Abstract
A growing increase in the number of serious infections caused by multidrug resistant bacteria (MDR) is challenging our society. Despite efforts to discover novel therapeutic options, few antibiotics targeting MDR have been approved by the Food and Drug Administration (FDA). Lactic acid bacteria have emerged as a promising therapeutic alternative due to their demonstrated ability to combat MDR pathogens in vitro. Our previous co-culture studies showed Lacticaseibacillus rhamnosus CRL 2244 as having a potent killing effect against carbapenem-resistant Acinetobacter baumannii (CRAB) strains. Here we report that cell-free conditioned media (CFCM) samples obtained from Lcb. rhamnosus CRL 2244 cultures incubated at different times display antimicrobial activity against 43 different pathogens, including CRAB, methicillin-resistant Staphylococcus aureus (MRSA) and carbapenemase Klebsiella pneumoniae (KPC)-positive strains. Furthermore, transwell and ultrafiltration analyses together with physical and chemical/biochemical tests showed that Lcb. rhamnosus CRL 2244 secretes a <3 kDa metabolite(s) whose antimicrobial activity is not significantly impaired by mild changes in pH, temperature and various enzymatic treatments. Furthermore, sensitivity and time-kill assays showed that the bactericidal activity of the Lcb. rhamnosus CRL 2244 metabolite(s) enhances the activity of some current FDA approved antibiotics. We hypothesize that this observation could be due to the effects of Lcb. rhamnosus CRL 2244 metabolite(s) on cell morphology and the enhanced transcriptional expression of genes coding for the phenylacetate (PAA) and histidine catabolic Hut pathways, metal acquisition and biofilm formation, all of which are associated with bacterial virulence. Interestingly, the extracellular presence of Lcb. rhamnosus CRL 2244 induced the transcription of the gene coding for the CidA/LgrA protein, which is involved in programmed cell death in some bacteria. Overall, the findings presented in this report underscore the promising potential of the compound(s) released by Lcb. rhamnosus CRL2244 as an alternative and/or complementary option to treat infections caused by A. baumannii as well as other MDR bacterial pathogens.
Introduction
Antibiotic resistance in bacteria constitutes a prominent and escalating menace to public health, jeopardizing healthcare systems, food security, and global progress. In recent years a significant surge in infections caused by highly antibiotic-resistant bacteria has been reported, leading to a substantial increase in healthcare expenses and necessitating the development of novel therapeutic agents [1, 2]. In this context, carbapenem-resistant Acinetobacter baumannii (CRAB) has earned a critical priority status from both the World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) [3, 4]. The global prevalence of A. baumannii strains resistant to multiple classes of antibiotics underscores the pressing demand for novel antimicrobial therapies. Despite concerted efforts by various research groups and pharmaceutical companies during the past decade, the approval of new drugs tailored to combat A. baumannii has been limited [5–7]. Presently, only two drugs, cefiderocol and sulbactam-durlobactam, have secured approval from the U.S. Food and Drug Administration (FDA) for treating A. baumannii infections [8, 9].
The lack of significant progress in the development of novel and efficacious antibiotics emphasizes the urgency of exploring innovative methods to address CRAB infections. Current research is directed toward therapeutic alternatives, such as the application of lactic acid bacteria (LAB) to treat bacterial infections [10–13]. The growing interest in LAB can be attributed to their Generally Regarded as Safe (GRAS) status, intrinsic antimicrobial attributes, and the myriad of health advantages they provide to the human host, particularly in terms of enhancing the equilibrium of the intestinal microbiota [14]. Our group has recently reported that the Lacticaseibacillus rhamnosus CRL 2244 strain significantly inhibits the growth of both susceptible and CRAB strains of A. baumannii [15]. We also observed changes at both the transcriptomic level as well as in A. baumannii morphology when it was co-cultured in the presence of Lcb rhamnosus CRL 2244 [15].
Based on these prior observations, the aim of this study is to investigate whether Lcb. rhamnosus CRL 2244 secretes substances capable of triggering or contributing to the death of CRAB cells. To achieve this goal, we isolated cell-free conditioned medium (CFCM) from cultures of Lcb. rhamnosus CRL 2244 and performed a comprehensive analysis, including the evaluation of its antimicrobial activity against various CRAB strains and other multidrug resistant (MDR) pathogens. In addition, we also characterized some of its physicochemical properties, explored potential synergistic effects with other antibiotics, and the determined of how A. baumannii responds to CFCM, including its growth rate and the differential expression of virulence-related genes.
Materials and methods
Bacterial strains and culture conditions
The Lcb. rhamnosus CRL 2244 strain, which displays antimicrobial activity against A. baumannii [15], was used to prepare cell-free conditioning medium (CFCM) after two subcultures at 37°C in Man, Rogosa, and Sharpe (MRS) broth (Oxoid, Basingstoke, Hampshire, United Kingdom). Three A. baumannii model strains, the A118 antibiotic-susceptible strain [16, 17] and the CRAB AB5075 (blaOXA-23 and blaOXA-51) [18] and AMA3[19] clinical isolates were used in this work. In addition, a total of 40 strains, including MDR isolates (n = 34), and susceptible strains (n = 6) were used for the spot-on-the-lawn assays to test antimicrobial activity (Table 1). The strains were grown on Cystine-Lactose-Electrolyte-Deficient (CLED) medium (Beckton Dickinson, Franklin Lakes, NJ, USA) overnight at 37°C and used within 24 h.
Table 1. Antimicrobial activity of Lacticaseibacillus rhamnosus CRL 2244 CFCM against multidrug pathogens tested by the spot-on-the-lawn method.
| Strains | IDH (mm) | Phenotype | Key resistance mechanismsa | |||
|---|---|---|---|---|---|---|
| 48 h | 96 h | 120 h | ||||
| Acinetobacter baumannii | ||||||
| AMA2 | 20 | 15 | 10 | MDR / CR | bla NDM-1 | |
| AMA3 | 16 | 20 | 20 | MDR / CR | blaNDM-1, blaPER-7 | |
| AMA5 | 14 | 25 | 25 | MDR / CR | bla NDM-1 | |
| AMA9 | 0 | 17 | 0 | MDR / CR | bla NDM-1 | |
| AMA10 | 0 | 19 | 20 | MDR / CR | bla NDM-1 | |
| AMA14 | 0 | 17 | 24 | MDR / CR | blaNDM-1, blaPER-7 | |
| AMA16 | 21 | 20 | 14 | MDR / CR | blaNDM-1, blaPER-7 | |
| AMA18 | 0 | 15 | 0 | MDR / CR | blaNDM-1, blaPER-7 | |
| AMA22 | 0 | 17 | 21 | MDR / CR | bla NDM-1 | |
| AMA30 | 17 | 19 | 21 | MDR / CR | blaNDM-1, blaPER-7 | |
| AMA31 | 20 | 23 | 15 | MDR / CR | bla NDM-1 | |
| AMA33 | 0 | 17 | 21 | MDR / CR | bla NDM-1 | |
| AMA40 | 22 | 17 | 22 | MDR / CR | bla NDM-1 | |
| AMA43 | 24 | 17 | 25 | MDR / CR | bla NDM-1 | |
| AMA116 | 20 | 20 | 18 | MDR / CR | bla OXA-23 | |
| AMA122 | 20 | 25 | 21 | MDR / CR | bla NDM-1 | |
| AMA166 | 21 | 20 | 25 | MDR / CR | bla OXA-23 | |
| AMANO | 0 | 17 | 0 | MDR / CR | blaNDM-1, blaOXA-23 | |
| AB0057 | 15 | 23 | 0 | MDR / CR | blaOXA-23, blaTEM-1 | |
| AB5075 | 8 | 20 | 20 | MDR / CR | blaOXA-23, blaOXA-51, blaGES-14 | |
| A118 | 11 | 18 | 16 | S | ||
| AB ATCC 17978 | 0 | 17 | 0 | S | ||
| Escherichia coli | ||||||
| ECO 4 | 10 | 12 | 11 | MDR | aadA1, Inti1 | |
| ECO 16 | 13 | 15 | 13 | MDR | aadA1, Inti1 | |
| ECO 7499 | 15 | 19 | 20 | MDR / CR | bla KPC-2 | |
| ECO PNDM5 | 16 | 21 | 23 | MDR / CR | bla NDM-1 | |
| ECO ATCC 25922 | 20 | 21 | 20 | S | ||
| Serratia marcescens | ||||||
| SmP9 | 12 | 12 | 12 | MDR / CR | blaKPC-2, blaCTX-M-2 | |
| SmP7 | 11 | 10 | 10 | MDR / CR | blaKPC-2, blaCTX-M-2, blaSHV-2 | |
| Sm 371 | 16 | 20 | 13 | MDR / CR | blaKPC-2, blaCTX-M-2 | |
| Sm 909 | 14 | 13 | 11 | MDR | dfrA1, ant(3”)-Ia, ant(2")-Ia, aac(3)-Ia | |
| Sm 55 | 13 | 13 | 10 | MDR / CR | bla KPC-2 | |
| Klebsiella pneumoniae | ||||||
| KPN 1224 | 0 | 11 | 9 | MDR / CR | bla KPC-2 | |
| KPN 25979 | 0 | 11 | 18 | MDR / CR | blaKPC-2, blaNDM-5, blaCTX-M-2 | |
| KPN RB_NDM5 | 0 | 16 | 19 | MDR / CR | bla NDM-5 | |
| Salmonella | ||||||
| S. Schwarzengrund 1024 | 11 | 11 | 13 | MDR / CR | bla KPC-2 | |
| Staphylococcus aureus | ||||||
| SAU 96 | 17 | 18 | 15 | MDR | mecA | |
| USA 300 | 22 | 20 | 24 | MDR | mecA | |
| SAU RN4220 | 9 | 12 | 12 | S | ||
| SAU ATCC 25923 | 17 | 20 | 20 | S | ||
| Providencia stuartii | ||||||
| PS 848 | 18 | 19 | 21 | MDR / CR | blaNDM-1, blaTEM-1 | |
| Staphylococcus epidermidis | ||||||
| ATCC 25933 | 22 | 21 | 16 | S | ||
| Enterococcus faecalis | ||||||
| ATCC 29212 | 0 | 0 | 0 | S | ||
| Response summary | ||||||
| Range | 24–0 | 25–0 | 25–0 | |||
| Mean | 11.74 | 17.04 | 15.06 | |||
| IDH50 | 14 | 17 | 16 | |||
IDH, inhibition diameter halos; MDR, multidrug resistance (resistant at least to three different antibiotics families); CR, carbapenem resistant; S. aureus USA300, methicillin-resistant (MRSA) model strain.
akey relevant mechanisms of resistance, such as carbapenemases among others, are mentioned. In the susceptible strain no relevant mechanism of resistant in stated.
Preparation of Lcb. rhamnosus CRL 2244 CFCM and detection of antimicrobial activity by spot-on-the-lawn assays
The evaluation of the antimicrobial activity of Lcb. rhamnosus CRL 2244 CFCM samples was performed following the modified well diffusion method described by Halder et al. (2017) [20]. Briefly, Lcb. rhamnosus CRL 2244 MRS broth (2% v/v inoculum) was incubated at 37°C for 48 h, 96 h and 120 h and CFCM samples were prepared after centrifugation at 9,200 xg for 5 min at 4°C and filtering the supernatant with 0.22 μm filters (Millipore, Billerica, MA, USA). Bacterial strains were suspended in sterile physiological solution (0.85% NaCl, w/v) at a concentration of 0.5 McFarland units (1.5 x 108 CFU/ml). These inocula were then seeded onto CLED agar plates. A total of 25 μl of CFCMs from different incubation times were applied to the surface of the plates. Following 24 h of incubation at 37°C, the diameters of the growth inhibition halos (IDH) were measured and classified as less active (IDH ≤ 10 mm), moderately active (IDH = 11–14 mm), and very active (IDH ≥15 mm) [20]. The assays were performed using three independent biological samples.
Transwell assays
Transwell inserts were used to confirm the diffusion of antimicrobial compounds secreted by Lcb. rhamnosus CRL 2244. Briefly, lysogeny broth (LB) cultures of A118 or AB5075 (1 x 107 CFU/mL) were seeded onto transwell inserts (CELLTREAT Scientific Products, Pepperell, MA) with a permeable polyethylene terephthalate (PET) membrane with a pore size of 0.4 μm and exposed to undiluted and half-diluted overnight MRS broth cultures of Lcb. rhamnosus CRL 2244. After incubation at 37°C for 18 h, the cell density (OD600) and viability (CFU/mL) of A. baumannii cells exposed to Lcb. rhamnosus CRL 2244 cultures were determined by serial dilution and plating on CHROMagar™ Acinetobacter plates. The assays were performed using three independent biological samples.
Preliminary characterization of Lcb. rhamnosus CRL 2244 CFCM
The effect of trichloroacetic acid (TCA), pH, temperature, and enzymes on the antimicrobial activity of compound(s) secreted by Lcb. rhamnosus CRL 2244 were studied using 96-h CFCM samples. Briefly, CFCM samples were treated with 20% cold TCA for 1 h and then centrifuged at 9,200 xg for 10 min in a microcentrifuge. The precipitate was washed 3 times with an ice-cold solution of 0.01 M HCl/90% acetone and resuspended in 100 mM Tris-HCl, pH 8.5, 8 M urea. To evaluate the effects of pH, CFCM samples were adjusted to pH 5.5, 6.5 and 8.0 with a 4 N NaOH solution. The effect of temperature was examined by treating CFCM samples at 25°C, 37°C, 50°C, 75°C and 100°C for 30 min. CFCM samples were treated separately with enzymes including proteinase K, lipase, alpha-chymotrypsin, pepsin, and catalase (all from Sigma, St. Louis, MO, USA). The enzyme reactions were carried out at a final enzyme concentration of 0.05 mg/mL for 30 min for catalase and of 0.5 mg/mL and 4 h for all other tested enzymes. All samples were incubated at 37°C. Untreated CFCM samples (pH 3.9) were used as a control. The antimicrobial activity of untreated and treated CFCM samples was tested on the AB5075 strain using the spot-on-the-lawn method. Technical triplicates using three independent biological samples were used to collect experimental data.
Antimicrobial chemical extraction assays
Precipitation of active compound(s): An equal volume of ice-cold acetone was added to CFCM and vigorously stirred for 20 min. The mixture was refrigerated at 4°C for 2 h and the resulting precipitate was collected by centrifugation 8,000 xg and decanting of the supernatant. The procedure was repeated two more times with the supernatant and the combined precipitates were dissolved in water and then evaluated for antimicrobial properties.
Extraction of active compounds(s): In a separatory funnel, CFCM was mixed with ethyl acetate (10% volume of the CFCM). The ethyl acetate layer was separated from the aqueous phase and collected. The original aqueous layer was extracted two more times with ethyl acetate. The combined ethyl acetate extracts were washed with a saturated NaCl aqueous solution, dried over magnesium sulfate, filtered, and evaporated to remove the ethyl acetate and obtain crude extracts containing molecules of interest. The crude extract was dissolved in DSMO and evaluated for biological activity. The antibacterial activity of all CFCM extracts against AB5075 and AMA3 was determined by the spot-on-the-lawn method, calculating the mass of the extracts that were resuspended in DMSO, diluted to 1% DMSO, and adjusted to specific concentrations. An organic solvent control was included on the plate. Technical triplicates using three independent biological samples were used to collect experimental data for each tested extract.
Minimum inhibitory activity and synergy assay of CFCMs
The CFCM obtained from a 96-h Lcb. rhamnosus CRL 2244 culture was lyophilized, weighed, and resuspended in phosphate buffered saline (PBS) to determine the minimum inhibitory concentration (MIC) by the broth microdilution method and minimum bactericidal concentration (MBC) assays, which were done following CLSI standards (CLSI 2020), against the AB5075 and AMA3 CRAB strains, and the MRSA USA300 isolate.
The synergy assays were performed by adding lyophilized CFCM at sub-inhibitory concentration (4 mg/mL) to cation-adjusted Muller-Hinton agar (CAMHA) medium. The gradient diffusion method was used to determine the MIC values for the AB5075 and AMA3 CRAB strains, E. coli 7499 and MRSA USA 300 using commercially available strips (Liofilchem S.r.l., Italy). The CRAB strains were tested for their susceptibility to meropenem (MEM), imipenem (IMP), ceftazidime/avibactam (CZA), amikacin (AK) and cefiderocol (CFDC). MRSA was tested for susceptibility to cefoxitin (FOX), ampicillin (AMP), cefazolin (CFZ), gentamicin (CN), erythromycin (ERY), trimethoprim-sulfamethoxazole (SXT) and vancomycin (VAN). CAMHA plates without added lyophilized CFCM were used as control. For each antibiotic evaluated, the fractional inhibitory concentration (FIC) was calculated as follows:
A FIC of ≤ 0.5 indicates synergism, > 0.5–1 indicates additive effects, ≥ 2–4 indifference, and > 4 is considered to be antagonism [21].Technical triplicates using three indepndent biological samples were used to collect experimental data obtained using each tested condition.
Effect of CFCM on A. baumannii colony morphology
Phenotypic changes in macrocolonies of the AMA3 and AB5075 strains exposed to lyophilized CFCM were evaluated. Briefly, 5 μl of overnight cultures of bacteria exposed to 4 mg/mL of CFCM were inoculated onto 40 mL plates of cation-adjusted Müller Hinton agar (CAMHA) with 2% glycerol and supplemented with 40 μg/mL of Congo red (Sigma-Aldrich), which allows the detection of the poly-β-(1–6)-N-acetylglucosamine (PNAG) biofilm component. Cultures of AMA3 and AB5075 not exposed to CFCM were included as controls. After 24–48 h of incubation at 37°C, macrocolony changes were observed and registered.
Time-killing assay of CFCM
The viability of A. baumannii AB5075 and AMA3 in the presence of lyophilized CFCM at MIC, 1 sub-MIC and 2 sub-MIC concentrations was assessed following the protocol of Balouiri et al. [22], with modifications. Briefly, tubes with LB broth inoculated with a bacterial suspension of 5 x 108 CFU/mL and the different concentrations of CFCM were incubated at 37°C for 24 h. Cells not exposed to CFCM were included as controls. Aliquots taken at various time intervals were serially diluted and plated on LB agar to determine the CFU/mL values of each tested sample. Technical triplicates using three independent biological samples were used to collect experimental data.
Scanning electronic microscopy (SEM)
From overnight cultures of A. baumannii on LB agar incubated at 37°C, one colony was resuspended in 100 μl of physiological solution and inoculated into 2 mL of LB broth supplemented with 4 mg/mL of lyophilized CFCM. A. baumannii cultured without exposure to lyophilized CFCM was included as a control. After overnight incubation at 37°C, the samples were centrifuged at 2,700 xg for 4 min, resuspended in Karnovsky fixative solution and processed for observation in the ZeissSupra 55VP Scanning Electron Microscope (Germany) belonging to the Centro Integral de Microscopía Electrónica (CIME-CONICET—Universidad Nacional de Tucumán, Argentina) as described before [15].
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays
A. baumannii AB5075 and AMA 3 cells were grown in the presence and absence of 4 mg/mL (sub-MIC concentration) of lyophilized CFCM for 18 h at 37°C. Total RNA extractions were carried out in triplicate for each condition using a commercial kit (Direct-zol RNA Kit, Zymo Research). DNase-treated RNA was used for cDNA synthesis using the iScriptTM Reverse Transcription Supermix kit (Bio-Rad, Hercules, CA, USA) following the manufacturer’s protocol. The cDNA concentrations were adjusted to 50 ng/μL, and qPCR was performed using the qPCRBIO SyGreen Blue Mix Lo-ROX following the manufacturer’s instructions (PCR Biosystems, Wayne, PA, USA). Each qPCR assay included at least three biological replicates of cDNA and run in triplicate utilizing the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Data are presented as NRQ (Normalized relative quantities) calculated by the qBASE method [23, 24], using recA and rpoB genes as normalizers. Statistically significant differences were denoted with asterisks and determined through ANOVA followed by Tukey’s multiple comparison test (P < 0.05), employing GraphPad Prism (GraphPad Software, San Diego, CA, USA).
Results
Antimicrobial activity of CFCMs from Lcb. rhamnosus CRL 2244 against MDR pathogens
The antimicrobial activity of CFCMs obtained from Lcb. rhamnosus CRL 2244 cultures incubated for 48 h (CFCM-48), 96 h (CFCM-96), and 120 h (CFCM-120) was evaluated against a total of 43 pathogenic strains, 34 of which exhibited MDR and 31 also possessed carbapenemases coding genes (CR) (Table 1, S1 Fig). The CFCMs showed variable activity against the tested strains, with growth inhibitory halo diameters ranging from 0 to 24/25 mm. None of the tested CFCMs exhibited activity against Enterococcus faecalis ATCC 29212. The highest antimicrobial activity was achieved with CFCM-96, which was active against most of the tested strains (97.7%) with an inhibition diameter halo (IDH) mean of 16.95 mm and an IDH50 equal to 17 mm (Table 1, S1 Fig). CFCM-120 samples also showed antimicrobial activity, although less than CFCM-96 as evidenced by significantly smaller IDH (15.06 mm) and IDH50 (16 mm) means and a loss of activity against five of the tested strains. CFCM-48 demonstrated the lowest activity (IDH mean = 11.74 mm and IDH50 = 14 mm), and it failed to inhibit 11 of the tested strains (Table 1).
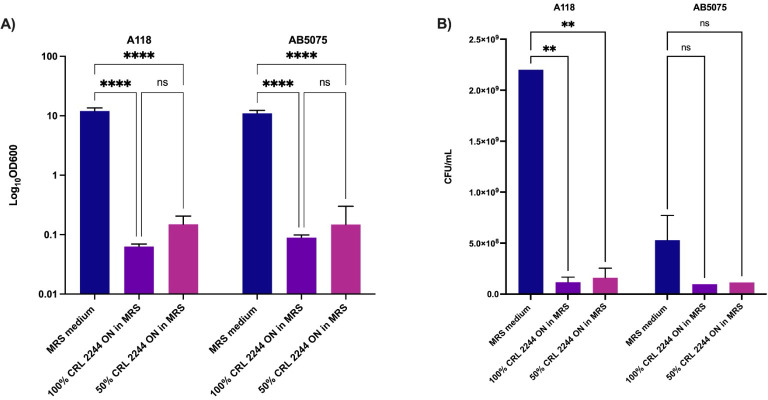
Transwell assays confirmed that the antimicrobial activity of Lcb. rhamnosus CRL 2244 on strains AB5075 and A118 is due to the production of a compound secreted into the culture medium (Fig 1). The cell density of A118 and AB5075 broth cultures was drastically reduced (OD600 values near 0.1) after 18 h exposure to pure (100%) and diluted (50%) Lcb. rhamnosus CRL 2244 CFCM samples collected after overnight (ON) incubation (Fig 1A) when compared to the control group, which was not exposed to CFCM. In these experiments, the number of surviving cells for strain A118 was less than 1x10-8, while no cells were detected for strain AB5075 (Fig 1B), indicating the potent lethal activity of the antimicrobial compound present in the CFCM samples. These results suggest that Lcb. rhamnosus CRL 2244 secretes one or more soluble and diffusible compounds that significantly inhibit A. baumannii cell growth.
Fig 1. Lcb. rhamnosus CRL 2244 secretes compounds that alter A. baumannii. growth.
A) Cell density (OD600), B) Surviving cells (CFU/mL). Transwell assays were performed using the A. baumannii clinical isolates A118 and AB5075. Bacteria were exposed to pure (100%) or diluted (50%) overnight Lcb. rhamnosus CRL 2244 cultures for 18 h at 37°C. Unexposed culture samples were used as controls. Technical triplicates using three independent biological samples were used to collect experimental data. Statistical significance (P < 0.05) was determined by two-way ANOVA followed by Tukey’s multiple comparison test. ****, P < 0.0001; ns, not significant. Error bars represents the standard deviation (SD).
To start the characterization of the active component(s) secreted by Lcb. rhamnosus CRL 2244, the CFCM-96 was subjected to different treatments. CFCM samples treated with TCA 20% displayed similar antimicrobial effects when compared to untreated samples (Table 2). The antimicrobial activity of CFCM remained relatively stable in the mild acidic to basic pH tested range, as well as after heat treatment (Table 2). Similarly, treatment with a variety of enzymes (protease, peptidase, lipase, and oxidoreductase) did not significantly affect antibacterial activity. An IDH = 19 was obtained for pepsin and α-chymotrypsin and 20 mm for proteinase K, lipase and catalase (IDH = 20 mm for the CFCM-non treated control).
Table 2. Stability of the antimicrobial activity of Lacticaseibacillus rhamnosus CRL 2244 CFCM subjected to different treatments.
| CFCM (IDH mm) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Protein-free | pH | Temperature | |||||||||
| TCA | Control | 5.5 | 6.5 | 8.0 | Control | 25° | 37° | 50° | 75° | 100° | Control | |
| AB5075 | 18 | 17 | 20 | 20 | 20 | 20 | 20 | 22 | 22 | 18 | 22 | 21 |
IDH, Inhibition diameter halo.
Control, CFCM non-treated, pH = 3.9 at room temperature.
The active compound(s) secreted by Lcb. rhamnosus CRL 2244 were extracted from the CFCM by precipitation with acetone and extraction with ethyl acetate. The compound(s) recovered from all extraction methods retained the growth inhibitory activity against AB5075, with IDH values similar to those observed for the crude CFCM sample (S1 Table). Furthermore, CFCM samples that underwent ultrafiltration using a 3-kDa molecular weight cutoff filter demonstrated antimicrobial effectiveness against the AB5075 and AMA3 CRAB strains comparable to that observed with untreated CFCM control. An IDH of 22 mm was obtained for both strains.
Taken together, these results indicate that the CFCM active component(s) is likely a secreted metabolite that is a non-peptide polar organic molecule soluble in organic solvents, smaller than 3 kDa in size, stable within a mild acidic-basic range, and relatively resistant to increased temperatures and various enzymes treatments.
Effect of Lcb. rhamnosus CRL 2244 CFCM on antibiotic susceptibility and colony morphology
Lyophilized Lcb. rhamnosus CRL 2244 CFCM samples were dissolved in PBS to determine MIC and MBC activities. The lyophilized CFCM MIC value was 8 mg/mL for both CRAB tested strains (AB5075 and AMA3), while the MIC value for the MRSA USA 300 strain was one dilution higher (15 mg/mL). The MBC values for the CRAB strains were consistent with the MIC values, whereas MBC value for USA 300 was 62 mg/mL (2 doubling dilution higher).
To evaluate the synergy of the antimicrobial compound(s) secreted by Lcb. rhamnosus CRL 2244 with clinically relevant antibiotics, a sub-inhibitory concentration of the lyophilized CFCM (4 mg/ml) was tested. For the AB5075 and AMA3 strains, a 4- and 5-fold dilution decrease in the MIC of meropenem was seen for each strain, respectively (Table 3, S2 Fig). Furthermore, strain AB5075 exhibited a reduction of 1-fold dilution for imipenem, ceftazidime/avibactam, and cefiderocol (Table 3, S2 Fig). MIC changes were not observed when AMA3 was tested for ceftazime/avibactam and cefiderocol. Similarly, synergy was not detected when amikacin was tested using the same experimental conditions. The same amikacin MIC of 128 mg/L was obtained for AB5075 with and without lyophilized CFCM. In addition, synergy for meropenem was also evaluated for E. coli 7499 (KPC producer) and, as expected, a synergistic effect was observed (Table 3, S2 Fig). The MRSA USA 300 MIC values for cefoxitin (MIC: 16 mg/L) and ampicillin (MIC: 1 mg/L) slightly decreased when the medium was supplemented with lyophilized CFCM (Table 3, S2 Fig). This strain also showed one-fold (MIC: 8 mg/L) and two-fold (MIC: 0.25 mg/L) dilution decreases for cefoxitin and ampicillin, respectively. In addition, the MRSA USA 300 strain exhibited a 2-dilution reduction for vancomycin in the presence of lyophilized CFCM (Table 3, S2 Fig).
Table 3. Synergy between Lacticaseibacillus rhamnosus CRL 2244 CFCM and antibiotics on multidrug resistant strains.
| Strains | MIC values (mg/L) | |||
|---|---|---|---|---|
| AB5075 | CAMHA | CAMHA + 4 mg/mL CFCM | FIC | Effects |
| MEM | 32 | 2 | 0.06 | synergism |
| IMP | 8 | 4 | 0.50 | synergism |
| CAZ/AVI | 128 | 64 | 0.50 | synergism |
| FDC | 0.5 | 0.25 | 0.50 | synergism |
| AMA3 | CAMHA | CAMHA + 4 mg/mL CFCM | FIC | Effects |
| MEM | 64 | 0.94 | 0.01 | synergism |
| IMP | >256 | 96 | 0.37 | synergism |
| CAZ/AVI | >256 | >256 | 1 | additive |
| FDC | 3 | 3 | 1 | additive |
| E. coli 7499 | CAMHA | CAMHA + 4 mg/mL CFCM | FIC | Effects |
| MEM | 1 | 0.19 | 0.19 | synergism |
| IMP | 4 | 8 | 2 | indifference |
| CAZ/AVI | 0.94 | 0.94 | 1 | additive |
| FDC | 0.19 | 0.19 | 1 | additive |
| S. aureus USA300 | CAMHA | CAMHA + 4 mg/mL CFCM | FIC | Effects |
| AMP | 1 | 0.25 | 0.25 | synergism |
| FOX | 16 | 8 | 0.50 | synergism |
| CFZ | 1 | 1 | 1 | additive |
| CN | 0.125 | 0.5 | 4 | indifference |
| ERY | 0.5 | 0.5 | 1 | additive |
| SXT | 0.064 | 0.064 | 1 | additive |
| VAN | 1 | 0.25 | 0.25 | synergism |
CAMHA, Cation-adjusted Mueller-Hinton Agar; MEM, meropenem; IMP, imipenem; CAZ/AVI, ceftazidime-avibactam; FDC, cefiderocol; AMP, ampicillin; FOX, cefoxitin; CFZ, cefazolin; CN, gentamicin; ERY, erythromycin; SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin.
In addition, the ability of the CRAB strains to produce poly-N-acetylglucosamine (PNAG), which is a major component of the biofilm extracellular matrix, in the presence or absence of a sub-inhibitory concentrations of CFCM was examined using the Congo red dye assay [25]. A notable decrease in the roughness of the edges and a slight lightening in the red color tone of the colonies were observed when these two CRAB strains were exposed to the lyophilized CFCM (S3 Fig).
Taken together, these findings suggest that CFCM acts synergistically with β-lactam antibiotics, increasing the susceptibility of MDR strains to these antibiotics. In addition, CFCM induces alterations in the morphological phenotype of macrocolonies that could influence biofilm formation. These responses would influence changes in antibiotic resistance and affect A. baumannii virulence.
Killing activity of Lcb. rhamnosus CRL 2244 CFCM on A. baumannii
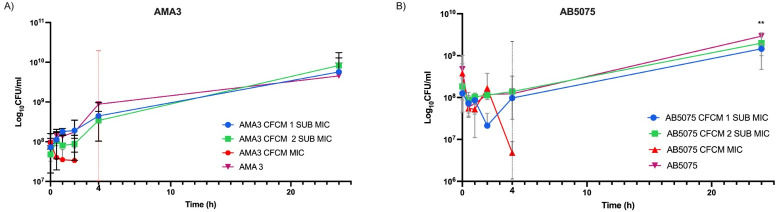
Time-kill assays were used to evaluate the bactericidal or bacteriostatic activity of lyophilized CFCM on AB5075 and AMA3 (Fig 2). At MIC concentrations, both CRAB strains showed a clear decrease in CFU/mL after short exposure to Lcb. rhamnosus CRL 2244 CFCM, and no cells were recovered after 24 h incubation. Concentrations below MIC affected the growth of AB5075 and AMA3 during the first 2 h, with slightly lower CFU values compared to the control. However, after 4 h, both strains showed an increase in CFU until reaching values similar to the control after 24 h incubation (Fig 2).
Fig 2. Lcb. rhamnosus CRL 2244 CFCM killing activity against A. baumannii.
The clinical isolates AMA3 (A) and AB5075 (B) were incubated at 37°C for 24 h in presence of lyophilized CFCM at MIC, 1 sub-MIC and 2 sub-MIC concentrations. CFU/mL were determined at different incubation times for a period of 24 h. All assays were carried out in technical triplicates. Statistical significance (P < 0.05) was determined by two-way ANOVA followed by Tukey’s multiple comparison test. * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P < 0.0001.
These results indicate that CFCM exhibits bactericidal behavior at concentrations equivalent to MIC. Whereas, at sub-MIC concentrations, CFCM induces less growth during the initial 8 hours of incubation.
Lcb. rhamnosus CRL 2244 CFCM exerts cell morphological changes
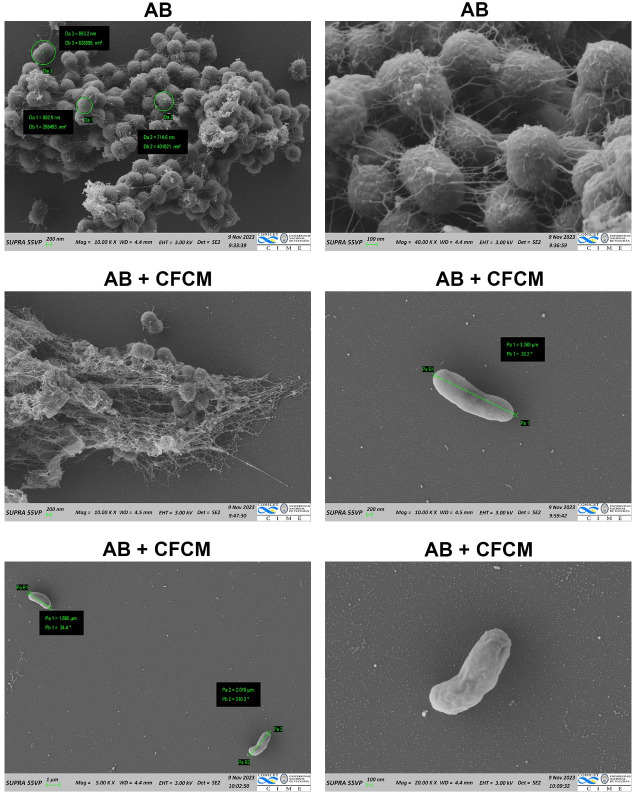
Scanning electron microscopy (SEM) was performed to assess the impact of lyophilized CFCM at a sub-MIC concentration on the morphology of A. baumannii cells (Fig 3). Microscopic images revealed a significant reduction in cell density and extracellular matrix when AB5075 cells were treated with lyophilized CFCM. Cells were observed as dispersed in single or paired formations, contrasting with the homogeneously distributed and biofilm-immersed control cells (Fig 3). Furthermore, exposure to lyophilized CFCM induced significant changes in cell length, with exposed cells being up to 5 times longer than non-exposed control cells (3,500 nm vs 700 nm). In addition, the cell surface of exposed cells displayed a smother texture when compared to the roughness observed in control cells (Fig 3).
Fig 3. Scanning electron microscopy of A. baumannii cells cultured in the presence of Lcb. rhamnosus CRL 2244 lyophilized CFCM and resuspended in PBS.
Micrographs were captured at a magnification of x5.000, ×10.000, x20.000 or x40.000. Bars, 1 μm, 100 nm or 200 nm.
Taken together, these results suggest that CFCM could influence the ability of AB5075 cells to form multicellular aggregates resembling biofilm structures on a solid surface and alter cell morphology.
Transcriptional response of A. baumannii to the presence of Lcb. rhamnosus CRL 2244 CFCM
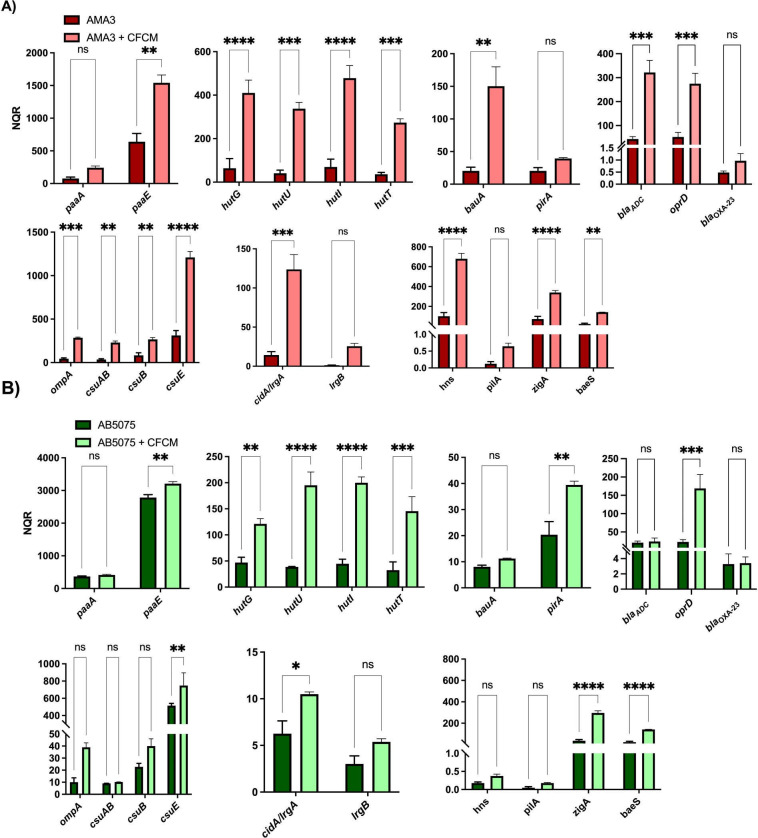
To explore how A. baumannii transcriptionally responds when exposed to lyophilized CFCM, we conducted quantitative RT-PCR (qRT-PCR) assays using the AMA3 (Fig 4A) and AB5075 (Fig 4B) strains. Both strains showed a significant overexpression of genes associated with A. baumannii virulence, including those involved in phenylacetate (PAA) catabolism, such as paaA, paaE; and the hutG, hutU, hutI, hutT genes, all of which are components of the histidine catabolic Hut pathway described in other bacteria (Fig 4). Furthermore, a significant increase in the expression of additional genes involved in the virulence of A. baumannii was observed, including the iron metabolism-related genes bauA and pirA genes in AMA3 and AB5075 respectively. Genes associated with biofilm formation such as ompA, csuAB, csuB and csuE were significantly upregulated in AMA3, while only csuE was seen significantly increased in AB5075 (Fig 4). In addition, genes associated with antibiotic resistance were also studied; oprD was significantly increased in both strains, while blaADC was increased only in AMA3 (Fig 4). We also noted the significant upregulation of genes related to zinc metabolism (zigA) and efflux functions (baeS) in both strains; however, the transcriptional expression of the hns global regulator was significantly increased only in AMA3 (Fig 4). Interestingly, the gene encoding the CidA/LgrA protein, which is involved in programmed cell death in some bacteria, was also found to be significantly overexpressed in both strains (Fig 4).
Fig 4. Transcriptional response of A. baumannii to Lcb. rhamnosus CRL 2244 CFCM.
The clinical isolates AMA3 (A) and AB5075 (B) were grown with and without sub-MIC concentration of lyophilized Lcb. rhamnosus CRL 2244 CFCM for 18 h at 37°C. The differential genes expression was determined by qRT-PCR of genes involved in the phenylacetic acid catabolic pathway (paaA and paaE), histidine (hutG, hutU, hutI and hutT) and iron metabolism (bauA and pirA), genes associated with antibiotic resistance (ampC, oprD and blaOXA-23), genes related to biofilm formation (ompA, csuAB, csuB and csuB), genes involved in programmed cell death (IrgA/cirA and IrgB), and genes associated with bacterial virulence, including those related to zinc metabolism (zigA), pili production (pilA), efflux functions (baeS), and the global transcriptional regulator hns. The data presented are the mean ± standard deviation (SD) of normalized relative quantities (NRQ) derived from transcript levels calculated using the qBASE method. Statistical significance (P < 0.05) was determined by one-way ANOVA followed by Tukey’s multiple-comparison test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. Technical triplicates using three independent biological samples were used to collect experimental data. Error bars represents the standard deviation (SD).
Taken together, these results indicate that the presence of Lcb. rhamnosus CRL 2244 CFCM in the extracellular medium results in the differential expression of genes associated with a variety of cellular functions such as cell metabolism, host pathogen-interactions and even bacterial death. However, this transcriptional response varies between AB5075 and AMA3, with the latter strain displaying a higher number of differentially transcribed genes.
Discussion
The lack of progress in the development of novel and effective antibiotics against MDR pathogens such as CRAB strains underscores the need for research into innovative approaches to address bacterial infections. Consequently, ongoing research efforts are focused on introducing therapeutic alternatives capable of slowing the evolution of antimicrobial resistance [26]. Probiotic LAB constitute a natural and safe therapeutic strategy. The antimicrobial activity of specific LAB strains or their extracellular products against pathogens is well documented in the literature [10, 12, 15]. LAB can produce numerous extracellular antimicrobial compounds because of their primary and secondary metabolisms, which can exert an antagonistic or antimicrobial action. These include the production of organic acids (mainly lactic and acetic acid), with a concomitant reduction in pH that limits the growth of other microorganisms [12, 27]; the production of other antimicrobial substances such as ethanol, diacetyl, hydrogen peroxide, reuterin and bacteriocins, among others [13, 27–30].
Nevertheless, limited research has been conducted on the potential use of LAB against extremely resistant strains such as CRAB clinical isolates [10, 13]. Published studies have indicated the antagonistic effects of different LAB against A. baumannii; however, there remains a need to identify the specific effectors and mechanisms responsible for their biological actions [10–13, 15]. Previously, in co-culture conditions, we observed a potent lethal activity of Lcb. rhamnosus CRL 2244 on A. baumannii strains [15]. Also, the transcriptomic analysis of A. baumannii cells co-cultured with Lcb. rhamnosus CRL 2244, revealed the differential expression of several genes mainly associated with metabolic pathways [15]. Our results indicated that A. baumannii actively responds to the stress imposed by LAB to enhance its survival strategies [15]. Based on these preliminary and published observations, we further evaluated in this present study whether the secretion of a substance(s) by Lcb. rhamnosus CRL2244 has the potential to induce or contribute to the death of CRAB cells as well as other MDR pathogens. Our findings revealed that the CFCM obtained after 96-h incubation presents a potent antimicrobial activity, with greater efficacy against Gram-negative bacilli, particularly CRAB strains. The transwell assay validated the antimicrobial effect observed with CFCM, indicating the presence of a compound(s) in the culture medium responsible for the killing activity. Notably, variations in pH, temperature, and exposure to different enzymes did not have a significant impact on the antimicrobial activity. Ladha et al. (2020) [11] identified a natural low molecular weight compound, containing a piperazine ring, produced by L. plantarum LJR13. This compound can inhibit the growth of A. baumannii, Listeria monocytogenes and S. aureus by forming vesicles and pores in the cell wall of sensitive cells. Other authors identified a common compound in the supernatant of twenty LAB strains, including species such as L. plantarum, L. paraplantarum, Pediococcus pentosaceus, P. acidilactici, Pediococcus spp., and Enterococcus spp. This compound, belonging to the diketopiperazine group, proved to be an effective agent against extensively drug-resistant A. baumannii [13]. Although the structure of the compound(s) secreted by Lcb. rhamnosus CRL 2244 remains unknown, the results presented in here suggest that it is a low molecular weight non-peptide organic compound that is actively stable after being exposed to changes in temperature, pH and enzymatic treatment.
Interestingly, the compound produced by Lcb. rhamnosus CRL2244 exhibits a synergistic interaction with the β-lactam antibiotic meropenem, restoring susceptibility to it in carbapenem-resistant strains. The synergistic interaction between natural compounds or other bioactive components, such as biogenic metallic nanoparticles, and existing antibiotics proves to be an effective approach in combating the resistance phenomenon, as they can facilitate the interaction between an antimicrobial agent and its target within the pathogen [21, 31–33]. Several authors have documented synergistic effects between natural products and antibiotics from different classes, employing diverse mechanisms against various MDR pathogens [21, 33–35]. However, understanding the mode of action of the Lcb. rhamnosus CRL2244 secreted compound(s) and its synergistic interaction with clinically relevant antibiotics requires further investigation. Nevertheless, we hypothesize that influencing the integrity of the cell membrane, as suggested by morphological changes observed in SEM images, such as elongation of cell length and a smoother bacterial surface appearance, is one of the mechanisms by which the Lcb. rhamnosus CRL2244 exerts it antibacterial activity.
The analysis of the transcriptional response A. baumannii exposed to a sub-MIC concentration of the CFCM from Lcb. rhamnosus CRL 2244 revealed an overexpression of genes coding for critical metabolic functions. These functions include the acquisition of the essential metals Zn and Fe, as well as the expression of key metabolic pathways, including the PAA catabolic pathway and the expression of the Hut system. It is well documented that the paa operon is one of the most differentially regulated A. baumannii pathways in response to numerous stress conditions, including the presence of antibiotics, and its role in antibiotic resistance; whereas the Hut histidine catabolic pathway provides carbon, nitrogen energy during infection [36, 37].
Another noteworthy finding is the significant overexpression of the cidA/lrgA genes, also consistent with our previous observations collected using a co-culture approach [15]. These genes are linked to programmed cell death in other bacteria, potentially through holin-antiholin activity or the regulation of cell death during overflow metabolism [38, 39]. Considering all these observations, we can speculate that the compound(s) produced and secreted by Lcb. rhamnosus CRL2244 modulate the A. baumannii transcriptional response to combat stress signals, which ultimately can affect cell integrity.
Conclusions
The present work revealed Lcb. rhamnosus CRL2244 releases a small compound(s) that has bactericidal activity against different MDR pathogens. In addition, a decrease of MIC values against selected MDR strains was detected when CFCM samples were combined with meropenem. We hypothesize that this response could be due to changes in cell morphology, the differential expression of genes coding for critical metabolic pathways and the expression of A. baumannii virulence functions including biofilm formation. Future experiments to further characterize and identify the nature and chemical structure of the released compounds(s) are needed. In sum, the results presented in this report, suggest the potential use of Lcb. rhamnosus CRL2244 released compound(s) as alternative and/or complementary options to treat infections caused by multi-drug resistant bacterial pathogens.
Supporting information
(DOCX)
(TIF)
The assays were performed in three independent technical and biological samples.
(TIF)
Bacterial cultures were gown on CAMHA supplemented with 2% glycerol and 40 μg/mL of Congo red. This is a representative image of assays performed in technical and biological triplicates.
(TIF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
NIH SC3GM125556 to MSR, R01AI100560, R01AI063517, R01AI072219 to RAB; PICT2018-03233 to RR and PIP 2020-817 and PICT 2021-00458 to CR. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to RAB from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 to RAB. There was no additional external funding received for this study.
References
- 1.(WHO). WHO. Global Priority List of Antibiotic-resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. [Google Scholar]
- 2.Cassini A, Hogberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. Epub 20181105. doi: 10.1016/S1473-3099(18)30605-4 ; PubMed Central PMCID: PMC6300481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. Antibiotic Resistance Threats in the United States. Atlanta, GA: US Department of Health and Human Services, CDC; 2019. 2019. [Google Scholar]
- 4.WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. News release by the WHO. 2017. [Google Scholar]
- 5.Abdul-Mutakabbir JC, Nguyen L, Maassen PT, Stamper KC, Kebriaei R, Kaye KS, et al. In Vitro Antibacterial Activity of Cefiderocol against Multidrug-Resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2021;65(9):e0264620. Epub 20210817. doi: 10.1128/AAC.02646-20 ; PubMed Central PMCID: PMC8370208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He S, He H, Chen Y, Chen Y, Wang W, Yu D. In vitro and in vivo analysis of antimicrobial agents alone and in combination against multi-drug resistant Acinetobacter baumannii. Front Microbiol. 2015;6:507. Epub 20150527. doi: 10.3389/fmicb.2015.00507 ; PubMed Central PMCID: PMC4444844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theuretzbacher U, Bush K, Harbarth S, Paul M, Rex JH, Tacconelli E, et al. Critical analysis of antibacterial agents in clinical development. Nature reviews Microbiology. 2020;18(5):286–98. Epub 20200309. doi: 10.1038/s41579-020-0340-0 . [DOI] [PubMed] [Google Scholar]
- 8.Andrei S, Droc G, Stefan G. FDA approved antibacterial drugs: 2018–2019. Discoveries (Craiova). 2019;7(4):e102. Epub 20191231. doi: 10.15190/d.2019.15 ; PubMed Central PMCID: PMC7086080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keam SJ. Sulbactam/Durlobactam: First Approval. Drugs. 2023;83(13):1245–52. doi: 10.1007/s40265-023-01920-6 . [DOI] [PubMed] [Google Scholar]
- 10.Stanbro J, Park JM, Bond M, Stockelman MG, Simons MP, Watters C. Topical Delivery of Lactobacillus Culture Supernatant Increases Survival and Wound Resolution in Traumatic Acinetobacter baumannii Infections. Probiotics Antimicrob Proteins. 2020;12(3):809–18. doi: 10.1007/s12602-019-09603-z . [DOI] [PubMed] [Google Scholar]
- 11.Ladha G, Jeevaratnam K. A novel antibacterial compound produced by Lactobacillus plantarum LJR13 isolated from rumen liquor of goat effectively controls multi-drug resistant human pathogens. Microbiol Res. 2020;241:126563. Epub 20200801. doi: 10.1016/j.micres.2020.126563 . [DOI] [PubMed] [Google Scholar]
- 12.Rastogi S, Mittal V, Singh A. Selection of Potential Probiotic Bacteria from Exclusively Breastfed Infant Faeces with Antagonistic Activity Against Multidrug-Resistant ESKAPE Pathogens. Probiotics Antimicrob Proteins. 2021;13(3):739–50. Epub 20201114. doi: 10.1007/s12602-020-09724-w . [DOI] [PubMed] [Google Scholar]
- 13.Yap PC, Ayuhan N, Woon JJ, Teh CSJ, Lee VS, Azman AS, et al. Profiling of Potential Antibacterial Compounds of Lactic Acid Bacteria against Extremely Drug Resistant (XDR) Acinetobacter baumannii. Molecules. 2021;26(6). Epub 20210319. doi: 10.3390/molecules26061727 ; PubMed Central PMCID: PMC8003687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cazorla SI, Maldonado-Galdeano C, Weill R, De Paula J, Perdigon GDV. Oral Administration of Probiotics Increases Paneth Cells and Intestinal Antimicrobial Activity. Front Microbiol. 2018;9:736. Epub 20180416. doi: 10.3389/fmicb.2018.00736 ; PubMed Central PMCID: PMC5911494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez C, Ramlaoui D, Georgeos N, Gasca B, Leal C, Subils T, et al. Antimicrobial activity of the Lacticaseibacillus rhamnosus CRL 2244 and its impact on the phenotypic and transcriptional responses in carbapenem resistant Acinetobacter baumannii. Sci Rep. 2023;13(1):14323. Epub 20230831. doi: 10.1038/s41598-023-41334-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traglia GM, Chua K, Centron D, Tolmasky ME, Ramirez MS. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome biology and evolution. 2014;6(9):2235–9. doi: 10.1093/gbe/evu176 ; PubMed Central PMCID: PMC4202317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez MS, Don M, Merkier AK, Bistue AJ, Zorreguieta A, Centron D, et al. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol. 2010;48(4):1488–90. Epub 2010/02/26. doi: 10.1128/JCM.01264-09 [pii] ; PubMed Central PMCID: PMC2849597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, et al. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. MBio. 2014;5(3):e01076–14. doi: 10.1128/mBio.01076-14 ; PubMed Central PMCID: PMC4045072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams MD, Pasteran F, Traglia GM, Martinez J, Huang F, Liu C, et al. Distinct mechanisms of dissemination of NDM-1 metallo- beta-lactamase in Acinetobacter spp. in Argentina. Antimicrob Agents Chemother. 2020. Epub 2020/03/04. doi: 10.1128/AAC.00324-20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halder D, Mandal M, Chatterjee SS, Pal NK, Mandal S. Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria. Biomedicines. 2017;5(2). Epub 20170616. doi: 10.3390/biomedicines5020031 ; PubMed Central PMCID: PMC5489817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Otibi FO, Yassin MT, Al-Askar AA, Maniah K. Green Biofabrication of Silver Nanoparticles of Potential Synergistic Activity with Antibacterial and Antifungal Agents against Some Nosocomial Pathogens. Microorganisms. 2023;11(4). Epub 20230404. doi: 10.3390/microorganisms11040945 ; PubMed Central PMCID: PMC10144991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: A review. J Pharm Anal. 2016;6(2):71–9. Epub 20151202. doi: 10.1016/j.jpha.2015.11.005 ; PubMed Central PMCID: PMC5762448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura B, Escalante J, Mezcord V, Tuttobene MR, Subils T, Actis LA, et al. Human serum albumin-induced modification of Ton-B-dependent receptor expression in cefiderocol-exposed carbapenem-resistant Acinetobacter baumannii. Int J Antimicrob Agents. 2023;62(5):106950. Epub 20230818. doi: 10.1016/j.ijantimicag.2023.106950 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mezcord V, Escalante J, Nishimura B, Traglia GM, Sharma R, Valle Q, et al. Induced Heteroresistance in Carbapenem-Resistant Acinetobacter baumannii (CRAB) via Exposure to Human Pleural Fluid (HPF) and Its Impact on Cefiderocol Susceptibility. International journal of molecular sciences. 2023;24(14). Epub 20230721. doi: 10.3390/ijms241411752 ; PubMed Central PMCID: PMC10380697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bentancor LV, O’Malley JM, Bozkurt-Guzel C, Pier GB, Maira-Litran T. Poly-N-acetyl-beta-(1–6)-glucosamine is a target for protective immunity against Acinetobacter baumannii infections. Infect Immun. 2012;80(2):651–6. Epub 20111121. doi: 10.1128/IAI.05653-11 ; PubMed Central PMCID: PMC3264292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma YX, Wang CY, Li YY, Li J, Wan QQ, Chen JH, et al. Considerations and Caveats in Combating ESKAPE Pathogens against Nosocomial Infections. Adv Sci (Weinh). 2020;7(1):1901872. Epub 20191205. doi: 10.1002/advs.201901872 ; PubMed Central PMCID: PMC6947519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biswas S, Turner L, Biswas I. Lactobacillus rhamnosus LRB mediated inhibition of oral streptococci. Mol Oral Microbiol. 2018;33(5):396–405. doi: 10.1111/omi.12242 . [DOI] [PubMed] [Google Scholar]
- 28.Rwubuzizi R, Carneiro KO, Holzapfel WH, Vaz-Velho M, Todorov SD. Bacteriocin and Antioxidant Production, a Beneficial Properties of Lactic Acid Bacteria Isolated from Fermented Vegetables of Northwest Bulgaria. Probiotics Antimicrob Proteins. 2023. Epub 20230817. doi: 10.1007/s12602-023-10140-z . [DOI] [PubMed] [Google Scholar]
- 29.Millette M, Cornut G, Dupont C, Shareck F, Archambault D, Lacroix M. Capacity of human nisin- and pediocin-producing lactic Acid bacteria to reduce intestinal colonization by vancomycin-resistant enterococci. Appl Environ Microbiol. 2008;74(7):1997–2003. Epub 20080201. doi: 10.1128/AEM.02150-07 ; PubMed Central PMCID: PMC2292579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Lira FM, Tanaka FYR, Rios EA, Carrilho SM, de Abreu SS, Ferreira GF, et al. Identification of lactic acid bacteria with anti-listeria activity. Characterization and application of a bacteriocinogenic strain in the control of Listeria monocytogenes in cheese. J Dairy Res. 2023;90(3):318–23. Epub 20231018. doi: 10.1017/S0022029923000584 . [DOI] [PubMed] [Google Scholar]
- 31.Oikonomou O, Liakopoulos A, Phee LM, Betts J, Mevius D, Wareham DW. Providencia stuartii Isolates from Greece: Co-Carriage of Cephalosporin (blaSHV-5, blaVEB-1), Carbapenem (blaVIM-1), and Aminoglycoside (rmtB) Resistance Determinants by a Multidrug-Resistant Outbreak Clone. Microb Drug Resist. 2016;22(5):379–86. Epub 2016/07/06. doi: 10.1089/mdr.2015.0215 . [DOI] [PubMed] [Google Scholar]
- 32.Cheesman MJ, Ilanko A, Blonk B, Cock IE. Developing New Antimicrobial Therapies: Are Synergistic Combinations of Plant Extracts/Compounds with Conventional Antibiotics the Solution? Pharmacogn Rev. 2017;11(22):57–72. doi: 10.4103/phrev.phrev_21_17 ; PubMed Central PMCID: PMC5628525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aljeldah MM, Yassin MT, Mostafa AA, Aboul-Soud MAM. Synergistic Antibacterial Potential of Greenly Synthesized Silver Nanoparticles with Fosfomycin Against Some Nosocomial Bacterial Pathogens. Infect Drug Resist. 2023;16:125–42. Epub 20230106. doi: 10.2147/IDR.S394600 ; PubMed Central PMCID: PMC9831080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coutinho HD, Costa JG, Lima EO, Falcao-Silva VS, Siqueira-Junior JP. Enhancement of the antibiotic activity against a multiresistant Escherichia coli by Mentha arvensis L. and chlorpromazine. Chemotherapy. 2008;54(4):328–30. Epub 20080812. doi: 10.1159/000151267 . [DOI] [PubMed] [Google Scholar]
- 35.Hemaiswarya S, Doble M. Synergistic interaction of phenylpropanoids with antibiotics against bacteria. Journal of medical microbiology. 2010;59(Pt 12):1469–76. Epub 20100819. doi: 10.1099/jmm.0.022426-0 . [DOI] [PubMed] [Google Scholar]
- 36.Lonergan ZR, Nairn BL, Wang J, Hsu YP, Hesse LE, Beavers WN, et al. An Acinetobacter baumannii, Zinc-Regulated Peptidase Maintains Cell Wall Integrity during Immune-Mediated Nutrient Sequestration. Cell Rep. 2019;26(8):2009–18 e6. Epub 2019/02/21. doi: 10.1016/j.celrep.2019.01.089 ; PubMed Central PMCID: PMC6441547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooppaw AJ, McGuffey JC, Di Venanzio G, Ortiz-Marquez JC, Weber BS, Lightly TJ, et al. The Phenylacetic Acid Catabolic Pathway Regulates Antibiotic and Oxidative Stress Responses in Acinetobacter. mBio. 2022;13(3):e0186321. Epub 20220425. doi: 10.1128/mbio.01863-21 ; PubMed Central PMCID: PMC9239106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ranjit DK, Endres JL, Bayles KW. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol. 2011;193(10):2468–76. Epub 20110318. doi: 10.1128/JB.01545-10 ; PubMed Central PMCID: PMC3133170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endres JL, Chaudhari SS, Zhang X, Prahlad J, Wang SQ, Foley LA, et al. The Staphylococcus aureus CidA and LrgA Proteins Are Functional Holins Involved in the Transport of By-Products of Carbohydrate Metabolism. mBio. 2021;13(1):e0282721. Epub 20220201. doi: 10.1128/mbio.02827-21 ; PubMed Central PMCID: PMC8805020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(TIF)
The assays were performed in three independent technical and biological samples.
(TIF)
Bacterial cultures were gown on CAMHA supplemented with 2% glycerol and 40 μg/mL of Congo red. This is a representative image of assays performed in technical and biological triplicates.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.