Abstract
In 1997, an H5N1 influenza virus outbreak occurred in chickens in Hong Kong, and the virus was transmitted directly to humans. Because there is limited information about the avian influenza virus reservoir in that region, we genetically characterized virus strains isolated in Hong Kong during the 1997 outbreak. We sequenced the gene segments of a heterogeneous group of viruses of seven different serotypes (H3N8, H4N8, H6N1, H6N9, H11N1, H11N9, and H11N8) isolated from various bird species. The phylogenetic relationships divided these viruses into several subgroups. An H6N1 virus isolated from teal (A/teal/Hong Kong/W312/97 [H6N1]) showed very high (>98%) nucleotide homology to the human influenza virus A/Hong Kong/156/97 (H5N1) in the six internal genes. The N1 neuraminidase sequence showed 97% nucleotide homology to that of the human H5N1 virus, and the N1 protein of both viruses had the same 19-amino-acid deletion in the stalk region. The deduced hemagglutinin amino acid sequence of the H6N1 virus was most similar to that of A/shearwater/Australia/1/72 (H6N5). The H6N1 virus is the first known isolate with seven H5N1-like segments and may have been the donor of the neuraminidase and the internal genes of the H5N1 viruses. The high homology between the internal genes of H9N2, H6N1, and the H5N1 isolates indicates that these subtypes are able to exchange their internal genes and are therefore a potential source of new pathogenic influenza virus strains. Our analysis suggests that surveillance for influenza A viruses should be conducted for wild aquatic birds as well as for poultry, pigs, and humans and that H6 isolates should be further characterized.
Influenza A viruses can infect various animal hosts, including avian and mammalian species (15). Serologic and genetic analyses have identified 15 different hemagglutinin (HA) and 9 different neuraminidase (NA) subtypes, indicating that the natural virus reservoirs contain a limited number of subtypes. The fact that all subtypes are found in wild aquatic birds underlines the importance of these species as the prime source of these viruses. Only 3 of the 15 HA subtypes found in wild and domestic aquatic birds (H1, H2, and H3) are known to have caused pandemics in humans. The catastrophic influenza pandemic of 1918, caused by an H1N1 virus, killed 20 to 40 million people. Pandemics caused by the Asian influenza A virus (H2N2) in 1957 and the Hong Kong virus (H3N2) in 1968 indicated that southern China is a hypothetical influenza epicenter (22).
In 1997, an outbreak of an H5N1 influenza virus in chickens was reported in Hong Kong, and 18 human influenza cases were confirmed (4, 28, 31). The death of an infected individual raised concerns about a possible pandemic (5). In that region, humans live in close proximity to domestic poultry, providing opportunities for interspecies virus transmission (23). The observation that avian-mammalian reassortants are not maintained in the avian reservoir suggests that avian-to-mammalian transmission is unidirectional (11). Genetic and phylogenetic analyses of avian influenza viruses isolated from domestic avian species in southern China from 1975 to 1980 revealed a vast number of subtypes (H3N8, H4N6, etc.). Most of the internal genes belonged to the Eurasian lineage, indicating that this reservoir is separable from the avian lineage in North America (11). However, the combinations of surface glycoproteins and internal genes that confer the ability to cross the species barrier are still not known. This information can be acquired only with increased knowledge about the gene pool of influenza A viruses in aquatic bird species.
There is evidence that the H5N1 virus was transmitted directly from poultry to humans (4, 24, 32). No additional cases of human H5N1 infections were reported after all birds in the Hong Kong poultry markets were killed, suggesting that the H5N1 strains may have been eliminated. However, the origin of these pathogenic viruses and the possible continued circulation of their genes in the Hong Kong area remain to be ascertained. Genetic characterization of the gene segments circulating in that region indicated that the H5N1 viruses were generated by reassortment. Guan et al. suggested that H9N2 viruses were the donors of the internal genes, including the three polymerase genes (PB2, PB1, and PA) and the nucleoprotein (NP), matrix (M), and nonstructural (NS) genes (8). Xu et al. (30) proposed that the H5 hemagglutinin was derived from the H5N1 virus A/goose/Guangdong/1/96, based on 99% nucleotide homology between the HA sequences of these H5N1 strains. However, the neuraminidase genes of A/goose/Guangdong/1/96 and H5N1 viruses isolated in 1997 were only 90% homologous, and the N1 protein of the goose virus lacked the 19-amino-acid stalk deletion that is characteristic of the pathogenic H5N1 viruses (30). These results suggest that subtypes other than H9N2 and H5N1 may have contributed genes to the H5N1 viruses.
To explore this possibility, influenza A viruses isolated in Hong Kong during December 1997 and January 1998 from various aquatic bird hosts, including ducks and geese, were characterized serologically and genetically. Our analysis revealed that multiple influenza A virus subtypes were cocirculating in these birds. We determined their evolutionary relationship to representative influenza A viruses and found evidence that gene segments of strains other than H5N1 and H9N2 are closely related to the pathogenic H5N1 virus isolates.
MATERIALS AND METHODS
Viruses and serological assays.
The viruses isolated in the Hong Kong region and the abbreviations used in this study are listed in Table 1. The viruses were collected in December 1997 and January 1998 during the H5N1 outbreak. Fecal and cloacal samples from the different bird species were grown in 9- to 11-day-old chicken embryos. Viruses were handled in a biosafety level 3+ facility at St. Jude Children's Research Hospital.
TABLE 1.
Influenza A viruses isolated from aquatic birds in Hong Kong and characterized in this study
| Isolate | Serotype | Abbreviation | Collection site | Sample | Health status |
|---|---|---|---|---|---|
| A/duck/Hong Kong/P151/97 | H3N8 | DKHK151-97 | Duck farm | Cloacal | Apparently healthy |
| A/duck/Hong Kong/P169/97 | H3N8 | DKHK169-97 | Duck farm | Cloacal | Apparently healthy |
| A/duck/Hong Kong/P185/97 | H3N8 | DKHK185-97 | Duck farm | Cloacal | Sick |
| A/teal/Hong Kong/W312/97 | H6N1 | TEALHK-97 | Live-poultry market | Tracheal + cloacala | Dead |
| A/duck/Hong Kong/Y264/97 | H4N8 | DKHK264-97 | Live-poultry market | Cloacal | Apparently healthy |
| A/goose/Hong Kong/W217/97 | H6N9 | GOHK-97 | Live-poultry market | Cloacal | Apparently healthy |
| A/aquatic bird/Hong Kong/M603/98 | H11N1 | ABHK-98 | Maipo Marshb | Fecal | Apparently healthy |
| A/duck/Hong Kong/P50/97 | H11N9 | DKHK50-97 | Live-poultry market | Fecal | Apparently healthy |
| A/duck/Hong Kong/P54/97 | H11N9 | DKHK54-97 | Live-poultry market | Fecal | Apparently healthy |
| A/duck/Hong Kong/T37/97 | H11N8 | DKHK37-97 | Live-poultry market | Fecal | Apparently healthy |
| A/duck/Hong Kong/T25/97 | H11N8 | DKHK25-97 | Live-poultry market | Fecal | Apparently healthy |
These samples were collected from a dead bird on an unloaded truck at the wholesale market.
Free-flying ducks were the predominant species present.
This study characterized influenza A viruses other than the H5N1 and H9N2 strains (25). All were isolated from aquatic birds. Where possible, cloacal samples were collected from individual birds so that the species could be identified. Fresh fecal samples were collected from “dropping trays” under cages of ducks in the live-poultry markets and from the banks of ponds in the Maipo Marsh Nature Reserve, where free-flying ducks were overwintering.
The samples collected from green-winged teal ducks at a poultry market require special note. These samples were collected on 29 December 1997, the day of depopulation of all poultry in Hong Kong. The birds were in bamboo cages that were still on the delivery truck and had not been moved to the market buildings. The truck contained mallard and green-winged teal ducks. These birds were probably caught wild and raised on a farm until they were transported to the market in Hong Kong. However, we cannot be sure if these ducks may have been raised on a farm. Because 3 of the 14 green-winged teal ducks in one cage were dead, tracheal and cloacal samples from each bird in that cage were collected. One dead duck yielded the virus designated A/teal/Hong Kong/W312/97 (H6N1). Hemagglutination titration and hemagglutination inhibition (HI) assays were performed in microtiter plates (18).
Mouse infection experiments.
Female BALB/c mice were maintained under specific-pathogen-free conditions until they were used at 8 to 12 weeks of age. To determine the dose lethal for 50% of mice (MLD50), groups of five mice were anesthetized by inhalation of metofane and inoculated intranasally with 100 μl of virus in different dilutions. Daily assessments of weight and mortality were made, and the MLD50 was calculated for each virus by the method of Reed and Muench (19). Mice were killed on day 3 to estimate the titer of virus in lungs and brain. The organs were removed under sterile conditions to make a 10% suspension and assayed for virus titer in embryonated chicken eggs. Titers of infectious virus are presented as log10 50% egg infectious doses (EID50).
RNA preparation, PCR, and sequencing.
Viral RNA was extracted from allantoic fluid using the commercially available RNA extraction kit RNeasy Mini-Kit (Qiagen, Valencia, Calif.) according to the manufacturer's protocol. Reverse transcriptase-PCR with segment-specific primers was performed to amplify the viral RNA. The sequences of the oligonucleotides used are available upon request. After purification with the QIAquick PCR purification kit (Qiagen), the PCR products were sequenced. The sequencing reactions were performed by the Center for Biotechnology at St. Jude Children's Research Hospital on template DNA with Prism BigDye Terminator Cycle Sequencing Ready Reaction kits with Ampli-Taq DNA polymerase FS (Perkin-Elmer/Applied Biosystems, Foster City, Calif.). Samples were electrophoresed, detected, and analyzed on Perkin-Elmer/Applied Biosystems model 373 and 377 DNA sequencers.
Sequence analysis and phylogenetic analysis.
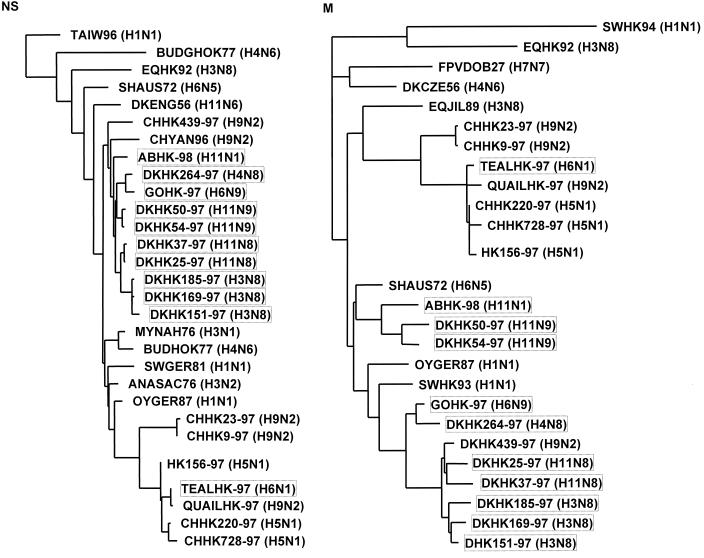
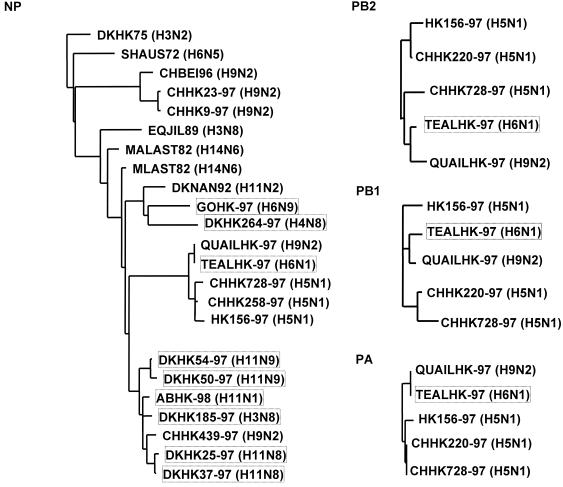
For sequence analysis and alignment, the Wisconsin Sequence Analysis Package, version 9.0 (Genetics Computer Group, Inc., Madison, Wis.), was used. The influenza virus nucleotide and amino acid sequences were obtained from the Influenza Sequence Database of the Los Alamos National Laboratory (http://www.flu.lanl.gov). Phylogenetic analysis was performed by using the maximum-likelihood method implemented by the software fastDNAml, which is derived from PHYLIP (6, 17). The TREEVIEW 1.5.2 software was used to draw phylogenetic trees in which horizontal distances are proportional to the number of differences. The trees presented in Fig. 2 and 3 are based on the nucleotide sequences of the gene segments NS (837 nucleotides [nts]), M (974 nts), NP (1,285 nts), PA (485 nts), PB1 (2,260 nts), and PB2 (2,150 nts).
FIG. 2.
Phylogenetic tree based on the nucleotide sequences of the NS and M gene segments of influenza viruses isolated in Hong Kong and on the comparable gene segments of other influenza virus isolates. The lengths of the horizontal lines are proportional to the number of nucleotide substitutions between branch points. The virus isolates shown in boxes were sequenced in this study.
FIG. 3.
Evolutionary relationship of influenza A virus polymerase complex genes (NP, PB2, PB1, and PA). The phylogenetic trees are based on the nucleotide sequences of the four genes.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been assigned GenBank accession numbers AF250470 to AF250502.
RESULTS
Serotypes of the viruses.
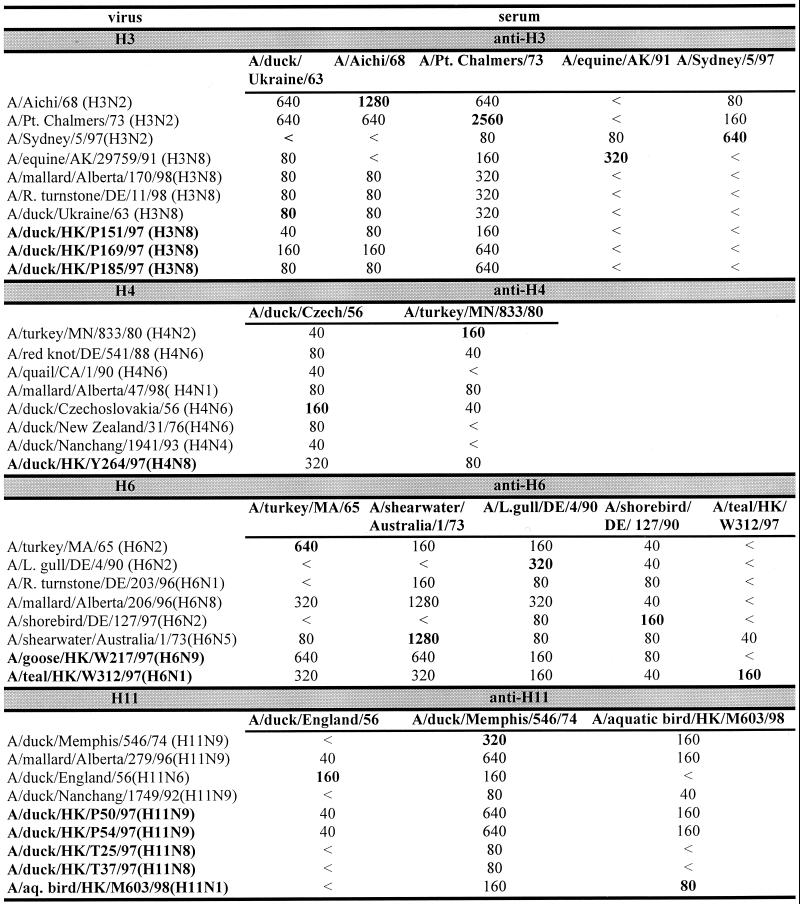
The influenza A viruses listed in Table 1 were serologically analyzed and found to contain different subtypes (Fig. 1). Of the isolates taken from ducks, three (DKHK151-97, DKHK169-97, and DKHK185-97) belonged to the H3N8 serotype, one (DKHK264-97) to H4N8, two (DKHK50-97 and DKHK54-97) to H11N9, and two (DKHK25-97 and DKHK37-97) to H11N8. The surface glycoproteins of the teal isolate (TEALHK-97) were identified as H6N1. The virus isolated from an aquatic bird (ABHK-98) was characterized as an H11N1 subtype, and the goose (GOHK-97) isolate was an H6N9 strain.
FIG. 1.
Comparison of influenza A viruses isolated from aquatic birds in HI assays. Titers in bold are titers to homologous sera. <, no inhibition detectable at a serum dilution of 1:40.
The HAs of the three H3N8 viruses were antigenically similar and were closely related to the HAs of the avian virus A/duck/Ukraine/1/63 (H3N8) and early human H3N2 influenza viruses (Fig. 1). Thus, the precursors of the pandemic human H3N2 influenza viruses continue to circulate in aquatic birds in southern China. Similarly, the HA of the H4N8 isolate was closely related antigenically to that of the prototype H4 influenza virus A/duck/Czech/56 (H4N6). The H6 hemagglutinins of the H6N1 and H6N9 viruses are antigenically similar to the prototype A/turkey/Mass/65 (H6N2) but could be distinguished from each other in the HI assay. The HAs of the five H11 influenza viruses are separable into two groups, H11N9 and others (H11N8 and H11N1). Each of the H11 isolates was distinguishable from the reference H11N6 strain A/duck/England/56. The HAs of the two H11N9 isolates show an antigenic similarity to that of the H11N1 isolate. These data show that a heterogeneous group of influenza A viruses were circulating in aquatic birds in the Hong Kong area during the time of the H5N1 outbreak. To characterize these virus isolates further, we performed sequence analysis of their internal genes. We analyzed these genes phylogenetically to determine the relationship between the isolates and to compare them with other influenza strains.
Sequence analysis of the NS and M gene segments.
We sequenced the NS and M gene segments of the serotypically different viruses. The sequence analysis of the NS gene segment revealed that the 11 strains characterized carry the A allele. Phylogenetic analysis was based on the nucleotide sequences of the NS and M gene segments of the isolated strains, of strains with similar HA or NA subtypes, and of strains isolated in Europe and Asia (Fig. 2).
The phylogenetic trees (Fig. 2) revealed that the 11 virus isolates can be divided into six separate groups: (i) the three H3N8 viruses (DKHK151-97, DKHK169-97, and DKHK185-97) which show high similarity to those originating at one root in the NS and in the M tree; (ii) isolates DKHK25-97 and DKHK37-97, with an H11N8 serotype; (iii) H11N9 viruses DKHK50-97 and DKHK54-97; (iv) the virus isolated from an aquatic bird (ABHK-98/H11N1); (v) the viruses isolated from a goose (GOHK-97/H6N9) and a duck (DKHK264-97/H4N8); and (vi) the H6N1 virus isolated from a teal (TEALHK-97), which formed a subgroup with QUAILHK97 (H9N2), H5N1 strains from chickens (CHHK220-97 and CHHK728-97), and the human strain HK156-97. Thus, the phylogenetic analysis of the M and NS segments reveals that these isolates form two different groups: the H9N2 (8), H5N1 (27, 28, 32), and H6N1 (this report) types and a different group distantly related to them that includes multiple subtypes.
Sequence analysis of the polymerase complex gene segments (NP, PB2, PB1, and PA).
The nucleoprotein (NP) is a component of the polymerase complex and a major factor in determining the host range of influenza A virus (20, 21). The phylogenetic tree of the NP sequences (Fig. 3) reveals six different subgroups among the isolates and shows an evolutionary relationship similar to that described for the M and NS segments. The H6N1 virus (TEALHK-97) shares a subgroup with an H9N2 virus (QUAILHK-97) and with the chicken (CHHK728-97) and human (HK156-97) H5N1 viruses. Thus, the NP segment of TEALHK-97 is closely related to that of the pathogenic H5N1 viruses isolated from poultry and humans.
The phylogenetic trees based on the PB2, PB1, and PA nucleotide sequences (Fig. 3) revealed that TEALHK-97 clustered in the same branch as QUAILHK97, CHHK220-97, CHHK728-97, and HK156-97. As shown in Table 2, the H6N1 virus has greater than 98% homology to the index human isolate A/156/97 in all six internal genes; no other isolate among the 11 analyzed showed a comparable close relationship. However, it is noteworthy that the H6N1 and the H9N2 viruses (8) both have an outgroup relationship with the H5N1 viruses, indicating that these viruses are similar but not identical.
TABLE 2.
Nucleotide homology between seven gene segments of A/teal/HK/W312 (H6N1) and the human virus A/HK/156 (H5N1)
| Segment | No. of nucleotides sequenced | % Homology to A/HK/156 (H5N1)
|
|
|---|---|---|---|
| Nucleotides | Amino acids (deduced) | ||
| PB2 | 2,280 | 98.6 | 99.7 |
| PB1 | 2,277 | 98.5 | 99.3 |
| PA | 2,146 | 98.4 | 98.8 |
| NP | 1,497 | 98.3 | 98.7 |
| M | 1,027 | 99.8 | 100 (M1) |
| 98.9 (M2) | |||
| NS | 890 | 98.8 | 99.1 (NS1) |
| 99.1 (NS2) | |||
| NAa | 1,350 | 97.1 | 97.1 |
Both NA proteins have the same 19-amino-acid stalk deletion.
Sequence analysis of the surface glycoproteins.
Because the origin of the N1 neuraminidase gene of the H5N1 viruses is not known (30), the N1 neuraminidase genes of our H11N1 and H6N1 isolates were sequenced. The N1 gene of the teal virus showed 97% nucleotide homology to the N1 gene of A/HK/156. The deduced amino acid sequence reveals that the N1 protein has a 19-amino-acid deletion in the stalk region. The N1 segment of the H11N1 virus (A/aquatic bird/HK/M603/98) is closely related to that of A/parrot/Ulster/73 (H7N1) and does not contain the deletion. These results show that the six internal gene segments as well as the neuraminidase gene of the H6N1 virus are closely related to those of the H5N1 viruses (Table 2).
The nucleotide sequence of the H6N1 isolate's HA segment had 88% homology to the HA of A/shearwater/Australia/1/72 (H6N5) (16). This segment contains an open reading frame encoding 566 amino acids that have 90% homology to the H6 protein of the H6N5 virus. It is noteworthy that the connecting peptide between the HA1 and HA2 parts of this molecule does not contain the additional basic amino acids that are found in the H5 and H7 hemagglutinins of highly pathogenic chicken influenza viruses (2, 4, 27, 29).
Pathogenicity of the H6N1 isolate in mice.
It is well established that A/Hong Kong/156/97 (H5N1)-like influenza viruses are lethal in mice without adaptation and that they spread to the brain (7, 9, 12, 24). After finding that seven of the eight gene segments of A/teal/Hong Kong/W312/97 (H6N1) were similar to those of A/Hong Kong/156/97 (H5N1), it was important to establish the pathogenicity of the teal H6N1 virus in mice.
High doses (8.5 EID50) of A/teal/Hong Kong/W312/97 (H6N1) killed BALB/c mice on initial inoculation and caused weight loss in those that survived inoculation; the virus replicated to high titers in the lungs (8.5 log10 EID50) but was not detected in the brain (Table 3). In comparison, the mouse lethal dose (MLD50) of human H5N1 virus [A/Hong Kong/156/97 (H5N1)] was 1.16 EID50, indicating that the H5N1 virus was more pathogenic in mice than A/teal/Hong Kong/W312/97 (H6N1). However, the pathogenicity of teal H6N1 increased as the virus was passaged in mice. Mice infected with the teal H6N1 virus had virus titers in lungs similar to those found with the human H5N1 influenza virus (Table 3). By the third passage, the MLD50 was 2.3 EID50 and the virus spread to the brain.
TABLE 3.
Replication of A/teal/Hong Kong/W312/97 (H6N1) influenza virus in mice
| Virusa | Virus titer
|
MLD50 (EID50)b | % wt loss on day 3 after infection | Titer of virus on day 3 after infection (log EID50/ml)
|
||
|---|---|---|---|---|---|---|
| In eggs (log EID50) | In mice (log MLD50) | Lungs | Brain | |||
| A/Hong Kong/156/97 | 9.0 | 7.7 | 1.16 | 15 | 8.5 | 2.0 |
| A/Teal/Hong Kong/W312/97 | ||||||
| P0 | 8.5 | 1.0 | 8.5 | 24 | 8.5 | None |
| P1 | 7.2 | Not studied | Not studied | Not studied | 7.5 | None |
| P2 | 8.0 | Not studied | Not studied | Not studied | 8.5 | 1.0 |
| P3 | 8.2 | 3.5 | 2.3 | 25 | 8.0 | 2.5 |
P0, original isolate; P1 to P3, infection with A/teal/Hong Kong W312/97 after passage 1, 2, or 3 times, respectively, in mouse lungs, with subsequent propagation in embryonated chicken eggs.
MLD50 (EID50), quantity of infectious virus units (EID50) in 1 MLD50.
DISCUSSION
The 1997 outbreak of H5N1 influenza in Hong Kong showed that influenza A viruses continue to evolve, introducing new subtypes from avian to mammalian species. The influenza A viral genome consists of eight negative-sense RNA segments that can generate new variants by genetic reassortment. Although each of the 15 hemagglutinin and 9 neuraminidase genes of influenza A virus has been identified in aquatic birds, only a few subtypes (i.e., H3N2 and H1N1) are circulating in humans and pigs. This observation suggests that a certain combination of genes is required for transmission from the avian to the mammalian species and for maintenance of a stable virus lineage in the new host (21). Because there is evidence that aquatic birds are the natural reservoir for influenza A viruses, we characterized virus subtypes isolated from these birds in Hong Kong during the H5N1 outbreak to gain information about the gene pool in this area and to investigate whether we can find viruses which are closely related to the pathogenic H5N1 viruses. We found that two groups of viruses are circulating in the Hong Kong area. One group contains H9N2 and H6N1 viruses that are closely related to each other and to the H5N1 viruses in their internal genes. Therefore, it is likely that reassortment can occur within this group. The phylogenetic data suggest that the other subtypes (H3, H4, H11, etc.) are too far removed from the H5N1-like group to allow reassortment between the two groups. This conclusion is consistent with the fact that none of our isolates apart from teal H6N1 were possible reassortants with the H5N1 viruses. Another explanation for the failure to find such reassortants would be that the these bird species were raised in separate areas and thus no transmission of virus could occur between them.
The host range of influenza A viruses is determined by multiple genes (10). By comparing avian and human influenza virus isolates, human virus-like amino acids were identified in the PB2, NP, and M2 proteins for the H5N1 strains (32). The influenza virus A/teal/HK/W312/97, which is closely related to the H5N1 viruses, has the human virus-like amino acids 199-Ala, 661-Thr, and 702-Arg in its PB2 protein and 136-Met in its nucleoprotein. The PA protein has a serine at amino acid 409 that is also found in A/chicken/Hong Kong/220/97; the human isolates have an asparagine at this position. The M2 protein of the teal H6N1 virus has an avian-like glutamic acid at position 16 instead of an human-like glycine. At amino acids 28 and 55, it has human-like valine and phenylalanine.
The N1 neuraminidase of the H6N1 isolate is closely related to the H5N1 neuraminidase and also has the 19-amino-acid deletion in its stalk region. The N1 stalk deletion is characteristic of the H5N1 viruses isolated from humans and from chickens (1, 4, 14, 27, 28). The stalk length is variable within and between neuraminidase subtypes, and the fact that viruses with different stalk lengths have different biological properties suggests that the sequence and length of the stalk region may affect the host range (3, 13, 14).
The high degree of homology between seven gene segments of the teal H6N1 virus and the pathogenic H5N1 virus raises the question whether the H6N1 virus donated its seven segments to the H5N1 virus or whether the exchange was in the opposite direction. The fact that the H6N1 and H5N1 viruses reside in different branches of the phylogenetic trees but originate at the same node supports the idea that both virus types are descendants of a common precursor virus. Thus, a new H5N1 virus could be generated by reassortment of the seven segments of an H6N1 precursor virus and the hemagglutinin of an H5 virus. The donor of this H5 glycoprotein could be the H5N1 virus A/goose/Guangdong/1/96 (or a closely related virus), isolated in the Guangdong province near Hong Kong (30). This virus caused disease in geese, and its hemagglutinin showed 99% homology to that of the pathogenic H5N1 viruses from Hong Kong. The remaining seven segments of this virus, including the N1 neuraminidase, showed no close relationship to those H5N1 viruses, supporting the idea that the N1 neuraminidase might have been acquired from a non-H5 subtype.
It was suggested that the internal genes of the H5N1 strains might have come from H9N2 viruses (8). Our phylogenetic analysis shows that the H6N1 from teal and the H9N2 from quail belong to the same subgroup and thus are closely related to each other and to the H5N1 isolates. These data indicate that in the influenza A virus gene pool in the Hong Kong region, closely related internal genes are distributed among multiple serotypes. It is likely that the H9N2 and H6N1 viruses can exchange these internal genes. Thus, we propose that the H6N1 and H9N2 strains, which were able to generate new variants by reassortment, were the source of the internal genes of the pathogenic H5N1 viruses. The fact that the H6N1 virus also has an H5N1-like neuraminidase gene, and therefore has seven H5N1-like segments, suggests that the H9N2 viruses were precursors of the H6N1 viruses. No H9 isolates were of the N1 subtype, indicating that the source of the neuraminidase gene of the H5N1 viruses was probably an H6N1 virus. However, it is also possible that an H6N1-teal-like virus was created by reassortment between an A/HK/156 (H5N1)-like virus and an H6 virus. Reassortment could have occurred on a farm, since one possibility is (although not known in this particular case) that teals were caught in the wild and then held on a farm until they were sold in the market. Future surveillance studies of H6 viruses should allow a more detailed characterization of the gene pool of this subtype in the Hong Kong region. The isolation of additional H6N1 viruses that are highly similar to the H5N1 viruses would support our proposal that the H6N1 viruses were the donors of the internal genes and the N1 neuraminidase gene and may have been the immediate precursors of the pathogenic H5N1 viruses.
Among the H5N1 influenza viruses isolated from humans in 1997 in Hong Kong, one group was highly pathogenic in mice and a second group was less pathogenic (7, 12). The molecular basis of these differences has not yet been established. The A/teal/Hong Kong/W312/97 (H6N1) virus initially behaved more like the less pathogenic H5N1 group, but it rapidly adapted to mice and behaved more like the highly pathogenic H5N1 group by the third passage. The H5N1 viruses contain a highly cleavable HA molecule that may contribute to their high pathogenicity in mice (26). The teal H6N1 does not contain a highly cleavable HA, which may contribute to the initial lower pathogenicity in the mice, but its rapid increase in pathogenicity suggests that the other gene segments also play an important role in mouse pathogenicity. Studies are in progress to identify the molecular changes in the gene segments of the teal H6N1 virus that allowed it to rapidly become highly pathogenic in mice.
ACKNOWLEDGMENTS
These studies were supported by Public Health Research grants AI95357 and AI29680 from the National Institute of Allergy and Infectious Diseases, by Cancer Center Support CORE grant CA-21765, and by the American Lebanese-Syrian Associated Charities.
We thank Lijuan Zhang and David Walker for excellent technical support. We also thank Sharon Naron for scientific editing.
REFERENCES
- 1.Bender C, Hall H, Huang J, Klimov A, Cox N, Hay A, Gregory V, Cameron K, Lim W, Subbarao K. Characterization of the surface proteins of influenza A (H5N1) viruses isolated from humans in 1997–1998. Virology. 1999;254:115–123. doi: 10.1006/viro.1998.9529. [DOI] [PubMed] [Google Scholar]
- 2.Bosch F X, Garten W, Klenk H D, Rott R. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology. 1981;113:725–735. doi: 10.1016/0042-6822(81)90201-4. [DOI] [PubMed] [Google Scholar]
- 3.Castrucci M R, Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claas E C, Osterhaus A D, van Beek R, de Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 5.De Jong J C, Claas E C, Osterhaus A D, Webster R G, Lim W L. A pandemic warning? Nature. 1997;389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 7.Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley A J, Kawaoka Y. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol. 1999;73:3184–3189. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan Y, Shortridge K F, Krauss S, Webster R G. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gubareva L V, McCullers J A, Bethell R C, Webster R G. Characterization of influenza A/Hong Kong/156/97 (H5N1) virus in a mouse model and protective effect of zanamivir on H5N1 infection in mice. J Infect Dis. 1998;178:1592–1596. doi: 10.1086/314515. [DOI] [PubMed] [Google Scholar]
- 10.Klenk H D, Rott R. The molecular biology of influenza virus pathogenicity. Adv Virus Res. 1988;34:247–281. doi: 10.1016/S0065-3527(08)60520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Y P, Shu L L, Wright S, Bean W J, Sharp G B, Shortridge K F, Webster R G. Analysis of the influenza virus gene pool of avian species from southern China. Virology. 1994;198:557–566. doi: 10.1006/viro.1994.1067. [DOI] [PubMed] [Google Scholar]
- 12.Lu X, Tumpey T M, Morken T, Zaki S R, Cox N J, Katz J M. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol. 1999;73:5903–5911. doi: 10.1128/jvi.73.7.5903-5911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo G, Chung J, Palese P. Alterations of the stalk of the influenza virus neuraminidase: deletions and insertions. Virus Res. 1993;29:141–153. doi: 10.1016/0168-1702(93)90055-r. [DOI] [PubMed] [Google Scholar]
- 14.Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy B R, Webster R G. Orthomyxoviruses. In: Fields B N, et al., editors. Fields Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1397–1445. [Google Scholar]
- 16.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 17.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 18.Palmer D F, Coleman M T, Dowdle W R, Schild G C. Advanced laboratory techniques for influenza diagnosis. U.S. Department of Health, Education and Welfare Immunology Series No. 6, Procedure Guide. Atlanta, Ga: Centers for Disease Control; 1975. [Google Scholar]
- 19.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 20.Scholtissek C, Burger H, Kistner O, Shortridge K F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 21.Scholtissek C, Ludwig S, Fitch W M. Analysis of influenza A virus nucleoproteins for the assessment of molecular genetic mechanisms leading to new phylogenetic virus lineages. Arch Virol. 1993;131:237–250. doi: 10.1007/BF01378629. [DOI] [PubMed] [Google Scholar]
- 22.Shortridge K F, Stuart-Harris C H. An influenza epicentre? Lancet. 1982;ii:812–813. doi: 10.1016/s0140-6736(82)92693-9. [DOI] [PubMed] [Google Scholar]
- 23.Shortridge K F. Pandemic influenza: a zoonosis? Semin Respir Infect. 1992;1:11–25. [PubMed] [Google Scholar]
- 24.Shortridge K F, Zhou N N, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti K G, Norwood M, Senne D, Sims L, Takada A, Webster R G. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- 25.Shortridge K F. Poultry and the influenza H5N1 outbreak in Hong Kong. 1997: abridged chronology and virus isolation. Vaccine. 1999;17(Suppl. 1):S26–S29. doi: 10.1016/s0264-410x(99)00102-4. [DOI] [PubMed] [Google Scholar]
- 26.Steinhauer D A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 27.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 29.Webster R G, Rott R. Influenza virus A pathogenicity: the pivotal role of hemagglutinin. Cell. 1987;50:665–666. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- 30.Xu X, Subbarao K, Cox N J, Guo Y. Genetic characterization of the pathogenic influenza A/goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999;261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 31.Yuen K Y, Chan P K, Peiris M, Tsang D N, Que T L, Shortridge K F, Cheung P T, To W K, Ho E T, Sung R, Cheng A F. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhou N N, Shortridge K F, Claas E C J, Krauss S L, Webster R G. Rapid evolution of H5N1 influenza viruses in chickens in Hong Kong. J Virol. 1999;73:3366–3374. doi: 10.1128/jvi.73.4.3366-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]