored at −20°C until use. On the day of injections, the ribonucleoprotein was assembled by incubating the dgRNA complexes (25 μM of total dgRNA) with the Cas9 Nuclease 3NLS (25 μM) (1074181, Integrated DNA Technologies) at 37 °C for 5 min 42. An injection cocktail of 5 μM dgRNA and 5 μM Cas9 protein and was injected into one-cell stage embryos.
Fragment analysis for capillary electrophoresis
To quantify the activity and efficiency of individual crRNAs, fragment analysis by capillary electrophoresis was used. Primers were designed to amplify an ~80 bp region surrounding the crRNA target site for ntn1a, ntn2, and ntn5. The forward primer was labeled with a 5’ 6-carboxyfluorescein tag (6-FAM, Integrated DNA Technologies). The following primers were used: Ntn1a_3 crRNA 56-FAM F: 5’-CGGATCCGTGTTACGACGAGAA-3’ (+6-FAM modification), Ntn1a_3 crRNA HRMA R: 5’-CTGGACGCGCGTACTTCTTTC-3’, Ntn2 crRNA 56-FAM F: 5’-AAGTGAAGACGCTCTCGGTG-3’ (+ 6-FAM), Ntn2 crRNA HRMA R: 5’-TCTCCCGGACACACCTAGAG-3’, Ntn5 crRNA 56-FAM F: 5’-GCTTCAGAGGGCTCCAGTG-3’ (+ 6-FAM), Ntn5 crRNA HRMA R: 5’-GGACTCAACCGAATCCACCT-3’. Genomic DNA was isolated from ptch2−/−; ntn1b−/− embryos either uninjected, or injected with ntn1a, ntn2, and ntn5 dgRNAs. Following PCR amplification, the fragments were diluted 10-fold with distilled water. Two microliters were further processed by the University of Utah DNA Sequencing Core Facility. Capillary electrophoresis was performed on an Applied BioSystems 3730 DNA analyzer (Applied Biosystems). Collected data were analyzed with GeneMapper Software (Applied Biosystems).
Analysis was adapted from previously published methods 43. Fragments <70 bp in size and peaks <150 in height were removed from the analysis. Six control uninjected ptch2−/−; ntn1b−/− samples were analyzed for each mutagenesis experiment, and the highest peak was identified as wild-type (if there was a second peak similarly substantially high, that was included), and a range of fragment sizes encompassing that peak +/− 0.5 bp made up the wild-type fragment range. For each control sample, the total height of all fragments and the total height of wild-type fragments were each determined and a ratio of wild-type/total was calculated. The average ratio was then acquired across all control samples. For injected ptch2−/−; ntn1b−/− samples, the same steps were followed. The total height of all fragments and the total height of wild-type fragments (defined using the control wild-type range) were determined, and a wild-type ratio was calculated. Each sample’s wild-type ratio was normalized using the wild-type ratio from the control samples (injected wild-type ratio/uninjected wild-type ratio). To determine the percent of transcripts in each sample that are edited (i.e. the mutant allele frequency), the equation: 1 - normalized ratio * 100 was used. Only injected samples with mutant allele frequencies for all targeted genes >70% were used in this study.
Imaging
For confocal imaging, both live and fixed, embryos were embedded in 1.6% low melting point agarose in E3 or PBS in PELCO glass bottom dishes (14027, Ted Pella). Images were acquired using either a Zeiss LSM710 or LSM880 laser-scanning confocal microscope. All imaging was performed with a 40x water immersion objective (1.2 NA). Datasets were acquired with the following parameters: 512×512; voxel size 0.69 × 0.69 × 2.1 μm3. All imaging and analyses were performed blinded to the genotype of each sample.
Image analysis: HCR quantification
ntn1a domain quantification:
Expression domain was quantified using volume data of HCR-stained embryos. 3D data sets were oriented in FluoRender 59 to achieve a lateral view. This orientation was captured in FluoRender and saved as a TIFF image. The domain of ntn1a expression was measured in FIJI/ImageJ using the angle tool; the vertex was positioned at the center of the lens with the rays of the angle projected to encompass the extent of the ntn1a signal expression domain (schematized in Fig. 2J).
ntn1a and ntn1b fluorescence intensity quantification:
HCR data (3D volume datasets with embryos mounted dorsally) were quantified as normalized fluorescence intensity measured in FIJI/ImageJ. For ntn1a, for which expression was observed in the anterior rim of the optic cup and nasal margin of the optic fissure, a maximum intensity projection was generated through the entire depth of the region. An ellipse (2019 μm2) was placed to encompass the anterior rim and nasal margin, and the mean fluorescence intensity measured. An ellipse of the same size (2019 μm2) was placed over the dorsal eye, an internal control region, and the mean fluorescence intensity again measured. Fluorescence intensity in the anterior rim and nasal margin was normalized to the dorsal eye for each embryo; the normalized values are plotted in Fig. 2K.
For ntn1b, quantification was carried out in a similar manner to ntn1a, except that the optic stalk, where signal was observed, was quantified. A maximum intensity projection through the entire depth of the optic stalk was generated. An ellipse (3031 μm2) was placed around the optic stalk, and the mean fluorescence intensity measured. An ellipse of the same size (3031 μm2) was placed over the dorsal eye, an internal control region, and the mean fluorescence intensity again measured. Fluorescence intensity in the optic stalk was normalized to the dorsal eye for each embryo; the normalized values are plotted in Fig. 2K.
Image analysis: Optic stalk volume
In FIJI/ImageJ, the segmentation editor was used to manually segment the optic stalk in 3D volume data sets of live embryos labeled for cell membranes (EGFP-CAAX). The optic stalk was outlined using the freehand selection tool, moving through the z-stack, slice by slice. Once the entire optic stalk was segmented, the stack labels were saved as a new tiff file. The total volume of the segmented region was measured in FluoRender using the Paint Brush tools. The entire volume was selected using the brush, and then the volume measured using the “Get Selection Size” function; output was in μm3, based on voxel size read in from the image data.
Image analysis: Optic fissure opening angle
Three-dimensional data sets of live embryos labeled for cell membranes (GFP-CAAX) were oriented in FluoRender 59 for a lateral view. Using the lateral cutaway tool, we cut to the lens midpoint. This orientation was captured in FluoRender and saved as a TIFF image. The opening angle of the optic fissure was measured in Fiji using the angle tool; the vertex was positioned at the center of the lens with the rays of the angle projected to each optic fissure margin.
Image analysis: Optic stalk cell roundness
Live embryos were mounted dorsally and TIFF images were captured containing a maximum intensity projection of 10 slices that contained one or more dispersed labeled optic cell in mCherry. An outline was drawn around the cell using the freehand tool in Fiji and roundness was manually calculated using the area and major axis measurement.
Box and whisker plots
Box and whisker plots were generated using the ggplot2 package in R Studio. The lower and upper hinges correspond to the first and third quartiles. The upper whisker extends from the hinge to the largest value no further than 1.5 * IQR from the hinge, and the lower whisker extends from the hinge to the smallest value at most 1.5 * IQR of the hinge. Data beyond the end of the whiskers are called “outlying” points and are plotted individually. The line in the box represents the median.
Statistics
For all quantifications, P-values were calculated using an unpaired Student’s t-test in which the means of the two comparison sets are considered statistically significant if P < 0.05. If the variance of the two comparison sets was significantly different, Welch’s correction was used. Throughout the manuscript, quantifications in the text are listed as mean ± standard error.
Key Findings:
Overactive Hedgehog signaling in the ptch2 mutant causes increased netrin expression
Spatiotemporally specific overexpression of netrin1a and netrin1b can cause coloboma
Spatiotemporally specific overexpression of netrin1a can disrupt optic fissure and stalk formation as well as optic stalk cell morphology, similar to the ptch2 mutant
Loss of netrin ligands in the ptch2 mutant does not rescue the phenotype
Acknowledgments
We are grateful to Puneet Dang and Jonathan Raper for generously sharing zebrafish lines. We are also grateful to David Grunwald and Kazuyuki Hoshijima; Mike Klein (University of Utah Genomics Core); and Natasha O’Brown for technical support and advice. Thanks to the Centralized Zebrafish Animal Resource and Kwan lab members past and present for useful discussion. This work was supported by the University of Utah Developmental Biology Training Grant (NIH T32 HD007491 to S. Lusk and S. LaPotin) and the National Eye Institute/National Institutes of Health (F31 EY030758 to S. Lusk, R01 EY025378 to K.M.K.).
Footnotes
The authors declare no conflicts of interest.
Ethics Statement:
All procedures and experiments with zebrafish were approved under Protocol #1647 by the University of Utah Institutional Animal Care and Use Committee and conformed to the ARVO guidelines for the use of animals in vision research.
References
- 1.ALSomiry AS, Gregory-Evans CY, Gregory-Evans K. An update on the genetics of ocular coloboma. Hum Genet. Sep 2019;138(8–9):865–880. 10.1007/s00439-019-02019-3. [DOI] [PubMed] [Google Scholar]
- 2.Chang L, Blain D, Bertuzzi S, Brooks BP. Uveal coloboma: clinical and basic science update. Curr Opin Ophthalmol. Oct 2006;17(5):447–70. 10.1097/01.icu.0000243020.82380.f6. [DOI] [PubMed] [Google Scholar]
- 3.Fitzpatrick DR, van Heyningen V. Developmental eye disorders. Curr Opin Genet Dev. Jun 2005;15(3):348–53. 10.1016/j.gde.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Gregory-Evans CY, Williams MJ, Halford S, Gregory-Evans K. Ocular coloboma: a reassessment in the age of molecular neuroscience. J Med Genet. Dec 2004;41(12):881–91. 10.1136/jmg.2004.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onwochei BC, Simon JW, Bateman JB, Couture KC, Mir E. Ocular colobomata. Surv Ophthalmol. Nov-Dec 2000;45(3):175–94. 10.1016/s0039-6257(00)00151-x. [DOI] [PubMed] [Google Scholar]
- 6.Patel A, Sowden JC. Genes and pathways in optic fissure closure. Semin Cell Dev Biol. Jul 2019;91:55–65. 10.1016/j.semcdb.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. Jun 14 1996;85(6):841–51. 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 8.Smyth I, Narang MA, Evans T, et al. Isolation and characterization of human patched 2 (PTCH2), a putative tumour suppressor gene inbasal cell carcinoma and medulloblastoma on chromosome 1p32. Hum Mol Genet. Feb 1999;8(2):291–7. 10.1093/hmg/8.2.291. [DOI] [PubMed] [Google Scholar]
- 9.Ragge NK, Salt A, Collin JR, Michalski A, Farndon PA. Gorlin syndrome: the PTCH gene links ocular developmental defects and tumour formation. Br J Ophthalmol. Aug 2005;89(8):988–91. 10.1136/bjo.2004.061390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, Willer JR, Willer GB, Smith K, Gregg RG, Gross JM. Zebrafish blowout provides genetic evidence for Patched1-mediated negative regulation of Hedgehog signaling within the proximal optic vesicle of the vertebrate eye. Dev Biol. Jul 1 2008;319(1):10–22. 10.1016/j.ydbio.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon HB, Lusk S, Carney KR, Wirick EO, Murray BF, Kwan KM. Hedgehog signaling regulates cell motility and optic fissure and stalk formation during vertebrate eye morphogenesis. Development. Nov 19 2018;145(22). 10.1242/dev.165068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yebra M, Montgomery AM, Diaferia GR, et al. Recognition of the neural chemoattractant Netrin-1 by integrins alpha6beta4 and alpha3beta1 regulates epithelial cell adhesion and migration. Dev Cell. Nov 2003;5(5):695–707. 10.1016/s1534-5807(03)00330-7. [DOI] [PubMed] [Google Scholar]
- 13.Mehlen P, Furne C. Netrin-1: when a neuronal guidance cue turns out to be a regulator of tumorigenesis. Cell Mol Life Sci. Nov 2005;62(22):2599–616. 10.1007/s00018-005-5191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai Wing Sun K, Correia JP, Kennedy TE. Netrins: versatile extracellular cues with diverse functions. Development. Jun 2011;138(11):2153–69. 10.1242/dev.044529. [DOI] [PubMed] [Google Scholar]
- 15.Boyer NP, Gupton SL. Revisiting Netrin-1: One Who Guides (Axons). Front Cell Neurosci. 2018;12:221. 10.3389/fncel.2018.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnold CR, Lamont RE, Walker JT, et al. Comparative analysis of genes regulated by Dzip1/iguana and hedgehog in zebrafish. Dev Dyn. Feb 2015;244(2):211–23. 10.1002/dvdy.24237. [DOI] [PubMed] [Google Scholar]
- 17.Lauderdale JD, Davis NM, Kuwada JY. Axon tracts correlate with netrin-1a expression in the zebrafish embryo. Mol Cell Neurosci. 1997;9(4):293–313. 10.1006/mcne.1997.0624. [DOI] [PubMed] [Google Scholar]
- 18.Park KW, Urness LD, Senchuk MM, et al. Identification of new netrin family members in zebrafish: developmental expression of netrin 2 and netrin 4. Dev Dyn. Nov 2005;234(3):726–31. 10.1002/dvdy.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strahle U, Fischer N, Blader P. Expression and regulation of a netrin homologue in the zebrafish embryo. Mech Dev. Mar 1997;62(2):147–60. 10.1016/s0925-4773(97)00657-6. [DOI] [PubMed] [Google Scholar]
- 20.Macdonald R, Scholes J, Strahle U, et al. The Pax protein Noi is required for commissural axon pathway formation in the rostral forebrain. Development. Jun 1997;124(12):2397–408. [DOI] [PubMed] [Google Scholar]
- 21.Lupo G, Gestri G, O’Brien M, et al. Retinoic acid receptor signaling regulates choroid fissure closure through independent mechanisms in the ventral optic cup and periocular mesenchyme. Proc Natl Acad Sci U S A. May 24 2011;108(21):8698–703. 10.1073/pnas.1103802108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norton WH, Mangoli M, Lele Z, et al. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphe neurones and cranial motoneurones. Development. Feb 2005;132(4):645–58. 10.1242/dev.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stacher Horndli C, Chien CB. Sonic hedgehog is indirectly required for intraretinal axon pathfinding by regulating chemokine expression in the optic stalk. Development. Jul 2012;139(14):2604–13. 10.1242/dev.077594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrasekhar A, Schauerte HE, Haffter P, Kuwada JY. The zebrafish detour gene is essential for cranial but not spinal motor neuron induction. Development. Jun 1999;126(12):2727–37. [DOI] [PubMed] [Google Scholar]
- 25.Vanderlaan G, Tyurina OV, Karlstrom RO, Chandrasekhar A. Gli function is essential for motor neuron induction in zebrafish. Dev Biol. Jun 15 2005;282(2):550–70. 10.1016/j.ydbio.2005.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert E, Coissieux MM, Laudet V, Mehlen P. Netrin-4 acts as a pro-angiogenic factor during zebrafish development. J Biol Chem. Feb 3 2012;287(6):3987–99. 10.1074/jbc.M111.289371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardy H, Prendergast JG, Patel A, et al. Detailed analysis of chick optic fissure closure reveals Netrin-1 as an essential mediator of epithelial fusion. Elife. Jun 4 2019;8. 10.7554/eLife.43877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson R, Owen N, Toms M, Young RM, Tracey-White D, Moosajee M. Transcriptome profiling of zebrafish optic fissure fusion. Sci Rep. Feb 7 2019;9(1):1541. 10.1038/s41598-018-38379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii N, Wadsworth WG, Stern BD, Culotti JG, Hedgecock EM. UNC-6, a laminin-related protein, guides cell and pioneer axon migrations in C. elegans. Neuron. Nov 1992;9(5):873–81. 10.1016/0896-6273(92)90240-e. [DOI] [PubMed] [Google Scholar]
- 30.Kang JS, Yi MJ, Zhang W, Feinleib JL, Cole F, Krauss RS. Netrins and neogenin promote myotube formation. J Cell Biol. Nov 8 2004;167(3):493–504. 10.1083/jcb.200405039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lejmi E, Leconte L, Pedron-Mazoyer S, et al. Netrin-4 inhibits angiogenesis via binding to neogenin and recruitment of Unc5B. Proc Natl Acad Sci U S A. Aug 26 2008;105(34):12491–6. 10.1073/pnas.0804008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Stein E, Oliver T, et al. Novel role for Netrins in regulating epithelial behavior during lung branching morphogenesis. Curr Biol. May 25 2004;14(10):897–905. 10.1016/j.cub.2004.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X, Le Noble F, Yuan L, et al. The netrin receptor UNC5B mediates guidance events controlling morphogenesis of the vascular system. Nature. Nov 11 2004;432(7014):179–86. 10.1038/nature03080. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasan K, Strickland P, Valdes A, Shin GC, Hinck L. Netrin-1/neogenin interaction stabilizes multipotent progenitor cap cells during mammary gland morphogenesis. Dev Cell. Mar 2003;4(3):371–82. 10.1016/s1534-5807(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 35.Choi HMT, Schwarzkopf M, Fornace ME, et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. Jun 26 2018;145(12). 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. Dec 2001;25(4):402–8. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Kwan KM, Fujimoto E, Grabher C, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. Nov 2007;236(11):3088–99. 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 38.Shen MC, Ozacar AT, Osgood M, et al. Heat-shock-mediated conditional regulation of hedgehog/gli signaling in zebrafish. Dev Dyn. May 2013;242(5):539–49. 10.1002/dvdy.23955. [DOI] [PubMed] [Google Scholar]
- 39.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. Jun 28 2012;9(7):676–82. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang P, Barnes DT, Cheng RP, et al. Netrins and Netrin Receptors are Essential for Normal Targeting of Sensory Axons in the Zebrafish Olfactory Bulb. Neuroscience. Jan 1 2023;508:19–29. 10.1016/j.neuroscience.2022.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobi AM, Rettig GR, Turk R, et al. Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes. Methods. May 15 2017;121–122:16–28. 10.1016/j.ymeth.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoshijima K, Jurynec MJ, Klatt Shaw D, Jacobi AM, Behlke MA, Grunwald DJ. Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev Cell. Dec 2 2019;51(5):645–657 e4. 10.1016/j.devcel.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrington B, Varshney GK, Burgess SM, Sood R. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. Dec 15 2015;43(22):e157. 10.1093/nar/gkv802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dominici C, Moreno-Bravo JA, Puiggros SR, et al. Floor-plate-derived netrin-1 is dispensable for commissural axon guidance. Nature. May 18 2017;545(7654):350–354. 10.1038/nature22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varadarajan SG, Butler SJ. Netrin1 establishes multiple boundaries for axon growth in the developing spinal cord. Dev Biol. Oct 1 2017;430(1):177–187. 10.1016/j.ydbio.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fricke C, Chien CB. Cloning of full-length zebrafish dcc and expression analysis during embryonic and early larval development. Dev Dyn. Nov 2005;234(3):732–9. 10.1002/dvdy.20492. [DOI] [PubMed] [Google Scholar]
- 47.Mawdsley DJ, Cooper HM, Hogan BM, Cody SH, Lieschke GJ, Heath JK. The Netrin receptor Neogenin is required for neural tube formation and somitogenesis in zebrafish. Dev Biol. May 1 2004;269(1):302–15. 10.1016/j.ydbio.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Suli A, Mortimer N, Shepherd I, Chien CB. Netrin/DCC signaling controls contralateral dendrites of octavolateralis efferent neurons. J Neurosci. Dec 20 2006;26(51):13328–37. 10.1523/JNEUROSCI.2858-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang B, Peng G, Gao J. Expression of unc5 family genes in zebrafish brain during embryonic development. Gene Expr Patterns. Dec 2013;13(8):311–8. 10.1016/j.gep.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Nikolopoulos SN, Giancotti FG. Netrin-integrin signaling in epithelial morphogenesis, axon guidance and vascular patterning. Cell Cycle. Mar 2005;4(3):e131–5. [PubMed] [Google Scholar]
- 51.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. Jul 1995;203(3):253–310. 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 52.Karlstrom RO, Trowe T, Klostermann S, et al. Zebrafish mutations affecting retinotectal axon pathfinding. Development. Dec 1996;123:427–38. [DOI] [PubMed] [Google Scholar]
- 53.Koudijs MJ, den Broeder MJ, Groot E, van Eeden FJ. Genetic analysis of the two zebrafish patched homologues identifies novel roles for the hedgehog signaling pathway. BMC Dev Biol. Feb 19 2008;8:15. 10.1186/1471-213X-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parant JM, George SA, Pryor R, Wittwer CT, Yost HJ. A rapid and efficient method of genotyping zebrafish mutants. Dev Dyn. Dec 2009;238(12):3168–74. 10.1002/dvdy.22143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. Aug 1993;4(2):403–10. 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 56.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 57.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. Jun 18 2002;3(7):RESEARCH0034. 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karra R, Knecht AK, Kikuchi K, Poss KD. Myocardial NF-kappaB activation is essential for zebrafish heart regeneration. Proc Natl Acad Sci U S A. Oct 27 2015;112(43):13255–60. 10.1073/pnas.1511209112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan Y, Otsuna H, Chien CB, Hansen C. FluoRender: An Application of 2D Image Space Methods for 3D and 4D Confocal Microscopy Data Visualization in Neurobiology Research. IEEE Pac Vis Symp. 2012:201–208. 10.1109/pacificvis.2012.6183592. [DOI] [PMC free article] [PubMed] [Google Scholar]