Abstract
Sequence analysis of the picornavirus echovirus 22 led to its classification as the first member of a new genus, Parechovirus, and renaming as human parechovirus type 1 (HPeV1). Although distinct from other genera in most of the genome, the 5′ untranslated region (5′UTR) shows similarities to that of cardio/aphthoviruses in some of its structural domains (A to L). The 5′UTR plays an important role in picornavirus translation initiation and in RNA synthesis. To investigate translation in HPeV1, we engineered an extensive range of mutations (including precise deletions and point mutations) into the 5′UTR. Their effects were studied both by in vitro transcription-translation using a bicistronic construct and by in vivo studies using an infectious, full-length HPeV1 cDNA. These approaches allowed the HPeV1 internal ribosome entry site (IRES) to be mapped. Deletions within the first 298 nucleotides had little impact in the in vitro system, while deletions of nucleotides 298 to 538 had a significant effect. Precise removal of domains H and L (nucleotides 287 to 316 and 664 to 682, respectively) did not significantly reduce translation efficiency in vitro, while domains I, J, and K (nucleotides 327 to 545, 551 to 661, and 614 to 645, respectively) appeared to have much more important roles. Mutation of a phylogenetically conserved GNRA motif (positions 421 to 424) within domain I severely reduced translation. We also confirmed the identity of the AUG (positions 710 to 712) which initiates the open reading frame, the positive identification of which has not been possible previously, as the N terminus of the polyprotein is blocked and not amenable to sequence analysis. This is therefore important in understanding parechovirus genome organization. Mutation of the AUG or an upstream polypyrimidine tract leads to aberrant translation, suggesting they both form part of the parechovirus Yn-Xm-AUG motif. In vivo experiments confirmed the importance of domains I, J, and K, the conserved GNRA motif, polypyrimidine sequences, and AUG, as mutations here were lethal. These features are also important in the IRES elements of cardio/aphthoviruses, but other features reported to be part of the IRES of some members of these genera, notably domains H and L, do not appear to be critical in HPeV1. This adds weight to the idea that there may be functional differences between the IRES elements of different picornaviruses, even when they share significant structural similarity.
Picornaviridae is a large family of single-stranded positive-sense RNA viruses, which includes a number of important human and animal pathogens. Until recently, picornaviruses were classified into five genera, Aphthovirus, Cardiovirus, Enterovirus, Hepatovirus, and Rhinovirus, but two former enteroviruses (echovirus 22 and 23) have now been placed in a new genus, Parechovirus, and renamed human parechovirus types 1 and 2 (HPeV1 and -2), respectively (68). Human parechoviruses had previously been shown to exhibit several atypical properties, including unusual cytopathology (65, 73) and failure to shut off host cell protein synthesis (8, 9). Complete nucleotide sequence determination showed that they form a genetically distinct group among picornaviruses (17, 30, 67). Characteristic molecular properties include an atypical form of 2A; lack of VP0 cleavage into VP4 and VP2, giving a virus particle with only three capsid proteins rather than four; lack of myristoylation of VP0; and a unique N-terminal extension to VP3 containing several basic amino acids (17, 27, 30, 61, 67–69). Subsequent partial nucleotide sequence analysis of Ljungan virus (a rodent pathogen) showed this to be highly related to human parechoviruses (53).
Like that of other picornaviruses, the HPeV1 5′ untranslated region (5′UTR) is long (712 nucleotides [nt]) and contains several AUGs which are probably not functional. Analysis of its secondary structure indicates the presence of 12 stem-loops (A to L), several of which are similar to structural domains of cardio/aphthoviruses (17, 68). Much of the picornavirus 5′UTR makes up the internal ribosome entry site (IRES), needed for the cap-independent translation exhibited by these viruses. In vitro and in vivo translation has been used to define IRES activity for representatives of the Aphthovirus (3, 43), Cardiovirus (13, 34, 35, 74), Enterovirus (22, 50, 55), Hepatovirus (6, 7, 18), and Rhinovirus (29, 71) genera. An IRES element also occurs in the genome of the hepatitis C virus and in some cellular mRNAs (45, 48).
Picornaviruses can be divided into three groups on the basis of IRES structure: entero/rhinoviruses, cardio/aphthoviruses, and hepatoviruses (32, 33, 44, 59, 60, 72). One common feature of picornavirus IRES elements is the presence at the 3′ border of a conserved, cis-acting element, the Yn-Xm-AUG motif, where Yn is a polypyrimidine tract and Xm is a random spacer sequence preceding an AUG triplet (37, 58). In cardio/aphthoviruses, as well as in hepatoviruses, translation is initiated usually at this AUG, whereas in entero/rhinoviruses this is not the case and the initiation codon is reached by ribosomal scanning from the Yn-Xm-AUG motif. Cardio/aphthovirus RNAs are translated efficiently in reticulocyte lysates, but the translation of entero/rhinovirus RNAs requires additional, trans-acting factors, obtained from cell extracts, and this correlates with the observed differences in IRES structure (5, 21).
Picornavirus IRES function seems to require canonical translation initiation factors, together with some cellular proteins that are not involved in cap-dependent translation (2). The initiation factors eIF-2, eIF-3, eIF-4A, and eIF-4B and the central domain of eIF-4G, but not eIF-4E, are required for the formation of 48S initiation complexes with the cardiovirus IRES (56, 57, 62). The cellular protein La has a role in poliovirus IRES function (49), while pyrimidine tract binding protein (PTB) is involved in IRES-mediated initiation in the cardiovirus encephalomyocarditis virus (EMCV) but not in another cardiovirus, Theiler's virus (TMEV) (36). PTB has a less significant role in foot-and-mouth disease virus (FMDV) (47, 51, 52) and poliovirus (24) translation, although it has a functionally significant interaction with the human rhinovirus IRES, which is closely related to that of enteroviruses such as poliovirus (29). In addition, poly(rC) binding protein 2 is involved in the switch between translation and replication of enteroviruses, by being able to bind to both the 5′ cloverleaf and the IRES. However, it does not seem to be required for translation directed by cardioviruses and aphthoviruses (72). More recently, Unr, a cytoplasmic RNA binding protein containing five cold shock domains, has been identified as binding to a region between stem-loop V and VI of the rhinovirus IRES (28).
Although the parechovirus 5′UTR shows some structural similarity to cardio/aphthoviruses at its extreme 5′ end and toward its 3′ end, the degree of functional identity is not clear. Moreover, there is little or no conservation of structure in the central part of the 5′UTR, which forms part of the IRES in at least some cardio/aphthoviruses (17, 70). To further analyze this newly recognized picornavirus genus and explore the conservation of translational determinants between genera, we have constructed an extensive range of HPeV1 5′UTR mutants. These include six where individual structural domains have been deleted precisely and five with point mutations. These have been tested both in vitro, using a bicistronic construct and in vivo with an infectious HPeV1 cDNA clone. Both approaches indicate the importance of structures and sequences toward the 3′ end of the 5′UTR to the parechovirus IRES and suggest that the core IRES resembles that of cardio/aphthoviruses. However, some structural domains reported to be part of the cardio/aphthovirus IRES are not necessary for parechovirus IRES activity.
MATERIALS AND METHODS
Cells and tissue culture.
Green monkey kidney (GMK) and human lung carcinoma (A549) cells were grown in Dulbecco modified Eagle medium (Gibco BRL) supplemented with 10% fetal calf serum as previously described (26).
Construct to produce bicistronic mRNAs.
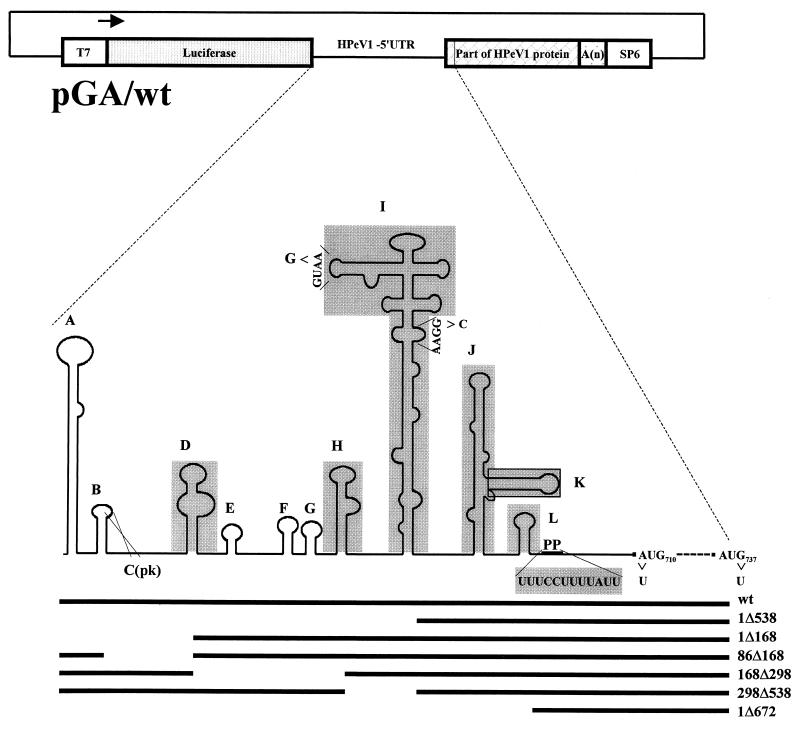
A DNA fragment containing the luciferase gene from the pGL3-control vector (Promega) was excised with BamHI and XbaI and was ligated into the pGEM-4Z vector (Promega), giving pGEM-4Z/T7-luciferase. A ClaI/BglII digest was performed on the complete HPeV1 cDNA clone, pHPeV1, excising material from positions 1653 to 7161. The DNA was end repaired and ligated, giving construct pHP5′UTR/VP, which has cDNA representing the 5′UTR and part of the capsid-encoding region (positions 1 to 1653), joined in frame to part of the 3D-encoding region, 3′UTR (position 7161 to end), and poly(A) tract. pHP5′UTR/VP and pGEM-4Z/T7-luciferase DNAs, following complete digestion with SacI and EcoRI, were ligated together, giving the product shown in Fig. 1. This has a T7 promoter to express a bicistronic mRNA of the form luciferase (control cistron)-HPeV1 5′UTR-partial coding sequence-3′UTR-poly(A) tract (second cistron) and was called pGA/wt.
FIG. 1.
Schematic representation of the bicistronic construct, pGA/wt, containing the HPeV1 5′UTR. Stem-loop domains (A to L) of the 5′UTR are labeled in accordance with those of the cardiovirus 5′UTR (68). Domain C(pk) is a pseudoknot involving the loop of domain B. The part of the 5′UTR remaining in deletion mutants produced using restriction enzymes (pGA/1Δ538, pGA/1Δ168, pGA/86Δ168, pGA/168Δ298, pGA/298Δ538, and pGA/1Δ672) is indicated by a horizontal black line. Specific domains deleted in mutations pGA/ΔD, pGA/ΔH, pGA/ΔI, pGA/ΔJ+K, pGA/ΔK, pGA/ΔpolyP, and pGA/ΔL are shaded grey. Point mutations are shown as arrows (pGA/GNRA424, pGA/GNRA499, pGA/AUG710, and pGA/AUG737).
Introduction of 5′UTR mutations into DNA subclones.
Several deletion mutants were constructed by using a KpnI subclone from pHPeV1 containing the first 575 nt of the 5′UTR. This was digested with a range of restriction enzymes; the ends of the DNA fragments were made blunt and then ligated (Fig. 1; SstI-BstBI [1Δ168], XcmI-BstBI [86Δ168], BstBI-BstXI [168Δ298], BstXI-SpeI [298Δ538], and SstI-SpeI [1Δ538]).
Precise deletion of domains D (36 nt), H (33 nt), I (220 nt), K (35 nt), and L (17 nt) was achieved by overlap PCR, to make a PCR fragment that lacked the element to be deleted. The primers used (Table 1) include the general outer primers OL240, OL537, OL566 and OL731, together with specific mutagenesis primers. Deletion of domains J and K together (J+K; 112 nt) and the polypyrimidine region was done by single-step PCR (Table 1). PCR-mediated site directed mutagenesis was also carried out to introduce the following single mutations into the 5′UTR: A→G at position 424 and G→C at position 499, both of which disrupted GNRA tetraloop motifs within domain I; A→U at position 710, which disrupted the putative initiator codon, AUG; and G→U at position 739, which disrupts a downstream AUG (Table 1). PCR-amplified DNA fragments were ligated into pGEMT-Easy vector (Promega) and sequenced to confirm the presence of only the desired mutation. All mutated fragments were then ligated into a subclone (pHP5′UTR/VP) containing cDNA representing the 5′UTR, part of the viral protein, 3′UTR, and poly(A) tract.
TABLE 1.
Oligonucleotides used for the construction of mutants
| Mutant construct(s) | Oligonucleotide name | Sequence (5′—3′) |
|---|---|---|
| pGA/ΔD | OL678 | GCCTGGTCCAACAAGGATGCTTAAAGCATAG |
| OL679 | GCATCCTTGTTGGACCAGGCATAGGGTTG | |
| pGA/ΔH | OL729 | TGGCGTGCCATAAATCTATTAACAGCCATCCTCTAGTAAG |
| OL730 | TGTTAATAGATTTATGGCACGCCA | |
| pGA/ΔI | OL768 | TGCCCACACAGCCATCCTTTGTAAGGCCCAC |
| OL769 | CTTACAAAGGATGGCTGTGTGGGCA | |
| pGA/ΔJ+K | OL736 | TGCAAACACTAGTTGTAAAACCCAGGGGGGA |
| pGA/ΔK | OL734 | ACGTTTCCCAGATCAGATCCACA |
| OL735 | TGTGGATCTGATCTGGGAAACGTCTAGTGGGCCAA | |
| pGA/ΔL | OL746 | CGTCTAGTGGGCCAAACTTTTCCTTTTATTGTTAATATTGACA |
| OL747 | GAAAGTTTGGCCCACTAGACG | |
| pGA/ΔpolyP | OL737 | GGATCCTGGTGTTAATATTGACATTA |
| pGA/GNRA424 | OL759 | CATCTGGTAGCAGATGCCTCT |
| OL760 | AGAGGCATCTGCTACCAGATG | |
| pGA/GNRA499 | OL758 | ACTAGTGTTTGCACCACTACCAGATTGATAGGTACTGAATATCTTCGAC |
| pGA/AUG710 | OL738 | GGATCCCTGGTTTCCTTTTATTGTTAATATTGACATTTTGGA |
| pGA/AUG737 | OL861 | GAGTATTGCAGATATTGCTACTGGTG |
| OL862 | CACCAGTAGCAATATCTGCAATACTC | |
| General primers | OL240 | GCCTCTG(G/C)GGCCAAAAGC |
| OL537 | CAGGCGAGCTCTTTGAAAGGGGTCTCC | |
| OL566 | CCAACCCTATGCCTGGTC | |
| OL731 | CAGTTGTGATGTCATAAGCCA |
Construction of a full-length cDNA of HPeV1 containing mutated fragments.
In the case of the deletions generated by restriction enzyme digestion, the mutated KpnI subclone was used to replace the wild-type fragment of HPeV1, following KpnI digestion and phosphatase treatment. In the case of other mutants, it was more convenient to ligate the SstI/SalI fragment (positions 1 to 1001) from pHP5′UTR/VP mutants into SstI/SalI-cut pHPeV1 cDNA.
Insertion of mutated fragments into the bicistronic construct.
An SstI/SalI fragment (positions 1 to 1001) from the full-length cDNA mutants or from the pHP5′UTR/VP mutant subclones was ligating into SstI/SalI-cut pGA/wt. All constructs were confirmed by both restriction enzyme digestion and nucleotide sequencing. The constructs are summarized in Fig. 1.
Coupled in vitro transcription-translation.
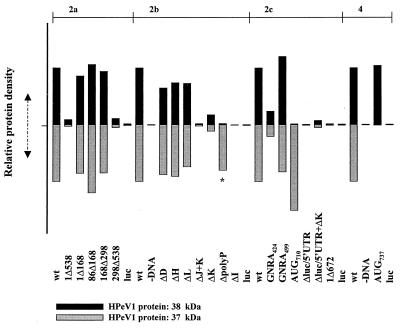
Coupled transcription-translation (system programmed with plasmid DNA) of the bicistronic construct DNA, under the control of the T7 RNA polymerase promoter, was carried out using the TNT-T7 Quick reticulocyte lysate system (Promega), containing [35S]methionine. Standard reaction conditions suggested by the manufacturer were used in all cases, and a total reaction volume of 25 μl was used. Incubation was for 90 min at 30°C. A 5-μl aliquot was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 10% polyacrylamide gel and visualized by autoradiography. Band intensities were measured using a model 300A densitometer (Molecular Dynamics) with ImageQuant software, scanning four times, in different directions, for each band. The HPeV1 protein in each sample was normalized according to the intensity of the corresponding luciferase band. Intensities were then expressed relative to the protein products of the wild-type construct in each case (Fig. 3).
FIG. 3.
Densitometric analysis of coupled in vitro transcription-translation reactions. The translation efficiency for each sample in Fig. 2 was measured by scanning four times in different directions and taking the mean value. The two HPeV1 proteins in each sample were normalized according to the intensity of the corresponding luciferase cistron. Intensities were then expressed relative to the intensity of the protein products of the wild-type construct loaded onto each gel. 2a, 2b, 2c, and 4 refer to figures showing the gels from which the data were derived. Black bars represent the relative intensity of the 38-kDa protein compared to that in the wild-type track, while the grey bars refer to the relative intensity of the 37-kDa protein. The ΔpolyP bar is labeled ∗ to indicate that although similar in size to the 37-kDa product, it is slightly smaller.
In vitro transcription, transfections, and analysis of viruses.
To study the effects of mutations on HPeV1, T7-mediated in vitro transcription of full-length, mutant cDNA, which had been linearized at the end of the poly(A) tract with MluI, was performed. Transfection of the RNA into GMK and A549 cells was performed using Lipofectin (Gibco BRL) as previously described (26). Partial sequence data from viruses recovered from infected cells were obtained by isolating RNA and performing reverse transcription-PCR on the region under investigation, followed by cloning into the pGEMT-Easy vector system or sequencing the PCR product directly (15).
RESULTS
Internal initiation in HPeV1.
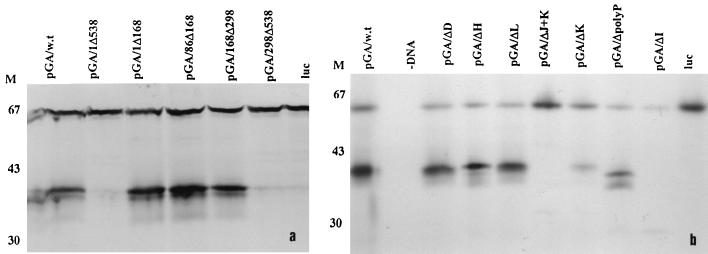
Picornavirus 5′UTR function has frequently been analyzed by in vitro translation of bicistronic RNAs containing the picornavirus 5′UTR inserted between two coding regions. Ribosomes can translate the first cistron, but the second cistron is translated only when a functional IRES is present. Modification of the 5′UTR therefore enables the determinants of IRES-mediated internal initiation to be analyzed. To determine if the 5′UTR of HPeV1, like that of other picornaviruses studied, possesses an IRES domain, it was used as an intercistronic spacer between two coding sequences. In the construct used (pGA/wt), a luciferase cDNA was used as the upstream cistron, while part of the HPeV1 viral protein was used as the downstream cistron (Fig. 1). This should generate bands of ≅63 kDa (luciferase) and ≅41 kDa (HPeV1 protein) when both cistrons are translated. pGA/wt was used to program a commercial in vitro transcription-translation system, and protein production was monitored by [35S]methionine incorporation, detected by SDS-PAGE. It can be seen (Fig. 2, lanes 1) that both cistrons are translated efficiently when an intact HPeV1 5′UTR is present, since bands of 59 and 38 kDa, close to the expected size, are produced. In addition, a smaller (37-kDa) band possibly due to initiation at a downstream AUG, appears close to the HPeV1 protein. In contrast, translation of construct pGA/1Δ672, in which only the 3′-most 30 nt of the 5′UTR remain, gives a single band, corresponding to that generated from the luciferase gene alone (Fig. 2). Thus, removal of HPeV1 5′UTR from the construct abolishes translation of the second cistron, indicating that the HPeV1 5′UTR does have IRES activity. For all pGA constructs described in this study, results obtained by visual inspections of the gels were confirmed by scanning and measuring intensities of the two HPeV1-specific bands. These were normalized to the luciferase band in each track and then expressed relative to the intensities of the corresponding band (size of 38 or 37 kDa) produced from the wild-type construct pGA/wt (Fig. 3).
FIG. 2.
Coupled transcription-translation analysis of HPeV1 5′UTR mutations within the bicistronic construct pGA/wt. Translation efficiency was examined by [35S]methionine incorporation as described in the text, and samples were resolved by SDS-PAGE on a 10% gel. M represents the position of protein markers; numbers to the left indicate approximate molecular masses in kilodaltons.
Mapping the HPeV1 IRES.
To define the borders of the IRES, mutant constructs were initially produced by restriction enzyme digestion and then tested in the coupled in vitro transcription-translation system (Fig. 2). These constructs were pGA/1Δ538, pGA/1Δ168, pGA/86Δ168, pGA/168Δ298, and pGA/298Δ538 (Fig. 1), where numbers indicate the start and end positions of the RNA deleted. Deletions of nt 1 to 168 and 86 to 168 did not reduce significantly the translational activity of the second cistron, and the pGA/86Δ168 deletion may have had a slight stimulatory effect (Fig. 2a). Deletion of nt 168 to 298 also had little effect (Fig. 2a). In contrast, constructs pGA/1Δ538 and pGA/298Δ538 exhibited very little translation of the second cistron, indicating that internal initiation had been abolished (Fig. 2a). Thus, these mutations served to narrow down the 5′ limit of the IRES, which must lie downstream of nt 298 and upstream of nt 538. The fact that nt 298 is in domain H suggests that domains A to H are not essential for IRES activity in this system.
Domains I, J, and K, but not H and L, are essential for internal initiation of translation.
Next, precise 5′UTR deletions were made in which stem-loop domains D, H, I, J+K, K, and L were individually removed. All of these domains show a high level of conservation of secondary structure with corresponding sequences in cardio/aphthoviruses, and there is considerable primary sequence identity at the top of domain I and in J and K (17). Several of these corresponding structures have been shown to be important in the aphtho/cardiovirus IRES (70). Analysis of the in vitro transcription-translation products showed that deletion of domains I and J+K effectively abolished translation of the second cistron (Fig. 2b). Deletion of domain K alone had a clear effect, but a significant albeit low level of translation remained (Fig. 2b and 3). To investigate whether this reduction in efficiency was due to a lower affinity for protein factors within the lysate, pGA/ΔK was translated in an extract to which pGA/Δluc/5′UTR had been added. This construct contained only the HPeV1 5′UTR and had no coding sequence. It could therefore itself generate no translation product and was added to act as a competitor to pGA/ΔK, by sequestering factors required in trans. There was some further depression of translation levels, suggesting that inefficient translation of pGA/ΔK may be due to lower affinity for such factors (Fig. 3). In contrast to the significant effect of removing domains I, J, and K, removal of domains D, H, and L had an undetectable effect on translation efficiency of the second cistron (Fig. 2b and 3).
The polypyrimidine region and downstream AUG.
In addition to 5′UTR structural elements, a key element in the IRES of other picornaviruses is the Yn-Xm-AUG motif, where Yn is a pyrimidine tract, followed by an AUG located 5 to 25 nt downstream (69). The HPeV1 pyrimidine-rich region (UUUCCUUUUAU) (nt 686 to 697) which is followed by an AUG triplet after a spacer region of 19 nt was therefore of particular interest. Precise removal of this pyrimidine-rich region resulted in a complete loss of the usual 38-kDa product seen upon analysis of the wild-type construct (Fig. 2b). Instead, significant levels of smaller products were observed, presumably due to incorrect initiation of the second cistron (Fig. 2b). One of these was similar in size to the second translation product (37 kDa) observed for the wild-type and other mutant constructs, but appeared to be slightly smaller (Fig. 3). These findings suggest that the pyrimidine-rich region is required for accurate initiation.
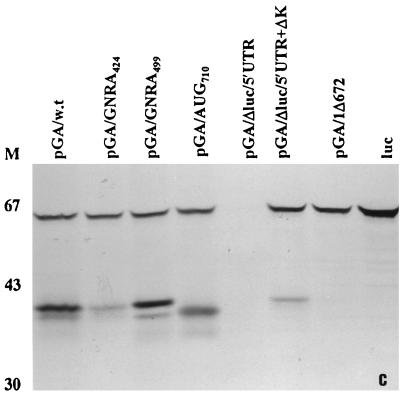
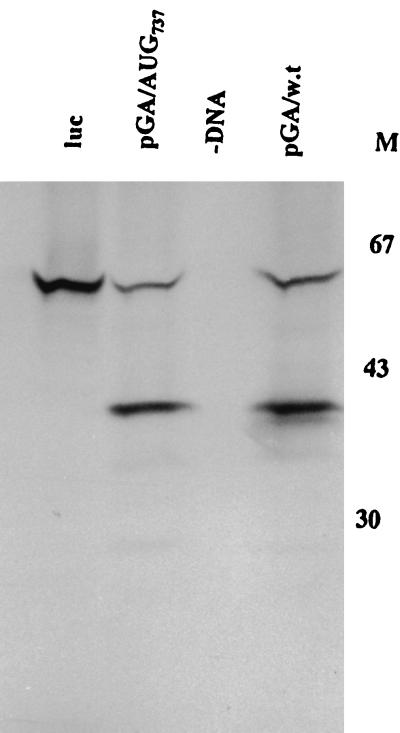
The mutation of AUG710 (the other element of the putative HPeV1 Yn-Xm-AUG motif) to UUG also removed the usual translation product, but an enhanced level of the second translation product was observed, suggesting that a downstream AUG (most likely AUG737) can function efficiently in translation initiation in vitro (Fig. 2c and 3).
A conserved GNRA motif is critical for HPeV1 IRES function.
Since GNRA tetraloops have been observed to be important in the IRES of other viruses, a search for these motifs was carried out in the HPeV1 5′UTR (11). Within domain I, the largest domain of the HPeV1 IRES, three GNRA motifs (a GUAA and two GGAAs) were found in tetraloops (Fig. 1). The GUAA (nt 421 to 424), located in the domain I hammerhead, is analogous to GNRA motifs in the cardio/aphthovirus 5′UTR. The two GGAA sequences form bulges opposite one another in the main domain I stem (nt 374 to 378 and 499 to 502), and TMEV has a GNRA motif in a location similar to the latter. We therefore performed mutagenesis of the GUAA and GGAA (nt 499 to 502) sequences to investigate possible roles in HPeV1 IRES activity. The efficiency of in vitro translation of a mutant containing a single substitution at the fourth position of the GUAA motif (A→G) was significantly reduced, suggesting that this motif is highly important for IRES activity (Fig. 2c). In contrast, mutation of the GGAA sequence (to CGAA) resulted in no apparent decrease in translation efficiency (Fig. 2c).
Effect of mutations in the 5′UTR on HPeV1 growth.
In addition to using in vitro translation, the possible functional role of sequences and structural elements in the IRES domain was also addressed by in vivo experiments. Here, several of the same mutations were introduced into an infectious cDNA clone of HPeV1 (Table 2). The wild-type cDNA clone and the various mutant clones were transcribed in vitro, and the RNA was used to transfect monolayers of GMK cells. The development of viral plaques was monitored for 3 to 6 days after application of plaquing overlay. The results are summarized in Table 2.
TABLE 2.
Approximation of growth ability of recombinant HPeV1 produced in monolayers of GMK cells
| Construct | Time when plaques were visible (days after transfection) | Relative virus growth in GMK cellsa |
|---|---|---|
| pHPeV1 | 3 | ++++++ |
| pHPeV1/1Δ168 | —b | −−−−−− |
| pHPeV1/86Δ168 | — | −−−−−− |
| pHPeV1/168Δ298 | — | −−−−−− |
| pHPeV1/298Δ538 | — | −−−−−− |
| pHPeV1/ΔD | 4 | +++−−− |
| pHPeV1/ΔH | 5 | ++−−−− |
| pHPeV1/ΔK | — | −−−−−− |
| pHPeV1/ΔL | 3 | ++++++ |
| pHPeV1/GNRA424 | —c | −−−−−− |
| pHPeV1/GNRA499 | 3 | ++++++ |
| pHPeV1/AUG710 | —c | −−−−−− |
| pHPeV1/AUG737 | 3 | ++++++ |
Ranging from ++++++ (wild-type growth) to −−−−−− (no growth), based on plaque size and period after transfection required for visible plaques to be observed.
—, no plaques or cytopathic effect visible after 6 days.
Revertants with wild-type sequence were obtained following blind passage.
When nt 1 to 168, 86 to 168, and 168 to 298 were deleted, there were no clear signs of viral growth. Although they have no effect on translation in vitro, such large deletions, encompassing more than one structural domain, probably adversely affect other aspects of virus replication. This is particularly the case for nt 1 to 168 and 86 to 168, which perturb 5′ proximal sequences known to play a key role in RNA replication in other picornaviruses (70). Deletion of domain K is lethal to HPeV1; this deletion did not completely abrogate translation in vitro in the bicistronic system (Fig. 3), but it substantially reduced its efficiency. This direct effect on translation probably accounts for the lethal phenotype, although other steps in replication may also be affected. Despite the phylogenetic conservation of domain L, its precise deletion gave a virus with wild-type growth properties in cultured cells, indicating that it does not have an important role in IRES activity or in other aspects of HPeV1 infection of these cells (Table 2). Although having no effect on in vitro translation, deletion of domain H gave a virus with highly impaired growth, evidenced by less extensive cytopathic effect which was slow to develop. The viability of this virus shows that domain H does not have an essential role in the IRES, but its impaired growth indicates that H is necessary for efficient virus replication (Fig. 4). Domain D was another structural element which could be deleted, albeit again giving a highly impaired virus. We also analyzed the effects of the GGAA→CGAA mutation (position 499) within the domain I main stem GNRA motif. This mutation did not alter viral growth properties (Table 2; Fig. 4). In contrast, a single mutation in the phylogenically conserved GNRA motif (GUAA→GUAG) was lethal, but infectious viruses could be obtained by blind passage of transfected cell supernatants. Upon sequencing, these were found to have reverted to the wild-type sequence. Mutation of AUG710 was also lethal, but revertant viruses with the wild-type sequence were recovered.
FIG. 4.
Coupled transcription-translation analysis of pGA/AUG737 compared to that of pGA/wt. Translation efficiency was examined by [35S]methionine incorporation as described in the text, and samples were resolved by SDS-PAGE on a 10% gel. M represents the position of protein markers; numbers to the right indicate approximate molecular masses in kilodaltons.
In vitro translation of HPeV1 RNA initiates at two AUGs, but this is not significant in vivo.
Picornaviruses differ in the precise role of the Yn-Xm-AUG motif. In FMDV, translation initiation occurs not only at this AUG but, at a higher frequency, at the next downstream AUG (1). In cardioviruses, ribosomes initiate translation at this AUG (38), while in entero/rhinoviruses virtually all initiation is at the next downstream AUG, presumably following scanning from the original entry site (14, 32). Sequence analysis of parechoviruses suggests that the AUG of the Yn-Xm-AUG motif (AUG710) initiates the open reading frame, but there is an AUG (AUG737) downstream (17, 30). The observation of two HPeV1-specific products seen in most translations shown in Fig. 2 suggests that HPeV1 translation in vitro can occur at two AUGs, the first one of these (presumably, AUG710) being preferred. Translation initiation from AUG710 was inhibited by an A→U mutation, but recognition of the second one, AUG737, was not affected (Fig. 2c and 3). To determine the relationship between these two AUGs, we analyzed the effect of mutation of AUG737 (to AUU). In the coupled in vitro translation system, this mutation prevented production of the smaller protein, confirming that this was indeed produced by initiation from AUG737 (Fig. 4). However, a HPeV1 infectious clone containing this mutation gave a virus with wild-type properties (Table 2), in contrast to the lethal effect of mutation of AUG710. The results suggest that the only significant translation is from AUG710 and that in vitro translation from AUG737 is not reflected in vivo.
DISCUSSION
This work represents the first molecular analysis of the newly recognized picornavirus genus, Parechovirus (68, 69), which includes the frequent human pathogen HPeV1. This genus is a distinct molecular entity among picornaviruses and exhibits a low overall degree of identity to any other genus. It is therefore important to have a fuller understanding of its molecular biology, which will also contribute to our understanding of functional conservation and diversity of picornaviruses. In view of its overall genetic distance from other picornaviruses, it is interesting that part of the 5′UTR, including what is probably the core IRES (located towards the 3′ end of the 5′UTR), exhibits primary and predicted secondary structural similarity to the corresponding region from cardioviruses and aphthoviruses (17). In contrast, much of the rest of the 5′UTR is quite distinct from that of cardioviruses and aphthoviruses. There are frequently differences in IRES structure and function even among viruses which exhibit extensive overall identity; for instance, the coxsackievirus B3 IRES is smaller and located closer to the initiation codon than that of poliovirus (46). A study of the parechovirus IRES was therefore undertaken to analyze how the pattern of conservation to cardioviruses and aphthoviruses was reflected in IRES function. We constructed a panel of 5′UTR mutants containing an extensive range of mutations, including large deletions, precise excision of individual structural domains, and point mutations. These were introduced into both a bicistronic construct and an infectious, full-length cDNA. Many virus mutants obtained showed defective properties, or the RNA produced from mutant cDNA was not infectious, which correlated well with reduced expression of viral protein observed during in vitro experiments.
Taken together, the in vitro and in vivo results allowed the IRES to be mapped and the contribution of specific elements within it to be assessed. Deletions within the first 298 nt of the 5′UTR were of little importance, while deletions of nt 298 to 538 had a significant effect on translation. The 5′ boundary of the core IRES must therefore be somewhere downstream of position 298 and contains maximally domains H to L. However, precise removal of domains H and L did not clearly reduce translation in vitro, and so domains I, J and K appear to constitute the structural core of the IRES. Separate removal of these large, complex regions of secondary structure completely destroyed IRES activity in vitro, with the exception of deletion of domain K, which reduced IRES activity substantially but not completely. Deletion of core IRES domains was lethal to HPeV1. These structural features are particularly well conserved between cardio/aphthoviruses and parechoviruses, consistent with their importance to the IRES core. Domains I, J, and K also appear to be the most critical for translation initiation in cardio/aphthoviruses (13, 25, 74). Although translation in picornaviruses may require other regions of the 5′UTR participating in a subtle interplay between translation and RNA replication (16), the structural domains defined here seem to represent the irreducible minimum for IRES activity.
The apparent lack of involvement of domain H in the HPeV1 IRES was unexpected, as in EMCV and FMDV it forms part of the binding site for PTB, reported to play a role in IRES function of several picornaviruses (29, 36, 39). However, the precise role of PTB in translation is not clear-cut. EMCV seems to require PTB binding, but another cardiovirus, TMEV, has PTB-independent IRES activity (36). Furthermore, a single point mutation in the EMCV IRES eliminates that dependence (39). Thus, PTB dependence in cardioviruses is not absolute or necessarily exhibited by all members (2). Mutation of some of the PTB binding sites selectively reduces the translation and replication efficiencies of poliovirus in neuronal cells but not in HeLa cells (21), while rhinoviruses, which are closely related to the enterovirus poliovirus, have a functional interaction with PTB (24, 29). In addition, the IRES of the flavivirus hepatitis C virus does not require PTB. Thus, PTB dependence of IRES activity is variable, even between closely related IRESs, and minor differences of sequence and structure may be responsible for different protein binding profiles. Therefore, it is perhaps not surprising that parechoviruses differ from some picornaviruses in terms of the significance of domain H, as this domain is poorly conserved between parechoviruses and other picornaviruses (17). It is also possible that the in vitro system based on rabbit reticulocytes, used here to study the parechovirus IRES, does not accurately reflect the requirements for protein-RNA interactions present in a normal infection. Moreover, it should be noted that complete removal of domain H gave a virus with severely impaired growth properties. It is possible that this is related to factors other than translation, such as RNA replication or encapsidation. Indeed, specific mutations in the IRES have been reported to have a dramatic effect on RNA replication, even when production of viral nonstructural proteins is under the control of a second IRES, and this therefore does not result from translation inhibition (4, 16, 31, 66). It is, nevertheless, possible that the impaired growth of the ΔH mutant is indicative of a role for stem-loop H in translation.
It has been reported that in TMEV, the insertion of 3 nt into domain H reduced IRES activity following transfection into BHK-21 cells, while little effect was seen in rabbit reticulocyte lysates (70). Virus containing this mutation had impaired growth properties and reduced neurovirulence. This observation suggests that the reticulocyte lysate system may not be very sensitive for determining the significance of noncritical IRES components. Thus, it would be interesting to explore the effects of deletion of domain H in other cellular systems. However, the fact that a viable ΔH virus was recovered at all suggests, in agreement with the in vitro results, that H is not an absolute requirement for internal initiation, although it could have an influence on its efficiency in vivo.
Deletion of domain D has a result similar to that seen upon deletion of domain H; i.e., there is no effect on in vitro translation, but the corresponding mutant virus grows poorly. Domain D is related to a corresponding feature in cardioviruses, which is absent in aphthoviruses (17), and lies well upstream of the core parechovirus IRES. It has been little studied but seems to lie at, or beyond, the 5′ border of the IRES elements of cardioviruses (70). Again, the fact that a viable ΔD mutant can be produced indicates that it is not necessary for core IRES activity. The defective nature of this mutant could be due to a more subtle lesion in translation in vivo or to an effect on other replication steps. In contrast to the defective growth properties of the ΔH and ΔD mutants, the ΔL mutant was indistinguishable from wild type. There is strong phylogenetic support for the presence of domain L in all cardio/aphtho/parechoviruses and in Ljungan virus, an animal picornavirus closely related to human parechoviruses (17, 53). Moreover, domain L appears to be part of the IRES of cardioviruses and aphthoviruses, and in the latter is another domain implicated in an interaction with PTB (32, 47). The apparent lack of effect, both in vitro and in vivo, of deleting this domain in HPeV1 is hence surprising, but it is possible that domain L has a cell-type-specific role and may be more important in natural infections.
In addition to the analysis of these structural elements, site-directed mutagenesis of four sequence motifs was also performed: two GNRA tetraloops and two elements of the putative Yn-Xm-AUG motif. Mutation of the latter motif had a critical effect on HPeV1 translation initiation in vitro and was lethal to HPeV1. Extensive previous work has demonstrated the importance of the pyrimidine-rich region in other picornaviruses (7, 19, 23, 36, 43, 50). The AUG located about 20 nt downstream from this pyrimidine-rich region differs functionally among picornaviruses. In cardio/aphthoviruses, it is an initiation codon and following ribosome binding translation is initiated directly, although initiation from a downstream AUG also takes place in aphthoviruses (1). In entero/rhinoviruses, ribosomes are translocated by an unknown mechanism, possibly scanning, to an initiator AUG, located further downstream (33, 50). Alteration of the AUG forming part of the putative Yn-Xm-AUG motif in HPeV1 severely reduced in vitro translation, and following transfection of the mutated RNA, no plaques were recovered. Following transfections without plaquing overlay, revertant viruses, containing the wild-type AUG, were observed. The product obtained from the AUG710 mutation in vitro was similar in size to that observed as a minor component upon translation of many of the constructs, probably due to initiation from the in-frame methionine codon, located 27 nt downstream of the putative Yn-Xm-AUG motif. Deletion of the Yn sequence gave a still smaller product of uncertain origin. These observations, together with sequence and structural similarities with cardio/aphthovirus IRESs, imply that the HPeV1 initiation codon is at position 710 (AUG710) as previously predicted and that the polypyrimidine sequence and AUG analyzed do form part of a functional Yn-Xm-AUG motif (17, 30, 67). As well as being significant to understanding parechovirus translation, this finding is important because amino acid sequencing studies could not give a definitive indication of the location of the N terminus of the parechovirus polyprotein, as the N-terminal amino acid of VP0 is blocked (67). These mutagenesis studies therefore shed important light on genome organization in parechoviruses.
In aphthovirus, about one-third of the internal initiation events occur at the Lab initiation site at the Yn-Xm-AUG motif; the rest occur at the next downstream AUG codon, the Lb site (1, 63). In cardioviruses, internal initiation at the Yn-Xm-AUG motif AUG predominates, but there is limited use of a second AUG, 12 nt downstream (38, 40). This different behavior could be explained by the context of the AUG at the 3′ end of the IRES (20, 41, 42). It has also been reported recently that other upstream sequences influence AUG selection (54). In vitro translation of the HPeV1 constructs showed that a second AUG (AUG737) could also serve as an initiation codon, as a significant level of a smaller product was obtained. Mutation of this second AUG demonstrated that it was indeed the origin of the smaller in vitro product, as this was no longer present (Fig. 4). However, it also proved that the second AUG was not functionally important, since its mutation had no effect on the properties of the virus. This second AUG certainly cannot substitute for AUG710 in vivo, as mutation of AUG710 is lethal, even though a translation product was obtained in vitro. Notwithstanding any effect on translation efficiency, presumably loss of the N-terminal amino acids of VP0 would have a severe effect on the virus. Thus, in parechoviruses there seems to be little if any role for AUG737.
GNRA tetraloops occur frequently in RNA molecules, possibly because they increase hairpin stability (10, 12). There is perfect conservation of one GNRA tetraloop (positions 421 to 424 in parechoviruses), located toward the top of domain I, among all aphtho/cardio/parechoviruses (64). The role of this well-conserved GNRA motif was analyzed in HPeV1 by in vitro translation of mutated RNAs and by introduction into a complete cDNA copy of the virus genome. It was demonstrated that alteration from GUAA to GUAG not only reduced translation in vitro but also was lethal to HPeV1. Transfection experiments led only to revertants of wild-type sequence. Mutagenesis of this motif has recently been performed on the FMDV and EMCV IRESs, and translation activities of mutated bicistronic constructs were analyzed following transfection (11, 64). This also indicated a significant effect of mutations that disrupt the GNRA motif. In contrast, the mutation of a second GNRA sequence, in the main domain I stem, had no effect on in vitro translation or growth, suggesting that it has no major significance. This GNRA sequence forms a 4-nt bulge in the domain I stem, rather than a terminal loop, and thus may be expected to be less important.
The results presented here, derived from experiments using an extensive panel of mutants, provide strong evidence for the requirement for structural elements and conserved sequence motifs in the HPeV1 IRES. The data show that the parechovirus IRES is similar in several functional as well as structural respects to that of cardio/aphthoviruses, but that there are some differences, notably the lack of involvement of domains H and L. As in cardio/aphthoviruses, domains I, J, and K are the main structural elements of the IRES, and the Yn-Xm-AUG motif is also critical. This seems to direct the production of protein from AUG710, which is part of the Yn-Xm-AUG motif. The data therefore contribute significantly to our understanding of the functional diversity and similarity within picornaviruses.
ACKNOWLEDGMENTS
A.S.N. thanks the Ministry of Higher Education of the Islamic Republic of Iran for the generous provision of a studentship. This work was supported by the Wellcome Trust, grant 046462.
REFERENCES
- 1.Belsham G J. Dual initiation sites of protein synthesis on FMDV RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 1992;11:1106–1110. doi: 10.1002/j.1460-2075.1992.tb05150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belsham G J, Brangwyn J K. A region of the 5′ noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J Virol. 1990;64:5389–5395. doi: 10.1128/jvi.64.11.5389-5395.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman A M, Deliat F G, Kean K M. Sequences within the poliovirus internal ribosome entry segment control viral RNA synthesis. EMBO J. 1994;13:3149–3157. doi: 10.1002/j.1460-2075.1994.tb06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borman A, Howell M T, Patton J C, Jackson R. The involvement of a sliceosome component in internal initiation of human rhinovirus RNA translation. J Gen Virol. 1993;74:1775–1788. doi: 10.1099/0022-1317-74-9-1775. [DOI] [PubMed] [Google Scholar]
- 6.Brown E A, Day S P, Jansen R W, Lemon S M. The 5′ nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J Virol. 1991;65:5828–5838. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown E A, Zajac A J, Lemon S M. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J Virol. 1994;68:1066–1074. doi: 10.1128/jvi.68.2.1066-1074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coller B A G, Chapman N M, Beck M A, Pallansch M A, Gauntt C J, Tracy S M. Echovirus 22 is an atypical enterovirus. J Virol. 1990;64:2692–2701. doi: 10.1128/jvi.64.6.2692-2701.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coller B A G, Tracy S M, Etchison D. Cap-binding complex protein p220 is not cleaved during echovirus 22 replication in HeLa cells. J Virol. 1991;65:3903–3905. doi: 10.1128/jvi.65.7.3903-3905.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa M, Michel F. Rules for recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.deQuinto S L, MartinezSalas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draper D E. Strategies for RNA folding. Trends Biochem Sci. 1996;21:145–149. [PubMed] [Google Scholar]
- 13.Duke G M, Hoffman M A, Palmenberg A C. Sequence and structural elements that contribute to efficient encephalomyocarditis RNA translation. J Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrenfeld E, Semler B L. Anatomy of the poliovirus internal ribosome entry site. Curr Top Microbiol Immunol. 1995;203:65–83. doi: 10.1007/978-3-642-79663-0_3. [DOI] [PubMed] [Google Scholar]
- 15.Gama R E, Hughes P J, Bruce C B, Stanway G. Polymerase chain reaction amplification of rhinovirus nucleic acids from clinical materials. Nucleic Acids Res. 1988;16:9346. doi: 10.1093/nar/16.19.9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghazi F, Hughes P J, Hyypiä T, Stanway G. Molecular analysis of human parechovirus type 2 (formerly echovirus 23) J Gen Virol. 1998;79:2641–2650. doi: 10.1099/0022-1317-79-11-2641. [DOI] [PubMed] [Google Scholar]
- 18.Glass M J, Jia X Y, Summers D F. Identification of the hepatitis A virus internal ribosome entry site: in vivo and in vitro analysis of bicistronic RNAs containing the HAV 5′ noncoding region. Virology. 1993;193:842–852. doi: 10.1006/viro.1993.1193. [DOI] [PubMed] [Google Scholar]
- 19.Gmyl A P, Pilipenko E V, Maslova S V, Belov G A, Agol V I. Functional and genetic plasticities of the poliovirus genome: quasi-infectious RNAs modified in the 5′ untranslated region yield a variety of pseudorevertants. J Virol. 1993;67:6309–6316. doi: 10.1128/jvi.67.10.6309-6316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunert S, Jackson R J. The immediate downstream codon strongly influences the efficiency of utilisation of eukaryotic translation initiation codons. EMBO J. 1994;13:3618–3630. doi: 10.1002/j.1460-2075.1994.tb06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez A L, Denova-Ocampo M, Racaniello V R, del Angel R M. Attenuating mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J Virol. 1997;71:3826–3833. doi: 10.1128/jvi.71.5.3826-3833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haller A A, Nguyen J H, Semler B L. Minimum internal ribosome entry site required for poliovirus infectivity. J Virol. 1993;67:7461–7471. doi: 10.1128/jvi.67.12.7461-7471.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haller A A, Semler B L. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J Virol. 1992;66:5075–5086. doi: 10.1128/jvi.66.8.5075-5086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellen C U T, Witherell G W, Schmid M, Shinm S H, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman M A, Palmenberg A C. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis IRES. J Virol. 1995;69:4399–4406. doi: 10.1128/jvi.69.7.4399-4406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes P J, Horsnell C, Hyypiä T, Stanway G. The coxsackievirus A9 RGD motif is not essential for virus infectivity. J Virol. 1995;69:8035–8040. doi: 10.1128/jvi.69.12.8035-8040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes P J, Stanway G. The 2A proteins of three picornaviruses are related to each other and to the H-rev 107 family of proteins involved in the control of cell proliferation. J Gen Virol. 2000;81:201–207. doi: 10.1099/0022-1317-81-1-201. [DOI] [PubMed] [Google Scholar]
- 28.Hunt S L, Hsuan J J, Totty N, Jackson R J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt S L, Jackson R J. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyypiä T, Horsnell C, Maaronen M, Khan M, Kalkkinen N, Auvinen P, Kinnunen L, Stanway G. A distinct picornavirus group identified by sequence analysis. Proc Natl Acad Sci USA. 1992;89:8847–8851. doi: 10.1073/pnas.89.18.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii T, Shiroki K, Iwai A, Nomoto A. Identification of a new element for RNA replication within the IRES of poliovirus RNA. J Gen Virol. 1999;80:917–920. doi: 10.1099/0022-1317-80-4-917. [DOI] [PubMed] [Google Scholar]
- 32.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson R J, Howell M T, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 34.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang S K, Wimmer E. Cap independent translation of EMCV RNA: structural elements of the IRES and involvement of a cellular 57kDa RNA-binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 37.Jang S K, Pestova T V, Hellen C U T, Witherell G W, Wimmer E. Cap-independent translation of picornavirus RNAs: structures and function of the internal ribosomal entry site. Enzyme. 1990;44:292–309. doi: 10.1159/000468766. [DOI] [PubMed] [Google Scholar]
- 38.Kaminski A, Belsham G J, Jackson R J. Translation of EMCV RNA: parameters influencing the selection of the internal initiation site. EMBO J. 1994;13:1673–1681. doi: 10.1002/j.1460-2075.1994.tb06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaminski A, Jackson R J. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–663. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong W P, Roos R P. Alternative translation initiation site in the DA strain of Theiler's murine encephalomyelitis virus. J Virol. 1991;65:3395–3399. doi: 10.1128/jvi.65.6.3395-3399.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulate translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 42.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhn R, Luz N, Beck E. Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J Virol. 1990;64:4625–4631. doi: 10.1128/jvi.64.10.4625-4631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le S Y, Zuker M. Common structures of the 5′ non-coding RNA in enteroviruses and rhinoviruses: thermodynamical stability and statical significance. J Mol Biol. 1990;216:729–741. doi: 10.1016/0022-2836(90)90395-3. [DOI] [PubMed] [Google Scholar]
- 45.Lemon S M, Honda M. Internal ribosome entry sites within the RNA genome of hepatitis C virus and other flaviviruses. Semin Virol. 1997;8:274–288. [Google Scholar]
- 46.Liu Z W, Carthy C M, Cheung P, Bohunek L, Wilson J E, McManus B M, Yang D C. Structural and functional analysis of the 5′ untranslated region of coxsackievirus B3 RNA: in vivo translational and infectivity studies of full-length mutants. Virology. 1999;265:206–217. doi: 10.1006/viro.1999.0048. [DOI] [PubMed] [Google Scholar]
- 47.Luz N, Beck E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J Virol. 1991;65:6486–6494. doi: 10.1128/jvi.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macejak D J, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular messenger RNA. Nature. 1991;335:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 49.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholson R, Pelletier J, Le S Y, Sonenberg N. Structural and functional analysis of the ribosome-landing pad of poliovirus type 2: in vivo studies. J Virol. 1991;65:1602–1609. doi: 10.1128/jvi.65.11.5886-5894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niepmann M. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and mouth disease virus. FEBS Lett. 1996;388:39–42. doi: 10.1016/0014-5793(96)00509-1. [DOI] [PubMed] [Google Scholar]
- 52.Niepmann M, Petersen A, Meyer K, Beck E. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J Virol. 1997;71:8330–8339. doi: 10.1128/jvi.71.11.8330-8339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niklasson B, Kinnunen L, Hornfeldt B, Benemar C, Horling J, Hedlund O, Matskova L, Hyypiä T, Winberg G. A new picornavirus isolated from bank voles (Clethrionomys glareolus) Virology. 1999;255:86–93. doi: 10.1006/viro.1998.9557. [DOI] [PubMed] [Google Scholar]
- 54.Ohlmann T, Jackson R J. The properties of chimeric picornavirus IRESes show that discrimination between internal translation initiation sites is influenced by the identity of the IRES and not just the context of the AUG codon. RNA. 1999;5:764–778. doi: 10.1017/s1355838299982158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Percy N, Belsham G, Brangwyn J K, Sulivan M, Stone D M, Almond J W. Intracellular modification induced by poliovirus reduce the requirement for structural motifs in the 5′ noncoding region of the genome involved in internal initiation of protein synthesis. J Virol. 1992;66:1695–1701. doi: 10.1128/jvi.66.3.1695-1701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pestova T V, Hellen C U, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate translation. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pilipenko E V, Gmyl A P, Maslova S V, Svitkin Y V, Sinyakov A N, Agol V I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- 59.Pilipenko E V, Blinov V, Chernov B K, Dmitrieva T M, Agol V I. Conservation of the secondary structure elements of the 5′ untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 1989;17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pöyry T, Kinnunen L, Hovi T. Genetic variation in vivo and proposed functional domains of the 5′ noncoding region of poliovirus RNA. J Virol. 1992;66:5313–5319. doi: 10.1128/jvi.66.9.5313-5319.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pulli T, Koivunen E, Hyypiä T. Cell-surface interactions of echovirus 22. J Biol Chem. 1997;272:21176–21180. doi: 10.1074/jbc.272.34.21176. [DOI] [PubMed] [Google Scholar]
- 62.Rust R C, Ochs K, Meyer K, Beck E, Niepmann M. Interaction of eukaryotic initiation factor eIF4B with the internal ribosome entry site of foot-and-mouth disease virus is independent of the polypyrimidine tract-binding protein. J Virol. 1999;73:6111–6113. doi: 10.1128/jvi.73.7.6111-6113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sangar D V, Newton S E, Rowlands D J, Clarke B E. All foot-and-mouth-disease serotypes initiate protein synthesis at two separate AUGs. Nucleic Acids Res. 1987;15:3305–3315. doi: 10.1093/nar/15.8.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seamons R A, Belsham G J. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shaver D N, Barron A L, Karzon D T. Distinctive cytopathology of ECHO viruses types 22 and 23. Proc Soc Exp Biol Med. 1961;106:648–652. [Google Scholar]
- 66.Shiroki K, Ishii T, Aoki T, Kobashi M, Ohka S, Nomoto A. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J Virol. 1995;69:6825–6832. doi: 10.1128/jvi.69.11.6825-6832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanway G, Kalkkinen N, Roivainen M, Ghazi F, Khan M, Smyth M, Meurman O, Hyypiä T. Molecular and biological characteristics of echovirus 22, a representative of a new picornavirus group. J Virol. 1994;68:8232–8238. doi: 10.1128/jvi.68.12.8232-8238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stanway G, Hyypiä T. Parechoviruses. J Virol. 1999;73:5249–5254. doi: 10.1128/jvi.73.7.5249-5254.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanway G, Joki-Korpla P, Hyypiä T. Human parechoviruses—biological and clinical significance. Rev Med Virol. 2000;10:57–69. doi: 10.1002/(sici)1099-1654(200001/02)10:1<57::aid-rmv266>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 70.Stewart S R, Semler B L. RNA determinants of picornavirus cap-independent translation initiation. Semin Virol. 1997;8:242–255. [Google Scholar]
- 71.Todd S, Towner J S, Semler B L. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology. 1997;229:90–97. doi: 10.1006/viro.1996.8416. [DOI] [PubMed] [Google Scholar]
- 72.Walter B L, Nguyen J H C, Ehrenfeld E, Semler B L. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wigand R, Sabin A B. Properties of ECHO types 22, 23 and 24 viruses. Arch Gesante Virusforsch. 1961;11:224–247. doi: 10.1007/BF01241688. [DOI] [PubMed] [Google Scholar]
- 74.Witherell G W, Schultz-Witherell C S, Wimmer E. Cis-acting elements of the EMCV internal ribosomal entry site. Virology. 1995;214:660–663. doi: 10.1006/viro.1995.0081. [DOI] [PubMed] [Google Scholar]