Highlights
-
•
The present study has identified TMSB10 as a cancer-promoting protein and a serum tumor marker in CRC diagnosis.
-
•
We proposes here a potential regulating strategy of TMSB10 which works via regulating the phosphorylation of p38 on the cell cycle and apoptosis of human CRC cells.
-
•
The multiple roles of TMSB10 in cell cycle progression and apoptosis suggest that TMSB10 could be used as a valuable biomarker for as well as a target to improve the clinical post-surgery efficacy of radio- as well as chemotherapy of CRC chemotherapy.
Keywords: Colorectal cancer, TMSB10, Serum tumor marker, p38
Abstract
Thymosin beta 10 (TMSB10) overexpression is a general characteristic in human carcinogenesis. It is involved in the malignant process of generating multiple cancers. However, there are only a few reports about TMSB10 in colorectal cancer (CRC) and the mechanism of its carcinogenetic effect is still poorly understood. The present study intends to clarify the biological roles and carcinogenic mechanism of TMSB10 in CRC and to explore the possibility whether TMSB10 might be useful as a non-invasive serum tumor biomarker in detecting CRC. Immunohistochemical results showed that TMSB10 protein expression in CRC tissues was generally higher than that in adjacent tissues, and the TMSB10 contents in serum of CRC patients was significantly elevated compared to that of healthy controls. Knockdown-TMSB10 increased apoptosis and induced S-cell cycle arrest, and finally inhibited cell proliferation in vitro and in vivo. Transcriptome sequencing and western blotting analysis revealed that knockdown-TMSB10 increased phosphorylation of p38 and activated the p38 pathway that blocked cell cycle and promoted apoptosis. Taken together, our study indicated that TMSB10 could serve as a minimally invasive serum tumor marker in detecting CRC. At the same time it demonstrates an effective regulatory capacity of TMSB10 on cell proliferation of CRC, suggesting that TMSB10 and downstream effector molecules regulated by TMSB10 could further be applied as an appealing target in clinical post-surgery chemotherapy.
Introduction
Colorectal cancer (CRC) causes high morbidity and mortality. There were about 1.93 million new cases diagnosed with CRC worldwide in 2020, and the CRC incidences are rising among younger adults [1,2]. During the past several decades, the five-year survival rate of the early CRC patients has been greatly increased to about 80 % with the progress of screening and systematic treatment of tumors, while the cases of advanced CRC patients was reduced to 14 % [3]. The ideal and primary treatment for CRC patients is the tumor resection of primary and metastatic cancers through surgical treatment at an early stage. However, most CRC patients are diagnosed at an advanced stage, and the effectiveness of surgical treatment on advanced CRC patients is poor. In recent years, molecular targeted therapy is playing an increasingly important role in improving therapeutic effects and survival rates of advanced cancer patients. Although targeted therapies gained great progress, they still face difficulties due to tumor resistance. It is, therefore, critical to better understand carcinogenic mechanisms of CRC to explore potential tumor markers and therapeutic targets for the treatment of CRC [4].
β-thymosins are a subfamily of thymosins secreted by the thymus gland, and the β-thymosin subfamily includes thymosin beta 4 (TMSB4), thymosin beta 10 (TMSB10), and thymosin beta 15 (TMSB15) [[5], [6], [7]]. These actin sequestering proteins are involved in the control of the cytoskeletal microfilament system, and play key physiological roles in the development of the central nervous system and the regulation of inflammation. Studies also found that β-thymosins might be positively correlated with the occurrence and development of multiple cancers, and the up-regulation of β-thymosins is a general event in human carcinogenesis [[8], [9], [10]]. Wirsching et al. [11] reported that TMSB4 could regulate cell invasiveness and that it was negatively correlated with overall survival. The up-regulation of TMSB15A was observed in breast cancer and prostate cancer, and TMSB15A was involved in cell growth and migration through the induction of transforming growth factor and epidermal growth factor in prostate cancer cells [12]. Up-regulated TMSB10 also have been reported in breast cancer and non-small cell lung cancer, and TMSB10 was proven as a key regulator of tumorigenesis and metastasis [9,13]. However, relevant studies on TMSB10 in CRC are less reported, and the mechanism of TMSB10 in CRC is still unknown. The aim of the present study is to understand the role of TMSB10 on malignant cell proliferation, and to further clarify the molecular mechanisms behind CRC development, which are imperative for further development of anti-cancer therapy of CRC patients.
Materials and methods
Clinical samples
Ethical approval of the present study was provided by the Human Ethics Committee of Shanxi provincial people's Hospital (Approval number: 2019–91), and written informed consent from each patient and healthy people was obtained after a detailed explanation. This study was performed in accordance with the standards of the Declaration of Helsinki. Paraffin-embedded CRC tumor tissues (n = 173 cases) were retrieved from the department of colorectal anorectum and pathology in Shanxi provincial people's Hospital. All cases were chosen from CRC patients without treatment of chemoradiotherapy treatment before operation, and the tumor-node-metastasis (TNM) staging in CRC was guided by the 8th edition of the American Joint Committee on Cancer and the International Union for Cancer Control update the TNM cancer staging system. Serum samples (n = 105) of CRC patients were also collected, and 69 serum samples of healthy people were randomly considered as the control group. The clinicopathological characteristics of CRC patients and healthy participants used for the present study are provided as additional files (Table S1 and S2).
TMSB10 mRNA expression analysis in CRC
Based on the data from Genotype-Tissue Expression projects (GTEx) and Cancer Genome Atlas (TCGA), TMSB10 mRNA expression in patients with colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) were obtained through Gene Expression Profiling Interactive Analysis (GEPIA) [14].
Protein expression analysis by tissue microarray technology (TMA) and immunohistochemistry (IHC).
Fabrication of tissue microarray: The histopathological characteristics of all samples were confirmed by pathologists from the Department of Colorectal Anorectum in Shanxi provincial people's Hospital, and the tumor and normal colorectal tissues were identified by hematoxylin-eosin staining. The marked tissues in paraffin wax were removed with a 2.0 mm diameter drilling needle to the blank paraffin blocks (30 mm × 25 mm × 10 mm). The TMA paraffin blocks were arranged in 8 columns of 4 rows, and then cut into 2-mm-thick slices for immunoassay.
Protein detection: The constructed TMA was deparaffinized in xylene, dehydrated with gradient ethanol, washed with distilled water (twice, 8 min/time), treated with 3 % H2O2-methanol at room temperature for 20 min, washed with distilled water (twice, 5 min/time), incubated in 0.01 mol/L sodium citrate buffer. After washing with PBS (twice, 5 min/time), the slides were sealed with 5 % BSA at 37 ℃ for 20 min, then incubated with anti-TMSB10 antibody (ab14338, Abcam, UK) or anti-p38 (phosphoT180, ab178867, Abcam, UK) or Ki67 (ab15580, Abcam, UK) at 4 ℃ overnight and Max Vision Mouse/Rabbit (MXB biotechnology, KIT-5020) at 37 ℃ for 1 h, visualized with 1,3-dimethylbarbituric acid (DBA) for 10 min, quickly washed with distilled water, counterstained with hematoxylin for 2 min. The image and data were collected using the ScanScope slide scanning system (Aperio Technologies Inc., Vista, CA, USA).
Enzyme linked immunosorbent assay (ELISA)
TMSB10 contents in serum of CRC patients were detected using double antibody sandwich ELISA. Firstly, a polystyrene microtiter plate was coated with anti-TMSB10 (sc-514,309, Santa Cruz, CA, USA). After overnight incubation at 4 °C, the coated liquid was discarded, and the plate was washed with PBST. Different concentrations of TMSB10 standard ranging from 10 to 300.00 ng/mL or serum samples were added, and the plate was incubated for 2 h at 37 °C. After the plates were incubated for 1 h with goat antimouse IgG-HRP (Sigma, St. Louis, MO, USA), TMB was added to the plate for 30 min followed by 50 μL of 2 M H2SO4 addition to plots to stop the reaction. The absorbance was measured at 450 nm on a spectrophotometer.
Cell culture and knockdown of TMSB10
Human colorectal cell lines (HT29 and SW480) used in the present study were purchased from Procell Life Sciences (CL-0118 and CL-0223, Wuhan, China), and were authenticated using STR analysis. Cells were cultured in RMPI 1640 medium containing 10 % calf serum and 1 % streptomycin-penicillin in 5 % CO2 at 37 ℃. The culture medium was replaced every two days. Slow virus vector sequence 5′- GATCCGCTGAAGAAAACGGAGACGCTTCAAGAGAGCGTCTCCGTTTTCTTCAGTTTTTTG-3′ (named shTMSB10) and 5′- GATCCGTTCTCCGAACGTGTCACGTAATTCAAGAGATTACGTGACACGTTCGGAGA ATTTTTTC-3′ (named shcon) were applied for the knockdown of the endogenous expression of TMSB10. The effectivenes of knockdown were determined via Western blot.
Cell cycle, apoptosis, and cell proliferation assays
By flow cytometry, cell cycle and apoptosis analysis were performed using PI staining and annexin V-FITC/PI double staining according to the manufacturer's protocol, respectively. Cell proliferation was examined by 3- (4, 5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT) assay and soft-agar colony forming assay. For an MTT assay, cells were placed in 96 well plates. 20 µL MTT (5 mg/mL in DMEM) were added to each well for 4 h, then dimethyl sulphoxide (DMSO) was added for 10 min. The absorbance at 490 nm of each well was measured by Multimode Reader (Varioskan Flash, Thermo Electron Co. US). For the soft agar colony forming assay, each agarose disk added to cell culture medium and inoculated with 1000 cells. After 14 days, cells were fixed with 5 % trioxane and dyed with crystal violet, and the number of colony forming units was counted.
Animal experiments
A mouse xenografted assay was performed to analyze the effects of TMSB10 on tumorigenesis in vivo. Female BALB/c nude mice (Strain NO.D000521) were purchased from GemPharmatech (Nanjing, China), and all mice were accomodated in appropriate pathogen-free conditions. About 4 × 106 TMSB10-knockdown HT29 cells were subcutaneously injected into the right oxter of 4-week-old nude mice and HT29 cells were used as a control. Tumor size was measured every 7 days using caliper and calculated using the following formula: tumor volume (mm3) = 1/2 × length (mm) × [width (mm)]2. After 5 weeks, the mice were euthanized with an overdose of sodium pentobarbital (150 mg/kg) via an intraperitoneal injection, and tumors were removed and measured. Mice experiments were approved (approval no. 2021–101) by the Animal Care and Use Committee of Shanxi Medical University (Taiyuan, China), and were proceeded according to the Laboratory Animals guide of Shanxi Medical University.
Transcriptome survey and expression analysis
Total RNA samples were isolated according to the protocols of Trizol (Invitrogen, Carlsbad, CA, USA), and the sequencing library was performed using Illumina hiseq 2500 sequencing platform. After that the clean reads were de novo assembled using Trinity software, the differentially expressed genes (DEGs) were calculated through reads per KB per million reads (RPKM) method with a false discovery rate P < 0.05 and a multiple difference ≥ 2. Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes pathway database (KEGG) pathway enrichment analysis were carried out to further clarify the biological functions of DEGs using the KOBAS2.0 and Blast2GO programs, respectively.
Western blotting
About 3 × 106 cells lysed in 200 μL RIPA buffer containing 10 μL Tris/HCl (50 mM, pH 7.4), 20 μL NaCl (150 mM), 4 μL EDTA (5 mM), 20 μL Triton X-100 (0.5 %), 2 μL PMSF (1 mM), and 4 μL cooktail (50 X) at 4℃ for 1 h were centrifuged at 10, 000 g for 15 min at 4 ℃. Total proteins were loaded into 10 % SDS-PAGE and transferred onto PVDF membranes which were incubated with PBST buffer (pH 8.0) containing 5 % nonfat dry milk to stand overnight at room temperature. After washing with PBST, the separated proteins were incubated with the special antibodies including anti-p38 (14,064–1-AP, ProteinTech, Chicago, USA), anti-p38 (phosphoT180, ab178867, Abcam, Cambridge, UK), anti-p53 (10,442–1-AP, ProteinTech, Chicago, USA), anti-p53 (phospho S15, ab1431, Abcam, Cambridge, UK), anti-p21 (ab109520, ProteinTech, Chicago, USA), anti-Rb (17,218–1-AP, ProteinTech, Chicago, USA), anti-Rb (phospho-S807, 8180T, Cell Signaling Technology, Frankfurt, Germany), and anti-Caspase3 (ab13847, Abcam, Cambridge, UK) at 4 ℃ overnight. Finally, these proteins were incubated with IRDye 680 goat anti-rabbit or anti-mouse antibodies (925–68,071 or 68,080, Li-Cor Biosciences, NE, USA), and was detected with Odyssey Imaging System (Li-Cor, Lincoln, NE, USA). The intensity of the bands was quantified using ImageJ software (National Institutes of Health, Maryland, USA).
Detection of oxidative stress in CRC cell lines
Reactive oxygen species (ROS), an index of oxidative stress, was detected by a chemical fluorescence method using a reactive oxygen species assay kit (Nanjing Jiancheng Bioengineering Institute, China). Dichloro-dihydro-fluorescein diacetate (DCFH-DA, 10 µM) was added to cells in darkness at 37 ℃ for 15 min. Cells were harvested and washed twice with PBS buffer at 4 ℃, and were suspended in PBS buffer for fluorescence detection using a flow cytometer.
Statistical analysis
Data were presented with mean ± standard deviation (S.D.) from at least three independent experiments. All statistical analyses were performed using SPSS software (version 15.0, SPSS Inc., Chicago, IL, USA), and data graphs were generated using GraphPad Prism 5 Software (GraphPad Software Inc., La Jolla, CA, USA). Data homogeneity of variance and normality were tested using the Levene tests and Kolmogorov–Smirnov tests, respectively. Student's t-test and one-way ANOVA analysis were for significant differences between groups. The receiver operating characteristic (ROC) curve was used to measure the best dividing values between high expression and low expression of the TMSB10 protein. The correlation analysis between TMSB10 expression with clinical information and Phos-p38 were carried out using Chi square test and Pearson correlation coefficients, respectively. Survival curves were constructed using the Kapla-Meier method and differences in survival were evaluated using the log-rank test. A p value < 0.05 was regarded as statistically significant, and statistical significances were denoted by asterisks * between different groups.
Results
Up-regulated TMSB10 expression in CRC tissues
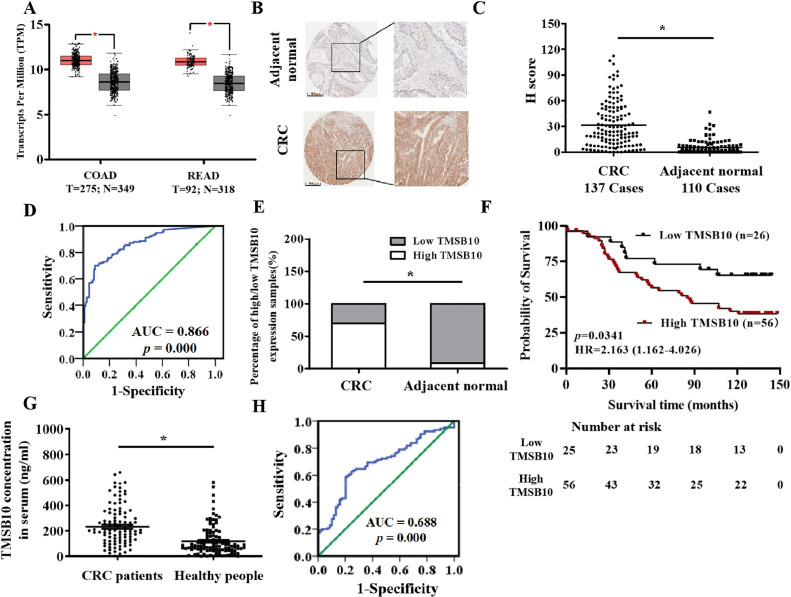
An elevation of TMSB10 mRNA in both COAD and READ tissues was observed based on the analyses by the GEPIA online database (Fig. 1A). TMSB10 protein expression from 173 human CRC tissue samples and matched adjacent non-tumor tissues was detected through TMA and immunohistochemical analysis, and 137 cases of colorectal cancer tissues and 110 cases of adjacent tissues were finally counted. The immunohistochemical outcomes showed that the TMSB10 protein in CRC tissues was significantly higher compared to these in normal tissues (Fig. 1B-C). According to the ROC curve (AUC = 0.866, p = 0.000, Fig. 1D), a H-value (13.9904) was identified as cut-off value, and all CRC tissue samples were divided into a low TMSB10 expression group (H-value ≤ 13.9904) and a high TMSB10 expression group (H-value > 13.9904). A rank sum test showed significant differences in the distribution between the high TMSB10 expression and low TMSB10 expression in CRC tissues and adjacent normal tissues (Fig. 1E). A Chi-square test discovered that TMSB10 expression was statistically associated with N-classification and T-classification (Table 1). Univariate and multivariate Cox regression analyses showed that TMSB10 expression, T classification, N classification, and AJCC stage was significantly associated with overall survival (Table 2). A significantly negative correlation between the TMSB10 and survival time was observed, and CRC patients with high TMSB10 exhibited a worse prognosis. A higher of TMSB10 level was observed in serum of CRC patients compared to that in controls (Fig. 1G). ROC analysis of CRC patients showed an area under the curve with 0.688 (95 % CI = 0.609 to 0.767, p < 0.05, Fig. 1H).
Fig. 1.
High expression of TMSB10 in CRC tissue. (A) TMSB10 mRNA expression analysis in human colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) clinical samples from GEPIA online database. (B) Representative images of TMSB10 protein expression in CRC tissue and normal colorectal tissue vis immunohistochemical analysis (Scale bar: 500 μm). (C) Histochemical score of TMSB10 protein expression in CRC and matched paracancerous tissue samples (*: p < 0.05). (D) Receiver operating characteristic (ROC) curve for TMSB10 protein expression in CRC tissue and normal colorectal tissue. (E) Proportion of high and low TMSB10 protein expression in CRC and normal tissue samples based on the cutoff value from ROC curves. (F) Kaplan–Meier survival analysis of overall survival of CRC patients. (G) Comparative analysis of TMSB10 concentration in the serum of CRC patients and healthy people. (H) ROC curve for TMSB10 content in serum of healthy individuals and CRC patients.
Table 1.
Relationship between TMSB10 expression and clinicopathological characteristics of CRC patients was tested by chi-square test.
| Parameters | Number of cases | TMSB10 expression |
chi-square test |
||
|---|---|---|---|---|---|
| Low | High | Χ2 values | p values | ||
| Overall | 137 | 41 | 96 | ||
| Age (years) | |||||
| < 60 | 56 | 18 | 38 | 0.222 | 0.638 |
| ≥ 60 | 81 | 23 | 58 | ||
| Gender | |||||
| Male | 82 | 25 | 57 | 0.031 | 0.861 |
| Female | 55 | 16 | 39 | ||
| Pathological grade | |||||
| G1 | 3 | 2 | 1 | 2.438 | 0.296 |
| G2 | 128 | 38 | 90 | ||
| G3 | 6 | 1 | 5 | ||
| Smoking history | |||||
| Yes | 47 | 15 | 32 | 0.135 | 0.713 |
| No | 90 | 26 | 64 | ||
| Drinking history | |||||
| Yes | 29 | 10 | 19 | 0.364 | 0.546 |
| No | 108 | 31 | 77 | ||
| T classification | |||||
| T1 | 3 | 2 | 1 | 4.255 | 0.235 |
| T2 | 30 | 8 | 22 | ||
| T3 | 23 | 4 | 19 | ||
| T4 | 81 | 27 | 54 | ||
| M classification | |||||
| M0 | 134 | 40 | 94 | 0.017 | 0.896 |
| M1 | 3 | 1 | 2 | ||
| N classification | |||||
| N0 | 77 | 18 | 59 | 6.141 | 0.046 |
| N1 | 40 | 18 | 22 | ||
| N2 | 20 | 5 | 15 | ||
| T classification | |||||
| I + II | 75 | 17 | 58 | 4.166 | 0.041 |
| III + IV | 62 | 24 | 38 | ||
| IHC status of Ki67 | |||||
| High | 43 | 5 | 31 | 9.733 | 0.002 |
| Low | 26 | 16 | 17 | ||
*p < 0.05.
Table 2.
Univariate and multivariate analysis for overall survival.
| Parameters | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95 % CI | p | HR | 95 % CI | P | |
| TMSB10 expression (Low vs High) | 1.668 | 1.015–2.742 | 0.043* | 1.407 | 1.016–2.427 | 0.035* |
| Age (< 60 vs ≥ 60) | 0.916 | 0.570–1.472 | 0.716 | 1.365 | 0.811–2.298 | 0.241 |
| Gender (Femal vs Male) | 1.067 | 0.660–1.726 | 0.791 | 1.1 | 0.593–2.039 | 0.763 |
| Smoking History (Yes vs No) | 0.848 | 0.514–1.399 | 0.519 | 0.972 | 0.420–2.248 | 0.946 |
| Drinking History (Yes vs No) | 0.914 | 0.516–1.620 | 0.758 | 0.996 | 0.410–2.419 | 0.994 |
| T classification (T1+2 vs T3+4) | 3.341 | 1.654–6.751 | 0.001* | 2.561 | 1.223–5.365 | 0.013* |
| N classification (N1+2 vs N0) | 0.209 | 0.126–0.348 | 0.000* | 0.116 | 0.022–0.608 | 0.011* |
| AJCC stage (III+IV vs I+II) | 0.183 | 0.109–0.308 | 0.000* | 0.025 | 0.004–0.136 | 0.000* |
p < 0.05.
Knockdown of TMSB10 inhibits cell proliferation by promoting the apoptosis and inducing G1/S arrest in human CRC
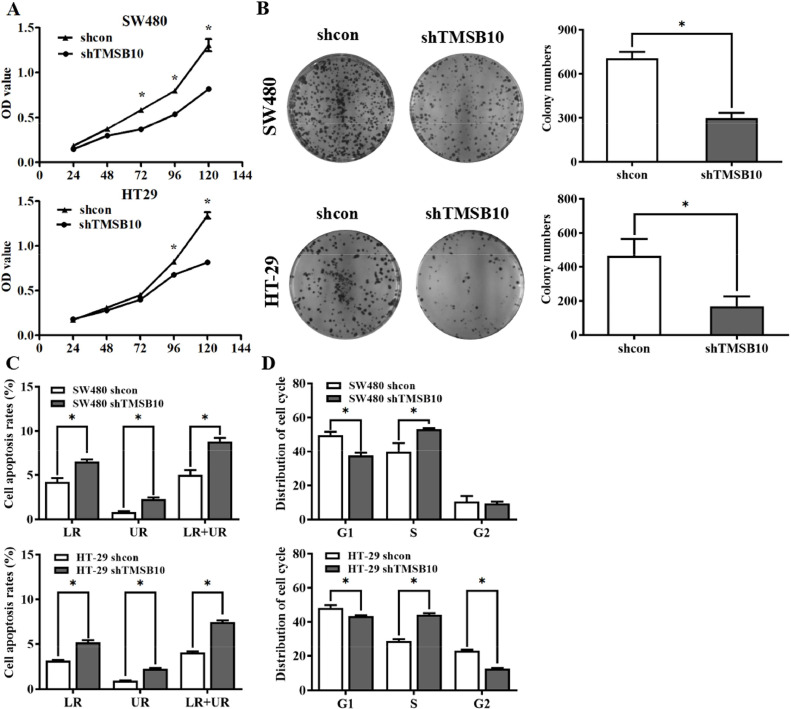
To explore the biological roles of TMSB10 in CRC, the TMSB10-knockdown HT-29 and SW480 cells were made by lentiviral vector transfection, and the efficiencies in TMSB10-knockdown cells were also confirmed by western blotting. MTT assays found significantly inhibited cell proliferation in both TMSB10-knockdown cells (Fig. 2A), and colony formation assay confirmed that the knockdown-TMSB10 reduced the colony-forming abilities of CRC cells (Fig. 2B). To clear the role of TMSB10 on apoptosis in CRC, comparative analysis was carried out between TMSB10-knockdown and control cells using an annexin V-FITC/PI double staining by flow-cytometry. As shown in Fig. 2C and Fig. S1, apoptosis in both TMSB10-knockdown cells was significantly up-regulated. The cell cycle of CRC cells was detected using propidium iodide (PI) staining by flow cytometry, and the S-phase in both TMSB10-knockdown HT29 and SW480 cell lines was significantly increased compared to those in the negative control cells (Fig. 2D, Fig. S2), but the G1 phase was significantly reduced, indicating that cell cycle progression was blocked in the G1/S phase. Next, the effect of TMSB10 on tumorigenesis in vivo was evaluated by a xenografted mouse model. The tumor volumes and weight in the TMSB10-knockdown group were significantly reduced compared to those in the control group (Fig. S4A-C). Immunohistochemical detection of Ki67 in TMSB10-knockdown tumor tissues found reduced numbers of Ki67-positive cells, and clinical analyses showed a positive correlation between TMSB10 and Ki67 (Table 1). The inhibited cell proliferation, induced apoptosis and blocked cell cycle in TMSB10-knockdown cells suggest that TMSB10 acts as a cancer-promoting gene to promote cell proliferation in CRC.
Fig. 2.
Knockdown of TMSB10 inhibits cell proliferation in colorectal cancer. (A) Cell proliferation assay was performed in TMSB10-knockdown cells and correspondence control cells, respectively. (B) Colony formation assay was performed in TMSB10-knockdown cells and correspondence control cells, respectively. (C) Analysis of apoptosis rate from TMSB10-knockdown cells and correspondence control cells. (D) Changes in each stage of cell cycle in TMSB10-knockdown cells were observed by flow cytometry. Data were representative of three independent experiments and were presented as means ± S.D., and a p value < 0.05 was regarded as statistically significant.
RNA sequencing and western blotting reveals that knockdown-TMSB10 alters p38 pathways in human CRC
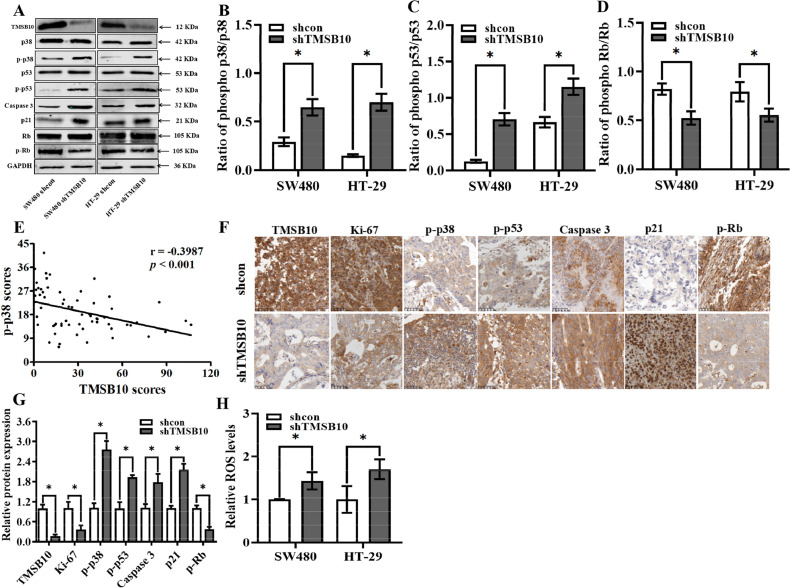
To further explore the regulatory mechanism of TMSB10 in CRC, the TMSB10-knockdown cells and negative control cells were sequenced using the transcriptome, and found significantly up-regulated mRNA expressions of p21 and Caspase 3. Western blotting further confirmed the proteins of p21 and Caspase 3 were highly expressed in TMSB10-knockdown cells (Fig. 3A). The higher p21 reduced the phosphorylation of Rb (Fig. 3D) resulting in cell cycle arrest in the G1/S phase, and the activated Caspase 3 in TMSB10-knockdown cells promoted apoptosis. The p38 and p53 plays a pivotal role in transcriptional activation of p21 and Caspase 3, and western blot analysis found up-regulated phosphorylation of p53 and p38 in TMSB10-knockdown cells (Fig. 3A and B). Immunohistochemical analysis also revealed a significant negative correlation between TMSB10 and phospho-p38 (p-p38) (Fig. 3E). Moreover, the p-p38, phospho-p53 (p-p53), p21, and phospho-Rb (p-Rb) expression in mouse tumor tissues was detected through immunohistochemical analysis. The results of the in vivo experiment were consistent with those of the in vitro study (Fig. 3F-G). The p38 pathways are closely related to intracellular reactive oxygen species (ROS), and results showed that the ROS levels were up-regulated in both TMSB10 knocked-down cells (Fig. 3H).
Fig. 3.
Knockdown of TMSB10 alters p38 pathways in human CRC. (A) TMSB10, p38, p-p38, p53, p-p53, caspase 3, p21, Rb and p-Rb were examined in both TMSB10-knockdown cells. (B) Ratio of p-p38/p38. (C) Ratio of p-p53/p53. (D) Ratio of p-Rb/Rb. (E) The correlation analysis between TMSB10 and p-p38 was carried out by Pearson correlation coefficient. (F and G) The expressions of TMSB10, Ki67, p-p38, p-p53, caspase 3, p21, p-Rb expression in tumor from mice injected with TMSB10-knockdown cells and negative control cells was were examined by IHC. All data are expressed as mean ± standard deviation (SD; three independent experiments), and statistical significance is denoted by * between different groups (p < 0.05).
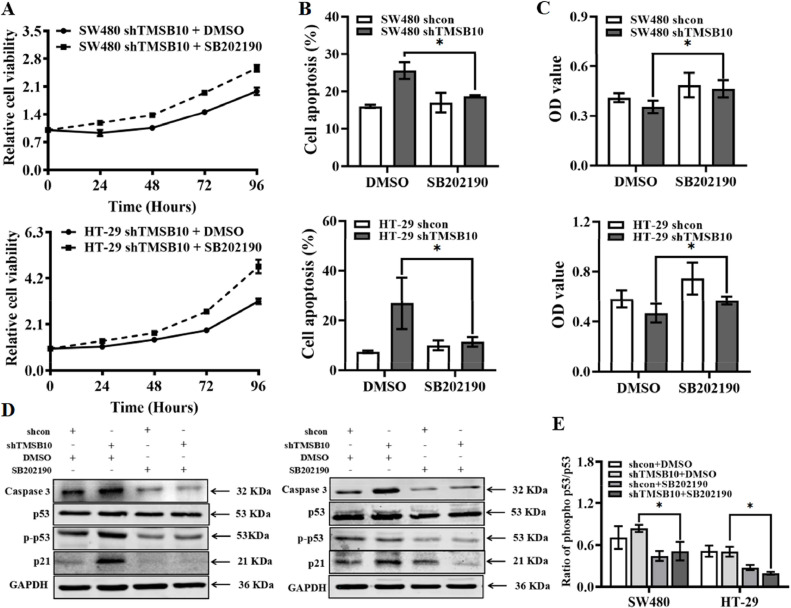
Based on these findings, we hypothesized that knockdown of TMSB10 increased the phosphorylation of p38 and the activated p38 pathway to block the cell cycle and promote apoptosis. To test this possibility, we treated TMSB10-knockdown and negative control cells with SB202190, a highly selective inhibitor of p38. After this, apoptosis, and proliferation in both group cells were examined (Fig. 4A-C, Fig. S3). Inhibition of p38 activation by SB202190 in TMSB10-knockdown cells reduced phosphorylation of p53 leading to a down-regulated transcription of p21 and caspase3. Finally, apoptosis was reduced and then a reactivated cell cycle provided cell proliferation. Collectively, these results revealed that TMSB10 was negatively correlated with the phosphorylation of p38, and possibly acted as a critical oncogene through activating the p38 pathway in CRC.
Fig. 4.
Analysis of cell viability after treatment with TMSB10 activity inhibitor (SB202190). (A) Effect of SB202190 on the proliferation of TMSB10-knockdown cells. (B) Effect of SB202190 on apoptosis of TMSB10-knockdown cells. (C) Cell proliferation of TMSB10-knockdown cells after treatment with SB202190 for 48 h was detected by MTT assay. (D and E) The expression of caspase-3, p53, p-p53 and p21 in TMSB10 knocked-down cells with SB202190 treatment for 48 h.
Discussion
TMSB10 is involved in the malignant process of multiple cancers, and is closely related to cancer cell functions including proliferation, migration, invasion, apoptosis, and cell division [13,[15], [16], [17]]. Zhang et al. [9] reported that the up-regulated TMSB10 contributed to the migration and invasion of breast cancer cells, and promoted cell proliferation of breast cancer by accelerating the cell cycle progression of breast cancer cells. In contrast, a down-regulated expression of TMSB10 was observed in kidney chromophobe, acute myeloid leukemia, and brain lower grade glioma, and the knockdown of TMSB10 in ovarian cancer cells promoted tumor growth and proliferation [18]. These findings revealed differences of TMSB10 expression in different cancers. However, the clinical expression and biological functions of TMSB10 in CRC remains unclear. The analysis of the GEPIA online database found that the expression of TMSB10 gene was generally up-regulated in human COAD and READ. Through the TMA and immunohistochemical analysis, we further confirmed an up-regulation of TMSB10 in CRC tissues. Importantly, our study found elevated TMSB10 in the serum of CRC patients, indicating that the TMSB10 could serve as a cancer biomarker to be used for micro-invasive diagnosis. MTT assays and colony formation assays were carried out and found inhibited cell proliferation and colony-forming abilities in TMSB10-knockdown cells. The annexin V-FITC/PI double stainings by flow cytometry revealed up-regulated apoptosis in TMSB10-knockdown cells, and propidium iodide staining by flow cytometry showed that cell cycle progression in TMSB10-knockdown cells was blocked during the G1/S phase. Cell proliferation inhibition, apoptosis induction and cell cycle arrest in TMSB10-knockdown cells suggest that TMSB10 acts as a cancer-promoting gene to promote cell proliferation in CRC.
Santra et al. [19] found that both thymosin β4 and thymosin α1 were involved in the regulation of cell function by the MAPK pathway, indicating the close relation between thymosin family members and the MAPK pathway. In the present study, the phosphorylation of p38 in TMSB10-knockdown cells was increased. The p-p38 works as a protein kinase involved in a variety of cellular processes including cell division and apoptosis through activating the intracellular signaling pathways [20,21]. The p53 is one of p38-targeted proteins, and p-p38 is involved in the phosphorylation of p53 [22]. Western blot analysis showed a significantly increased expression of p-p53 in TMSB10-knockdown cells. p-53 plays a major role in regulating of the cell cycle machinery and apoptosis, and more p-p53 could enhance the transcription of intracellular p21 and caspase 3 [[23], [24], [25]]. Importantly, higher levels of p21 could inhibit the activation of Cdk2/cyclinE complex and reduce the phosphorylation of Rb which could inactivate E2F1 to weaken transcription of genes related to cell cycle that finally resulted in the blocking of mitotic cell division [26,27]. Activated caspase 3 in TMSB10-knockdown cells could result in increased apoptosis. The above regulatory pathways might be mediated by ROS, and there are reports that mitochondrial ROS can act as a signaling molecule to initiate the phosphorylation of p38 MAPkinase to regulate cell cycles and apoptosis [28], and Liu et al. [29] also confirmed that the elevated ROS could activated p38 MAPK signaling pathway. Moreover, the study from Liu et al. [30] amd Jia et al. [31] showed that the up-regulation of ROS was involved in apoptosis of human colorectal cancer cells and lung cancer cells through the p38/p53 signaling pathway. Effects and regulatory mechanism of TMSB10 on ROS will be the focus of future research. Based on these findings, we hypothesized that knockdown of TMSB10 could inhibit cell proliferation via the p38 pathway. This possibility was strengthened by results of our drug intervention experiments where TMSB10-knockdown and negative control cells were treated with a highly selective p38 inhibitor (SB202190). The results showed the reduced phospho-p53, p21 and Caspase3 in TMSB10-knockdown cells where SB202190 was added. This led to reduced apoptosis and reactivated cell division.
Conclusions
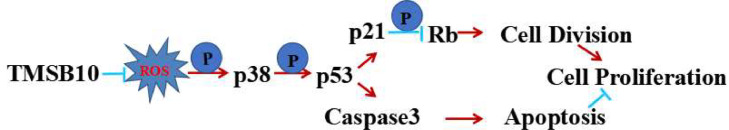
To conclude, the present study has identified TMSB10 as a cancer-promoting protein and a serum tumor biomarker for CRC diagnosis. We propose here a regulatory strategy of TMSB10 regulating phosphorylation of p38 to affect the cell cycle and apoptosis in human CRC cells as shown in Fig. 5. The multiple roles of TMSB10 in cell cycle progression and apoptosis suggest that TMSB10 could be used as a useful biomarker for CRC diagnosis as well as a target to improve the clinical post-surgery efficacy of CRC radio- as well as chemotherapy.
Fig. 5.
Regulation model of TMSB10 on cell proliferation in CRC. Knockdown- TMSB10 promotes the phosphorylation of p38 and p53. The up-regulated p-p53 inhibits cell cycle progression and promotes apoptosis via the p21/Rb pathway and caspase3 pathway, respectively. Arrows (→) and truncated lines (—|) indicate promoting and inhibiting effects, respectively.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Gene expression analysis was performed using through the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn).
CRediT authorship contribution statement
Jian Yang: Conceptualization, Data curation, Formal analysis, Funding acquisition, Writing – original draft, Writing – review & editing. Xiaolong Yang: Data curation, Formal analysis, Methodology, Validation. Tianyi Guo: Data curation, Formal analysis, Methodology, Validation. Lingxiao Wang: Data curation, Formal analysis, Methodology, Validation. Zhenxiang Zhao: Data curation, Formal analysis, Validation. Zhen Hu: Data curation, Formal analysis, Funding acquisition, Methodology, Validation. Yaoping Li: Conceptualization, Funding acquisition, Resources, Supervision.
Declaration of competing interest
All authors have participated in the research and/or article preparation and agree with submission of this manuscript to Translational Oncology. The material has not been published previously, and will not be submitted for publication elsewhere. None of the authors have potential conflicts of interest with regard to this manuscript to disclose.
Funding
This research was supported by funding from the National Natural Science Foundation of China (82103175) and the Natural Science Foundation of Shanxi Province (20210302123316, 202103021224379, and 202203021221185).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.102026.
Contributor Information
Jian Yang, Email: jiany@sxmu.edu.cn.
Yaoping Li, Email: liyaoping1600@sina.com.
Appendix. Supplementary materials
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Vuik F.E., Nieuwenburg S.A., M.Bardou I.Lansdorp-Vogelaar, Dinis-Ribeir M., Bento M.J., Zadnik V., Pellisé M., Esteban L., Kaminski M.F. Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut. 2019;68:1820–1826. doi: 10.1136/gutjnl-2018-317592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W., Zhang Y., Zhang X., Teng S., Yao H., Li X., Hu Z. Prognosis factor of patients with colorectal cance:a retrospective study of surveillance, epidemiology, and end results population-based data. Chin. J. Colorec. Dis. (Electr. Edit.) 2017;6:21–27. [Google Scholar]
- 4.Huang L., Zhang Y., Li Z., Zhao X., Xi Z., Chen H., Shi H., Xin T., Shen R., Wang T. MiR-4319 suppresses colorectal cancer progression by targeting ABTB1. United. European. Gastroenterol. J. 2019;7:517–528. doi: 10.1177/2050640619837440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watts J.D., Cary P.D., Sautiere P., Crane-Robinson C. Thymosins: both nuclear and cytoplasmic proteins. Eur. J. Biochem. 1990;192:643–651. doi: 10.1111/j.1432-1033.1990.tb19271.x. [DOI] [PubMed] [Google Scholar]
- 6.Inzitari R., Cabras T., Pisano E., Fanali C., Manconi B., Scarano E., Fiorita A., Paludetti G., Manni A., Nemolato S., Faa G., Castagnola M., Messana I. HPLC-ESI-MS analysis of oral human fluids reveals that gingival crevicular fluid is the main source of oral thymosins beta(4) and beta(10) J. Sep. Sci. 2009;32:57–63. doi: 10.1002/jssc.200800496. [DOI] [PubMed] [Google Scholar]
- 7.Padmanabhan K., Grobe H., Cohen J., Soffer A., Mahly A., Adir O. Thymosin β4 is essential for adherens junction stability and epidermal planar cell polarity. Development. 2020;147 doi: 10.1242/dev.193425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santelli G., Califano D., Chiappetta G., Vento M.T., Bartoli P.C., Zullo F., Trapasso F., Viglietto G., Fusco A. Thymosin beta-10 gene overexpression is a general event in human carcinogenesis. Am. J. Pathol. 1999;155:799–804. doi: 10.1016/s0002-9440(10)65178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X., Ren D., Guo L., Wang L., Wu S., Lin C., Ye L., Zhu J., Li J., Song L., Lin H., He Z. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast. Cancer Res. 2017;19:15. doi: 10.1186/s13058-016-0785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An H.W., Kim S.Y., Kwon J.W., Seok S.H., Woo S.H., Kim D.Y. In vivo CRISPR-Cas9 knockout screening using quantitative PCR identifies thymosin beta-4 X-linked that promotes diffuse-type gastric cancer metastasis. Mol. Carcinog. 2021;60:597–606. doi: 10.1002/mc.23326. [DOI] [PubMed] [Google Scholar]
- 11.Wirsching H.G., Krishnan S., Florea A.M., Frei K., Krayenbühl N., Hasenbach K., Reifenberger G., Weller M., Tabatabai G. Thymosin β 4 gene silencing decreases stemness and invasiveness in glioblastoma. Brain. 2014;137:433–448. doi: 10.1093/brain/awt333. [DOI] [PubMed] [Google Scholar]
- 12.Banyard J., Barrows C., Zetter B.R. Differential regulation of human thymosin beta 15 isoforms by transforming growth factor beta 1. Genes Chromos. Cancer. 2009;48:502–509. doi: 10.1002/gcc.20659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X.J., Y.R.Su D.Liu, Xu D.B., Zeng M.S., Chen W.K. Thymosin beta 10 correlates with lymph node metastases of papillary thyroid carcinoma. J. Surg. Res. 2014;192:487–493. doi: 10.1016/j.jss.2014.05.066. [DOI] [PubMed] [Google Scholar]
- 14.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic. Acids. Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng J., Yang X., Yang L., Li W., Zheng Y. Thymosin β 10 promotes tumor-associated macrophages M2 conversion and proliferation via the PI3K/Akt pathway in lung adenocarcinoma. Respir. Res. 2020;21:328. doi: 10.1186/s12931-020-01587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W., W.Chu Q.Liu, Coates D., Shang Y., Li C. Deer thymosin beta 10 functions as a novel factor for angiogenesis and chondrogenesis during antler growth and regeneration. Stem Cell Res. Ther. 2018;9:166. doi: 10.1186/s13287-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maelan A.E., Rasmussen T.K., Larsson L.I. Localization of thymosin beta10 in breast cancer cells: relationship to actin cytoskeletal remodeling and cell motility. Histochem. Cell Biol. 2007;127:109–113. doi: 10.1007/s00418-006-0208-z. [DOI] [PubMed] [Google Scholar]
- 18.Lee S.H., Son M.J., Oh S.H., Rho S.B., Park K., Kim Y.J., Park M.S., Lee J.H. Thymosin {beta}(10) inhibits angiogenesis and tumor growth by interfering with Ras function. Cancer Res. 2005;65:137–148. [PubMed] [Google Scholar]
- 19.Santra M., Chopp M., Zhang Z.G., Lu M., Santra S., Nalani A., Santra S., Morris D.C. Thymosin β4 mediates oligodendrocyte differentiation by upregulating p38 MAPK. Glia. 2012;60:1826–1838. doi: 10.1002/glia.22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono K., Han J. The p38 signal transduction pathway: activation and function. Cell Signal. 2000;12:1–13. doi: 10.1016/s0898-6568(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 21.Lu Z.W., Wen D., Wei W.J., Han L.T., Xiang J., Wang Y.L. Silencing of PPM1D inhibits cell proliferation and invasion through the p38 MAPK and p53 signaling pathway in papillary thyroid carcinoma. Oncol. Rep. 2020;43:783–794. doi: 10.3892/or.2020.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Y., Ye F., Li X., Yang Q., Zhou J., Xu W. 3,3′-diindolylmethane exerts antiproliferation and apoptosis induction by TRAF2-p38 axis in gastric cancer. Anticancer Drugs. 2021;32:189–202. doi: 10.1097/CAD.0000000000000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies L., Gray D., Spiller D., White M.R., Damato B., Grierson I., Paraoan L. P53 apoptosis mediator PERP: localization, function and caspase activation in uveal melanoma. J. Cell Mol. Med. 2009;13:1995–2007. doi: 10.1111/j.1582-4934.2008.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreis N.N., Louwen F., Yuan J. Less understood issues: p21(Cip1) in mitosis and its therapeutic potential. Oncogene. 2015;34:1758–1767. doi: 10.1038/onc.2014.133. [DOI] [PubMed] [Google Scholar]
- 25.Savaryn J.P., Reitsma J.M., Bigley T.M., Halligan B.D., Qian Z., Yu D. Human cytomegalovirus pUL29/28 and pUL38 repression of p53-regulated p21CIP1 and caspase 1 promoters during infection. J. Virol. 2013;87:2463–2474. doi: 10.1128/JVI.01926-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heo S.Y., Jeong M.S., Lee H.S., Park W.S., Choi I.W., Yi M M. Dieckol induces cell cycle arrest by down-regulating CDK2/cyclin E in response to p21/p53 activation in human tracheal fibroblasts. Cell Biochem. Funct. 2022;40:71–78. doi: 10.1002/cbf.3675. [DOI] [PubMed] [Google Scholar]
- 27.Huang S.Y., Hsieh M.J., Chen C.Y., Chen Y.J., Chen J.Y., Chen M.R. Epstein-Barr virus Rta-mediated transactivation of p21 and 14-3-3σ arrests cells at the G1/S transition by reducing cyclin E/CDK2 activity. J. Gen. Virol. 2012;93:139–149. doi: 10.1099/vir.0.034405-0. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Ma H.L., Han L., Liu W.Y., Zhao B.X., Zhang S.L. Novel ferrocenyl derivatives exert anti-cancer effect in human lung cancer cells in vitro via inducing G1-phase arrest and senescence. Acta Pharmacol. Sin. 2013;34:960–968. doi: 10.1038/aps.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Wei Y., Jia W., Can C., Wang R., Yang X. Chenodeoxycholic acid suppresses AML progression through promoting lipid peroxidation via ROS/p38 MAPK/DGAT1 pathway and inhibiting M2 macrophage polarization. Redox. Biol. 2022;56 doi: 10.1016/j.redox.2022.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B., Yuan B., Zhang L., Mu W., Wang C. ROS/p38/p53/Puma signaling pathway is involved in emodin-induced apoptosis of human colorectal cancer cells. Int J Clin Exp. 2015;15:15413–15422. [PMC free article] [PubMed] [Google Scholar]
- 31.Jia X.B., Zhang Q., Xu L., Yao W.J., Wei L. Lotus leaf flavonoids induce apoptosis of human lung cancer A549 cells through the ROS/p38 MAPK pathway. Biol. Res. 2021;54:7. doi: 10.1186/s40659-021-00330-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Gene expression analysis was performed using through the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn).