Abstract
Expression of the latency-associated transcript (LAT) gene is a hallmark of alphaherpesvirus latency, and yet its control and function remain an enigma. Resolution of this problem will require verification and subsequent elimination or disabling of elements regulating LAT gene transcription so that the influence of the resultant RNA can be evaluated. Toward this end, we generated a novel pseudorabies virus (PrV) recombinant in which a 282-bp region containing the LAP1 (first latency-active promoter) consensus sequence was replaced by a reporter cassette. Despite this substitution, replication of the recombinant was comparable to that of the parental and rescuant viruses both in cultured mammalian cells and in the natural host, swine. Furthermore, production of the LAT gene-associated 2.0- and 8.0-kb RNAs during an in vitro lytic infection of cultured neuronal cells was unaffected. However, the otherwise constitutively produced and processed 8.4-kb LAT was not detected in porcine trigeminal ganglia latently infected with this novel recombinant, although the viral genome was shown to be present. Therefore, LAP1 is apparently the basal promoter for PrV LAT gene expression during viral latency but is not required for such activity during an in vitro lytic infection of neuronal cells. More importantly, the ability of PrV to persist in a latent state in the absence of LAT suggests that other factors are responsible for this event in the natural host.
Like many alphaherpesviruses, pseudorabies virus (PrV) can establish a latent infection characterized by limited expression of the viral genome, which results in the production of latency-associated transcripts (LATs). During PrV latency in the trigeminal nerve ganglia (TG) of its natural host, swine (9, 10), LATs are generated from an inverted repeat region in a manner similar to that of herpes simplex virus type 1 (HSV-1) (3, 4, 5, 17, 18, 23, 25). However, unlike HSV-1, PrV expresses a different-sized, spliced transcript from this region during an in vitro productive (lytic) infection of cultured mammalian cells (2.0 kb) compared to an in vivo latent infection (8.4 kb) (3, 12). The lytic cycle 2.0-kb RNA lacks the same intron as the 8.4-kb LAT even though its 5′ initiation site is 243 bp downstream from that of the 8.4-kb RNA (12). Moreover, an 8.0-kb RNA species, whose sequence overlaps the second exon of the 2.0-kb RNA and 8.4-kb LAT, also is present in PrV-infected, cultured neuronal cells (12).
How PrV LAT gene expression is regulated during latency remains unknown. Based on the available data (7, 12), two different promoters may be involved, one active during a productive infection and the other active during a latent infection. In this regard, two distinct TATA sequences are in the proximity of the 5′ transcriptional initiation site of the 8.4-kb LAT (5, 18). The first is located 34 nucleotides upstream, while the second is positioned 143 bp downstream, of this initiation site. Three CAAT boxes are present at 171, 149, and 21 nucleotides upstream of the first TATA region, which is also flanked by three possible consensus simian virus 40 Sp1 sites (GC boxes) (18). Due to this topography, the sequence containing these seven promoter consensus elements has been designated the first latency-active promoter (LAP1). Since its consensus element distribution is very similar to that of the HSV-1 LAT gene promoter, it is not surprising that the PrV genomic sequence between −420 to +66 bp relative to the first TATA box can functionally replace its HSV-1 counterpart (11). However, when the PrV LAP1 was transferred to a site upstream of the native gC gene, the promoter failed to direct any transcription in infected cultured cells (11). This suggests that LAT gene expression is most likely mediated by both the LAT promoter and cis elements external to this designated region. The area defined by the two GC boxes located 77 nucleotides upstream and 150 nucleotides downstream of the second TATA sequence is thought to act as a second regulator (LAP2). Using a transient reporter gene expression assay, the LAP2 was shown to be active in both neuronal and nonneuronal cells (7), whereas LAP1 activity was detectable only in neuronal cells (11). Thus, LAP1 may be responsible for the production of LATs during latency.
To investigate the role of the LAP1 in PrV LAT gene expression, the delineated region extending from approximately 274 bp upstream to 8 bp downstream of the first TATA box was eliminated. The effect of this deletion on the synthesis of LATs by PrV during a lytic infection of cultured mammalian cells and a latent infection of pigs was then examined.
MATERIALS AND METHODS
Viruses and cells.
The Becker strain of PrV (PrV-Be) was used as the parental virus for the generation of the LAP1 deletion mutant (M5′). Both viruses and a rescue mutant (R5′) were propagated in Crandall-Rees feline kidney (CRFK) cells as described elsewhere (12). The murine neuroblastoma cell line N1E was used for investigating LAT production during an in vitro lytic infection as previously reported (12).
Plasmid construction.
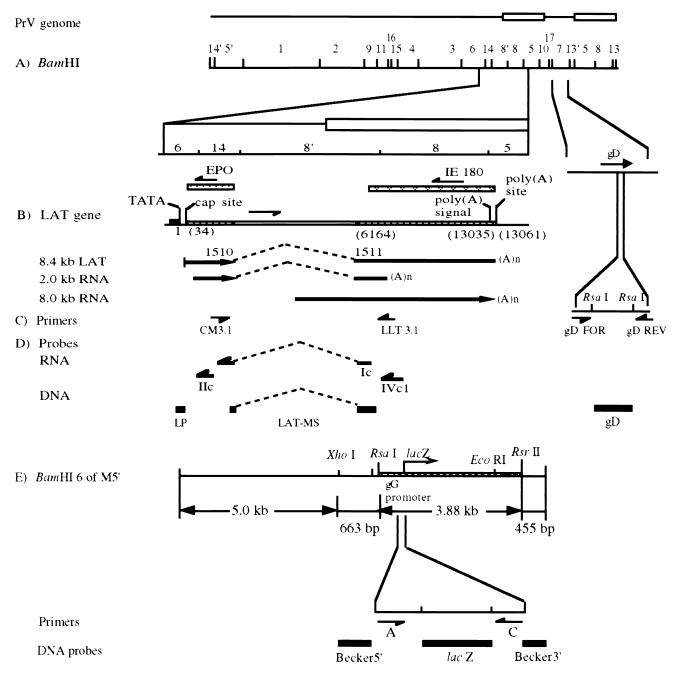
Plasmid pBS was constructed by ligating a 2.6-kb BamHI-SalI fragment from the PrV genomic BamHI 6 region into the respective sites in pUC19. A 1.4-kb subfragment of the viral DNA insert in pBS, containing the LAP1 region of the LAT gene, was excised by digestion with BamHI and XhoI and then inserted between the PvuII and RsaI sites of pUC19 to produce pBX22. This plasmid was then digested with RsaI and RsrII to release the 282-bp sequence containing the TATA, CAAT, and GC boxes located upstream of the 5′ end of the LAT gene. The plasmid was then ligated with a 3.88-kb transcriptional unit (Escherichia coli lacZ coding sequence fused to a simian virus 40 polyadenylation signal and under the control of the PrV gG gene promoter), which was released from pXSB110 (22) by digestion with SalI and SmaI. The resultant plasmid, pRR12, has the LAP1 sequence replaced with the lacZ reporter gene, which is transcribed in the same orientation as the LAT gene.
Plasmids pB14 and pB8 were generated by inserting the respective PrV BamHI 14 or BamHI 8 fragment in the BamHI site of pBluescript SKII(+) (Stratagene, La Jolla, Calif.). Plasmid pAC38 (provided by A. K. Cheung, National Animal Disease Center, Ames, Iowa) contains a 656-bp insert derived from a cDNA of the sequences flanking the intron of the in vivo PrV 8.4-kb LAT.
Generation of recombinant viruses.
The LAP1 deletion/E. coli lacZ insertion (PrV LP−/lacZ+) genotype mutant, M5′, and the rescue mutant, R5′, were respectively generated by homologous recombination between PrV-Be DNA and pRR12 or M5′ genome and pBX22. In each case, viral DNA (approximately 2 to 3 μg/ml), isolated as described by Scherba et al. (21), and linearized plasmid (10 to 15 μg/ml) were cotransfected by the calcium phosphate method (13) into monolayers of CRFK cells. Expression of β-d-galactosidase, encoded by the lacZ gene, or lack thereof was used to identify the deletion or rescue mutants, respectively (19). Recombinant viruses (two M5′ isolates and one R5′ isolate) were subjected to at least three rounds of plaque purification and stored at −70°C.
Primers.
Oligonucleotides were synthesized by the standard phosphoramidite procedure on an automated synthesizer (Operon Technologies, Inc., Alameda, Calif.). The DNA sequence data necessary for PrV LAT gene-specific primer construction were obtained from published sequences (5, 26). As shown in Fig. 1, primer LLT3.1 extends from nucleotides (numbered with reference to the transcriptional start site) 1578 to 1558 on the LAT gene anticoding strand (6, 12). Primer CM3.1 extends from nucleotides 1326 to 1346 on the LAT gene coding strand (6, 12). Primers gD FOR (CACGGAGGACGAGCTGGGGCT) and gD REV (GTCCACGCCCCGCTTGAAGCT) extend from nucleotides 433 to 454 and 651 to 630 downstream of the initiation codon on the gD gene coding and anticoding strands, respectively (26). Primer A (TCCACAGCTCAACAATGAAGTGGG) spans nucleotides −14 to 10 of the gG gene sequence relative to its transcriptional start site (21) (Fig. 1). Primer C (TTACGTTGGTGTAGATGGGCGCAT) corresponds to nucleotides (numbered from the ninth codon of the lacZ gene) 310 to 287 on the anticoding strand (21).
FIG. 1.
Schematic of the PrV parental and LP−/lacZ+ (M5′) recombinant genomes, with locations of PCR and RT-PCR primers and viral DNA probes. (A) PrV genomic BamHI restriction endonuclease fragment map and the expanded BamHI 6, 14, 8′, and 8 regions and gD gene. (B) Location of the LAT gene: its intron boundaries (dashed line), map units before (in parentheses) and after intron splicing, and the resultant in vivo (8.4-kb) and in vitro (2.0-kb) spliced transcripts (arrows showing the direction of transcription). (C) Locations of primers LLT3.1 and CM3.1 (used for amplification of the processed in vivo 8.4-kb LAT and in vitro 2.0-kb transcript), primers gD FOR and gD REV (used for amplification of the gD gene sequence), and endonuclease restriction sites used to remove primer sequences from the ends of the gD DNA probe. (D) Locations of ssRNA probes (Ic, IIc, and IVc1, used for Northern hybridization [the “c” denotes complementarity to the LAT sequences]) and DNA probes (LP, used for genotype verification; LAT-MS and gD, used for verification of the LAT RT-PCR and gD PCR products, respectively). All probes are drawn in an approximate scale with respect to their genomic locations shown in panel D. (E) Expanded BamHI 6 region of the M5′ recombinant virus showing the location and orientation of the gG-lacZ transcriptional unit insert and positions of the PCR primers (A and C) used for amplification of the specific gG-lacZ insert and the DNA probes (lacZ, Becker5′, and Becker3′) used for genomic analysis. The endonuclease restriction sites used to remove primer sequences from the ends of the probes are also shown.
Probes.
DNA probes Becker5′, Becker3′, and LP were obtained by digestion of approximately 1 μg of pBS with XhoI plus RsaI, BamHI plus RsrII, and RsrII plus RsaI, respectively (Fig. 1). Probes gD, lacZ, and LAT-MS were generated from PrV-Be DNA, pRR12, and pAC38, respectively, by PCR amplifications (described below) using primers gD FOR and gD REV (probe gD), A and C (probe lacZ), or CM3.1 and LLT3.1 (probe LAT-MS) followed by digestion with RsaI, MaeIII, or SmaI and MaeIII, respectively, to remove the primer sequences present in the products (Fig. 1). The restricted plasmid fragments and PCR products were then labeled with [α-32P]dCTP (Amersham Corp., Arlington Heights, Ill.) by using a random primer DNA labeling system (Life Technologies, Inc.) and following the manufacturer's recommendations.
The single-stranded RNA (ssRNA) probes, Ic, IIc, and IVc1 (Fig. 1), used portions of the LAT gene coding strand as templates and were labeled with [α-32P]UTP as previously described (12). Probe Ic is complementary to 656 nucleotides flanking the intron of the spliced 8.4-kb LAT. Probe IIc was derived from about 400 bp of the PrV BamHI 14 fragment located between an internal XhoI site and the BamHI site at the BamHI 6-14 junction. Probe IVc1 was transcribed from about 600 bp of the BamHI 8 sequence between an internal SmaI site and the BamHI site adjacent to the BamHI 8′ fragment (12).
Isolation of viral genomes and RNA.
Genomes from viruses propagated in cell culture were purified from nucleocapsids prepared by nonionic detergent treatment of infected cells as previously described (21). Viral DNA was isolated from porcine tissues as described by Scherba et al. (21). Total and poly(A) RNAs were isolated as previously reported (12).
Genomic analysis.
The predicted genotypes of the parental and recombinant viruses were verified by using restriction endonuclease analysis and Southern hybridization. Approximately 1 μg of the M5′, R5′, and PrV-Be viral DNAs was separately digested with either BamHI or BamHI and EcoRI. The resultant fragments were electrophoresed in an 1% agarose gel and then transferred to a nylon membrane (Zeta-Probe; Bio-Rad Laboratories, Richmond, Calif.) in the presence of 0.4 N NaOH for a period of at least 4 h. Conditions for prehybridization and hybridization have been previously described (21). Probes Becker5′, Becker3′, and LP were used separately and sequentially. Hybridization with a subsequent probe was performed after removal of the previous probe by boiling the membrane in 0.1% sodium dodecyl sulfate–0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 1 to 2 min. The sizes of the restricted DNA were estimated by comparison of their mobilities to those of commercially available lambda DNA markers (Life Technologies).
In vitro virus replication.
To compare the in vitro replication kinetics of the recombinant and rescuant to that of the parental virus, separate confluent monolayers of CRFK cells in 10-cm2 plates were infected with virus at 0.1 PFU/cell. At various times between 0 to 12 h postinfection (p.i.), infected cultures were harvested and the cells were lysed by one cycle of freezing and thawing. The infectious virus yields were titered in a plaque assay using fresh CRFK cell monolayers (20).
PCR.
PCR reagents were supplied as a commercially available kit, GeneAmp (Perkin-Elmer Cetus, Norwalk, Conn.). Amplification of the PrV gD gene was performed in 50 μl of reaction mixture consisting of 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, 200 μM each deoxynucleoside triphosphate, 1.0 μM each primer (gD FOR and gD REV), 1× Q-Solution (Qiagen, Inc., Chatsworth, Calif.), 1.0 U of Taq polymerase, and 5 μg of tissue DNA. The PCR was performed in a simplified hot start program by loading samples in a thermocycler (MJ Research, Inc., Watertown, Mass.) set at 94°C and then cycling 30 times at 94°C for 1 min, 63°C for 40 s, and 72°C for 45 s and once at 94°C for 1 min, 63°C for 40 s, and 72°C for 5 min. Amplification of the lacZ gene with primers A and C was done with the same reaction mixture as the gD PCR except that each primer was diluted to 0.8 μM. The reaction was initially cycled 5 times at 94°C for 1 min, 58°C for 2 min, and 72°C for 2 min; then 30 times at 94°C for 1 min, 58°C for 1 min, and 72°C for 1.5 min; and once at 94°C for 1 min, 58°C for 1 min, and 72°C for 5 min. Then 10 μl (one-fifth) of each PCR sample was analyzed in an 1.5% agarose gel in TBE buffer (50 mM Tris-HCl [pH 7.4], 50 mM boric acid, 1 mM EDTA) and then visualized by UV illumination after staining with ethidium bromide (1 μg/ml). Commercially available HaeIII-digested øX174 replicative-form DNA fragments (Life Technologies) served as molecular weight markers.
RT-PCR.
First-strand cDNA was synthesized from total or poly(A) RNA with random hexamers in the presence of Superscript reverse transcriptase II (Life Technologies) according to the manufacturer's recommendations. PCR was performed in a volume of 40 μl as previously reported (12) except that 0.8 μM each primer (LLT3.1 and CM3.1), 1× Q-Solution (Qiagen), and 0.8 U of Taq polymerase (Perkin-Elmer Cetus) were used. The reverse transcription (RT)-PCR products were analyzed as described for PCR amplimers.
Verification of PCR and RT-PCR products.
To verify the fidelity of the PCR and RT-PCR, the amplimers in the stained agarose gels were transferred to a Zeta-Probe membrane (Bio-Rad Laboratories) for Southern hybridization as previously described (12, 21). The PCR products generated by amplification of the gD and lacZ gene sequences were verified by hybridization with probes gD and lacZ, respectively. The RT-PCR products amplified from the processed LAT (intronless) were verified using probe LAT-MS.
Northern hybridization analysis.
Conditions for electrophoresis and transfer of total and poly(A) RNAs and for subsequent prehybridization and hybridization reactions have been previously described (12). The sizes of the RNAs were estimated by comparison of their mobilities to those of commercially available RNA markers (Ambion, Inc., Austin, Tex.).
In vitro LAT gene expression.
PrV LAT gene expression during a productive infection was investigated in N1E cells infected with 5 to 6 PFU/cell. Total RNA and poly(A) RNA, isolated from infected N1E cells at 6 h p.i., were examined for the presence of LAT gene transcripts by Northern hybridization using ssRNA probes.
Primary in vivo virus infection.
Sixteen 4- to 5-week-old pigs were acquired from the University of Illinois College of Veterinary Medicine PrV-free herd. The pigs were vaccinated intramuscularly with 2 ml of an inactivated PrV product (Porcilis Aujeszky; USO Veterinario-Intervet Mexico, S.A. de C.V. Mexico) to ensure their survival during the acute infection. Ten days after immunization, four pigs per group were inoculated intranasally with either M5′ (105 50% tissue culture infective dose [TCID50]/ml), R5′ (4 × 105 TCID50/ml), or PrV-Be (4 × 104 TCID50/ml). These viral titers are equivalent to or greater than a parental virus lethal dose in the absence of immunization. Four pigs not infected with virus served as controls. To monitor viral replication in and shedding from the host, the nasal cavities of the individual animals were swabbed daily for 3 or 4 days after infection and at 35 days p.i. The swabs were then placed in 1 ml of minimal essential medium containing penicillin (200 U/ml), streptomycin (200 μg/ml), and gentamicin (0.5 mg/ml). Virus released from the swabs was titered in CRFK cells (20).
Assays to evaluate establishment of latency.
At 35 days p.i., tonsil, brain, and TG were harvested and examined for evidence of a lingering productive infection by attempted virus isolation and a direct fluorescent antibody (FA) test. Samples for direct virus isolation were prepared by homogenization in 5 ml of minimal essential medium supplemented with 1,000 U of penicillin, 1,000 U of streptomycin, and 2.5 mg of gentamicin per g of tissue. Homogenates were clarified by low-speed centrifugation and then filtered through 0.45-μm-pore-size disposable sterile syringe filters (Corning Laboratories, Corning, N.Y.). The filtered tissue supernatants were screened for the presence of infectious virus by titration in CRFK monolayers. For FA testing, fresh tissue samples were processed into 8-μm cryostatic sections, fixed in cold 100% acetone, incubated at room temperature with anti-PrV antibody conjugated to fluorescein isothiocyanate (National Animal Disease Laboratories, Ames, Iowa), washed in phosphate-buffered saline, and then coverslipped using mounting fluid (1:1, glycerol:phosphate-buffered saline).
RESULTS
Genomic analysis.
To verify that the LAP1 region in the genome of M5′ had been replaced by the gG gene promoter-lacZ gene transcriptional (gG-lacZ) unit and that the parental genotype had been restored in R5′, their DNAs were compared to that of the parental virus.
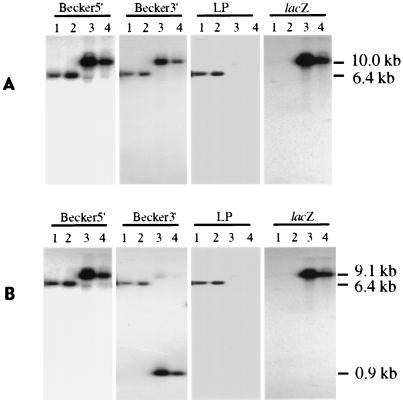
When the BamHI-digested viral genomes were hybridized with probes containing the BamHI 6 sequence upstream (Becker5′) or downstream (Becker3′) of the LAP1 region (Fig. 1), only a 6.4-kb fragment (BamHI 6) in the parental and R5′ viral genomes (Fig. 2A, lanes 1 and 2) and a 3.6-kb-larger fragment (10.0 kb) in the DNAs from two different M5′ virus isolates (lanes 3 and 4) were recognized. The increased size of the BamHI 6 fragment in the M5′ genome corresponded to replacement of the 282-bp LAP1 sequence with the 3.88-kb gG-lacZ unit.
FIG. 2.
Southern hybridization analysis of the PrV parental, rescuant, and recombinant viral genomes. BamHI (A)- and BamHI-EcoRI (B)-restricted viral DNA fragments were separated by gel electrophoresis, transferred to a nylon membrane, and then hybridized sequentially with 32P-labeled DNA probes (Becker5′, Becker3′, LP, and lacZ) as indicated above each autoradiogram. The parental and rescue viral DNAs were resolved in lanes 1 and 2, respectively; genomes of two distinct recombinant M5′ isolates were resolved in lanes 3 and 4. Sizes of the viral DNA fragments complementary to the various probes are indicated on the right.
The correctly placed gG-lacZ insert should be about 5.6 kb downstream from the 5′ end and about 0.45 kb upstream from the 3′ end of the BamHI 6 region. The unique EcoRI site present in the insert about 450 bp upstream from its 3′ end, but absent in the parental BamHI 6 fragment, was used to demonstrate the orientation of the insert in the M5′ viral genome. As anticipated, EcoRI in conjunction with BamHI digestion did not alter the size of the parental and rescuant viral genomic fragment annealing to both Becker5′ and Becker3′ probes (Fig. 2B, lanes 1 and 2). Likewise, as expected, the 10.0-kb M5′ DNA fragment partitioned into 9.1- and 0.9-kb portions complementary to Becker5′ and Becker3′ probes, respectively (lanes 3 and 4). When the same restricted viral DNAs were hybridized with the lacZ probe, which comprises a region upstream of the lacZ gene EcoRI site (Fig. 1), only the correctly sized fragment of 10.0 or 9.1 kb was detected in the M5′ viral genomes digested with either BamHI (Fig. 2A, lanes 3 and 4) or BamHI and EcoRI (Fig. 2B, lanes 3 and 4), respectively. As anticipated, the lacZ probe did not hybridize to either the parental or rescuant viral genomes (Fig. 2A and B, lanes 1 and 2). Thus, the insertion was correctly oriented within the predicted site.
To verify removal of the LAP1 region from the genome of the M5′ viruses, the PrV DNAs were hybridized with the LP probe, which is composed of only that portion of the PrV genome targeted for deletion (Fig. 1). As with the Becker5′ and Becker3′ probes, annealing only to a 6.4-kb fragment of the parental and rescuant viral genomes (Fig. 2A and B, lanes 1 and 2) was observed. In contrast, the LP probe did not hybridize with any fragment derived from the M5′ viral DNA (Fig. 2A and B, lanes 3 and 4). Thus, the putative LAT promoter region had been eliminated from the M5′ DNA and restored in the R5′ genome.
To determine if the M5′ virus retained its genetic modification after replication in the host animal, virus was isolated from each infected animal at 2 to 3 day p.i. via nasal swabs. The same hybridization patterns as previously observed for each virus were obtained when using the Becker5′, Becker3′, lacZ, and LP probes (data not shown). Therefore, the M5′ genome was stable during in vivo replication.
In vitro virus replication.
To assess any effect of LAP1-deletion on in vitro virus propagation, virus replication kinetics and yields of the recombinant and rescuant compared to that of the parental virus were determined in the PrV-permissive CRFK cell line. The M5′ mutant replicated as efficiently as the parental and rescuant viruses in that all three exhibited similar growth kinetics and had comparable titers at 12 h p.i. (data not shown). Furthermore, the mutant viral plaques were indistinguishable in size and appearance from those produced by the parental and rescuant viruses. Therefore, in vitro viral replication and yields were not measurably affected by replacement of the LAP1 sequence with a gG-lacZ cassette.
In vitro LAT expression.
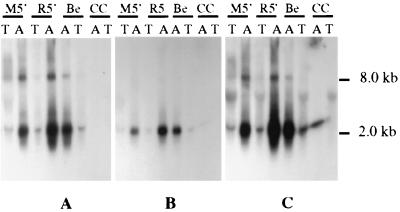
To determine whether removal of the LAP1 region affected in vitro lytic cycle LAT production, total and poly(A) RNAs isolated from M5′, R5′, and parental virus-infected N1E cells at 6 h p.i. were examined by Northern hybridization using ssRNA probes complementary to transcripts originating from the LAT gene (12) (Fig. 1). Probe Ic, specific for the sequences flanking the LAT gene intron, annealed to both 2.0- and 8.0-kb poly(A) RNAs obtained from parental, M5′, and R5′ virus-infected N1E cells (Fig. 3A). A similar pattern was obtained when probe IVc1, specific for the second exon of the LAT gene, was used (Fig. 3C). When the RNAs were hybridized with the first exon-specific probe, IIc, only the 2.0-kb and not the 8.0-kb (which originates within the intron [Fig. 1 and reference 12]) transcript was detected regardless of the source of virus-infected cells (Fig. 3B). Specific annealing to the RNA fractions from uninfected N1E cells was not observed for any of the probes. Thus, the PrV LAP1 does not appear to be essential for LAT gene expression during a productive infection in cultured neuronal cells, as removal of this promoter did not affect in vitro LAT gene-directed transcription.
FIG. 3.
Northern hybridization analysis of total and poly(A) RNAs from virus-infected and uninfected N1E cells at 6 h p.i. Uninfected control (CC), parental virus-infected (Be), recombinant virus-infected (M5′), and rescuant-infected (R5′) cellular sources of RNA are indicated above each autoradiogram. The RNAs were separated in a 2.2 M formaldehyde–1% agarose gel and then transferred to a nylon membrane. The membrane was sequentially hybridized with [32P]UTP-labeled ssRNA probes Ic (A), IIc (B), and IVc1 (C). As indicated above each lane, either total RNA (T) or poly(A) RNA (A) was used. The sizes of the RNAs complementary to the various probes are indicated on the right.
Primary in vivo virus infection.
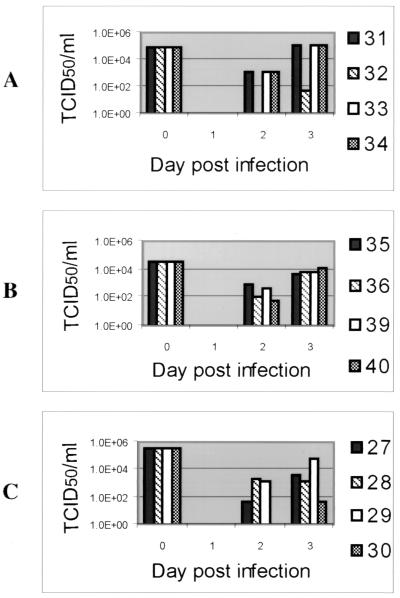
Within the first 7 days after receiving either the parental or rescuant viruses, the prevaccinated pigs developed severe clinical signs (pyrexia, anorexia, dullness, and sneezing) characteristic of an acute PrV infection. Two of the animals that received the R5′ virus and one infected with the parental virus exhibited severe neurological signs (incoordination, opisthotonus, and epileptiform convulsions, as well as muscular trembling). Animals infected with the M5′ virus had only mild disease (pyrexia, anorexia, and dullness). All animals manifested a high fever within the first 7 days after infection. As shown in Fig. 4, comparable amounts of each virus were excreted from their hosts within 2 to 3 day p.i. All of the infected pigs had recovered by 7 to 8 days p.i. The uninfected control pigs remained clinically normal throughout the study period.
FIG. 4.
Titer of infectious virus recovered from virus-infected pigs. The amount of excreted infectious virus (TCID50/milliliter) was determined from nasal swab samples obtained from each animal (designated by number in the key) infected with recombinant M5′ (A), parental (B), and rescuant R5′ (C) viruses. At day 0 p.i., the bar represents the titer of each virus inoculum given to the pigs. Starting at 2 days p.i., each bar represents the titer of virus excreted from the animals at the specified time points.
Establishment of latency.
The latent infection status of the pigs was determined at 35 days p.i. At this time, attempts to detect the excretion of infectious virus from the nasal cavities of infected pigs were unsuccessful. Likewise, evidence of virus replication in other tissues (tonsil, TG, and brain) could not be shown through direct virus isolation or FA testing. Such results indicate that an in vivo productive infection, if present, was not measurable by standard methodologies.
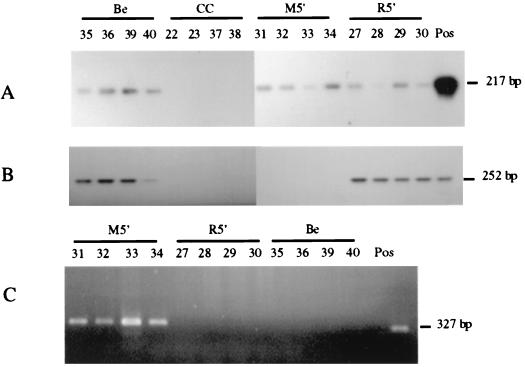
To verify PrV latency, the presence of viral genome in the TG should be demonstrable. To this end, total DNA was isolated from the TG of all pigs and screened for the presence of both parental and mutant viral genomes by a PCR for the unaltered PrV gD gene. Products of the correct size and composition were amplified from all of the infected pigs regardless of the inoculum used (Fig. 5A). Moreover, a similar specific amplification of the lacZ gene was successful only when TG DNA from the M5′ virus-infected animals was used (Fig. 5C). In contrast, attempts to detect gD-encoded mRNA expression, an event indicative of a productive PrV infection, in the TG by RT-PCR were unsuccessful (data not shown). These results demonstrate that all three viruses established a latent infection in their host animals and that the latent M5′ recombinant had retained the lacZ gene insertion.
FIG. 5.
Analysis of the PrV gD and E. coli lacZ gene PCR amplicons and the processed PrV LAT RT-PCR products generated from the porcine TG tissue preparations. Individual pigs are identified by number and represent uninfected negative controls (CC), parental virus-infected (Be), recombinant virus-infected (M5′), and rescuant-infected (R5′) animals. (A) Autoradiogram of gD gene amplicons using DNA probe gD. The positive control (Pos) uses PrV-Be DNA as the template and shows the anticipated 217-bp product. (B) Autoradiogram of LAT RT-PCR products using DNA probe LAT-MS. The positive control (Pos) uses pAC38 as the template and shows the predicted 252-bp product. (C) Electrophoregram of lacZ PCR products generated using primers A and C. The positive control (Pos) uses the lacZ insert in pRR12 as the template and shows the expected 327-bp product.
In vivo LAT expression.
To investigate the necessity of the LAP1 region for in vivo LAT expression during latency, total and poly(A) RNA extracts from the TG of the virus-infected pigs were examined for the presence of the processed 8.4-kb LAT by RT-PCR. Primers CM3.1 and LLT3.1 (Fig. 1) were selected so as to enable the amplification of portions of the two exon sequences flanking the intron in the spliced 8.4-kb LAT. A processed LAT was detected in all ganglionic total (data not shown) and poly(A) RNAs (Fig. 5B) from pigs infected with the parental and rescuant viruses. Amplification of this specific 252-bp LAT sequence did not occur when TG RNA from the uninfected or the M5′ virus-infected pigs was used (Fig. 5B). Likewise, a product of this size was not detected when the RT reaction was performed in the absence of reverse transcriptase. These results indicate that the LAP1 sequence removed from the genome of the M5′ mutant may be the in vivo basal promoter for the PrV LAT gene or, more specifically, for production of the 8.4-kb LAT in the virus's natural host.
DISCUSSION
The PrV LAP1 was initially predicted to be a LAT gene promoter based on its relative location to the 8.4-kb LAT transcriptional start site and the presence of consensus TATA, CAAT, and GC boxes (5, 18). Later, this preliminary attribution was confirmed by the demonstrated ability of LAP1 to direct HSV-1 LAT gene transcription when substituted for the homologous regulatory sequence (11). To determine the relevance of LAP1 in the PrV genome, we created a novel mutant virus whose DNA lacks a 282-bp sequence encompassing this predicted promoter. The modification was engineered so as not to affect the termination of the nearby and oppositely transcribed early EP0 gene, whose polyadenylation signal sequence is 180 nucleotides further downstream. Our study demonstrated that this deletion did not affect in vitro viral replication or the ability of the virus to enter an in vivo latent state in the natural host, swine.
Since the LAT gene of the M5′ virus was transcriptionally quiescent during latency, this inactivity confirms that the LAP1 is the basal promoter for in vivo regulation of this gene. In agreement with the findings on in vivo HSV LAT gene expression in animal models (1, 8, 15, 16, 24), the production of the in vivo 8.4-kb LAT is not required for the establishment of PrV latency in its natural host. However, it has yet to be determined whether the 2.0- and 8.0-kb viral RNAs detected during lytic cycle viral replication (12) participate in this event. Our data also confirmed a previous finding (14) that the TG is the preferred site of PrV latency since the viral genome was easily detected in this tissue during latency, but not in the tonsils or brain (data not shown). Although these two tissues also have been found to harbor PrV genomes (6), the detection frequency of viral DNA in them is always lower than that for the TG site.
Our study shows that the PrV LAP1 sequence, presumably essential for the production of the 8.4-kb LAT during a latent infection, is not required for LAT gene expression during an in vitro lytic (productive) infection of cultured neuronal cells. This result indicates that a different promoter may regulate lytic cycle viral LAT gene transcription. As previously reported, a second TATA box (LAP2) is located within the 8.4-kb LAT coding sequence 177 nucleotides downstream from the first one (5). By using an in vitro transient reporter gene expression assay, the LAP2 but not the LAP1 region was shown to be active in both neuronal and nonneuronal cells (7). Likewise, we have found that the LAP1 sequence fails to direct transcription from a fused reporter gene in cultured nonneuronal cells (unpublished data). Our previous in vitro LAT gene expression study demonstrated that transcription of the lytic cycle 2.0-kb viral RNA is initiated from a site about 200 bp downstream of the 8.4-kb LAT transcriptional start site (12). Therefore, it is probable that by virtue of proximity, consensus promoter elements, and demonstrated activity, this second TATA region is responsible for lytic cycle PrV LAT gene expression. In that instance, the PrV LAT gene would have dual regulatory promoters, as is the case for HSV-1 (2).
ACKNOWLEDGMENTS
We thank A. K. Cheung for providing plasmid pAC38.
This work was supported in part by USDA Animal Health and Disease grant ILLU-70-0989 and in part by University of Illinois Campus Research Board grant 95126.
REFERENCES
- 1.Block T M, Spivack J G, Steiner I, Deshmane S, McIntosh M T, Lirette R P, Fraser N W. A herpes simplex virus type 1 latency-associated transcript mutant reactivates with normal kinetics from latent infection. J Virol. 1990;64:3417–3426. doi: 10.1128/jvi.64.7.3417-3426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, Schmidt M C, Goins W F, Glorioso J C. Two herpes simplex virus type 1 latency-acting promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections. J Virol. 1995;69:7899–7908. doi: 10.1128/jvi.69.12.7899-7908.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung A K. Detection of pseudorabies virus transcripts in trigeminal ganglia of latently infected swine. J Virol. 1989;63:2908–2913. doi: 10.1128/jvi.63.7.2908-2913.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung A K. The BamHI J fragment (0.76 to 0.737 map units) of pseudorabies virus is transcriptionally active during viral infection. J Virol. 1990;64:977–983. doi: 10.1128/jvi.64.3.977-983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung A K. Cloning of the latency gene and the early protein 0 gene of pseudorabies virus. J Virol. 1991;65:5260–5271. doi: 10.1128/jvi.65.10.5260-5271.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung A K. Investigation of pseudorabies virus DNA and RNA in trigeminal ganglia and tonsil tissues of latently infected swine. Am J Vet Res. 1995;56:45–50. [PubMed] [Google Scholar]
- 7.Cheung A K, Smith T A. Analysis of the latency-associated transcript/UL1-3.5 gene cluster promoter complex of pseudorabies virus. Arch Virol. 1999;144:381–391. doi: 10.1007/s007050050511. [DOI] [PubMed] [Google Scholar]
- 8.Fareed M U, Spivack J G. Two open reading frames (ORF1 and ORF2) within the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1 are not essential for reactivation from latency. J Virol. 1994;68:8071–8081. doi: 10.1128/jvi.68.12.8071-8081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutekunst D E. Latent pseudorabies virus infection in swine detected by RNA-DNA hybridization. Am J Vet Res. 1979;40:1568–1572. [PubMed] [Google Scholar]
- 10.Gutekunst D E, Pirtle E C, Miller L D, Stewart W C. Isolation of pseudorabies virus from trigeminal ganglia of a latently sow. Am J Vet Res. 1980;41:1315–1316. [PubMed] [Google Scholar]
- 11.Huang C J, Rice M K, Devi-Rao G B, Wagner E K. The activity of the pseudorabies virus latency-associated transcript promoter is dependent on its genomic location in herpes simplex virus recombinants as well as on the type of cell infected. J Virol. 1994;68:1972–1976. doi: 10.1128/jvi.68.3.1972-1976.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin L, Scherba G. Expression of pseudorabies virus latency-associated transcript gene during productive infection of cultured cells. J Virol. 1999;73:9781–9788. doi: 10.1128/jvi.73.12.9781-9788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kingston R E. Calcium phosphate transfection. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1990. pp. 9.1.4–9.1.11. [Google Scholar]
- 14.Maes R K, Sussman M D, Vilnis A, Thacker B J. Recent developments in latency and recombination of Aujeszky's disease (pseudorabies) virus. Vet Microbiol. 1997;55:13–27. doi: 10.1016/s0378-1135(96)01305-3. [DOI] [PubMed] [Google Scholar]
- 15.Maggioncalda J, Mehta A, Fraser N W, Block T M. Analysis of a herpes simplex virus type 1 LAT mutant with a deletion between the putative promoter and the 5′ end of the 2.0-kilobase transcript. J Virol. 1994;68:7816–7824. doi: 10.1128/jvi.68.12.7816-7824.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maggioncalda J, Mehta A, Bagasra O, Fraser N W, Block T M. A herpes simplex virus type 1 mutant with a deletion immediately upstream of the LAT locus establishes latency and reactivates from latently infected mice with normal kinetics. J Neurovirol. 1996;2:268–278. doi: 10.3109/13550289609146890. [DOI] [PubMed] [Google Scholar]
- 17.Priola S A, Gustafson D P, Wagner E K, Stevens J G. A major portion of the latent pseudorabies virus genome is transcribed in trigeminal ganglia of pigs. J Virol. 1990;64:4755–4760. doi: 10.1128/jvi.64.10.4755-4760.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Priola S A, Stevens J G. The 5′ and 3′ limits of transcription in the pseudorabies virus latency associated transcription unit. Virology. 1991;182:852–856. doi: 10.1016/0042-6822(91)90628-o. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 16:66–16:67. [Google Scholar]
- 20.Schang L M, Kutish G F, Osorio F A. Correlation between precolonization of trigeminal ganglia by attenuated strains of pseudorabies virus and resistance to wild-type virus latency. J Virol. 1994;68:8470–8476. doi: 10.1128/jvi.68.12.8470-8476.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherba G, Jin L, Schnitzlein W M, Vodkin M H. Differential polymerase chain reaction for detection of wild-type and a vaccine strain of Aujeszky's disease. J Virol Methods. 1992;38:131–144. doi: 10.1016/0166-0934(92)90176-e. [DOI] [PubMed] [Google Scholar]
- 22.Schnitzlein W M, Winans R, Ellsworth S, Tripathy D N. Generation of thymidine kinase-deficient mutants of infectious laryngotracheitis virus. Virology. 1995;209:304–314. doi: 10.1006/viro.1995.1262. [DOI] [PubMed] [Google Scholar]
- 23.Spivack J G, Fraser N W. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987;61:3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steiner I, Spivack J G, Lirette R P, Brown S M, MacLean A R, Subak-Sharpe J H, Fraser N W. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 1989;8:505–511. doi: 10.1002/j.1460-2075.1989.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 26.Wheeler J G, Osorio F A. Investigation of sites of pseudorabies virus latency, using polymerase chain reaction. Am J Vet Res. 1991;52:1799–1803. [PubMed] [Google Scholar]