Abstract
Rotavirus infectivity is dependent on the proteolytic cleavage of the VP4 spike protein into VP8* and VP5* proteins. Proteolytically activated virus, as well as expressed VP5*, permeabilizes membranes, suggesting that cleavage exposes a membrane-interactive domain of VP5* which effects rapid viral entry. The VP5* protein contains a single long hydrophobic domain (VP5*-HD, residues 385 to 404) at an internal site. In order to address the role of the VP5*-HD in permeabilizing cellular membranes, we analyzed the entry of o-nitrophenyl-β-d-galactopyranoside (ONPG) into cells induced to express VP5* or mutated VP5* polypeptides. Following IPTG (isopropyl-β-d-thiogalactopyranoside) induction, VP5* and VP5* truncations containing the VP5*-HD permeabilized cells to the entry and cleavage of ONPG, while VP8* and control proteins had no effect on cellular permeability. Expression of VP5* deletions containing residues 265 to 474 or 265 to 404 permeabilized cells; however, C-terminal truncations which remove the conserved GGA (residues 399 to 401) within the HD abolished membrane permeability. Site-directed mutagenesis of the VP5-HD further demonstrated a requirement for residues within the HD for VP5*-induced membrane permeability. Functional analysis of mutant VP5*s indicate that conserved glycines within the HD are required and suggest that a random coiled structure rather than the strictly hydrophobic character of the domain is required for permeability. Expressed VP5* did not alter bacterial growth kinetics or lyse bacteria following induction. Instead, VP5*-mediated size-selective membrane permeability, releasing 376-Da carboxyfluorescein but not 4-kDa fluorescein isothiocyanate-dextran from preloaded liposomes. These findings suggest that the fundamental role for VP5* in the rotavirus entry process may be to expose triple-layered particles to low [Ca]i, which uncoats the virus, rather than to effect the detergent-like lysis of early endosomal membranes.
Rotaviruses are nonenveloped icosahedral viruses with eleven double-stranded RNA (dsRNA) gene segments inside a 70-nm triple-layered particle (TLP) (49). The capsids of infectious TLPs are formed by four structural proteins, VP2, VP6, VP7, and VP4 (16). TLPs are converted to transcriptionally active, noninfectious double-layered particles (DLPs) following exposure to low calcium ion concentrations which mimic cytoplasmic levels (7, 39, 54). VP6-encapsidated DLPs contain a dsRNA-dependent RNA polymerase complex and are transcriptionally active, extruding RNAs into the cytoplasm through pores in the DLP (7, 35). Baculovirus coexpression of rotavirus structural proteins results in the self-assembly of rotavirus proteins into TLP and DLP virus-like particles (VLPs) (9).
The rotavirus outer capsid is comprised of viral proteins VP4 and VP7 (16). VP7 is the major structural protein on the surface of TLPs, while 60 dimeric VP4 spikes project from the rotavirus surface (16, 50). Antibodies to VP4 and VP7 proteins neutralize rotaviruses, and immune responses to these proteins protect animals from disease (16). The VP4 and VP8* proteins of some rotaviruses hemagglutinate and mediate viral attachment to sialic-acid-containing cell surface components (16, 19). Rotaviruses also reportedly attach to cellular α4β1 and α2β1 integrins, and integrin interactions may represent the primary means of cellular attachment for human rotaviruses which lack hemagglutinin activity (5, 8, 43).
Protease treatment of rotaviruses is required for viral infectivity (1, 16, 17). On the virion, trypsin cleaves VP4 (86-kDa) into VP8* (28-kDa) and VP5* (60-kDa) fragments, activating the virus for infection (1, 17, 22). Proteolytically activated rotaviruses enter cells with a t1/2 of 5 to 10 min, and rotaviruses which are not proteolytically activated are endocytosed and degraded but do not infect cells (28, 29, 39, 58). Rotavirus entry does not require endosome acidification since viral entry is not inhibited by lysosomotropic agents, which basicify endosomes or macrolide antibiotics, bafilomycin A1, and concanamycin A, which selectively inhibit the endosome-specific [H+]ATPase (10, 28, 29, 37, 39).
There is relatively little information available on the means by which nonenveloped viruses cross the plasma membrane during entry (18, 20, 23, 28, 44, 52, 53, 56, 60, 61). Activated rotavirus TLPs, but not DLPs, permeabilize liposomes or vesicles preloaded with carboxyfluorescein (CF), indicating that the outercapsid is crucial to the viral interaction with cellular membranes (44, 53). Rotaviruses or complete VLPs also permeabilize cells permitting cell-cell fusion, ethidium bromide entry, the coentry of cellular toxins, and an increase in permeability to divalent cations (18, 24, 37, 46, 52, 53). In each case membrane permeability requires trypsinized TLPs, is temperature dependent, occurs at neutral pH, and is inhibited by neutralizing monoclonal antibodies (MAbs).
We have recently reported that purified VP5* and VP5* truncations which contain a single internal hydrophobic domain (HD) permeabilize model membranes (12, 40). As with virus, VP5*-induced permeability occurs at neutral pH and is dose and temperature dependent (12). VP5* permeability is blocked by neutralizing MAbs which recognize a large conformationally determined epitope on the VP5* protein (residues 248 to 474) and which select for viral variants with mutations in the HD (40, 41). The VP5* HD also shares homology with the fusion domain of the E1 proteins of enveloped Sindbis virus and Semliki Forest virus (SFV) (40). Collectively, these findings suggest that following cleavage of VP4, the VP5*-HD may interact with cell membranes and play an essential role in the rotavirus entry process.
In order to investigate the role of the VP5*-HD in membrane permeability, we generated a series of VP5* mutants and evaluated their function by using a bacterial permeability assay originally developed to investigate the function of viral fusion and pore-forming proteins (26, 32). Our findings demonstrate that VP5* proteins which permeabilize liposomes and cells contain the HD (residues 385 to 404) and that permeability is abolished by C-terminal truncations which remove a conserved GGA motif (residues 399 to 401) or by site-directed mutagenesis of the HD. Our findings further demonstrate the importance of conserved glycine residues within the VP5*-HD and point to a required random coiled structure of the HD for membrane permeabilization. In contrast to other membrane interactive proteins, VP5* did not inhibit bacterial growth or lyse bacteria following induction. VP5* also permeabilized liposomes to 376-Da CF but not to 4-kDa fluorescein isothiocyanate (FITC)-dextran, suggesting that VP5* permeabilization is size selective. These findings suggest a specific role for the VP5*-HD in the rotavirus entry process.
MATERIALS AND METHODS
Reagents.
Egg yolk phosphatidylcholine (l-α-lecithin, 760 Da) in chloroform was obtained from Avanti Polar Lipids, Inc. (Alabaster, Ala.). CF (376 kDa) was purchased from Molecular Probes (Eugene, Oreg.), and FITC-dextran was from Sigma. Isopropyl-β-d-thiogalactopyranoside (IPTG) and o-nitrophenyl-β-d- galactopyranoside (ONPG) were from Lab Scientific, Inc. (Livingston, N.J.).
Plasmids.
Oligonucleotide primers to the 5′ and 3′ ends of rhesus rotavirus (RRV) VP4 and VP4 fragments were synthesized with 5′ BamHI and 3′ SstI or XhoI sites. Products containing VP4(1–776), VP8(1–231), VP5*(248–776), VP5(248–475), and VP5*(265–475) and other truncations were generated by PCR and ligated directionally into pET-6HIS plasmids by standard methods. The pET-6HIS vector is a modified version of pET30a (Novagen) containing a substitution of the NdeI-to-BamHI fragment with a methionine and six histidines in frame with the BamHI site (12). Constructs were verified by restriction enzyme digestion, sequencing, and protein expression (12).
Protein expression and purification.
pET-6HIS plasmids containing RRV VP4, VP8, VP5*, VP5(248–474), or VP5*(265–474) were transformed into BL21(DE3) cells (Novagen) and grown in Luria broth (LB) with 50 μg of kanamycin per ml (40). Overnight cultures were diluted, grown to an OD600 of 0.2 to 0.4, and induced with IPTG for 3 h. Cells were harvested by centrifugation at 3,000 × g for 15 min and sonicated in 10 mM Tris (pH 8.0)–0.1 M NaH2PO4–2 M urea. Supernatants were discarded, and pellets were solubilized in 10 mM Tris (pH 8.0)–0.1 M NaH2PO4–8 M urea (buffer B). Lysates were pelleted in a microfuge for 10 min, purified by affinity chromatography on Nickel NTA-Agarose (Qiagen), and sequentially dialyzed as previously described (12).
Site-directed mutagenesis.
Single amino acid substitution mutants of VP5*(265–474) were generated by site-directed mutagenesis (QuikChange; Stratagene). Briefly, pairs of complementary oligonucleotide primers containing each desired mutation were synthesized. CsCl-purified VP5*(265–474) pET-6HIS plasmid (0.05 μg) was incubated with 0.125 μg of each primer and Pfu DNA polymerase for 16 cycles with the following cycle parameters: 95°C for 30 s, 55°C for 1 min, and 68°C for 14 min. PCR products were treated with DpnI at 37°C for 1 h and transformed into Escherichia coli strain XL-1 Blue. Mutations were confirmed by DNA sequencing.
ONPG entry assay.
The ONPG bacterial permeabilization assay was performed as previously described (26, 34). pET-6HIS clones were transformed into BL21(DE3) cells (Novagen) and grown overnight at 37°C in LB containing 50 μg of kanamycin per ml. Overnight cultures were diluted 1:20 and grown to an OD600 of 0.2 to 0.4. Cultures were split, and half the culture was induced with IPTG (1 mM). At given times postinduction, 1 ml of induced or uninduced culture was removed and the OD600 was measured. Additional aliquots (0.5 ml) were added to 0.5 ml of LB containing 50 μg of streptomycin per ml to stop translation. Cells were pelleted (1 min at 14,000 rpm) and resuspended in 1 ml of LB-streptomycin. ONPG (2 mM) was added to resuspended pellets and incubated at 30°C for 10 min. Cells were pelleted, and supernatants (1 ml) were added to 0.4 ml of 1 M sodium carbonate buffer (pH 9.5) to stop the reaction. The absorbance at 420 nm was read to measure ONPG cleavage within permeabilized cells. Recombinant protein present in 1 OD600 of IPTG-induced culture was purified by nickel affinity chromatography, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%), and stained with Coomassie blue (12). Bands from the scanned gel were quantitated densitometrically using NIH Image 1.5 and compared to protein standards, and ONPG readings were adjusted to comparable levels of expressed protein. For equivalently expressed VP5*(265–474) mutants, ONPG cleavage was measured by quantitating increases in OD420 compared to controls. A420 readings were standardized to the bacterial growth (OD600) following subtraction of OD420 values of uninduced controls (<0.1 OD420).
Preparation of liposomes.
Liposomes of phosphotidylcholine (PC) containing CF were prepared by an extrusion method (44). Briefly, 2 mg of PC was dissolved in chloroform, dried under nitrogen, and further dried under vacuum. Lipids were resuspended in 20 μl of CF (70 mM in 10 mM Tris [pH 7.35]) by vortexing. Following five cycles of freezing and thawing, lipid unilamellar vesicles (LUVs) or liposomes were prepared by extrusion through a 0.1-μm (pore-size) membrane (Mini-Extruder; Avanti Polar Lipids). Extravesicular fluorophore was eliminated by size exclusion chromatography on Sephadex G-50 (Pharmacia) in 10 mM Tris-HCl (pH 7.35)–140 mM NaCl (TN Buffer) (44). Fractions were collected (150 μl), and 3 μl was assayed in 1.6 ml of TN buffer for fluorescence dequenching following the addition of 0.125% Triton X-100. Fluorescence dequenching was monitored in the Perkin-Elmer Luminescence Spectrometer LS-5B at 520 nm (490-nm excitation) (Molecular Probes) (27). Fractions with a fluorescence change ratio of >15 were pooled and used in the CF release assays.
CF release assays.
The ability of purified VP5* to permeabilize LUVs and cause CF release was assayed by measuring the fluorescence dequenching of the released fluor as previously described (12, 44, 53). CF-LUVs (5 to 10 μl) were equilibrated in 1.6 ml of TN buffer in fluorimeter cuvettes at 37°C for 3 min with constant stirring prior to protein addition. Fluorescence dequenching was monitored in the Perkin-Elmer Luminescence Spectrometer LS-5B at 520 nm (490-nm excitation). Experiments were reproduced several times with different preparations of liposomes and VP5* protein. As previously described, VP5*-induced permeability was abolished by VP5*-specific neutralizing MAbs which recognize a large conformationally determined epitope requiring residues 265 to 474 of VP4 (12, 40, 41). At the conclusion of each experiment LUVs were lysed with 0.1% Triton X-100 to determine the maximum CF release within each sample. Samples with control proteins (VP8* or bovine serum albumin) were incubated and similarly treated to control for spontaneous CF release from liposomes (12). Results are expressed as a percentage of total fluorescence dequenching resulting from Triton X-100 addition. The percent fluorophore dequenching was calculated according to the following formula: percent release = [(Ft − F0)/(FT − F0)] × 100, where F0 is the background fluorescence, Ft is the fluorescence at time t, and FT is the total fluorescence of the sample (12, 44, 53).
Release of FITC-dextrans.
LUVs were prepared as described above, except that a 125-mg/ml solution of FITC-dextran of 4,000 Da (FD-4) was used in place of CF for resuspension of dried PC essentially as previously described (3, 12, 31, 40–42, 57). FD-4 LUVs (5 to 10 μl) were equilibrated in 1.6 ml of TN buffer in fluorimeter cuvettes at 37°C for 3 min prior to the addition of VP5* (4 μg, 44 nM) or melittin (0.03 nM) (Sigma). Samples were incubated at 37°C for 30 min and assayed for FD-4 release. Released FD-4 was separated from the LUVs by centrifugation through a Centricon-100 microseparator (Amicon). The flowthrough (free FD-4) was assayed directly, and the retentate (LUV-associated FD-4) was assayed after resuspension in 0.125% Triton X-100 in 1.6 ml of TN buffer and centrifugation. Fluorescence was monitored in a Perkin Elmer Luminescence Spectrometer (LS-5B) at 520 nm (490-nm excitation). FD-4 release was compared to the total FITC-dextran fluorescence of the sample (the sum of the flowthrough and the retentate) (31, 42).
RESULTS
VP5* permeabilizes BL21(DE3) cells.
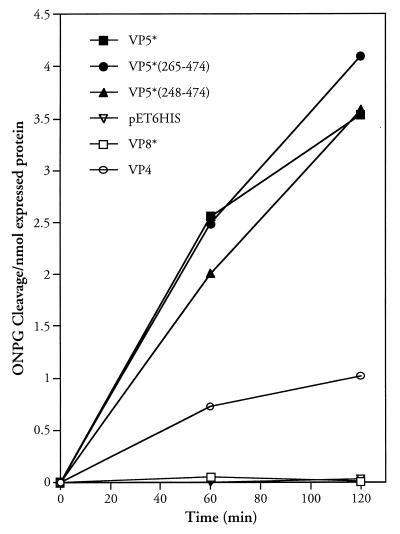
We previously demonstrated that purified expressed VP5* and VP5* truncations containing residues 265 to 474 permeabilized liposomes and that this activity was blocked by VP5*-specific neutralizing MAbs (40, 41). In order to assay the function of VP5* mutants, we used a bacterial membrane permeability assay previously used to study fusion and channel-forming proteins (26, 32). VP5*, VP5* truncations as well as VP8*, VP4, and the empty pET-6HIS plasmid were assayed for their ability to permeabilize E. coli following IPTG induction. In Fig. 1 the ability of induced proteins to permeabilize cells to ONPG was compared by measuring ONPG cleavage within cells as previously described (26, 32). To account for differences in the expression of VP4, VP8*, and VP5* proteins from pET6HIS, ONPG cleavage was standardized to recombinant protein expression levels using NIH image software analysis of induced proteins. VP8*(1–247), VP5*(248–474) or VP5*(265–474) truncations are expressed at comparable levels while VP5*(248–776) and VP4 are expressed at two- to fourfold-lower levels, respectively (12). BL21(DE3) cells were not permeabilized by IPTG induction of the recombinant VP8* protein or the pET-6HIS vector alone. In contrast, VP5* and VP5* truncations containing residues 265 to 474 permeabilized BL21(DE3) cells upon IPTG induction. VP4 also permeabilized cells at low but reproducible levels. Since VP8* lacks the ability to permeabilize cells, these findings suggest that VP4 either has some activity in permeabilizing membranes or that degradation products containing membrane interactive portions of VP5* permeabilize cells following VP4 induction. These findings are nearly identical to studies performed using purified VP8*, VP4, and VP5* proteins with CF-loaded liposomes and indicate the utility of the bacterial permeabilization assay in defining requirements for VP5*-induced membrane permeability (12).
FIG. 1.
VP5* induction permeabilizes bacteria. Bacterial permeabilization following IPTG induction was measured as previously described (12). E. coli BL21(DE3) cells containing pET-6HIS plasmids expressing VP4, VP8*, VP5*, VP5*(248–474), VP5*(265–474), or empty controls were grown to an OD600 of 0.2 to 0.4 and induced by the addition of IPTG (1 mM). At the indicated times cells were pelleted, resuspended in LB containing 50 μg of streptomycin per ml and 2 mM ONPG and incubated at 30°C for 10 min (26). Cleaved ONPG in supernatants was assayed spectrophotometrically (OD420), and the absorbance was standardized to nanomoles of recombinant expressed protein. This experiment was repeated six times with similar results.
Permeability induced by VP5* truncations.
To evaluate requirements for VP5*-induced membrane permeability, we generated a series of C- and N-terminal truncations of the functional VP5*(265–474) protein. We evaluated the ability of VP5* truncations to permeabilize cells using the ONPG cleavage assay and compared new truncations to VP5*s containing residues 248 to 474 or 265 to 474 of the RRV VP4 protein. N-terminal truncations containing residues 301 to 474 or 322 to 474 were unable to permeabilize cells (Fig. 2). However, a C-terminal truncation of residues 248 to 404 permeabilized cells with approximately 65% of the activity of the VP5*(248–474) positive control. Deleting three additional residues (residues 248 to 401) still permitted some membrane permeability (25% of residues 248 to 474) (Fig. 2). In contrast, induction of VP5*(248–398) which lacks a conserved GGA sequence (residues 399 to 401) from the VP5*-HD abolished permeability by the expressed VP5* protein. Further C-terminal truncations of the VP5*-HD were similarly unable to permeabilize cells following IPTG induction (Fig. 2).
FIG. 2.
Permeabilizing activity of VP5* truncations. E. coli BL21(DE3) containing pET-6HIS plasmids expressing the indicated VP5* truncations were grown and induced as previously described (12). Bacterial permeabilization was determined by using the ONPG cleavage assay as in Fig. 1 and standardized to OD600 of the culture at each time point. Uninduced controls resulted in ONPG cleavage of <0.1/OD600. This experiment was repeated at least three times with similar results.
Truncations which removed any residues between positions 380 and 401 of the HD abolished VP5*-mediated membrane permeability, and VP5*(265–380) lacks a hydrophobic potential membrane-spanning domain. N-terminal truncations which remove the cysteine at position 318 also abolished the membrane-permeabilizing function of VP5*. This finding suggests that the disulfide bond between cysteines at positions 318 and 380 may hold VP5* in a membrane-interactive conformation. This does not explain why the VP5*(301–474) fragment, which contains the disulfide bond and the HD, does not permeabilize membranes. However, residues 265 to 317 may similarly contribute to VP5* conformations required for HD interactions with membranes (40, 41).
Mutagenesis of residues in the VP5*-HD abolishes membrane permeability.
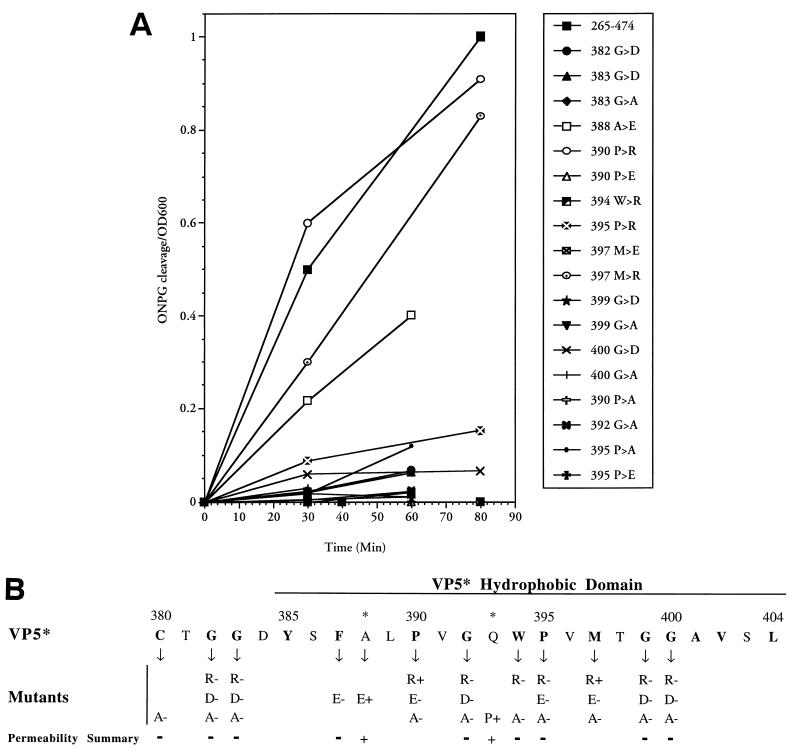
From an analysis of truncated VP5* proteins, membrane permeabilization requires the presence of the VP5*-HD, and deletions of the conserved GGA within the HD abolish this activity. In order to demonstrate a functional requirement of the VP5*-HD within VP5*(265–474), we generated a series of alanine scanning and charge inserted mutants of the VP5* hydrophobic domain. Since C-terminal truncations which removed the GGA (residues 399 to 401) abolished permeability by VP5* (see Fig. 4), mutagenesis was focused on the region between 380 and 400 and was further directed to residues which are highly conserved among all group A rotavirus VP4 proteins. Mutants were sequenced and analyzed by nickel affinity purification to demonstrate that expression levels were comparable to that of the control (data not shown). The functions of a representative number of VP5* mutants in the ONPG permeability assay are presented and compared to the VP5*(265–474) control (Fig. 3A).
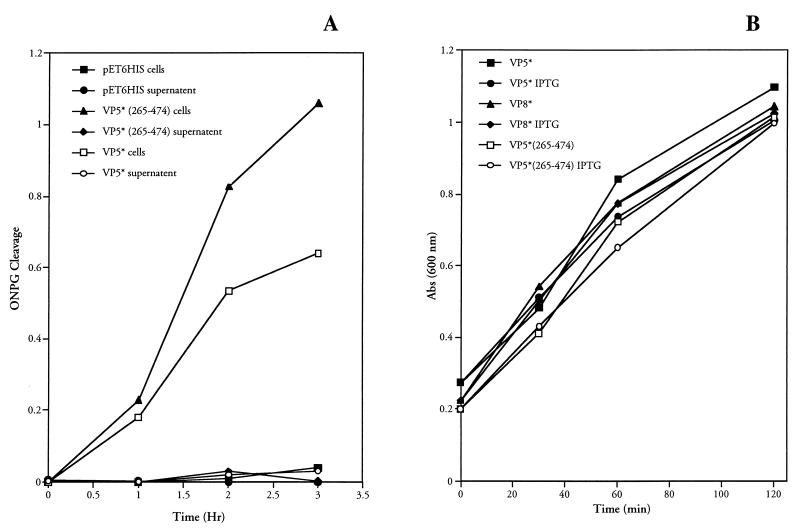
FIG. 4.
Bacteria are not lysed by VP5* expression. (A) β-Galactosidase is not released following VP5* expression. E. coli BL21(DE3)/pLysS containing the pET-6HIS plasmid with no insert or expressing either VP5*(265–474) or VP5*(248–776) were assayed for their ability to cause the release of β-galactosidase into the culture medium as measured by ONPG cleavage. Bacteria were grown to an OD600 of 0.2 to 0.4 and induced by the addition of IPTG (1 mM). Samples of each culture were removed at the indicated times after IPTG induction, and the OD600 was measured. Cells were pelleted, and both resuspended cells and supernatants were analyzed for ONPG cleavage as in Fig. 1. This experiment was repeated two times with similar results. (B) VP5* expression does not alter bacterial growth. E. coli BL21(DE3)/pLysS transformed with pET-6HIS plasmids expressing either VP5*(265–474), VP5*, or VP8* were grown to an OD600 of 0.2 to 0.4 and split into separate tubes. One tube was induced by the addition of IPTG (1 mM), and another was grown in parallel as a control. At given times postinduction, samples of each culture were removed and the OD600 was measured; growth curves of induced and uninduced cultures are shown. This experiment was repeated three times with similar results.
FIG. 3.
Mutagenesis of the VP5* HD inhibits membrane permeability. (A) VP5*(265–474) or indicated VP5*(265–474) point mutants were assayed for their ability to permeabilize E. coli as measured by the ONPG cleavage assay (Fig. 1). Bacteria were grown, IPTG induced, and assayed for ONPG cleavage as in Fig. 1. Cleaved ONPG in the supernatants was assayed as in Fig. 2. Each mutant was tested at least three times with similar results. (B) Summary of VP5* mutants. The permeability induced by oligo-nucleotide-directed VP5* mutants is summarized. Residues and introduced changes within the VP5* hydrophobic domain mutants are shown along with their ability (+) or inability (−) to permeabilize cells. Residues highly conserved among rotavirus strains are indicated in boldface. ∗, residue change in neutralizing MAb escape mutants.
VP5* mutants fell into two groups: mutations with little or no effect and mutations with little or no ability to permeabilize cells (Fig. 3A). A summary of mutagenesis findings is presented in Fig. 3B along with the sequence of the VP5*-HD and conserved residues from the Cys-380 to the end of the HD at Leu-404. Of the 30 mutations tested thus far, only three changes of highly conserved residues permitted VP5*-mediated permeability. Two were positively charged residue substitutions, and one was the same as a previously selected RRV neutralization escape mutant (A388→E) and occurs at a nonconserved residue. Alanine scanning mutagenesis of conserved residues abolished membrane permeability, indicating that the ability to permeabilize membranes is not strictly due to the hydrophobic nature of the domain. Although substitution of positively charged residues also abolished membrane permeability at most sites, Arg substitution for conserved Pro-390 or Met-397 residues had no affect on VP5*-mediated permeability. In contrast, any substitution of glycine residues within or adjacent to the HD abolished permeability, while introduction of a proline at residue 393 (Q→P) permitted VP5* function.
VP5* induction does not alter bacterial growth or lyse cells.
Bacterial permeabilization could release β-galactosidase from cells which causes ONPG cleavage. In order to determine if VP5* expression induced the release of β-galactosidase from cells, we determined whether ONPG cleavage was effected by supernatants of cells expressing VP5* or by ONPG entry into the cells themselves. In Fig. 4A, the release of β-galactosidase from cells was assessed following IPTG induction of VP5*, VP5*(265–474), or control empty pET-6HIS containing BL21(DE3)/pLysS cells. At each point postinduction cells were pelleted, and both supernatant and pellets were assayed for β-galactosidase activity using the ONPG cleavage assay. Neither supernatants nor cells from pET-6HIS-induced plasmids demonstrated β-galactosidase activity. As shown above, cells from IPTG-induced VP5* or VP5* truncations cleaved ONPG from 1 to 3 h postinduction. However, supernatants from induced cells had no β-galactosidase activity at any time point tested, indicating that the 86-kDa β-galactosidase protein is not released from cells and instead that ONPG cleavage occurs within the permeabilized bacteria.
Expressed fusion proteins are often deleterious to bacteria and affect bacterial growth kinetics (25, 26, 32, 47, 55). However, during the course of these experiments it was noted that bacterial growth following VP5* induction was unaltered. Figure 4B demonstrates that bacterial growth curves are nearly identical with or without IPTG induction of VP5*, VP5* truncations, or VP8. Collectively, these findings indicate that bacterial membranes were not lysed by VP5* expression and suggest that a transient, size-selective permeabilization might be effected by VP5* membrane interactions.
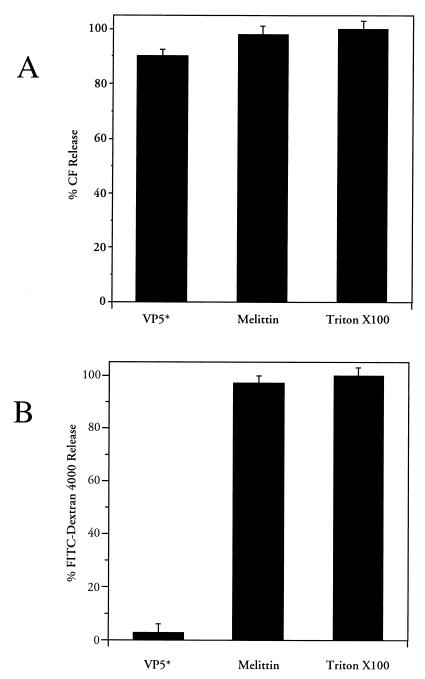
VP5* permeabilization of liposomes is size selective.
Normal bacterial growth curves and the absence of the 86-kDa β-galactosidase in the media suggested that VP5* lacks the ability to lyse membranes but may instead selectively permeabilize membranes. In order to determine whether VP5*-induced permeability is size selective, we tested the ability of purified VP5*(265–474) to release 376-Da CF or a 4-kDa FITC-dextran from preloaded liposomes (i.e., LUVs) (12, 31, 42). In Fig. 5 VP5*-induced fluorophore release was compared to that of the pore-forming protein melittin, as previously described, for 30 min at 37°C. Figure 5A demonstrates that both VP5 and melittin permeabilize liposomes and permit the release of CF to nearly the same extent as addition of a membrane-disruptive detergent, Triton X-100. Melittin also permitted the release of the 4-kDa FITC-dextran from liposomes. However, VP5* which permitted CF release from liposomes did not permeabilize liposomes to the 4-kDa fluor (Fig. 5B). Collectively, these findings indicate that VP5* does not lyse or generate large pores within membranes and instead suggest that VP5* is likely to cause a transient, size-selective permeabilization of membranes.
FIG. 5.
VP5* selectively permeabilizes liposomes. Purified LUVs containing either CF (70 mM) or FITC-dextran 4000 (FD-4) (30 mM) were incubated in 1.6 ml of TN buffer at 37°C for 30 min with nickel affinity-purified VP5* (4 μg, 44 nM) or melittin (0.03 nM) in duplicate (31, 42). (A) To measure CF release, fluorescence dequenching of samples was measured in a fluorimeter at an excitation wavelength of 490 nm and an emission wavelength of 520 nm. The total CF content of each sample was assessed by the addition of Triton X-100 (0.125%) and quantitated as described in Materials and Methods. (B) FD-4 is not present at self-quenching concentrations, and release was determined after centrifuging samples in a Centricon-100 microseparator (Amicon) and measuring released FD-4 in the effluent using a Perkin-Elmer Luminescence Spectrometer (LS-5B) at 520 nm (490-nm excitation). Liposome-associated FD-4 was assayed after resuspension in 1.6 ml of TN buffer plus 0.125% Triton X-100. Released FD-4 is expressed as a percentage of the total fluorescence (the sum of the effluent and the retentate). This experiment was repeated twice with similar results.
DISCUSSION
Once attached to cells, viruses are faced with the task of crossing the plasma membrane. Although the fusion of two lipid bilayers is relatively easy to envision and a commonplace event of intracellular trafficking, it is far less clear how nonenveloped viruses cross lipid bilayers. Proteolytic activation of rotaviruses is required for infectivity and is a key step in the virus' ability to cross the plasma membrane (17, 22) and permeabilize membranes (18, 22, 24, 44, 52, 53). Proteolytically activated rotaviruses enter cells with a t1/2 of 5 to 10 min while uncleaved virus is degraded in acidified endosomes (28, 29, 39, 58). It is now clear that proteases in the proximal small intestine cleave the rotavirus VP4 spike protein into VP8* and VP5* fragments (38). We have demonstrated that the VP5* cleavage product permeabilizes model membranes, and this provides a means for rotavirus interactions with cell membranes and a rationale for viral entry following proteolytic activation (12). The findings presented here define elements of the VP5*-HD required for permeabilizing membranes and further suggest a specific role for VP5* membrane interactions in the rotavirus entry process.
VP5* and two VP5* truncations, VP5*(248–474) and VP5*(265–474), were previously shown to permeabilize model membranes (12). However, E. coli-based membrane permeabilization assays have also been used to monitor the function of proteins which fuse or permeabilize membranes and are well suited for analyzing mutagenized proteins (33). The ability of VP5* to permeabilize bacterial membranes permitted us to define domains and residues of VP5* which are required for this function. The only potential membrane-spanning domain of VP5* occurs in residues 385 to 404 and is highly conserved (40). Both N- and C-terminal truncations of VP5* were identified that abolished membrane permeabilization. However, N-terminal deletions past residue 265 (residues 301 to 474 or 322 to 474) abolished the VP5* permeabilizing function even though no potential membrane-spanning regions of the protein were removed by this deletion. Deletion of the conserved cysteine, Cys-318, disrupts the disulfide bond with Cys-380 and alters the secondary structure of the protein required for function (45). We previously demonstrated that N-terminal deletions (residues 280 to 474) also abolished binding of neutralizing MAbs to VP5* which select for escape mutants within the HD (40, 41). These findings suggest that this region forms a large, conformationally determined epitope which may structurally constrain interactions of the HD with membranes and antibodies. It is possible that residues 265 to 317 are required to hold the HD in a membrane-interactive conformation, perhaps through interactions with the region between the cysteines. At present it is unclear whether further N-terminal truncations alleviate the block to membrane permeabilization or whether peptides containing the VP5* HD alone are capable of permeabilizing membranes.
C-terminal truncations including residues 265 to 404 were completely functional in permeabilizing membranes and contain the VP5* HD (residues 385 to 404) just downstream of Cys-380. Removal of three to six additional residues from the C terminus reduced or abolished permeabilizing activity, respectively, and shortened the HD. Additional C-terminal truncations of the VP5* HD lacked permeabilizing activity. These findings demonstrate that the VP5* HD is required for membrane permeabilizing function and that deleting even a few residues from this region abolishes permeability.
In order to assay the role of the HD in permeability and to define residues required for function, we mutated conserved residues within the VP5* HD. Interestingly, nearly any change introduced into conserved residues of the HD abolished membrane permeabilizing function. In addition, mutagenesis of any of the glycine residues in or around the HD abolished permeability. This included changes to a diglycine motif, homologous to the SFV E1 protein and required for E1-induced membrane fusion (30, 36). Our findings suggest that most conserved residues within the HD are absolutely required for the permeabilizing function. The inability of alanine residues to substitute and confer protein function further suggests that simple hydrophobic amino acid substitutions do not compensate for conserved residue functions. Although substitution of negatively charged residues at conserved HD sites is not tolerated for permeabilization, positively charged residues are permitted at two positions. Perhaps at these sites the positive charge facilitates HD interactions with negatively charged polar head groups of the lipid bilayer.
Glycines within the vesicular stomatitis virus G protein fusion domain are essential for fusion and have been suggested to be a common element of membrane fusion domains (6, 36). Glycine and proline residues are helix breaking, and their conservation in many fusion proteins may prevent helix formation or form hinges within the domain which permit cis-trans isomerization. Rotavirus VP5* proteins contain five glycine and two proline residues in and around the HD, with a diglycine motif at either end, and impart a random coiled structure to the domain (Fig. 3B). In fact, a Q→P substitution at residue 393 is tolerated in both neutralization escape mutants (40) and VP5* permeability mutants, suggesting that additional helix-breaking residues are permitted for VP5* function. Similar to what has been suggested for fusion or pore-forming proteins, these residues play an essential role in VP5*'s ability to permeabilize membranes.
In contrast to other membrane permeabilizing proteins (25, 26, 32, 47, 55), VP5* induction did not lyse bacteria or reduce bacterial viability, and this suggested a transient pore-forming function for VP5* membrane interactions. When we tested the ability of VP5* to permeabilize liposomes to 376- or 4,000-Da fluors, we similarly found that membranes were not lysed by VP5* but instead that a size-selective permeabilization was effected by VP5* which permitted efflux of only the 376-Da CF. Release of CF from liposomes has been reported to require a pore with a diameter of ∼10 Å, based on the radius of the CF molecule (51). The pore-forming peptide melittin was used as a control in these studies since it forms 25- to 30-Å pores in membranes, as measured by the release of 4-kDa but not 70-kDa FITC-dextrans (31). These findings suggest that VP5 may generate pores in membranes at least 10 Å in diameter which permit the transit of low-molecular-weight compounds.
Insight into VP5* permeabilizing functions may be provided by pore-forming protein interactions with membranes. Melittin, bee venom, is a 29-residue peptide which contains a series of glycine and proline residues (three glycines and one proline) within its 19-residue HD (11, 42). These residues facilitate cis-trans isomerization of the peptide, permit it to assume a water-soluble conformation, and are suggested to be important for membrane insertion (6). Melittin also forms a voltage-gated ion conductance channel in membranes which is size selective and contains a basic domain that presumably permits its association with polar head groups of the membrane (11, 59). The influenza M2 protein also forms voltage-gated ion channels in membranes which facilitate virus entry and exit from cells, and M2 pores are blocked by the anti-influenza drug amantadine (26, 48, 63). The diphtheria toxin B subunit also permeabilizes membranes by forming 8-Å diameter channels in membranes and may deliver the toxic A subunit through these pores or by forming a hydrophobic cleft in the membrane which permits the subunit to enter (2, 62). Although it is not known whether VP5* forms voltage-gated channels in membranes, a study of VP5* pores may further define the selectivity of VP5*-membrane interactions and may permit the identification of inhibitors of VP5* permeability.
There have been several hypotheses on how rotaviruses enter cells. All of these hypotheses must take into consideration that (i) rotaviruses enter cells rapidly; (ii) entry does not require endosome acidification; (iii) rotaviruses permit the coentry of exogenously provided compounds, such as the 28-kDa toxin alpha sarcin; (iv) rotavirus TLPs uncoat, forming DLPs, within a low-calcium environment; and (v) rotavirus entry results in the delivery of transcriptionally active 50-nm DLPs into the cytoplasm of cells (10, 17, 18, 21, 24, 28, 29, 37, 52, 54). Our findings suggest an additional element which must be considered in the delivery of rotavirus DLPs into the cytoplasm: that VP5* selectively rather than lytically permeabilizes membranes.
Although a direct-entry mechanism has not been totally discounted, the inability of rotaviruses to uncoat in high-extracellular-[Ca]i environments and the inability to lower [Ca]i surrounding the virions in the absence of a membrane-bound compartment suggest that infectious rotaviruses rapidly enter into early endosomal vesicles (7, 37, 54). A recent study also suggests that rotavirus particles are present in vesicles immediately following infection (52). This accounts for the coentry of large toxins along with the 70- or 50-nm particle, but it does not establish whether virion uncoating occurs before or after membrane lysis. It was suggested that early endosomal [Ca2+]i is reduced from 1 mM to the intracellular 100 nM level by diffusion or by endocytic channels (13, 52). It was further suggested that the concomitant uncoating of virions releases outer capsid proteins which lyse early endosomal membranes (52). However, our findings suggest that VP5*-mediated permeability is size selective and not lytic and instead suggests that VP5* may play a role in lowering the [Ca]i within endosomes rather than in disrupting the endosomal membrane.
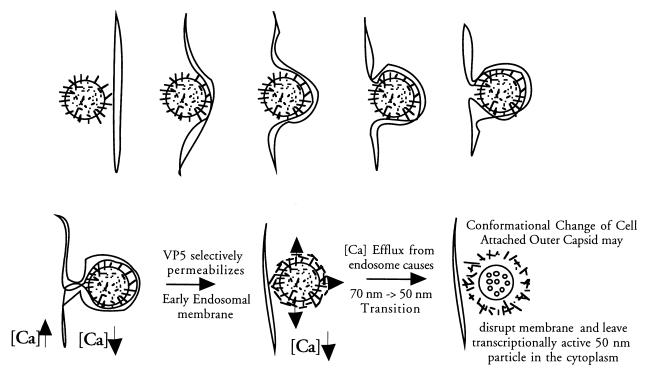
The TLP-to-DLP transition is a dramatic conformational change resulting from the loss of 60 VP4 spikes and 260 calcium-dependent VP7 trimers from the virion's surface (14, 15, 49). The inability of VP5* to lyse membranes and instead facilitate uncoating suggests that membrane disruption could be effected by the energy of the uncoating process itself while the outer capsid proteins are still bound to cellular receptors in the membrane (Fig. 6). It is also possible that VP5* lowers endocytic [Ca]i and that virion uncoating releases a second permeabilizing polypeptide, perhaps a lytic function of the protease digested VP7 (4). Although there are many points that need to be addressed to more fully understand the role of VP5* in the rotavirus entry process, proteolytic cleavage of VP4 appears to trigger VP5* to selectively permeabilize membranes and suggests that VP5* may function as a membrane pore which facilitates virion uncoating during viral entry.
FIG. 6.
Role of VP5* in rotavirus entry. The role of VP5* in the rotavirus entry process is hypothesized in light of its ability to selectively permeabilize membranes. Rotavirus is drawn as a circle with protruding VP4 spikes initially entering an early endosome. The outer capsid proteins on the virion are presumed to maintain contacts with receptors, while fusogenic domains of VP5* are exposed following VP4 cleavage in order to selectively permeabilize the early endosomal membrane. Permeabilization of early endosomes presumably lowers the [Ca]i surrounding the virions and permits virion uncoating.
ACKNOWLEDGMENTS
We are grateful to Erwin London for insightful discussions and to Yildiz Farooqui and Jignesh Patel for technical assistance.
This work was supported by a Merit Award from the Veterans Administration and by NIH grants R01-AI31016 and RO3-AI42150 to E.R.M.
REFERENCES
- 1.Arias C F, Romero P, Alvarez V, Lopez S. Trypsin activation pathway of rotavirus infectivity. J Virol. 1996;70:5832–5839. doi: 10.1128/jvi.70.9.5832-5839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaumelle B, Bensammar L, Bienvenue A. Selective translocation of the A chain diphtheria toxin across the membrane of purified endosomes. J Biol Chem. 1992;267:11525–11531. [PubMed] [Google Scholar]
- 3.Butko P, Huang F, Pusztai-Carey M, Surewicz W K. Membrane permeabilization induced by cytolytic delta-endotoxin CytA from Bacillus thuringiensis var. israelensis. Biochemistry. 1996;35:11355–11360. doi: 10.1021/bi960970s. [DOI] [PubMed] [Google Scholar]
- 4.Charpilienne A, Abad M J, Michelangeli F, Alvarado F, Vasseur M, Cohen J, Ruiz M C. Solubilized and cleaved VP7, the outer glycoprotein of rotavirus, induces permeabilization of cell membrane vesicles. J Gen Virol. 1997;78:1367–1371. doi: 10.1099/0022-1317-78-6-1367. [DOI] [PubMed] [Google Scholar]
- 5.Ciarlet M, Estes M K. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J Gen Virol. 1999;80:943–948. doi: 10.1099/0022-1317-80-4-943. [DOI] [PubMed] [Google Scholar]
- 6.Cleverley D Z, Lenard J. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc Natl Acad Sci USA. 1998;95:3425–3430. doi: 10.1073/pnas.95.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J, Laporte J, Charpilienne A, Scherrer R. Activation of rotavirus RNA polymerase by calcium chelation. Arch Virol. 1979;60:177–186. doi: 10.1007/BF01317489. [DOI] [PubMed] [Google Scholar]
- 8.Coulson B S, Londrigan S L, Lee D J. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc Natl Acad Sci USA. 1997;94:5389–5394. doi: 10.1073/pnas.94.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford S E, Labbe M, Cohen J, Burroughs M H, Zhou Y J, Estes M K. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5945–5922. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuadras M A, Arias C F, Lopez S. Rotaviruses induce an early membrane permeabilization of MA104 cells and do not require a low intracellular Ca2+ concentration to initiate their replication cycle. J Virol. 1997;71:9065–9074. doi: 10.1128/jvi.71.12.9065-9074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dempsey C E. The actions of melittin on membranes. Biochim Biophys Acta. 1990;1031:143–161. doi: 10.1016/0304-4157(90)90006-x. [DOI] [PubMed] [Google Scholar]
- 12.Denisova E, Dowling W, LaMonica R, Shaw R, Scarlata S, Ruggeri F, Mackow E R. Rotavirus capsid protein VP5* permeabilizes membranes. J Virol. 1999;73:3147–3153. doi: 10.1128/jvi.73.4.3147-3153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz R, Wileman T E, Anderson S J, Stahl P. The use of permeabilized cells to study the ion requirements of receptor-ligand dissociation in endosomes. Biochem J. 1989;260:127–134. doi: 10.1042/bj2600127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dormitzer P, Both G, Greenberg H B. Presentation of neutralizing epitopes by engineered rotavirus VP7s expressed by recombinant vaccinia viruses. Virology. 1994;204:391–402. doi: 10.1006/viro.1994.1543. [DOI] [PubMed] [Google Scholar]
- 15.Dormitzer P R, Greenberg H B. Calcium chelation induces a conformational change in recombinant herpes simplex virus-1-expressed rotavirus VP7. Virology. 1992;189:828–832. doi: 10.1016/0042-6822(92)90616-w. [DOI] [PubMed] [Google Scholar]
- 16.Estes M K. Rotaviruses and their replication. In: Knipe D M, Fields B N, editors. Virology. New York, N.Y: Raven Press; 1996. pp. 1329–1352. [Google Scholar]
- 17.Estes M K, Graham D Y, Mason B B. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falconer M M, Gilbert J M, Roper A M, Greenberg H B, Gavora J S. Rotavirus-induced fusion from without in tissue culture cells. J Virol. 1995;69:5582–5591. doi: 10.1128/jvi.69.9.5582-5591.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiore L, Greenberg H B, Mackow E R. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology. 1991;181:553–563. doi: 10.1016/0042-6822(91)90888-i. [DOI] [PubMed] [Google Scholar]
- 20.Fricks C E, Hogle J M. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukuhara N, Yoshie O, Kitaoka S, Konno T, Ishida N. Evidence for endocytosis-independent infection by human rotavirus. Arch Virol. 1987;97:93–99. doi: 10.1007/BF01310737. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert J, Greenberg H. Cleavage of rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J Virol. 1998;72:5323–5327. doi: 10.1128/jvi.72.6.5323-5327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert J, Greenberg H. Virus-like particle-induced fusion form without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J Virol. 1997;71:4555–4563. doi: 10.1128/jvi.71.6.4555-4563.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert J, Greenberg H. Virus-like particle-induced fusion from without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J Virol. 1997;71:4555–4563. doi: 10.1128/jvi.71.6.4555-4563.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez M E, Carrasco L. The human immunodeficiency virus type 1 Vpu protein enhances membrane permeability. Biochemistry. 1998;37:13710–13719. doi: 10.1021/bi981527f. [DOI] [PubMed] [Google Scholar]
- 26.Guinea R, Carrasco L. Influenza virus M2 protein modifies membrane permeability in E. coli cells. FEBS Lett. 1994;343:242–246. doi: 10.1016/0014-5793(94)80564-4. [DOI] [PubMed] [Google Scholar]
- 27.Haugland R P. Polar tracers. In: Spence M T Z, editor. Handbook of fluorescent probes and research chemicals. 6th ed. Portland, Oreg: Molecular Probes Inc.; 1990. pp. 331–356. [Google Scholar]
- 28.Kaljot K T, Shaw R D, Rubin D H, Greenberg H B. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J Virol. 1988;62:1136–1144. doi: 10.1128/jvi.62.4.1136-1144.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keljo D J, Kuhn M, Smith A. Acidification of endosomes is not important for the entry of rotavirus into the cell. J Pediatr Gastroenterol Nutr. 1988;7:257–263. doi: 10.1097/00005176-198803000-00016. [DOI] [PubMed] [Google Scholar]
- 30.Kielian M, Klimjack M R, Ghosh S, Duffus W A. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J Cell Biol. 1996;134:863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladokhin A S, Selsted M E, White S H. Sizing membrane pores in lipid vesicles by leakage of co-encapsulated markers: pore formation by melittin. Biophys J. 1997;72:1762–1766. doi: 10.1016/S0006-3495(97)78822-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lama J, Carrasco L. Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J Biol Chem. 1992;267:15932–15937. [PubMed] [Google Scholar]
- 33.Lama J, Carrasco L. Mutations in the hydrophobic domain of poliovirus protein 3AB abrogate its permeabilizing activity. FEBS Lett. 1995;367:5–11. doi: 10.1016/0014-5793(95)00523-c. [DOI] [PubMed] [Google Scholar]
- 34.Lama J, Carrasco L. Screening for membrane-permeabilizing mutants of the poliovirus protein 3AB. J Gen Virol. 1996;77:2109–2119. doi: 10.1099/0022-1317-77-9-2109. [DOI] [PubMed] [Google Scholar]
- 35.Lawton J A, Estes M K, Prasad B V. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 36.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liprandi F, Moros Z, Gerder M, Ludert J E, Pujol F H, Ruiz M C, Michelangeli F, Charpilienne A, Cohen J. Productive penetration of rotavirus in cultured cells induces coentry of the translation inhibitor alpha-sarcin. Epidemiol Infect. 1997;119:227–230. doi: 10.1006/viro.1997.8803. [DOI] [PubMed] [Google Scholar]
- 38.Ludert J E, Krishnaney A A, Burns J W, Vo P T, Greenberg H B. Cleavage of rotavirus VP4 in vivo. J Gen Virol. 1996;77:391–395. doi: 10.1099/0022-1317-77-3-391. [DOI] [PubMed] [Google Scholar]
- 39.Ludert J E, Michelangeli F, Gil F, Liprandi F, Esparza J. Penetration and uncoating of rotaviruses in cultured cells. Intervirology. 1987;27:95–101. doi: 10.1159/000149726. [DOI] [PubMed] [Google Scholar]
- 40.Mackow E R, Shaw R D, Matsui S M, Vo P T, Dang M N, Greenberg H B. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci USA. 1988;85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackow E R, Yamanaka M Y, Dang M N, Greenberg H B. DNA amplification-restricted transcription-translation: rapid analysis of rhesus rotavirus neutralization sites. Proc Natl Acad Sci USA. 1990;87:518–522. doi: 10.1073/pnas.87.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuzaki K, Yoneyama S, Miyajima K. Pore formation and translocation of melittin. Biophys J. 1997;73:831–838. doi: 10.1016/S0006-3495(97)78115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendez E, Lopez S, Cuadras M A, Romero P, Arias C F. Entry of rotaviruses is a multistep process. Virology. 1999;263:450–459. doi: 10.1006/viro.1999.9976. [DOI] [PubMed] [Google Scholar]
- 44.Nandi P, Charpilienne A, Cohen J. Interaction of rotavirus particles with liposomes. J Virol. 1992;66:3363–3367. doi: 10.1128/jvi.66.6.3363-3367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patton J T, Hua J, Mansell E A. Location of intrachain disulfide bonds in the VP5* and VP8* trypsin cleavage fragments of the rhesus rotavirus spike protein VP4. J Virol. 1993;67:4848–4855. doi: 10.1128/jvi.67.8.4848-4855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez J F, Ruiz M C, Chemello M E, Michelangeli F. Characterization of a membrane calcium pathway induced by rotavirus infection in cultured cells. J Virol. 1999;73:2481–2490. doi: 10.1128/jvi.73.3.2481-2490.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez M, Garcia-Barreno B, Melero J A, Carrasco L, Guinea R. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology. 1997;235:342–351. doi: 10.1006/viro.1997.8696. [DOI] [PubMed] [Google Scholar]
- 48.Pinto L H, Holsinger L J, Lamb R A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 49.Prasad B V, Chiu W. Structure of rotavirus. Curr Top Microbiol Immunol. 1994;185:9–29. doi: 10.1007/978-3-642-78256-5_2. [DOI] [PubMed] [Google Scholar]
- 50.Prasad B V, Wang G J, Clerx J P, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 51.Rex S, Schwarz G. Quantitative studies on the melittin-induced leakage mechanism of lipid vesicles. Biochemistry. 1998;37:2336–2345. doi: 10.1021/bi971009p. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz M C, Abad M J, Charpilienne A, Cohen J, Michelangeli F. Cell lines susceptible to infection are permeabilized by cleaved and solubilized outer layer proteins of rotavirus. J Virol. 1997;71:9458–9465. doi: 10.1099/0022-1317-78-11-2883. [DOI] [PubMed] [Google Scholar]
- 53.Ruiz M C, Alonso-Torre S R, Charpilienne A, Vasseur M, Michelangeli F, Cohen J, Alvarado F. Rotavirus interaction with isolated membrane vesicles. J Virol. 1994;68:4009–4016. doi: 10.1128/jvi.68.6.4009-4016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruiz M C, Charpilienne A, Liprandi F, Gajardo R, Michelangeli F, Cohen J. The concentration of Ca2+ that solubilizes outer capsid proteins from rotavirus particles is dependent on the strain. J Virol. 1996;70:4877–4883. doi: 10.1128/jvi.70.8.4877-4883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanz M A, Perez L, Carrasco L. Semliki Forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J Biol Chem. 1994;269:12106–12110. [PubMed] [Google Scholar]
- 56.Seth P. Mechanism of adenovirus-mediated endosome lysis: role of the intact adenovirus capsid structure. Biochem Biophys Res Commun. 1994;205:1318–1324. doi: 10.1006/bbrc.1994.2809. [DOI] [PubMed] [Google Scholar]
- 57.Shaw R D, Vo P T, Offit P A, Coulson B S, Greenberg H B. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology. 1986;155:434–451. doi: 10.1016/0042-6822(86)90205-9. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki H, Kitaoka S, Konno T, Sato T, Ishida N. Two modes of human rotavirus entry into MA104 cells. Arch Virol. 1985;85:25–34. doi: 10.1007/BF01317003. [DOI] [PubMed] [Google Scholar]
- 59.Tosteson M T, Alvarez O, Hubbell W, Bieganski R M, Attenbach C, Caporales L H, Levy J J, Nutt R F, Rosenblatt M, Tosteson D C. Primary structure of peptides and ion channels. Role of amino acid side chains in voltage gating of melittin channels. Biophys J. 1990;58:1367–1375. doi: 10.1016/S0006-3495(90)82483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tosteson M T, Chow M. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J Virol. 1997;71:507–511. doi: 10.1128/jvi.71.1.507-511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tosteson M T, Nibert M L, Fields B N. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc Natl Acad Sci USA. 1993;90:10549–10552. doi: 10.1073/pnas.90.22.10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Umata T, Mekada E. Diphtheria toxin translocation across endosome membranes. A novel cell permeabilization assay reveals new diphtheria toxin fragments in endocytic vesicles. J Biol Chem. 1998;273:8351–8359. doi: 10.1074/jbc.273.14.8351. [DOI] [PubMed] [Google Scholar]
- 63.Wang C, Takeuchi K, Pinto L H, Lamb R A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]