Abstract
Objective:
The aim of this literature review is to yield a comprehensive and exhaustive overview of the existing evidence and up-to-date applications of artificial intelligence for knee osteoarthritis.
Methods:
A literature review was performed by using PubMed, Google Scholar, and IEEE databases for articles published in peer-reviewed journals in 2022. The articles focusing on the use of artificial intelligence in diagnosis and prognosis of knee osteoarthritis and accelerating the image acquisition were selected. For each selected study, the code availability, considered number of patients and knees, imaging type, covariates, grading type of osteoarthritis, models, validation approaches, objectives, and results were reviewed.
Results:
395 articles were screened, and 35 of them were reviewed. Eight articles were based on diagnosis, six on prognosis prediction, three on classification, three on accelerated image acquisition, and 15 on segmentation of knee osteoarthritis. 57% of the articles used MRI, 26% radiography, 6% MRI together with radiography, 6% ultrasonography, and 6% only clinical data. 23% of the articles made the computer codes available for their study, and 26% used clinical data. External validation and nested cross-validation were used in 17% and 14% of articles, respectively.
Conclusions:
The use of artificial intelligence provided a promising potential to enhance the detection and management of knee osteoarthritis. Translating the developed models into clinics is still in the early stages of development. The translation of artificial intelligence models is expected to be further examined in prospective studies to support clinicians in improving routine healthcare practice.
Keywords: Knee osteoarthritis, Artificial intelligence, Machine learning, Deep learning, Literature review
1. Introduction
Osteoarthritis is one of the most common forms of arthritis which affects many joints, yet is widely located in weight-bearing ones and mostly occur in the knee [1]. It is a multifactorial disease leading to joint deformations and severe disability. The increased number of patients having Knee Osteoarthritis (KOA) highlights the importance of early diagnosis of the disease. KOA is mainly diagnosed by using clinical and radiographic changes that emerged from structural deformations. However, current treatment methods including both pharmacological and non-pharmacological ones have limited efficacy for preventing Osteoarthritis (OA) progression [2]. The main radiographic findings of KOA are osteophyte formation, Joint Space Narrowing (JSN), and subchondral sclerosis [3,4]. As the severity of these findings approaches advanced stages, nonsurgical interventions become increasingly ineffective. The disease leads to excruciating pain and mostly a Total Knee Replacement (TKR) is required. Therefore, early diagnosis of the disease could play a crucial role in postponing the TKR [1,3].
The diagnosis of KOA is based on clinically reported symptoms and reports from imaging modalities. The most common clinical symptoms include inflammation, debilitating pain, and functional limitations [5]. Different imaging modalities such as radiography or X-ray, Magnetic Resonance Imaging (MRI), and ultrasonography are used for KOA diagnosis [1,3,6]. Radiography is used to detect structural changes in bone [7], ultra- sonography enables evaluation of the superficial soft tissues [8], while MRI is used to detect soft-tissue alterations, bone marrow lesions, and early osteophytic changes [9]. The diagnosis of KOA is studied by using different imaging modalities together with various Artificial Intelligence (AI) approaches [7,8,10,1].
Deep Learning (DL), a branch of AI, has been widely used in the field of KOA for the last decade [1,11,12]. The performance of DL in medical imaging field has outperformed human readers in many tasks [3]. DL employs automatic feature extraction considering different Convolutional Neural Network (CNN) models [2,11]. Recently, CNN approaches have been used with Transfer learning (TL) scheme, since it reuses the information obtained through pre-trained models as a starting point for a new task, hence decreases the training time [13]. Additionally, using CNN with TL enhances model performance compared to the ones trained without pre-trained information [13].
Using AI applications on KOA has been increased significantly in the last decade [1–3,11,14–20]. In [1], different imaging modalities for traditional OA diagnosis and recent image-based machine learning algorithms were reviewed to discover KOA imaging features for diagnosis and prognosis. Existing literature on KOA imaging modalities, knee joint localization, classification of OA severity, and prediction of disease progression was yielded in a detailed way. However, the study mostly provided the papers published before 2022 in which novel and updated approaches on KOA were investigated and it reviewed only 3 papers from 2022. In [11], the use of machine learning in the clinical care of OA was reviewed. Instead of reviewing each article in detail, the number of patients, ML algorithms, type of data, validation methods, and data availability of the 46 reviewed articles were reported together. While [18] reviewed 23 articles using AI, machine learning, and DL algorithms in Total Knee Arthroplasty (TKA), [14] reviewed 23 articles using AI in diagnosis of KOA and prediction of arthroplasty outcomes providing the prediction outcome, algorithm, statistical performance, strengths, weaknesses, and clinical significance of the papers. The [2] reviewed 31 articles with AI applications in OA including OA classification tasks through imaging and non-imaging-based ML models, analysis of both radiographs for automatic detection of OA severity, and MR images for detection of cartilage/meniscus lesions and cartilage segmentation for automatic T2 quantification by using DL approaches. In [15], 36 articles using DL applications in knee joint imaging were reviewed using a checklist for AI in medical imaging, divided by imaging modality, and characterized by imaging task, data source, algorithm type, and outcome metrics. In [3], segmentation methods allowing the estimation of articular cartilage loss rate ranging from traditional to DL algorithms were reviewed from 30 articles.
Differently from the existing literature, in this review paper, a comprehensive study on AI approaches in KOA was performed. The study was based on 35 papers, 34 of which were published in 2022. It included knee joint detection and segmentation, OA severity classification and disease progression prediction with different imaging modalities, and image acquisition with accelerated MRI. Since the AI field is moving fast compared to traditional fields of research, a review paper including up-to-date algorithms used in the field helps researchers steer future research directions.
2. Methods
A literature search was performed by using PubMed, Google Scholar, and IEEE databases for articles published in peer-reviewed journals, MICCAI 2022, and MIDL 2022 conferences between the 1st of January 2022 and the 31st of December 2022. The keywords of ((knee) AND ((osteoarthritis) OR (arthroplasty)) AND ((artificial intelligence) OR (deep learning) OR (machine learning))) were used for searching the articles. We selected studies focusing on the use of AI in the diagnosis of KOA, prediction of the need for TKR, segmentation and localization of knee structures, and accelerating the image acquisition. Non-English language articles and articles with no English translation were excluded. A reviewer screened the articles for their relevance to the strength of the evidence. Detailed search strategies and selection criteria can be found in Fig. 1. The majority of the articles reviewed in this paper used Osteoarthritis Initiative (OAI) database for KOA prediction, segmentation, and classification [16,11,3]. The Multicenter Osteoarthritis Study (MOST) and Prevention of Knee Osteoarthritis in Overweight Females (PROOF) are the other considered databases [21]. Some articles used these databases separately or ensemble them to compare the performance of their proposed methods [22,23,21]. The number of papers using different database are given in Fig. 2.
Fig. 1.
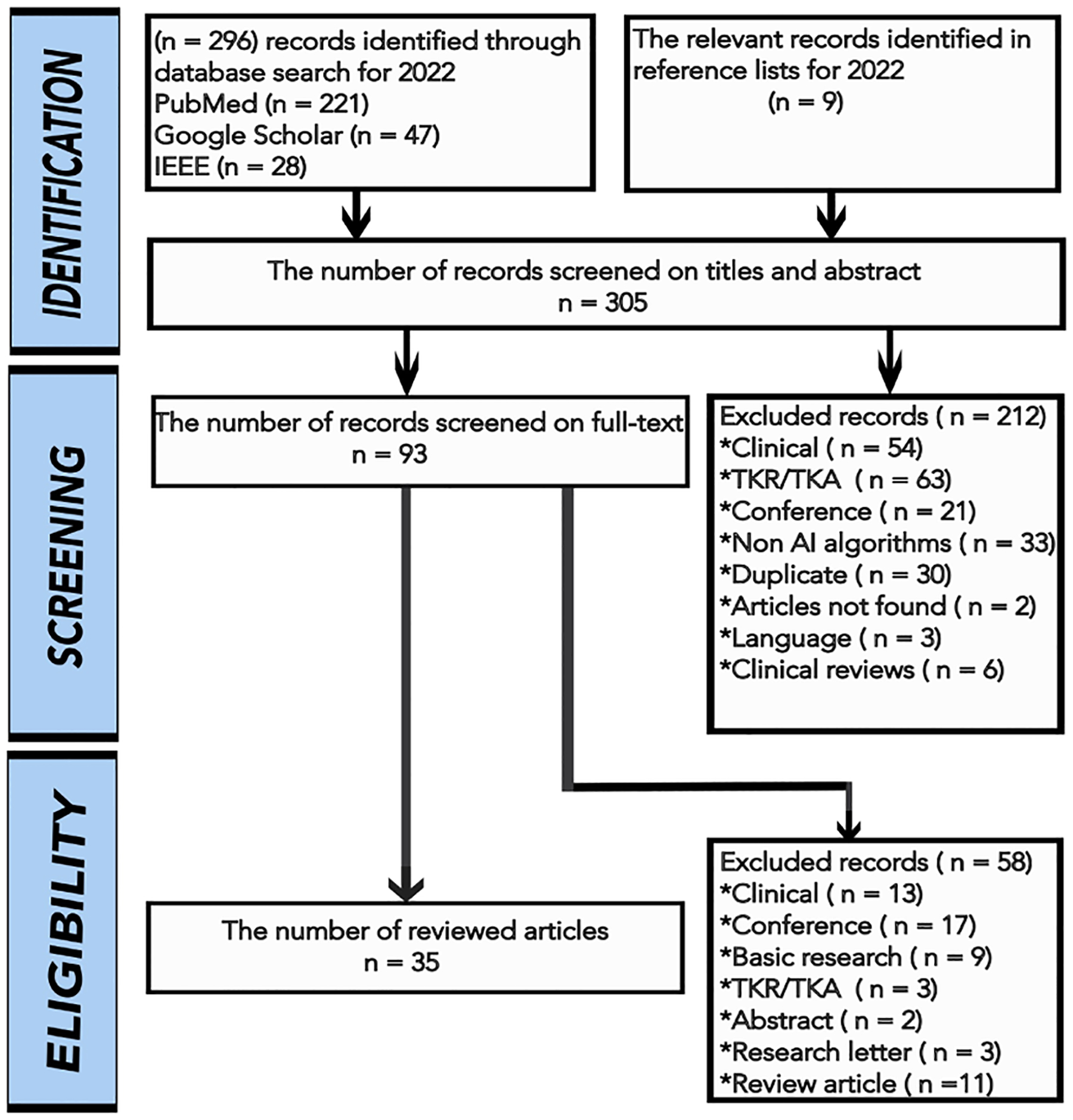
The article selection flowchart with exclusion criteria.
Fig. 2.
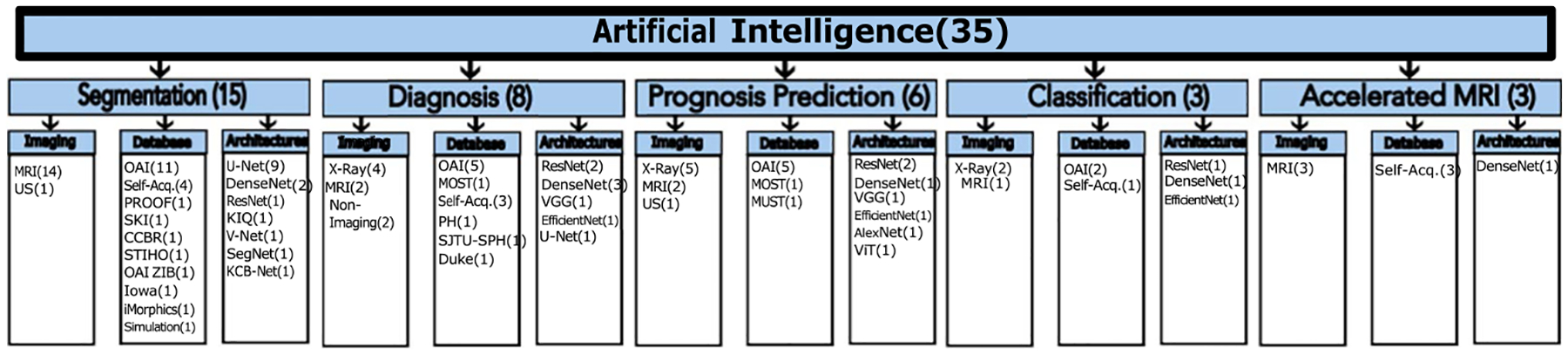
The reviewed articles on KOA prediction using AI algorithms.
3. Imaging of knee osteoarthritis
The knee joint includes various tissues, such as bones, muscles, cartilage, meniscus, tendons, and ligaments which might have complex pathological changes by time [3,15]. KOA is a degenerative joint disease with symptoms of osteophyte formation, cartilage degeneration, JSN, and subchondral sclerosis. Radiograph and MRI of the knee are the most commonly used knee imaging techniques in literature [1].
The knee joint structures are analyzed using different KOA imaging modalities such as radiography, MRI, and ultrasonography [1,24]. Radiography or X-ray is mostly used for the prediction of KOA and the classification of KOA severity, while MRI is the mostly considered for segmentation of knee joints to develop predictive models for KOA (Tables 1 and 2).
Table 1.
The reviewed studies on the diagnosis, prognosis prediction, and classification of KOA using AI algorithms.
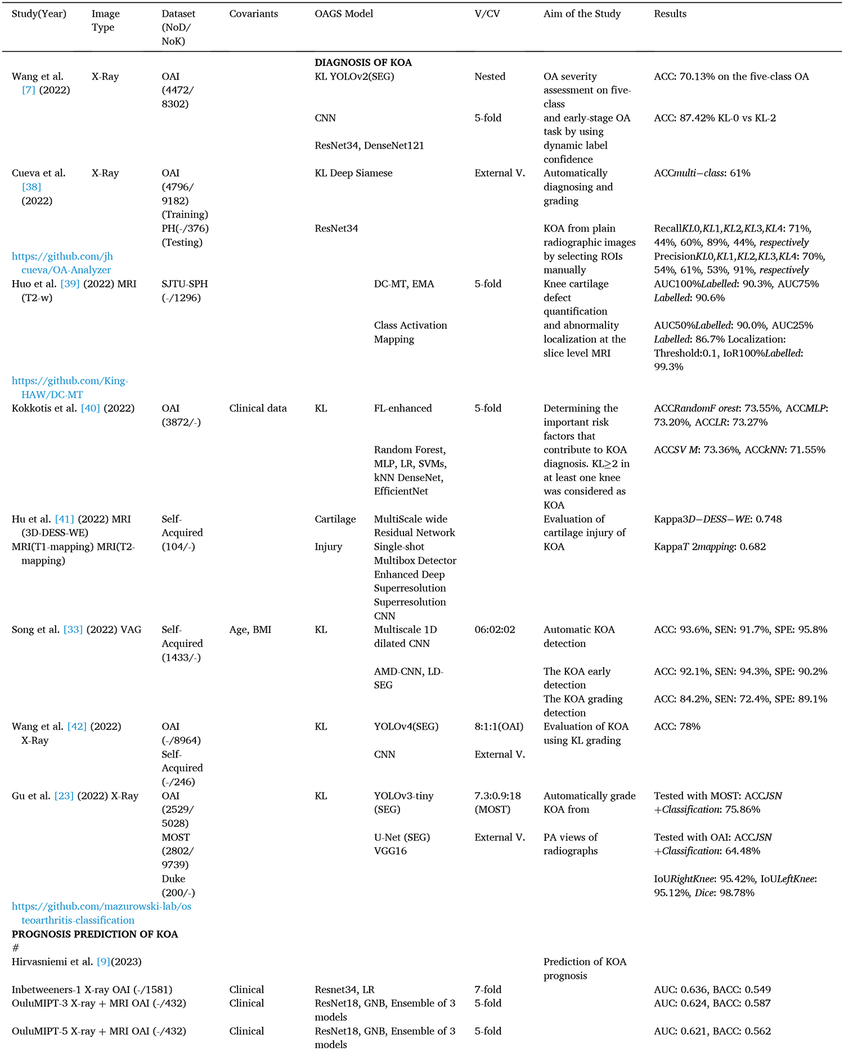

|
Table 2.
The reviewed studies on KOA segmentation/localization and accelerated MRI using AI algorithms.
| Study(Year) | Image Type | Dataset(NoD/NoK) | Model | V/CV | Aim of the Study | Results |
|---|---|---|---|---|---|---|
| SEGMENTATION/LOCALIZATION OF KOA | ||||||
| Desai et al. [45](2021) | MRI (3D DESS) | OAI(88/176) | 6.8:1.6:1.6 | Knee MRI segmentation | ||
| Team1: https://github.com/denizlab/2019_IWOAI_Challenge | 3D U-Net | Dicefemoral: 0.88, Dicetibial: 0.87, Dicepatellar: 0.83, Dicemeniscus: 0.84 | ||||
| Team2: | 3D V-Net+Dropout(30,31)+3D V-Net+Dropout | Dicefemoral: 0.90, Dicetibial: 0.89, Dicepatellar: 0.86, Dicemeniscus: 0.88 | ||||
| Team3: https://github.com/perslev/MultiPlanarUNet | 2D V-Net+Dropout+2D Multiplanar U-Net | Dicefemoral: 0.90, Dicetibial: 0.89, Dicepatellar: 0.85, Dicemeniscus: 0.88 | ||||
| Team4: | 2D DeeplabV3+DenseNet | Dicefemoral: 0.90, Dicetibial: 0.89, Dicepatellar: 0.86, Dicemeniscus: 0.87 | ||||
| Team5: http://github.com/ali-mor/IWOAI_challenge | 2D Encoder-Decoder | Dicefemoral: 0.87, Dicetibial: 0.85, Dicepatellar: 0.81, Dicemeniscus: 0.83 | ||||
| Team6: http://github.com/ad12/DOSMA | 2D U-Net | Dicefemoral: 0.90, Dicetibial: 0.88, Dicepatellar: 0.86, Dicemeniscus: 0.8 | ||||
| Perslev et al. [21](2022) | MRI(3D DESS) | OAI(88/176) | MPUnet | Nested LOO | Evaluating the crosscohort performance | MDCCBRof MPUnet: 0.83, KIQ : 0.81, and U-Net : 0.82 |
| MRI(Turbo 3D T1w) | CCBR(140/140) | 2D U-Net | 25-fold | and robustness of MPUnet, KIQ, and | MDOAIof MPUnet: 0.86, KIQ : 0.84, and U-Net : 0.85 | |
| MRI(3D DESS) | PROOF(25/25) | KIQ | 2D U-Net on three clinical cohorts. | MDPROOFOf MPUnet: 0.78, KIQ : 0.77, and U-Net : 0.73 | ||
| https://github.com/perslev/MultiPlanarUNet | ||||||
| Toit et al. [8](2022) | 3D Ultrasound | Self-Acquired(25/-) | U-Net | 8:2 | Automatically segment the | Dice: 73.1%, Recall: 74.8%, Precision: 72.0% |
| Self-Acquired(4/-) | FAC in 3D ultrasound images | Volume Percent Difference: 10.4, Mean Surface Distance: 0.3 | ||||
| of the knee | Hausdorff Distance: 1.6 | |||||
| Schmidt et al. [46](2022) | MRI(qDESS) | OAI(59/82) | DL(qDESS-trained) | To generalize the automated MRI | DiceqDESS:·0·79–0·93, DiceOAI—DESS: 0·59–0·79 | |
| DL(OAI-DESS-trained) | cartilage segmentation models | CSS(T2)qDESS: 0.75–0–0.98, CSS(T 2)OAI–DESS: 0.35–0.90 | ||||
| across four independent datasets | CSS(Volume)qDESS: 0.47–0.95, CSS(Volume)OAI–dess: 0.13–0.84 | |||||
| Yang et al. [47](2022) | MRI(3D Sagittal DESS) | OAI(-/176) | GAN with TL, UNet | Nested | Automatically segment heterogeneous | GAN with TL, 5-fold CV: Dice: 0.819, Hausdorff: 1.463mm, Distsymmetricsurface: 0.350mm |
| MRI(Sagittal PDw) | Cleveland Clinic (-/30) | GAN without TL | 5-fold | clinical MRI scans | GAN with TL, 5 random Holdout: Dice: 0.801, Hausdorff: 1.746mm, DistSymmetricSurface: 0.364mm | |
| 5 random holdout | ||||||
| Chadoulos et al. [48](2022) | MRI(3D DESS) | OAI(76/-) | SegNet, VoxelMorph | 6:2:2 | Segmentation of knee cartilage using | HyLP: Dice: 88.89%, Precision: 89.86%, Recall: 88.12% |
| RegLP, HyLP | LOO | a two-stage multi-atlas approach built | 5-class: DiceFemoral: 88.22%, DiceTibial: 85.84% | |||
| SSL | under SSL scheme | |||||
| Felfeliya et al. [49](2022) | MRI(3T DESS) | OAI(-/500) | Improved-Mask Rb-CNN | Segmentation and identification of | Mask Rb-CNN: DiceFemur: 95%, DiceTibial: 95%, DiceFemoral:·71%, DiceTibial:81% | |
| MRI(3T STIR) | STIHO (76/-) | ResNet, DenseNet | OA-related tissues and pathologies | Improved-Mask Rb-CNN: DiceFemur: 98%, DiceTibial: 97%, DiceFemoral: 80%, DiceTibial: 82% | ||
| without having imbalanced class problem | Effusion D: DiceRCNN: 71%, DiceMask-RCNN: 72% | |||||
| Peng et al. [50](2022) | MRI(3D) | SKI10(-/150) | KCB-Net | 5-fold | Segmentation 3D knee cartilage | SKI10: DiceFemur: 98.41%, DiceFemoral: 81.67%, DiceTibia: 97.97%, DiceTibial: 78.19% |
| MRI(3D) | OAI ZIB(-/507) | UNet | and bone via sparse annotation | OAI ZIB: DiceFemur:·98.62%, DiceFemoral:·90.24%, DiceTibia: 98.76%, DiceTibial: 87.16% | ||
| MRI(3T DESS) | Iowa(248/−1462-) | using KCB-Net model | Iowa: DiceFemur: 96.47%, DiceFemoral: 86.73%, DiceTibia: 96.49%, DiceTibial: 84.34% | |||
| MRI(3D) | iMorphics(-/176) | iMorphics: DiceFemur: 92.35%, DiceFemoral: 91.27%, DiceTibia: 90.58%, DiceTibial: 89.31% | ||||
| Khan et al. [51](2022) | MRI(3D DESS) | OAI(88/176) | MoBilenet Convolutional | 5-fold | Segmentation precise knee tissue from | DiceFemoral: 91%. DiceTibial: 92% |
| Self-Acquired(−/−) | Unet | MRI using deep collaborative network with alpha matte | SS(Femoral+Tibial+LateralTibial+MedialTibial+Patellar) Cartilage+(Lateral+Medial)Meniscus: 89% | |||
| Li et al. [52](2022) | MRI(3D DESS) | OAI(-/507 | nnU-Net | 5:5 | Automatic articular cartilage segmentation | DiceCombinedHip: 92.1% |
| MRI(dGEMRIC Hip) | Self-acquired(25/-) | 2-fold | using an nnU-Net based two-stage approach for knee and hip datasets | DiceFemoral: 89.8%, DiceTibial: 86.4% | ||
| Zaman et al. [53](2022) | MRI(3D DESS) | OAI(248/1462) | CNN | 4-fold | Segmentation quality assessment by automated | SENKnee:·0.96, SENCalf:·0.96 |
| MRI(3D Calf Muscle) | Self-Acquired(93/175) | U-Net | detection of erroneous surface regions in knee OA and calf muscle datasets | PPVKnee: 0.87, PPVcalf:·0.93 | ||
| Kessler et al. [54](2022) | MRI(3D DESS) | OAI(507/-) | CNN, 3D-CaSM | 9.2:0.4:0.4 | Automatic segmentations for surface-based | 2D U-Net: DiceFemur: 0.97, DiceTibia: 0.97, CartilageThicknessBias:·−0.12 to 0.33mm |
| 2D U-Net | analysis of cartilage morphology | 3D U-Net: DiceFemur: 0.98, DiceTibia: 0.98, CartilageThicknessBias: 0.07 to 0.14mm | ||||
| 3D U-Net | and composition on knee MRI | 2D U-Net: T2 Bias: −0.16 to 1.32ms, 3D U-Net: T2 Bias: −0.05 to 0.46ms | ||||
| Huang et al. [55](2022) | MRI | OAI(4349/21229)(Right) OAI(4343/19915)(Left) Simulations (160/624) |
DADP | LOO | Proposing a DADP framework for longitudinal knee MRI analysis | Dice: 0.81, SEN: 0.98, SPE: 0.95 |
| ACCELERATED MRI | ||||||
| Tolpadi et al. [61](2022) | MRI(Knee) | (244/446) | recurrent U-Net | 6:2:2 | ROI-specific optimization of accelerated | Knee: ErrorR=2 < 10, ErrorR=10 < 9 |
| MRI(Hip) | (67/89) | CNN | 5.8:2.2:2 | acquisitions to improve T2 quantification | Hip: ErrorR=2 < 10, ErrorR=10 < 9 | |
| MRI(LumbarSpine) | (21/24) | DenseNet121(CLS) | 6.2:1.9:1.9 | in knee, hip and lumbar spine MRI | IVD: ErrorR=2toR=6 < 12 | |
| Kim et al. [62](2022) | MRI | (33/-) | SMS | Evaluating diagnostic performance of | κInterreadrr: 0.574–0.838, κIntermethod: 0.755–1.000 | |
| PI | different combination of PI and SMS | Decreased Acquisition Timecomparing2–foldPI: P 2S2: 47%, P 3S2: 62%, P 4S2: 71 | ||||
| acceleration reconstruction in knee MRI | ||||||
| Wang et al. [63](2022) | MRI(Coronal) | (20/-) | 1D CNN, ODLS | 6:0.5:3.5 | Making the deep network much easier | RLNEAF =3 = 0.08, PSNRAF =3 = 36.20dB, SSIMAF=3 = 0.93 |
| MRI(Sagittal) | (20/-) | GRAPPA | to be trained and generalized for accelerated | RLNEAF =4 = 0.10, PSNRAF =4 = 33.60dB, SSIMAF =4 = 0.90 | ||
| MR image reconstruction | RLNEAF =6 = 0.13, PSNRAF =6 = 30.57dB, SSIMAF =6 = 0.85 | |||||
| Qin et al. [56](2022) | MRI(3D DESS) | OAI-ZIB(-/507) | iSegFormer (Transformer Combines | 8:1:1 | Transformer-based interactive image segmentation and its application | Dice3DKneeFemoral: 82.2% |
| https://github.com/uncbiag/iSegFor | Swin Transformer+MLP decoder) | to 3D images | ||||
| Liang et al. [57](2022) | MRI(3D DESS) | OAI-ZIB(-/507) | nnU-Net | 2-fold | Ensuring accurate continuous knee | DiceFemoral: 89.35%, DiceTibial: 86.14% |
| PCAM | cartilage segmentation using a combination | |||||
| https://github.com/LeongDong/PCAMNet | of segmentation network and PCAM | |||||
Different imaging modalities have various KOA grading systems to evaluate the severity of the disease. The Kellgren-Lawrence (KL) and Osteoarthritis Research Society International (OARSI) are mostly used grading systems for radiography to investigate the OA features such as osteophyte, JSN, percentage of JSN, bone end deformity, and subchondral sclerosis [1]. The KL grading system uses five grades to classify KOA severity from the radiographs, where Grade 0 corresponds to the normal knee, and the other grades correspond to the progression of the disease. Grade 0 is normal, Grade 1 shows doubtful signs of OA, Grade 2 is mild OA, Grade 3 is moderate OA, and Grade 4 is severe OA [7]. The Whole-Organ Magnetic Resonance Imaging Scores (WORMS) scoring method incorporates 14 features for evaluating the KOA using MRI [10]. The Knee injury and Osteoarthritis Outcome Score (KOOS) is a self-reported 42-item questionnaire with 5 subscales that evaluates knee health [25]. The MRI Osteoarthritis Knee Score (MOAKS) scoring tool has 14 subregions and individual features for KOA [1]. These are mostly used grading systems for MR imaging modalities to examine cartilage, meniscal tears, bone marrow lesions, anterior cruciate ligament tears, subchondral cysts, bone attrition, effusion synovitis, osteophytes, ligaments, and synovial thickening.
3.1. Radiography (X-Ray)
In a radiograph of an advanced OA joint, a decrease in joint space width is typically observed between the bones of the joint, indicating cartilage degeneration [26,27]. Therefore, the bone starts to rub against the bone which could lead to cysts or fluid-filled cavities [28]. In addition, the radiograph enables visualizations of sclerosis, increased bone density, and bone spurs that are emerged due to joint surface misalignment. Even though radiography has a low cost and it allows bony structure visualization, it is mostly limited to 2D visualization and does not provide soft tissue information, which is important because research shows OA degeneration starts from soft tissues [1,29].
3.2. Magnetic resonance imaging
MR imaging helps to evaluate the structural changes in KOA and to visualize the affected tissues due to OA pathologies, such as soft tissue, subchondral bone, meniscus, and cartilage [30,31]. MRI has been shown to have high specificity for the diagnosis of cartilage deformation, yet low sensitivity for investigation of small abnormalities and injuries [32]. It is widely used as a non-invasive technique for assessing cartilage injuries and allows both 2D and 3D visualizations. However, the diagnostic performance of MRI highly depends on the level of readers expertise. Additionally, knee implants affect the quality of images and it is an expensive modality.
3.3. Ultrasonography
Ultrasonography is a non-invasive diagnostic tool that uses high-frequency sound waves to produce real-time images of the knee joints and other internal structures within the body, including muscles, tendons, blood vessels, and organs [33]. It enables immediate evaluation of soft tissues, such as cartilage and menisci [34,35]. Differently from radiographs, it facilitates a 3D evaluation of the joint without ionizing radiation exposure [36]. Ultrasonography is a non-invasive, fast, operator-dependent, and low-cost imaging modality. It allows the real-time evaluation of the synovium and ligaments. The main limitations of ultrasonography in assessing large joints include its inability to pass through bony structures, difficulty in visualizing deeper structures, and its inability to estimate bone erosion depth [37].
4. Applications of artificial intelligence in knee osteoarthritis
Current diagnosis of KOA mostly relies on evaluation of medical images using KL grading system. Recently, there has been a growing interest in the use of AI for automatic diagnosis, prognosis, segmentation, and classification of KOA to enhance diagnostic accuracy and efficiency [3,14]. AI can help in automatic diagnosis by analyzing large amounts of data to provide diagnoses. The following reviewed studies have represented that AI helps to enhance the performance of KOA diagnosis and prognosis in terms of accuracy and reproducibility compared to human readers.
4.1. Diagnosis of KOA
In [7], to address the ambiguity between adjacent KL grades, particularly in early-stage OA, an investigation was conducted on OA severity assessment utilizing a two-step scheme that employs an object detection CNN to segment knee joint areas. The study focused on the label uncertainty and proposed an approach that enabled the model to learn from the highly confident samples. These samples were characterized by considering an integrated learning technique that fuses label confidence estimation, which focuses on the probability of correctly labeled samples. This estimation process involves preserving the predicted label distribution for all samples in the validation set, calculating the self-confidence for each class, and using this as a threshold to distinguish between highly and low confident samples within the validation set based on their reliability. In order to help CNN learn from two sets accordingly, a hybrid loss function which enhances the effects of highly confident samples and controls the impacts of low confident ones with a weight parameter was proposed. The early-stage OA assessment includes KL-0 vs. KL-1, KL-1 vs. KL-2, and KL-0 vs. KL-2 classifications. A mean accuracy of 70.13% and a Matthews correlation coefficient of 0.5864 were obtained on the five-class OA assessment task. However, the study did not include an examination of whether the use of high-confidence labels introduces bias into the system and whether its accuracy is comparable to that of human readers.
Detection of the OA lesions in the two knees simultaneously was studied by using a deep Siamese CNN model [38]. The last Fully Connected (FC) layer was modified and two extra FC layers were added to employ the transfer learning. The Deep Siamese network was studied in order to identify the similarity metrics between the lateral and medial parts of a cropped knee image. The medial side was horizontally flipped to maintain symmetry with the lateral one. The multi-class accuracy of 61% was obtained by using the proposed model. The precision of 70%, 54%, 61%, 53%, and 91% as well as recall of 71%, 44%, 60%, 89%, and 44% were obtained for KL0, KL1, KL2, KL3, and KL4, respectively. The classification of KL-0, KL-3, and KL-4 classes yielded better precision and recall results than KL-1 and KL-2. It was explained in the study that this is due to the slight differences between KL-1 and KL-2, making it challenging for even specialists to distinguish between KL-1 and KL-0 or KL-2. However, using private data limits the fair comparison of the results with the existing literature ones.
A semi-supervised scheme based on Dual-Consistency Mean Teacher (DC-MT) classification model was proposed to utilize unlabeled data effectively for better assessment of knee cartilage defect grading at the slice level of MR images [39]. An attention loss function was employed to force the network focus on and lie within the cartilage regions, which may provide accurate attention masks and enhance classification performance, simultaneously. An aggregation scheme was employed to ensembles slice-level classification outcomes for identifying the final subject-level diagnosis. The student model was subject to both supervised and unsupervised losses. The supervised loss included classification and attention losses, while the unsupervised loss included classification and attention consistency losses. The teacher model was updated using an exponential moving average strategy based on the student model. The two models were fed with different noise perturbations of the input MR slice and encouraged to be consistent. The class activation mapping method was employed to generate the attention masks that emphasizes the particular regions for the classification network. The study determined the severity of knee cartilage defect and yielded the attention masks for cartilage region localization simultaneously. The proposed method demonstrated an improvement in both the classification and localization of knee cartilage defects when compared to a fully-supervised baseline network trained with labeled data. This improvement was especially notable when using limited labeled data, with the Area Under Curve (AUC) increasing from 81.5% to 86.7% under training with 25% labeled data and 75% unlabeled data.
In order to determine the important risk factors that contribute to KOA diagnosis, a fuzzy logic-based ensemble feature selection method aggregating the output of different feature selection algorithms with several ML models was used [40]. The multidimensional clinical data was utilized. The participants with KL ≥ 2 in at least one of the two knees or in both at baseline were considered as KOA, whereas the ones with KL0 or KL1 grade at baseline were considered as Non-KOA. The impact of risk factors on the diagnosis of KOA was evaluated using a SHapley additive explanations approach. This method, based on game theory’s Shapley Values and local explanations, allowed for the determination of the influence of selected features and the understanding of the decision-making process of the best-performing model. Among the studied risk factors, knee symptoms, risk factors, or both, history of knee surgery, age, Body Mass Index (BMI), and KOOS score were found to be the most important ones in KOA diagnosis, respectively. It was concluded that considering heterogeneous risk factors from various feature categories was required for the effective diagnosis of KOA. However, the study only focused on the clinical parameters and ignored the effects of imaging techniques on KOA diagnosis.
In [41], the cartilage injury assessment of KOA was studied by using an image superresolution algorithm based on an improved multiscale wide residual network model together with several MRI sequences. The performance of the proposed model was compared with the single-shot multibox detector, superresolution CNN, and enhanced deep superresolution algorithms. The proposed algorithm outperformed the compared ones in overall quality, with quantitative analysis showing significantly higher Peak Signal- to-Noise Ratio (PSNR) (38.87 dB) and Structural Similarity Index Measure (SSIM) (0.956) values compared to superresolution CNN (PSNR = 30.41 dB, SSIM = 0.892), single-shot multibox detector (PSNR = 26.11 dB, SSIM = 0.749), and enhanced deep superresolution (PSNR = 27.84 dB, SSIM = 0.788) algorithms. Arthroscopic analysis provided that grade I and grade II lesions concentrated on patella and femoral trochlear and grade III and grade IV lesions gradually developed into the medial and lateral articular cartilage. Among different MRI sequences, the 3D-DESS-WE one yielded the highest diagnostic accuracy of over 95% in grade IV lesions. The Kappa consistency test values were 0.748 and 0.682 for the 3D-DESS-WE and T2 mapping sequences, respectively. The study stated that DL together with MRI could clearly demonstrates the cartilage lesions of KOA. However, the data is self-acquired and is not available with an open-access, but from the corresponding author upon request.
A KOA Computer-Assisted Diagnosis (CAD) using multivariate information, namely Vibroarthrographics (VAGs) signal emitted by human knee joints, age, and BMI, based on a DL model was investigated for the automatic KOA detection, the KOA early detection, and the KOA grading detection [33]. By placing an accelerometer at the articular surface, a VAG signal can be obtained which contains pathological information related to the roughness, softening, and breakdown of cartilage. An Aggregated Multiscale Dilated (AMD)-CNN framework having two parallel pathways was designed to extract features from multivariate information of KOA patients. The features from VAG signals were captured in the first pathway using a multi-scale 1D dilated CNN and the ones from age and BMI signals were obtained in the second pathway using a FC Neural Network (NN). A Laplace distribution-based scheme was used to classify the extracted featFures from the AMD-CNN approach.
The KOA evaluation using KL grading in real-life knee radiographs was investigated by using a CNN model [42]. To assess the model’s performance and interobserver agreement, two orthopedic surgeons and a musculoskeletal radiologist evaluated the OAI testing set and external validation images for KL classification. The quadratic kappa values of the model–surgeon 1, model–surgeon 2, and model–radiologist were 0.80, 0.84, and 0.86, respectively. The model provided the accuracy of 78% which was consistent with interobserver agreement for the OAI knees and external validation knees. A lower interSobserver agreement for the images misclassified by the model was obtained. The surgical candidate detection performance of the model for KL3 and KL4 grades was better than that of specialists with an F1 score of 0.923.
A DL based framework was proposed in [23] to automatically grade KOA from PA views of radiographs. The proposed model has five steps: image preprocessing, localization of knees joints considering the YOLO v3-tiny model, initial evaluation of the severity of OA using a VGG16 based CNN classifier, segmentation of the joints and calculation of the JSN taking into account U-net architecture for bone segmentation, and a combination of the JSN and the initial evaluation to predict the KL grade. When MOST data was used as a test set, the accuracy of 58.86%, 71.93%, and 75.86% were obtained for using only segmentation based JSN without classification model, only classification model without the JSN component, and segmentation based JSN together with classification model, respectively. The accuracy of 64.48% were obtained for segmentation based JSN together with classification model when the OAI data was used as a test set.
4.2. Prognosis prediction for KOA
In [9], the results of seven teams with 23 entries submitted to KNOAP2020 challenge to predict the incident symptomatic radiographic KOA within 78 months on a test set. The ResNet-34 model trained with 1581 knees from the OAI to predict TKR surgery from radiographs together with age, BMI, and KL considering Logistic Regression (LR) prediction method provided the highest AUC of 0.64. Using automatically extracted radiograph and MRI features together with clinical variables yielded the highest Balanced Accuracy (BACC) of 0.59 for the ensemble of three different models, namely model 1 using radiograph, MRI, and clinical variable as modalities, joint shape and space features for radiograph and automatically extracted cartilage features for MRI as feature extraction, and gradient boosting machine as prediction model, model 2 using radiograph as modality, ResNet18 as feature extraction and prediction model, model 3 using radiograph and clinical variable as modalities, joint shape and space features for radiograph as feature extraction, and gradient boosting machine as prediction model.
An Adversarial Evolving Neural Network (A-ENN) with an adversarial training scheme was examined to predict KL grading based on the baseline radiograph observation without using the scans from follow-up visits [43]. As disease level progresses, the Evolving Neural Network (ENN) involves the progression of KOA patterns for accurately addressing the disease through comparing a set of template images of different KL grades. An adversarial training scheme with a discriminator provided how the input image changes to/from templates of each KL grades. A mean square error was used to measure the distance between an estimated template and each ground-truth template in a pixel-wise manner. By using multiple evolving traces and the original input image, the classification of raw longitudinal KOA grading probabilities was performed. A maximum pooling was applied on the KL grade probabilities and they were fused with the input image to predict a longitudinal KL grade. An overall accuracy of 62.7% was obtained.
In [10], an ML-based prediction model was developed for incident radiographic OA of the right knee over 8 years using MRI, demographics, and clinical predictors including muscle strength and symptoms. The radiographic images of participants with KL grade of 0–1 at baseline from OAI were analyzed. Subjects having the same KL grade over 8 years were accepted to be false, whereas subjects having KL grade of 2–4 were defined to be a true set. The prediction performance of three different models was compared: Model 1 with 112 predictors based on OA risk factors and MRI imaging features, namely cartilage T2 and WORMS; Model 2 with top ten predictors based on feature importance score from Model 1 and clinical relevance, and Model 3 with Model 2 without the imaging predictors. Among these three models, the Model 1 performed best with the highest AUC of 0.792.
A DL risk evaluation model analyzing the baseline PA knee radiograph was investigated for predicting the pain progression in subjects with or at risk of KOA [44]. Pain progression was identified as a 9-point or greater increase in Western Ontario and Mc- Master Universities Osteoarthritis (WOMAC) Pain Score (PS) between the baseline and two or more follow-up time points over 48 months. The cascaded two stage DL model consisted of two deep CNNs was considered. The EfficientNet DL model with AUC of 0.77 yielded better diagnostic performance than the DenseNet with AUC of 0.75. The traditional model considering the baseline clinical and imaging risk factors resulted in an AUC of 0.69. A combined joint training model developed by using DL analysis of baseline radiographs with clinical, demographic, and radiographic risk factors provided the highest AUC of 0.81.
The effect of different acquisition modalities and image qualities on the performance of the combined Trabecular Bone Texture (TBT)-Siamese CNN prediction model was investigated and compared to LR technique for prediction the medial JSN progression [22]. The TBT was defined as an imaging descriptor providing information on trabecular bone changes owing to KOA and calculated by a variogram-based method. The Siamese CNN calculated the probability distribution of the KL grades of baseline radiographs. The KOA progression prediction was employed by using of the same KOA progression prediction model validated on independent OAI and MOST datasets, namely training the model with one dataset and testing it with the other, and vice versa. The radiographs were automatically segmented by BoneFinder to identify 16 Region of Interests (ROIs). The KOA progression was evaluated based on an increase in the OARSI medial-JSN grades over 48 months in OAI and 60 months in MOST. The proposed TBT-CNN model predicted JSN progression with and AUC of 0.75 and 0.81 in OAI and MOST, respectively. The prediction ability of TBT-CNN was invariant from the acquisition modality or image quality when the model was trained and tested on the same cohort.
In [35], the prediction of TKR was examined. The predictive performance of ultrasound features such as osteophytes, meniscal extrusion, synovitis in the suprapatellar recess, femoral cartilage thickness, and quality for future KOA surgery were investigated. In the multivariate analysis, five multivariate predictive models were considered, namely Model1 using age, sex, BMI, knee injury, familial OA, and occupational load clinical variables, Model2 using Model1 features and KL grade from radiographic images, Model3 using Model1 features and ultrasound features, Model4 using Model1 features together with ultrasound features and KL grade, Model5 or ensemble model using ensemble of Model2 and Model3 which averaged their output probabilities. The study stated that considering ultrasound features along with clinical variables could predict KOA surgery almost as good as using the KL grading based radiographic images.
4.3. Segmentation and localization
The cross-cohort performance and robustness of the different architectures on three clinical cohorts without manual adaptation of model or optimization of hyperparameters were studied in a retrospective cohort study [21]. Four OA segmentation models were compared. The MPUnet and Knee Imaging Quantification (KIQ) frameworks were applied with default settings across all cohorts. The MPUnet was trained using only the sagittal view, which corresponds to training a 2D U-Net with the MPUnet’s augmentation strategy and training pipeline. The impact of incorporating multiple views was examined. The KIQ method is a validated automatic segmentation technique based on task-specific knowledge. It aligns scans to a reference knee MRI model using rigid multi-atlas registration and computes Gaussian derivative features within regions of interest. Voxel-wise classifications were performed using compartment-specific classifiers and largest connected component analysis selects final segmentation volumes for each compartment. The results of the study represented that the MPUnet matched or exceeded the performance of KIQ and 2D U-Net on all compartments across three cohorts and no manual tuning was required for MPUnet.
In [45], the results of six networks to address the semantic and clinical efficacy of automatic segmentation models for identifying OA progression was provided. Different DL models were assessed and compared for segmenting articular (femoral, tibial, and patellar) cartilage and the meniscus. The segmentation metrics of all tissues (P=.99) were similar for the four top-performing networks. The Dice coefficient correlations between network pairs were high (>0.85), per-scan thickness errors were negligible among networks Team1–Team4 (P=.99), and longitudinal changes represented minimal bias (<0.03mm).
The correlation between segmentation metrics and thickness error was low (p<0.41). The result of majority-vote ensemble and top-performing networks was similar (P=.99). Empirical upper-bound performances were similar for both combinations (P=.99).
A 3D reconstruction was analyzed to develop and validate an automatic Femoral Articular Cartilage (FAC) segmentation framework for 3D ultrasound knee data of KOA patients and healthy volunteers was analyzed in [8]. In order to monitor the femoral trochlea cartilage, the proposed scheme included the DL model predictions on the 2D ultrasound slices sampled in the transverse plane followed by reconstruction into a 3D surface. Considering 3D reconstruction decreased the prediction performance comparing to using 2D predictions only, yet the difference was not statistically significant. The effects of using DL approaches to systematically evaluate generalizability of auto- mated MRI cartilage segmentation models across four independent datasets differ in MR vendor and model, subject population, and image acquisition parameters were examined [46]. The clinically related cartilage morphometry and relaxometry metrics besides the traditional segmentation ones were used in the study. It is concluded that quantitative Double-Echo Steady-State (qDESS)-trained models generalize well to independent qDESS datasets regardless of MR scanner type, MR scan parameters, and subject population.
In [47], a conditional Generative Adversarial Network (GAN) based DL model was studied. The segmentation of heterogeneous clinical MRI scans was performed by optimizing a pre-trained model built from a homogeneous research dataset with a TL algorithm. The study considered the TL model with a U-Net architecture as the generator and a CNN as the discriminator. The model with TL outperformed that of without TL and performed similarly as the manual segmentation.
The knee cartilage segmentation using a two-stage multi-atlas segmentation approach together with a Semi-Supervised Learning (SSL) scheme was investigated [48]. The target voxels were labeled by using the spectral information of globally sampled images from the target data and their spatially correspondent images obtained from the atlases. The proposed framework consisted of sparse reconstructions of voxels from linear neighbor- hoods, HOG feature descriptors of regions, and label propagation through sparse graph constructions. The performance of the proposed segmentation approach outperformed the existing patch-based methods and the average execution time computational load of the propose method decreased by more than 70% compared to the other considered patch-based ones. Additionally, using a 5-class problem which split the bone and cartilage components of 3-class one into their respective femoral and tibial parts yielded that the proposed approach provided comparable performance to the existing state-of-the-art knee cartilage segmentation methods.
In [49], an improved Mask Rb-CNN approach, which is an instance segmentation framework that distinguishes individual objects of interest such as different anatomical structures (e.g. bone and cartilage) was studied. The Mask Rb-CNN model was modified to enhance the segmentation accuracy around the instance edges. It could be applicable for various tissue scales, pathologies, and MRI sequences related to OA, overcoming the imbalanced class problem. The improved-mask Rb-CNN model consisted of an extra ROIAligned block, an additional decoder block in the segmentation header, and connecting them considering a skip connection. A high agreement between the two readers were obtained for both Mask Rb-CNN and improved-mask Rb-CNN models.
A framework called KCB-Net was introduced to segment cartilages and bones in 3D knee joint MR images with sparse annotation [50]. Each 2D slice in an unlabeled training set of 3D images was initially encoded into a feature vector in an unsupervised manner. Subsets of diverse image slices were selected for expert annotations by ranking the most information-contributing slices highest. This allowed image segmentation models to be trained using annotations with a high-sparsity ratio. In the proposed framework, an ensemble of three 2D segmentation modules and one 3D module that incorporated features from multiple scales with edge-sensitive branches was utilized to produce pseudo-labels for the un-annotated slices. These pseudo-labels were subsequently employed to re-train the 3D model for several iterations. The final segmentation results were produced by post-processing the feature maps generated by the ensemble model. The primal–dual internal point method was used for fine-tuning the segmentation results. The study stated that the framework on full annotations outperformed the state-of-the-art methods and provided high quality outputs for small annotation ratios down to 10%.
The knee tissue segmentation model using a deep collaborative method including an encoder and decoder-based segmentation network together with a low rank tensor- reconstructed one was examined [51]. The segmentation network was built in two paths where the first one might be unsuccessful and overlook the boundary patches, the second one having low rank reconstructed input mitigated the impacts of this failure. The features from both the source and their low rank reconstruction images were extracted by using the proposed method.
An automatic articular cartilage segmentation method using an nnU-Net based two- stage approach was examined in [52]. The outputs of the first stage were considered to calculate the intermediate features for cartilage segmentation refinement at the second stage. At the first stage, nnU-Net was used to capture hard tissue and articular cartilage segmentation, and distance and entropy maps were calculated to encode uncertainty in the initial cartilage segmentation. These maps were then concatenated with the original image at the second stage. A sub-volume around the cartilage was cropped using the initial segmentation approach and used as input for another nnU-Net for segmentation refinement. In [53], the erroneous regions on the boundary surfaces of segmented objects were addressed by using a DL framework for the quality control and segmentation evaluation. A CNN model was examined to capture the image features on the boundaries of multi-objects which may be considered to determine the location-based inaccurate segmentation.
The performance of a surface-based examination on cartilage morphology was studied [54]. To capture the precise regional analyses of cartilage morphology and relaxometry, a U-Net based CNN generated knee MRI segmentation in the 3D Cartilage Surface Mapping (3D-CaSM) method was considered. The performance of segmentation from 3D U-Net provided higher accuracy in the extracted thickness and T2 features using 3D-CaSM compared to that from 2D U-Net.
In [55], a Dynamic Abnormality Detection and Progression (DADP) framework for longitudinal knee MRI analyzes was studied to overcome the lack of building spatial- temporal correspondences and correlations in cartilage thickness as well as the spatio-temporal heterogeneity in cartilage lesions. The 2D cartilage thickness maps were extracted from 3D images and the spatial correspondences were built on these 2D flattened data across the time within each data and across all data. A dynamic functional mixed- effects model was proposed to simultaneously differentiate individual cartilage lesions on MRI data at baseline, 12 months, 24 months, and 48 months for dynamic abnormality detection and progression. The study concluded that the proposed DADP model detected the subject-specific dynamic cartilage regions, effectively and yielded population-level statistical disease mapping and subgroup analysis.
The development of Transformer-based interactive image segmentation approaches for 3D MR images was examined in [56]. The study aimed to address the limited memory capacity of computationally demanding Transformers by proposing a memory-efficient Transformer named iSegFormer. This Transformer combined a Swin Transformer with a lightweight Multi-Layer Perception (MLP) decoder. To overcome the limited availability of labels for 3D MR images, the iSegFormer was pre-trained using a large amount of unlabeled data before being fine-tuned with a limited number of segmented 2D slices. A pre-existing segmentation propagation model, which was pretrained on videos, was considered to propagate the 2D segmentations obtained by iSegFormer to unsegmented slices in 3D images. The iSegFormer was trained with 507 3D MR images from OAI-ZIB dataset. Three segmented slices per image (40, 80, and 120) resulted in 1521 training, 150 validation, and 150 testing slices. A Dice score of 82.2% was obtained for 3D knee cartilage segmentation when the 2D interactive segmentations of 5 slices were extended to other unprocessed ones within the same 3D volume.
To ensure accurate segmentation of knee cartilage on a continuous basis, a Position- prior Clustering-based Self-attention Module (PCAM) was proposed [57]. The self-attention method was used to collect long-range contextual information inaccurate dis- continuous segmentation caused by the limited receptive field in CNNs. To estimate class centers, the clustering-based method was considered, which promoted intra-class consistency and improved segmentation accuracy. The position-prior enhanced the precision of center estimation through excluding false positives from the side-output. The proposed PCAM can be integrated into any segmentation NN with an encoder-decoder structure. The experiments were performed using 507 3D MR images from OAI-ZIB dataset. The Dice score of 89.35% and 86.14% were obtained for femoral and tibial cartilage, respectively.
4.4. Classification
The viability of a knee arthroplasty prediction model using 3 different radiograph views (anterior-posterior, lateral, and sunrise) was evaluated to determine whether a patient of KOA requires a TKA, Unicompartmental Knee Arthroplasty (UKA), or No Surgery [58]. The study suggested that the machine learning approach represented viability for predicting which patients are candidates for a UKA, TKA, or no surgical intervention. An accuracy of 87.8% on the holdout test set and a quadratic Cohen’s kappa score of 0.81 were obtained.
In [59], two GANs that can generate an unlimited number of KOA radiographs at different KL grades was proposed. The KL0 and KL1 grade images were merged into the KL01 class, whereas KL2, KL3, and KL4 ones were merged into the KL234 class. In [59], 320.000 DeepFake images were generated synthetically from 5.556 knee joints radiographs with varying osteoarthritis severity using the GAN approach. The experts were asked to classify 30 real and 30 DeepFake images. The rate of classifying DeepFakes images as real ones was higher than classifying real images as DeepFakes ones.
The performance of ResNet-50, DenseNet-121, and Convolutional Variational AutoEncoder (CVAE) DL models were investigated to predict KOA incidence within 24 months [60]. To capture the ROI containing the knee joint from the IW-TSE sequence, a DL model was used to segment bone on a SAG/3D/DESS/WE sequence. After IW-TSE and DESS sequences registration, the DESS segmentations were applied to the related IW-TSE scans. Combining the patient data with MRI-based features improved the performance.
4.5. Accelerated MRI
Accelerated quantitative imaging using AI is a novel topic with great promise in MSK imaging field. Recent reconstruction approaches using Parallel Imaging (PI) and compressed sensing have been shown to yield decreased acquisition time while maintaining acceptable image quality at a low acceleration rate compared to MRI. In [61], a ROI- specific optimization of accelerated acquisitions was examined. The considered DL model yielded T2 maps in knee and hip cartilage, and lumbar spine IVDs from accelerated T2- prepared snapshot gradient-echo acquisitions and InterVertebral Disc (IVD) performance with a 4-component loss function. The gray level co-occurrence matrix-based approach provided better results than the considered state-of-the-art models for the knee and hip pipelines and retained smooth textures for the most acceleration factors R as well as sharper ones through moderate acceleration factors. The study concluded that the proposed approach provided robust T2 maps and better preservation of small and clinically related features.
The diagnostic performance of different combinations of Simultaneous Multi-Slice (SMS) and PI approaches considering DL models for reconstruction was compared in [62]. The edge rise distance and noise power were calculated for quantitative evaluations of image sharpness and noise, respectively. The diagnostic performance of overall image quality and internal knee derangement were similar for 2-fold PI with 2-fold SMS acceleration (P2S2), P3S2, and P4S2 protocols. While the edge rise distance values were similar for DL protocols and conventional imaging, the noise power ones considering the DL protocols were importantly lower than conventional imaging. The study stated that using DL-enhanced 8-fold acceleration imaging, 4-fold PI with 2-fold SMS, provided comparable results with considering conventional 2-fold PI for the assessment of internal knee derangement, with a 71% decrease in acquisition time.
In [63], a 1D convolution was used to make the deep network memory-efficient, easier to be trained and generalized. The 1D CNN model was built through unrolling the iteration procedure of a low-rank and sparse reconstruction model that searches the coil correlation and image sparsity, respectively. The experimental results provided that One-dimensional Deep Low-rank and Sparse Network (ODLS) is robust to different undersampling scenarios and some mismatches between the training and test data. The Relative L2 Norm Error (RLNE) of 0.08 was obtained for Acceleration Factors (AF)=3.
5. Discussions and future directions
AI has been widely used for segmentation, prediction, and classification of KOA using radiographs and MRI imaging modalities. These models aim to diagnose the KOA at its early stages and prevent or postpone the TKR. Radiograph was used to classify the KOA and predict KOA progression based on KL grading system, and MRI was used to localize the knee joints and predict knee pain. Clinical and demographic variables, pain level, gait performances were also used for KOA prediction. Recent studies have started to consider the simultaneous analysis of radiographs or MRI, along with clinical and demographic variables, to predict the progression of KOA [9]. In order to develop an automated tool for assisting clinicians in evaluating the severity of KOA, it is expected to be more informative to facilitate all available patient data coherently.
The preliminary results of DL on fully-automated segmentation of knee cartilage and bone have shown reasonable potential in terms of generalizability and robustness. Using DL methods for segmentation reduced the segmentation time compared to model- and atlas-based approaches. Moreover, DL has been shown to identify OA phenotypes, predict KOA severity, and identify the KOA progression from mild to severe. In the near future, DL models that incorporate more than a single imaging modality and patient data from multiple time points are expected to be studied broadly to improve the performance of DL in KOA.
DL image reconstruction has been used to accelerate MRI by significantly reducing the scan time required for MRI examinations without compromising image quality or diagnostic accuracy. Reducing the duration of MRI acquisition might decrease discomfort of a patient and the costs of patient care. The existing studies have shown a high degree of interchangeability between standard and accelerated images.
DL models have gained popularity owing to their ability to accurately model complex data and make predictions. Currently, many users viewing DL models as a black box, a model whose inputs and outputs can be observed, yet whose underlying processes are not well understood. This lack of understanding can limit interpretability and explainability. Additionally, DL models also have high generalization power when trained on large datasets, raising questions about performance and data acquisition. Recent advancements in computer science that involve the use of visualization tools open new avenues to better understand the underlying learning behavior of DL models. Even though these visualization tools provide general information about the decision task, they have not been able to provide one-to-one correspondence on how a decision was performed based on the inputs. This is an active area of research in the AI field. Further studies need to be performed to better understand how the decision process takes place for predicting KOA-related tasks.
A large portion of the articles reviewed did not validate developed models in an external validation set. External validation is a process that evaluates the performance of an AI model using a test dataset that was not part of the training data. Without external validation, it remains uncertain how well the model will perform on new data from different populations or healthcare settings. Hence, external validation sets should be used to identify the generalizability of the developed models. Considering external validation can enhance the reliability of AI models used for KOA. Moreover, some of the articles did not provide source code, and the information about cohorts or how to access the private dataset was missing. In order to compare the performance of a predictive model with previous approaches, it is important to share cohorts, trained model weights, model parameters and source codes with other researchers. Future studies need to ensure the reproducibility of the presented approaches. This will enable fast validation of the original results and fast progress to the next phase of KOA research.
With the help of observational datasets, such as OAI and MOST, model development for KOA has been studied extensively with well-structured data. Due to a lack of new data, the researchers have not been able to study the effect of current improvements on imaging devices and changes in the imaging parameters on the DL model performance effectively. In order to incorporate DL models in clinics, the generalizability of these models need to be validated on the current imaging protocols/devices as well as on the patient cohorts understudied in the available datasets. Currently, the development of effective AI models may require the use of large datasets to obtain high generalization power. However, acquisition and annotation of these datasets and the availability of high-quality data for training AI models may pose significant challenges. Generating synthetic data, augmentation, or leveraging existing datasets through transfer learning have been used as solutions to address these challenges [47,38,39]. Moreover, new era of DL approaches using self/un-supervised pretraining opens new venues for improving the generalization of DL models in limited data regime [64].
Acknowledgments
This study is supported in part by the National Institutes of Health (R01 AR074453).
Abbreviations:
- A-ENN
adversarial evolving neural network
- AF
acceleration factors
- AI
artificial intelligence
- AMD
aggregated multiscale dilated
- AP
average precision
- AUC
area under curve
- BACC
balanced accuracy
- BMI
body mass index
- CAD
computer-assisted diagnosis
- CLS
classification
- CNN
convolutional neural network
- CV
cross-validation
- CVAE
convolutional variational autoEncoder
- DADP
dynamic abnormality detection and progression
- DC-MT
dual-consistency mean teacher
- dGEMRIC
delayed gadolinium-enhanced MRI of cartilage
- DL
deep learning
- EMA
exponential moving average
- ENN
evolving neural network
- FAC
femoral articular cartilage
- FC
fully connected
- FLASH
fast low-angle shoot
- FSE/TSE
fast or turbo spin echo
- GAN
generative adversarial network
- GNB
gaussian naive Bayesian
- HyLP
hybrid label propagation
- IoR
intersection over detected region
- IoU
intersection over union
- IVD
interVertebral disc
- JSN
joint space narrowing
- KIQ
knee imaging quantification
- KL
Kellgren-lawrence
- kNN
k-nearest neighbors
- KOA
Knee Osteoarthritis
- KOOS
Knee injury and Osteoarthritis Outcome Score
- LOO
leave-one-out
- LR
logistic regression
- MLP
multilayer perception
- MOAKS
MRI osteoarthritis knee score
- MOST
multicenter osteoarthritis study
- MRI
Magnetic Resonance Imaging
- MSK
MusculoSKeletal
- MUST
musculoskeletal pain in ullensaker study
- NN
neural network
- NoD
Number of data/image
- NoK
number of knee
- OA
osteoarthritis
- OAGS
osteoarthritis grading system
- OAI
osteoarthritis initiative
- OARSI
osteoarthritis research society international
- ODLS
one-dimensional deep low-rank and sparse network
- PCAM
position-prior clustering-based self-attention module
- PH
private hospital
- PI
Parallel imaging
- PROOF
prevention of knee osteoarthritis in overweight females
- PS
pain score
- PSNR
peak signal-to-noise ratio
- PPV
positive predictive values
- P2S2
2-fold PI with 2-fold SMS acceleration
- qDESS
quantitative double-echo steady-State
- Rb-CNN
region-based convolutional neural networks
- RegLP
regional label propagation
- RLNE
relative L2 norm error
- ROI
region of interest
- SEG
segmentation
- SJTU-SPH
Shanghai Jiao Tong University Affiliated Sixth People’s Hospital
- SMS
simultaneous multi-slice
- SSIM
structural similarity index measure
- SSL
semi-supervised learning
- SVM
support-vector machines
- TBT
trabecular bone texture
- TKA
total knee arthroplasty
- TKR
total knee replacement
- TL
transfer learning
- UKA
unicompartmental knee arthroplasty
- VAG
VibroArthroGraphic
- VGG
visual geometry group
- ViT
visual transformer
- WOMAC
western ontario and McMaster universities osteoarthritis
- ZIB
Zuse Institute Berlin
- 3D-CaSM
3D cartilage surface Mapping.
Footnotes
Declaration of Competing Interest
No conflict of interest to declare for any of the authors.
References
- [1].Teoh YX, Lai KW, Usman J, Goh SL, Mohafez H, Hasikin K, Qian P, Jiang Y, Zhang Y, Dhanalakshmi S, Discovering knee osteoarthritis imaging features for diagnosis and prognosis: review of manual imaging grading and machine learning approaches, J. Healthc. Eng 2022 (2022), 4138666, 10.1155/2022/4138666. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [2].Joseph GB, McCulloch CE, Sohn JH, Pedoia V, Majumdar S, Link TM, Ai msk clinical applications: cartilage and osteoarthritis, Skeletal Radiol. 51 (2) (2022) 331–343, 10.1007/s00256-021-03909-2. [DOI] [PubMed] [Google Scholar]
- [3].Ahmed SM, Mstafa RJ, A comprehensive survey on bone segmentation techniques in knee osteoarthritis research: from conventional methods to deep learning, Diagnostics 12 (3) (2022), 10.3390/diagnostics12030611. https://www.mdpi.com/2075-4418/12/3/611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tang X, et al. , Fully automatic knee joint segmentation and quantitative analysis for osteoarthritis from magnetic resonance (mr) images using a deep learning model, Med. Sci. Monit. Int. Med. J. Exp. Clin. Res (2022). https://pubmed.ncbi.nlm.nih.gov/35698440/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hunter DJ, Bierma-Zeinstra S, Osteoarthritis, Lancet 393 (10182) (2019. April 27) 1745–1759, 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- [6].Eckstein F, Chaudhari AS, Fuerst D, Gaisberger M, Kemnitz J, Baumgartner CF, Konukoglu E, Hunter DJ, Wirth W, Detection of differences in longitudinal cartilage thickness loss using a deep-learning automated segmentation algorithm: data from the foundation for the national institutes of health biomarkers study of the osteoarthritis initiative, Arthr. Care Res. (Hoboken) 74 (6) (2022) 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang Y, Bi Z, Xie Y, Wu T, Zeng X, Chen S, Zhou D, Learning from highly confident samples for automatic knee osteoarthritis severity assessment: Data from the osteoarthritis initiative, IEEE J. Biomed. Health Inform 26 (3) (2022) 1239–1250, 10.1109/JBHI.2021.3102090. [DOI] [PubMed] [Google Scholar]
- [8].du Toit C, Orlando N, Papernick S, Dima R, Gyacskov I, Fenster A, Automatic femoral articular cartilage segmentation using deep learning in three-dimensional ultrasound images of the knee, Osteoarthr. Cartil. Open 4 (3) (2022), 100290, 10.1016/j.ocarto.2022.100290. https://www.sciencedirect.com/science/article/pii/S2665913122000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hirvasniemi J, Runhaar J, van der Heijden R, Zokaeinikoo M, Yang M, Li X, Tan J, Rajamo- han H, Zhou Y, Deniz C, Caliva F, Iriondo C, Lee J, Liu F, Martinez A, Namiri N, Pedoia V, Panfilov E, Bayramoglu N, Nguyen H, Nieminen M, Saarakkala S, Tiulpin A, Lin E, Li A, Li V, Dam E, Chaudhari A, Kijowski R, Bierma-Zeinstra S, Oei E, Klein S, The knee osteoarthritis prediction (knoap2020) challenge: an image analysis challenge to predict incident symptomatic radiographic knee osteoarthritis from mri and x-ray images, Osteoarthr. Cartil 31 (1) (2023) 115–125, 10.1016/j.joca.2022.10.001. https://www.sciencedirect.com/science/article/pii/S1063458422008640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Joseph G, McCulloch C, Nevitt M, Link T, Sohn J, Machine learning to predict incident radiographic knee osteoarthritis over 8 years using combined mr imaging features, demographics, and clinical factors: data from the osteoarthritis initiative, Osteoarthr. Cartil 30 (2) (2022) 270–279, 10.1016/j.joca.2021.11.007. https://www.sciencedirect.com/science/article/pii/S1063458421009675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Binvignat M, Pedoia V, Butte AJ, Louati K, Klatzmann D, Berenbaum F, Mariotti- Ferrandiz E, Sellam J, Use of machine learning in osteoarthritis research: a systematic literature review, RMD Open 8 (1) (2022), 10.1136/rmdopen-2021-001998 arXiv: h*t*t*p*s://rmdopen.bmj.com/content/8/1/e001998.full.pdf, https://rmdopen.bmj.com/content/8/1/e001998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brejnebøl MW, Hansen P, Nybing JU, Bachmann R, Ratjen U, Hansen IV, Lenskjold A, Axelsen M, Lundemann M, Boesen M, External validation of an artificial intelligence tool for radiographic knee osteoarthritis severity classification, Eur. J. Radiol 150 (2022), 110249, 10.1016/j.ejrad.2022.110249. https://www.sciencedirect.com/science/article/pii/S0720048X22000997. [DOI] [PubMed] [Google Scholar]
- [13].LeCun Y, Bengio Y, Hinton G, Deep learning, Nature 521 (7553) (2015) 436–444, 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- [14].Lee LS, Chan PK, Wen C, Fung WC, Cheung A, Chan VWK, Cheung MH, Fu H, Yan CH, Chiu KY, Artificial intelligence in diagnosis of knee osteoarthritis and prediction of arthroplasty outcomes: a review, Arthroplasty 4 (1) (2022) 16, 10.1186/s42836-022-00118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Si L, Zhong J, Huo J, Xuan K, Zhuang Z, Hu Y, Wang Q, Zhang H, Yao W, Deep learning in knee imaging: a systematic review utilizing a checklist for artificial intelligence in medical imaging (claim), Eur. Radiol 32 (2) (2022) 1353–1361, 10.1007/s00330-021-08190-4. [DOI] [PubMed] [Google Scholar]
- [16].Banjar M, Horiuchi S, Gedeon DN, Yoshioka H, Review of quantitative knee articular cartilage MR imaging, Magn. Reson. Med. Sci 21 (1) (2022) 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Buchlak QD, Clair J, Esmaili N, Barmare A, Chandrasekaran S, Clinical outcomes associated with robotic and computer-navigated total knee arthroplasty: a machine learning-augmented systematic review, Eur. J. Orthopaedic Surg. Traumatol 32 (5) (2022) 915–931, 10.1007/s00590-021-03059-0. [DOI] [PubMed] [Google Scholar]
- [18].Rodŕıguez-Merchán EC, The current role of the virtual elements of artificial intelligence in total knee arthroplasty, EFORT Open Rev 7 (7) (2022) 491–497, 10.1530/EOR-21-0107. https://eor.bioscientifica.com/view/journals/eor/7/7/EOR-21-0107.xml. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Almhdie-Imjabbar A, Toumi H, Lespessailles E, Radiographic biomarkers for knee osteoarthritis: a narrative review, Life 13 (1) (2023), 10.3390/life13010237. https://www.mdpi.com/2075-1729/13/1/237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hinterwimmer F, Lazic I, Suren C, Hirschmann MT, Pohlig F, Rueckert D, Burgkart R, von Eisenhart-Rothe R, Machine learning in knee arthroplasty: specific data are key—a systematic review, Knee Surg. Sports Traumatol. Arthrosc 30 (2) (2022) 376–388, 10.1007/s00167-021-06848-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Perslev M, Pai A, Runhaar J, Igel C, Dam EB, Cross-cohort automatic knee mri seg- mentation with multi-planar u-nets, J. Magn. Reson. Imaging 55 (6) (2022) 1650–1663, 10.1002/jmri.27978, arXiv:h*t*t*p*s://onlinelibrary.wiley.com/doi/pdf/10.1002/jmri.27978, https://onlinelibrary.wiley.com/doi/abs/10.1002/jmri.27978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Almhdie-Imjabbar A, Nguyen KL, Toumi H, Jennane R, Lespessailles E, Prediction of knee osteoarthritis progression using radiological descriptors obtained from bone texture analysis and siamese neural networks: data from oai and most cohorts, Arthr. Res. Ther 24 (1) (2022) 66, 10.1186/s13075-022-02743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gu H, Li K, Colglazier RJ, Yang J, Lebhar M, O’Donnell J, Jiranek WA, Mather RC, French RJ, Said N, Zhang J, Park C, Mazurowski MA, Knee arthritis severity measurement using deep learning: a publicly available algorithm with a multi-institutional validation showing radiologist-level performance (2022). doi: 10.48550/ARXIV.2203.08914. https://arxiv.org/abs/2203.08914. [DOI] [Google Scholar]
- [24].Jerban S, Oei EHG, Ding J, Editorial: cartilage assessment using quantitative mri, Front. Endocrinol 13 (2022), 10.3389/fendo.2022.1092354. https://www.frontiersin.org/articles/10.3389/fendo.2022.1092354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Roemer FW, Jansen M, Marijnissen ACA, Guermazi A, Heiss R, Maschek S, Lalande A, Blanco FJ, Berenbaum F, van de Stadt LA, Kloppenburg M, Haugen IK, Ladel CH, Bacardit J, Wisser A, Eckstein F, Lafeber FPJG, Weinans HH, Wirth W, Structural tissue damage and 24-month progression of semi-quantitative mri biomarkers of knee osteoarthritis in the imi-approach cohort, BMC Musculoskelet. Disord 23 (1) (2022) 988, 10.1186/s12891-022-05926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Swagerty DL Jr, Hellinger D, Radiographic assessment of osteoarthritis, Am. Fam. Phys 64 (2) (2001) 279–286. [PubMed] [Google Scholar]
- [27].Pelsma ICM, Kroon HM, van Trigt VR, Pereira AM, Kloppenburg M, Biermasz NR, Claessen KMJA, Clinical and radiographic assessment of peripheral joints in controlled acromegaly, Pituitary 25 (4) (2022) 622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chitnavis J, Maghsoudi D, Grewal H, Bilaterally symmetrical ganglion and subchondral cysts of the knee: a case report, J. Surg. Case Rep (3) (2022), 10.1093/jscr/rjac064 rjac064 (032022). arXiv: https://academic.oup.com/jscr/article-pdf/2022/3/rjac064/42972621/rjac064.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Steenkamp W, Rachuene PA, Dey R, Mzayiya NL, Ramasuvha BE, The correlation between clinical and radiological severity of osteoarthritis of the knee, SICOT J. 8 (2022) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gill J, Sayre EC, Guermazi A, Nicolaou S, Cibere J, Association between statins and progression of osteoarthritis features on magnetic resonance imaging in a predominantly pre-radiographic cohort: the vancouver longitudinal study of early knee osteoarthritis (VALSEKO): a cohort study, BMC Musculoskelet. Disord 23 (1) (2022) 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Aghdam YH, Moradi A, Großterlinden LG, Jafari MS, Heverhagen JT, Daneshvar K, Accuracy of magnetic resonance imaging in assessing knee cartilage changes over time in patients with osteoarthritis: a systematic review, North. Clin. Istanb 9 (4) (2022) 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huang B, Huang Y, Ma X, Chen Y, Intelligent algorithm-based magnetic resonance for evaluating the effect of platelet-rich plasma in the treatment of intractable pain of knee arthritis, Contrast Media & Mol. Imaging 2022 (2022), 9223928, 10.1155/2022/9223928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Song J, Zhang R, A novel computer-assisted diagnosis method of knee osteoarthritis based on multivariate information and deep learning modelimage 1, Digital Signal Process. (2022), 103863, 10.1016/j.dsp.2022.103863. https://www.sciencedirect.com/science/article/pii/S1051200422004808. [DOI] [Google Scholar]
- [34].Abraham AM, Goff I, Pearce MS, Francis RM, Birrell F, Reliability and validity of ultrasound imaging of features of knee osteoarthritis in the community, BMC Musculoskelet. Disord 12 (1) (2011) 70, 10.1186/1471-2474-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tiulpin A, Saarakkala S, Mathiessen A, Hammer HB, Furnes O, Nordsletten L, Englund M, Magnusson K, Predicting total knee arthroplasty from ultrasonography using machine learning, Osteoarthr. Cartil. Open 4 (4) (2022), 100319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ishibashi K, Sasaki E, Chiba D, Oyama T, Ota S, Ishibashi H, Yamamoto Y, Tsuda E, Sawada K, Jung S, Ishibashi Y, Effusion detected by ultrasonography and overweight may predict the risk of knee osteoarthritis in females with early knee osteoarthritis: a retrospective analysis of iwaki cohort data, BMC Musculoskelet. Disord 23 (1) (2022) 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Oo WM, Bo MT, Role of ultrasonography in knee osteoarthritis, JCR: J. Clin. Rheumatol 22 (6) (2016). https://journals.lww.com/jclinrheum/Fulltext/2016/09000/Role_of_Ultrasonography_in_Knee_Osteoarthritis.7.aspx. [DOI] [PubMed] [Google Scholar]
- [38].Cueva JH, Castillo D, Espinós-Morató H, Durán D, Díaz P, Lakshminarayanan V, Detection and classification of knee osteoarthritis, Diagnostics 12 (10) (2022), 10.3390/diagnostics12102362. https://www.mdpi.com/2075-4418/12/10/2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huo J, Ouyang X, Si L, Xuan K, Wang S, Yao W, Liu Y, Xu J, Qian D, Xue Z, Wang Q, Shen D, Zhang L, Automatic grading assessments for knee mri cartilage defects via self-ensembling semi-supervised learning with dual-consistency, Med. Image Anal 80 (2022), 102508, 10.1016/j.media.2022.102508. https://www.sciencedirect.com/science/article/pii/S1361841522001554. [DOI] [PubMed] [Google Scholar]
- [40].Kokkotis C, Ntakolia C, Moustakidis S, Giakas G, Tsaopoulos D, Explainable machine learning for knee osteoarthritis diagnosis based on a novel fuzzy feature selection methodology, Phys. Eng. Sci. Med 45 (1) (2022) 219–229, 10.1007/s13246-022-01106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hu Y, Tang J, Zhao S, Li Y, Deep learning-based multimodal 3 t mri for the diagnosis of knee osteoarthritis, Comput. Math. Methods Med 2022 (2022), 7643487, 10.1155/2022/7643487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang CT, Huang B, Thogiti N, Zhu WX, Chang CH, Pao JL, Lai F, Successful real-world application of an osteoarthritis classification deep-learning model using 9210 knees-an orthopedic surgeon’s view, J. Orthop. Res (2022). Jul. [DOI] [PubMed] [Google Scholar]
- [43].Hu K, Wu W, Li W, Simic M, Zomaya A, Wang Z, Adversarial evolving neural network for longitudinal knee osteoarthritis prediction, IEEE Trans. Med. Imaging 41 (11) (2022) 3207–3217, 10.1109/TMI.2022.3181060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Guan B, Liu F, Mizaian AH, Demehri S, Samsonov A, Guermazi A, Kijowski R, Deep learning approach to predict pain progression in knee osteoarthritis, Skeletal Radiol. 51 (2) (2022) 363–373, 10.1007/s00256-021-03773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Desai AD, Caliva F, Iriondo C, Mortazi A, Jambawalikar S, Bagci U, Perslev M, Igel C, Dam EB, Gaj S, Yang M, Li X, Deniz CM, Juras V, Regatte R, Gold GE, Hargreaves BA, Pedoia V, Chaudhari AS, IWOAI Segmentation Challenge Writing Group, The international workshop on osteoarthritis imaging knee MRI segmentation challenge: A multi-institute evaluation and analysis framework on a standardized dataset, Radiol. Artif. Intell 3 (3) (2021), e200078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schmidt AM, Desai AD, Watkins LE, Crowder HA, Black MS, Mazzoli V, Rubin EB, Lu Q, MacKay JW, Boutin RD, Kogan F, Gold GE, Hargreaves BA, Chaudhari AS, Generalizability of deep learning segmentation algorithms for automated assessment of cartilage morphology and MRI relaxometry, J. Magn. Reson. Imaging (2022). Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yang M, Colak C, Chundru KK, Gaj S, Nanavati A, Jones MH, Winalski CS, Subhas N, Li X, Automated knee cartilage segmentation for heterogeneous clinical mri using generative adversarial networks with transfer learning, Quant. Imaging Med. Surg 12 (5) (2022). https://qims.amegroups.com/article/view/89910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chadoulos CG, Tsaopoulos DE, Moustakidis S, Tsakiridis NL, Theocharis JB, A novel multi-atlas segmentation approach under the semi-supervised learning framework: application to knee cartilage segmentation, Comput. Methods Programs Biomed 227 (2022), 107208, 10.1016/j.cmpb.2022.107208. https://www.sciencedirect.com/science/article/pii/S0169260722005892. [DOI] [PubMed] [Google Scholar]
- [49].Felfeliyan B, Hareendranathan A, Kuntze G, Jaremko JL, Ronsky JL, Improved-mask r-cnn: Towards an accurate generic msk mri instance segmentation platform (data from the osteoarthritis initiative), Comput. Med. Imaging Graph 97 (2022), 102056, 10.1016/j.compmedimag.2022.102056. https://www.sciencedirect.com/science/article/pii/S0895611122000295. [DOI] [PubMed] [Google Scholar]
- [50].Peng Y, Zheng H, Liang P, Zhang L, Zaman F, Wu X, Sonka M, Chen DZ, Kcb-net: A 3d knee cartilage and bone segmentation network via sparse annotation, Med. Image Anal 82 (2022), 102574, 10.1016/j.media.2022.102574. https://www.sciencedirect.com/science/article/pii/S1361841522002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Khan S, Azam B, Yao Y, Chen W, Deep collaborative network with alpha matte for precise knee tissue segmentation from mri, Comput. Methods Programs Biomed 222 (2022), 106963, 10.1016/j.cmpb.2022.106963. https://www.sciencedirect.com/science/article/pii/S0169260722003455. [DOI] [PubMed] [Google Scholar]
- [52].Li Z, Chen K, Liu P, Chen X, Zheng G, Entropy and distance maps-guided segmentation of articular cartilage: data from the osteoarthritis initiative, International Journal of Computer Assisted Radiology and Surgery 17 (3) (2022) 553–560, 10.1007/s11548-021-02555-2. [DOI] [PubMed] [Google Scholar]
- [53].Zaman FA, Zhang L, Zhang H, Sonka M, Wu X, Segmentation quality assessment by automated detection of erroneous surface regions in medical images (May 2022). doi: 10.36227/techrxiv.19767661.v1. https://www.techrxiv.org/articles/preprint/Segmentation_Quality_Assessment_by_Automated_Detection_of_Erroneous_Surface_Regions_in_Medical_Images/19767661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kessler DA, MacKay JW, McDonnell SM, Janiczek RL, Graves MJ, Kaggie JD, Gilbert FJ, Segmentation of knee mri data with convolutional neural networks for semi-automated three-dimensional surface-based analysis of cartilage morphology and composition, Osteoarthr. Imaging 2 (2) (2022), 100010, 10.1016/j.ostima.2022.100010. https://www.sciencedirect.com/science/article/pii/S2772654122000046. [DOI] [Google Scholar]
- [55].Huang C, Xu Z, Shen Z, Luo T, Li T, Nissman D, Nelson A, Golightly Y, Niethammer M, Zhu H, Dadp: Dynamic abnormality detection and progression for longitudinal knee magnetic resonance images from the osteoarthritis initiative, Med. Image Anal 77 (2022), 102343, 10.1016/j.media.2021.102343. https://www.sciencedirect.com/science/article/pii/S1361841521003881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Liu Q, Xu Z, Jiao Y, Niethammer M, isegformer: Interactive segmentation via transformers with application to 3d knee mr images, in: Wang L, Dou Q, Fletcher PT, Speidel S, Li S (Eds.), Medical Image Computing and Computer Assisted Intervention – MICCAI 2022, Springer Nature Switzerland, Cham, 2022, pp. 464–474. [Google Scholar]
- [57].Liang D, Liu J, Wang K, Luo G, Wang W, Li S, Position-prior clustering-based self-attention module for knee cartilage segmentation, in: Wang L, Dou Q, Fletcher PT, Speidel S, Li S (Eds.), Medical Image Computing and Computer Assisted Intervention – MICCAI 2022, Springer Nature Switzerland, Cham, 2022, pp. 193–202. [Google Scholar]
- [58].Houserman DJ, Berend KR, Lombardi AV, Fischetti CE, Duhaime EP, Jain A, Crawford DA, The viability of an artificial intelligence/machine learning prediction model to determine candidates for knee arthroplasty, J. Arthroplasty (2022), 10.1016/j.arth.2022.04.003. https://www.sciencedirect.com/science/article/pii/S0883540322003941. [DOI] [PubMed] [Google Scholar]
- [59].Prezja F, Paloneva J, Polonen I, Niinimäki E, Ayramo S, Deepfake knee osteoarthritis x-rays from generative adversarial neural networks deceive medical experts and offer augmentation potential to automatic classification, Sci. Rep 12 (1) (2022) 18573, 10.1038/s41598-022-23081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Alexopoulos A, Hirvasniemi J, Tümer N, Early detection of knee osteoarthritis using deep learning on knee magnetic resonance images (2022). doi: 10.48550/ARXIV.2209.01192. https://arxiv.org/abs/2209.01192. [DOI] [Google Scholar]
- [61].Tolpadi AA, Han M, Caliva F `, V. Pedoia, S. Majumdar, Region of interest-specific loss functions improve t2 quantification with ultrafast t2 mapping mri sequences in knee, hip and lumbar spine, Sci. Rep 12 (1) (2022) 22208, 10.1038/s41598-022-26266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kim M, Lee SM, Park C, Lee D, Kim KS, Jeong HS, Kim S, Choi MH, Nickel D, Deep learning-enhanced parallel imaging and simultaneous multislice acceleration reconstruction in knee MRI, Invest. Radiol 57 (12) (2022) 826–833. [DOI] [PubMed] [Google Scholar]
- [63].Wang Z, Qian C, Guo D, Sun H, Li R, Zhao B, Qu X, One-dimensional deep low-rank and sparse network for accelerated mri, IEEE Trans. Med. Imaging 42 (1) (2023) 79–90, 10.1109/TMI.2022.3203312. [DOI] [PubMed] [Google Scholar]
- [64].Balestriero R, Ibrahim M, Sobal V, Morcos A, Shekhar S, Goldstein T, Bordes F, Bardes A, Mialon G, Tian Y, Schwarzschild A, Wilson AG, Geiping J, Garrido Q, Fernandez P, Bar A, Pirsiavash H, LeCun Y, Goldblum M, A cookbook of self-supervised learning (2023). arXiv:2304.12210.


