Abstract
We have studied the role of an RNA region at nucleotides (nt) +200 to +233, just downstream of the 5′ long terminal repeat, in encapsidation of human immunodeficiency virus type 1 genomic RNA. Three deletion mutations, namely, BH-D0, BH-D1, and BH-D2, were generated to eliminate sequences at positions nt +200 to +219, +200 to +226, and +200 to +233. The result in each case was decreased levels of packaging of viral RNA into the mutated viruses, with the BH-D2 virus being the most severely affected. Consistently, all three deletions resulted in impaired viral infectiousness and the BH-D2 mutation showed the most dramatic impact in this regard. Further analysis revealed additional defects in Gag precursor processing and in the extension efficiency of the tRNA3Lys primer in reverse transcription reactions performed with these mutated viruses. To shed further light on the function of these deleted sequences in viral replication, the mutated viruses were cultured in MT-2 cells over prolonged periods to enable them to reacquire wild-type replication kinetics. Sequencing of the reverted viruses revealed point mutations in both the noncoding region and the gag gene. In the case of the BH-D0 revertant, two mutations were observed at positions G112A in the U5 region, termed M1, and T24I in the nucleocapsid protein, termed MNC, respectively. Either of these two mutations was able to confer wild-type replication capacity on BH-D0. In the case of BH-D1, each of the M1 mutations, a mutation termed M2, i.e., C227T, just downstream of the primer binding site, a mutation termed MP2 (T12I) in the p2 protein, and the MNC mutation were observed. A combination of either M1 and M2 or MP2 and MNC was able to rescue BH-D1. In the case of the BH-D2 deletion-containing viruses, three point mutations, i.e., M1, MP2, and MNC, were observed and the presence of all three was required to restore viral replication to wild-type levels.
Human immunodeficiency virus type 1 (HIV-1) packages two identical copies of the full-length viral RNA that are noncovalently linked at the 5′ end of the genome to form a dimer (6). Viral RNA packaging is a specific process involving specific recognition between viral proteins and viral RNA elements in the cytoplasm and recruitment of viral RNA into virus particles. Nucleocapsid protein (NCp) is the major domain in Gag protein that recognizes a stretch of approximately 140-nucleotide (nt) RNA sequences at the 5′ end of the viral genome, termed encapsidation (E/Ψ) signals. NCp contains two zinc finger motifs, as well as a number of basic amino acid residues, all of which contribute to specific encapsidation of viral RNA (1, 7–9, 15, 16, 18, 20, 45). Four RNA stem-loop structures, termed SL1 to SL4, constitute the E/Ψ site, among which SL1 and SL3 are the major elements that bind NCp and recruit viral RNA into virus particles (3, 10, 21, 38, 39). Interestingly, SL1 is located upstream of the 5′ splice donor site and yet stimulates packaging of the full-length viral RNA (26, 35, 38, 39). It is thought that SL1 may serve as the initiation site of viral RNA dimerization, i.e., the dimerization initiation site (DIS), and that the latter may be a prerequisite for viral RNA packaging (2, 5, 12, 19, 25, 30–32, 37, 40–44, 48).
Other viral RNA sequences that include segments in the env gene and the 5′ R region have also been reported to affect viral RNA encapsidation (17, 46). One stretch of RNA sequences located between the 5′ long terminal repeat (LTR) and SL1 is thought to help stabilize secondary structures of the E/Ψ signals (21, 22). One example is a CT-rich sequence in this region that is predicted to bind to a GA-rich sequence downstream of SL3 and to help to hold SL1 to SL3 in a large RNA complex. However, the role of this RNA segment in viral RNA packaging and viral replication is still unclear.
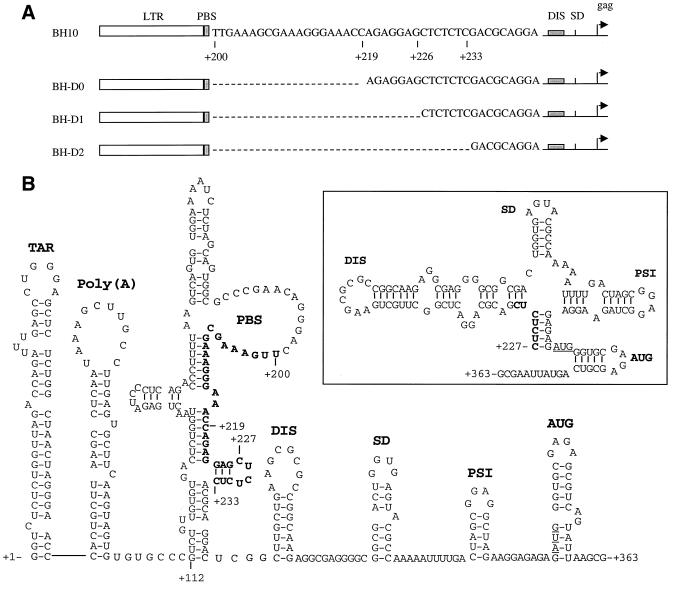
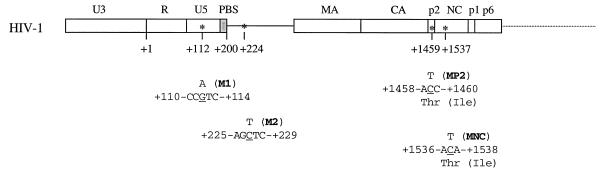
We have previously eliminated sequences in the SL1 motif and have identified point mutations in the Gag protein that are able to restore impaired viral infectiousness of the deletion-containing viruses to wild-type levels, an observation that strongly suggested functional relationships between the SL1 RNA motif and the Gag protein (33, 35, 36). In the present studies, we have used the same strategy of “forced evolution” to pursue the function of RNA sequences just downstream of the 5′ LTR in viral RNA packaging and replication. Toward this end, three nested deletion mutations, i.e., BH-D0, BH-D1, and BH-D2, were constructed to eliminate sequences at nt +200 to +219, +200 to +226, and +200 to +233, respectively (Fig. 1A). The rationale whereby these deletions were chosen is the presence of GA repeats at nt +220 to +225 and CT repeats at nt +226 to +233. We now show that RNA sequences at nt positions +200 to +233 affect viral RNA packaging and that deletion of the above sequences can be rescued by compensatory mutations in both the U5 region and several different Gag proteins.
FIG. 1.
(A) Schematic illustration of deletions of sequences at nt +200 to +219, +200 to +226, and +200 to +233 in the region just downstream of the primer-binding site (PBS). The deleted sequences are indicated by dashed lines. Nucleotide numbers refer to the initiation site of gene transcription. (B) Secondary structures of the HIV-1 5′ viral RNA segment from nt +1 to nt +363 on the basis of published models (4, 14, 21). A different model with regard to the organization of the DIS, SD, and PSI stem-loop structures in a large RNA complex is shown in the insert. Deleted sequences are in boldface letters. The DIS, SD, PSI, and AUG regions have been named SL1, SL2, SL3, and SL4, respectively, in various publications. TAR, transactivation response element; SD, splice donor site; PSI, RNA encapsidation site.
MATERIALS AND METHODS
Construction of mutant HIV-1 cDNA clones.
The BH10 clone of infectious HIV-1 cDNA was employed as the starting material for these mutagenesis studies. Viral DNA segments at nt +200 to +219, +200 to +226, and +200 to +233 were deleted by PCR through the use of primer pairs pD0 (5′-AGCAGTGGCGCCCGAACAGGGACAGAGGAGCTCTCTCGACGC-3′ [+177 to +238])–pApa-A (5′-CCTAGGGGCCCTGCAATTTCTG-3′ [+1599 to +1538]), pD1 (5′-AGCAGTGGCGCCCGAACAGGGACCTCTCTCGACGCAGGAC-3′ [+177 to +243])–pApa-A, and pD2 (5′-AGCAGTGGCGCCCGAACAGGGACGACGCAGGACTCGGCTTG-3′ [+177 to +251])–pApa-A, respectively, to generate the BH-D0, BH-D1, and BH-D2 mutant constructs (Fig. 1A). Cloning was facilitated by the following restriction sites located within the primer sequences; ApaI in primer pApa-A and BssHII in primers pD0, pD1, and pD2. Generation of the MP2 and MNC point mutations has been described previously (35). The M1 and M2 point mutations were engineered by PCR through the use of primers pM1 (5′-GTGCCCATCTGTTGTGTGAC-3′ [+106 to +125]) and pM2 (5′-AGCAGTGGCGCCCGAACAGGGACTTCTCTCGAC-3′ [+177 to +236]), respectively. All viral constructs and the presence of mutations were confirmed by sequencing.
Transfection and infection assays.
MT-2 and COS-7 cells were maintained in RPMI 1640 medium and Dulbecco modified Eagle medium (DMEM), respectively, supplemented with 10% fetal calf serum. Transfection of COS-7 cells was performed in 100-mm-diameter dishes through the use of either calcium phosphate or Lipofectamine (GIBCO BRL, Montreal, Québec, Canada) (47). Progeny viruses were collected at 48 h after transfection and clarified on a Beckman GS-6R bench centrifuge at 3,000 rpm for 30 min at 4°C. The amount of virus was determined by measuring the p24 antigen (Ag) level using an enzyme-linked immunosorption assay (Abbott Laboratories, Abbott Park, Ill.).
The infectiousness of the mutated and wild-type viruses was examined by infection of MT-2 cells. An amount of virus equivalent to 3 ng of capsid protein (CA) p24 Ag was used to infect 5 × 105 MT-2 cells in 2 ml of RPMI 1640 medium. Cells were washed twice at 2 h after infection and cultured in 10 ml of complete RPMI 1640 medium. Culture fluids were collected at various times, and reverse transcriptase (RT) activity was measured to monitor viral replication (35). Multinuclear activation of a galactosidase indicator (MAGI) assays were performed using HeLa cells stably transfected with retroviral vectors expressing both CD4 and a truncated HIV-1 LTR–β-galactosidase plasmid (i.e., HeLa–CD4–LTR–β-gal [NIH AIDS Research and Reference Reagent Program; reagent supplied by Michael Emerman]) (26a). Toward this end, cells were prepared at a concentration of 4 × 104/well in a 24-well plate at 1 day before infection. Wild-type virus was diluted to determine the appropriate concentration for use in infection studies (the optimal concentration of virus produced 100 to 200 blue-stained cells per well). Forty-eight hours after infection, cells were fixed with a solution containing 1% formaldehyde and 0.2% glutaraldehyde in phosphate-buffered saline (PBS) for 5 min. After extensive washing with PBS, cells were incubated in staining solution (4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal] at 0.4 mg/ml) for 50 min. The number of blue-stained cells was scored by microscopy. For each viral preparation, three independent infections were performed and the average number of blue-stained cells was determined.
Viral protein analysis by either Western blotting or immunoprecipitation.
Culture fluids from transfected COS-7 cells were clarified on a Beckman GS-6R bench centrifuge at 3,000 rpm for 30 min at 4°C. Virus particles were then purified through a 20% sucrose cushion at 40,000 rpm for 1 h at 4°C using an SW41 rotor in a Beckman L8-M ultracentrifuge. Virus pellets were suspended in NP-40 lysis buffer and subjected to Western blotting through the use of an anti-HIV-1 CA (p24) immunoglobulin G monoclonal antibody (MAb) (ID Labs Inc., Toronto, Ontario, Canada). Transfected COS-7 cells were washed twice with cold PBS and lysed in 200 μl of NP-40 lysis buffer. Ten-microliter volumes of cell lysates were analyzed by Western blotting as described above.
Viral proteins in the cells were also analyzed by short-term radiolabeling and immunoprecipitation experiments. Transfected COS-7 cells were starved at 37°C for 30 min in DMEM without methionine (Met) and cysteine (Cys). Radiolabeling was performed with [35S]Met and [35S]Cys at a concentration of 100 μCi/ml for 30 min at 37°C, after which the cells were thoroughly washed with complete DMEM and cultured for 1 h. Cells were washed twice with cold PBS and lysed in buffer containing 0.1% NP-40. Cell lysates were incubated with the MAb against HIV-1 CA at 4°C for 30 min, and the resultant Ag-antibody complexes were harvested through 30 min of incubation with protein A–Sepharose CL-4B (Amersham Pharmacia Biotech, Montreal, Québec, Canada). The recovered viral proteins were fractioned by 12% polyacrylamide gel electrophoresis and exposed to X-ray film.
Viral RNA analysis.
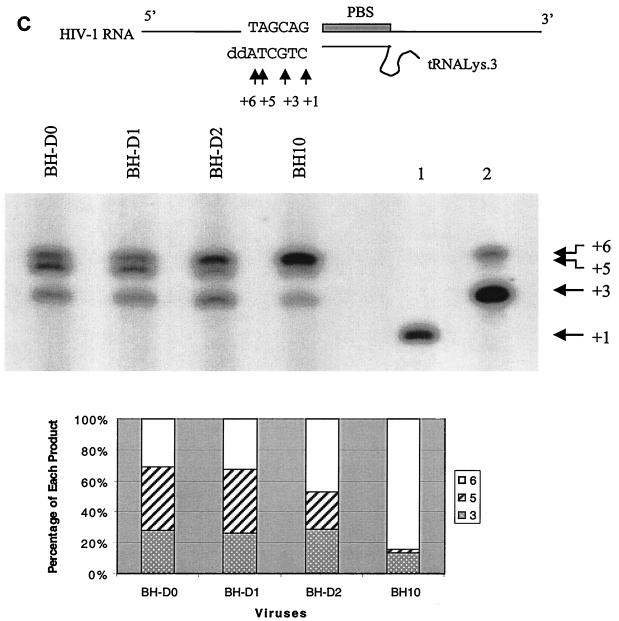
Viral particles were purified as described above and treated with Trizol (GIBCO, Montreal, Québec, Canada) to extract viral RNA. Viral RNA samples from viral preparations equivalent to 2 ng of p24 Ag were analyzed by RT-PCR as previously described (35). Extension reactions from primer tRNA3Lys, which had been annealed onto the viral RNA genome within virus particles, were performed at 37°C for 15 min in a system containing 50 ng of HIV-1 recombinant RT, dCTP (α-32P labeled), and 100 μM each dTTP, dGTP, and ddATP. Inclusion of ddATP terminated the extension of tRNA3Lys at the sixth nucleotide (see Fig. 3C). The reactions were stopped by addition of loading buffer containing 50% formamide, and the reaction mixtures were boiled for 5 min before being analyzed on 8% denaturing polyacrylamide gels.
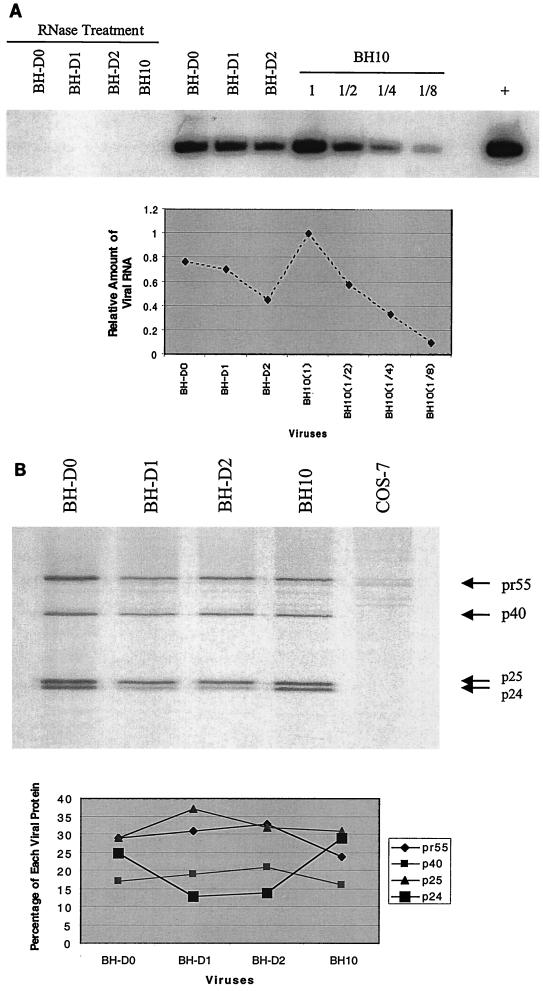
FIG. 3.
Deficits in viral RNA packaging, Gag protein processing, and reverse transcription as a result of the BH-D0, BH-D1, and BH-D2 deletions. (A) Levels of viral RNA in wild-type and mutated viruses as determined by RT-PCR and use of the primer pair pGAG1-pST (35). RNA samples were treated with RNase as a negative control to exclude the possibility of DNA contamination. The wild-type BH10 RNA sample was diluted 1:2, 1:4, and 1:8 to show the linear range of RT-PCR. BH10 DNA was used in the RT-PCR as a positive control (+). The amounts of PCR product were quantified through the use of the NIH Image Program, and the wild-type level was arbitrarily set at 1. (B) Examination of processing of Gag precursor protein by radiolabeling of transfected COS-7 cells and immunoprecipitation of viral proteins with antibodies against the HIV-1 p24 (CA) Ag. The positions of viral proteins are shown on the right. The amount of each viral protein as a percentage of the total viral proteins was calculated using the NIH Image Program. (C) In vitro extension by the tRNA3Lys primer of wild-type and mutated viruses. Three extension products, namely, nt +3, +5, and +6, were observed. Lanes 1 and 2 are controls performed with a tRNA3Lys-viral RNA complex formed by heat annealing. The reaction mixture in lane 1 included only dCTP (α-32P labeled) to indicate the position of the nt +1 product. Lane 2 contained a reaction mixture that included dCTP (α-32P labeled), dGTP, dTTP, and ddATP and shows the positions of the nt +3 and +6 products. Relative amounts of extension products in each virus were calculated using the NIH Image Program and plotted. PBS, primer-binding site.
RESULTS
Deletion of sequences downstream of the 5′ LTR results in impaired viral infectiousness.
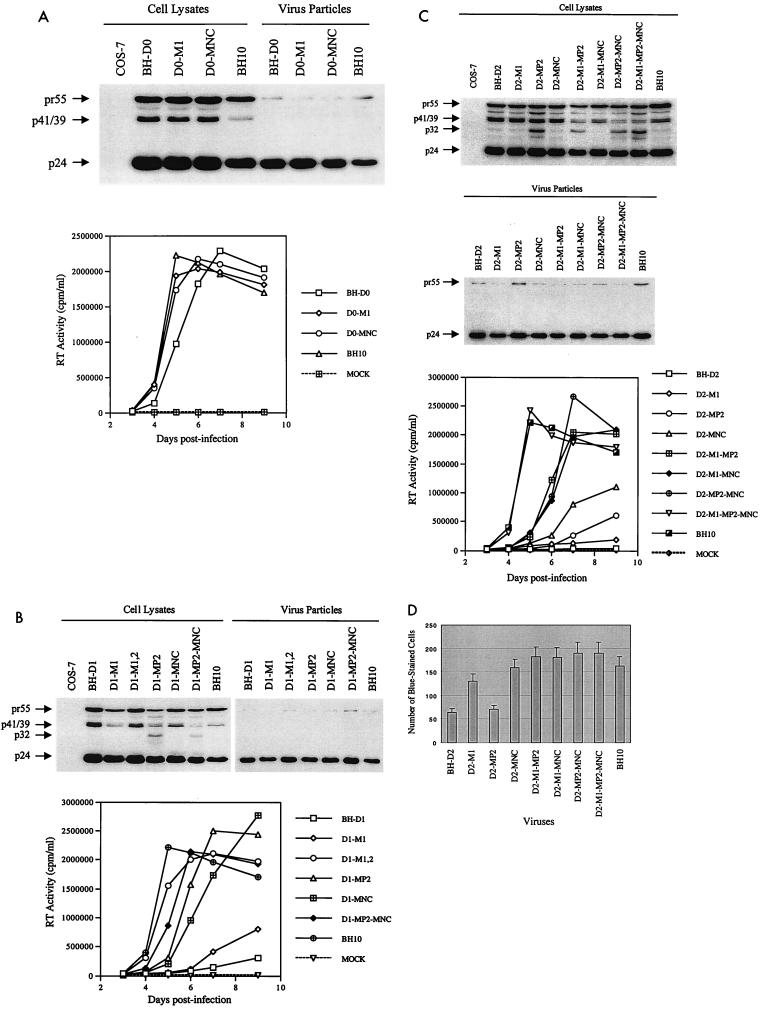
Sequences at nt +200 to +219, +200 to +226, and +200 to +233 were deleted to create the BH-D0, BH-D1, and BH-D2 constructs, respectively (Fig. 1A). The wild type and these various mutated HIV-1 cDNA constructs were transfected into COS-7 cells, and levels of both intracellular and extracellular viral proteins were analyzed by Western blotting through the use of a MAb against the HIV-1 p24 (CA) Ag. Figure 2A shows that all of these constructs produced HIV-1 Gag precursor proteins that were further processed to mature CA. The infectiousness of the progeny viruses was further tested by infecting MT-2 cells with the same amount of virus on the basis of CA levels. The results in Fig. 2B show that the BH-D0 deletion moderately diminished viral infectiousness while both the BH-D1 and BH-D2 deletions resulted in severe attenuation of viral replication, with BH-D2 being the most impaired in this regard. Therefore, we concluded that viral RNA sequences at nt positions +200 to +233 play important roles in viral replication.
FIG. 2.
Effects of BH-D0, BH-D1, and BH-D2 deletions on viral infectiousness. (A) Protein analysis of transfected COS-7 cells and virus particles by Western blotting through the use of a MAb against the HIV-1 p24 (CA) Ag. The positions of viral proteins are on the left. (B) Replication kinetics of wild-type and mutated viruses in MT-2 cells on the basis of RT activity in culture fluids. MOCK, negative control in which MT-2 cells were exposed to heat-inactivated wild-type virus.
RNA sequences at nt positions +200 to +233 are necessary for viral RNA packaging, processing of Gag precursor protein, and reverse transcription.
To shed light on the deficits that might result from these deletions, we first analyzed the efficiency of viral RNA packaging in the mutated viruses by RT-PCR as previously described (35). Primer pair pGAG1-pST was employed to amplify a region of the gag gene to provide information on the packaging of viral RNA into virus particles. The results in Fig. 3A show that the BH-D0 and BH-D1 deletions decreased viral RNA packaging to 77 and 70% of the wild-type level, respectively, while the BH-D2 deletion resulted in a decrease to 40% of the wild-type level. Thus, the sequence at nt positions +200 to +233 presumably participate in viral RNA packaging, which may be achieved either directly or indirectly through pairing with other sequences to facilitate the folding and presentation of other signals involved in viral RNA encapsidation (Fig. 1B).
We next examined the processing of Gag precursor proteins in the wild-type and mutated viruses through short-term radiolabeling and immunoprecipitation assays. Gag precursor Pr55, intermediate proteins p40 and p25 (CA-p2), and the mature p24 product (CA) can be observed on the gel (Fig. 3B). The amount of each protein was quantified by densitometry, and for each virus, the percentage of a particular protein versus the total viral proteins was plotted. The results show that BH-D0 and wild-type BH10 gave rise to similar proportions of these four proteins. In contrast, both the BH-D1 and BH-D2 deletions resulted in accumulation of the Pr55, p40, and p25 proteins and diminished levels of p24 (CA). Therefore, deletion of sequences at nt positions +220 to +233 caused delayed processing of the Gag precursor protein while sequences at nt +200 to +219 may not have been important in this regard.
Since the RNA sequences at nt +200 to +233 are just downstream of the primer-binding site, it is highly likely that they affect the priming of reverse transcription. To analyze this, we studied the ability of primer tRNA3Lys, which had already been annealed onto viral genomic RNA within viruses, to be extended. The antiviral drug ddATP was included in the reaction mixtures to cause chain termination of reverse transcription at the sixth nucleotide position. In the case of wild-type virus, two major bands can be observed, at nt positions +3 and +6, with the +6 product being in the majority (Fig. 3C). The +3 band indicates a strong pause site during initiation of reverse transcription, as previously shown (34), while the +6 band represents the chain termination site. With the mutated viruses, an additional strong band at nt position +5 was observed; this was particularly intense in the cases of BH-D0 and BH-D1. When relative amounts of extension products were calculated for each virus, it was found that more of the +3 and +5 products accumulated with the mutated than with the wild-type virus; this indicates a block in reverse transcription before the +5 nt site. Since deletion of the nt sequence from +200 to +219 alone caused the accumulation of the nt +3 and +5 products, it follows that this segment is required for efficient reverse transcription.
Long-term culture of mutated viruses in MT-2 cells and emergence of revertants with wild-type replication kinetics.
The BH-D0, BH-D1, and BH-D2 mutated viruses were cultured in MT-2 cells for prolonged periods until wild-type replication kinetics were observed. At this time, cellular DNA from the infected MT-2 cells was extracted and PCR was performed to amplify a long viral DNA fragment, termed HA, at nt −454 to +1546, through use of the primer pair pHpa-s–pApa-A (35). Sequencing analysis showed that each of the three revertant viruses maintained the original deletions, i.e., BH-D0, BH-D1, and BH-D2, respectively. Therefore, other parts of the viral genome must have been altered in order to rescue viral replication.
We then replaced the equivalent fragment in the wild-type BH10 cDNA with the above-described HA PCR product to generate constructs D0-HA, D1-HA, and D2-HA. Each of these recombinant DNA clones was transfected into COS-7 cells; we found that the progeny that had been generated were as infectious for MT-2 cells as were the wild-type viruses (data not shown). Therefore, novel mutations must have been present in the HA DNA fragment that were able to rescue the BH-D0, BH-D1, and BH-D2 deletions. Sequencing of the HA region revealed point mutations in both the U5 region and the gag gene in BH-D0, BH-D1, and BH-D2 (Fig. 4).
FIG. 4.
Illustration of compensatory mutations identified in mutated viruses. Nucleotide numbers are relative to the initiation site of RNA transcription. Mutated nucleotides are underlined, and names of mutations are in boldface letters. PBS, primer-binding site; MA, matrix protein; NC, nucleocapsid.
The decreased infectiousness of BH-D0 is restored to wild-type levels by either the M1 or the MNC point mutation.
Two point mutations, M1 (G112A) in the U5 region and MNC (T24I) in the NCp, were identified in BH-D0 (Fig. 4) in revertant viruses at day 31 postinfection. By day 61, most of the viruses that were sequenced harbored these two point mutations (Table 1). To determine whether either the M1 or the MNC mutation or both could compensate for the BH-D0 deletion, each of them was inserted separately into the BH-D0 clone to generate constructs D0-M1 and D0-MNC. Each of these two constructs yielded normal patterns of viral proteins when transfected into COS-7 cells, as shown by Western blotting (Fig. 5A). The results of infection studies also indicated that the recombinant viruses D0-M1 and D0-MNC were as infectious as wild-type BH10 in MT-2 cells (Fig. 5A). Therefore, either M1 or MNC alone can compensate for the decreased infectiousness of BH-D0.
TABLE 1.
Compensatory mutations accumulated with long-term culture of the BH-D0, BH-D1, and BH-D2 mutants in MT-2 cellsa
| Deletion and no. of days after infection | No. of mutations/no. of clones sequenced
|
|||
|---|---|---|---|---|
| M1 | M2 | MP2 | MNC | |
| BH-D0 | ||||
| 12 | ||||
| 31 | 1/6 | 1/6 | ||
| 46 | 3/6 | 2/6 | ||
| 61 | 4/6 | 4/6 | ||
| BH-D1 | ||||
| 28 | 2/6 | 1/6b | 1/6b | |
| 40 | 2/6 | 4/6 | 1/6b | 1/6b |
| 47 | 6/6 | 6/6 | 2/6b | 2/6b |
| BH-D2 | ||||
| 28 | 6/6 | 1/6 | ||
| 42 | 2/6 | 6/6 | 1/6 | |
| 57 | 2/6 | 6/6 | 2/6 | |
| 64 | 6/6 | 6/6 | 4/6 | |
Six clones were sequenced per passage, and the number of clones containing a particular mutation is shown.
The MP2 and MNC point mutations were identified in different clones.
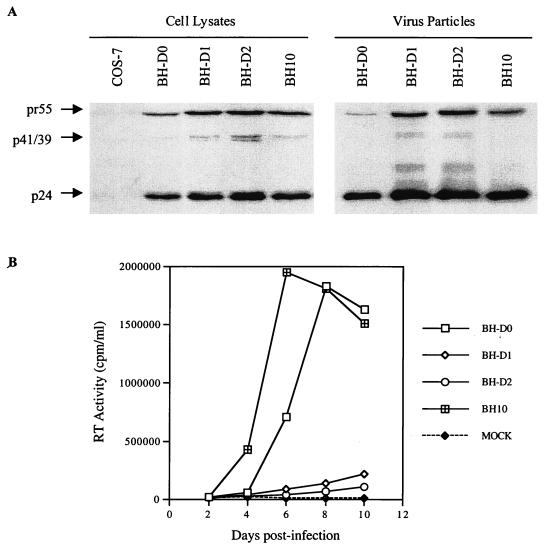
FIG. 5.
Roles of various point mutations in rescuing the decreased infectiousness of mutated viruses. (A) Rescue of BH-D0 by the M1 and MNC point mutations. Viral proteins in cell lysates and virus particles were analyzed by Western blotting through the use of antibodies against the HIV-1 p24 (CA) Ag. Infectiousness of viruses generated by transfection of COS-7 cells was examined in MT-2 cells by monitoring RT activity in the culture fluids. (B) Compensation for BH-D1 by the M1, M2, MP2, and MNC point mutations, as analyzed by Western blotting and infection studies. (C) Compensation for BH-D2 by the M1, MP2, and MNC point mutations as shown by Western blotting and infection studies. (D) MAGI assays of the BH-D2 mutated virus and various recombinant viruses containing compensatory mutations. HeLa–LTR–β-gal cells were infected with these viruses; after 48 h, cells were fixed and stained as described in Materials and Methods. Numbers of blue-stained cells were scored and plotted. Three independent infections were performed for each virus studied. Results are expressed as averages ± standard deviations. MOCK, negative control in which MT-2 cells were exposed to heat-inactivated wild-type virus.
BH-D1 mutated viruses can be compensated by point mutations in either the noncoding region or the gag gene.
In the case of BH-D1, four point mutations were identified, i.e., M1, M2 (C227T, just downstream of the 5′ LTR), MP2 (T12I) in p2, and MNC (Fig. 4). Interestingly, all of the clones analyzed contained both the M1 and M2 point mutations at day 47 postinfection while only two of six clones contained either the MP2 or MNC mutation; this suggests that the M1 and M2 mutations may be more important with regard to compensation for the BH-D1 deletion (Table 1). To evaluate the roles of the above-described substitutions in the compensation for BH-D1, constructs were generated to recombine these various mutations with BH-D1, i.e., D1-M1, D1-M1,2 (containing both M1 and M2), D1-MP2, D1-MNC, and D1-MP2-MNC. These constructs were transfected into COS-7 cells, and production of viral proteins was detected in Western blots (Fig. 5B). Interestingly, the presence of the MP2 point mutation within either D1-MP2 or D1-MP2-MNC yielded a novel protein band of approximately 32 kDa in cell lysates. The results of infection experiments showed that the D1-M1 recombinant virus was barely infectious, while both D1-M1,2 and D1-MP2-MNC possessed wild-type replication kinetics. The D1-MP2 and D1-MNC viruses both showed increased replication capacity in comparison to BH-D1 (Fig. 5B). Therefore, both M1 and M2 acting together and MP2 and MNC were able to rescue the BH-D1 deletion.
The impaired replication kinetics of BH-D2 can be restored to wild-type levels only when all of the M1, MP2, and MNC point mutations are present.
The M1, MP2, and MNC point mutations were also identified in BH-D2 (Fig. 4). Indeed, the majority of clones contained all three point mutations by day 64 postinfection (Table 1). Sequencing results of earlier passages had also shown that the MP2 point mutation had become dominant by day 28. To examine whether the above point mutations were sufficient and necessary to rescue BH-D2, the following constructs were generated: D2-M1, D2-MP2, D2-MNC, D2-MP2-MNC, D2-M1-MP2, D2-M1-MNC, and D2-M1-MP2-MNC. Each of these constructs was able to produce viral proteins when transfected into COS-7 cells (Fig. 5C). Again, a protein band of around 32 kDa was observed with constructs containing the MP2 mutation (i.e., D2-MP2, D2-M1-MP2, D2-MP2-MNC, and D2-M1-MP2-MNC). The results of infection studies with MT-2 cells showed that D2-M1, D2-MP2, and D2-MNC all produced virus particles with marginally higher replication capacity than BH-D2; therefore, neither the M1, the MP2, nor the MNC point mutation alone could confer wild-type infectiousness on BH-D2 (Fig. 5C). In contrast, the D2-M1-MP2, D2-M1-MP2, and D2-MP2-MNC recombinant viruses were moderately infectious and the D2-M1-MP2-MNC virus showed wild-type replication kinetics (Fig. 5C). Therefore, all three point mutations were needed to restore the impaired infectiousness of BH-D2 to wild-type levels. The infectiousness of our various mutated viruses was further analyzed in a MAGI cell assay. The results in Fig. 5D show that the BH-D2 and D2-MP2 mutants generated far fewer blue-stained cells than did the wild-type BH10 virus. The other mutated viruses studied yielded numbers of blue-stained cells similar to those yielded by BH10.
Either the M1 or the MNC point mutation can correct defective viral RNA packaging, and the MP2 point mutation facilitates the processing of the Gag precursor in the mutated viruses.
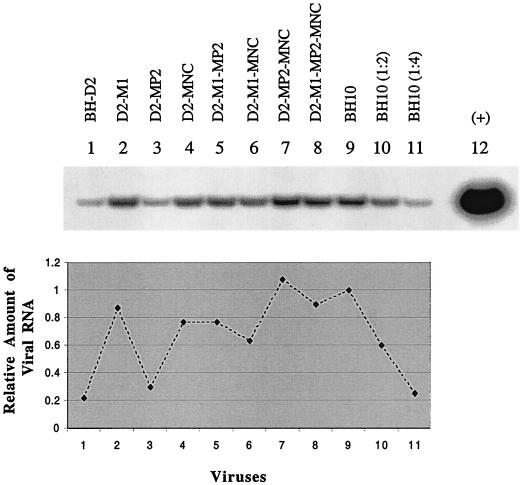
To shed light on the mechanisms by which the above-described point mutations compensate for the deletions, levels of viral RNA packaging in the BH-D2 viruses containing the M1, MP2, or MNC point mutation were examined by RT-PCR as described above (Fig. 6). The D2-MP2 virus packaged low levels of viral RNA similar to those packaged by BH-D2, while the D2-M1 and D2-MNC viruses showed markedly increased amounts of viral RNA. Consistently, D2-M1-MP2, D2-M1-MNC, D2-MP2-MNC, and D2-M1-MP2-MNC possessed much higher levels of viral RNA than did either BH-D2 or D2-MP2. Therefore, the M1 and MNC point mutations helped to compensate for defects in viral RNA packaging in the BH-D2 viruses while the MP2 mutation did not.
FIG. 6.
Levels of viral RNA packaging in BH-D2 mutated viruses containing the M1, MP2, or MNC point mutation. BH10 RNA samples were used undiluted and diluted 1:2 and 1:4 (lanes 10 and 11) for RT-PCR to ensure the linear range of the PCR. Lane 12 served as a positive control performed with BH10 plasmid DNA. The intensity of each RT-PCR product on the gel was quantified with the NIH Image Program, and the level of viral RNA in BH10 was arbitrarily set at 1.0. RNA samples were treated with RNase before RT-PCR as negative controls to rule out the possibility of DNA contamination (data not shown).
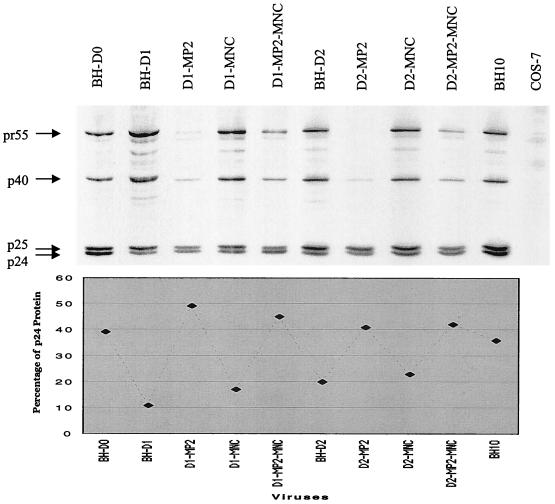
The MP2 point mutation results in a change in one of the amino acids at the protease cleavage site between p2 and NCp. Therefore, it follows that MP2 may affect this cleavage to modify the processing of Gag proteins. To test this hypothesis, processing of Pr55gag in the mutated viruses, i.e., D1-MP2, D1-MNC, D1-MP2-MNC, D2-MP2, D2-MNC, and D2-MP2-MNC, was studied by radiolabeling of viral proteins and immunoprecipitation experiments. Relative levels of mature CA were calculated in each virus recovered from COS-7 cells to evaluate the efficiency of Gag protein processing. The results in Fig. 7 show that the MP2 mutation restored the decreased levels of CA in the various mutated viruses to wild-type levels (i.e., D1-MP2, D1-MP2-MNC, D2-MP2, and D2-MP2-MNC), while the MNC point mutation had no effect in this regard (i.e., D1-MNC and D2-MNC).
FIG. 7.
Effects of the MP2 and MNC point mutations on Gag protein processing in the mutated BH-D1 and BH-D2 viruses. For details, see the legend to Fig. 3B.
DISCUSSION
The HIV-1 RNA genome contains a long 5′ noncoding leader sequence spanning nt positions +1 to +335. Computer modeling and biochemical analysis have revealed the presence of complex secondary structures in this region that perform important functions in viral replication (Fig. 1B). A transactivation response element (TAR) structure, formed by the sequences at the 5′ end, serves as the binding site for viral transactivation protein (Tat); the poly(A) stem-loop is the binding site for cleavage factors to terminate gene transcription (13). The U5 region and parts of the sequences downstream of the primer-binding site form complex structures, and different models for these regions have been proposed (Fig. 1B) (4, 14, 21). Four stem-loop structures, namely, DIS, SD, PSI, and AUG, have been predicted to exist with the following characteristics. (i) DIS promotes viral RNA dimerization and packaging. (ii) SD contains signals for viral RNA splicing. (iii) PSI dictates specific viral RNA encapsidation. (iv) AUG contains the AUG initiation codon for Gag protein synthesis (3, 11). A more complex structure has also been proposed that comprises the above four stem-loop structures (Fig. 1B, insert) (21). Our BH-D2 deletion mutation eliminated a CTCTC sequence that is assumed to bind to a GAGAG sequence on the basis of the above model and, hence, might have resulted in the opening of this large RNA complex. Since the BH-D2 construct led to decreased viral RNA encapsidation, it is speculated that formation of the large RNA complex under natural conditions may help to present the DIS (SL1) and PSI (SL3) RNA structures, i.e., the major viral RNA encapsidation signals, to viral proteins and therefore may facilitate viral RNA packaging.
Extension via reverse transcription from the cognate primer tRNA3Lys of HIV involves two stages, initiation and elongation (23, 24, 27–29, 34). This concept has been developed on the basis of cell-free RT reactions that use natural tRNA3Lys as a primer and in vitro-transcribed viral RNA templates. The initiation stage is defined by early pausing events, especially at nt +1, +3, and +5, and is further distinguished from the subsequent elongation stage by different RT dissociation constants. We purified tRNA3Lys-viral RNA complexes from the various wild-type and mutated virus particles employed in this study. An nt +3 extension product was observed in these reaction mixtures, duplicating results obtained earlier with cell-free RT reactions (Fig. 3C) (34). This supports the existence of a specific initiation stage of reverse transcription in vivo. Yet, the amount of in vivo nt +3 product was lower than that observed in in vitro reaction mixtures (lane 2, Fig. 3C). This suggests that the in vivo tRNA3Lys-viral RNA complex is more processive than that formed by heating in vitro. Interestingly, when sequences at nt +200 to +219, just downstream of the primer-binding site, were deleted in BH-D0, an intense band at nt position +5, representing an extension product, was observed. Thus, the BH-D0 deletion led to a higher probability that reactions would stop during initiation than with the wild-type virus. According to published secondary structures of the tRNA3Lys-viral RNA complex, the sequences at nt +200 to +219 do not directly contribute to interactions between tRNA3Lys and viral RNA (24). However, this segment can bind to sequences in the U5 region (Fig. 1B). This may help to organize the highly ordered secondary structure of the tRNA3Lys-viral RNA complex that has been shown, in a three-dimensional model, to be important for RT to properly bind to the above RNA complex and to initiate reverse transcription.
Deletions of the SL1 RNA sequences have been previously shown to affect the efficiency of Gag protein processing (33). We observed similar effects on the processing of Pr55gag with deletions of RNA sequences at nt +200 to +233, just upstream of the SL1 region. This suggests that the above two RNA regions interact with Gag proteins in a similar way to help Pr55gag to adopt certain conformations that facilitate protease (PR)-mediated cleavages. Although the sequences at nt +200 to +233 were found to affect viral RNA packaging, reverse transcription, and Gag protein processing, the defects in the latter may primarily account for the impaired viral infectiousness of relevant constructs. This is suggested by the fact that the BH-D0 deletion at nt +200 to +219 resulted in both decreased levels of viral RNA encapsidation and reduced efficiency of reverse transcription, yet viruses containing this deletion retained normal processing of Gag proteins, as well as a moderately high viral replication capacity. In contrast, the BH-D1 deletion at nt +200 to +226 caused similar defects in viral RNA packaging and reverse transcription but defective Gag processing and dramatically decreased viral infectiousness were observed in this situation.
We have previously reported point mutations in the gag gene that were able to rescue deletions of the SL1 (DIS) sequence (35, 36). Interestingly, some of these mutations, i.e., MP2 and MNC, were observed to compensate for deletions of sequences at nt +200 to +233, just upstream of SL1. The MNC point mutation alone can confer wild-type replication capacity on BH-D0. In terms of BH-D1, both MP2 and MNC are needed to restore wild-type replication kinetics. When a 34-nt RNA segment was deleted in BH-D2, i.e., nt +200 to +233, both the MP2 and MNC mutations were able to increase the impaired infectiousness to higher levels. Seemingly, the deletion of longer sequences may increase the dependence of the mutated viruses on the MP2 and MNC point mutations to gain wild-type replication kinetics. Since both MP2 and MNC can also rescue deletions within the SL1 region, this suggests that RNA sequences at nt +200 to +233 may form a complex with SL1 sequences that is important in viral replication. The fact that BH-D0 and BH-D1 can also be rescued by point mutations in noncoding RNA sequences, which has not been observed with deletions of SL1, suggests that the sequences deleted in BH-D0 and BH-D1 interact with regions in U5 and fulfill functions distinct from those of SL1.
The MP2 point mutation is important in rescue of the deletion of the sequence including nt +200 to +233 because of its ability to restore diminished efficiency of Gag processing in mutated viruses to wild-type levels (Fig. 7). MP2 changes the amino acids at the viral protease cleavage site between p2 and NCp; this presumably enhanced Gag processing. However, the observation of a 32-kDa intermediate product in Western blots of cell lysates in samples containing the MP2 point mutation renders the situation more complex (Fig. 5B and C). Obviously, the 32-kDa protein might contain CA (p24), p2, and NCp (p7) in order to possess a size of 32 kDa. The accumulation of the 32-kDa protein therefore indicates a lowered rate of cleavage between p2 and NCp; this leads to the conclusion that MP2 decreased the efficiency of p2-NCp cleavage. Therefore, MP2 may facilitate the use by PR of other cleavage sites at MA-CA and NCp-p6 to increase the efficiency of Gag protein processing in the mutated viruses. Indeed, MP2 might simultaneously result in an increase in cleavage at the p2-CA junction while diminishing that at p2-NC7.
Both the BH-D2 and D2-MP2 viruses were defective in the MAGI assay, consistent with their inability to package normal levels of full-length viral RNA. Since the generation of blue-stained cells in this assay depends on levels of Tat protein that are synthesized after viral entry into cells, the numbers of such cells reflect the efficiency of initial viral replication events, including entry, reverse transcription, integration, and early viral gene expression. Deficits in levels of viral RNA packaging could result in the crippled production of proviral DNA after infection, leading ultimately to lower levels of Tat protein. As a result, fewer blue-stained cells should be observed than in the case of the wild-type virus. Both the D-M1 and D2-MNC constructs showed numbers of blue-stained cells in the MAGI assay similar to that of the wild-type virus, perhaps because of their correction of the viral RNA packaging deficit. Yet, both of these viruses were poorly infectious in MT-2 cells. Therefore, other defects must also be present in these viruses that maintain a state of attenuated replication.
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council of Canada.
We thank Mervi Detorio and Maureen Oliveira for technical assistance.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 3.Baudin G, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz R D, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J L, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J L, Wong M L, Parslow T G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 14.Damgaard C K, Dyhr-Mikkelsen H, Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type 1 Gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 17.Das A T, Klaver B, Berkhout B. The 5′ and 3′ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison G P, Miele G, Hunter E, Lever A M L. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J Virol. 1998;72:5886–5896. doi: 10.1128/jvi.72.7.5886-5896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isel C, Lanchy J M, Le Grice S F, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 24.Isel C, Westhof E, Massire C, Le Grice S F, Ehresmann B, Ehresmann C, Marquet R. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J. 1999;18:1038–1048. doi: 10.1093/emboj/18.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaddrick M, Lear A L, Cann A J, Heaphy S. Evidence that a kissing loop structure facilitates genomic RNA dimerization in HIV-1. J Mol Biol. 1996;259:58–68. doi: 10.1006/jmbi.1996.0301. [DOI] [PubMed] [Google Scholar]
- 26.Kim H J, Lee K, O'Rear J J. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 26a.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanchy J M, Ehresmann C, Le Grice S F, Ehresmann B, Marquet R. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J. 1996;15:7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 28.Lanchy J M, Isel C, Ehresmann C, Marquet R, Ehresmann B. Structural and functional evidence that initiation and elongation of HIV-1 reverse transcription are distinct processes. Biochimie. 1996;78:1087–1096. doi: 10.1016/s0300-9084(97)86734-x. [DOI] [PubMed] [Google Scholar]
- 29.Lanchy J M, Keith G, Le Grice S F, Ehresmann B, Ehresmann C, Marquet R. Contacts between reverse transcriptase and the primer strand govern the transition from initiation to elongation of HIV-1 reverse transcription. J Biol Chem. 1998;273:24425–24432. doi: 10.1074/jbc.273.38.24425. [DOI] [PubMed] [Google Scholar]
- 30.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus type 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 31.Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensible for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 32.Laughrea M, Jette L. HIV-1 genomic dimerisation: formation kinetics and thermal stability of dimeric HIV-1 LAI RNAs are not improved by the 1–232 and 292–790 regions flanking the kissing loop domain. Biochemistry. 1996;35:9366–9374. doi: 10.1021/bi960395s. [DOI] [PubMed] [Google Scholar]
- 33.Liang C, Rong L, Cherry E, Kleiman L, Laughrea M, Wainberg M A. Deletion mutagenesis within the dimerization initiation site of human immunodeficiency virus type 1 results in delayed processing of the p2 peptide from precursor proteins. J Virol. 1999;73:6147–6151. doi: 10.1128/jvi.73.7.6147-6151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang C, Rong L, Gotte M, Li X, Quan Y, Kleiman L, Wainberg M A. Mechanistic studies of early pausing events during initiation of HIV-1 reverse transcription. J Biol Chem. 1998;273:21309–21315. doi: 10.1074/jbc.273.33.21309. [DOI] [PubMed] [Google Scholar]
- 35.Liang C, Rong L, Laughrea M, Kleiman L, Wainberg M A. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initiation site can restore viral replication. J Virol. 1998;72:6629–6636. doi: 10.1128/jvi.72.8.6629-6636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang C, Rong L, Quan Y, Laughrea M, Kleiman L, Wainberg M A. Mutations within four distinct Gag proteins are required to restore replication of human immunodeficiency virus type 1 after deletion mutagenesis within the dimerization initiation site. J Virol. 1999;73:7014–7020. doi: 10.1128/jvi.73.8.7014-7020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marquet R, Paillart J C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muriaux D, Girard P M, Bonnet-Mathoniere B, Paoletti J. Dimerization of HIV-1 LAI at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 41.Paillart J C, Berthoux L, Ottmann M, Darlix J-L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paillart J C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 43.Paillart J C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paillart J C, Westhof E, Ehresmann C, Ehresmann B, Marquet R. Noncanonical interactions in a kissing loop complex: the dimerisation initiation site of HIV-1 genomic RNA. J Mol Biol. 1997;270:36–49. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- 45.Poon D T K, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson J H, Child L A, Lever A M L. Packaging of human immunodeficiency virus type 1 RNA requires cis-acting sequences outside the 5′ leader region. J Virol. 1993;67:3997–4005. doi: 10.1128/jvi.67.7.3997-4005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]