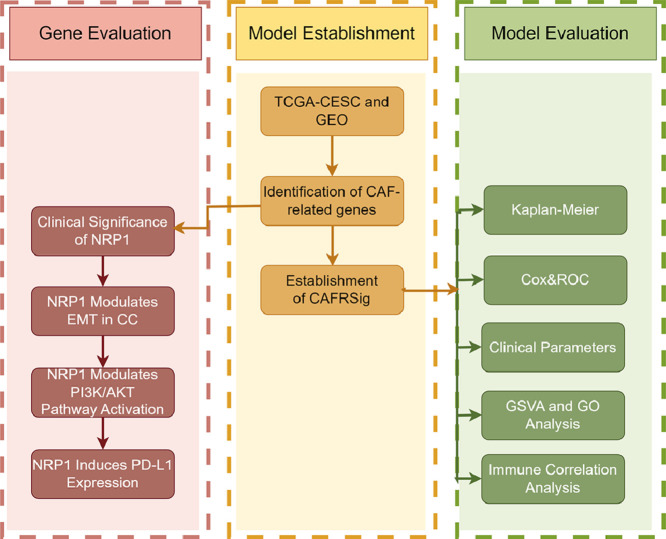
Highlights
-
•
This study introduces a pioneering model predicting cervical cancer outcomes using cancer-associated fibroblasts (CAFs) genes.
-
•
The CAF-related risk score (CAFRSig) effectively distinguishes survival rates and immune responses in cervical cancer patients.
-
•
Multiple bioinformatics tools and diverse datasets were utilized to robustly validate the CAFRSig model.
-
•
CAFRSig's potential in identifying cervical cancer patients likely to respond to immune checkpoint therapies is highlighted.
-
•
NRP1, key in CAFRSig, is crucial in cervical cancer progression, affecting cell proliferation and EMT, linked to the PI3K/AKT pathway.
Keywords: Cancer-associated fibroblasts, Cervical cancer, Immune response, Risk score signature, Neuropilin 1
Abstract
This study developed a prognostic signature for cervical cancer using transcriptome profiling and clinical data from The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), and TISCH database, focusing on cancer-associated fibroblasts (CAFs). Through LASSO Cox regression and integrated bioinformatics analyses, we identified 144 differentially expressed genes (DEGs) related to CAFs, from which an 11-gene CAF-related signature (CAFRSig) was constructed. The CAFRSig effectively stratified patients into high- and low-risk categories, demonstrating significant prognostic capability in predicting overall survival. Gene ontology (GO) and gene set variation analysis (GSVA) linked the DEGs to crucial pathways in tumor malignancy, immune response, and fatty acid metabolism. The immune landscape analysis, utilizing the TIMER platform and CIBERSORT algorithm, revealed a positive correlation between immune cell effector functions and CAFRSig scores, highlighting the model's potential to identify patients likely to respond to immune checkpoint blockade (ICB) therapies. Furthermore, neuropilin 1 (NRP1), a key gene in the CAFRSig, was upregulated in cervical cancer tissues and associated with disease progression and differentiation. The downregulation of NRP1 curbed cell proliferation and influenced the epithelial-mesenchymal transition (EMT), implicating the PI3K/AKT pathway and modulating PD-L1 expression. This comprehensive analysis establishes a robust prognostic signature based on CAF-related genes, offering valuable insights for optimizing therapeutic strategies in cervical cancer management.
Graphical abstract
Introduction
In 2020, cervical cancer (CC) accounted for approximately 604,000 new cases and 342,000 deaths globally, ranking as the fourth most common type of cancer and the fourth highest contributor to cancer-related mortality among women [1]. A notable proportion, about 13%, of CC patients receive their diagnosis in the advanced stages of the disease [2]. Presently, there is no curative treatment approach for individuals with advanced, recurrent, or metastatic CC [[3], [4], [5]]. The success rate of existing therapeutic options, such as the conventional chemotherapy regimen of cisplatin/paclitaxel and targeted therapies like bevacizumab, falls below 50% in these patient groups [4,6].
CC's heterogeneity significantly impacts drug resistance and oncological outcomes [7]. The tumor microenvironment (TME), which includes a diverse array of cells like tumor cells, immune cells, and cancer-associated fibroblasts (CAFs), plays a pivotal role in tumor progression [8]. Cancer-associated fibroblasts (CAFs) within the TME play a vital role in tumor dynamics by producing growth factors, remodeling the ECM, and promoting angiogenesis [9]. These activities contribute to cell invasion, therapy resistance, and tumor recurrence. CAFs also regulate immune cell infiltration in the TME, aiding immune evasion [[10], [11], [12]]. Their diversity, stemming from various cell types like adipocytes and stem cells, leads to distinct functional subtypes [[13], [14], [15]]. While studies targeting CAFs and their related molecules as tumor biomarkers have been explored in other cancers [16,17], research on CC is limited. Understanding CAFs' molecular characteristics and roles in the TME could clarify CC heterogeneity and enhance immunotherapy strategies.
Advances in high-throughput sequencing analysis across multiple cancer types, coupled with the evolution of bioinformatics methodologies, have facilitated the discovery of critical targets and novel predictive signatures [[18], [19], [20]]. In this study, eleven CAF-related prognostic genes were identified to develop a CAF-related signature (CAFRSig), offering valuable information for utilizing the CAF landscape in predicting clinical outcomes and treatment effectiveness in CC.
Materials and methods
Data collection and processing
Information regarding the gene expression profiles and pertinent clinical information for cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) was acquired from the cancer genome atlas (TCGA). This information was compiled using Perl scripts, incorporating data from the GSE44001 cohort in the Gene Expression Omnibus (GEO), along with transcriptome and clinical data, as well as single-cell sequencing data from the GSE168652 cohort in the TISCH database. Inclusion criteria for TCGA-CESC and GSE44001 cohort study cases were the inclusion of both transcriptome and survival information. The ComBat function of the R package 'SVA' was conducted to eliminate batch processing effects in different datasets. Genes associated with CAF were acquired from The Human Gene Database [21].
Identification of CAF-related genes in CESC
R (version 4.2.1) was used to extract the mRNA expression data for CAF-associated genes from the TCGA-CESC cohort. Genes showing differential expression (DEGs) were pinpointed in normal and tumor tissues based on a |fold change| (FC) exceeding 2 and a false discovery rate (FDR) below 0.05. The R package 'pheatmap' was utilized to create heatmaps that illustrate the expression of genes that are differentially expressed (DEGs). Analysis of differentially expressed genes (DEGs) in the TCGA-CESC and GSE44001 groups was conducted with the assistance of the 'limma' and 'sva' software packages.
Establishment of a CAFRSig in CESC
Univariate Cox analysis identified survival-related DEGs in the TCGA-CESC cohort. The CAFRSig was created using LASSO regression analysis to pinpoint the most appropriate genes. Each patient's risk score was determined by assessing the expression levels of the genes associated with CAFRSig and their respective regression coefficient. Patients were grouped into high and low risk categories based on the median risk score. Kaplan-Meier curves were used to analyze the GSE168652 cohort for genes linked to the signature, and their expression was examined.
Validation of the CAFRSig in CESC
Risk heatmaps, curves, survival status maps, and Kaplan-Meier curves were generated using the TCGA-CESC and GSE44001 cohorts. The R packages 'pheatmap', 'survival', and 'survminer' were utilized to generate these visual representations. The predictive importance of the CAFRSig was assessed using both univariate and multivariate Cox analyses. Additionally, the predictive significance of the CAFRSig was evaluated through the creation of receiver operating characteristic (ROC) curves utilizing the 'survminer', 'survival', and 'timeROC' R packages. The size of the area under the curves (AUC) was utilized for evaluation.
Correlation analysis of the CAFRSig with clinical parameters in CESC
To evaluate the CAFRSig, patients were categorized into two groups according to their age, stage, and grade. Kaplan-Meier curves were utilized to analyze the variation in survival rates among groups with different risk levels based on a range of clinical factors. Heatmaps displaying different clinical parameters in each risk subgroup were generated using the 'ComplexHeatmap' package.
GO and GSVA analysis
DEGs with significant changes (|FC| > 2, FDR < 0.05) between various risk groups were identified using the 'limma' software. The DEGs underwent analysis using gene ontology (GO) to explore their roles in cellular component, molecular function, and biological process. The examination made use of resources such as 'org.Hs.eg.db', ‘GOplot’, and ‘clusterProfiler’.
The Gene Set Variation Analysis (GSVA) was conducted to assess alterations in pathway activities among the subgroups [22]. This required analyzing the enrichment of KEGG pathways in both high-risk groups and exploring the relationship between these pathways and the expression of key genes. This comprehensive analysis utilized R packages including ‘GSVA’, ‘pheatmap’, and ‘ggplot2’.
Correlation analysis of the CAFRSig and immune landscape in CESC
The study utilized the TIMER platform to evaluate the extent of immune cell infiltration in tumors [23,24]. The system incorporates a variety of algorithms such as MCPCOUNTER, TIMER, CIBERSORT, XCELL, QUANTISEQ, and EPIC to evaluate the association between CAFRSig risk scores and different degrees of immune cell infiltration, a Spearman correlation analysis was performed. Data analysis and visualization were performed using various R packages including 'ggplot2′, 'ggtext', 'scales', 'tidyverse', and 'ggpubr'.
GSEA, or gene set enrichment analysis, is a method utilized to discover genomes that share similar biological traits [25]. The study utilized the ssGSEA algorithm, available through the 'GSEABase' and 'GSVA' packages, to assess the presence of immune cells in tumor samples from the TCGA-CESC dataset. This method produced results concerning the presence of immune cells and the function of the immune system in each individual. Differential box plots were created in R with the 'ggpubr' and 'reshape2′ packages to illustrate the differences in scores among high and low-risk groups.
The CIBERSORT algorithm, leveraging machine learning, was applied for high-throughput analysis of various cell types like tumor-infiltrating immune cells (TIICs) [26]. This analysis, using tools such as ‘limma’, ‘CIBERSORT’, ‘preprocessCore’, ‘e1071’, and ‘parallel’, focused on quantifying 22 distinct types of TIICs and comparing their distribution across the two risk subgroups.
ESTIMATE, an algorithm for assessing tumor purity through gene expression analysis, was used in this study [27]. Employing the ‘ESTIMATE’ package, stromal and immune scores within tumor tissues were computed. These scores were then visualized for each risk group using box plots, created with the ‘ggpubr’ package.
Correlation analysis of the CAFRSig with immunotherapy response
The efficacy of immunotherapy was examined in this research, focusing on the influence of immune checkpoint activation, which hinders the immune system's capacity to eliminate cancerous cells [28]. The research involved analyzing the expression of various immune checkpoint genes in different risk subgroups. In order to evaluate the survival outcomes among these categories, Kaplan-Meier curves were generated by utilizing information from the IMvigor210 immunotherapy cohort.
Patient information and tissue specimens
The ongoing research encompasses 103 cervical squamous-cell carcinoma specimens collected from the First Affiliated Hospital of Anhui Medical University between 2015 and 2017. From October 2022 to May 2023, we procured eight sets of early-stage CC and adjacent healthy tissue from surgical patients at our facility. To be eligible, participants must have undergone RH without prior chemotherapy, radiotherapy, or hormonal therapy, have a diagnosis of FIGO stage IB–IIA according to the 2009 criteria, show no evidence of distant metastasis, provide complete clinical data, and have follow-up records for at least 5 years, with a deadline of January 2014. Table 2 provides a concise overview of the patient's clinical information. The main CC group was followed up for a period ranging from 5.8 to 89.2 months, with a median follow-up duration of 64.8 months.
Table 2.
Correlation between NRP1 expression and the clinicopathological features of CC.
| Characteristics | Total |
NRP1 Expression |
χ2 | P value | |
|---|---|---|---|---|---|
| No or Weak | Moderate or Strong | ||||
| Age(years) | |||||
| <45 | 38 | 20(0.53) | 18(0.47) | 1.437 | 0.231 |
| ≥45 | 65 | 42(0.65) | 23(0.35) | ||
| HPV Infection | |||||
| Yes | 31 | 19(0.61) | 12(0.39) | 0.220 | 0.881 |
| No | 72 | 43(0.60) | 29(0.40) | ||
| FIGO Stage | |||||
| IB | 59 | 40(0.68) | 19(0.32) | 4.204 | 0.040 |
| IIA | 44 | 21(0.48) | 23(0.52) | ||
| Squamous Cell Carcinoma Antigen | |||||
| <1.5 ng/mL | 66 | 38(0.58) | 28(0.42) | 0.526 | 0.468 |
| ≥1.5 ng/mL | 37 | 24(0.65) | 13(0.35) | ||
| Tumor Size | |||||
| <4 cm | 61 | 35(0.57) | 26(0.43) | 0.495 | 0.481 |
| ≥4 cm | 42 | 27(0.64) | 15(0.36) | ||
| Differentiation Grade | |||||
| Well | 27 | 19(0.31) | 8(0.20) | 7.930 | 0.019 |
| Moderately | 40 | 28(0.45) | 12(0.29) | ||
| Poorly | 36 | 15(0.24) | 21(0.51) | ||
IHC staining and evaluation
Immunohistochemistry (IHC) was employed to analyze NRP1 expression in the study's tissue samples. Initially, the specimens were treated with a 4% solution of formaldehyde, then encased in paraffin, and finally sliced into sections measuring 4 μm in thickness. After deparaffinization in xylene and dehydration through graded alcohols, antigen retrieval was achieved using microwave heating. Afterward, the segments were treated with a primary antibody against NRP1 and PD-L1 from Abcam in Cambridge, USA, then a secondary antibody conjugated with peroxidase, and finally diaminobenzidine was used for development. The immunoreactive score (IRS) was determined by two pathologists who independently assessed the staining intensity and the proportion of positive cells, utilizing a semi-quantitative scoring system.
Cell culture and cell transfection
The Siha and Hela cervical cell lines were grown in DMEM medium (Gibco) supplemented with 10% fetal bovine serum, in a 5% CO2 environment at 37 °C. To assess the effects of NRP1, cells were treated with NRP1-targeting siRNAs (Genomeditech, Shanghai, China) using Lipofectamine 2000 (Invitrogen, California, USA) according to the provided instructions. Western blot analysis confirmed the efficiency of the transfections.
Cell proliferation assay
Cell proliferation was monitored by utilizing a Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan). A density of 1000 cells per well was used to plate cells in 96-well plates. Cell viability was evaluated daily for a week by introducing CCK-8 solution to each well and measuring absorbance following a 4-hour incubation at 37 °C.
Cell migration assay
Cell migration ability was assessed using wound-healing and transwell assays. For the transwell experiment, 10,000 cells were added to the upper chambers of transwell inserts (Labselect, China) with 200 μL of serum-free medium. These inserts were then placed in 24-well plates with 800 μL of medium containing 10% FBS. Following a 24-hour incubation period, cells that migrated through the membrane were fixed using 4% paraformaldehyde (PFA) and stained with crystal violet. The migrated cells were then imaged and counted to assess migration ability. CC cells that were transfected were grown in six-well plates for the wound-healing test, reaching around 80% confluence. Subsequently, a 200 μL pipette tip was utilized to make a straight scrape through the cell layer. Following the removal of dislodged cells using PBS, a new culture medium was introduced, allowing the cells to migrate into the wound area for a period of 12 h. To quantify the migration, the average distance between the wound's edges was measured at various points throughout its length.
Western blotting
Proteins were extracted from the cells and quantified using the Coomassie Protein Assay (Thermo Fisher Scientific, USA). After undergoing separation using SDS-PAGE, the samples were then transferred to PVDF membranes. After a 60-minute block with skim milk, the membranes were incubated overnight at 4 °C with primary antibodies. The following day, a second antibody (diluted 1:5000, sourced from Cell Signaling Technology in the United States) was administered and allowed to incubate for 2 h at ambient temperature. β-actin served as the internal control. The Bio-Rad ChemiDoc XRS+ system from Bio-Rad Laboratories in Hercules, California, USA, was used for detecting and capturing images. Protein quantities were measured with Image J program and adjusted based on β-actin levels. The primary antibodies used included NRP1 (Abcam, Cambridge, USA), PD-L1 (Abcam, Cambridge, USA), E-cadherin (Abcam, Cambridge, USA), α-SMA (Abmart, Shanghai, China), Vimentin (Abclonal, Wuhan, Hubei, China), Snail (Abclonal, Wuhan, Hubei, China), p-AKT (CST, Danvers, MA, USA), AKT (CST, Danvers, MA, USA), p-PI3K (CST, Danvers, MA, USA), and PI3K (CST, Danvers, MA, USA).
Statistical analysis
SPSS 22.0 software (SPSS, Chicago, IL, USA) was utilized for all statistical analyses. The correlation between NRP1 expression levels and clinicopathological characteristics in patients with CC was assessed through the χ2 test. Survival rates were calculated using the Kaplan-Meier technique. A statistical significance was determined with a significance level below 0.05.
Results
Identification of CAF-related genes in CESC
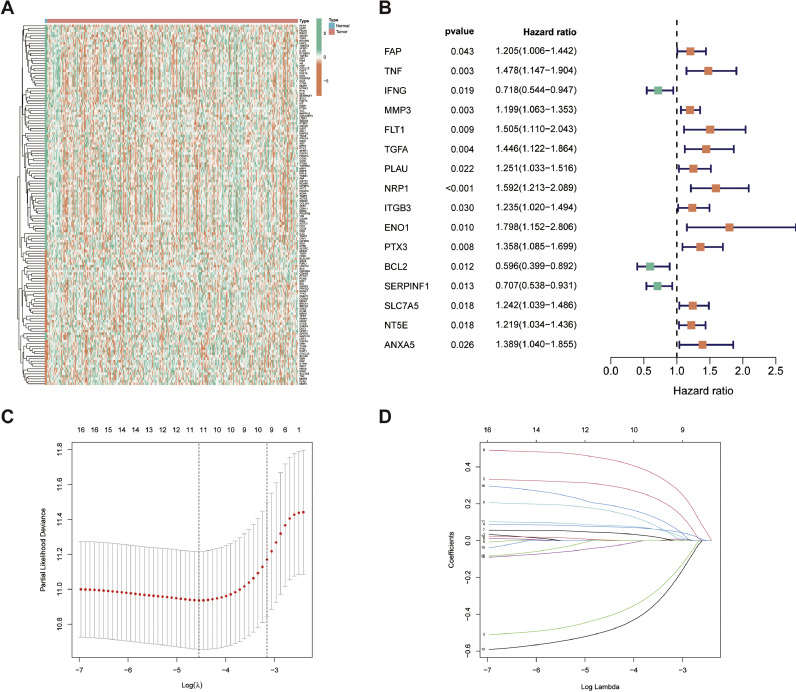
From GeneCards, 401 protein-coding genes associated with CAFs were identified, each with a relevance score above 5 (Appendix 1). Among these, 144 genes showed differential expression between CESC tumors and normal tissues. Their expression patterns are depicted in a heatmap (Fig. 1A). Cox regression analysis revealed 16 of these genes as significant for CESC survival in the TCGA-CESC cohort (Fig. 2B).
Fig. 1.
CAF-related genes in CESC. (A) Heat map of differentially expressed CAF-related genes in the TCGA-CESC cohort. (B) Risk forest plot showing that 16 CAF-related genes are associated with CESC prognosis in the TCGA-CESC cohort. (C-D) Coefficient and partial likelihood deviance of the prognostic signature.
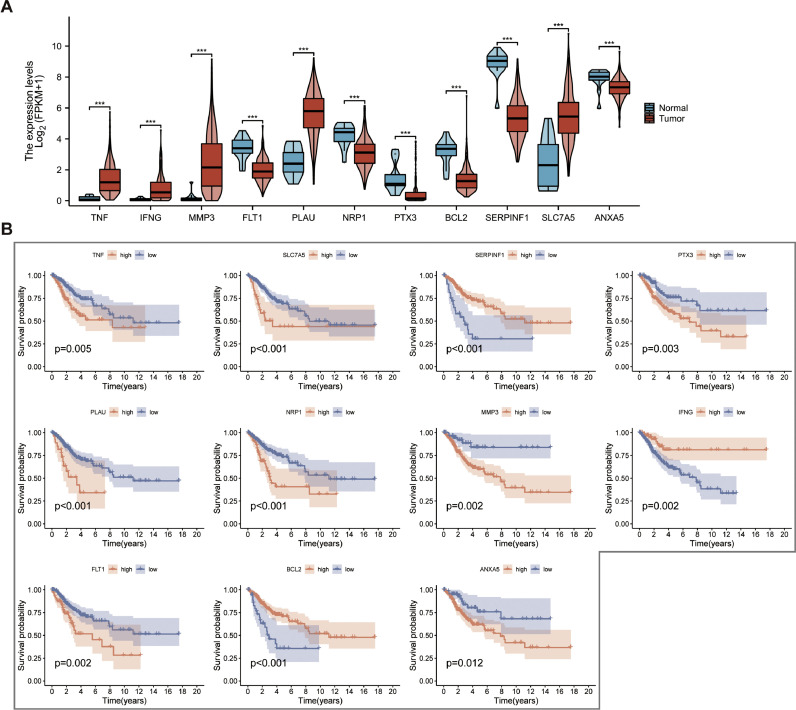
Fig. 2.
CAFRSig-related genes in CESC. (A) Violin diagram displaying the expression of CAFRSig-related genes in CESC tumor and normal tissues. (B) Kaplan-Meier curves analyzing the correlation between the expression of the CAFRSig-related genes and the prognosis of CESC.
Construction and validation of the CAFRSig in CESC
The LASSO algorithm was utilized to reduce overfitting, leading to the selection of 11 critical genes for the CAF-related signature (CAFRSig) (Fig. 1C-D). These genes, which include six down-regulated and five up-regulated ones, were found to be differentially expressed in CESC tumor versus normal tissues (Fig. 2A). Survival analysis indicated a correlation between high expression of genes such as SERPINF1, BCL2, and IFNG and improved survival rates, contrasting with other genes that had an adverse impact on survival (Fig. 2B). The risk score for each patient was calculated using a formula that multiplies the expression levels of these genes by their respective risk coefficients (Table 1). The risk score equation is: Risk score = Expression levels of genes × (corresponding regression coefficients).
Table 1.
CAFs-related signature in CESC.
| CAFs-related genes | Coef |
|---|---|
| TNF | 0.300650993622782 |
| IFNG | −0.413099491372077 |
| MMP3 | 0.0745949051737052 |
| FLT1 | 0.163600855835463 |
| PLAU | 0.0429657212298335 |
| NRP1 | 0.443691502689255 |
| PTX3 | 0.0828054282243763 |
| BCL2 | −0.0333116920947764 |
| SERPINF1 | −0.490981840464332 |
| SLC7A5 | 0.00340557287936776 |
| ANXA5 | 0.194906012392434 |
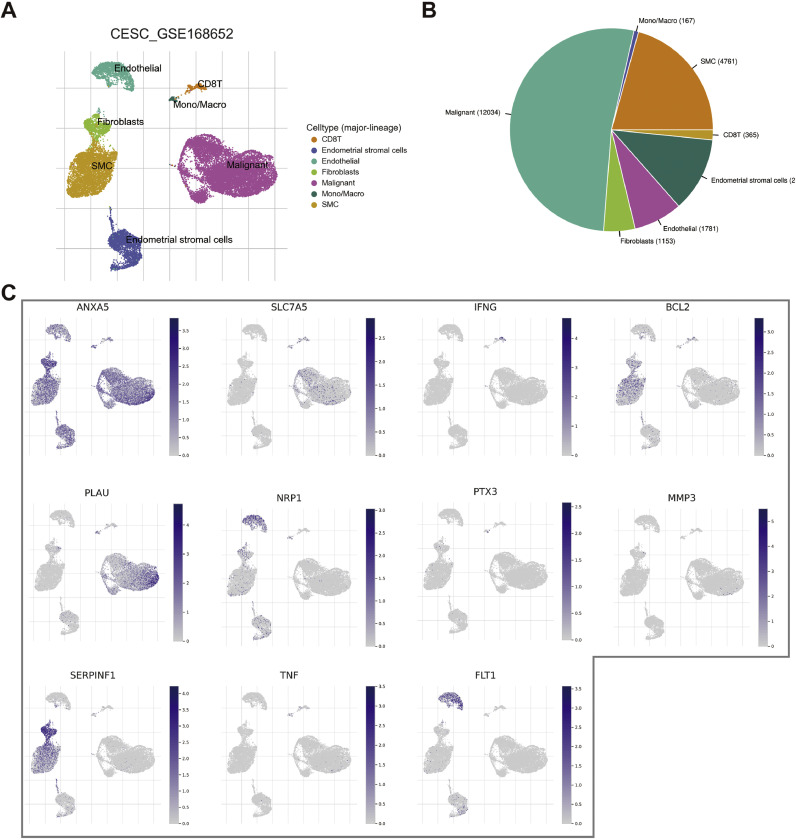
Furthermore, we analyzed the expression of signature-related genes in different cell types of the GSE168652 cohort (Fig. 3A, B). The results showed that ANXA5 and SERPINF1 were expressed at high levels in fibroblasts (Fig. 3C).
Fig. 3.
CAFRSig-related genes in different cell types of the GSE168652 cohort. (A) Annotation of cell clusters into 7 cell categories. (B) Proportion of different cell types. (C) Expression of 11 CAFRSig-related genes in different cells of the GSE168652 cohort.
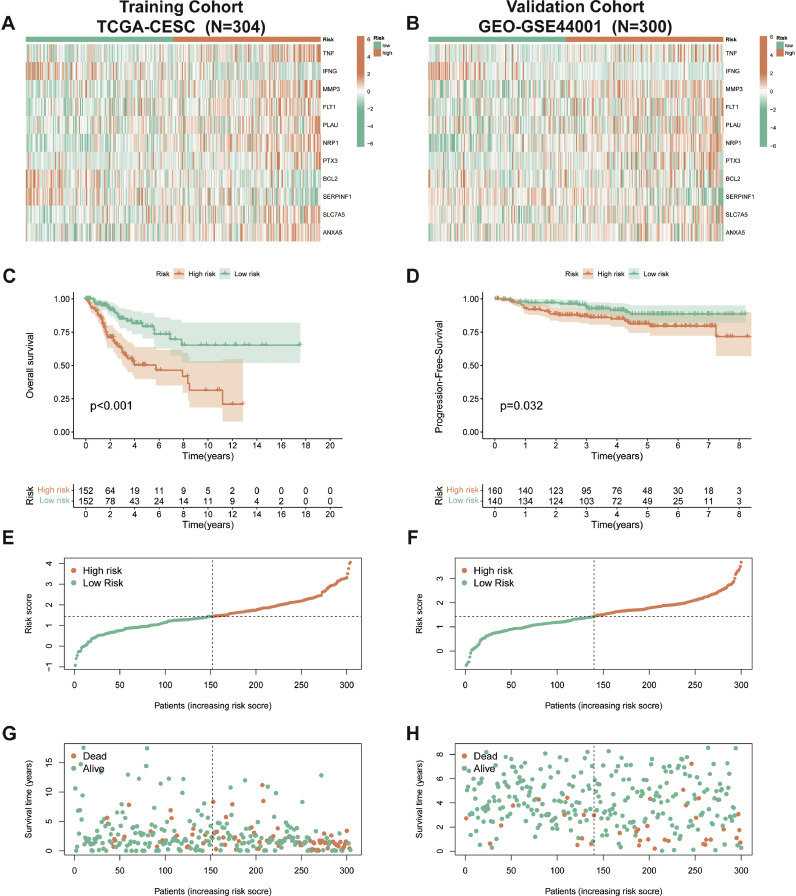
The validation of CAFRSig was also confirmed in the TCGA-CESC and GSE44001 groups. The cohorts were analyzed using heatmaps (Fig. 4A, B) to illustrate the expression of the 11 genes associated with CAFRSig. Kaplan-Meier analyses showed that patients in the high-risk subgroup had poorer outcomes (Fig. 4C and D). Additionally, analyses of risk score distributions and survival status maps further supported the association between higher risk scores and increased mortality rates (Fig. 4E-H).
Fig. 4.
Validation of the CAFRSig in the TCGA-CESC and GSE44001 cohorts. (A-B) Heat maps showing the expression of the 11 CAFRSig-associated genes in the TCGA-CESC and GSE44001 cohorts. (C-D) Kaplan-Meier curves for the CAFRSig in the TCGA-CESC and GSE44001 cohorts. (E-H) Risk score distribution curves and survival status plots for the CAFRSig in the TCGA-CESC and GSE44001 cohorts.
Assessment of the CAFRSig in CESC
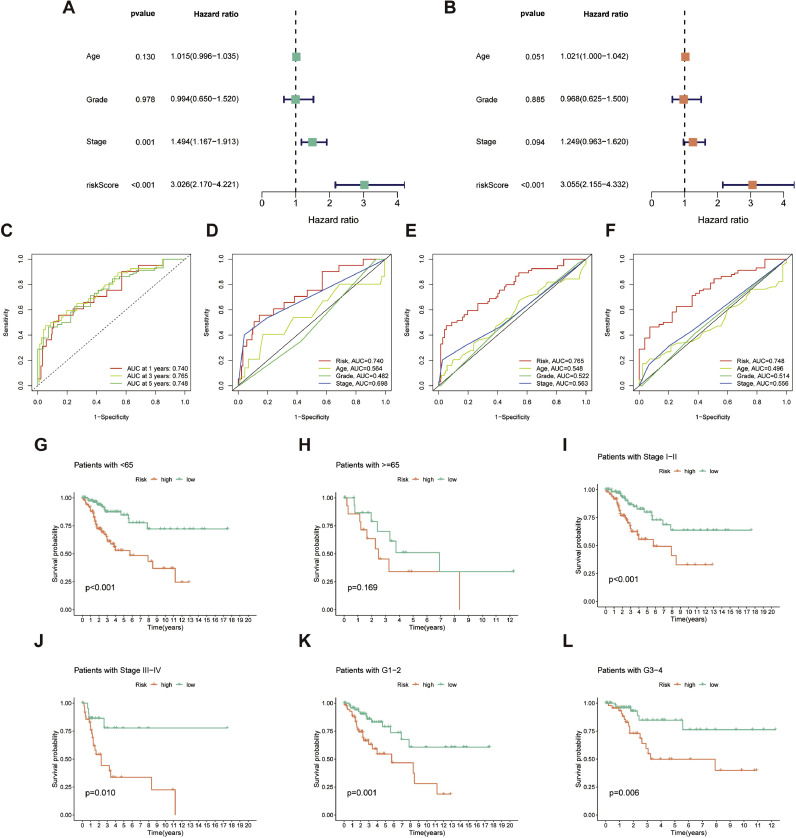
Significant hazard ratios (HR) demonstrated that risk scores derived from CAFRSig were independent prognostic indicators, as determined by univariate and multivariate Cox regression analysis (Fig. 5A and B). ROC curves in Fig. 5C-F assess the predictive precision of CAFRSig for 1-, 3-, and 5-year overall survival (OS) in CESC prognosis, demonstrating a strong level of specificity and sensitivity. Upon further analysis of patient subgroups categorized by gender, age, and tumor stage, it was found that the high-risk group consistently had a poor prognosis, as indicated by subsequent survival analysis (Fig. 5G-L).
Fig. 5.
Assessment of the CAFRSig in the TCGA-CESC cohort. (A-B) Forest plots for univariate and multivariate Cox regression analyses. (C) ROC curves for 1-, 3-, and 5-year survival predictions using the CAFRSig. (D-F) Comparison of the prediction accuracy of the CAFRSig with age, stage, and grade at 1-, 3-, and 5- years. (G-L) Kaplan-Meier curves of low- and high-risk subgroups sorted by age, stage, and grade.
GSVA and go analysis
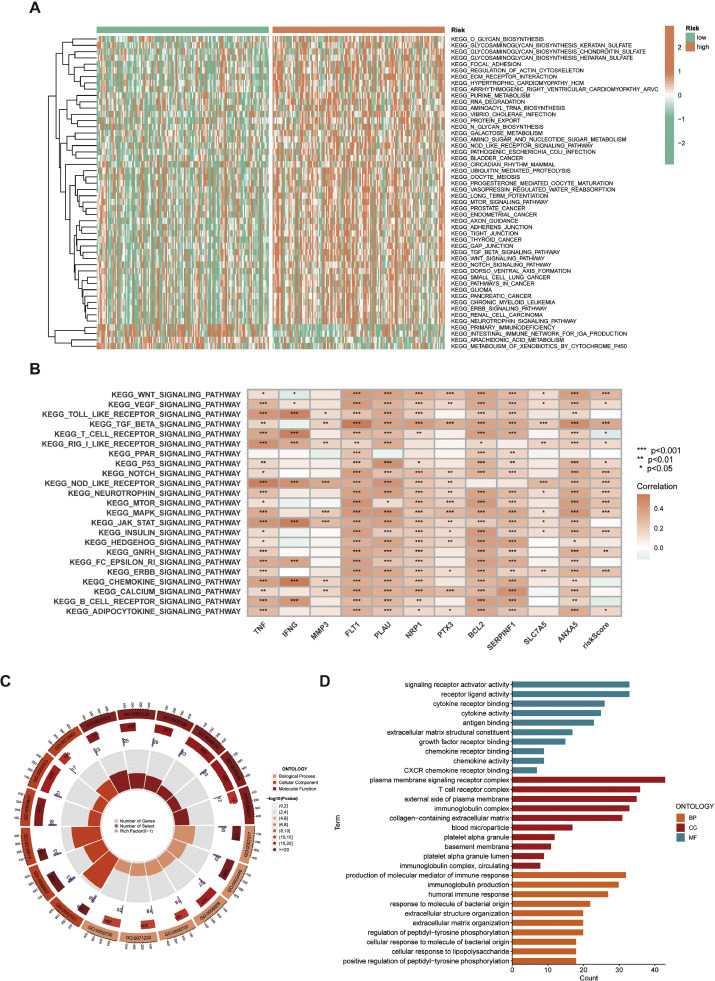
GSVA revealed that the high-risk subgroup in the study was characterized by a significant engagement in tumor-related pathways including the Wnt, TGF-beta, Notch, mTOR, and ERBB signaling pathways. Furthermore, the low-risk subgroup exhibited enrichment in pathways associated with primary immunodeficiency, intestinal immune network for IgA synthesis, arachidonic acid metabolism, and metabolism of xenobiotics by cytochrome P450 (Fig. 6A). The heatmap indicated a robust association between the 11 genes' expression in the CAFRSig and signaling pathways associated with tumor malignancies (Fig. 6B).
Fig. 6.
GSVA and GO analysis in the TCGA-CESC cohort. (A) Heat map of functional pathway enrichment differences between the two risk groups. (B) Heat map of the correlation between the expression of signature genes and signaling pathways. (C, D) GO analysis displaying the enrichment of DEGs between the high- and low-risk subgroups.
Moreover, the study investigated the physiological functions of DEGs in these susceptible subgroups. The analysis showed that DEGs in biological processes were mainly associated with the synthesis of immune response molecules, production of immunoglobulins, and humoral immune responses. Molecular functions of these DEGs included signaling receptor activation, receptor-ligand interactions, and cytokine receptor binding, emphasizing their involvement in crucial cellular signaling complexes related to T cell receptor function and immune regulation. (Fig. 6C, D).
Correlation of the CAFRSig with immune landscape in CESC
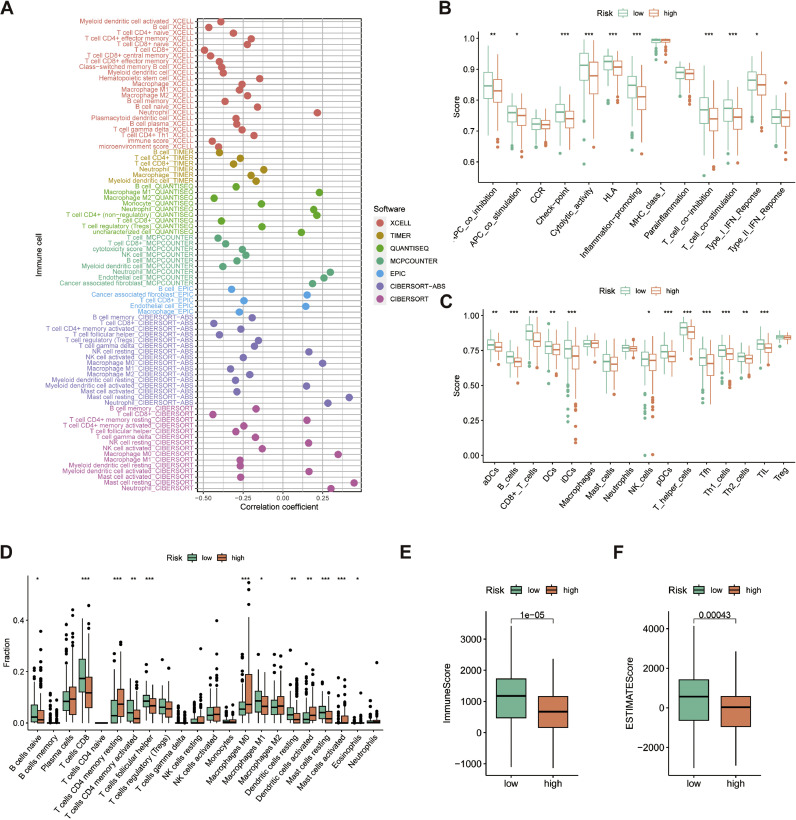
Building on the discovery of DEGs related to immune functions in GO analysis, the study proceeded to assess the connection between CAFRSig and the Tumor Immune Microenvironment (TIME) in the TCGA-CESC cohort. Using the TIMER platform, a positive correlation was found between most immune cells, including key effector cells, and CAFRSig risk scores (Fig. 7A). Furthermore, ssGSEA results revealed heightened immune-related activities in the low-risk category, such as APC co-stimulation, cytolytic function, immune checkpoints, human leukocyte antigens, processes that enhance inflammation, T cell co-stimulation and co-inhibition, and type I IFN response (Fig. 7B). The subset showed a notable presence of various immune cells, such as dendritic cells (both activated and immature), CD8+ T cells, B cells, natural killer cells, follicular helper T cells, and helper T cells. Additionally, there was an elevated presence of tumor-infiltrating lymphocytes (Fig. 7C). To further substantiate the link between the CAFRSig and TIME, CIBERSORT analysis was performed. The results of the analysis revealed a significantly higher percentage of CD8T cells in the low-risk group compared to the high-risk group, as illustrated in Fig. 7D. The examination of the ESTIMATE algorithm confirmed the existence of increased ESTIMATE and immune scores in the low-risk category (Fig. 7E-F).
Fig. 7.
Association of the CAFRSig with the immune microenvironment in the TCGA-CESC cohort. (A) Bubble plot of correlation coefficients between immune cells and risk scores. (B) Box plot showing differences in immune-related functions between the high- and low-risk groups using the ssGSEA algorithm. (C) Box plot showing differences in immune cells between the high- and low-risk groups using the ssGSEA algorithm. (D) CIBERSORT algorithm analysis of immune cell differences between high- and low-risk groups. (E-F) Immune score and ESTIMATE score in the two risk subgroups. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. NC.
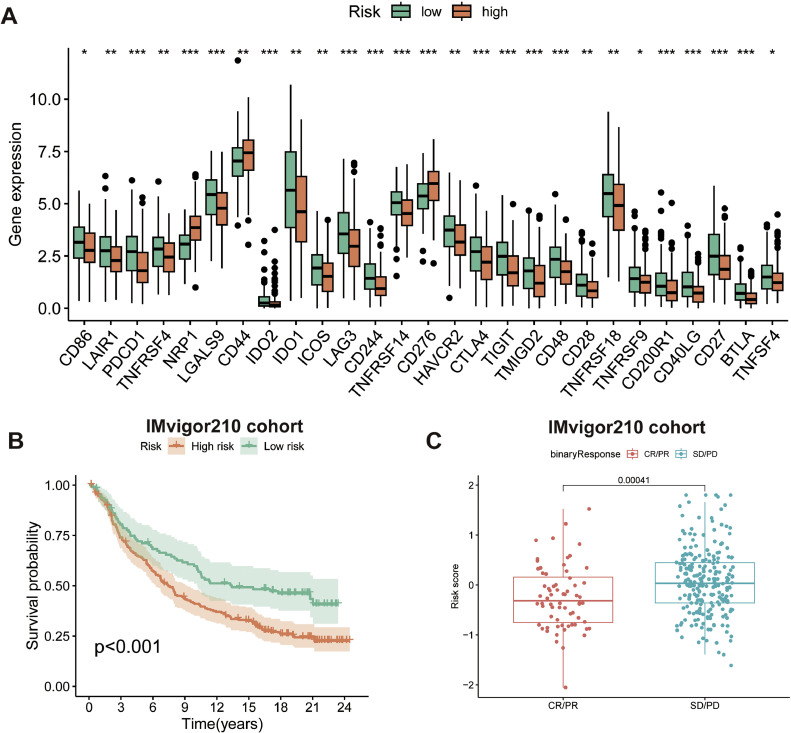
Correlation analysis of the CAFRSig with immunotherapy response
Additionally, the investigation examined the correlation between CAFRSig and the effectiveness of immune checkpoint blockade (ICB) treatments in CC. Analysis indicated that immune checkpoints were more highly expressed in the CAFRSig low-risk subgroup (Fig. 8A), suggesting potential responsiveness to ICB treatments in these patients. To confirm this, participants in the IMvigor210 immunotherapy cohort were categorized as high or low risk based on CAFRSig. Fig. 8B demonstrated that the low-risk cohort achieved superior survival results when administered with ICBs in comparison to the high-risk cohort. In addition, individuals who experienced a complete or partial response after receiving ICB treatment had considerably lower risk scores compared to those who had stable or progressive disease (Fig. 8C).
Fig. 8.
Correlation analysis of the CAFRSig with immunotherapy response. (A) Differences in the expression of immune checkpoints across the risk subgroups. (B) Kaplan-Meier curves for the high and low-risk groups of the IMvigor210 cohort. (C) Differences in risk scores between patients with CR/PR and patients with SD/PD after treatment with ICIs.
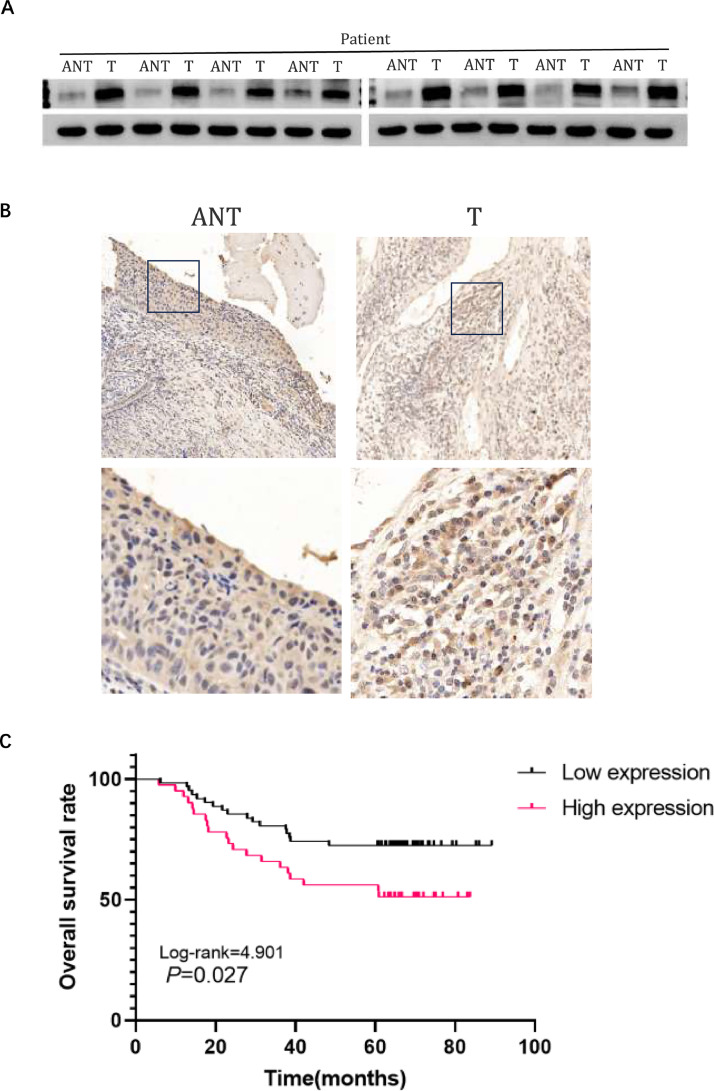
NRP1 is upregulated in CC tissues
Analysis of 8 paired cervical cancer (T) and neighboring noncancerous tissues (ANT) showed a rise in NRP1 protein levels in the cancerous specimens (Fig. 9A). This was consistent with IHC analysis results, confirming NRP1′s upregulation in cervical CC tissues (Fig. 9B).
Fig. 9.
NRP1 Upregulation in Cervical Cancer Tissues Correlates with Poor Prognosis. (A) Representative Western blots of NRP1 protein expression in 8 matched pairs of cervical cancer tissues (T) and adjacent noncancerous tissues (ANT); (B) Immunohistochemical (IHC) assay of NRP1 protein expression in matched cervical cancer tissues. (C) Kaplan–Meier analysis of CC patient survival rates in relation to NRP1 protein expression.
Clinicopathological significance of NRP1 overexpression and high NRP1 expression indicates poor prognosis in CC
To assess the link between NRP1 protein levels and the clinicopathological characteristics of cervical cancer, immunohistochemical analysis was performed on 103 cervical squamous cell carcinoma (SCC) samples(Table 2). This set included FIGO stage IB (59 cases, 38.1%) and stage IIA1 (44 cases, 28.5%) diseases. Notably, 41 samples (39.8%) exhibited high NRP1 protein expression, in contrast to the 62 samples (60.2%) that showed weak or negative staining. Table 2 summarizes that high NRP1 protein expression tends to strongly correlate with clinical stage and differentiation. Specifically, the rate of strong NRP1 protein positivity was significantly higher in stage IIA cervical SCCs (52.3%, 23/44) compared to stage IB (43.2%, 19/44) (P = 0.004). Similarly, high rates of NRP1 positivity were observed in poorly differentiated cervical SCCs (51.2%, 21/41) and moderately differentiated cervical SCCs (29.3%, 12/41), significantly surpassing those in well-differentiated cases (19.5%, 8/41) (P = 0.019). Nevertheless, there was no notable association observed between NRP1 expression and patient age, HPV infection, squamous cell carcinoma antigen, or tumor size. The importance of NRP1 in clinical settings was emphasized by the Kaplan-Meier analysis of OS in 103 cases of cervical SCC, showing that patients with high NRP1 expression had a lower OS compared to those with low expression (P = 0.027) (Fig. 9C).
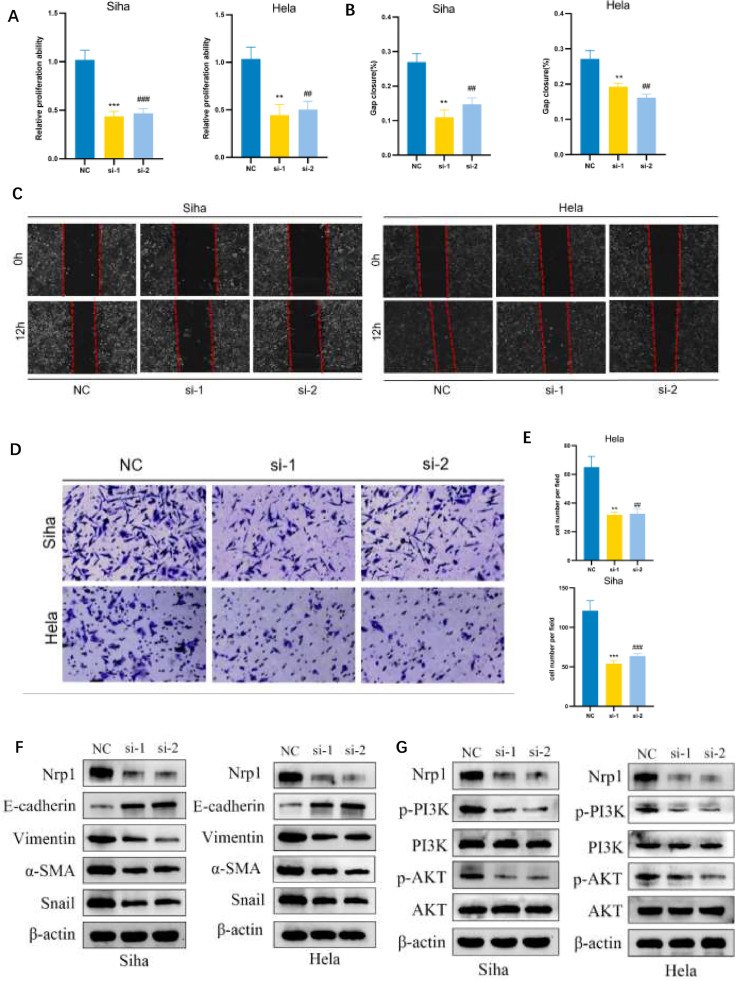
NRP1 promotes CC progression by modulating EMT and PI3K/AKT pathway activation
In our study, the CKK8 assay results revealed that NRP1 overexpression notably diminished the proliferation of CC cells (Fig. 10A). This effect was complemented by a significant increase in cell migration, as evidenced in transwell and wound-healing assays (Fig. 10B-E). Significantly, blocking NRP1 led to an increase in E-cadherin expression and a simultaneous decrease in α-SMA, Snail, and Vimentin levels, which are important markers in the epithelial–mesenchymal transition (EMT) process (Fig. 10F). These findings indicate a pivotal role of NRP1 in modulating EMT, potentially altering the typical metastatic pathway in CC cells. Furthermore, our examination revealed a decrease in levels of phosphorylated AKT and PI3K in cells where NRP1 was knocked down in comparison to the control group (Fig. 10G), suggesting that the regulatory effect of NRP1 on EMT may be mediated through the PI3K/AKT signaling pathway.
Fig. 10.
NRP1 Promotes CC Progression by Modulating EMT and PI3K/AKT Pathway Activation. (A) CCK8 assay reveals that NRP1 depletion attenuates cell viability in Siha and Hela cells. (B-C) Wound-healing assay and corresponding statistical chart showing that NRP1 depletion inhibits cell migration in Siha and Hela cells. (D-E) Transwell assay and corresponding statistical chart demonstrating that NRP1 depletion inhibits cell migration in Siha and Hela cells. (F) Immunoblot assay showing EMT-related proteins in Siha and Hela cells after NRP1 depletion. (G) Immunoblot assay showing PI3K/AKT pathway-related proteins in Siha and Hela cells after NRP1 depletion. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. NC. #P < 0.05, ##P < 0.01, and ###P < 0.001 vs. NC.
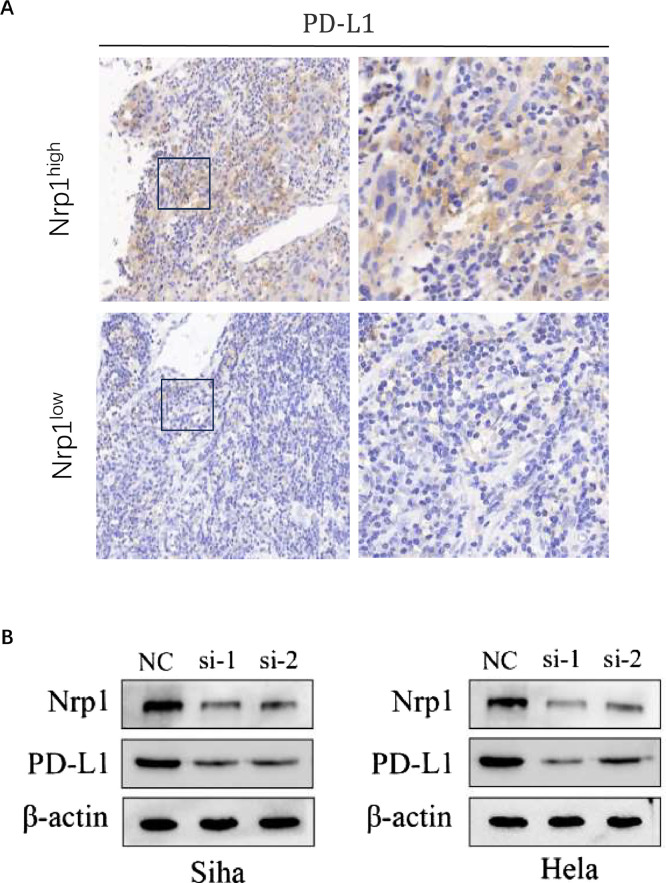
NRP1 induces PD-L1 expression in CC
IHC analysis of samples from 103 cervical SCC patients revealed a significant positive correlation between NRP1 and PD-L1 expression (r = 0.195, P = 0.014) (Fig. 11A, Table 3). Subsequent studies on NRP1-suppressed cells showed a decrease in PD-L1 levels compared to the control group (Fig. 11B). This data implies that NRP1 may play a role in modulating PD-L1 expression within CC, potentially engaging in the tumor's immune evasion strategies.
Fig. 11.
NRP1 induces PD-L1 expression in CC. (A) IHC assay of PD-L1 protein expression in cervical cancer tissues with high and low NRP1 expression. (B) Western blot assay demonstrating PD-L1 protein levels in Siha and Hela cells following NRP1 depletion.
Table 3.
Correlation between NRP1 and PD-L1 expression of CC.
| PD-L1 |
NRP1 |
χ2 | p-value | r | p-value | |
|---|---|---|---|---|---|---|
| Negative | Positive | |||||
| Negative | 50 | 30 | 6.057 | 0.014 | 0.195 | 0.014 |
| Positive | 12 | 11 | ||||
Discussion
Emerging research highlights the significant role of CAFs in CC progression. Yugawa et al. [29] revealed that CAFs may promote CC proliferation by disrupting hsa-miR-324–3p through circZNF208. Sheng and his team also found that cancer-related fibroblasts can activate M2 polarized macrophages, increasing CC's resistance to radiation therapy through the CCL2 pathway [30].
However, most existing studies focus on individual CAF-associated genetic factors in CC, leaving the collective impact of numerous CAF-associated genes less understood [31,32]. This study addresses this by evaluating the roles of CAF-related genes in CC progression, potentially refining therapeutic strategies. This is the first exploration into the prognostic significance of these genes in CC patients.
Using gene sets from the TCGA database, a predictive model was constructed using univariate Cox and LASSO regression, followed by multivariate analysis to confirm their prognostic value. This model, based on 11 CAF-related genes, effectively predicts overall survival (OS) in CC patients, with validation from TCGA-CESC and GSE44001 databases indicating that individuals with lower risk scores show significantly better OS outcomes. Enrichment analysis underscored the influence of CAFs on tumor malignancy-related pathways, involving key pathways such as Wnt, TGF-beta, Notch, mTOR, and ERBB signaling pathways, and functional pathways related to fatty acid metabolism and immune response were also closely associated with CAF and immune landscape.
Additionally, the study explored the correlation between the CAFRSig and the immune landscape, identifying a positive association between immune cell functions and risk scores. The study analyzed the correlation between immune therapy response and risk scores, revealing that patients with low-risk scores who received ICBs treatment experienced improved survival outcomes. The prognosis model was capable of identifying patients relatively more sensitive to ICBs treatment, thereby improving ICBs therapy effectiveness and prognosis.
In our study, using the LASSO algorithm to identify 11 hub genes for CAFRSig development, the four genes with higher risk scores were TNF, IFNG, NRP1, and SERPINF1. Notably, TNF and IFNG are recognized for their inflammatory roles and their involvement in oncogenesis. SERPINF1, also known as Pigment Epithelium-Derived Factor, is noted for its anti-angiogenic properties and its role in nerve growth, which are vital for cancer inhibition and nerve tissue support [[32], [33], [34], [35], [36]]. Research by Murata et al. [37] highlights the synergistic effects of HB-EGF and PDGF in promoting cancer cell growth via paracrine interaction with surrounding fibroblasts in CC. Neuropilins (NRPs), which are transmembrane glycoprotein receptors, play a diverse role in binding with semaphorins and different members of the vascular endothelial growth factor family. NRP1 encodes a specific NRP, involved in axon guidance and angiogenesis [38,39]. High expression of NRP1 has been observed in various tumor types, including neuroblastoma and cancers of the ovary, stomach, pancreas, lung, prostate, breast, and colon [[40], [41], [42], [43], [44], [45]]. However, the mechanisms by which NRP1 influences cervical cancer progression remain to be fully elucidated. Therefore, in subsequent validation experiments, we investigated NRP1′s role and underlying mechanisms in cervical cancer, confirming its upregulation in earlier stages (IB-IIA) and strong correlation with clinical severity and differentiation. These findings underscore NRP1′s potential as a biomarker for aggressive cervical cancer. Experiments showed NRP1 suppression reduces cell proliferation and may reverse the epithelial-mesenchymal transition, influencing cell mobility through the PI3K/Akt pathway. Further, a significant correlation between NRP1 and PD-L1 suggests NRP1′s involvement in tumor immune evasion, highlighting its dual role as a predictive and therapeutic target in cervical cancer progression.
However, this study has limitations, including an incomplete understanding of the mechanistic roles of CAF-related genes, necessitating additional research to deepen our knowledge. Further clinical trials are essential to validate the prognostic model based on these 11 CAF-related genes. The reliance on public databases also raises the possibility of selection bias in the study's findings. Additionally, although we validated the constructed signature in the IMvigor210 immunotherapy cohort, there is still a lack of real-world validation of the CESC immunotherapy cohort. We will further validate the predictive ability of the signature in future prospective studies.
Conclusion
This study investigated the prognostic role of CAFs in CC, identifying key associated genes. A risk score model, based on these genes, was developed to improve both prognostic accuracy and immunotherapy response prediction in CC, potentially advancing personalized treatment approaches. Additionally, it demonstrates that NRP1, by modulating EMT and influencing the PI3K/AKT pathway, contributes to CC progression and is associated with poor prognosis and immune evasion.
Funding
Funding for this research was provided by the Anhui Medical University Postgraduate Innovation Research and Practice Program (No.YJS20230019).
Data availability
All original data in this research are available upon reasonable request from the corresponding authors.
Approval of ethics and agreement to participate
The procedures of this study were approved by the Ethics Committee at the First Affiliated Hospital of Anhui Medical University, in accordance with the principles outlined in the Declaration of Helsinki.Due to the study being retrospective, explicit written consent from patients was not required, and all patient data was kept confidential.
CRediT authorship contribution statement
Zhongxuan Gui: Writing – original draft, Methodology. Yingquan Ye: Validation. Yu Li: Software. Zhengting Ren: Visualization. Nan Wei: Writing – original draft. Li Liu: Writing – original draft. Hua Wang: Writing – review & editing. Mei Zhang: Writing – review & editing, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Hua Wang, Email: qq296901523@163.com.
Mei Zhang, Email: zhangmei@ahmu.edu.cn.
References
- 1.Sung H., Ferlay J., Siegel R.L. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Li H., Wu X. Advances in diagnosis and treatment of metastatic cervical cancer. 2016; 27(4): e43. [DOI] [PMC free article] [PubMed]
- 3.Arbyn M., Weiderpass E., Bruni L., de Sanjosé S., Saraiya M., Ferlay J. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob. Health. 2020;8(2):e191–e203. doi: 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen P.A., Jhingran A., Oaknin A., Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 5.Canfell K., Kim J.J., Brisson M., Keane A., Simms K.T., Caruana M. Mortality impact of achieving WHO cervical cancer elimination targets: a comparative modelling analysis in 78 low-income and lower-middle-income countries. Lancet. 2020;395(10224):591–603. doi: 10.1016/S0140-6736(20)30157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kossaï M., Leary A., Scoazec J.Y., Genestie C. Ovarian cancer: a heterogeneous disease. Pathobiology. 2018;85(1–2):41–49. doi: 10.1159/000479006. [DOI] [PubMed] [Google Scholar]
- 7.Rojas V., Hirshfield K.M., Ganesan S., Rodriguez-Rodriguez L. Molecular characterization of epithelial ovarian cancer: implications for diagnosis and treatment. Int. J. Mol. Sci. 2016;17(12) doi: 10.3390/ijms17122113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baghban R., Roshangar L., Jahanban-Esfahlan R., Seidi K., Ebrahimi-Kalan A., Jaymand M. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020;18(1):59. doi: 10.1186/s12964-020-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X., Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019;18(2):99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 10.Sahai E., Astsaturov I. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer. 2020;20(3):174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiori M.E., Di Franco S., Villanova L., Bianca P., Stassi G., De Maria R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol. Cancer. 2019;18(1):70. doi: 10.1186/s12943-019-0994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra Jena B., Kanta Das C., Banerjee I., Das S., Bharadwaj D., Majumder R. Paracrine TGF-β1 from breast cancer contributes to chemoresistance in cancer associated fibroblasts via upregulation of the p44/42 MAPK signaling pathway. Biochem. Pharmacol. 2021;186 doi: 10.1016/j.bcp.2021.114474. [DOI] [PubMed] [Google Scholar]
- 13.Strong A.L., Pei D.T., Hurst C.G., Gimble J.M., Burow M.E., Bunnell B.A. Obesity enhances the conversion of adipose-derived stromal/stem cells into carcinoma-associated fibroblast leading to cancer cell proliferation and progression to an invasive phenotype. Stem Cells Int. 2017;2017 doi: 10.1155/2017/9216502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raz Y., Cohen N., Shani O., Bell R.E., Novitskiy S.V., Abramovitz L. Bone marrow-derived fibroblasts are a functionally distinct stromal cell population in breast cancer. J. Exp. Med. 2018;215(12):3075–3093. doi: 10.1084/jem.20180818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeon J.H., Jeong H.E., Seo H., Cho S., Kim K., Na D. Cancer-derived exosomes trigger endothelial to mesenchymal transition followed by the induction of cancer-associated fibroblasts. Acta Biomater. 2018;76:146–153. doi: 10.1016/j.actbio.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Zheng H., Liu H., Ge Y., Wang X. Integrated single-cell and bulk RNA sequencing analysis identifies a cancer associated fibroblast-related signature for predicting prognosis and therapeutic responses in colorectal cancer. Cancer Cell Int. 2021;21(1):552. doi: 10.1186/s12935-021-02252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye Y., Zhang S., Jiang Y., Huang Y., Wang G., Zhang M. Identification of a cancer associated fibroblasts-related index to predict prognosis and immune landscape in ovarian cancer. Sci. Rep. 2023;13(1):21565. doi: 10.1038/s41598-023-48653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Meng H., Nie S., Sun Y., Jiang P., Li S. Identification of a prognostic signature of epithelial ovarian cancer based on tumor immune microenvironment exploration. Genomics. 2020;112(6):4827–4841. doi: 10.1016/j.ygeno.2020.08.027. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Geng R., Ni S., Cai L., Yang S., Shao F. Pyroptosis-related lncRNAs are potential biomarkers for predicting prognoses and immune responses in patients with UCEC. Mol. Ther. Nucleic Acids. 2022;27:1036–1055. doi: 10.1016/j.omtn.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J., Chen C., Wang Y., Qian C., Wei J., Xing Y. Comprehensive of N1-methyladenosine modifications patterns and immunological characteristics in ovarian cancer. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.746647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y., Zhao Q., Wu Y., Wang G., Huang Y., Sun W. Construction of a cancer-associated fibroblasts-related long non-coding RNA signature to predict prognosis and immune landscape in pancreatic adenocarcinoma. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.989719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucl. Acids Res. 2020;48(W1):W509–w514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman A.M., Liu C.L., Green M.R. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi M., Niu M., Xu L., Luo S., Wu K. Regulation of PD-L1 expression in the tumor microenvironment. 2021; 14(1): 10. [DOI] [PMC free article] [PubMed]
- 29.Sun W., Hang D., Han S., Fu S., Ma H., Dong X. Construction of circRNA-associated ceRNA network reveals the regulation of fibroblast proliferation in cervical cancer. Gene. 2022;844 doi: 10.1016/j.gene.2022.146824. [DOI] [PubMed] [Google Scholar]
- 30.Sheng Y., Zhang B., Xing B., Liu Z., Chang Y., Wu G. Cancer-associated fibroblasts exposed to high-dose ionizing radiation promote M2 polarization of macrophages, which induce radiosensitivity in cervical cancer. Cancers (Basel) 2023;15(5) doi: 10.3390/cancers15051620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ou Z., Lin S., Qiu J., Ding W., Ren P., Chen D., et al. Single-nucleus RNA sequencing and spatial transcriptomics reveal the immunological microenvironment of cervical squamous cell carcinoma. 2022; 9(29): e2203040. [DOI] [PMC free article] [PubMed]
- 32.Belkacemi L., Zhang S.X. Anti-tumor effects of pigment epithelium-derived factor (PEDF): implication for cancer therapy. A mini-review. J. Exp. Clin. Cancer Res. 2016;35:4. doi: 10.1186/s13046-015-0278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simonovic M., Gettins P.G., Volz K. Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor. Proc. Natl. Acad. Sci. U S A. 2001;98(20):11131–11135. doi: 10.1073/pnas.211268598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagdadi N., Sawaied A., AbuMadighem A. The expression levels and cellular localization of pigment epithelium derived factor (PEDF) in mouse testis: its possible involvement in the differentiation of spermatogonial cells. Int. J. Mol. Sci. 2021;22(3) doi: 10.3390/ijms22031147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robl B., Botter S.M., Boro A., Meier D., Neri D., Fuchs B. Evaluation of F8-TNF-α in models of early and progressive metastatic osteosarcoma. Transl. Oncol. 2017;10(3):419–430. doi: 10.1016/j.tranon.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye Z.H., Jiang X.M., Huang M.Y., Xu Y.L., Chen Y.C., Yuan L.W. Regulation of CD47 expression by interferon-gamma in cancer cells. Transl. Oncol. 2021;14(9) doi: 10.1016/j.tranon.2021.101162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murata T., Mizushima H., Chinen I., Moribe H., Yagi S., Hoffman R.M. HB-EGF and PDGF mediate reciprocal interactions of carcinoma cells with cancer-associated fibroblasts to support progression of uterine cervical cancers. Cancer Res. 2011;71(21):6633–6642. doi: 10.1158/0008-5472.CAN-11-0034. [DOI] [PubMed] [Google Scholar]
- 38.Geretti E., Shimizu A., Klagsbrun M. Neuropilin structure governs VEGF and semaphorin binding and regulates angiogenesis. Angiogenesis. 2008;11(1):31–39. doi: 10.1007/s10456-008-9097-1. [DOI] [PubMed] [Google Scholar]
- 39.Plein A., Fantin A., Ruhrberg C. Neuropilin regulation of angiogenesis, arteriogenesis, and vascular permeability. Microcirculation. 2014;21(4):315–323. doi: 10.1111/micc.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu C., Somasundaram A., Manne S., Gocher A.M., Szymczak-Workman A.L., Vignali K.M., et al. Neuropilin-1 is a T cell memory checkpoint limiting long-term antitumor immunity. 2020; 21(9): 1010–1021. [DOI] [PMC free article] [PubMed]
- 41.Huang X., Ye Q., Chen M., Li A., Mi W., Fang Y., et al. N-glycosylation-defective splice variants of neuropilin-1 promote metastasis by activating endosomal signals. 2019; 10(1): 3708. [DOI] [PMC free article] [PubMed]
- 42.Xin Y., Li J., Wu J., Kinard R., Weekes C.D., Patnaik A. Pharmacokinetic and pharmacodynamic analysis of circulating biomarkers of anti-NRP1, a novel antiangiogenesis agent, in two phase I trials in patients with advanced solid tumors. Clin. Cancer Res. 2012;18(21):6040–6048. doi: 10.1158/1078-0432.CCR-12-1652. [DOI] [PubMed] [Google Scholar]
- 43.Vescarelli E., Gerini G., Megiorni F., Anastasiadou E., Pontecorvi P., Solito L., et al. MiR-200c sensitizes Olaparib-resistant ovarian cancer cells by targeting Neuropilin 1. 2020; 39(1): 3. [DOI] [PMC free article] [PubMed]
- 44.Chuckran C.A., Liu C., Bruno T.C., Workman C.J., Vignali D.A. Neuropilin-1: a checkpoint target with unique implications for cancer immunology and immunotherapy. J. Immunother. Cancer. 2020;8(2) doi: 10.1136/jitc-2020-000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mei B., Chen J., Yang N., Peng Y. The regulatory mechanism and biological significance of the Snail-miR590-VEGFR-NRP1 axis in the angiogenesis, growth and metastasis of gastric cancer. Cell Death Dis. 2020;11(4):241. doi: 10.1038/s41419-020-2428-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All original data in this research are available upon reasonable request from the corresponding authors.