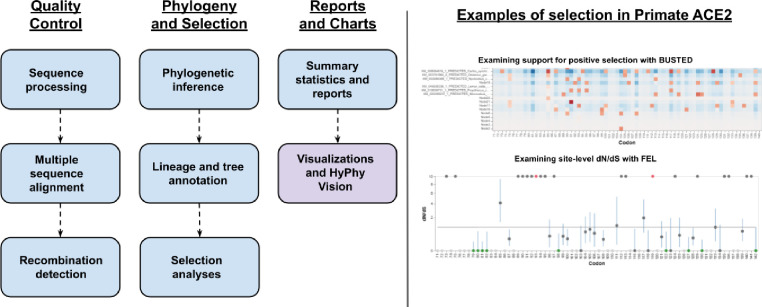
Figure 1. A flowchart diagram of the AOC workflow and an example using Primate ACE2 data.
The workflow consists of three parts, the first of which does quality control, and converts input transcript and protein files from the NCBI ortholog database into codon-aware alignments and checks for phylogenetic evidence of genetic recombination. The second part performs full maximum-likelihood phylogenetic inference and lineage annotation based on NCBI Taxonomy and runs a full suite of selection detection methods using HyPhy. The last part consists of summarizing results into useful tables and visualizations that can be used for post-hoc interpretation and interactions.