Abstract
During infection with herpes simplex virus type 1 (HSV-1), VP16 serves multiple functions, including transcriptional activation of viral immediate early genes and downregulation of the virion host shutoff protein vhs. Furthermore, VP16 has been shown to be involved in some aspect of virus assembly and/or maturation. Experiments with a VP16 null virus, 8MA, suggested that VP16 plays a direct role in virion assembly, since removal of VP16 from the HSV-1 genome results in reduced levels of encapsidated DNA and a failure to produce extracellular enveloped particles. However, VP16 null mutants display a severe translational arrest due to unrestrained vhs activity, thus complicating interpretation of these data. We examine here the role of VP16 in virion assembly and egress in the context of a vhs null background, using the virus 8MA/ΔSma (VP16− vhs−). Comparison of 8MA and 8MA/ΔSma with respect to viral DNA accumulation and encapsidation and accumulation of the major capsid protein, VP5, revealed that the 8MA lethal phenotype is only partially due to uncontrolled vhs activity, indicating that VP16 is required in HSV-1 virion formation. Electron microscopy confirmed these results and further showed that VP16 is required for HSV-1 egress beyond the perinuclear space. In addition, we describe the isolation and characterization of an 8MA derivative capable of propagation on Vero cells, due to second site mutations in the vhs and UL53 (gK) genes. Taken together, these results show that VP16 is required for viral egress downstream of the initial envelopment step and further underscore the importance of VP16 in controlling vhs activity within an infected cell.
Herpes simplex virus type 1 (HSV-1) is a large DNA virus consisting of an icosahedral capsid surrounded by an amorphous protein layer termed the tegument and bounded by an envelope derived from host membranes (50). The tegument contains important regulatory proteins that are released into the newly infected cell following fusion of the virion envelope with the host cell plasma membrane. While the functions of many of the tegument proteins have yet to be precisely defined, several have been shown to aid in the initiation of the viral replicative cycle (50). Among these are VP16 (also known as Vmw65, ICP25, UL48, or α-TIF) (5, 11, 44, 46) and the virion host shutoff protein (vhs or UL41 gene product) (21, 34, 48, 52).
VP16 is an abundant 65-kDa virion phosphoprotein that is synthesized late in infection and subsequently packaged into virions (37–39). VP16 delivered by the infecting virion acts during the earliest stages of infection to stimulate transcription of the viral immediate-early (IE) genes, thereby facilitating the onset of the lytic program of viral gene expression (reviewed in references 41 and 50). Intensive studies have shown that the C-terminal portion of VP16 is a potent transcriptional activation domain and that VP16 is targeted to the TAATGARATTC consensus sequence found in IE promoters through interactions with the host factors Oct-1 and HCF. Mutations that inactivate the transcriptional activation function of VP16 result in reduced levels of IE gene expression during low-multiplicity infection and a greatly increased particle-to-PFU ratio, indicating that transactivation by VP16 increases the probability that cells infected with a single virus particle will enter the lytic cycle (2, 53, 56). Such transactivation-deficient mutants can, however, be propagated in tissue culture, demonstrating that the activation function of VP16 is not essential for virus replication and assembly. In contrast, certain VP16 mutations prevent production of infectious progeny virus: for example, Ace et al. demonstrated that a ts mutation in HSV-2 VP16 (ts2203) is lethal, as are some in-frame linker insertion mutations in the HSV-1 VP16 gene (1, 2). The ts2203 mutation blocks virus assembly, arguing that VP16 plays an essential role in this process. More recently, Weinheimer et al. provided additional evidence supporting a role for VP16 in virion maturation, by demonstrating that an HSV-1 VP16 null mutant (8MA) displays a severe defect in virus assembly during infection of noncomplementing cells (58). The 8MA mutant failed to produce extracellular enveloped virions and exhibited a marked reduction in the efficiency of packaging viral DNA into capsids.
Previous studies from our laboratory have uncovered an additional regulatory function of VP16 that potentially complicates the interpretation of the foregoing findings. We found that VP16 binds to the virion host shutoff (vhs) protein encoded by HSV gene UL41 and provided evidence that this interaction modulates vhs activity (36, 51). vhs is a tegument protein that triggers shutoff of host protein synthesis and accelerated degradation of both cellular and viral mRNAs (21, 34, 48, 52). Although the mechanism of vhs action has yet to be precisely defined, vhs displays significant amino acid sequence similarity to the fen-1 family of nucleases (15, 20), and current evidence strongly suggests that it is either an endo-RNase or a required subunit of an endo-RNase that also includes one or more cellular subunits (17, 18). The vhs-dependent RNase displays little if any sequence specificity and targets both viral and cellular mRNAs in vivo and in vitro (17, 18, 22, 33, 34, 36, 42, 43, 62). Lam et al. have shown that vhs activity is greatly enhanced during infection with the 8MA VP16 null mutant, leading to greatly exaggerated mRNA turnover and essentially complete translation arrest midway through the infective cycle (36). These observations suggested that VP16 downregulates vhs activity at intermediate and late times postinfection, thereby allowing the maintenance of viral protein synthesis. Consistent with this hypothesis, the translational arrest phenotype of 8MA was suppressed by a transcriptionally incompetent mutant form of VP16 which retained the ability to bind vhs and was reversed by inactivating the vhs gene (36). Taken in combination, these results demonstrated that VP16 acts posttranscriptionally to stimulate viral protein synthesis at intermediate and late times postinfection.
The severe decline in viral protein synthesis observed during infection with 8MA raises the possibility that the previously described effects of VP16 loss-of-function mutations on virus assembly might stem, at least in part, from reduced synthesis of one or more virion components rather than the absence of functional VP16 per se. Indeed, the VP16 homologue of varicella-zoster virus is dispensable for virus replication in tissue culture, demonstrating that VP16 does not play an essential role in the assembly of all alphaherpesviruses (12). We examine here the phenotype of a VP16 null mutant in which the vhs gene has been inactivated. We confirm that VP16 is essential for virus maturation, and we present evidence that VP16 is required for one or more steps in the egress pathway downstream of the initial envelopment event. In addition, our studies provide additional evidence that VP16 downregulates vhs activity.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were propagated in Dulbecco modified Eagle medium (DMEM) with 5% fetal bovine serum (FBS). 16-8 cells (kindly provided by S. Weinheimer) were maintained in DMEM with 5% FBS supplemented with 0.45 mg of G418 (Geneticin; Gibco-BRL) per ml. QL10 cells were propagated in histidine-deficient DMEM with 10% FBS supplemented with 1 mM histidinol (Sigma). 16-8 cells and QL10 cells are Vero cell derivatives that are stably transfected with the HSV-1 VP16 gene (36, 58). Both cell lines complement the VP16 null mutant 8MA; QL10 cells support efficient plaque formation by 8MA, while 16-8 cells generate a larger yield of infectious virus.
HSV-1 strains KOS, 8MA-R, and ΔSma were grown and titrated on Vero cells. Mutants encoding altered forms of VP16 (8MA, 8MA/ΔSma, and 8MA-V) were grown on 16-8 cells and titrated on QL10 cells.
Single-step growth analysis.
Vero or 16-8 cells (106 cells) were infected at a multiplicity of infection (MOI) of 5. After 1 h, unabsorbed virus was removed, the cell monolayer was extensively washed with phosphate-buffered saline, and DMEM–5% FBS was added. Cells were harvested into 1 ml at various times postinoculation, and virus titers were determined by plaque assay on QL10 cells. All growth analyses were performed in triplicate.
DNA encapsidation assay.
Encapsidation assays were performed as previously described (58). In brief, DNA from untreated and DNase I-treated cell extracts was cleaved with BamHI and then analyzed by Southern blot hybridization using the L-S junction-spanning BamHI K fragment as a probe. The efficiency of encapsidation was then deduced from the fraction of the total hybridization signal that was present in terminal fragments.
Immunoblotting analyses.
A total of 5 × 105 Vero cells were inoculated with virus at an MOI of 5, and infected cell lysates were harvested directly into sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) lysis buffer at the indicated times postinfection. Lysates were run on 9% polyacrylamide gels, transferred to nitrocellulose, and probed with the anti-VP5 antibody NC-1 (1:20,000 dilution), kindly provided by G. H. Cohen and R. J. Eisenberg.
Electron microscopy.
Cells in 100-mm dishes were infected with virus at the indicated MOI for 24 h and fixed for 1 h with 1% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3), followed by several washes in the buffer. The samples were postfixed with 1% osmium tetroxide and dehydrated in a graded series of ethanol baths. Prior to embedding in 100% LX112, samples were transferred to propylene oxide followed by a 1:1 mix of propylene oxide and LX112. Ultrathin sections were cut and stained with uranyl acetate and lead citrate and then examined using a Philips model 410 transmission electron microscope.
Metabolic labeling.
Vero cells (4 × 105) were inoculated with virus at an MOI of 10. Complete medium was replaced with medium minus methionine supplemented with 50 μCi of [35S]methionine for 1 h prior to harvesting of infected monolayers. Lysates were subjected to SDS-PAGE analysis, treated with EN3HANCE according to the manufacturer's specifications, and visualized by autoradiography.
Mapping of second-site mutations in 8MA-V.
Initial Southern blot analysis of the vhs locus of 8MA-V revealed a ca. 800-nucleotide deletion which removed both internal SmaI sites (residues 442 and 1030 in the vhs open reading frame), while maintaining the PvuI and EcoRV restriction sites at residues 246 and 1380, respectively. Primers external to these latter sites were generated, and the resulting PCR fragment was sequenced in order to define the deletion endpoints. Mutations in the UL53 gene were mapped by sequencing overlapping PCR fragments spanning the entire UL53 open reading frame. Products of three independent PCR reactions were sequenced for each interval in order to avoid fidelity problems associated with Taq polymerase. All PCR reactions contained 30 mM Tricine (pH 8.4), 2 mM MgCl2, 5 μM β-mercaptoethanol, 0.1% Thesit, 0.2 mM concentrations of each deoxynucleoside triphosphate, 1 μM concentrations of each primer, 10% dimethyl sulfoxide, and 1 U of Taq polymerase. Sequencing was performed on an ABI373 DNA sequencer with Taq cycling to >5-fold redundancy.
Construction of recombinant viruses.
The BamHI L fragment containing the intact UL53 gene from 8MA-V was cloned into pUC19 creating the plasmid pKM1. To construct 8MA/UL53syn, 8MA/ΔSma/UL53syn, and KOS/UL53syn, 4 μg of infectious 8MA, 8MA/ΔSma, and KOS DNA, respectively, were cotransfected into 16-8 cells with 0.5 μg of HindIII-linearized pKM1 using Lipofectamine (Gibco-BRL). Recombinant viruses displaying syncytial lesions were subsequently plaque purified three times on QL10 monolayers. The UL53 gene of each recombinant was amplified by PCR and sequenced to confirm that both point mutations present in the UL53 gene of 8MA-V were present.
Propagation of syncytial viruses.
Syncytial viruses were serially propagated on Vero cells by allowing infected monolayers to proceed to a stage where all cells were rounded and floating in the culture medium. An aliquot of this medium was then added to the culture medium of a fresh Vero monolayer. Successive passages were performed in this manner.
RESULTS
The lethal defect of a VP16 null mutant is not overcome by inactivating vhs.
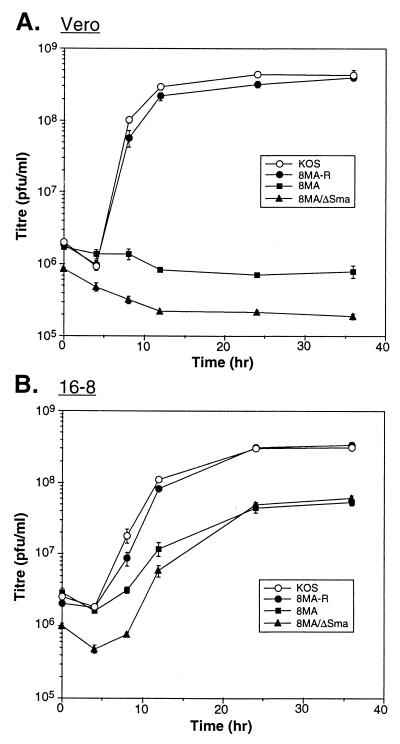
As reviewed in the introduction, the VP16-null mutant 8MA undergoes a severe vhs-induced decline in viral protein synthesis midway through infection of noncomplementing cells. This finding raised the possibility that the lethal defect displayed by 8MA and other VP16 loss-of-function mutants might stem (at least in part) from unrestrained vhs action rather than the absence of functional VP16 per se. Specifically, it seemed possible that reduced expression of one or more virion structural components might contribute to the defects in DNA packaging and virion assembly observed by Weinheimer et al. (58). The translational arrest phenotype of 8MA is eliminated when the vhs gene is inactivated in the double mutant 8MA/ΔSma (36). To determine if inactivation of vhs detectably alleviates the lethal defect of the 8MA mutation, single-step growth experiments were performed. Noncomplementing Vero cells and complementing 16-8 cells were infected with 8MA (VP16 null), 8MA/ΔSma (VP16 null-vhs inactivated), 8MA-R (a VP16+ rescue product of 8MA), and wild-type HSV-1 KOS at an MOI of 5, and infected cultures were assayed for production of infectious progeny virus by titration on complementing QL10 cells. KOS and 8MA-R replicated efficiently in both Vero and 16-8 cells, while 8MA and 8MA/ΔSma replicated only in 16-8 cells (Fig. 1). The yield of 8MA and 8MA/ΔSma was consistently lower than that of KOS and 8MA-R on 16-8 cells, indicating that this cell line only partially complements the VP16 mutation under our conditions of infection. We conclude from this experiment that, although uncontrolled vhs activity may contribute to the lethal phenotype of 8MA (see below), VP16 serves at least one additional function that is required for the formation of infectious progeny virus.
FIG. 1.
Single-step growth assay. Triplicate monolayers of Vero (A) or 16-8 (B) cells were infected with KOS, 8MA-R, 8MA, or 8MA/ΔSma at an MOI of 5. At the indicated times postinfection, monolayers were harvested and titers were determined on QL10 cells.
Inactivation of vhs partially relieves the DNA encapsidation defect of 8MA.
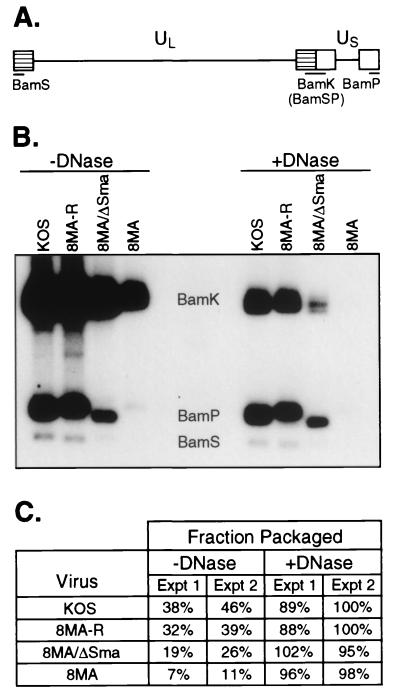
Weinheimer et. al. reported that 8MA displays a 50 to 75% reduction in the efficiency of DNA encapsidation (58). We asked if this defect was detectably alleviated by inactivating vhs. HSV DNA replication produces large “endless” concatamers which are cleaved to unit length during packaging into capsids (29, 35). We examined the ratio of unit length to concatameric viral DNA by Southern blot analysis as a measure of the encapsidation efficiency. As illustrated in Fig. 2A, the BamHI K, P, and S fragments are derived from the inverted repeat sequences that flank the L and S components of viral DNA. BamHI K arises from the L-S junction, while BamHI S and P are derived from the L and S termini, respectively. Thus, BamHI digestion of unit length viral DNA gives rise to equimolar yields of the terminal BamHI S and P fragments and the joint-spanning BamHI K fragment. In contrast, concatamers yield predominantly BamHI K due to the paucity of free genomic ends.
FIG. 2.
DNA encapsidation assay. (A) Schematic diagram of the HSV-1 genome. The unique long (UL) and unique short (US) regions are flanked by the long and short inverted repeats (hatched and open boxes, respectively). Digestion by BamHI within these repeats results in the indicated BamHI restriction fragments. (B) Southern blot analysis of HSV-1 DNA. Infected Vero cell lysates were harvested, either treated or left untreated with DNase I, and subjected to SDS and proteinase K treatment. DNA was subsequently extracted for BamHI digestion. The locations of the BamHI K, S, and P restriction fragments are indicated. (C) Quantitation of the fraction of packaged DNA. The intensities of the BamHI K, S, and P fragments from panel B (experiment [Expt] 1) were quantitated by phosphorimager analyses, and the fraction of packaged viral DNA was determined. Quantitation of an independent experiment not illustrated in panel B is also shown (experiment 2).
Vero cells were infected with KOS, 8MA-R, 8MA/ΔSma, and 8MA. Viral DNA was then extracted from untreated and DNase-treated cell extracts at 24 h postinfection, cleaved with BamHI, and analyzed by Southern blot hybridization with a radiolabeled BamHI K probe (Fig. 2B). Signal intensities were quantified by phosphorimager analysis, and the fraction of the total signal present in terminal fragments was determined (i.e., [P + S]/[K + P + S]). This value was then multiplied by 2 to yield the proportion of packaged genomes (Fig. 2C). The DNase I-treated samples yielded estimates of packaging efficiency that closely approximated the predicted value of 100% for DNase I-resistant (i.e., encapsidated) DNA. Approximately 38 to 46% of the DNA detected in untreated samples derived from KOS-infected cells was packaged into virions. In contrast, only 7 to 11% of 8MA DNA was packaged. The packaging defect of 8MA was substantially corrected in 8MA-R, demonstrating that it stems (either directly or indirectly) from the loss of VP16. 8MA/ΔSma displayed intermediate values of 19 to 26%. These data confirm the earlier report that 8MA displays a packaging defect and indicate that deletion of vhs partially corrects this phenotype. Quantitation of the total yield of DNA in untreated samples demonstrated that, compared to KOS, the amounts of 8MA and 8MA/ΔSma DNA are approximately one-third and one-half, respectively. A similar reduction in the total yield of viral DNA was previously noted by Weinheimer et al. (58). Whether this stems from a reduced rate of viral DNA synthesis and/or instability of unpackaged DNA in the infected cell remains to be determined.
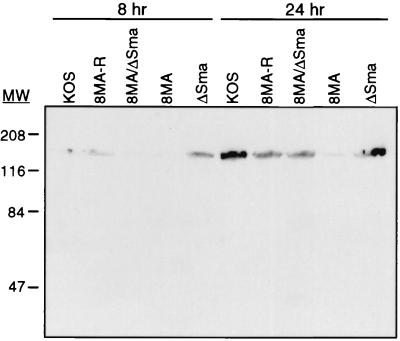
It seemed plausible that the decline in the rate of viral protein synthesis observed midway through infection with 8MA could lead to a significant reduction in the accumulated levels of virion components, thereby potentially explaining (at least in part) the encapsidation defect. As one approach to examining this possibility, we monitored the accumulation of the major capsid protein VP5 by Western blot analysis (Fig. 3). The results indicated that 8MA accumulated greatly reduced levels of VP5 compared to KOS and 8MA-R by 24 h postinfection. As predicted, this defect was substantially corrected by inactivating vhs in 8MA/ΔSma. These data document that vhs limits the accumulation of an essential structural component of the virion during infection with 8MA and are consistent with the hypothesis that this effect contributes to the reduced levels of DNA encapsidation.
FIG. 3.
Western blot analysis of HSV-1-infected Vero cells. Vero cells were infected with the indicated viruses at an MOI of 5 and harvested into SDS-PAGE lysis buffer at 8 or 24 h postinfection. The major capsid protein, VP5, was visualized by using a 1:20,000 dilution of NC-1. The location of molecular mass markers are indicated in kilodaltons.
VP16 is required for viral egress at a step after initial envelopment.
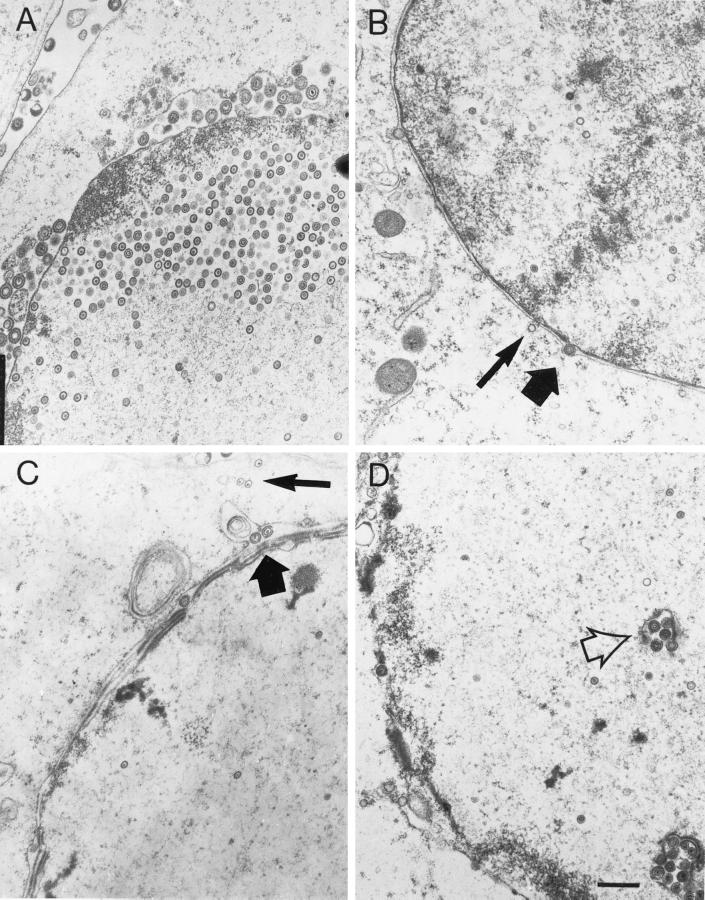
The foregoing results indicated that vhs contributes to reduced accumulation of VP5 and a lower efficiency of DNA encapsidation during infection with 8MA. We therefore reevaluated the role of VP16 in virion assembly and egress in a situation where these potentially confounding secondary effects were eliminated. To this end, we used transmission electron microscopy (TEM) to characterize virion assembly and egress during infection of noncomplementing cells with 8MA and 8MA/ΔSma (Fig. 4 and Table 1). Vero cell cultures were fixed and prepared for TEM 24 h postinfection with 8MA, 8MA/ΔSma, and KOS. As a control, 16-8 cells were also infected with the same viral strains, and no significant differences were observed other than a slightly higher rate of infection with KOS relative to 8MA and 8MA/ΔSma (data not shown).
FIG. 4.
TEM of HSV-1-infected Vero cells. Vero cells were infected with KOS (A), 8MA (B), or 8MA/ΔSma (C and D) at an MOI of 5 for 24 h. Within 8MA- and 8MA/ΔSma-infected cells, empty and full capsids are observed within the cytoplasm (thin arrows), while enveloped capsids accumulate within the perinuclear space (thick arrows). 8MA/ΔSma-infected cell nuclei often harbor enveloped virions within nuclear vacuoles believed to result from cross-sectioning of invaginated nuclear membranes (open arrow). Bar, 0.5 μm (panel D).
TABLE 1.
Quantitative analysis of Vero cells infected with various strains of HSV-1
| Parameter | Strain
|
||
|---|---|---|---|
| KOS | 8MA/ΔSma | 8MA | |
| No. of cells counted | 20 | 30 | 30 |
| No. of cells with visible viral particles | 19 | 19 | 12 |
| No. of capsids/cell counted in: | |||
| Nucleus | 55.6 | 8.3 | 2.2 |
| Cytoplasm | 6.1 | 0.9 | 0.2 |
| No. of enveloped particles/cell counted in: | |||
| Perinuclear space | 6.5 | 2.7 | 0.2 |
| Cytoplasm | 1.1 | 0 | 0 |
| Extracellular space | 23 | 0 | 0 |
| Total no. of viral particles/cell counted | 92.3 | 11.9 | 2.6 |
| Total no. of viral particles relative to KOS (%) | 100 | 13 | 3 |
| % Packaged DNAa | 100 | 31 | 4 |
Amount of packaged DNA relative to KOS as calculated from the total signal of samples treated with DNase (see Fig. 2, experiment 1).
As expected, Vero cells infected with wild-type HSV-1 KOS showed abundant nuclear capsids, the majority of which appeared to contain viral DNA (Fig. 4A). In a small number of cells, naked cytoplasmic capsids were also observed, and the majority of these appeared to be full (Table 1 and data not shown). Enveloped capsids were routinely observed between the inner and outer nuclear membranes, on the cell surface, and in the extracellular space and were occasionally observed within the cytoplasm, predominantly within membrane-bound structures. Virtually every cell within the KOS-infected sample harbored recognizable HSV-1 capsids and enveloped virions. In contrast, numerous cells within the 8MA- or 8MA/ΔSma-infected cultures were void of visible viral particles (Table 1). Moreover, individual 8MA-infected cells contained substantially fewer HSV particles than those infected with KOS or 8MA-R. For example, the cell depicted in Fig. 4B represents the greatest abundance of viral particles observed following 8MA infection. Compared to KOS, a higher percentage of the nuclear 8MA capsids appeared to be empty, a finding consistent with the encapsidation results, and individual capsids observed within the cytoplasm were predominantly empty (Fig. 4B, indicated by the thin arrow). Enveloped particles were rare, and those that were observed were located within the perinuclear space (Fig. 4B, thick arrow).
Several interesting features were observed in Vero cells infected with 8MA/ΔSma (Fig. 4C and D and Table 1). First, the total number of capsids (naked plus enveloped) was increased substantially relative to 8MA but did not approach the levels obtained with wild-type virus (Table 1). While this finding parallels the effects of deleting vhs on the total levels of packaged DNA (Table 1) and VP5 accumulation (Fig. 3), the magnitude of the increase in capsid accumulation was somewhat less than the increase in packaged DNA (13 and 31% of KOS, respectively [Table 1]) and markedly less than the enhancement of VP5 production. Although the basis for these differences remains unclear, we note that the efficiencies of capsid assembly and DNA packaging are not necessarily linear functions of the levels of viral protein production.
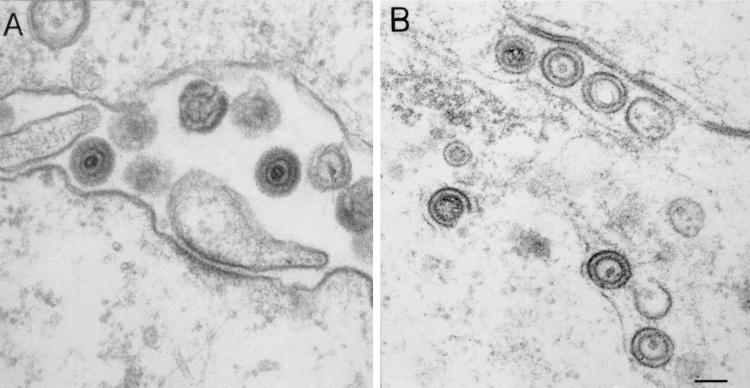
A second feature of 8MA/ΔSma is that naked capsids were observed within the cytoplasm (Fig. 4C, thin arrow), and the majority of these appeared to contain DNA. Third, enveloped particles were readily observed between the inner and outer nuclear membranes (Fig. 4C, thick arrow) and in membrane-bound intranuclear structures (Fig. 4D, open arrow) but never within the cytoplasm or extracellular space. Nuclear vacuoles harboring HSV-1 particles have been reported previously and are believed to be continuous with the perinuclear cisterna, resulting from cross-sectioning of nuclear membrane invaginations (13). Although we occasionally observed enveloped KOS particles within such nuclear structures, the incidence was much greater in cells infected with 8MA/ΔSma (data not shown). The significance of this observation is unknown. Fourth, the morphology of the enveloped 8MA/ΔSma particles differed in two respects from that of KOS virions (Fig. 5). Enveloped KOS virions were heterogeneous in diameter, reflecting a variable distance between the capsid and the envelope (Fig. 5A). In contrast, 8MA/ΔSma virions displayed a smaller and more uniform diameter, with the envelope tightly wrapped around the capsid (Fig. 5B). In addition, 8MA/ΔSma envelopes appeared as sharp thin lines and lacked the fuzzy appearance of KOS envelopes. Although such “thin” envelopes are occasionally observed in KOS infected cells (data not shown), their composition and significance remain unknown.
FIG. 5.
Morphology of KOS and 8MA/ΔSma enveloped virions. Enveloped virions from KOS- (A) or 8MA/ΔSma (B)-infected Vero cells are shown at a high magnification to illustrate the difference in morphology of the envelopes. Bar, 0.1 μm.
Taken in combination, these data indicate that both 8MA and 8MA/ΔSma virions are able to acquire an envelope by budding through the inner nuclear membrane. However, no enveloped particles could be detected in the cytoplasm or extracellular space. These findings argue that VP16 is required for one or more steps in the virion egress pathway, downstream of the initial envelopment step. Inasmuch as no infectious virus can be detected within cells infected with 8MA/ΔSma (Fig. 1), the enveloped particles observed between the nuclear membranes are almost certainly defective. However, we have not yet been able to purify sufficient quantities of these particles to characterize the nature of their defect further.
Second-site mutations allow propagation of VP16-null virus on noncomplementing cells.
The data outlined above suggest that deletion of VP16 impairs the production of infectious progeny virions in at least two distinct ways. First, unrestrained vhs action reduces the production of virion components and limits DNA packaging. Second, loss of VP16 appears to block viral egress at a step downstream of the initial envelopment event at the inner nuclear membrane and prevents acquisition of infectivity. Independent evidence supporting these conclusions emerged through the analysis of a derivative of 8MA that is capable of limited propagation on noncomplementing cells through cell-cell fusion.
During the routine characterization of a stock of 8MA, we plated 106 PFU onto 5 × 107 Vero cells. As expected, no plaques were observed after 3 days, demonstrating that the stock was devoid of VP16+ contaminants. The flask was then returned to the incubator, and 2 weeks later a single large syncytial plaque (ca. 4 cm in diameter) was observed in the monolayer. No infectious virus was detected in the extracellular medium by plaque assay on complementing QL10 cells. The syncytial area of the monolayer was detached by manual agitation and inoculated onto uninfected Vero cells. The monolayer gradually fused into a single large syncytium over the course of 2 weeks. Although no infectious virus was present in the medium or in cell extracts, we were able to serially passage the infection by transferring infected cells onto fresh cells. Unfortunately, no record was kept of the number of subcultures. After approximately 4 months, the rate of propagation significantly increased. At this point, an aliquot of the infected cells was added to complementing 16-8 cells to rescue the resident virus as an infectious virus stock. The resulting stock was plaque purified on complementing QL10 cells to yield isolate 8MA-V, which was then expanded into a high-titer stock on 16-8 cells. Stocks of 8MA-V were subsequently maintained on 16-8 cells. Although 8MA-V forms syncytial plaques on Vero cells (see Fig. 8C), no infectious virus capable of growth on complementing cells could be detected in the medium or in extracts of the infected cells. However, 8MA-V could be passaged on Vero cells by serially inoculating infected cells onto monolayers of uninfected cells.
FIG. 8.
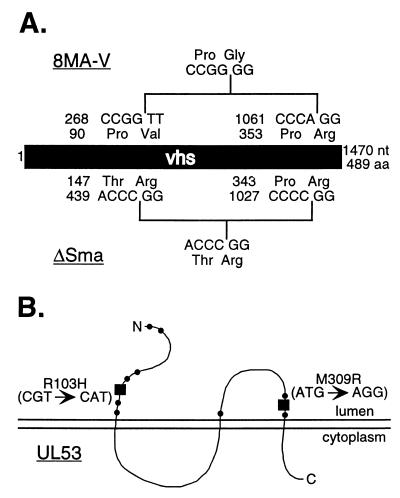
Mapping of lesions within the 8MA-V genome. (A) An in-frame deletion within the vhs gene of 8MA-V removes residues 91 to 354. The location of the in-frame SmaI deletion of ΔSma is shown for comparison. Both the nucleotide and the resulting amino acid sequences are indicated. (B) Two independent point mutations within the UL53 gene of 8MA-V were found that altered residues 103 and 309 (■). Each altered residue is located within an ectodomain where known syncytial mutations reside (●).
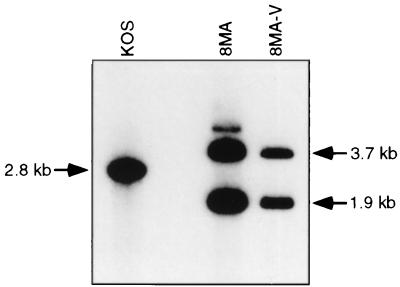
We used Southern blot analysis to demonstrate that 8MA-V retained the VP16 null mutation present in the parental 8MA mutant (Fig. 6). Vero cells were infected with KOS, 8MA, and 8MA-V, and total cellular DNA extracted 24 h postinfection was digested with PstI and XhoI and hybridized to a 2.8-kb PstI/XhoI fragment spanning the VP16 locus. KOS produced the expected 2.8-kb VP16 fragment, while 8MA-V displayed the 3.7- and 1.9-kb fragments predicted for 8MA VP16 deletion-lacZ substitution mutation (58). The 8MA mutation removes all but eight carboxy-terminal residues of VP16 (58). Therefore, these data implied that 8MA-V harbors one or more second-site mutations in other genes that allow limited propagation in the absence of VP16.
FIG. 6.
Southern blot analysis of 8MA-V DNA. Total DNA harvested from Vero cells infected with KOS, 8MA, or 8MA-V was cleaved with PstI and XhoI. The resulting Southern blot was hybridized with a 2.8-kb PstI/XhoI fragment spanning the VP16 locus. 8MA-V displayed the 3.7- and 1.9-kb VP16 fragments indicative of the 8MA VP16 deletion-lacZ substitution mutation.
8MA-V bears a large in-frame deletion at the vhs locus and two point mutations in the UL53 gene.
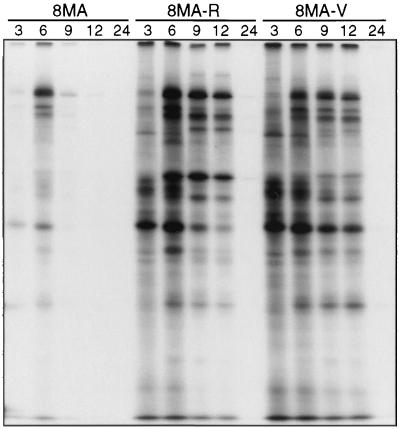
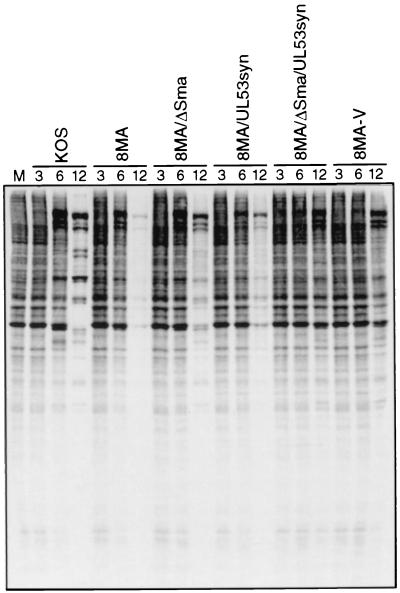
We asked if 8MA-V undergoes the severe vhs-induced reduction in the rate of viral protein synthesis characteristic of 8MA. Vero cells were infected with 8MA, 8MA-R, and 8MA-V at an MOI of 10 PFU/cell, labeled with [35S]methionine for 1 h at various times throughout the lytic cycle, and then analyzed by SDS-PAGE (Fig. 7). As expected, cells infected with 8MA displayed a severe reduction in the rate of viral and cellular protein synthesis at 9 and 12 h postinfection. In marked contrast, 8MA-V (and 8MA-R) sustained high levels of protein synthesis for up to 12 h postinfection. Inasmuch as 8MA-V displayed a pattern similar to that previously observed with 8MA/ΔSma, these results raised the possibility that the vhs gene had been inactivated during the evolution of the 8MA-V isolate. This hypothesis was confirmed by Southern blot analysis, which revealed a ∼800-nucleotide deletion of vhs coding sequences (data not shown). We then amplified the relevant segment of 8MA-V DNA by PCR and determined the nucleotide sequence across the vhs deletion. The results (summarized in Fig. 8A) revealed an in-frame deletion of DNA sequences encoding vhs amino acid residues 91 to 354. The 8MA-V deletion encompasses all of the sequences deleted by the inactivating ΔSma mutation (47) and, as such, is predicted to eliminate vhs function. These data explain the ability of 8MA-V to sustain viral protein synthesis at late times postinfection and suggest that inactivation of vhs represents one of the second-site mutations that allow propagation of 8MA-V.
FIG. 7.
Metabolic labeling of HSV-1-infected Vero cells. Vero cells infected with 8MA, 8MA-R, or 8MA-V were labeled with 50 μCi of [35S]methionine for 1 h prior to harvesting at 3, 6, 9, 12 or 24 h postinfection. Lysates were subjected to SDS-PAGE analysis and visualized by autoradiography.
The lethal phenotype exhibited by 8MA/ΔSma demonstrates that inactivation of vhs does not suffice to allow propagation of 8MA on noncomplementing cells (Fig. 1). Moreover, 8MA-V displays a syncytial plaque morphology while 8MA/ΔSma does not. These considerations suggested that 8MA-V contains at least one additional second-site mutation in addition to the deletion at the vhs locus. Syncytial mutations have been documented in at least four distinct HSV-1 genes. However, the majority of these lesions map to the UL53 gene, which encodes gK (6, 7, 14, 16, 45). We therefore cloned and sequenced the UL53 gene from the 8MA-V genome. This analysis revealed that 8MA-V contains two separate point mutations that alter the predicted amino acid sequence of gK relative to KOS and 8MA (R103→H, M309→R; Fig. 8B). Both of these mutations reside within the predicted ectodomains of gK, where all other known gK syncytial mutants can be mapped (40). Marker rescue experiments revealed that each of these mutations is individually sufficient to confer a syncytial plaque morphology to HSV-1 KOS (data not shown). Further dissection of the 8MA-V genome has not yet been completed, and therefore it is entirely possible that additional mutations exist which contribute to the ability of 8MA-V to be serially propagated on Vero cells.
We transferred both of the UL53 mutations present in 8MA-V into the genomes of 8MA, 8MA/ΔSma, and KOS in order to determine whether or not these lesions were sufficient to produce VP16-deficient recombinants capable of propagation on Vero cells through cell-cell fusion. These recombinants (8MA/UL53syn, 8MA/ΔSma/UL53syn, and KOS/UL53syn) were isolated on complementing QL10 cells on the basis of their syncytial phenotype and then expanded on 16-8 cells. DNA sequence analysis confirmed the presence of both UL53 mutations in all three recombinants. Unlike 8MA and 8MA/ΔSma, 8MA/UL53syn and 8MA/ΔSma/UL53syn formed small (and syncytial) plaques on Vero cells similar to those produced by 8MA-V (Fig. 9). Moreover, 8MA/UL53syn and 8MA/ΔSma/UL53syn could be serially passaged on Vero cells by transferring infected cells onto uninfected monolayers, although no infectious virus was produced (data not shown). As expected, KOS/UL53syn formed large syncytial plaques and gave rise to high levels of infectious virus.
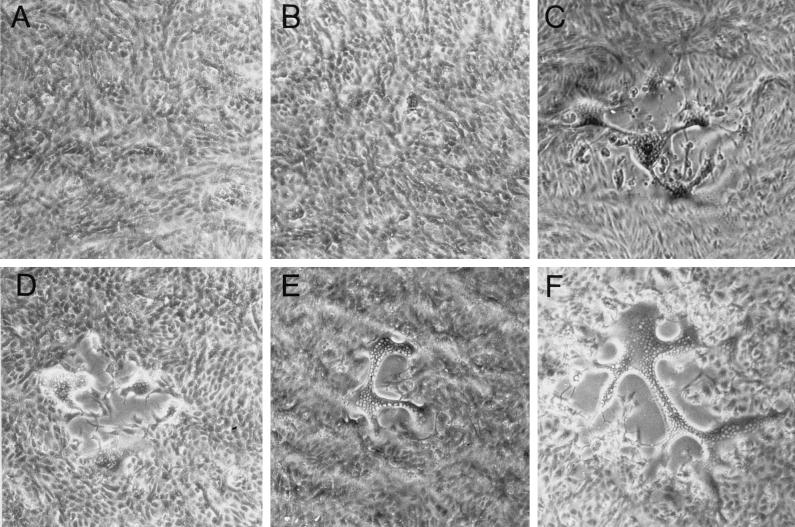
FIG. 9.
Plaque morphology of HSV-1 recombinants. Vero cells were infected with 8MA (A), 8MA/ΔSma (B), 8MA-V (C), 8MA/UL53syn (D), 8MA/ΔSma/UL53syn (E), and KOS/UL53syn (F) under limiting dilutions in order to visualize individual plaques. Monolayers were photographed at ×40 magnification 2 days postinfection. 8MA and 8MA/ΔSma fail to plaque on Vero cells, while the remaining recombinant viruses form syncytial plaques where individual nuclei can be visualized within a syncytium.
The observation that 8MA/UL53syn displayed growth properties similar to those of 8MA/ΔSma/UL53syn and 8MA-V was surprising in view of the fact that the original 8MA-V isolate had sustained a large deletion at the vhs locus. We therefore asked if 8MA/UL53syn was able to maintain viral protein synthesis throughout the course of infection in Vero cells. The results indicated that 8MA/UL53syn did not undergo the late decline in protein synthesis characteristic of 8MA (Fig. 10). Perhaps the UL53 mutations in this isolate somehow temper vhs activity; alternatively, it is possible that the 8MA/UL53syn isolate suffered an inactivating mutation at the vhs locus during isolation or propagation. Further experiments are required to distinguish between these possibilities. In addition, 8MA/ΔSma/UL53syn failed to display the delayed shutoff of host protein synthesis that was observed during infection with 8MA/ΔSma and KOS at a late time postinfection (12 h), perhaps indicating that this isolate progresses through the lytic cycle more slowly than the other viruses analyzed.
FIG. 10.
Metabolic labeling of HSV-1 wild type and 8MA-based recombinant viruses. Vero cells mock infected (M) or infected with the indicated virus were labeled with 50 μCi of [35S]methionine for 1 h prior to harvesting at 3, 6, or 12 h postinfection. Lysates were subjected to SDS-PAGE analysis and visualized by autoradiography.
The UL53 mutations cause fusion of outer nuclear membranes.
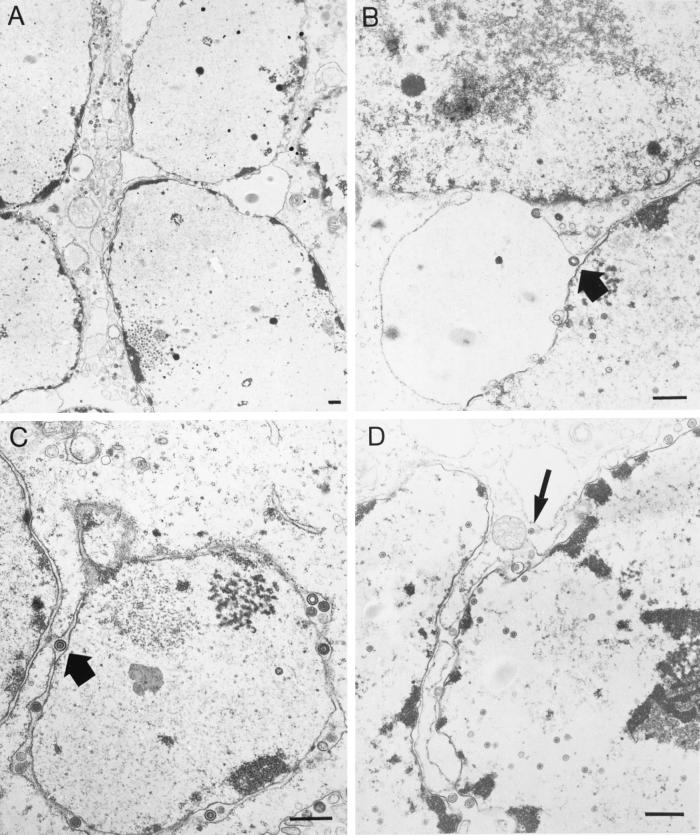
We examined cells infected with 8MA-V and 8MA/ΔSma/UL53syn by TEM to determine if the defects in virion assembly and egress previously noted for 8MA/ΔSma were detectably altered by the additional mutations present in these isolates (Fig. 11B, C, and D). 8MA-V and 8MA/ΔSma/UL53syn were indistinguishable and, with one exception, maintained all of the features observed with 8MA/ΔSma. In particular, enveloped virions were observed between the inner and outer nuclear membranes and within intranuclear vesicles but were absent from the cytoplasm and extracellular space. In addition, naked and apparently full capsids were observed in the cytoplasm. As expected, and in marked contrast to infection with nonsyncytial isolates, multiple nuclei were often observed within a common cytoplasm. Strikingly, the outer nuclear membranes of adjacent nuclei were often fused, in many cases producing large perinuclear “pockets” that approached half the size of a nucleus. In some cells these appeared to pinch off and detach, forming perinuclear vesicles. Fusion of the outer membranes of adjacent nuclei was also observed with KOS/UL53syn (Fig. 11A), demonstrating that the UL53 mutations are sufficient to produce this effect.
FIG. 11.
TEM of HSV-1 viruses harboring the UL53 gene from 8MA-V. Vero cells were infected with KOS/UL53syn (A), 8MA-V (B and C), and 8MA/ΔSma/UL53syn (D) at an MOI of 0.1 and harvested 24 h postinfection. Alteration of gK results in the formation of both large syncytia and perinuclear “pockets” caused by fusion of adjacent outer nuclear membranes. 8MA-V and 8MA/ΔSma/UL53syn enveloped virions are blocked within the perinuclear space, often adjacent to areas of nuclear membrane fusion (thick arrows). In addition, full capsids are often viewed within the cytoplasm of infected cells (thin arrow). Bars, 0.5 μm.
These results indicate that the UL53 mutations present in 8MA-V do not overcome the defect in virion assembly and egress induced by the 8MA mutation. As discussed further below, it seems likely that these mutations help propagate the infection by recruiting uninfected nuclei into infected cells through cell-cell fusion.
DISCUSSION
Previous studies have clearly established that VP16 serves multiple functions during HSV infection: it activates transcription of the viral IE genes, binds to the virion host shutoff protein vhs and downregulates its activity, and forms a complex with the tegument protein VP22 (19, 36, 41). In addition, VP16 is one of the most abundant components of the virion tegument (26, 49). The transcriptional activation function of VP16 can be inactivated without eliminating virus replication in tissue culture (1, 2, 53, 56); in contrast, other VP16 mutations are lethal, and viral mutants harboring such lesions fail to produce infectious progeny virions under nonpermissive conditions (1, 58). These observations demonstrate that VP16 is required, either directly or indirectly, for one or more steps in virus assembly and/or maturation. Weinheimer et al. investigated the nature of this requirement in some detail and showed that the VP16-null mutant 8MA displays markedly reduced levels of encapsidated DNA and does not produce extracellular enveloped particles (58). Taken in combination, these data are compatible with the hypothesis that VP16 plays a direct role in virion assembly. However, as outlined in the Introduction, VP16-null mutants undergo essentially complete translational arrest midway through the lytic cycle due to unrestrained vhs action (36). This finding raised the possibility that at least some of the defects observed during infection with VP16 null mutants might stem from reduced levels of one or more virion components, rather than loss of VP16 per se. We therefore evaluated the effects of deleting VP16 on virion assembly and maturation in a vhs-null background.
Our results provide strong evidence that the exaggerated vhs activity produced by deleting VP16 contributes to some, but not all, of the defects in virion assembly previously described by Weinheimer et al. for the 8MA virus (58). Thus, the DNA packaging defect of 8MA was substantially reversed by deleting the vhs gene in 8MA/ΔSma (Fig. 2), and 8MA/ΔSma accumulated greatly increased amounts of the major capsid protein VP5 compared to 8MA (Fig. 3). The simplest interpretation of these observations is that in the absence of VP16, vhs activity limits accumulation of some or all capsid components (and elements of the DNA cleavage-packaging machinery), thereby indirectly reducing the efficiency of DNA encapsidation. Consistent with this view, we also observed that deleting vhs increased the proportion of infected cells displaying recognizable HSV-1 capsids and/or virions. Although our results demonstrate that deletion of vhs has a major effect on the efficiency of encapsidation, it is important to stress that 8MA/ΔSma still displays a defect relative to wild-type HSV-1. Whether this defect is due to the vhs mutation or stems from loss of a vhs-independent function of VP16 remains to be determined.
8MA/ΔSma produced substantially more full capsids than 8MA, allowing us to search for other potential defects in the virion assembly and egress pathway using TEM. Before discussing the results of this analysis, it is useful to review the prevailing models for HSV egress. It is generally agreed that mature HSV capsids bud through modified patches of the inner nuclear membrane, delivering the now-enveloped virion into the space between the inner and the outer nuclear membranes. The subsequent steps in the egress pathway have yet to be clearly defined and are currently the subject of much debate in the literature (23, 60). One model, popularized by Johnson and Spear (31), holds that the enveloped virion transits the endoplasmic reticulum and Golgi network through the secretory apparatus. According to this view, the envelope of the mature infectious virus particle is derived from the inner nuclear membrane, and the tegument components and envelope glycoproteins are incorporated into virions as they bud out of the nucleus. In contrast, the envelopment-deenvelopment-reenvelopment model postulates that the initial virion envelope fuses with the outer nuclear membrane, delivering a naked capsid into the cytoplasm (9, 24, 25, 32, 54, 57, 59, 60). The capsid then acquires its final envelope and tegument as it buds into cytoplasmic (likely post-Golgi) vesicles. We found that cells infected with 8MA/ΔSma (and 8MA) displayed readily detectable numbers of enveloped virions located in the space between the inner and the outer nuclear membranes (Fig. 4). However, we were unable to detect any enveloped virions in the cytoplasm or extracellular space with either of these VP16-deficient viruses: all of the cytoplasmic capsids were naked. The simplest interpretation of these data is that VP16 is required for one or more steps in the virus egress pathway, downstream of the initial envelopment event at the inner nuclear membrane.
The observation that 8MA/ΔSma displays naked capsids in the cytoplasm can be reconciled with either of the current models for HSV egress. In the Johnson and Spear model such cytoplasmic capsids are interpreted as dead-end products resulting from inappropriate fusion of the virion envelope with the outer nuclear membrane. It seems possible that the frequency of such aberrant membrane fusion events might increase if the normal egress pathway through the secretory apparatus was blocked by loss of VP16. In contrast, cytoplasmic capsids are viewed as bona fide functional intermediates in the envelopment-deenvelopment-reenvelopment model of egress. Under this scenario, the simplest interpretation of the 8MA/ΔSma phenotype is that VP16 acts after delivery of capsid into the cytoplasm and prior to reenvelopment.
The enveloped particles detected between the inner and the outer nuclear membranes in cells infected with 8MA/ΔSma are noninfectious and appear to be qualitatively distinct from wild-type HSV-1 particles. Their envelopes appear to be devoid of spikes, which have been attributed in part to clustering of glycoproteins gB and gD (55). In this context, it may be relevant that gB and gD have been reported to interact with VP16 (65). It is possible that VP16 is required for the assembly and integration of an intact tegument and incorporation of membrane glycoproteins at the inner nuclear membrane. Consistent with this possibility, a mutant form of vhs that cannot bind VP16 is not incorporated into HSV-1 virions (47). Moreover, VP16 binds the tegument protein VP22 (19), and the tegument proteins VP11/12 and VP13/14 have been reported to modulate VP16 function (50, 63, 64), possibly indicating a physical interaction. Determining the polypeptide composition of the enveloped 8MA/ΔSma particles will be an important step in clarifying the role of VP16 in virion assembly.
Previous studies have shown that mutations within the genes encoding gD, UL11, UL20, and ICP34.5 affect the egress of enveloped perinuclear virions (3, 4, 8, 10). The defect in egress exhibited by 8MA/ΔSma could reflect physical or functional interactions with one or more of these proteins. Alternatively, it could indicate that only particles with a proper polypeptide composition proceed beyond the initial stages of envelopment. Consistent with the latter possibility, we occasionally observe enveloped KOS particles that are similar in appearance to those produced by 8MA and 8MA/ΔSma; when present, these are seen exclusively in the perinuclear space and never within cytoplasmic vesicles, on the cell surface, or in the extracellular space.
Our analysis of 8MA and 8MA/ΔSma has led us to draw two major conclusions regarding the role of VP16 in virion assembly and egress. First, deletion of VP16 results in greatly exaggerated vhs activity, which in turn severely reduces the accumulation of at least one virion component (VP5) and markedly limits the efficiency of genome encapsidation. Second, loss of VP16 independently blocks virus egress downstream of initial envelopment event and prevents the acquisition of infectivity. Additional evidence supporting these conclusions was obtained through the isolation and characterization of a spontaneous derivative of 8MA (8MA-V) that can be propagated on noncomplementing cells. 8MA-V bears a large inactivating in-frame deletion at the vhs locus and two point mutations in the UL53 gene that cause a syncytial plaque phenotype. Consistent with its vhs-null genotype, 8MA-V failed to display the translational arrest characteristic of its parent, 8MA. The selection of a vhs-null mutation during the isolation of 8MA-V supports our previous conclusion that VP16 downregulates vhs activity and suggests that inactivation of vhs is strongly selected for during propagation of VP16-null mutants on noncomplementing cells. 8MA-V exhibited the same defect in virus assembly and egress as had 8MA/ΔSma. Thus, no infectious virus was produced, and enveloped virions were confined to the space between the two nuclear membranes. However, the 8MA-V infection could be serially propagated by inoculating infected cells onto uninfected monolayers. The isolate harbors two point mutations in gK that are each individually sufficient to confer a syncytial plaque morphology. The simplest interpretation is that the 8MA-V infection spreads through cell-cell fusion induced by these gK mutations. Consistent with this view, derivatives of 8MA and 8MA/ΔSma bearing these mutations were able to form syncytial plaques on noncomplementing cells and could be serially propagated in the same fashion as the original 8MA-V isolate. Presumably, the defect in egress resulting from the VP16 mutation is overcome by cell-cell fusion events that recruit uninfected cells (and nuclei) into infected cells (see below).
The two point mutations in the UL53 gene of 8MA-V alter residues located in the predicted ectodomains of gK. These results are consistent with those of previous studies, where 10 UL53syn mutants were shown to contain from one to three missense mutations resulting in residue changes within the two predicted ectodomains (16, 40). The two amino acid changes recovered in 8MA-V are each individually sufficient to produce a syncytial phenotype (data not shown), suggesting strong selection for this phenotype during the evolution of the 8MA-V isolate. gK localizes almost exclusively to the perinuclear and nuclear membranes, fails to reach the Golgi apparatus and cell surface, and is not incorporated into HSV-1 virions (28). Earlier studies with two gK-null mutants suggest that gK plays an important role in efficient virion envelopment and translocation to the extracellular space (27, 30), possibly by preventing inappropriate fusion of virion and cellular membranes, while a recent report with a number of gK recombinant viruses implicates gK as a multifunctional protein whose different domains are required for a variety of functions (23). The exclusively intracellular location of gK raises interesting questions about how gK syn mutants initiate cell-cell fusion. It has been suggested that the mutant forms of gK syn somehow alter the transport of a fusion complex or other factors to the cell surface, possibly through interactions between the ectodomains of gK and the ectodomains of other viral glycoproteins (27, 30). Our results indicate that at least some mutant forms of gK also induce fusion events involving the outer nuclear membrane. Thus, adjacent nuclei in cells infected with isolates bearing the mutant gK gene derived from 8MA-V often shared a common outer nuclear membrane, which occasionally detached to form large perinuclear “pockets.” This effect, which to our knowledge has not been previously described, provides strong evidence that gK can influence membrane fusion events involving the outer nuclear envelope.
Our ability to serially propagate the 8MA-V infection through multiple passages through cell-cell fusion implies that progeny viral genomes produced in one nucleus must somehow be transmitted to at least some of the previously uninfected nuclei that are newly recruited into the syncytium. Although we frequently detect enveloped particles within the shared space located between the inner and the outer membranes of adjacent nuclei, it seems highly unlikely that this represents a means of transmitting the infection between nuclei. While the precise mechanism of nuclear import of HSV-1 genomes has yet to be defined, it is generally accepted that the unenveloped capsids delivered into the cytoplasm by infection migrate to the nuclear envelope, dock with the nuclear pore complex, and inject their DNA through the nuclear pore (61). It is improbable, therefore, that the 8MA-V infection is transmitted between adjacent nuclei by enveloped virions that we observe in the common perinuclear space. Such a mode of transmission would involve fusion of the virion envelope with the inner nuclear membrane of the second nucleus and therefore require a previously undescribed mechanism for controlled release of viral DNA from intranuclear capsids. It seems more probable that unenveloped cytoplasmic capsids produced in one nucleus can deliver progeny genomes to previously uninfected nuclei. It would be interesting to determine whether syncytial mutations in other genes could also rescue the cell-associated infectivity of 8MA/ΔSma.
Taken together, these data show that VP16 is involved in HSV-1 maturation and egress at a point downstream of the initial envelopment event. While the intricacies of this involvement remain to be determined, it is clear that controlling the activity of vhs is one important function of VP16, in order to allow the accumulation of key structural proteins such as VP5. It is also clear that while mutations in vhs and UL53 allow for the propagation of a VP16-null virus, they do not negate the block in egress. These findings demonstrate that VP16 is a multifunctional protein that functions throughout the life cycle of HSV-1.
ACKNOWLEDGMENTS
We thank Steve Weinheimer for 8MA, 8MA-R, and pVP16KOS; G. H. Cohen and R. J. Eisenberg for the NC-1 antiserum; and Steve Rice for generously providing lab space and critical comments to K.L.M. during the initial stages of this work.
This research was supported by a grant from the Medical Research Council of Canada (MT-12172). J.R.S. was a Terry Fox Senior Scientist of the National Cancer Institute of Canada, and K.L.M. holds postdoctoral fellowships from the Medical Research Council of Canada and the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Ace C I, Dalrymple M A, Ramsay F H, Preston V G, Preston C M. Mutational analysis of the herpes simplex virus type 1 trans-inducing factor Vmw65. J Gen Virol. 1988;69:2595–2605. doi: 10.1099/0022-1317-69-10-2595. [DOI] [PubMed] [Google Scholar]
- 2.Ace C I, McKee T A, Ryan J M, Cameron J M, Preston C M. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines J D, Roizman B. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1993;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines J D, Ward P L, Campadelli-Fiume G, Roizman B. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batterson W, Roizman B. Characterization of the herpes simplex virion-association factor responsible for the induction of α genes. J Virol. 1983;46:371–377. doi: 10.1128/jvi.46.2.371-377.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond V C, Person S. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology. 1984;132:368–376. doi: 10.1016/0042-6822(84)90042-4. [DOI] [PubMed] [Google Scholar]
- 7.Bond V C, Person S, Warner S C. The isolation and characterization of mutants of herpes simplex virus type 1 that induce cell fusion. J Gen Virol. 1982;61:245–254. doi: 10.1099/0022-1317-61-2-245. [DOI] [PubMed] [Google Scholar]
- 8.Brown S M, MacLean A R, Aitken J D, Harland J. ICP34.5 influences herpes simplex virus type 1 maturation and egress from infected cells in vitro. J Gen Virol. 1994;75:3679–3686. doi: 10.1099/0022-1317-75-12-3679. [DOI] [PubMed] [Google Scholar]
- 9.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campadelli-Fiume G, Poletti L, Dall'Olio D, Serafini-Cessi F. Origin of unenveloped plasmids in the cytoplasm of cells infected with herpes simplex virus I. J Virol. 1991;65:1589–1595. doi: 10.1128/jvi.65.3.1589-1595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell M E M, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate-early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 12.Cohen J I, Seidel K. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J Virol. 1994;68:7850–7858. doi: 10.1128/jvi.68.12.7850-7858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darlington R W, Moss L H., III The envelope of herpesviruses. Prog Med Virol. 1969;11:16–45. [PubMed] [Google Scholar]
- 14.Debroy C, Pederson N, Person S. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology. 1985;145:36–48. doi: 10.1016/0042-6822(85)90199-0. [DOI] [PubMed] [Google Scholar]
- 15.Doherty A J, Serpell L C, Ponting C P. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolter K E, Ramaswamy R, Holland T C. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J Virol. 1994;68:8277–8281. doi: 10.1128/jvi.68.12.8277-8281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elgadi M M, Hayes C E, Smiley J R. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J Virol. 1999;73:7153–7164. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elgadi M M, Smiley J R. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J Virol. 1999;73:9222–9231. doi: 10.1128/jvi.73.11.9222-9231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott G, Mouzakitis G, O'Hare P. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everly D N, Read G S. Mutational analysis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): characterization of HSV type 1 (HSV-1)/HSV-2 chimeras. J Virol. 1997;71:7157–7166. doi: 10.1128/jvi.71.10.7157-7166.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenwick M L, Everett R D. Inactivation of the shutoff gene (UL41) of herpes simplex virus types 1 and 2. J Gen Virol. 1990;71:2961–2967. doi: 10.1099/0022-1317-71-12-2961. [DOI] [PubMed] [Google Scholar]
- 22.Fenwick M L, Owen S A. On the control of immediate early (alpha) mRNA survival in cells infected with herpes simplex virus. J Gen Virol. 1988;69:2869–2877. doi: 10.1099/0022-1317-69-11-2869. [DOI] [PubMed] [Google Scholar]
- 23.Foster T P, Kousoulas K G. Genetic analysis of the role of herpes simplex virus type I glycoprotein K in infectious virus production and egress. J Virol. 1999;73:8457–8468. doi: 10.1128/jvi.73.10.8457-8468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granzow H, Weiland F, Jons A, Klupp B G, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heine J W, Honess R W, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutchinson L, Johnson D C. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69:5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutchinson L, Roop C, Johnson D C. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J Virol. 1995;69:4556–4563. doi: 10.1128/jvi.69.7.4556-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatamers in nuclei of infected cells and their role in the generation of the four isomeric arrangement of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayachandra S, Baghian A, Kousoulas K G. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J Virol. 1997;71:5012–5024. doi: 10.1128/jvi.71.7.5012-5024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komuro M, Tajima M, Kato K. Transformation of Golgi membrane into the envelope of herpes simplex virus in rat anterior pituitary cells. Eur J Cell Biol. 1989;50:398–406. [PubMed] [Google Scholar]
- 33.Kwong A D, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ladin B F, Ihara S, Hampl H, Ben-Porat T. Pathway of assembly of herpesvirus capsids: an analysis using DNA+ temperature-sensitive mutants of pseudorabies virus. Virology. 1982;116:544–561. doi: 10.1016/0042-6822(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 36.Lam Q, Smibert C A, Koop K E, Lavery C, Capone J P, Weinheimer S P, Smiley J R. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 1996;15:2575–2581. [PMC free article] [PubMed] [Google Scholar]
- 37.Lemaster S, Roizman B. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. J Virol. 1979;35:798–811. doi: 10.1128/jvi.35.3.798-811.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marsden H S, Stow N D, Preston V G, Timbury M C, Wilkie N M. Physical mapping of herpes simplex virus-induced polypeptide. J Virol. 1978;28:624–642. doi: 10.1128/jvi.28.2.624-642.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLean G, Rixon F, Langeland N, Haarr L, Marsden H. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J Gen Virol. 1990;71:2953–2960. doi: 10.1099/0022-1317-71-12-2953. [DOI] [PubMed] [Google Scholar]
- 40.Mo C, Holland T C. Determination of the transmembrane topology of herpes simplex virus type 1 glycoprotein K (gK) J Biol Chem. 1997;272:33305–33311. doi: 10.1074/jbc.272.52.33305. [DOI] [PubMed] [Google Scholar]
- 41.O'Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 42.Oroskar A A, Read G S. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oroskar A A, Read G S. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. J Virol. 1987;61:604–606. doi: 10.1128/jvi.61.2.604-606.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellett P E, McKnight J L C, Jenkins F J, Roizman B. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing α genes. Proc Natl Acad Sci USA. 1985;82:5870–5874. doi: 10.1073/pnas.82.17.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pogue-Geile K L, Lee G T, Shapira S K, Spear P G. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology. 1984;136:100–109. doi: 10.1016/0042-6822(84)90251-4. [DOI] [PubMed] [Google Scholar]
- 46.Post L E, Conley A J, Mocarski E S, Roizman B. Regulation of α genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with α gene promoters. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 47.Read G S, Bradley B M, Knight K. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptides. J Virol. 1993;67:7149–7160. doi: 10.1128/jvi.67.12.7149-7160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of a (immediate early) viral polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roizman B, Furlong D. The replication of herpesviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. Vol. 3. New York, N.Y: Plenum Press; 1974. pp. 229–403. [Google Scholar]
- 50.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1043–1107. [Google Scholar]
- 51.Smibert C A, Popova B, Xiao P, Capone J P, Smiley J R. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J Virol. 1994;68:2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smibert C A, Smiley J R. Differential regulation of endogenous and transduced β-globin genes during infection of erythroid cells in herpes simplex virus type 1 recombinant. J Virol. 1990;64:3882–3894. doi: 10.1128/jvi.64.8.3882-3894.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smiley J R, Duncan J. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the in1814 linker insertion mutation. J Virol. 1997;71:6191–6193. doi: 10.1128/jvi.71.8.6191-6193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stackpole C W. Herpes-type virus of the frog renal adenocarcinoma. J Virol. 1969;4:75–93. doi: 10.1128/jvi.4.1.75-93.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stannard L M, Fuller A O, Spear P G. Herpes simplex virus glycoproteins associated with different morphological entities projecting from the virion envelope. J Gen Virol. 1987;68:715–725. doi: 10.1099/0022-1317-68-3-715. [DOI] [PubMed] [Google Scholar]
- 56.Tal-Singer R, Pichyangkura R, Chung E, Lasner T M, Randazzo B P, Trojanowski J Q, Fraser N W, Triezenberg S J. The transcriptional activation domain of VP16 is required for efficient infection and establishment of latency by HSV-1 in the murine peripheral and central nervous systems. Virology. 1999;259:20–33. doi: 10.1006/viro.1999.9756. [DOI] [PubMed] [Google Scholar]
- 57.van Genderen I, Brandimarti R, Torrisi M, Campadelli-Fiume G, van Meer G. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology. 1994;200:831–836. doi: 10.1006/viro.1994.1252. [DOI] [PubMed] [Google Scholar]
- 58.Weinheimer S P, Boyd B A, Durham S K, Resnick J L, O'Boyle D R. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J Virol. 1992;66:258–269. doi: 10.1128/jvi.66.1.258-269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whiteley A, Bruun B, Minson T, Browne H. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J Virol. 1999;73:9515–9520. doi: 10.1128/jvi.73.11.9515-9520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whittaker G R, Helenius A. Nuclear import and export of viruses and virus genomes. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 62.Zelus B D, Stewart R S, Ross J. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, McKnight J L C. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J Virol. 1993;67:1482–1492. doi: 10.1128/jvi.67.3.1482-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Sirko D A, McKnight J L. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J Virol. 1991;65:829–841. doi: 10.1128/jvi.65.2.829-841.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu Q, Courtney R J. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology. 1994;204:590–599. doi: 10.1006/viro.1994.1573. [DOI] [PubMed] [Google Scholar]