Abstract
Prurigo nodularis (PN) is a chronic inflammatory skin disease characterized by intense pruritus and skin nodules. Beyond the skin, PN involves circulating blood inflammation that may contribute to systemic disease comorbidities. Dupilumab was recently approved for treatment of PN, but its effects on systemic inflammation are unknown. Thus, we aimed to characterize changes in plasma concentrations of inflammatory proteins after dupilumab treatment. In this exploratory study, plasma samples were collected from 3 patients with moderate-to-severe PN before and after ≥6 months of dupilumab treatment. All patients exhibited clinically significant improvements after treatment. Of the 2569 proteins tested, 186 were differentially expressed after treatment (q < 0.1, fold change > 1.3). Downregulated proteins included cytokines associated with T helper (Th) 1 (IFN-γ, TNF-α), Th2 (IL-4, IL-13), and Th17/Th22 (IL-6, IL-22) signaling. Markers of innate immunity (IL-19, toll-like receptor 1, nitric oxide synthase 2), immune cell migration (CCL20, CD177), and fibrosis (IL-11, IL-22) were also decreased (q < 0.1). Gene set variation analysis of Th2, Th17, and epithelial–mesenchymal transition gene sets showed reduced pathway expression in the post-treatment cohort (P < .05). Plasma cytokine levels of IL-11, nitric oxide synthase 2, IL-13, IL-4, and IFNG (R2 > 0.75, q < 0.10) showed the strongest correlations with pruritus severity. Dupilumab may reduce systemic inflammatory proteins associated with multiple immune and fibrosis pathways in patients with PN, potentially modulating the development of systemic disease comorbidities.
Keywords: Biologics, Immunomodulatory therapy, Proteomics, Pruritus
Introduction
Prurigo nodularis (PN) is a chronic inflammatory skin disorder characterized by severe pruritus and fibrotic nodules, predominantly located on the limbs and trunk. PN significantly impairs QOL, driving heightened sleep disturbances and psychological distress (Whang et al, 2022).
Similar to other chronic inflammatory skin diseases such as psoriasis (Reich, 2012), PN also exhibits evidence of systemic inflammation. Patients with PN, for example, report itching in both affected and healthy-appearing skin areas, and systemic inflammation markers have been observed in the blood (Belzberg et al, 2021). Plasma studies in patients indicate elevated levels of IL-4, IL-22, and TNF-α, along with cutaneous dysregulation of T helper (Th) 1, Th17, and Th22 pathways (Sutaria et al, 2022). These findings, coupled with the notable prevalence of comorbidities such as kidney disease, chronic obstructive pulmonary disease, and congestive heart failure among patients with PN, indicate a potential link between cutaneous inflammation and systemic disease.
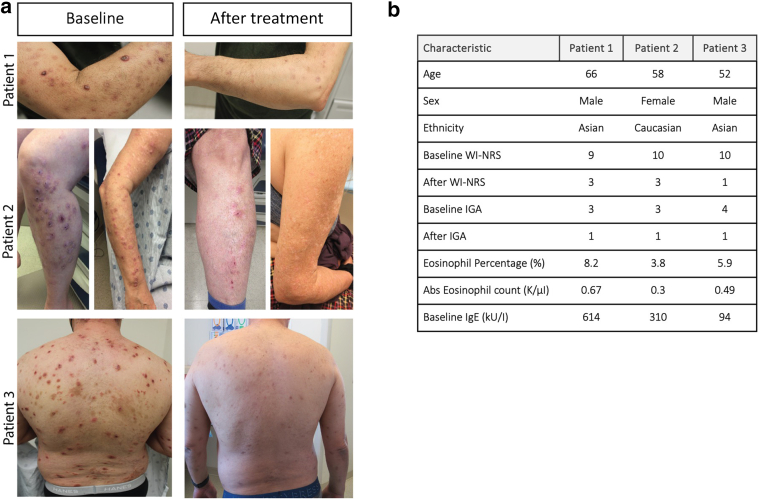
Dupilumab, a mAb that targets IL-4/IL-13 signaling by blocking the IL-4Rα, received regulatory approval for the treatment of PN after successful phase 3 clinical trials (Yosipovitch et al, 2023). However, the impact of dupilumab on systemic inflammation in PN remains unexplored. To address this knowledge gap, we conducted a preliminary study to analyze plasma obtained from 3 patients with PN before and after at least 6 months of sustained dupilumab therapy (Figure 1a and b).
Figure 1.
Clinical response to dupilumab treatment. (a) Baseline and post-treatment (≥6 months of dupilumab therapy) photographs. (b) Patient demographic and relevant clinical information. All participants provided consent for the publication of the images presented. IGA, Investigator’s Global Assessment; WI-NRS, Worst Itch Numeric Rating Scale.
Results
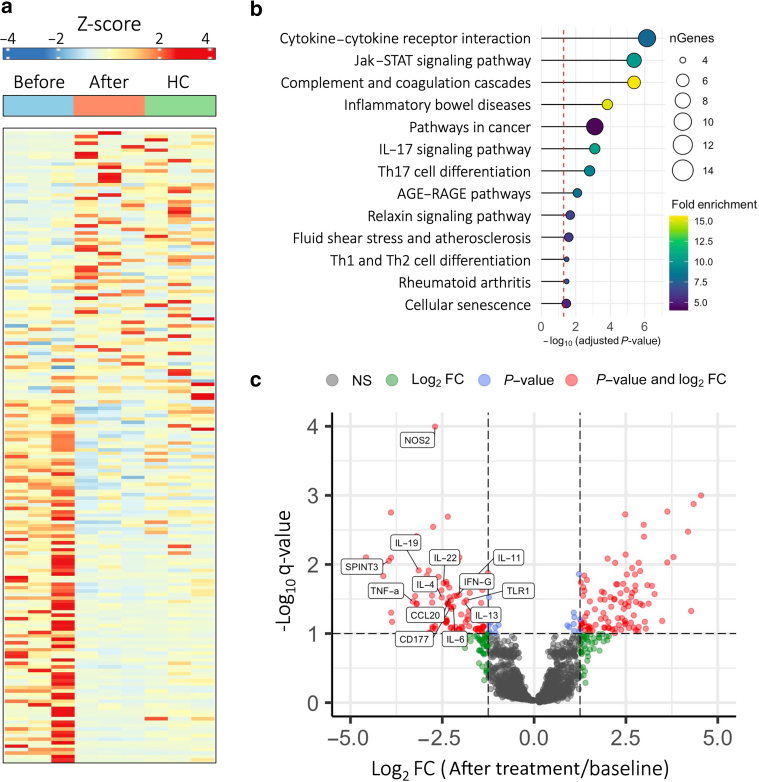
Of the 2569 plasma proteins tested, 184 plasma proteins (7%) exhibited significant changes in expression after dupilumab therapy (Figure 2a). A total of 126 plasma proteins (68%) were significantly upregulated, whereas 58 plasma proteins (32%) were significantly downregulated after treatment. Pathway enrichment analysis (Ge et al, 2020) revealed enrichment of distinct Kyoto Encyclopedia of Genes and Genomes pathways, including cytokine–cytokine receptor interactions, Jak–signal transducer and activator of transcription signaling, and IL-17 signaling (P < .001, fold enrichment > 7) (Figure 2b). Pathways related to cancer and inflammatory bowel disease were also found to be enriched (P < .05, fold enrichment > 5).
Figure 2.
Systemic immunomodulatory effects of dupilumab in prurigo nodularis. (a) Heatmap depicting relative plasma expression levels (Z-Score) of differentially expressed proteins at baseline (before), after treatment (After), and in matched HCs. Color intensity reflects relative expression levels. (b) Pathway enrichment analysis displaying top enriched pathways in the differentially expressed protein set. Bubble size corresponds to number of differentially expressed genes, and color intensity demonstrates fold enrichment. (c) Volcano plot illustrating fold change (after treatment/baseline) versus adjusted q-value obtained from moderated t-tests. Higher absolute magnitude signifies greater changes in expression, and negative and positive FCs represent decreased and increased expression after treatment, respectively. Differentially expressed and immune-related proteins are highlighted. FC, fold change; HC, healthy control; STAT, signal transducer and activator of transcription; Th, T helper.
Differential analysis of individual plasma proteins revealed elevations in key markers of Th1 (IFN-γ, TNF-α), Th2 (IL-4, IL-13), and Th17/22 (IL-6/IL-22) signaling at baseline compared with those in matched healthy controls (fold change > 1.3, q < 0.1). After dupilumab therapy, these markers displayed significant reductions in plasma concentration (Figure 2c). Proteins related to innate immunity (IL-19, toll-like receptor 1, nitric oxide synthase 2), wound repair (IL-11, SPINT3, IL-22), and immune cell migration (CCL20, CD177) exhibited a similar trend.
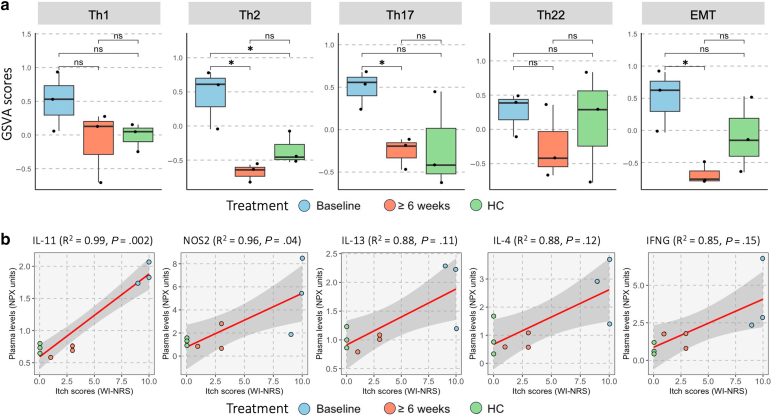
Gene set variation analysis (GSVA) of established immune pathways (Hänzelmann et al, 2013) revealed reduced post-treatment expression of gene sets associated with Th2, Th17, and epithelial–mesenchymal transition (P < .05). The Th2-associated gene set was also significantly enriched at baseline relative to that in healthy controls (P < .05). Th1- and Th22-associated gene sets also displayed a nonstatistically significant reduction in mean GSVA score after treatment (Figure 3a).
Figure 3.
GSVA and cytokine–itch correlation. (a) GSVA scores of each patient group using 5 distinct gene sets representing Th1, Th2, Th17, Th22, and EMT pathways. ∗P < .05. (b) Plasma cytokine levels (NPX) and pruritus severity (WI-NRS) were correlated with patient cohort (3 baseline, 3 after dupilumab treatment) and 3 healthy controls. EMT, epithelial–mesenchymal transition; GSVA, gene set variation analysis; NPX, normalized protein unit; ns, not significant; Th, T helper; WI-NRS, Worst Itch Numeric Rating Scale.
To identify cytokines potentially driving itch sensations, we correlated healthy control and patient Worst Itch Numeric Rating Scale scores with plasma protein levels at baseline and after treatment. Among these, IL-11, nitric oxide synthase 2, IL-13, IL-4, and IFNG exhibited the strongest positive correlation trends (R2 > 0.85 and P < .15) (Figure 3b).
Discussion
Our study outcomes suggest that systemic impact of dupilumab may extend beyond a reduction in Th2 inflammation. We observed significant downregulation of the Th17 pathway, alongside alterations in key Th1- and Th22-associated proteins. These findings could be attributed to the role of Th2 signals in activating dendritic cells and promoting subsequent Th1/Th17 differentiation (Segura et al, 2013). Prior research has also established the importance of Th2/Th22 coregulatory cytokines in the pathogenesis of atopic dermatitis, emphasizing the frequent involvement of multiple immune axes in inflammatory skin conditions (Lou et al, 2017).
Furthermore, we identified a broader impact of dupilumab on innate immunity–related (IL-19, TNF-α, toll-like receptor 1, nitric oxide synthase 2) and chemokine (CCL20, CD177) proteins. These findings suggest a suppression of the inflammatory cascade and immune cell recruitment, emphasizing the potential of dupilumab to attenuate both local and systemic immune-mediated pathologies in PN.
Our study also highlights the involvement of fibroproliferative pathways in PN pathogenesis. Baseline upregulation of proteins associated with keratinocyte proliferation (IL-22), TGF-β signaling (SPINT3), and profibrotic processes (IL-11) demonstrated a notable reduction after treatment (Fung et al, 2022; Zhuang et al, 2020). Particularly, IL-11 exhibited the strongest correlation with patient itch scores in our study, and elevated expression has been linked to chronic inflammatory conditions such as chronic obstructive pulmonary disease, rheumatoid arthritis, and psoriasis (Fung et al, 2022). This trend in fibroproliferative cytokines aligned with reduced GSVA scores for the epithelial–mesenchymal transition gene set, comprised of markers relevant to wound healing, fibrosis, and metastasis.
Our plasma cytokine findings corroborate recent single-cell RNA-sequencing studies demonstrating the unique upregulation of extracellular matrix organization, collagen synthesis, and stromal remodeling (Alkon et al, 2023) in the skin of patients with PN. Our study expands this understanding by demonstrating the potential reversibility of the systemic fibrotic phenotype with dupilumab, paralleling single-cell observation studies demonstrating normalization of immune–stromal crosstalk after nemolizumab treatment (Ma et al, 2024). The potential normalization of fibrotic pathways with dupilumab is clinically relevant, given the high prevalence of comorbid fibroproliferative diseases such as renal sclerosis and pulmonary fibrosis in PN (Patel et al, 2024). These antifibrotic effects provide a compelling mechanism through which dupilumab could mitigate systemic comorbidities in PN. Our preliminary results contribute to the characterization of the systemic fibrotic profile of PN and offer insights into its potential reversibility with targeted biologic therapy.
This study has several limitations worth noting. Most critically, the small sample size restricts the generalizability of our findings. Although statistically feasible for microarray analyses (Lin et al, 2010), small sample sizes carry tradeoffs between power and sensitivity, increasing the potential of false positives in both our differential expression and GSVA results. To address this risk, we implemented adjustments for false discoveries at a pre-established threshold (false discovery rate < 0.1 or a false positive rate of 10%), thus increasing the reliability of the reported significant changes in protein expression. Another limitation arises from the variable duration of dupilumab treatment at the time of the after blood draw, stemming from the real-world nature of the study. Diverse participant follow-up patterns led to early blood draws for some, whereas others, facing barriers to care, had later blood draws. Finally, different ethnic backgrounds may have underlying immunological differences. Efforts were made to address this limitation and maximize local statistical power through a pairwise study design and addition of matched controls. Given the substantial fold changes before and after treatment and the pilot nature of the study, these limitations were deemed tolerable a priori.
Nevertheless, our study provides preliminary insights into the systemic immune dysregulation in PN, marked by the modulation of proinflammatory cytokines associated with Th2, Th17, and fibroproliferative pathways. The reduction of these systemic biomarkers after clinical improvement of PN with dupilumab therapy supports a connection between PN’s cutaneous and systemic manifestations and suggests potential for mitigation of PN-associated comorbidities linked to chronic systemic inflammation. To substantiate the broader applicability of our observations, further cohort studies with larger sample sizes and longitudinal designs are imperative. These future investigations will be vital in delineating the temporal dynamics of cytokine changes during therapy and confirming the generalizability of our initial findings. In conclusion, our study lays the groundwork for a preliminary hypothesis regarding how dupilumab modulates systemic inflammatory networks implicated in PN pathogenesis and comorbidities, paving the way for more extensive and nuanced research on the subject.
Materials and Methods
Cohort selection
Participants were recruited from a tertiary academic medical center. Inclusion criteria encompassed a diagnosis of PN by a board-certified dermatologist; the presence of ≥20 bilateral nodules; and a Worst Itch Numeric Rating Scale score ≥7, with 0 and 10 representing no itch and the most severe imaginable itch, respectively. In addition, patients must have received ≥6 consecutive months of dupilumab therapy (600 mg loading dose, followed by 300 mg biweekly) and exhibited clinically significant improvements in itch and cutaneous disease, defined by a ≥4-point reduction in Worst Itch Numeric Rating Scale scores and an endpoint Investigator’s Global Assessment score ≤1 (Figure 1a and b). Topical corticosteroids were concurrently used with dupilumab as the exclusive systemic therapy. Demographics and clinical characteristics of patients were recorded at the initial blood draw (Figure 1a and b). Healthy patients were matched on the basis of age, sex assigned at birth, and race. Within the control cohort, there were no major comorbidities, and patient laboratory values were in normal range. All participants provided consent for the publication of the images presented.
Sample processing
Plasma samples were isolated from the whole blood by centrifugation for 10 minutes at 1960g. Plasma was then carefully separated, avoiding the buffy coat, and transferred into separate tubes to create 1 ml aliquots, with at least 4 aliquots isolated. The aliquots were slowly frozen at a temperature of −80 °C for 24–72 hours and transferred into liquid nitrogen vapor phase for long-term storage.
Differentially expressed proteins
Plasma samples underwent testing using proteomic immunoassays (Olink Explore 3072). A total of 372 proteins with a missing data frequency exceeding 80% were excluded from analysis. The limma package (version 3.58.1) in R (version 4.3.2) facilitated moderated pairwise 2-tailed t-tests for comparing cytokine levels at baseline with those after treatment. False discovery rates were applied to nominal P-values to obtain adjusted q-values. Similar to criteria in prior studies, proteins were deemed differentially expressed if they met the criteria of an absolute log2 fold change exceeding 1.3 and a false discovery rate–adjusted q-value below 0.1 (Al-Nesf et al, 2022).
Pathway enrichment, GSVA, and itch correlation analysis
Pathway enrichment analysis utilized ShinyGo (version 0.77), employing all differentially expressed genes as input, with the total panel genes as background. GSVA was conducted using the GSVA package (version 1.50.0), and gene sets were obtained from the Gene Set Enrichment Analysis Molecular Signatures Database. Similar to the analysis of differentially expressed proteins, statistical differences in GSVA enrichment scores between groups were obtained through moderated pairwise 2-tailed t-tests using limma (He et al, 2022). Itch correlation analysis involved a correlation test for repeated measures between Worst Itch Numeric Rating Scale and cytokine plasma expression levels for a total of 9 sample points (3 baseline, 3 after treatment, and 3 healthy controls) using the rmcorr package (version 0.6.0).
Disclaimer
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Ethics Statement
The study was conducted at Johns Hopkins Medicine and received approval from the Johns Hopkins Medicine Institutional Review Board. Written, informed consent was obtained from all patients and control participants. Adhering to ethical principles of human research regulations, the research team prioritized the protection of participants’ rights, welfare, and privacy, employing secure measures for storing and handling identifiable patient information. The study, exclusively focused on human plasma samples, excluded the use of animal subjects. The institutional review board approval encompassed all aspects of the research, spanning data collection, analysis, and dissemination of results. The authors affirm their compliance with necessary approvals and documentation, with the corresponding author ready to furnish proof of institutional review board approval in response to potential inquiries.
Data Availability Statement
The prurigo nodularis data are not publicly available owing to patient wishes but are available upon reasonable request to the corresponding author at kwatra.shawn@gmail.com.
ORCIDs
Aaron Bao: http://orcid.org/0000-0002-2069-9985
Hannah Cornman: http://orcid.org/0000-0003-1462-2479
Emily Ma: http://orcid.org/0000-0001-6022-6659
Anusha Kambala: http://orcid.org/0000-0002-0350-8622
Jaya Manjunath: http://orcid.org/0000-0002-4455-4764
Alexander L. Kollhoff: http://orcid.org/0000-0003-2026-6781
Brenda Umenita Imo: http://orcid.org/0009-0008-8481-4031
Madan M. Kwatra: http://orcid.org/0000-0002-6547-8852
Shawn G. Kwatra: http://orcid.org/0000-0003-3736-1515
Conflict of Interest
SGK is an advisory board member/consultant for Abbvie, Aslan Pharmaceuticals, Arcutis Biotherapeutics, Celldex Therapeutics, Galderma, Genzada Pharmaceuticals, Incyte, Johnson & Johnson, Novartis Pharmaceuticals, Pfizer, Regeneron Pharmaceuticals, and Sanofi and has served as an investigator for Galderma, Pfizer, Incyte, and Sanofi. All other authors declare they have no competing interests.
Acknowledgments
SGK is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number K23AR077073-01A1.
Author Contributions
Conceptualization: AB, HC, SGK; Data Curation: AB, HC, AK, SGK, MMK; Formal Analysis: AB, HC, MMK; Methodology: AB, HC, SGK; Supervision: SGK; Visualization: AB, HC, SGK; Writing - Original Draft Preparation: AB, HC; Writing - Review and Editing: AB, HC, EM, AK, JM, ALK, BUI
Declaration of Artificial Intelligence (AI)/Large Language Models (LLMS) Use
The author(s) did not use AI/LLM in any part of the research process and/or manuscript preparation.
accepted manuscript published online XXX; corrected published online XXX
References
- Alkon N., Assen F.P., Arnoldner T., Bauer W.M., Medjimorec M.A., Shaw L.E., et al. Single-cell RNA sequencing defines disease-specific differences between chronic nodular prurigo and atopic dermatitis. J Allergy Clin Immunol. 2023;152:420–435. doi: 10.1016/j.jaci.2023.04.019. [DOI] [PubMed] [Google Scholar]
- Al-Nesf M.A.Y., Abdesselem H.B., Bensmail I., Ibrahim S., Saeed W.A.H., Mohammed S.S.I., et al. Prognostic tools and candidate drugs based on plasma proteomics of patients with severe COVID-19 complications. Nat Commun. 2022;13:946. doi: 10.1038/s41467-022-28639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzberg M., Alphonse M.P., Brown I., Williams K.A., Khanna R., Ho B., et al. Prurigo nodularis is characterized by systemic and cutaneous T helper 22 immune polarization. J Invest Dermatol. 2021;141:2208–2218.e14. doi: 10.1016/j.jid.2021.02.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung K.Y., Louis C., Metcalfe R.D., Kosasih C.C., Wicks I.P., Griffin M.D.W., et al. Emerging roles for IL-11 in inflammatory diseases. Cytokine. 2022;149 doi: 10.1016/j.cyto.2021.155750. [DOI] [PubMed] [Google Scholar]
- Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Fan B., Du P., Jin Y. Construction and validation of two hepatocellular carcinoma-progression prognostic scores based on gene set variation analysis. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.806989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.J., Hsueh H.M., Chen J.J. Power and sample size estimation in microarray studies. BMC Bioinformatics. 2010;11:48. doi: 10.1186/1471-2105-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H., Lu J., Choi E.B., Oh M.H., Jeong M., Barmettler S., et al. Expression of IL-22 in the skin causes Th2-biased immunity, epidermal barrier dysfunction, and pruritus via stimulating epithelial Th2 cytokines and the GRP pathway. J Immunol. 2017;198:2543–2555. doi: 10.4049/jimmunol.1600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Plazyo O., Billi A.C., Tsoi L.C., Xing X., Wasikowski R., et al. Single cell and spatial sequencing define processes by which keratinocytes and fibroblasts amplify inflammatory responses in psoriasis. Nat Commun. 2023;14:3455. doi: 10.1038/s41467-023-39020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J.R., Joel M.Z., Lee K.K., Kambala A., Cornman H., Oladipo O., et al. Single-cell RNA sequencing reveals dysregulated POSTN+WNT5A+ fibroblast subclusters in prurigo nodularis [e-pub ahead of print] J Invest Dermatol. 2024 doi: 10.1016/j.jid.2023.12.021. [DOI] [PubMed] [Google Scholar]
- Reich K. The concept of psoriasis as a systemic inflammation: implications for disease management. J Eur Acad Dermatol Venereol. 2012;26:S3–S11. doi: 10.1111/j.1468-3083.2011.04410.x. [DOI] [PubMed] [Google Scholar]
- Segura E., Touzot M., Bohineust A., Cappuccio A., Chiocchia G., Hosmalin A., et al. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. 2013;38:336–348. doi: 10.1016/j.immuni.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Sutaria N., Alphonse M.P., Marani M., Parthasarathy V., Deng J., Wongvibulsin S., et al. Cluster analysis of circulating plasma biomarkers in prurigo nodularis reveals a distinct systemic inflammatory signature in African Americans. J Invest Dermatol. 2022;142:1300–1308.e3. doi: 10.1016/j.jid.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang K.A., Le T.K., Khanna R., Williams K.A., Roh Y.S., Sutaria N., et al. Health-related quality of life and economic burden of prurigo nodularis. J Am Acad Dermatol. 2022;86:573–580. doi: 10.1016/j.jaad.2021.05.036. [DOI] [PubMed] [Google Scholar]
- Yosipovitch G., Mollanazar N., Ständer S., Kwatra S.G., Kim B.S., Laws E., et al. Dupilumab in patients with prurigo nodularis: two randomized, double-blind, placebo-controlled phase 3 trials. Nat Med. 2023;29:1180–1190. doi: 10.1038/s41591-023-02320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L., Ma W., Yan J., Zhong H. Evaluation of the effects of IL-22 on the proliferation and differentiation of keratinocytes in vitro. Mol Med Rep. 2020;22:2715–2722. doi: 10.3892/mmr.2020.11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The prurigo nodularis data are not publicly available owing to patient wishes but are available upon reasonable request to the corresponding author at kwatra.shawn@gmail.com.