Abstract
Squalene is a highly sought-after triterpene compound in growing demand, and its production offers a promising avenue for circular economy practices. In this study, we applied metabolic engineering principles to enhance squalene production in the nonconventional yeast Yarrowia lipolytica, using waste cooking oil as a substrate. By overexpressing key enzymes in the mevalonate pathway — specifically ERG9 encoding squalene synthase, ERG20 encoding farnesyl diphosphate synthase, and HMGR encoding hydroxy-methyl-glutaryl-CoA reductase — we achieved a yield of 779.9 mg/L of squalene. Further co-overexpression of DGA1, encoding diacylglycerol acyltransferase, and CAT2, encoding carnitine acetyltransferase, in combination with prior metabolic enhancements, boosted squalene production to 1381.4 mg/L in the engineered strain Po1g17. To enhance the supply of the precursor acetyl-CoA and inhibit downstream squalene conversion, we supplemented with 6 g/L pyruvic acid and 0.7 mg/L terbinafine, resulting in an overall squalene titer of 2594.1 mg/L. These advancements underscore the potential for sustainable, large-scale squalene production using Y. lipolytica cell factories, contributing to circular economy initiatives by valorizing waste materials.
Keywords: Squalene, Y. lipolytica, Metabolic engineering, Waste cooking oil
Graphical abstract
Highlights
-
•
Up-cycling of waste cooking oil into squalene using engineered Y. lipolytica.
-
•
Overexpression of pathway genes directed metabolic flux towards squalene biosynthesis.
-
•
Co-overexpressing CAT2 and DGA1 boosted the squalene titers in Y. lipolytica.
1. Introduction
Squalene, a highly unsaturated triterpene with a chemical formula of C30H50, was first isolated from the liver oil of deep-sea sharks (Squalus spp). At the molecular level, squalene holds the important biological role as a precursor in the biosynthesis of cholesterol, hormones, and vitamins, and thereby regulates cell division and growth (Berghoff et al., 2021). Furthermore, squalene has direct uses in anti-cancer, anti-oxidation, maintaining cell membrane integrity and providing protective and moisturizing advantages in the food, cosmetic and pharmaceutical sectors (Paramasivan and Mutturi, 2022; Brown et al., 2019; Garcia-Bermudez et al., 2019; Tateno et al., 2020).
In 2005, responding to global marine wildlife conservation efforts against harvesting squalene from shark livers, cosmetic and pharmaceutical companies started exploring phytoextraction as an alternative squalene source. Nevertheless, this method faces inefficiency due to extensive organic solvent use and the relatively low squalene content in plants (Chang and Keasling, 2006). A substantial quantity of plant biomass is required for relatively meager squalene yields, rendering the process economically impractical. In recent years, systems metabolic engineering has opened new avenues for producing plant natural products using microbial cell factories. Yarrowia lipolytica, a marine yeast valued for its biological safety and metabolic benefits, adeptly stores lipids and lipid-derived substances. These reserves can be converted into acetyl-CoA precursors, essential for powering squalene biosynthesis (Chattopadhyay et al., 2021). Y. lipolytica also exhibits good tolerance to harsh environments, making it suitable for various industrial applications (Barth and Gaillardin, 1997; Madzak C, 2021).
Numerous strategies have been employed to boost squalene production in engineered yeast cell factories. These includes:
-
1)
Up-regulation of squalene synthesis pathways: Increasing the expression of genes in the mevalonate (MVA) pathway and sterol synthesis pathway has been shown to enhance squalene production significantly (Li et al., 2020). Notably, the overexpression of the HMG-CoA reductase enzyme (HMGR), a rate-limiting enzyme in the squalene synthesis pathway, has proven effective in boosting squalene production (Huang et al., 2018).
-
2)
Weakening of downstream degradation pathways: Strategies involving the suppression of squalene degradation pathways have been implemented. One approach is to modify the promoter of the ERG1 gene, responsible for squalene degradation, leading to increased squalene production (Hull et al., 2014).
-
3)
Enzyme engineering: Mutations in enzymes such as ERG1 have been used to increase squalene production (Zhou et al., 2021).
-
4)
Increasing squalene storage: Enhancing lipid droplet formation in yeast cells has been achieved through overexpression of the DGA1 gene, resulting in significantly increased squalene production (Wei et al., 2018).
-
5)
Compartmentalization engineering: Strategies that involve compartmentalization in mitochondria and peroxisomes have been used to increase squalene production (Zhu et al., 2021).
-
6)
Bioprocess optimization: Various carbon sources, including sugarcane molasses and xylose, have been utilized to boost squalene production (Kwak et al., 2017). Additionally, the addition of specific substances during fermentation, like terbinafine and methyl jasmonate, has been found to enhance squalene production (Tang et al., 2021).
A review of literatures published before December 2023 revealed that squalene production in metabolically engineered Y. lipolytica typically yielded less than 1 g/L in shake flask cultures, a benchmark often regarded as the threshold for initiating industrial fermentation processes (Huang et al., 2018; Liu et al., 2020; Tang et al., 2021; Wei et al., 2021). In recent studies led by Ma et al. and Ning et al., Yarrowia strains capable of producing squalene titers of 3820 mg/L and 2549 mg/L within 72 h of shake flask cultures were reported (Ma et al., 2024; Ning et al., 2024). These advancements were accomplished by compartmentalizing key enzymes in the peroxisome and utilizing glucose as the primary carbon source.
In our previous research, we illustrated Y. lipolytica's capability to produce acetyl-CoA-derived biochemical from lipids. This capability is mainly due to its robust beta-oxidation capacity, breaking down fatty acids into acetyl-CoA, and its expanded array of extracellular lipases (Kong et al., 2022). Supplementary to the abovementioned works, in this study, we explored the synergistic combination of various individual methods recognized for their potential to enhance squalene production in Y. lipolytica, as illustrated in Fig. 1. Firstly, we verified that waste cooking oil (WCO), as the carbon source, provided greater benefits compared to glucose and glycerol in stimulating endogenous squalene biosynthesis using a parental strain of Y. lipolytica. We then enhanced squalene production by individually and collectively overexpressing key genes in the mevalonate and β-oxidation pathways. We further studied the effect of enhancing the transmembrane transport rate of acetyl-CoA and increasing squalene storage capacity on squalene titers. Finally, with the optimization of fermentation media, we attained 2.5 g/L squalene titer and a productivity of 21.6 mg/L/h in shake flask cultures. These results hold great promise for scaling up squalene production in a sustainable circular economy, where the fermentation process primarily utilizes WCO as the carbon source.
Fig. 1.
Squalene biosynthesis in Y. lipolytica on various carbon sources, such as glucose, glycerol, and free fatty acids (FAs) from waste cooking oil (WCO). Black arrows indicate the endogenous squalene biosynthetic pathway metabolic pathway; target genes denoted in red were overexpressed in this study. The enzymatic activity of squalene oxidase ERG1 denoted in blue can be inhibited by terbinafine. Genes: HMGR, HMG-CoA reductase; ERG20, farnesyl pyrophosphate synthase; ERG9, squalene synthase; MFE1, multifunctional enzyme; POT1, peroxisomal thiolase; DGA1, diacylglycerol acyltransferase; CAT2, peroxisomal carnitine acetyltransferases; ERG1, squalene epoxidase. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2. Materials and methods
2.1. Strains, media, and reagents
Escherichia coli DH5α served as the host for plasmid construction and amplification. Bacterial cells were cultured in Luria-Bertani (LB) medium, composed of 0.5% w/v yeast extract, 1% w/v tryptone, and 1% w/v NaCl, and maintained at 37 °C, supplemented with 100 μg/mL of ampicillin when required. The parental strain for this study was Y. lipolytica Po1gΔKU70, created by disrupting the KU70 gene in the original Po1g strain (Yu et al., 2016). Cultivation of Y. lipolytica strains was carried out at 30 °C in either Yeast Peptone Dextrose (YPD) medium, consisting of 1% w/v yeast extract, 2% w/v peptone, and 2% w/v glucose, Yeast peptone waste cooking oil medium (YPO), containing 1% w/v yeast extract, 2% w/v peptone, and 1.18% w/v WCO, or Yeast peptone glycerol (YPG) medium, consisting of 1% w/v yeast extract, 2% w/v peptone, and 2% w/v glycerol. WCO was sourced from a domestic kitchen in the university campus and filter sterilized with a 0.22 μm filter before blending into YPO medium. When using YPO as a fermentation medium, 0.2% Tween-80 was supplemented. The amount of WCO was adjusted to provide an equivalent number of carbon atoms as in the YPD medium. The WCO's fatty acid composition was analyzed by Qingdao Sci-tech Innovation Quality Testing Co., Ltd. (Table S1).
Yeast transformants were screened in SD-Leu medium, a synthetic complete drop-out medium containing 2% D-glucose and lacking leucine. Polymerase chain reaction (PCR) amplification was carried out using 2 × Phanta® Flash Master Mix from Vazyme Biotech, Nanjing, China. PCR fragments were purified using the FastPure® Gel DNA Extraction Kit from the same supplier. Plasmid extraction was performed using the FastPure Plasmid Mini Kit, also from Vazyme Biotech. The linear plasmid containing the expression cassette was obtained by restriction endonuclease digestion and subsequently integrated into the yeast genome.
2.2. Plasmid construction
All plasmids used in this study are listed in Table 1. To facilitate the construction of the gene expression cassette, two types of plasmids were initially prepared: the blank pYLEX1 plasmid and the pYLEX1H plasmid (Lu et al., 2023). The genes involved in this study derived from the Po1gΔKU70 genome were amplified by PCR. The PCR primers used were synthesized by Genewiz (Jiangsu, China) and are detailed in Table S2. Subsequently, the desired gene fragments were purified using the FastPure® Gel DNA Extraction Kit. The assembly of recombinant plasmids was executed using the ClonExpress® II One Step Cloning Kit from Vazyme Biotech, following the principles of homologous recombination. Plasmid extraction was conducted using the FastPure Plasmid Mini Kit, also from Vazyme Biotech, while linearized plasmids were obtained through the enzymatic digestion process employing restriction endonucleases from New England Biolabs.
Table 1.
Plasmids used in this study.
| Plasmids | Properties | Source |
|---|---|---|
| DH5α | supE44ΔlacU169φ80lacZΔM15 hsdR17 recAl endAl gyrA96 thi1 relA | laboratory |
| pYLEX1 | Y. lipolytica-integrative plasmid, Php4d-TXPR2, leu2, Apr | laboratory |
| pYLEX1H | Y. lipolytica-integrative plasmid, Php4d-TXPR2, hyg, Apr | laboratory |
| pYLE9 | Php4d-erg9-TXPR2, leu2, Apr | this study |
| pYLE20 | Php4d-erg20-TXPR2, leu2, Apr | this study |
| pYLH | Php4d-hmgr-TXPR2, leu2, Apr | this study |
| pYL2E | Php4d -erg9-TXPR2, Php4d -erg20-TXPR2, leu2, Apr | this study |
| pYL9H | Php4d-erg9-TXPR2, Php4d -hmgr-TXPR2, leu2, Apr | this study |
| pYL20H | Php4d -erg20- TXPR2, Php4d -hmgr- TXPR2, leu2, Apr | this study |
| pYL2EH | Php4d -erg9- TXPR2, Php4d -erg20- TXPR2, Php4d -hmgr- TXPR2, leu2, Apr | this study |
| pYLM | Php4d -mfe1- TXPR2, leu2, Apr | this study |
| pYLP | Php4d -pot1- TXPR2, leu2, Apr | this study |
| pYL-AXP-M | Php4d -mfe1- TXPR2, hyg, Apr | this study |
| pYL-AXP-P | Php4d -pot1- TXPR2, hyg, Apr | this study |
| pYL-AXP-MP | Php4d -mfe1- TXPR2, Php4d -pot1- TXPR2, hyg, Apr | this study |
| pYLC | Php4d -cat2- TXPR2, leu2, Apr | this study |
| pYLD | Php4d -dga1- TXPR2, leu2, Apr | this study |
| pYL2EHC | Php4d -erg9- TXPR2, Php4d -erg20- TXPR2, Php4d -hmgr- TXPR2, Php4d -cat2- TXPR2, leu2, Apr | this study |
| pYL2EHCD | Php4d -erg9- TXPR2, Php4d -erg20- TXPR2, Php4d -hmgr- TXPR2, Php4d -cat2- TXPR2, Php4d -dga1- TXPR2, leu2, Apr | this study |
2.3. Yeast construction
All the strains employed in this investigation are detailed in Table 2. The recombinant plasmids, utilizing the empty plasmid pYLEX1 as the carrier, were rendered linear through Spe I digestion and then integrated into the pBR322 site within the Y. lipolytica Po1gΔKU70 genome, employing the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (Yu et al., 2016). In contrast, the recombinant plasmids, employing the empty plasmid pYLEX1H as the vector, were linearized with Not I and subsequently integrated into the AXP site of the yeast strain's genome. Following selection on SD-Leu medium, the engineered Y. lipolytica strains were successfully created. Genomic DNA PCR and genome sequencing were performed to verify the integration of the target genes into the genome.
Table 2.
Strains used in this study.
| Y. lipolytica strains | Genotype | Source |
|---|---|---|
| Po1g0 (Po1g ΔKU70) | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2 | laboratory |
| Po1g1 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg9 | this study |
| Po1g2 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg20 | this study |
| Po1g3 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, hmgr | this study |
| Po1g4 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg9, erg20 | this study |
| Po1g5 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg9, hmgr | this study |
| Po1g6 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg20, hmgr | this study |
| Po1g7 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg9, erg20, hmgr | this study |
| Po1g8 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, mfe1 | this study |
| Po1g9 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, pot1 | this study |
| Po1g10 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, mfe1, pot1 | this study |
| Po1g11 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg9, erg20, hmgr, mfe1 | this study |
| Po1g12 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg9, erg20, hmgr, pot1 | this study |
| Po1g13 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg9, erg20, hmgr, mfe1, pot1 | this study |
| Po1g14 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, cat2 | this study |
| Po1g15 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, dga1 | this study |
| Po1g16 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg9, erg20, hmgr, cat2 | this study |
| Po1g17 | MATA, xpr2-332, leu2-270, ku70-, ura3-302::URA3, Axp-2, erg9, erg20, hmgr, cat2, dga1 | this study |
2.4. Shake flask cultivation of engineered strains
For the cultivation of an engineered yeast strain, a 250 mL shake flask containing 50 mL of YPO medium was employed. A loopful of yeast culture was inoculated onto three portions of SD-Leu agar plates and allowed to incubate for 12–16 h. The following day, a single colony of Y. lipolytica was selected from one of the SD-Leu agar plates and transferred into a 5 mL tube filled with YPD medium. This tube was then placed in an incubator at 30 °C with continuous shaking at 225 rpm for 24 h to establish a primary seed culture. Afterward, 1% of the primary seed culture was transferred from the tube to a 25 mL YPD fermentation medium. The resulting mixture was incubated at 30 °C with shaking at 220 rpm for 16 h to generate a secondary seed culture. Subsequently, an initial optical density of 0.1 at 600 nm (OD600 nm) was used to inoculate the 50 mL YPO medium in the shake flask. The flask fermentation was carried out at 30 °C with shaking at 220 rpm and continued for 120 h.
2.5. Extraction and quantification of squalene
After 120 h of shake flask fermentation, a 15 mL portion of the culture broth was transferred into a 50 mL centrifuge tube. The tube was subsequently subjected to centrifugation at 5000 rpm for 6 min, leading to the removal of the resulting supernatant. The pellet was then reconstituted by suspending it in 5 mL of sterile water, and this process was repeated twice. Following this, an equivalent volume of acid-washed glass beads, sized between 0.4 and 0.6 mm, and 2 mL of chromatographic-grade n-hexane were introduced to the pellet. The mixture was vigorously agitated with a vortex mixer for 20–30 min and then centrifuged at 7500 rpm for 6 min. The supernatant obtained was collected and transferred to a 2 mL centrifuge tube. Subsequently, an appropriate amount of anhydrous sodium sulfate was added, and the tube was gently mix by inversion before being centrifuged at 12000 rpm for 1 min. The resulting supernatant was filtered with a 0.22 μm hydrophopic filter and aliquoted into a 2 mL brown sample vial for downstream analysis.
For the detection of squalene, an Agilent Technologies gas chromatography-mass spectrometry (GC-MS) system was utilized, comprising a 5970B mass spectrometer and a 7890B gas chromatograph. The capillary column employed in this process was an HP-5 column (30 m × 0.25 mm, with a 0.25 μm film thickness). The ion source temperature was set to 280 °C, the interface temperature was maintained at 280 °C, and the quadrupole temperature was held at 200 °C. Helium gas served as the carrier gas with a flow rate of 1 mL/min. The mass spectrometer operated in full-scan mode, with a solvent delay of 3 min. The initial temperature was established at 200 °C and held for 1 min, after which the temperature was incrementally increased at a rate of 20 °C/min until reaching 310 °C, where it was then maintained for 5 min. A 1 μL sample injection was used, and the split ratio was set at 10:1.
3. Results and discussion
3.1. WCO was the preferred carbon source for squalene biosynthesis in Y. lipolytica
Y. lipolytica, despite its marine heritage, is a lipophilic yeast with robust oxidative metabolic capabilities (Liu et al., 2021). It efficiently metabolizes lipids via β-oxidation, yielding large pools of acetyl-CoA as precursors for downstream assimilation into long-chain terpenoids, such as squalene. At elevated levels, squalene sequesters within lipid droplets, and its concentration is positively linked to the overall cellular lipid content, partly due to its cytotoxic effects (Valachovic et al., 2016). On this premise, we theorized that lipid-rich WCO (in YPO) could potentially outperform commonly used carbon sources like glucose (in YPD) and glycerol (in YPG) for squalene production in native yeast. Our hypothesis was confirmed using a parental strain of Y. lipolytica (Po1gΔKU70, referred to as Po1g0), where we achieved a squalene yield of 6.6 mg/L after 120 h of fermentation with WCO, while glucose and glycerol yielded only 1.9 mg/L and 2.8 mg/L of squalene, respectively (Fig. 2).
Fig. 2.
Squalene titers and OD600 data were measured in wild-type Y. lipolytica strains after 120 h of shake flask cultivation using glucose, WCO, and glycerol as carbon sources. The carbon atom contributions were standardized at the same level for all three media types. GLU stands for glucose, WCO for waste cooking oil, and GLY for glycerol. Error bar represents the standard deviation of biological triplicates.
3.2. Co-expression of key genes in the mevalonate pathway drove metabolic flux towards squalene biosynthesis
With WCO affirmed as the preferred carbon source for squalene production in Y. lipolytica, we next sought to improve squalene titers by directing metabolic flux towards the squalene node. Besides HMGR serving as a primary bottleneck in the mevalonate pathway, ERG20 is responsible for encoding farnesyl diphosphate synthase, a pivotal enzyme involved in the synthesis of geranyl diphosphate (GPP) and farnesyl diphosphate (FPP). FPP, in its role as a precursor, contributes to squalene synthesis. Studies in Saccharomyces cerevisiae have confirmed that the co-expression of ERG20 and the squalene synthase gene ERG9 provides benefits for improving squalene production (Dai et al., 2013; Guo et al., 2018). In our previous attempt to produce α/β-amyrin, a terpenoid compound in which squalene plays an intermediary role, we found that overexpressing key genes in the mevalonate pathway, such as HMGR (HMG1), ERG9, and ERG20, resulted in elevated squalene levels in Y. lipolytica (Kong et al., 2022). Building on this knowledge, we proceeded to overexpress the target genes either individually or in various combinations using the synthetic pHP4D promoter. This effort resulted in the development of seven strains, identified as Po1g1 to Po1g7, as detailed in Table 3. The pHP4D promoter primarily activates gene expression during the early stages of the stationary phase, facilitating efficient resource allocation to support both cell growth and bioproduction.
Table 3.
Squalene titers and productivities of Y. lipolytica following 120 h of shake flask fermentation using WCO as the primary carbon source. Values represent mean of biological triplicates ± standard deviation.
| Strains (Genotype) | Titers (mg/L) | OD600 | Productivity (mg/L/h) |
|---|---|---|---|
| Po1g0 (Po1gΔKU70; wild-type) | 6.6 ± 1.33 | 26.8 ± 3.89 | 0.06 ± 0.01 |
| Po1g1 (↑ERG9) | 62.3 ± 3.59 | 29.5 ± 2.83 | 0.52 ± 0.03 |
| Po1g2 (↑ERG20) | 58.2 ± 4.81 | 28.6 ± 0.14 | 0.49 ± 0.04 |
| Po1g3 (↑HMGR) | 498.7 ± 12.87 | 29.4 ± 2.83 | 4.20 ± 0.11 |
| Po1g4 (↑ERG9, ↑ERG20) | 89.2 ± 4.53 | 36.2 ± 4.17 | 0.74 ± 0.04 |
| Po1g5 (↑ERG9, ↑HMGR) | 680.3 ± 24.32 | 37.6 ± 2.97 | 5.45 ± 0.20 |
| Po1g6 (↑ERG20, ↑HMGR) | 564.9 ± 19.23 | 37.5 ± 0.28 | 4.70 ± 0.16 |
| Po1g7 (↑ERG9, ↑ERG20, ↑HMGR) | 779.9 ± 21.92 | 37.6 ± 1.27 | 6.49 ± 0.18 |
| Po1g8 (↑MFE1) | 9.9 ± 0.57 | 30.7 ± 3.11 | 0.08 ± 0.01 |
| Po1g9 (↑POT1) | 10.5 ± 0.42 | 29.8 ± 3.11 | 0.08 ± 0.01 |
| Po1g10 (↑MFE1, ↑POT1) | 15.9 ± 2.26 | 30.0 ± 1.56 | 0.15 ± 0.02 |
| Po1g11 (↑ERG9, ↑ERG20, ↑HMGR, ↑MFE1) | 595.4 ± 1.84 | 29.8 ± 2.12 | 4.96 ± 0.02 |
| Po1g12 (↑ERG9, ↑ERG20, ↑HMGR, ↑POT1) | 664.5 ± 0.57 | 29.4 ± 1.41 | 5.53 ± 0.01 |
| Po1g13 (↑ERG9, ↑ERG20, ↑HMGR, ↑MFE1, ↑POT1) | 753.9 ± 19.09 | 33.5 ± 0.28 | 6.28 ± 0.16 |
| Po1g14 (↑CAT2) | 63.1 ± 1.98 | 31.7 ± 1.70 | 0.53 ± 0.02 |
| Po1g15 (↑DGA1) | 155.0 ± 3.96 | 34.9 ± 1.70 | 1.29 ± 0.03 |
| Po1g16 (↑ERG9, ↑ERG20, ↑HMGR, ↑CAT2) | 1199.5 ± 8.88 | 36.2 ± 1.70 | 9.99 ± 0.07 |
| Po1g17 (↑ERG9, ↑ERG20, ↑HMGR, ↑CAT2, ↑DGA1) | 1381.4 ± 34.66 | 39.7 ± 0.85 | 11.50 ± 0.29 |
As shown in Tables 3 and in the context of individual gene overexpression within the mevalonate pathway, the up-regulation of HMGR (strain Po1g3) had the most notable effect, resulting in a squalene titer of 498.7 mg/L using WCO. This marked an impressive 75-fold increase when compared to the wild-type strain. Up-regulating ERG9 (strain Po1g1) and ERG20 (strain Po1g2) also yielded higher squalene titers, although their impact was not as substantial, yielding an increase of no more than 9-fold compared to the parental strain. Similarly, in strain Po1g4, the overexpression of both ERG9 and ERG20 led to only a modest 13-fold increase in squalene titer compared to the parental strain, up to 89.2 mg/L. Among the seven tested strains, strain Po1g7, featuring the overexpression of all three target genes, exhibited the highest squalene titer at 779.9 mg/L, corresponding to a productivity of 6.49 mg/L per hour.
We next sought to increase the cytoplasmic acetyl-CoA pool, an important precursor for both mevalonate and downstream terpenoid products. WCO-derived free fatty acids are absorbed by Y. lipolytica and converted into fatty acyl-CoA. These acyl-CoA molecules then enter the peroxisome, undergoing a four-reaction cycle within the β-oxidation pathway, encompassing oxidation, hydration, dehydrogenation, and thiolysis, culminating in the formation of acetyl-CoA (Braga and Belo, 2016). The hydration and dehydrogenation reactions are orchestrated by the multifunctional enzyme MFE1 (Black et al., 2000; Dulermo et al., 2013). Thiolysis, on the other hand, is mediated by peroxisomal thiolase, POT1 (Wang et al., 2020).
In a prior study conducted with Y. lipolytica cultivated on WCO, we showcased that overexpressing either MFE1 or POT1 redirected metabolic flux towards the Krebs cycle, highlighting an increased availability of peroxisomal acetyl-CoA (Rong et al., 2022). To verify if either of these enzymes would play a major role in the squalene biosynthesis from WCO, we overexpressed MFE1, POT1 and both genes in combination in wild-type Y. lipolytica and strain Po1g7. This resulted in the development of six strains, identified as Po1g8 to Po1g13. Surprisingly, when MFE1 and POT1 were expressed individually and in combination in Y. lipolytica, there were no notable alterations in squalene titers. The squalene levels ranged from a 1.5-fold increase (in strains Po1g8 and Po1g9) to a 2.4-fold increase (in strain Po1g10) compared to the wild-type strain. This result sharply contrasts with the outcomes observed for the other examined gene targets in this study (Table 3). Similarly, overexpressing these genes in Po1g7 did not yield improved squalene titers; in fact, squalene titers decreased in strainPo1g11, where MFE1 was up-regulated, and in Po1g12 and Po1g13, where POT1 was up-regulated, by 14.8% and 23.6%, respectively. Considering that β-oxidation occurs within peroxisomes and acetyl-CoA must traverse across cellular membranes to be used in the cytoplasm, it's plausible that peroxisomal acetyl-CoA shuttling might be the metabolic constraint impacting squalene production.
3.3. Overexpression of Peroxisomal CAT2 and DGA1 significantly boosted squalene titer in Y. lipolytica
Carnitine acetyltransferase in Candida yeast shuttles acetyl groups between different cellular compartments, converting acetyl carnitine to acetyl-CoA and vice versa (Strijbis et al., 2008, 2010). Xu et al. previously reported the expression of the peroxisomal CAT2 gene in Y. lipolytica to shuttle peroxisomal acetyl-CoA (Xu et al., 2016). Hence, to redirect acetyl-CoA from peroxisomes to the cytoplasm for squalene synthesis, we overexpressed peroxisomal CAT2 in wild-type and Po1g7 to create strains Po1g14 and Po1g16, respectively. Impressively, the squalene titer rose to 63.1 mg/L in strain Po1g14 and a striking 1199.5 mg/L in Po1g16 (Table 3), affirming peroxisomal CAT2's role in governing the acetyl-CoA translocation across the peroxisome. Squalene, being a hydrophobic compound, exhibits a close relationship with the lipid content within yeast cells. In a study conducted elsewhere, it was shown that hydrophobic compounds may preferentially accumulate within lipid droplets (LDs), with triacylglycerol (TAG) constituting the major component of these lipids (Ma et al., 2019). DGA1 is the predominant gene, in addition to DGA2 and LRO1, which governs TAG synthesis in Yarrowia (Beopoulos et al., 2012). We postulated that enhancing DGA1 expression could lead to a simultaneous increase in intracellular squalene levels in engineered Y. lipolytica. As anticipated, squalene levels were elevated to 155 mg/L in strain Po1g15 with DGA1 overexpression alone, and up to 1381.4 mg/L in strain Po1g17 when ERG9, ERG20, HMGR, peroxisomal CAT2, and DGA1 were co-overexpressed (Table 3). The cell growth curve of this strain is provided in the supplementary file (Fig. S1). This resulted in a squalene productivity of 11.5 mg/L per hour, a remarkable 191-fold increase compared to the wild-type.
3.4. Supplementation of acetyl-CoA precursor and terbinafine boosted squalene titers
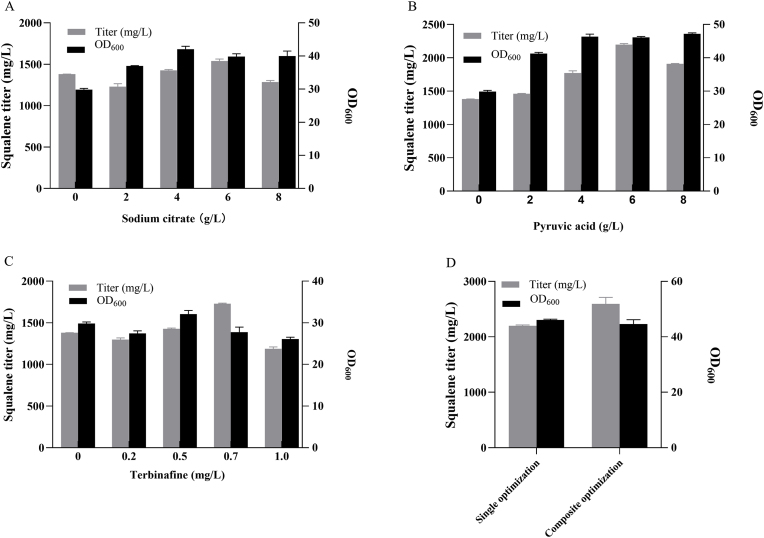
Addition of auxiliary carbon sources linked to pyruvate (pyruvic acid) metabolism and the citric acid cycle could boost the abundance of cytoplasmic acetyl-CoA, potentially enhancing the production of terpenoid products (Huang et al., 2018; Li et al., 2020). Briefly, pyruvate is transformed into acetyl-CoA by the mitochondrial pyruvate dehydrogenase complex. This acetyl-CoA is then converted into citrate and exported to the cytoplasm. There, citrate is broken down by ATP-citrate lyase into cytoplasmic acetyl-CoA and oxaloacetate. To test this hypothesis, we introduced varying quantities of sodium citrate and pyruvic acid to the YPO media at the onset of fermentation. As depicted in Fig. 3A and B, among the two options, the exogenous addition of pyruvic acid up to 6 g/L proved to be the more effective choice for enhancing squalene levels and Y. lipolytica growth in the YPO medium. The squalene titer saw a substantial increase of nearly 58%, reaching 2187.2 mg/L when 6 g/L was introduced. In contrast, the squalene titer only experienced a 10% increase when 6 g/L of sodium citrate was applied. Squalene is enzymatically converted into 2,3-epoxysqualene by squalene epoxidase. Terbinafine, at sub-inhibitory concentrations, can suppress the activity of this enzyme and enable the intracellular accumulation of squalene (Nowosielski et al., 2011; Petranyi et al., 1984). To determine the optimal terbinafine concentration, strain Po1g17 was grown on YPO medium with terbinafine concentrations varying from zero to 1 mg/L. Our findings indicate that the addition of terbinafine up to 0.7 mg/L increased intracellular squalene levels, reaching a maximum of 1724.4 mg/L. Beyond this point, cell growth was adversely affected (Fig. 3C). By combining the supplementation of both pyruvic acid and terbinafine at 6 g/L and 0.7 mg/L, the engineered strain Po1g17 attained an improved squalene titer of 2594.1 mg/L on WCO (Fig. 3D), which represented a remarkable 392-fold increase in titer compared to the wild-type.
Fig. 3.
Effects of auxiliary carbon supplementation and terbinafine on squalene titers and cell growth in Yarrowia strain Po1g17 after 120 h of shake flask fermentation on WCO. Varied concentrations of (A) sodium citrate, (B) pyruvic acid and (C) terbinafine. (D) Squalene titer and cell growth in strain Po1g17 with single supplementation of pyruvic acid and composite supplementation of both pyruvic acid and terbinafine. Error bars indicate the standard deviation from biological triplicates.
4. Conclusion
In this study, we harnessed Y. lipolytica as our host organism and utilized WCO as the primary carbon source for squalene production in shake flask experiments. Firstly, by overexpressing ERG9, ERG20, and HMGR, we enhanced the metabolic flux within the mevalonate pathway, significantly boosting the squalene production capacity of our engineered strain. Next, we focused on the transmembrane transport of acetyl-CoA and increased squalene storage capacity by overexpressing peroxisomal CAT2 and DGA1, which could synergistically enhance squalene production. We then conducted experiments involving the introduction of varying concentrations of pyruvic acid, citrate salts, and terbinafine into the fermentation medium. Notably, our engineered strain Po1g17 achieved a high squalene titer of 2594.1 mg/L under shake flask fermentation conditions. Comparing between different microbial hosts, metabolic engineering applications with Y. lipolytica had shown higher squalene titers than the conventional cell factories such as S. cerevisiae and E. coli (Table S3). While recent studies have demonstrated the production of squalene at grams per liter levels in engineered Yarrowia cell factories using glucose, our research presents a distinctive edge by harnessing WCO as the primary carbon source. By concentrating on repurposing WCO to generate squalene, our study not only reduces the cost of the bioprocess but also aligns seamlessly with the principles of a sustainable circular economy. This innovative approach holds promise for expanding squalene production in a more environmentally conscious and economically efficient manner.
Declarations
This manuscript does not involve any studies with human participants or animals. The authors declare no competing interests.
Consent for publication
All authors give consent to publish the research in Metabolic Engineering Communications.
Availability of data and material
All relevant data generated or analyzed during this study were included in this published article. Strains and plasmids developed in this study may be provided upon requests.
CRediT authorship contribution statement
Shuhui Wang: Writing – original draft, Visualization, Validation, Project administration, Methodology, Investigation, Formal analysis, Data curation. Xu Sun: Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Yuqing Han: Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Zhuo Li: Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Xiaocong Lu: Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Hongrui Shi: Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Cui-ying Zhang: Writing – review & editing, Supervision, Funding acquisition, Conceptualization. Adison Wong: Writing – review & editing, Writing – original draft, Supervision, Funding acquisition, Conceptualization. Aiqun Yu: Writing – original draft, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
AQY expresses gratitude for research funding from National Key R&D Program of China (No. 2023YFA0914500), the 2023 Provincial Scientific and Technological Innovation Strategy Special Project (Major Project + Task List) of Qingyuan City (No. 2023DZX012), the Natural Science Foundation of Tianjin, China (No. 22JCYBJC00170), Startup Fund for Haihe Young Scholars of Tianjin University of Science and Technology, the Thousand Young Talents Program of Tianjin, China. AW acknowledges funding from the Lee Foundation, Singapore (T-LEE-T201-A001).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2024.e00240.
Contributor Information
Cui-ying Zhang, Email: cyzhangcy@tust.edu.cn.
Adison Wong, Email: adison.wong@singaporetech.edu.sg.
Aiqun Yu, Email: yuaiqun@tust.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Barth G., Gaillardin C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 1997;19(4):219–237. doi: 10.1111/j.1574-6976.1997.tb00299.x. [DOI] [PubMed] [Google Scholar]
- Beopoulos A., Haddouche R., Kabran P., Dulermo T., Chardot T., Nicaud J.M. Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeastYarrowia lipolytica. New insights into the storage lipid metabolism of oleaginousyeasts. Appl. Microbiol. Biotechnol. 2012;93(4):1523–1537. doi: 10.1007/s00253-011-3506-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghoff S.A., Spieth L., Sun T., Hosang L., Schlaphoff L., Depp C., Düking T., Winchenbach J., Neuber J., Ewers D., Scholz P., van der Meer F., Cantuti-Castelvetri L., Sasmita A.O., Meschkat M., Ruhwedel T., Möbius W., Sankowski R., Prinz M., Saher G. Microglia facilitate repair of demyelinated lesions via post-squalene sterol synthesis. Nat. Neurosci. 2021;24(1):47–60. doi: 10.1038/s41593-020-00757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black P.N., Færgeman N.J., DiRusso C.C. Long-chain acyl-CoA–dependent regulation of gene expression in bacteria, yeast and mammals. J. Nutr. 2000;130(2):305S–309S. doi: 10.1093/jn/130.2.305S. [DOI] [PubMed] [Google Scholar]
- Braga A., Belo I. Biotechnological production of γ-decalactone, a peach like aroma, by Yarrowia lipolytica. World J. Microbiol. Biotechnol. 2016;32(10):169. doi: 10.1007/s11274-016-2116-2. [DOI] [PubMed] [Google Scholar]
- Brown A.J., Chua N.K., Yan N. The shape of human squalene epoxidase expands the arsenal against cancer. Nat. Commun. 2019;10(1):888. doi: 10.1038/s41467-019-08866-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M.C.Y., Keasling J.D. Production of isoprenoid pharmaceuticals by engineered microbes. Nat. Chem. Biol. 2006;2(12):674–681. doi: 10.1038/nchembio836. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A., Mitra M., Maiti M.K. Recent advances in lipid metabolic engineering of oleaginous yeasts. Biotechnol. Adv. 2021;53 doi: 10.1016/j.biotechadv.2021.107722. [DOI] [PubMed] [Google Scholar]
- Dai Z., Liu Y., Zhang X., Shi M., Wang B., Wang D., Huang L., Zhang X. Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab. Eng. 2013;20:146–156. doi: 10.1016/j.ymben.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Dulermo T., Tréton B., Beopoulos A., Kabran Gnankon A.P., Haddouche R., Nicaud J.M. Characterization of the two intracellular lipases of Y. lipolytica encoded by TGL3 and TGL4 genes: new insights into the role of intracellular lipases and lipid body organisation. BBA-Mol. Cell Biol. L. 2013;1831(9):1486–1495. doi: 10.1016/j.bbalip.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Garcia-Bermudez J., Baudrier L., Bayraktar E.C., Shen Y., La K., Guarecuco R., Yucel B., Fiore D., Tavora B., Freinkman E., Chan S.H., Lewis C., Min W., Inghirami G., Sabatini D.M., Birsoy K. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature. 2019;567(7746):118–122. doi: 10.1038/s41586-019-0945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X.J., Xiao W.H., Wang Y., Yao M.D., Zeng B.X., Liu H., Zhao G.R., Yuan Y.J. Metabolic engineering of Saccharomyces cerevisiae for 7-dehydrocholesterol overproduction. Biotechnol. Biofuels. 2018;11(1):192. doi: 10.1186/s13068-018-1194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.Y., Jian X.X., Lv Y.B., Nian K.Q., Gao Q., Chen J., Wei L.J., Hua Q. Enhanced squalene biosynthesis in Yarrowia lipolytica based on metabolically engineered acetyl-CoA metabolism. J. Biotechnol. 2018;281(9):106–114. doi: 10.1016/j.jbiotec.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Hull C.M., Loveridge E.J., Rolley N.J., Donnison I.S., Kelly S.L., Kelly D.E. Co-production of ethanol and squalene using a Saccharomyces cerevisiae ERG1 (squalene epoxidase) mutant and agro-industrial feedstock. Biotechnol. Biofuels. 2014;7(1):133. doi: 10.1186/s13068-014-0133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J., Miao L., Lu Z.H., Wang S.H., Zhao B.X., Zhang C.Y., Xiao D.G., Teo D., Leong S.S.J., Wong A., Yu A.Q. Enhance production of amyrin in Yarrowia lipolytica using a combinatorial protein and metabolic engineering approach. Microb. Cell Factories. 2022;21(1):186. doi: 10.1186/s12934-022-01915-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S., Kim S.R., Xu H., Zhang G.C., Lane S., Kim H., Jin Y.S. Enhanced isoprenoid production from xylose by engineered Saccharomyces cerevisiae. Biotechnol. Bioeng. 2017;114(11):2581–2591. doi: 10.1002/bit.26369. [DOI] [PubMed] [Google Scholar]
- Li M., Hou F., Wu T., Jiang X., Li F., Liu H., Xian M., Zhang H. Recent advances of metabolic engineering strategies in natural isoprenoid production using cell factories. Nat. Prod. Rep. 2020;37(1):80–99. doi: 10.1039/c9np00016j. [DOI] [PubMed] [Google Scholar]
- Liu H., Wang F., Deng L., Xu P. Genetic and bioprocess engineering to improve squalene production in Yarrowia lipolytica. Bioresour. Technol. 2020;317 doi: 10.1016/j.biortech.2020.123991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang Z., Cui Z., Qi Q., Hou J. α-Farnesene production from lipid by engineered Yarrowia lipolytica. Bioresour. Bioprocess. 2021;8(1):78. doi: 10.1186/s40643-021-00431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z.H., Wang Y.P., Li Z., Zhang Y.H., He S.C., Zhang Z.Y., Leong S.S.J., Wong A., Zhang C.Y., Yu A.Q. Combining metabolic engineering and lipid droplet storage engineering for improved α-bisabolene production in Yarrowia lipolytica. J. Agric. Food Chem. 2023;71(30):11534–11543. doi: 10.1021/acs.jafc.3c02472. [DOI] [PubMed] [Google Scholar]
- Ma T., Shi B., Ye Z., Li X., Liu M., Chen Y., Xia J., Nielsen J., Deng Z., Liu T. Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metab. Eng. 2019;52:134–142. doi: 10.1016/j.ymben.2018.11.009. [DOI] [PubMed] [Google Scholar]
- Ma Y., Shang Y., Stephanopoulos G. Engineering peroxisomal biosynthetic pathways for maximization of triterpene production in Yarrowia lipolytica. Proc. Natl. Acad. Sci. U. S. A. 2024;121(5) doi: 10.1073/pnas.231479812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madzak C. Yarrowia lipolytica strains and their biotechnological applications: how natural biodiversity and metabolic engineering could contribute to cell factories improvement. J. Fungi. 2021;7(7):548. doi: 10.3390/jof7070548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y., Liu M., Ru Z., Zeng W., Liu S., Zhou J. Efficient synthesis of squalene by cytoplasmic-peroxisomal engineering and regulating lipid metabolism in Yarrowia lipolytica. Bioresour. Technol. 2024;395 doi: 10.1016/j.biortech.2024.130379. [DOI] [PubMed] [Google Scholar]
- Nowosielski M., Hoffmann M., Wyrwicz L.S., Stepniak P., Plewczynski D.M., Lazniewski M., Ginalski K., Rychlewski L. Detailed mechanism of squalene epoxidase inhibition by terbinafine. J. Chem. Inf. Model. 2011;51(2):455–462. doi: 10.1021/ci100403b. [DOI] [PubMed] [Google Scholar]
- Paramasivan K., Mutturi S. Recent advances in the microbial production of squalene. World J. Microbiol. Biotechnol. 2022;38(5):91. doi: 10.1007/s11274-022-03273-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranyi G., Ryder N.S., Stütz A. Allylamine derivatives: new class of synthetic antifungal agents inhibiting fungal squalene epoxidase. Science. 1984;224(4654):1239–1241. doi: 10.1126/science.6547247. [DOI] [PubMed] [Google Scholar]
- Rong L.X., Miao L., Wang S.H., Wang Y.P., Liu S.Q., Lu Z.H., Zhao B.X., Zhang C.Y., Xiao D.G., Pushpanathan K., Wong A., Yu A.Q. Engineering Yarrowia lipolytica to produce itaconic acid from waste cooking oil. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.888869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbis K., van Roermund C.W., Visser W.F., Mol E.C., van den Burg J., MacCallum D.M., Odds F.C., Paramonova E., Krom B.P., Distel B. Carnitine-dependent transport of acetyl coenzyme A in Candida albicans is essential for growth on nonfermentable carbon sources and contributesto biofilm formation. Eukaryot. Cell. 2008;7(4):610–618. doi: 10.1128/EC.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbis K., van Roermund C.W., van den Burg J., van den Berg M., Hardy G.P.M., Wanders R.J., Distel B. Contributions of carnitine acetyltransferases to intracellular acetyl unit transport in Candida albicans. J. Biol. Chem. 2010;285(32):24335–24346. doi: 10.1074/jbc.M109.094250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.Y., Wang D.P., Tian Y., Fan X., Wang C., Lu X.Y., Li P.W., Ji X.J., Liu H.H. Metabolic engineering of Yarrowia lipolytica for improving squalene production. Bioresour. Technol. 2021;323 doi: 10.1016/j.biortech.2020.124652. [DOI] [PubMed] [Google Scholar]
- Tateno M., Stone B.J., Srodulski S.J., Reedy S., Gawriluk T.R., Chambers T.M., Woodward J., Chappell J., Kempinski C.F. Synthetic biology-derived triterpenes as efficacious immunomodulating adjuvants. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-73868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valachovic M., Garaiova M., Holic R., Hapala I. Squalene is lipotoxic to yeast cells defective in lipid droplet biogenesis. Biochem. Biophys. Res. Commun. 2016;469(4):1123–1128. doi: 10.1016/j.bbrc.2015.12.050. [DOI] [PubMed] [Google Scholar]
- Wang J., Ledesma-Amaro R., Wei Y., Ji B., Ji X.J. Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica – a Review. Bioresour. Technol. 2020;313 doi: 10.1016/j.biortech.2020.123707. [DOI] [PubMed] [Google Scholar]
- Wei L.J., Kwak S., Liu J.J., Lane S., Hua Q., Kweon D.H., Jin Y.S. Improved squalene production through increasing lipid contents in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2018;115(7):1793–1800. doi: 10.1002/bit.26595. [DOI] [PubMed] [Google Scholar]
- Wei L.J., Cao X., Liu J.J., Kwak S., Jin Y.S., Wang W., Hua Q. Increased accumulation of squalene in engineered Yarrowia lipolytica through deletion of PEX10 and URE2. Appl. Environ. Microbiol. 2021;87(17) doi: 10.1128/AEM.00481-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Qiao K., Ahn W.S., Stephanopoulos G. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc. Natl. Acad. Sci. U. S. A. 2016;113(39):10848–10853. doi: 10.1073/pnas.1607295113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A.Q., Pratomo N., Ng T.K., Ling H., Cho H.S., Leong S.S.J., Chang M.W. Genetic engineering of an unconventional yeast for renewable biofuel and biochemical production. J. Vis. Exp. 2016;115 doi: 10.3791/54371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Li M., Lu S., Cheng Y., Guo X., He X., Wang Z., He X.P. Engineering of cis-element in Saccharomyces cerevisiae for efficient accumulation of value-added compound squalene via downregulation of the downstream metabolic flux. J. Agric. Food Chem. 2021;69(42):12474–12484. doi: 10.1021/acs.jafc.1c04978. [DOI] [PubMed] [Google Scholar]
- Zhu Z.T., Du M.M., Gao B., Tao X.Y., Zhao M., Ren Y.H., Wang F.Q., Wei D.Z. Metabolic compartmentalization in yeast mitochondria: burden and solution for squalene overproduction. Metab. Eng. 2021;68:232–245. doi: 10.1016/j.ymben.2021.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data generated or analyzed during this study were included in this published article. Strains and plasmids developed in this study may be provided upon requests.
Data will be made available on request.