Abstract
Replacement therapy for the salivary gland (SG) remains an unmet clinical need. Xerostomia (“dry mouth”) due to hyposalivation can result from injury or disease to the SG, such as salivary acinar death caused by radiation therapy (RT) for head and neck squamous cell carcinoma (HNSCC). Currently, only palliative treatments exist for xerostomia, and many patients endure deteriorated oral health and poor quality of life. Tissue engineering could offer a permanent solution for SG replacement by isolating healthy SG tissues prior to RT, expanding its cells in vitro, and recreating a functional salivary neogland for implantation post-RT. 3D bioprinting methods potentiate spatial cell deposition into defined hydrogel-based architectures, mimicking the thin epithelia developed during the complex branching morphogenesis of SG. By leveraging a microfluidics-based bioprinter with coaxial polymer and crosslinker streams, we fabricated thin, biocompatible, and reproducible hydrogel features that recapitulate the thin epithelia characteristics of SG. This flexible platform enabled two modes of printing: we produced solid hydrogel fibers, with diameters <100 μm, that could be rastered to create larger mm-scale structures. By a second method, we generated hollow tubes with wall thicknesses ranging 45–80 μm, total tube diameters spanning 0.6 – 2.2 mm, and confirmed tube patency. In both cases, SG cells could be printed within the thin hydrogel features, with preserved phenotype and high viability, even at high density (5.0 × 106 cells/mL). Our work demonstrates hydrogel feature control across multiple length scales, and a new paradigm for addressing SG restoration by creating microscale tissue engineered components.
Keywords: 3D bioprinting, salivary gland, tissue engineering, biomaterials, hydrogels, sodium alginate
1. Introduction
The salivary glands (SGs) are integral functional components of the oral cavity. Through their secretion of saliva, and its associated functions of lubrication, enzymatic digestion of food, and bacteriostasis, the SGs enable proper speech, digestion, and dental health. SG dysfunction accordingly impacts these aspects of daily living, and can be initiated by multiple factors, including aging, pharmaceutical side-effects, auto-immune conditions (e.g. Sjogren’s syndrome), and damage from common anti-cancer therapies [1–3]. As one example, head and neck squamous cell carcinoma (HNSCC) comprises a broad constellation of soft tissue-based tumors that arise within the oral cavity, nasal cavity, and the pharyngeal regions [4]. Radiation therapy (RT) is a preferred treatment for improving overall survival and preserving adjacent tissues, however, the nearby SG often are damaged significantly and permanently during that process, resulting in significant and persistent loss of salivary structures and function [5].
This RT-induced damage to the SG quickly leads to hyposalivation and the resultant perception of xerostomia (dry mouth). These dramatically impact oral health through the loss of salivary function, and current treatments are palliative [5], providing temporary relief at best. Given the >60,000 new HNSCC cases diagnosed in the U.S. annually [6], xerostomia is an increasingly relevant health concern with no effective treatment, and thus new methods are needed to either preserve or restore SG function. For these patients, a biopsy of healthy SG tissue, prior to RT, could enable the ex vivo development, and subsequent autologous re-implantation, of a tissue-engineered substitute to restore salivary function [7].
As a highly branched and secretory structure [8], the SGs represent challenging targets for replacement. SGs comprise multiple differentiated cell types, forming epithelial ducts and acini, surrounded by supportive myoepithelial cells, mesenchyme, endothelium, and nerve [9]. Many current paradigms for SG engineering employ SG-derived epithelial cells, encapsulated within a soft, supportive hydrogel matrix, or similar synthetic matrix mimic.[10–16] We have described extensively [17–24] the three-dimensional (3D) culture of primary human stem/progenitor cells (hS/PCs) as well-defined multicellular clusters within hyaluronic acid (HA) hydrogels that mimic the native connective tissue in which SGs form. By modifying gel porosity and biologically relevant tethered peptide signals, we can impact the cell-matrix interface, affecting the morphology of cluster peripheries, the distribution of activated integrins, and the deposition of basement membrane (BM) proteins around each cluster. However, spatial control is lacking in these bulk 3D culture methods, particularly when targeting the branched architecture and length scale of native SGs. In particular, the fine epithelial layers of the ducts and acini are exceptionally thin (1–2 cells thick, ~ 30–50 μm with associated matrix) [25] and challenging to recreate by synthetic means.
Improvements in biomanufacturing methods, such as bioprinting, have accelerated dramatically over the past decade, aiming toward precision fabrication of tissues and organs [26]. Bioprinting methods often exploit additive deposition of cells within pre-polymer solutions, via syringe extrusion, inkjet-style printing, or similar means, to enable cell deposition within soft, but structurally secure, hydrogel geometries. These methods enable blunt reassembly of tissue-like structures, but all strain to reach a practical filament resolution limit of ~100 μm, well beyond single-cell size [27]. At these small diameters, extrusion of thick polymer solutions through metal syringe needles can induce damaging shear stresses. Thus, for complex epithelial structures, with small cell size and monolayer-scale features, bioprinting faces innate restrictions: standard methods do not allow localized spatial deposition near single-cell resolution, nor do they permit controlled growth, after gelation, toward complex secondary structures.
In contrast, microfluidics-based bioprinting offers exceptional control over both spatial and temporal deposition of biomaterials, with reduced shear stress compared to other extrusion-based printing methods [27,28]. Such systems enable exquisite control over fluid flow, with fast switching among multiple inputs, control over mixing, and sub-microliter precision.[29] Additionally, coaxial microfluidics (CMF) enables concentric variations of inputs, with precise core-shell morphologies. Rapid crosslinking polymers, such as sodium alginate, an anionic polysaccharide obtained from seaweed, are commonly used for microfluidics-based bioprinting. Alginate has been one of the most extensively used materials due to its immediate crosslinking with divalent cations such as calcium or barium [30–32]. Microfiber or microtube structures made of alginate-based hydrogel have been developed for tissue engineering applications, such as stem cell expansion and lineage differentiation. Lee et al described early applications of microfluidic devices for alginate microfibers as a carrier of human fibroblasts and therapeutic materials,[33] as well as for vasculature.[34] Onoe et al studied extended CMF microfibers across multiple tissue types.[35] More recently, Abelseth et al used an RX1 modular bioprinter to generate neural tissues derived from human induced pluripotent stem cells using a fibrin-alginate bioink.[36] Within the SG field, Jorgensen et al made alginate microtubes with a needle-in-needle method, to co-culture mouse SG ductal epithelial cells with NIH 3T3 fibroblasts in an epithelial-stromal model of organization and interaction. [16]
In the present work, we demonstrate the utility of a CMF-based bioprinter, in printing hydrogel structures with variable dimensions that uniquely approximate the size and characteristics of salivary epithelia. The selected printheads enabled the generation of (1) solid alginate fibers that can be used to delineate the branching structure of SG; and (2) hollow alginate tubes, with a sacrificial liquid core and extremely thin hydrogel walls within the desired dimensions of salivary epithelial layers. Moreover, the ability to print cells across multiple densities within the hydrogel tubes permitted the fine tuning of the cellular parameters for obtaining the most viable system for 3D cell culture.
2. Results
2.1. Inlet solution pressures dictate fiber dimensions
The dimensions of mature SG structures serve as a template for targeting feature sizes in bioprinted structures (Figure 1(a) [37]). Optimally, a CMF printer would access features <100 μm, which are closest to the feature sizes of a native gland. As shown in Figure 1(b), two inlet solutions, Material (i.e., the 1.5% (w/v) alginate polymer solution) and Crosslinker (i.e., 125 mM CaCl2 solution), are aligned coaxially, to enable the printing of solid hydrogel fibers. Adjusting the pressures of each inlet allows for an array of accessible diameters. To explore the relationship between hydrogel fiber diameter and fluid pressures, first, we kept crosslinker pressures constant and varied Material inlet pressures (Figure 2(a)); then, we held material pressures constant and tested a range of crosslinker pressures (Figure 2(b)). The dispensed hydrogel fibers were stable, with no kinking or spiraling, and modest deviation among repeat measures (Figure 2(c)–(d)).
Figure 1. Physiological inspiration of bioprinting salivary gland ductal and epithelial features.
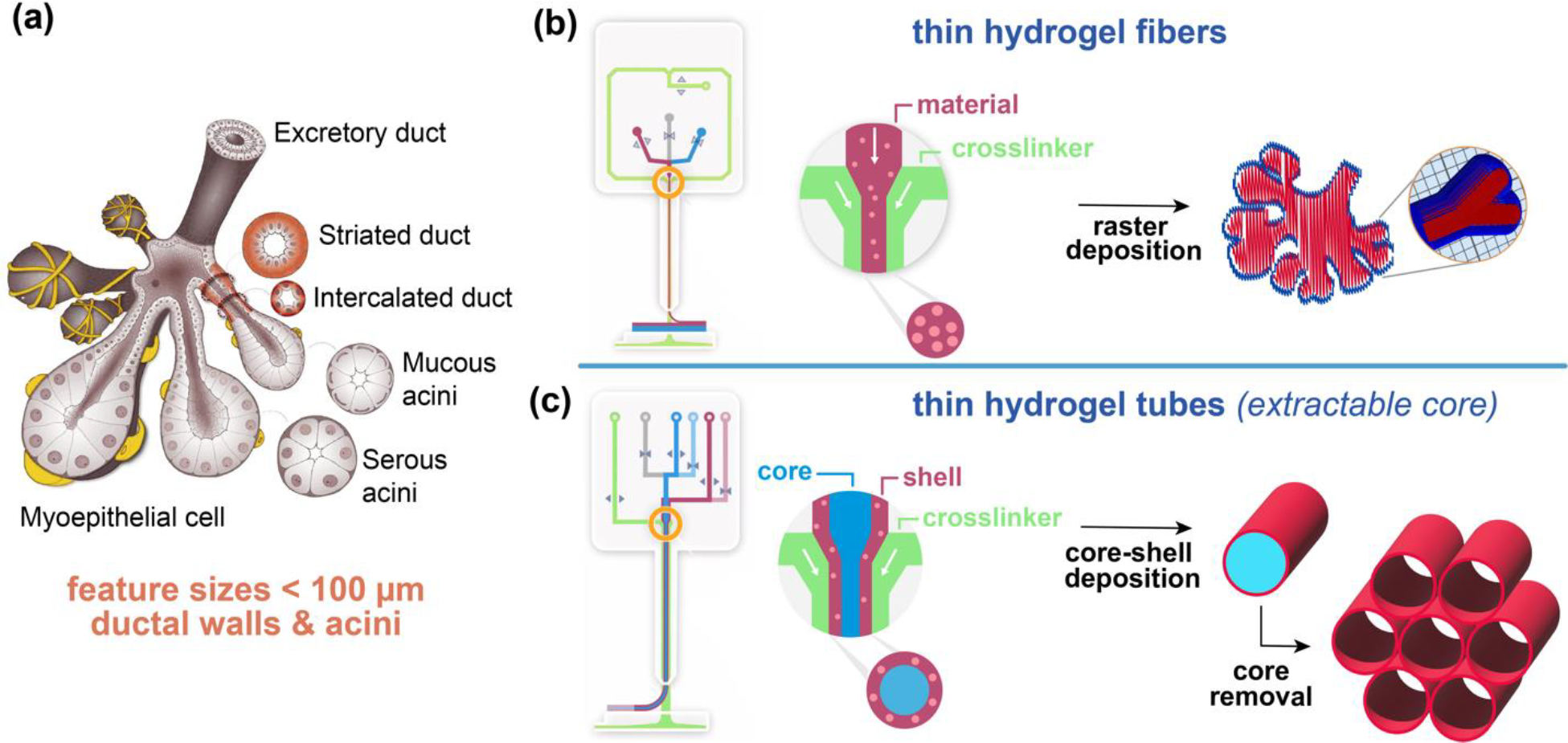
(a) Scheme of fully developed salivary gland (adopted from Ref [37]). (b) Scheme of DUO Printhead of RX1 bioprinter and the raster deposition method using solid fibers to print biomimetic structures. (c) Scheme of CENTRA Printhead of RX1 bioprinter and a core-shell deposition method to produce hollow tubes with thin walls.
Figure 2. Hydrogel fiber diameter can be controlled within a broad range by adjusting input pressures.
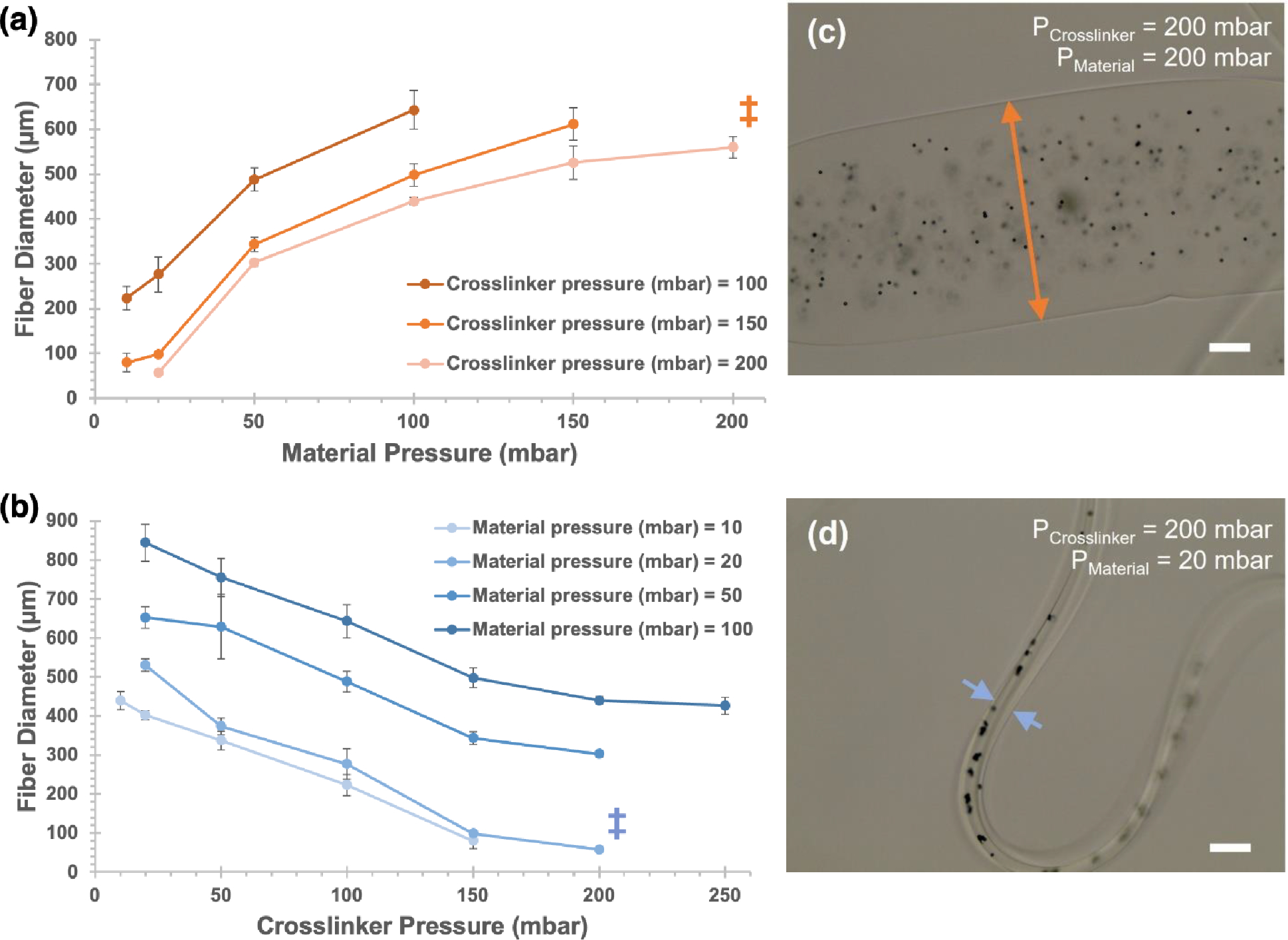
1.5% (w/v) alginate solutions were printed as hydrogel fibers, using 125 mM CaCl2 solution as a crosslinking agent. (a) For crosslinker pressures held constant at 100–200 mbar, varying material pressures yielded hydrogel fibers with diameters ranging from 57 μm to 642 μm. (b) For material pressures held constant at 10–100 mbar, varying crosslinker pressures yielded hydrogel fibers with diameters from 57 μm to 844 μm. (c) and (d) show representative brightfield micrographs of fibers, at pressure combinations indicated by ‡ (scale bar = 100 μm). Arrowheads show representative fiber widths at each condition. Opaque microbeads (ø = 6 μm) were added for visualization. Error bars indicate +/− standard deviation for n=3 measures.
2.2. Inlet solution pressures dictate tube dimensions, and core solution concentration influences tube diameter, but not wall thickness
As shown in Figure 1(c), the addition of a third inlet solution enables the printing of hydrogel tubes with thin walls and a comparably large interior diameter. A dilute aqueous solution of poly(vinyl alcohol) (PVA) was used in this third inlet, which coaxially prints as a sacrificial “core” that is readily cleared after printing to leave a hollow center. To explore the relationship between the inlet pressures and the resulting wall thickness of the hydrogel tubes, first, we maintained constant Shell (200 mbar) and Crosslinker (50 mbar) pressures and varied the Core inlet pressure (150 – 250 mbar) (Figure 3(a)). Then, we maintained constant Core (200 mbar) and Shell (200 mbar) pressures, and varied Crosslinker pressure (25 – 125 mbar) (Figure 3(b)). Lastly, we configured the Core (200 mbar) and Crosslinker (50 mbar) pressures to be held constant and altered the Shell pressure (150 – 250 mbar) (Figure 3(c)). After printing and clearance of the PVA solution, we incubated the tubes with a dilute aqueous solution of 2MDa FITC-dextran, which provided high contrast and facile assessment of tube edges (Figure 3(d)). The pressures shown in Figure 3(a)–(c) represent the ranges that produced stable tubes. For printing with 6% (w/v) PVA as a core solution, the relationships shown in Figure 3(a)–3(c) were observed for variations in each solution pressure: average wall thickness decreased with increasing core solution pressure, increased with increasing shell solution pressure, and changed minimally or decreased with increasing crosslinker solution pressure. In all cases, the average wall thickness spanned ~ 45 – 80 μm, within the desired range to mimic salivary epithelia. Inner diameters of the printed tubes measured ~ 400 μm.
Figure 3. Shell pressure defines wall thickness for printed tubes.
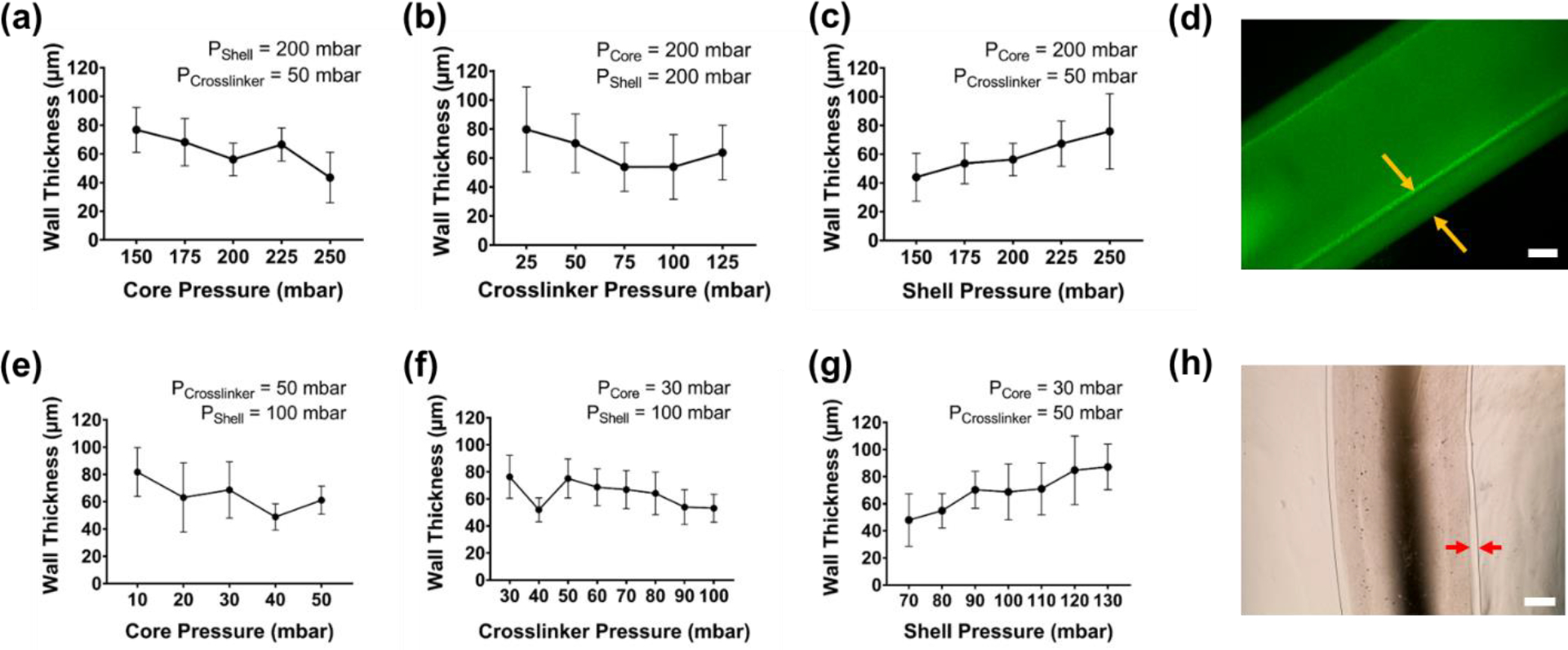
In (a) to (d), open alginate tubes were printed using 6% PVA solution in the Core inlet, 1.5% alginate in the Shell inlet, and 125 mM calcium chloride solution in the Crosslinker inlet. As Core (a) and Crosslinker (b) pressures were varied, minimal changes were observed in the wall thickness, as measured by brightfield microscopy. (c) In contrast, wall thickness was adjustable as a function of Shell pressure and could be tuned from ~45–75 μm across pressure ranges of 150–250 mbar. (d) Fluorescence micrograph demonstrates a representative printed tube. Yellow arrows indicate the tube wall being measured. Scale bar = 100 μm. Similarly, in (e) to (h), open alginate tubes were printed using 0.006% PVA solution in the Core inlet, 1.5% alginate in the Shell inlet, and 125 mM calcium chloride solution in the Crosslinker inlet. As Core (e) and Crosslinker (f) pressures were varied, minimal changes were observed in the wall width, as measured by brightfield microscopy. (g) In contrast, wall width was adjustable as a function of Shell pressure and could be tuned from ~50–90 μm across pressure ranges of 70–120 mbar. (h) Brightfield image demonstrates a representative printed tube. Red arrows indicate the tube wall being measured. Scale bar = 500μm. Error bars indicate +/− standard deviation for n = 3 measures.
To assess the range of achievable tube sizes, we diluted the PVA solution 1000-fold to 0.006% (w/v) and observed the pressure relationships noted in Figure 3(e)–3(g). For this dataset, wall thicknesses were measured using brightfield imaging (Figure 3(h)). Trends in the impact of solution pressures on wall thicknesses tracked closely with those observed for the 6% PVA core solution: wall thickness increased with increasing shell pressure, and changed minimally or decreased with increasing crosslinker pressure, within a similar range of target wall thicknesses. Notably, the necessary shell pressures to reproduce wall thicknesses of 45 – 80 μm were roughly half of those needed for the 6% (w/v) PVA used in Figure 3(a)–3(c). Crosslinker pressures remained relatively unchanged in both conditions. Core pressures were substantially reduced, ranging from 10 – 50 mbar. In these experiments, we focused on the wall thicknesses for each tube, but we also recorded the total (outer) tube diameters, which are presented in Figure S3 in SI. The trends for total tube diameter, as a function of Core, Crosslinker, and Shell pressures largely matched the same trends observed above for wall thickness.
2.3. Hydrogel tubes remain patent after printing
To confirm the creation and retention of open lumen of hydrogel tubes, microparticles were doped into the Core or Shell solutions and imaged by confocal microscopy after printing. A 3D reconstruction of the confocal Z-stack in Figure 4(a) demonstrates that red fluorescent microparticles doped into the Shell alginate solution were confined within the thin hydrogel tube walls and were not visible in the lumen after clearance of the sacrificial PVA solution. In contrast, Figure 4(b) shows a still frame from a video (Movie S1 in SI) of a hydrogel tube, immediately after printing into a PBS bath. Opaque microparticles were included within the Core PVA solution during printing. After the hydrogel tube was printed, the opaque microparticles were observed to flow slowly out of the open ends of the tubes (Figure 4(b’)), as they were displaced anddiluted by PBS.
Figure 4. Microparticles demonstrate tube patency.
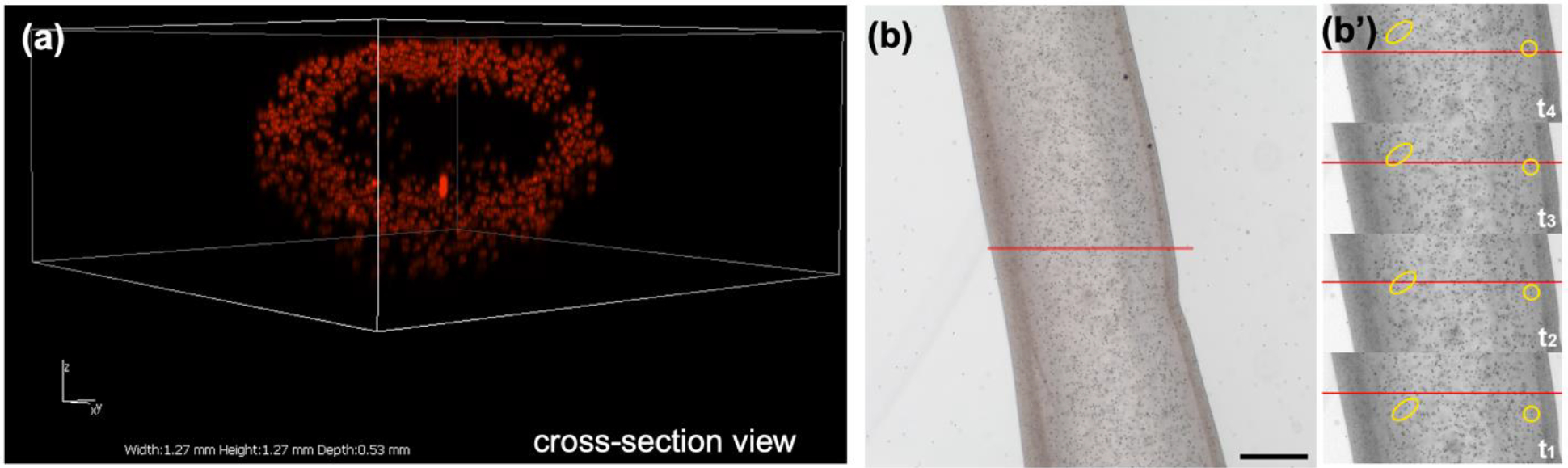
(a) Fluorescent microparticles (14 μm diameter) within the Shell alginate solution were readily printed within the tube walls and were exclusively retained there. Image is a 3D reconstruction of a confocal Z-stack, recorded throughout the sample depth. Axis dimensions: x-axis length = 1.27 mm; y-axis length = 1.27 mm; z-axis depth = 0.53 mm. (b) Opaque microparticles (6 μm diameter), doped within the Core (6% PVA) solution were printed within the center of a tube. After printing, microparticle flow out of the tube was recorded (single frame image at time zero; scale bar = 250 μm). (b’) Single greyscale frames at times t1-t4, spanning ~4 sec, show two arbitrary groups of particles (yellow) progressing past a reference point (red line). (Particle flow visible in Movie S1 in SI).
2.4. Bioprinting enabled xyz patterning and rapid material switching
Individual hollow tubes could be printed along xyz pathways to form complex 3D assemblies of stacked tubes in close-packed, honeycomb-like patterns (Figure 5(a)–5(c)). After bisecting the stack with a sharp razor blade, distinct core-shell separation was visible for the stacked tubes, as well as their retained patency (Figure 5(c)). Printer features enabled separate selection between two hydrogel “inks”, labeled with blue and red dyes, on opposite sides of the assembled structure (shown for hollow tubes in Figure 5(b) and solid fibers in 5(d)). Simple replicas of branched structures with 2, 3, or 4 lobes could be designed in computer-aided design (CAD) software, sliced, and printed at the centimeter scale using the fiber-based printhead (Figure 5(e)).
Figure 5. Bioprinting enabled xyz patterning and materials switching.
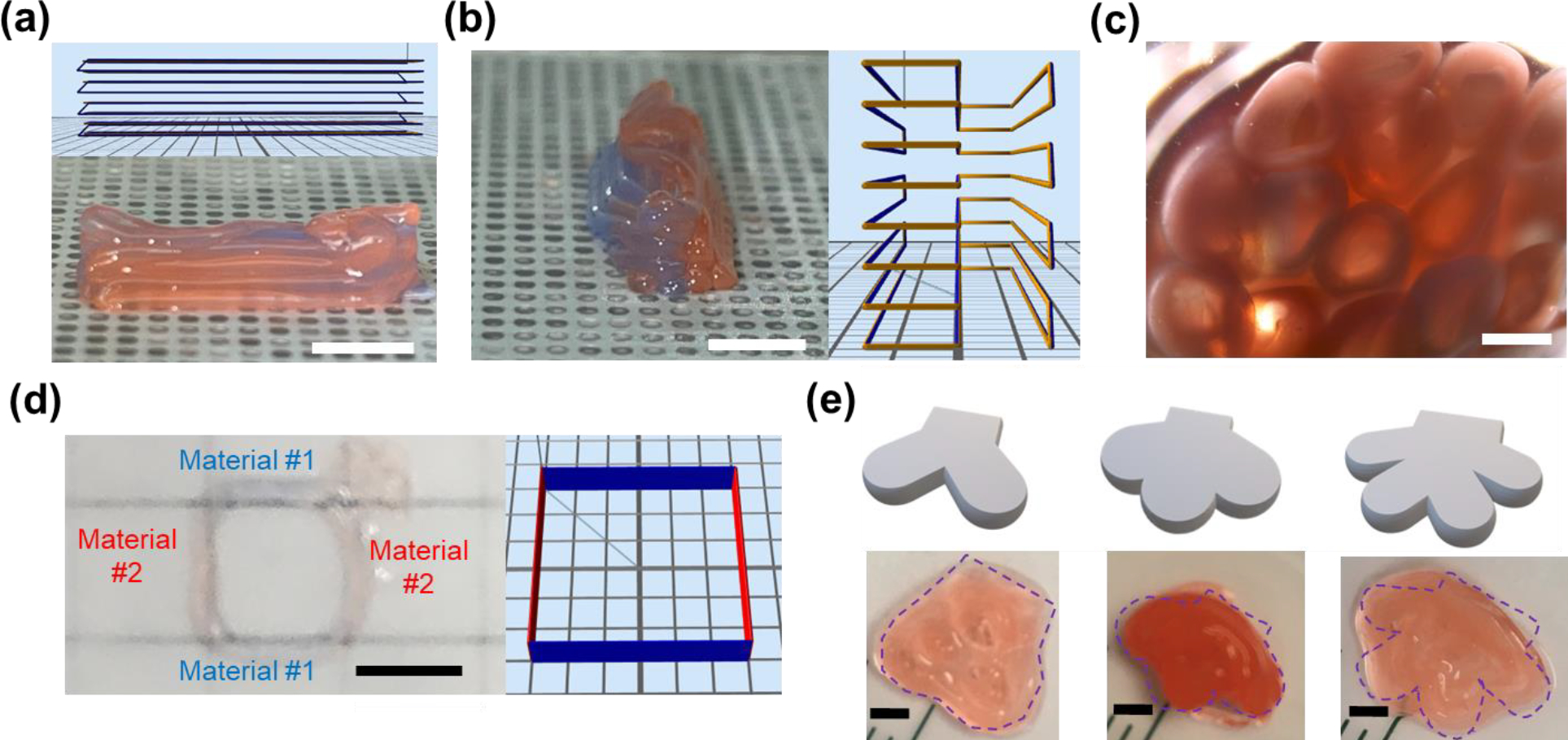
In Figure 5(a)–(c), alginate hollow tubes were printed in a parallel, repetitive pattern in the xy directions along the build platform and stacked in the z direction. Macroscopic photographs demonstrate this stacking along orthogonal xy directions in (a, b) (scale bar = 5 mm) and a zoomed view in cross section in (c) (scale bar = 1 mm). Insets in (a) and (b) show the building paths in the printing software. Hydrogel fibers were printed in a square pattern in (d) (scale bar = 5 mm). Additionally, (b) and (d) demonstrate the ability of both printheads to switch rapidly between two materials on opposite sides of the structures, shown as blue and red dyed components. Dyes in (d) appear fainter due to the magnification and fiber diameter. As shown in Figure 5(e), in contrast to printing hollow tubes demonstrated in (a)-(c), solid fibers were printed using the raster deposition mechanism to generate bioprinted structures mimicking two-/three-/four-branched salivary structures on the cm scale, based on computer-aided design (CAD) models. Scale bar = 2 mm.
2.5. Primary human salivary epithelial cells can be bioprinted at multiple alginate concentrations and cell densities with no significant impact on viability over time
Cells can be incorporated into any of the printing streams, to enable encapsulation, e.g., within the primary Material inlet for fiber production, or within the Shell inlet to produce cell-laden hydrogel tube walls. Viability after printing is a key consideration for this method. Given the extremely thin dimensions, the alginate hydrogel fibers and tubes can ensure efficient mass transport and exchange.
As an initial assessment of alginate concentrations, primary hS/PCs were isolated from human tissues, expanded in 2D culture, and printed within hydrogel fibers at 5 million cells/mL using either 0.5% (w/v) or 1.5% (w/v) alginate solutions through the Material channel, using established settings that yield fiber diameter of ~100 μm. After 3 days of culture in complete salivary media, the cell-laden hydrogel fibers were assessed for viability using confocal microscopy and quantitative fluorescence image analysis. Viability levels were indistinguishable between the two materials three days after bioprinting (Figure 6(a)).
Figure 6. Salivary epithelial cells can be printed with minimal impact on viability.
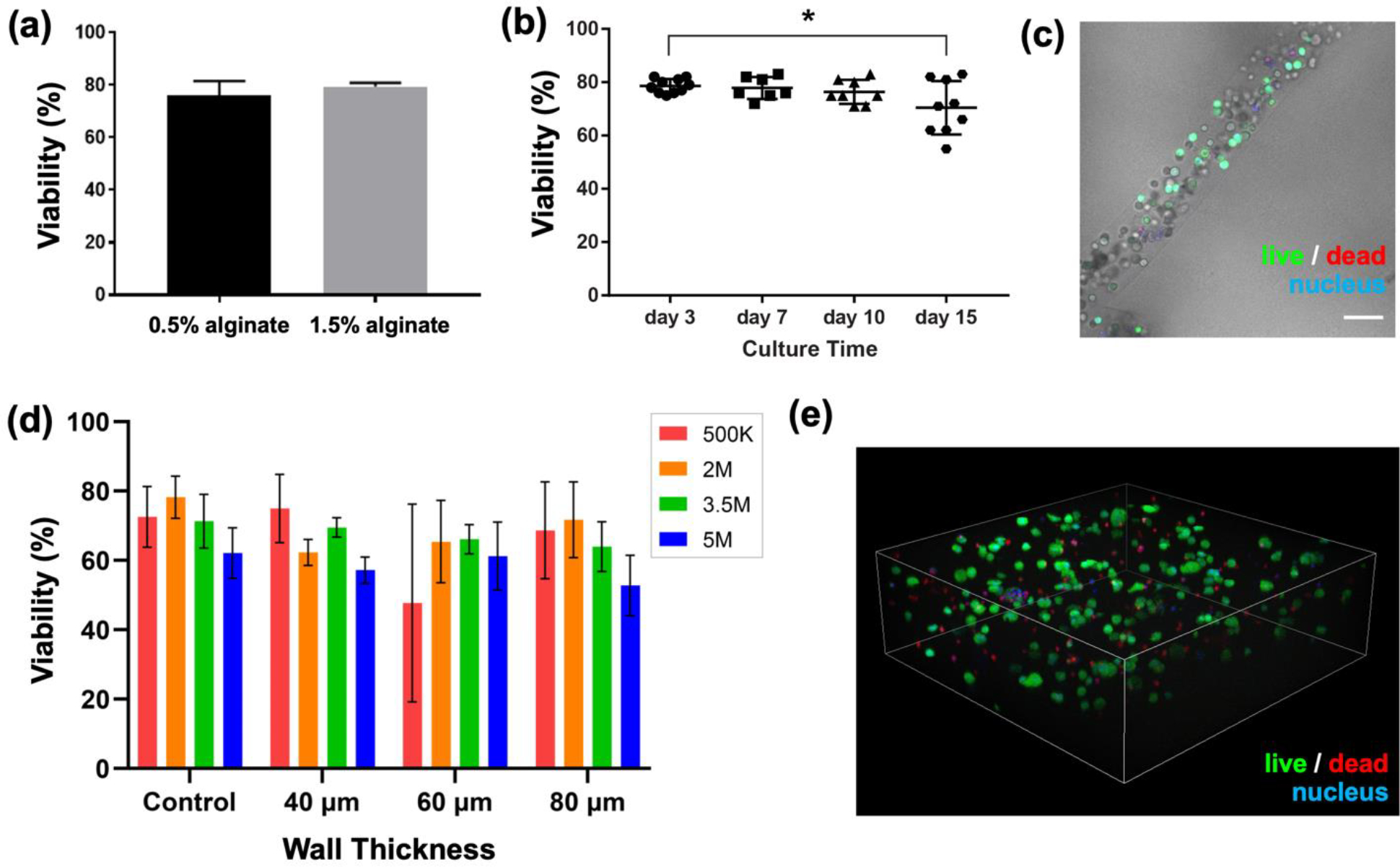
(a) Cells bioprinted in 0.5% or 1.5% (w/v) alginate fibers retained indistinguishable viability three days after bioprinting. (b) Viability of hS/PCs bioprinted in 1.5% (w/v) alginate fibers was monitored over time. Each data point represents one sample. Error bars indicate +/− standard deviation. * p< 0.05. (c) A representative bright field image overlaid with live/dead staining confocal image shows hS/PC distribution within the bioprinted fiber structure (from day 3 sample with 5.0 × 106 cells/mL density). Scale bar = 100 μm. (d) hS/PC viability within the printed hollow tubes, quantified as a function of both cell density (noted in legend as cells/mL) and wall thickness (x-axis) vs. bulk control (no printing). Error bars indicate +/− standard deviation for n=3 measures. (e) Representative 3D rendering of Z-stack images taken of bioprinted hS/PCs after being stained in a viability assay. Image shows 2E6 cells/mL density in 40 μm wall thickness hollow tube samples. Dimensions of Z-stack: width: 636.4 μm; depth: 636.4 μm; height: 201.4 μm.
In solid fiber structures, hS/PCs were printed at 5 million cells/mL with 1.5% (w/v) alginate and monitored for viability over a course of fifteen days. The viability of these bioprinted hS/PCs cultured in complete salivary media remained unchanged through day 10, with a slight drop by day 15 (Figure 6(b)). Figure 6(c) is a representative bright field image overlaid with live/dead staining confocal image to show hS/PC distribution within the bioprinted fiber structure (from day 3 sample with 5 million cells/mL density) and high viability.
A more extensive study of hS/PC viability was conducted for cells printed in the hollow tube geometry. hS/PCs were printed at cell concentrations varying from 0.5 – 5.0 million cells/mL within the 1.5% (w/v) alginate Shell stream solution, using established settings that yield wall thicknesses of 40, 60, and 80 μm. A complementary control experiment used the same cells and material, but cast as a bulk cell-gel suspension into defined static molds. After three days of culture, samples from each set were stained, imaged, and quantified, yielding the data in Figure 6(d). Figure 6(e) shows a representative 3D rendering of Z-stack images of bioprinted hS/PCs in a hydrogel tube (3.5 million cells/mL, 40 μm wall thickness) after being stained in a viability assay. Across the multiple cell concentrations and printing conditions tested, no statistical difference in viability was observed as compared to the controls (hS/PCs encapsulated directly in bulk hydrogel, without printing).
2.6. Bioprinted hS/PCs retain stemness markers
To investigate how hS/PCs behave in bioprinted hydrogel tubes, we conducted immunocytochemistry analyses. Just prior to encapsulation (i.e. “day 0”), nearly all hS/PCs express characteristic markers of stem/progenitor nature: keratin 5 (K5), keratin 14 (K14), and p63.[20] After 15 days in culture post-printing, hS/PCs encapsulated in 3D still retain these markers, matching 2D counterparts on glass substrates (Figure 7). As expected, nearly all hS/PCs stained positive for these markers, with appropriate morphology and intracelullar localization. K5/K14 staining was cytoplasmic and concentrated at cell periphery, and p63 staining was localized to nuclei, as is phenotypic for these cells in 3D. Phenotype staining for these markers was qualitatively equivalent between adherent hS/PC 2D monolayers on tissue culture substrates, and hS/PCs printed in 3D hydrogels for up to 15 days, demonstrating no loss of these markers over that culture period.
Figure 7. Bioprinted hS/PCs retain stemness markers.
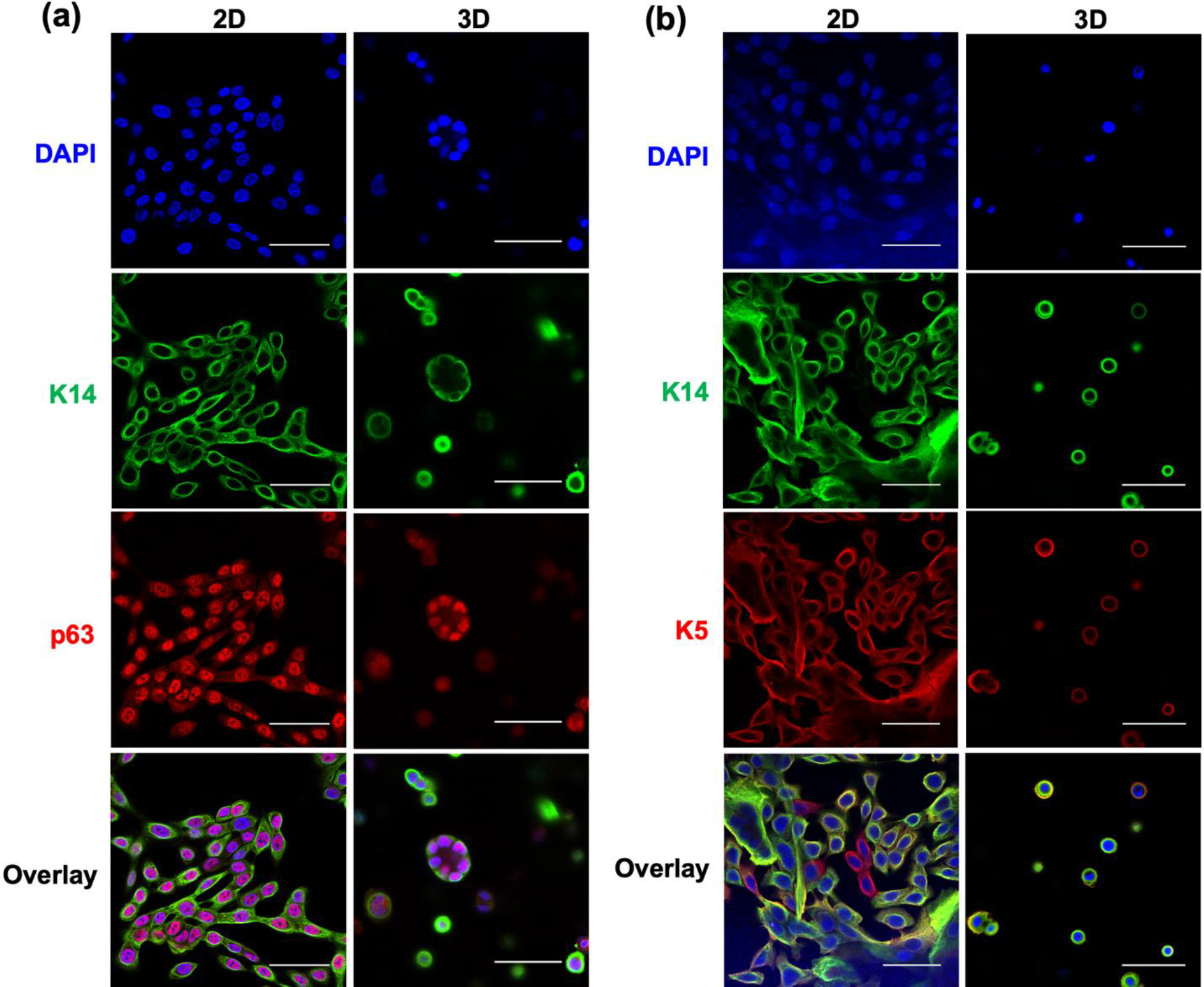
Immunocytochemistry characterizations revealed that hS/PCs in 3D bioprinted 1.5% (w/v) alginate hydrogel hollow tubes (60 μm wall thickness; 5 million cells/mL) after 15 days in culture still could express (a) basal stem cell marker p63 and (b) basal cell keratin pair K5/K14, indicating stemness was retained after bioprinting, and was comparable to hS/PCs on 2D glass substrates. For all images, scale bar = 50 μm.
3. Discussion
Unlike many other target tissues for bioprinting, glands uniquely rely on methods that can manufacture a thin epithelial layer. Leveraging a microfluidics-based bioprinter with coaxial polymer and crosslinker streams, we demonstrated parallel biocompatible printing strategies suitable for deposition of hydrogel layers <100 μm. Thin features like cell-laden microfibers or microtubes of physiologically relevant sizes can be successfully generated with ensured cell viability, retained stemness markers, and reliable reproducibility of feature sizes. CMF functionality provides this fine resolution at the individual layer-level, while preserving the ability to print larger hydrogel structures on the mm- and cm-scale. Because technologies at this scale have not been applied to SG engineering/replacement, this demonstration paves the way for fundamental studies of intercellular communication across cell types, spatial customization of ECM composition, and targeted cytokine delivery. Other elegant work [38] in SG fabrication has employed intracellular magnetic particles to promote cell assembly into multicellular aggregates; we see bioprinting as a complement to such efforts, by adding spatial deposition of cells within hydrogel matrices, with control across multiple length scales.
During development, gland formation begins as an epithelial bud, which branches progressively through interaction with adjacent stromal fibroblasts and nerve fibers, until it terminates in the secretory acini. Our ability to print multiple matrices (as in Figure 5(a,b,d)), either within a given fiber, or in multiple fiber layers, provides the toolkit to reconstruct spatial elements of this native gland arrangement in future work. The multiple material feeds shown in Figure 5 can be variations in gel composition, and/or associated cells. Other researchers in the field have demonstrated the interactions of human SG fibroblasts and SG epithelia in in vitro co-cultures, with broad utility in understanding and impacting phenotypic behavior.[16,39–41] Our own future efforts will integrate and organize these cell types and assess the impact of spatial scaling and density on their responses.
The complementary manufacturing methods demonstrated here, of fibers and tubes, offer separate advantages that may lead to different end uses. Fiber-based printing would lead to a raster-based workflow, that may yield higher resolution and precision, at the potential expense of speed or size. Conversely, tube-based printing closely resembles the native ductal structure, and with the availability of single-layered and double-layered (not shown here) CMF printheads, hydrogel tubes with complex radial structure and ultra-thin outer walls are possible. However, tube-based printing suffers from an inability to create connected, branched structures. Instead, this may require an alternate paradigm, e.g., constructing parallel tracks of single-outlet tubes, similar to Figure 5(c). In all cases, biomanufacturing methods will require tandem appreciation for the biological function of the gland, and the ultimate need for surgical integration.
For tissue engineering applications, alginate hydrogel substrates are often modified with adhesion motifs (e.g., RGD, IKVAV, or other peptides) to activate integrin signaling or other cell-matrix cues. In the present work, alginate was used without such modification, as SG-derived cells, particularly of epithelial character, are often able to survive well solely through cell-cell adhesions, and eventually through their own peripheral matrix deposition. Here, we found that hS/PC viability after bioprinting was consistent across print parameters, but slightly lower than observed in other matrices. We anticipate that viability could be further improved by using peptide-functionalized alginate. Printing pre-clustered hS/PCs, in which hS/PCs would be additionally stabilized by cell-cell adhesions, could be another strategy, although counter to our goal of a coherent mono-epithelial layer. Ultimately, alginate may serve as a fast-gelling, but temporary and extractable, scaffold for co-printing with other hydrogels, such as hyaluronic acid, which we have shown can support high hS/PC viability. The modularity of chemically functionalized HA enables an array of modifications, including adhesive ligands, enzyme-labile crosslinkers, and depots for cytokine retention/release.
This promising method of creating salivary epithelium-mimicking structures represents potential for future engineered salivary therapeutics that can restore salivary function. The printed structures can also possibly serve as models for studying the biology of secretory function, disease of exocrine tissues or high-throughput drug screening. This initial proof of concept establishes a platform for future complex combinations of cell types, material compositions, and biological cues to direct cell growth in a SG biomimic.
4. Conclusion
In summary, we exploited a microfluidics-based coaxial bioprinter with alginate as a model biomaterial for generating refined hydrogel structures with dimensions and morphologies inspired by salivary epithelia. We printed a broad range of accessible structure sizes, with reliable control over diameter for both fibers and hollow tubes, and wall thickness for tubes. Fiber diameters ranged from ~100–800μm, and tube diameters ranged from ~600–2000 μm, all within the range of utility for salivary structures. Primary human SG cell viability was preserved for bioprinted tubes, compared to control bulk encapsulation, across multiple cell densities (5.0 × 105 – 5.0 × 106 cells/mL) and variable wall thicknesses (40–80 μm). hS/PC marker expression was also preserved over 15 days after bioprinting. The ability to create structures with total size on the centimeter scale, but fine features of <100 μm, with minimal impact on hS/PC viability, makes this bioprinting system a promising tool for future efforts in SG tissue engineering. The diversity of available printing formats, as tubes or as hollow fibers, provides multiple avenues for constructing bioinspired mimics of SG structures, across multiple length scales, and defined spatial deposition of multiple cell types and hydrogel compositions.
5. Materials and Methods
5.1. Bioprinter and Bioinks
We used a RX1 bioprinter from Aspect Biosystems to print solid fibers using a disposable DUO™ microfluidic printhead (nozzle dimensions: 28 mm L × 2.2 mm outer diameter) or hollow tubes using a CENTRA™ printhead (nozzle dimensions: 25.8 mm L × 4.0 mm outer diameter) and maintained in a standard laminar flow cell culture hood throughout all printing procedures. Calcium chloride (Fisher Scientific, C70–500) (CaCl2)-based crosslinking solution (125 mM) was prepared in MilliQ water and sterile filtered by 0.22 μm PES syringe filter. Sodium alginate (Sigma-Aldrich®, 180947) was reconstituted with MilliQ water or 1X PBS (Corning®, 21–040-CV) to make 0.5% or 1.5% w/v solution and sterile filtered. Poly(vinyl alcohol) (PVA) (Sigma-Aldrich®, 363081–25G) was dissolved in MilliQ water to 6% w/v working concentration and autoclaved for sterilization. Fluorescent red microspheres with Nile Red excitation/emission spectra (Spherotech Inc., FP-15056–2, 10.0–14.0 μm, 1% w/v) were diluted prior to use. 2 MDa FITC-dextran (Sigma-Aldrich®, FD2000S-100MG) was diluted to 2.5 mg/mL using 1X PBS.
5.2. Human Salivary Stem/Progenitor Cell Isolation, Expansion, and Maintenance
Human salivary stem/progenitor cells (hS/PCs) were generated from salivary tissue explants, obtained from patients providing informed consent under IRB-approved protocols at respective institutions (IRB #HSC-DB-16–1060 for University of Delaware, PI R. Witt and Stanford University, PI Q. Le; HSC-DB-18–0057 for Houston Methodist Hospital, PI A. Farach). hS/PC isolation, expansion, and maintenance were performed in accordance with previously published protocols.[42] For passaging and collecting for bioprinting, hS/PC monolayers were exposed to trypsin for 3–4 minutes (Gibco™, 25–200-072, 0.25% trypsin-EDTA), incubated with trypsin inhibitor Glycine max (soybean) (Sigma-Aldrich®, T6522, 1 mg/mL) at a 1:1 (v/v) ratio to trypsin-EDTA, and centrifuged to a pellet. The supernatant was removed and the cell pellet was further processed for expansion, freezing, or bioprinting. hS/PCs (female, Caucasian, 52 years of age) at passage 3 were used for bioprinting in solid fibers and hS/PCs (female, Caucasian, 59 years of age) at passage 4 were used for bioprinting in hollow tubes. hS/PCs (female, Caucasian/Hispanic, 66 years of age) at passage 2 were used for immunocytochemistry. All hS/PCs were generated from human parotid gland tissues. The following culture medium formula[42] (the complete salivary medium) was used throughout: Williams’ E media (Sigma-Aldrich, W4128–500ML) supplemented with 1% (v/v) ITS (InVitria, 777ITS091100ML), 1 mg/mL (w/v) human serum albumin (Sigma-Aldrich, A1887–10G), 1% (v/v) GlutaMax™ (Gibco™, A1286001), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco™, 15140122), 10 μM dexamethasone (Sigma-Aldrich, St. Louis, MO), and 10 ng/mL human epidermal growth factor (hEGF) (Gibco™, PHG0311).
5.3. Bioprinted Feature Fidelity Measurements
Defined build paths were established in the Aspect Studio software (Aspect Biosystems, Canada) to direct the printhead to dispense hydrogel material onto a build platform. For ease of collection and measurement of feature sizes, fibers were printed as the periphery of a cylinder of 1 mm in height and 20 mm in diameter; hydrogel tubes were printed as the single-layer rectilinear infill of a cylinder of 25 mm in diameter. Structures were printed directly into a 35mm glass bottom dish (Cellvis, D35201.5N, 35mm).
Fiber diameters were varied by adjusting the pressures of the Material (i.e., 1.5% w/v alginate solution) and Crosslinker (i.e., 125 mM CaCl2 solution) feeds. Brightfield micrographs of printed fiber structures were captured by a EVOS™ XL Core microscope (ThermoFisher Scientific). Tube dimensions were varied by adjusting Core, Shell, and Crosslinker pressures and core solution concentrations (i.e., 6% w/v PVA and 0.006% w/v PVA). Printed tube structures were examined either under brightfield microscopy or confocal microscopy (A1R/MP Microscope, Nikon Instruments) after mixing the 6% w/v PVA core solution with FITC-dextran solution. Images were quantified using ImageJ2 [43].
5.4. Bioprinted Acellular Structures
To print extended 3D structures, build paths were established in Aspect Studio software, and hydrogel fibers and tubes were printed onto filter paper on the standard build platform, which applies light vacuum to aspirate excess crosslinker solution.
To demonstrate material switching, blue or red acrylic dyes (Artist’s Loft™, USA) were added in dilute amounts (~ 10 μL dye per 1 mL of polymer solution) to each of the two separate input feeds on the bioprinter system to be printed through Material 1 and Material 2 channels. Selection of Material channels and coordinates for switching between materials were designed in the Aspect Studio Software to preferentially print each material on specific sides of the final macroscopic object. Fibers were printed at Material and Crosslinker pressures of 25 mbar and 35 mbar respectively. Tubes were printed at Core, Shell, and Crosslinker pressures of 200, 200, and 50 mbar, respectively.
To demonstrate the ability to print tubes with cells encapsulated within the tubule walls, Nile Red microspheres of size similar to cells were added to the alginate shell solution at an approximate concentration of 1:5 (v/v) Nile Red microsphere solution to 1.5% (w/v) sodium alginate, and printed by the above means. Tube patency was visualized (1) by printing microspheres within the alginate solution and then imaged using confocal microscopy; and (2) by printing Polybead® Black Dyed Microspheres (Polysciences Inc., 24293–5, ø = 6.00 μm) in 6% PVA solution and then visualizing microsphere motion in the solid tube (SI Movie 1). Similarly, FITC-dextran accumulation within the tubes’ inner walls defined tube dimensions and demonstrated patency.
5.5. Bioprinted Cellularized Structures
hS/PCs were resuspended at varying cell densities (0.5 – 5.0 million cells/mL) within sterile alginate shell solution, and printed onto glass bottom dishes. Printing pressures were controlled to print ~100 μm fibers containing cells, or hollow tubes at wall thicknesses of 40, 60, and 80 μm with cells in the shell. The excess printing solutions (i.e., cross linker solution, printing buffer, and leached PVA core solutions) were replaced with fresh warm media and bioprinted structures were quickly passed to the humidified incubator (37°C / 5% CO2). The media was changed once again after one hour post printing to remove any remaining printing solutions. Control studies involved separate bulk cell encapsulations at equivalent densities, within 50 μL of 1.5% (w/v) alginate, crosslinked using 125 mM CaCl2 solution in a PDMS mold.
Cells were cultured with daily media change and assessed for viability after 3 days. Live/dead viability reagents, Calcein AM (Invitrogen, C3100MP), Ethidium Homodimer-III (EthD-III, Biotium, 40050), and Hoechst 33342 (Enzo, ENZ-52401), were all diluted in 1X HBSS (Gibco™, 14025134) to 2 μM, 4 μM, and 2 μg/mL, respectively. Samples were incubated in a 35mm glass bottom dish (Cellvis, D35201.5N, 35mm) in the dark at room temperature for 15 min before imaging. A Z-stack of ~200 μm was imaged for each of the cell-laden specimens, with 3 regions of interest (ROIs) per condition. Each Z-stack was recorded at 0.95 μm per step under resonant scanning mode using Nikon A1 Piezo Z Drive under Nikon 20X water lens, with NA = 0.95. Imaris 9.1 (Bitplane, UK) software was used to reconstruct each image stack. Spot function in Imaris was applied in each channel for objects above a minimum background signal and within a standard size range for cell features. The number of objects within each 3D structure were counted. Viability was calculated based on the following equation, which identifies the number of dead cells, as a percentage of total cells, corrected for aberrant co-staining:
In the above equation, “Dead” refers to all cells that stained red by EthD-III within the nucleus region; “Nuclei” counts all nuclei that stained blue using Hoechst 33342. “Dead_Nuclei_Colocalized” refers to spots that stained both blue and red. To avoid overcounting, this portion was subtracted from the denominator of the viability formula.
5.6. Statistical analyses
Statistical analyses were performed on the three repeated prints of each condition in Figure 6(b) using one-way analysis of variance (ANOVA) with Tukey’s post hoc tests and on each of the three repeated prints of each condition in Figure 6(d) using two-way analysis of variance (ANOVA) with Tukey’s post hoc tests in GraphPad Prism 5 Project. p<0.05 was regarded as statistically significant.
5.7. Immunocytochemistry
Immunocytochemistry of K5, K14, and p63-α were done following previously established protocols [25]. Briefly, samples were gently washed with pre-warmed 1X HBSS and placed in 4% (w/v) paraformaldehyde solution in 1X HBSS for 10 min under room temperature in a chemical fume hood. Samples were washed with 1X HBSS for three rounds of 10 minutes on shaker at 50 rpm. Permeabilization was done using 0.2% (v/v) Triton X-100 (Thermo Scientific™, A16046-AP) in 1X HBSS for three rounds of 10 minutes on shaker at 50 rpm. Blocking was completed using 10% (v/v) goat serum (Thermal Fisher, 5058835) in 0.2% (v/v) Triton X-100 in 1X HBSS, filtered using 0.22 μm PES syringe filter, for one-hour incubation at room temperature on shaker at 50 rpm. Primary antibodies, listed in Table S1 in SI, were diluted in filtered 10% (v/v) goat serum in 0.2% (v/v) Triton X-100 in 1X HBSS and incubated with samples overnight (12–24 hours) in 4°C cold room on shaker at 60 rpm. After removing primary antibody, samples were washed with 0.2% (v/v) Triton X-100 in 1X HBSS for four rounds of 5 minutes on shaker at 50 rpm. Meanwhile, secondary antibodies were centrifuged at 10,000G for 15 minutes and only the supernatant was used at a dilution ratio of 1:2000 (v/v) in 0.2% (v/v) Triton X-100 in 1X HBSS. HNA-Cy3 (Millipore, MAB1281) was used as a positive control to confirm antibody penetration, at 1:100 dilution in 0.2% (v/v) Triton X-100 in 1X HBSS. Secondary antibody incubation lasted for an hour at room temperature in the dark. Samples were then washed with 1X HBSS for four rounds of 5 minutes on shaker at 50 rpm. Finally, to counterstain the nuclei, 4’,6-diamidino-2-phenylindole (DAPI) (Biotium, 40043, 1 μg/mL in 1X HBSS) was used for 10 min at room temperature at 50 rpm. 3D bioprinted samples were imaged using Nikon A1R/MP confocal microscope with 20X water lens.
For comparison against bioprinted samples, hS/PCs were cultured on 2D in a 35mm tissue culture treated, #1.5 polymer coverslip microscopy dish (ibidi, ibiTreat 81156) to 50–70% confluency, and fixed, permeabilized, and stained by similar methods as described above (described as “2D” in Figure 7). 2D cell samples were imaged using Nikon A1R/MP confocal microscope using a 40X water lens, with NA = 1.15.
Supplementary Material
Highlights.
Microfluidic coaxial 3D bioprinting for ultra-thin hydrogel micro-fibers/tubes
Bioprinted alginate structures of near-epithelium size mimicking salivary gland
Bioprinted human salivary stem/progenitor cells with high viability and stemness
Acknowledgments
This work was supported by the National Science Foundation CAREER award 2240008 to D.A.H., National Institutes of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) grants R03DE028988 to D.A.H. and R56DE026530 to M.C.F-C; National Cancer Institute grant P01CA257907 to Dr. Quynh-Thu Le at Stanford University for tissue collection support; UTHSC-H startup funding to D.A.H; UTSTARS Award to M.C.F-C supporting the bioprinter and microscopy facilities. E.J.V-R. was supported by NIH RISE-E-BASE grant R25GM127191. The authors acknowledge Rachel Hildebrand, a research coordinator in Dr. Quynh-Thu Le’s group, who helped to consent patients for human parotid tissue collections. The authors thank Caitlynn Barrows for explanting parotid tissue from Houston Methodist Hospital and Dr. Nadia Mohyuddin at Houston Methodist Hospital for providing surgical specimens. Thanks to Dr. Erin Bedford at Aspect Biosystems for her technical support of the bioprinter.
Footnotes
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Daniel Harrington reports financial support was provided by National Institutes of Health and National Science Foundation. Mary C. Farach-Carson reports financial support was provided by National Institutes of Health. Ephraim J. Vazquez-Rosado reports financial support was provided by National Institutes of Health.
CRediT author statement
Yu Yin: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – Original draft preparation, Writing – Reviewing and Editing, Visualization
Ephraim J. Vazquez-Rosado: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – Original draft preparation, Writing – Reviewing and Editing, Visualization
Danielle Wu: Resources, Writing - Review & Editing
Andrew Farach: Resources
Vignesh Viswananthan: Resources
Mary C. Farach-Carson: Resources, Writing – Reviewing and Editing, Supervision, Funding acquisition
Daniel A. Harrington: Conceptualization, Methodology, Resources, Validation, Writing – Original draft preparation, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Saleh J, Figueiredo MAZ, Cherubini K, Salum FG, Salivary hypofunction: An update on aetiology, diagnosis and therapeutics, Arch. Oral Biol. 60 (2015) 242–255. 10.1016/j.archoralbio.2014.10.004. [DOI] [PubMed] [Google Scholar]
- [2].Turner MD, Hyposalivation and Xerostomia: Etiology, Complications, and Medical Management, Dent. Clin. North Am. 60 (2016) 435–443. 10.1016/j.cden.2015.11.003. [DOI] [PubMed] [Google Scholar]
- [3].Guggenheimer J, Moore PA, Xerostomia: Etiology, recognition and treatment, J. Am. Dent. Assoc. 134 (2003) 61–69. 10.14219/jada.archive.2003.0018. [DOI] [PubMed] [Google Scholar]
- [4].Chow LQM, Head and Neck Cancer, N. Engl. J. Med 382 (2020) 60–72. 10.1056/NEJMra1715715. [DOI] [PubMed] [Google Scholar]
- [5].Jasmer KJ, Gilman KE, Muñoz Forti K, Weisman GA, Limesand KH, Radiation-Induced Salivary Gland Dysfunction: Mechanisms, Therapeutics and Future Directions, J. Clin. Med. 9 (2020) 4095. 10.3390/jcm9124095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Head and Neck Cancers - NCI, (2021). https://www.cancer.gov/types/head-and-neck/head-neck-fact-sheet (accessed October 19, 2022).
- [7].Piraino LR, Benoit DSW, DeLouise LA, Salivary Gland Tissue Engineering Approaches: State of the Art and Future Directions, Cells. 10 (2021) 1723. 10.3390/cells10071723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang S, Matsumoto K, Lish SR, Cartagena-Rivera AX, Yamada Correspondence KM, Yamada KM, Budding epithelial morphogenesis driven by cell-matrix versus cell-cell adhesion, Cell. 184 (2021). 10.1016/j.cell.2021.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Holmberg KV, Hoffman MP, Anatomy, Biogenesis and Regeneration of Salivary Glands, Monogr. Oral Sci 24 (2014) 1–13. 10.1159/000358776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Miyajima H, Matsumoto T, Sakai T, Yamaguchi S, An SH, Abe M, Wakisaka S, Lee KY, Egusa H, Imazato S, Hydrogel-based biomimetic environment for in vitro modulation of branching morphogenesis, Biomaterials. 32 (2011) 6754–6763. 10.1016/j.biomaterials.2011.05.072. [DOI] [PubMed] [Google Scholar]
- [11].Soscia DA, Sequeira SJ, Schramm RA, Jayarathanam K, Cantara SI, Larsen M, Castracane J, Salivary gland cell differentiation and organization on micropatterned PLGA nanofiber craters, Biomaterials. 34 (2013) 6773–6784. 10.1016/j.biomaterials.2013.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shubin AD, Felong TJ, Graunke D, Ovitt CE, Benoit DSW, Development of Poly(Ethylene Glycol) Hydrogels for Salivary Gland Tissue Engineering Applications, Tissue Eng. Part A 21 (2015) 1733–1751. 10.1089/ten.tea.2014.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nam K, Jones JP, Lei P, Andreadis ST, Baker OJ, Laminin-111 Peptides Conjugated to Fibrin Hydrogels Promote Formation of Lumen Containing Parotid Gland Cell Clusters, Biomacromolecules. 17 (2016) 2293–2301. 10.1021/acs.biomac.6b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shin K, Koo KH, Jeong J, Park SJ, Choi DJ, Ko Y-G, Kwon H, Three-Dimensional Culture of Salivary Gland Stem Cell in Orthotropic Decellularized Extracellular Matrix Hydrogels, Tissue Eng. Part A. 25 (2019) 1396–1403. 10.1089/ten.tea.2018.0308. [DOI] [PubMed] [Google Scholar]
- [15].Fowler EW, Ravikrishnan A, Witt RL, Pradhan-Bhatt S, Jia X, RGDSP-Decorated Hyaluronate Hydrogels Facilitate Rapid 3D Expansion of Amylase-Expressing Salivary Gland Progenitor Cells, ACS Biomater. Sci. Eng. 7 (2021) 5749–5761. 10.1021/acsbiomaterials.1c00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jorgensen M, Ramesh P, Toro M, Evans E, Moskwa N, Zhang X, Sharfstein ST, Larsen M, Xie Y, Alginate Hydrogel Microtubes for Salivary Gland Cell Organization and Cavitation, Bioengineering. 9 (2022) 38. 10.3390/bioengineering9010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pradhan-Bhatt S, Harrington DA, Duncan RL, Jia X, Witt RL, Farach-Carson MC, Implantable three-dimensional salivary spheroid assemblies demonstrate fluid and protein secretory responses to neurotransmitters, Tissue Eng. - Part A. 19 (2013) 1610–1620. 10.1089/ten.tea.2012.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pradhan-Bhatt S, Harrington DA, Duncan RL, Farach-Carson MC, Jia X, Witt RL, A novel in vivo model for evaluating functional restoration of a tissue-engineered salivary gland, Laryngoscope. 124 (2014) 456–461. 10.1002/lary.24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ozdemir T, Fowler EW, Liu S, Harrington DA, Witt RL, Farach-Carson MC, Pradhan-Bhatt S, Jia X, Tuning Hydrogel Properties to Promote the Assembly of Salivary Gland Spheroids in 3D, ACS Biomater. Sci. Eng. 2 (2016) 2217–2230. 10.1021/acsbiomaterials.6b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Srinivasan PP, Patel VN, Liu S, Harrington DA, Hoffman MP, Jia X, Witt RL, Farach-Carson MC, Pradhan-Bhatt S, Primary Salivary Human Stem/Progenitor Cells Undergo Microenvironment-Driven Acinar-Like Differentiation in Hyaluronate Hydrogel Culture, STEM CELLS Transl. Med 6 (2017) 110–120. 10.5966/sctm.2016-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ozdemir T, Srinivasan PP, Zakheim DR, Harrington DA, Witt RL, Farach-Carson MC, Jia X, Pradhan-Bhatt S, Bottom-up assembly of salivary gland microtissues for assessing myoepithelial cell function, Biomaterials. 142 (2017) 124–135. 10.1016/j.biomaterials.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wu D, Witt RL, Harrington DA, Farach-Carson MC, Dynamic Assembly of Human Salivary Stem/Progenitor Microstructures Requires Coordinated α1β1 Integrin-Mediated Motility, Front. Cell Dev. Biol. 0 (2019) 224. 10.3389/FCELL.2019.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wu D, Lombaert IMA, DeLeon M, Pradhan-Bhatt S, Witt RL, Harrington DA, Trombetta MG, Passineau MJ, Farach-Carson MC, Immunosuppressed Miniswine as a Model for Testing Cell Therapy Success: Experience With Implants of Human Salivary Stem/Progenitor Cell Constructs, Front. Mol. Biosci 8 (2021) 1–15. 10.3389/fmolb.2021.711602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Martinez M, Witt RL, Farach-Carson MC, Harrington DA, Functionalized Biomimetic Hydrogels Enhance Salivary Stem/Progenitor Cell Organization, BioRxiv. (2021) 2021.08.05.455302. 10.1101/2021.08.05.455302. [DOI] [Google Scholar]
- [25].Wu D, Chapela PJ, Barrows CML, Harrington DA, Carson DD, Witt RL, Mohyuddin NG, Pradhan-Bhatt S, Farach-Carson MC, MUC1 and Polarity Markers INADL and SCRIB Identify Salivary Ductal Cells, J. Dent. Res. 101 (2022) 983–991. 10.1177/00220345221076122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Picollet-D’hahan N, Dolega ME, Liguori L, Marquette C, Le Gac S, Gidrol X, Martin DK, A 3D Toolbox to Enhance Physiological Relevance of Human Tissue Models, Trends Biotechnol. 34 (2016) 757–769. 10.1016/j.tibtech.2016.06.012. [DOI] [PubMed] [Google Scholar]
- [27].Ozbolat IT, Hospodiuk M, Current advances and future perspectives in extrusion-based bioprinting, Biomaterials. 76 (2015) 321–343. 10.1016/j.biomaterials.2015.10.076. [DOI] [PubMed] [Google Scholar]
- [28].Rojek KO, Ćwiklińska M, Kuczak J, Guzowski J, Microfluidic Formulation of Topological Hydrogels for Microtissue Engineering, Chem. Rev. 122 (2022) 16839–16909. 10.1021/acs.chemrev.1c00798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Davoodi E, Sarikhani E, Montazerian H, Ahadian S, Costantini M, Swieszkowski W, Willerth SM, Walus K, Mofidfar M, Toyserkani E, Khademhosseini A, Ashammakhi N, Extrusion and Microfluidic‐ Based Bioprinting to Fabricate Biomimetic Tissues and Organs, Adv. Mater. Technol. 5 (2020) 1901044. 10.1002/admt.201901044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rowley JA, Madlambayan G, Mooney DJ, Alginate hydrogels as synthetic extracellular matrix materials, Biomaterials. 20 (1999) 45–53. 10.1016/S0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- [31].Augst AD, Kong HJ, Mooney DJ, Alginate Hydrogels as Biomaterials, Macromol. Biosci 6 (2006) 623–633. 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- [32].Lee KY, Mooney DJ, Alginate: Properties and biomedical applications, Prog. Polym. Sci. Oxf 37 (2012) 106–126. 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shin S-J, Park J-Y, Lee J-Y, Park H, Park Y-D, Lee K-B, Whang C-M, Lee S-H, “On the Fly” Continuous Generation of Alginate Fibers Using a Microfluidic Device, Langmuir. 23 (2007) 9104–9108. 10.1021/la700818q. [DOI] [PubMed] [Google Scholar]
- [34].Lee KH, Shin SJ, Park Y, Lee S-H, Synthesis of Cell-Laden Alginate Hollow Fibers Using Microfluidic Chips and Microvascularized Tissue-Engineering Applications, Small. 5 (2009) 1264–1268. 10.1002/smll.200801667. [DOI] [PubMed] [Google Scholar]
- [35].Onoe H, Okitsu T, Itou A, Kato-Negishi M, Gojo R, Kiriya D, Sato K, Miura S, Iwanaga S, Kuribayashi-Shigetomi K, Matsunaga YT, Shimoyama Y, Takeuchi S, Metre-long cell-laden microfibres exhibit tissue morphologies and functions, Nat. Mater. 2013 126. 12 (2013) 584–590. 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- [36].Abelseth E, Abelseth L, De La Vega L, Beyer ST, Wadsworth SJ, Willerth SM, 3D Printing of Neural Tissues Derived from Human Induced Pluripotent Stem Cells Using a Fibrin-Based Bioink, ACS Biomater. Sci. Eng. 5 (2019) 234–243. 10.1021/acsbiomaterials.8b01235. [DOI] [PubMed] [Google Scholar]
- [37].de Paula F, Teshima THN, Hsieh R, Souza MM, Nico MMS, Lourenco SV, Overview of Human Salivary Glands: Highlights of Morphology and Developing Processes, Anat. Rec. 300 (2017) 1180–1188. 10.1002/ar.23569. [DOI] [PubMed] [Google Scholar]
- [38].Chansaenroj A, Adine C, Charoenlappanit S, Roytrakul S, Sariya L, Osathanon T, Rungarunlert S, Urkasemsin G, Chaisuparat R, Yodmuang S, Souza GR, Ferreira JN, Magnetic bioassembly platforms towards the generation of extracellular vesicles from human salivary gland functional organoids for epithelial repair, Bioact. Mater. 18 (2022) 151–163. 10.1016/j.bioactmat.2022.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Altrieth AL, O’Keefe KJ, Gellatly VA, Tavarez JR, Feminella SM, Moskwa NL, Cordi CV, Turrieta JC, Nelson DA, Larsen M, Identifying fibrogenic cells following salivary gland obstructive injury, Front. Cell Dev. Biol. 11 (2023) 1190386. 10.3389/fcell.2023.1190386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Moskwa N, Mahmood A, Nelson DA, Altrieth AL, Forni P, Larsen M, PDFGRα+ Stromal Cells Promote Salivary Gland Proacinar Differentiation Through FGF2-dependent BMP7 Signaling, (2021) 2021.11.19.469144. 10.1101/2021.11.19.469144. [DOI] [Google Scholar]
- [41].Yamamoto S, Fukumoto E, Yoshizaki K, Iwamoto T, Yamada A, Tanaka K, Suzuki H, Aizawa S, Arakaki M, Yuasa K, Oka K, Chai Y, Nonaka K, Fukumoto S, Platelet-derived Growth Factor Receptor Regulates Salivary Gland Morphogenesis via Fibroblast Growth Factor Expression*, J. Biol. Chem. 283 (2008) 23139–23149. 10.1074/jbc.M710308200. [DOI] [PubMed] [Google Scholar]
- [42].Wu D, Chapela P, Farach-Carson MC, Reassembly of Functional Human Stem/Progenitor Cells in 3D Culture, in: Epithel. Cell Cult, Humana Press, New York, NY, 2018: pp. 19–32. 10.1007/978-1-4939-8600-2_3. [DOI] [PubMed] [Google Scholar]
- [43].Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW, ImageJ2: ImageJ for the next generation of scientific image data, BMC Bioinformatics. 18 (2017) 529. 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


