Abstract
In recent years, there has been a growing interest in modulating the performance of probiotic, mainly Lactic Acid Bacteria (LAB), in the field of probiotic food. Attenuation, induced by sub-lethal stresses, delays the probiotic metabolism, and induces a metabolic shift as survival strategy. In this paper, RNA sequencing was used to uncover the transcriptional regulation in Lacticaseibacillus casei ATCC 393 after ultrasound-induced attenuation. Six (T) and 8 (ST) min of sonication induced a significant differential expression of 742 and 409 genes, respectively. We identified 198 up-regulated and 321 down-regulated genes in T, and similarly 321 up-regulated and 249 down-regulated in ST. These results revealed a strong defensive response at 6 min, followed by adaptation at 8 min. Ultrasound attenuation modified the expression of genes related to a series of crucial biomolecular processes including membrane transport, carbohydrate and purine metabolism, phage-related genes, and translation. Specifically, genes encoding PTS transporters and genes involved in the glycolytic pathway and pyruvate metabolism were up-regulated, indicating an increased need for energy supply, as also suggested by an increase in the transcription of purine biosynthetic genes. Instead, protein translation, a high-energy process, was inhibited with the down-regulation of ribosomal protein biosynthetic genes. Moreover, phage-related genes were down-regulated suggesting a tight transcriptional control on DNA structure. The observed phenomena highlight the cell need of ATP to cope with the multiple ultrasound stresses and the activation of processes to stabilize and preserve the DNA structure. Our work demonstrates that ultrasound has remarkable effects on the tested strain and elucidates the involvement of different pathways in its defensive stress-response and in the modification of its phenotype.
Keywords: Probiotic, Attenuation, RNA sequencing, Metabolic change
1. Introduction
Lactic Acid Bacteria (LAB) are well characterized bacteria for their technological [1], [2], [3] and beneficial properties [4], [5]. The primary and secondary metabolic pathways of LAB have been exploited to formulate foods with targeted rheological and sensory properties [6], increase the bioavailability of food components [7], formulate balanced dietary supplements for targeted human body functions [8], and bio-convert waste [9]. The wide range of applications of LAB also depends on their high adaptability and resistance to various environmental conditions. In fact, LAB exert high survival rate during the challenging conditions of food processing, storage, and digestion [10], [11], [12]. Depending on the specific application, the activities of these bacteria could be enhanced or reduced. Gene transfer and bioengineering are common approaches used for these purposes [13], [14]. However, the relatively high cost limits their applicability. Therefore, in recent years different strategies aiming at modulating the target microbial performance have been investigated, especially in the probiotics field.
Probiotics comprehend strains of different species and genera, although microorganisms belonging to the Lactobacillus genus (recently refined in 23 distinct genera and three emended genera) are the most used ones. Probiotics, as defined by the International Scientific Association for Probiotics and Prebiotics [15], are mostly supplemented in the diet through functional foods [16]. In such context, attenuation, that is the modulation of probiotics performances, helps in counteracting physico-chemical and sensorial deviations of food characteristics that may be induced by bacteria metabolites released in the food matrix [17]. However, attenuation implies both controlling of metabolism and retention of viability. Thermal shock is one of the most established technologies used to attenuate microbial metabolism, however it comes with a loss of viability [18]. This problem can be overcome by alternative non-thermal technologies which may also provide eco-friendly and less time-consuming solutions.
In this context, sonication has been shown to be successful in the food area for a wide range of food processing and quality control applications [19], [20], [21]. Focusing on quality control, ultrasound has been applied to preserve heat-sensitive compounds in water disinfection and beverage [22]. Ultrasound triggers in bacteria multiple damages to the cell membrane, nucleic acids, and proteins. An important aspect is that the efficacy of ultrasound in bacteria inactivation strictly depends on the target bacteria and on process conditions, especially in terms of treatment intensity [23]. High intensity treatments lead to irreversible damages causing the inactivation or death of bacteria. On the other hand, low intensity treatments lead to transient damages that modulate or delay bacteria metabolism [23]. Therefore, ultrasound can be exploited to induce stress, a relevant factor in the probiotic field for attenuation purposes.
Stress is defined as a transition from a normal to an abnormal condition that causes changes in the cell genome, transcriptome, proteome, and/or metabolome. Large literature has shown that microbial cells have decreased survival potential, slower or reduced growth and metabolic activity under stressful conditions [24]. Therefore, exposure of microorganisms to stressful environments or treatments is one of the approaches currently being evaluated to modulate their metabolism. However, challenges arise when ultrasound must be applied efficiently to probiotic attenuation since it is necessary to know and control the cell stress response. LAB employ various resistance strategies to face different abiotic and biotic stressors. Main stress response mechanisms include cell energy conservation, macromolecular defence, and cell envelope protection. In addition, stress response mechanisms can be categorized as specific or generic and together define the bacterial stressome. As a general stress response, bacteria shift to a metabolic pattern that ensures their survival [24]. In addition, LAB protect themselves through the mechanism of cross-protection [25], which is observed under the sequential exposure of bacteria to mild and severe stressors. Mild stressor activates stress response pathways, triggering the synthesis of specific protective proteins that also contribute to cell resistance against unrelated and unspecific stressors.
Response of LAB to stress can be evaluated using a multi-omics approaches [26]. Among them, RNA sequencing (RNA-Seq) is a powerful tool to uncover global changes in bacterial gene expression and the intricate gene regulatory networks associated with stress response mechanisms. It belongs to the Next Generation Sequencing (NGS) technologies and main outcome is represented by cell transcripts [27]. Such technology can be exploited to assess probiotics response to the detrimental effects of ultrasound stress, for which no studies have been performed in the literature.
The aim of this study was to elucidate mechanisms of action of ultrasound on probiotics. More specifically, we assessed response of the probiotic strain Lacticaseibacillus casei ATCC 393 to ultrasound attenuation and evaluated differential gene expression patterns through a transcriptomic analysis based on RNA-Seq. We also wanted to identify key differentially expressed genes to modulate the target metabolism in a controllable and reproducible manner.
2. Materials and Methods
2.1. Bacteria preparation
Lacticaseibacillus casei ATCC 393 was used in this study as a probiotic model, and it was purchased from American Type Culture Collection. The strain was kept at −20 °C in MRS broth (OXOID Ltd., Basingstoke, Hampshire, England) supplemented with glycerol (30 %). Before each assay, the strain was cultured in MRS broth for 18 h at 37 °C (9 Log CFU/ml). Before each sonication assay, Lc. casei ATCC 393 was cultured in 90 ml of MRS broth and then divided in 3 aliquots of 30 ml. Two aliquots were sonicated, and one was used as control (C). The cell pellet was collected by centrifugation (6000 x g, 15 min, 4 °C) and resuspended in equal volume of deionized water.
2.2. Ultrasonic exposure
The experiments were performed at the Institute of Agrochemistry and Food Technology (IATA) (Valencia, Spain) using the SONOPLUS HD 2200 ultrasonic homogenizer with a 12 mm diameter titanium probe (BANDELIN electronic GmbH & Co. KG, Germany). The operating frequency was 20 kHz, and the maximum net power of the ultrasound device was 200 W. The amplitude was set to 28.5 % to obtain a working power of 57 W. The energy density of the treatments was calculated by applying Formula 1, as described by Huang et al. [28]:
| (1) |
where P is the power applied (W), t is the time of the treatment (s), and V is the volume of the treated suspension (ml).
The probiotic water suspension in conical plastic tubes (Falcon®) was exposed to ultrasound in pulsed mode (50 % duty cycle) for 6 min (Treated, T) to achieve an ED of 684 J/ml and 8 min (Super-Treated, ST) to achieve an ED of 912 J/ml.
The probe was placed 2 cm from the bottom of the tube. The samples were kept in an ice box during the ultrasound treatments to dissipate the heat generated during the treatment. Three independent sonication experiments were performed.
2.3. Total RNA isolation
The pellet of 100 µl of probiotic water suspensions for each sample was collected by centrifugation, suspended in RNA later solution and rapidly frozen. The pellet was stored at −80 °C until use for RNA extraction.
Total DNA and RNA were extracted from each sample using the DNA/RNA Patho Gene-spin Extraction kit (INtRON Biotechnology, Korea) according to the standard protocol. Briefly, bacterial pellet was lysed with 300 µl lysis buffer, and the lysate was loaded onto a column. It was subjected to 3 washes and eluted with 30 µl elution buffer. The RNA was subjected to clean-up and digestion with DNAseI using the RNA Clean & Concentrator Kit (Zymo Research), following the manufacturer's recommendations. The quantity and quality of RNA was assessed through a capillary gel electrophoresis performed by the Fragment Analyzer. RNA Quality Number (RQN) was used as quantitative indication of RNA integrity. Integer RNA was declared for an RQN > 7.
2.4. cDNA library construction and sequencing
The cDNA library construction and sequencing were performed at the Seqplexing S.L. (Valencia, Spain). Bacterial rRNA was removed using the RiboCopTM for Bacteria (Lexogen GmbH, Vienna, Austria). A DNase treatment was also applied to purify the sample using DNA-free™ Kit DNase Treatment and Removal Reagents (Ambion®, Germany). The cDNA library quality was evaluated using the QIAxcel Advaned System (Qiagen, Germany). The final cDNA library was sequenced using 3′mRNA sequencing by use of PE 150x2 paired-end with 10 millions of reads/sample using NextSeq500 (Illumina Platform).
2.5. Bioinformatic data analysis
Bioinformatic data analysis was performed on the open-source web-based platform Galaxy. The raw data files from the RNA-Seq analysis were imported into the bioinformatics software and further analysed through RStudio tool. The data were filtered to remove low quality reads through fastqc and trimmed using cutadapt. We downloaded the latest reference genome of Lc. casei ATCC 393 (GCF_000829055.1) from the NCBI website) (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000829055.1/) with the associated annotation file. We performed RNA-Seq alignment using the STAR software and computed counts of aligned reads through the command featureCounts. https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000829055.1/Differential gene expression (DGE) analysis was performed using the edgeR tool. The gene expression fold changes (log2FC) were calculated by comparing treated vs control samples. False discovery rate (FDR) was applied on P-values to perform multiple hypothesis testing correction, and significant differential gene expression was declared for FDR < 0.05. The threshold for differentially expressed genes (DEGs) was log2FC < -1 for down regulated genes and log2FC > 1 for up regulated genes. Finally, DEGs functional categories were individuated based on Gene Ontology Enrichment analysis and consulting the Kyoto Encyclopedia of Genes and Genomes (KEGG). The genetic circular map displaying the distribution and expression patterns of DEGs was generated using the Circular Genomic Viewer available in BioCyc.
3. Results and discussion
3.1. Differential gene expression (DGE) analysis
Lc. casei ATCC 393 was sonicated to attenuate its metabolism and delay the metabolic activities related to the utilization of food components. Treated (T; 6 min of sonication) and Super-Treated (ST; 8 min of sonication) samples were compared with the non-treated strain used as a control reference (C). Three independent sonication experiments were performed and generated a total of 9 samples that were sequenced through RNA-Seq to perform DGE analysis between treated and untreated samples (see Methods).
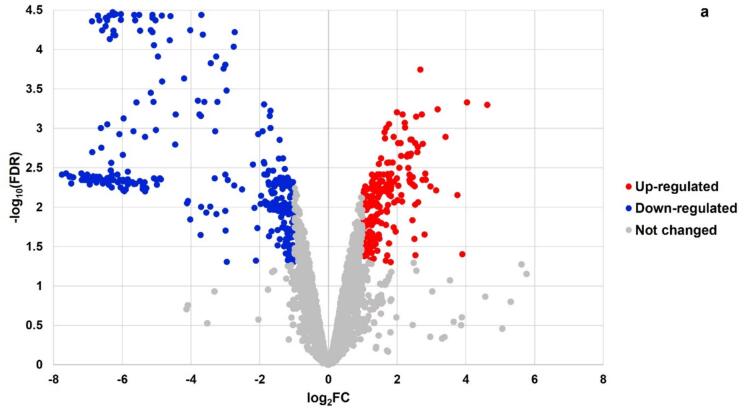
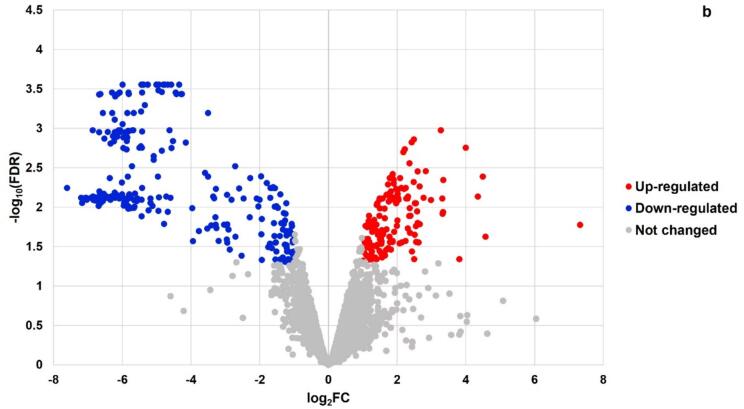
The Lc. casei ATCC 393 is composed by a total of 2882 annotated genes, and we identified hundreds of them as DEGs by RNA-Seq analysis in both sonicated samples (Table 1). More specifically, we detected 742 DEGs (FDR < 0.05) in T and 409 DEGs in ST. In addition, 6 min of sonication induced up-regulation (log2FC > 1) of 198 genes (Fig. 1a), and similarly down-regulation (log2FC < -1) of 321 genes (Fig. 1b). A similar pattern was observed after 8 min of sonication, with 140 up-regulated and 249 down-regulated genes. Moreover down-regulated genes were expressed with a higher fold change in both samples.
Table 1.
Summary of RNA-Seq results in terms of number of genes. T: Lc. casei ATCC 393 sonicated for 6 min; ST: Lc. casei ATCC 393 sonicated for 8 min.
| T | ST | |
|---|---|---|
| Total number of genes | 2882 | 2882 |
| DEGs (FDR < 0.05) | 742 | 409 |
| DEGs up-regulated (log2FC > 1) | 198 | 140 |
| DEGs down-regulated (log2FC < -1) | 321 | 249 |
Fig. 1a.
Volcano plot illustrating the landscape of Differentially Expressed Genes (DEGs) of Lc. casei ATCC 393 after 6 min of sonication. Each point represents a gene (N = 2882 genes in total), with colors highlighting up-regulated (in red) and down-regulated (in blue) genes.
Fig. 1b.
Volcano plot illustrating the landscape of Differentially Expressed Genes (DEGs) of Lc. casei ATCC 393 after 8 min of sonication. Each point represents a gene (N = 2882 genes in total), with colors highlighting up-regulated (in red) and down-regulated (in blue) genes.
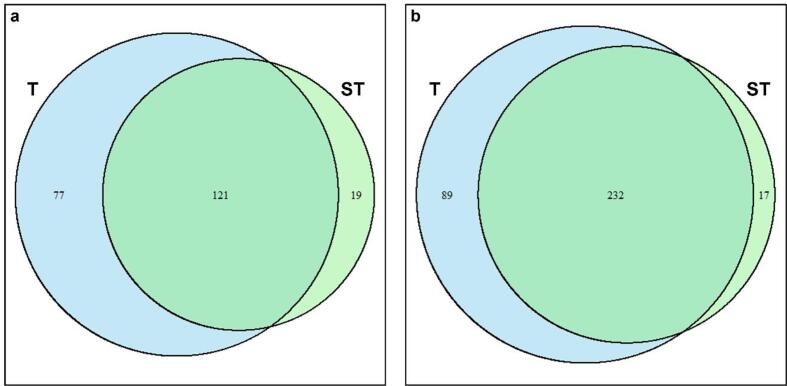
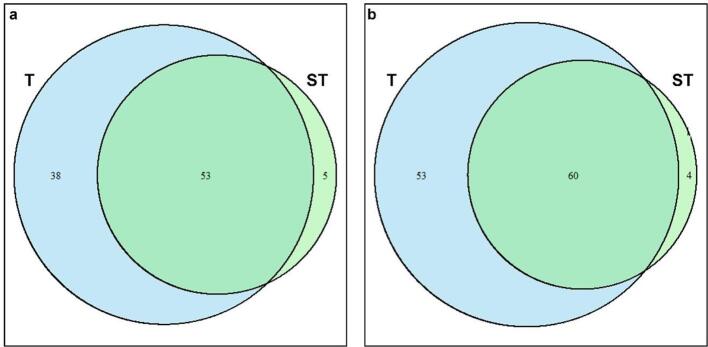
The number of DEGs shared between the two treated samples are visualized in the Venn diagram of Fig. 2, which also provides insights on the number of unique transcriptional responses. This visualization highlights the number of overlapping genes that may play critical roles in shared biological processes, a necessary step to clarify the complex interplay of gene expression patterns.
Fig. 2.
Venn Diagram illustrating the overlap of Differentially Expressed Genes (DEGs) between 6-min (T) and 8-min (ST) treated Lc. casei ATCC 393. Each circle represents the number of DEGs in the specific condition, with shared genes represented in the overlapping regions. The size of circles is proportional to the number of DEGs. a: up-regulated DEGs; b: down-regulated DEGs.
Among 217 DEGs up-regulated in at least one sample, the majority of them (121; 56 %) were found both in T and ST. On the other hand, 77 and 19 genes were up-regulated only in T and ST, respectively. Similarly, 232 of the DEGs down-regulated in T were also found in ST. Instead, only 89 and 17 down-regulated DEGs were specific to 6 min and 8 min of sonication, respectively. We speculate that a non-complete overlap of the up- and down-regulated DEGs across timepoints may be due to cross-protection and adaptation. The overlapping subset probably includes several genes related to general stress response. In addition, while the strain detects unfavorable conditions and activates a strong defensive response in the very first minutes of sonication, the cell seems to adapt to the new condition by decreasing the transcriptional response with prolonged exposure. A similar transcriptional adaptive response was observed by Fenouil et al. [29]. High hydrostatic pressure above the growth optimum range induced differential expression of several genes in Pseudothermotoga elfii DSM9442. The authors observed that the number of DEGs was higher under moderate pressure increase conditions than under extreme pressure increase conditions. This suggests that more intense stress conditions allow the bacteria to transcriptionally adapt to them.
3.2. Gene Ontology analysis of DEGs
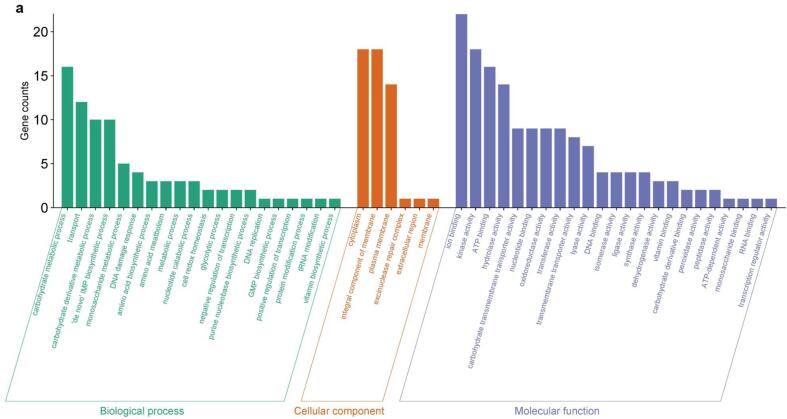
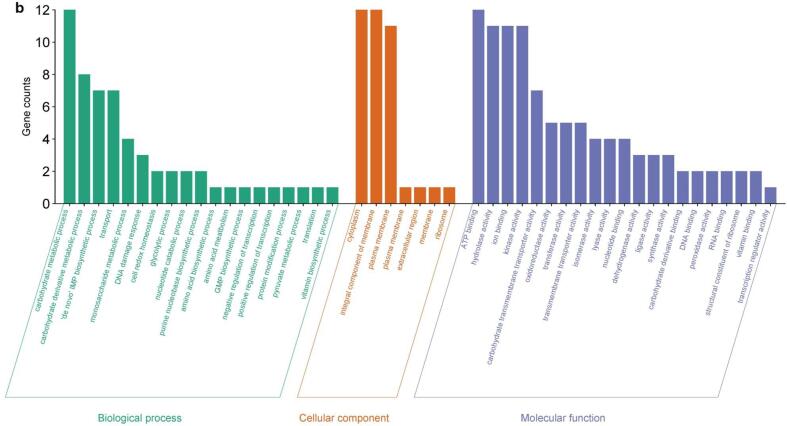
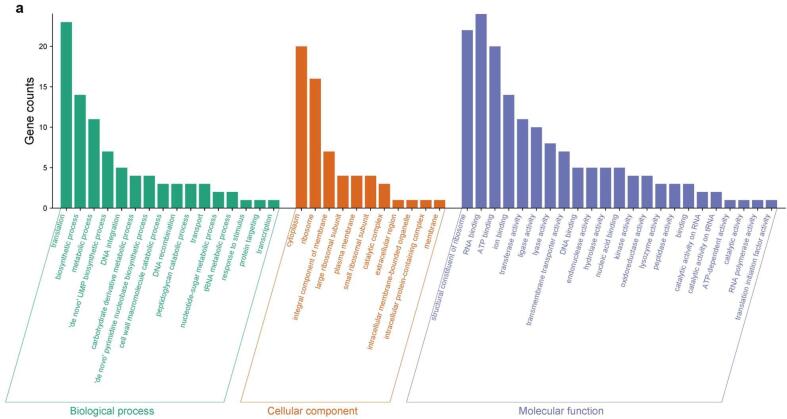
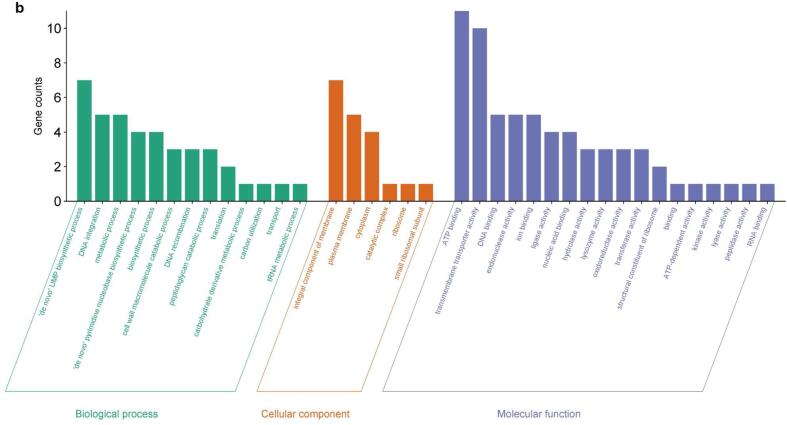
Annotated DEGs (see Methods) were grouped into three Gene Ontology (GO) categories: biological process (BP), cellular component (CC), and molecular function (MF). In each category, different GO terms (sub-categories) were enriched after sonication treatments. The number of DEGs across GO categories and sub-categories is shown in Fig. 3a, Fig. 3b and Fig. 4a, Fig. 4b for up-regulated and down-regulated genes, respectively.
Fig. 3a.
Grouping of the DEGs up-regulated in Lc. casei ATCC 393 after 6 min of sonication in three main GO categories: biological process (BP), cellular component (CC), and molecular function (MF).
Fig. 3b.
Grouping of the DEGs up-regulated in Lc. casei ATCC 393 after 8 min of sonication in three main GO categories: biological process (BP), cellular component (CC), and molecular function (MF).
Fig. 4a.
Grouping of the DEGs down-regulated in Lc. casei ATCC 393 after 6 min of sonication in three main GO categories: biological process (BP), cellular component (CC), and molecular function (MF).
Fig. 4b.
Grouping of the DEGs down-regulated in Lc. casei ATCC 393 after 8 min of sonication in three main GO categories: biological process (BP), cellular component (CC), and molecular function (MF.
This plot provides a comprehensive overview of the functional categories that are significantly affected by transcriptional changes and therefore helps to understand the molecular mechanisms shaping Lc. casei ATCC 393 transcriptome upon ultrasound exposure.
For up-regulated DEGs, the same GO subcategories were the most enriched in both T and ST: “carbohydrates metabolic process”, “transport”, “carbohydrate derivate metabolic process”, and “de novo IMP biosynthetic process” in BP; “cytoplasm”, “integral component of membrane”, and “plasma membrane” in CP; “ion binding”, “kinase activity”, “ATP binding”, and “hydrolase activity” in MF. More remarkable differences between T and ST were found in down-regulated DEGs. Here for BP, “translation”, “biosynthetic process”, “metabolic process”, “de novo UMP biosynthetic process”, and “DNA integration” were the most prevalent subcategories, with only the last three ones particularly enriched also in ST. For CP, “cytoplasm”, “ribosome”, “integral component of membrane”, and “large ribosomal subunit” were the main subcategories for T, while “integral component of membrane”, “plasma membrane” and “cytoplasm” were occurrent in ST. Finally, for MF the most enriched terms were “structural constituent of ribosome”, “RNA binding”, “ATP binding”, “ion binding”, and “transferase activity” in T, and “ATP binding”, “transmembrane transporter activity”, “DNA binding”, “endonuclease activity”, and “ion binding” in ST.
We also identified overlapping GO terms that are summarized as Venn Diagram in Fig. 5. The overlap ratio and the size of each subset were also observed after filtering the data. Interestingly, most of the GO terms identified in ST were also detected in T for both up-regulated (Fig. 5a) and down-regulated (Fig. 5b) genes, while a much larger number of terms were specific to T.
Fig. 5.
Venn Diagram illustrating the overlap of Differentially Expressed Genes (DEGs) belonging to the Gene Ontology database across 6-min (T) and 8-min (ST) treated Lc. casei ATCC 393. Each circle represents the number of DEGs in the specific condition, with shared genes represented in the overlapping regions. The size of circles is proportional to the number of DEGs. a: up-regulated DEGs; b: down-regulated DEGs.
3.3. Degs pathways
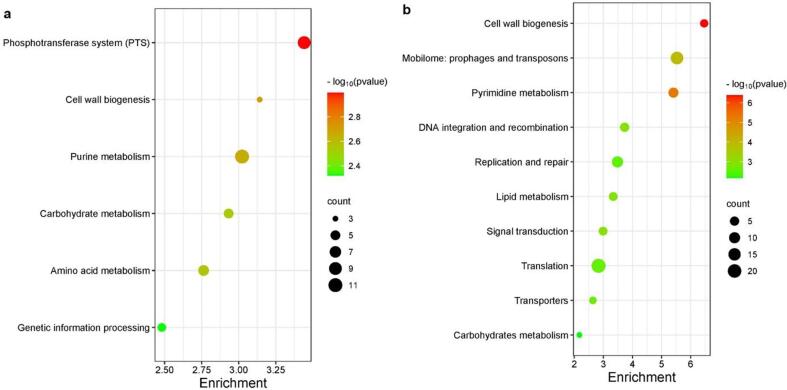
We further grouped the gene ontologies associated with DEGs to the main pathways in which they are involved using the KEGG database. The main pathways to which belong up- and down-regulated DEGs are shown in the bubble plot of Fig. 6a and Fig. 6b, respectively. This plot provides an overview of the major biological processes associated with the identified DEGs and highlights key pathways that play an important role in the molecular response of Lc. casei ATCC 393 to ultrasound attenuation.
Fig. 6.
Metabolic Pathway Analysis of Differentially Expressed Genes (DEGs) found in Lc. casei ATCC 393 upon sonication. The graphs were build based on the gene counts and the relative enrichment values (−log10(P)) for each category. Each circle represents a distinct biological pathway, and the circle size corresponds to the number of DEGs implicated in that particular pathway. The color differentiates between the enrichment value within each pathway. a: metabolic pathways of up-regulated DEGs; b: metabolic pathways of down-regulated DEGs.
Sonication induced up-regulation of genes involved in the phosphotransferase system (PTS), cell wall biogenesis, purine metabolism, amino acid metabolism, carbohydrate metabolism, and genetic information processing. More specifically, PTS and purine metabolism exhibited the highest gene counts and lowest P-values. Very different pathways were instead associated with down-regulated genes. Translation had the highest gene count, but with a low enrichment value. Cell wall biogenesis and pyrimidine metabolism were the pathways on which ultrasound had the major effect, and also the mobilome was characterized by a medium/high enrichment score. Other pathways detected at lower enrichment were DNA integration and recombination, replication and repair, lipid metabolism, signal transduction, transporter, and carbohydrates metabolism. These results underline that several pathways of Lc. casei ATCC 393 metabolism are affected by ultrasound.
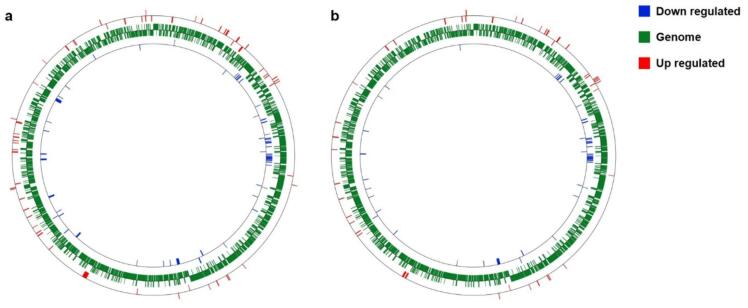
We also investigated the spatial distribution of transcriptional variations by building the circular genetic map (Fig. 7), which offers insights into genomic hotspots of differential expression and potential regulatory interactions. This circular layout facilitates the identification of regional gene clusters and aids in understanding the spatial context of DEGs within sonicated Lc. casei ATCC 393.
Fig. 7.
Genetic Circular Map displaying the distribution and expression patterns of Differentially Expressed Genes (DEGs) across 6-min (a) and 8-min (b) treated Lc. casei ATCC 393. The outer and the inner circle represents the genomic coordinates of up-regulated and down-regulated DEGs, respectively, across the Lc. casei ATCC 393 genome.
3.4. Investigation of the main pathways associated with DEGs
We list in Table 2 the up- and down-regulated DEGs found in each pathway with relative fold change, description of each gene function, and sample in which it was found. Here, we focus the discussion on providing insights on the most enriched and relevant pathways.
Table 2.
Main functional categories associated with Differentially Expressed Genes (DEGs) in ultrasound-treated Lc. casei ATCC 393. T: 6 min of sonication; ST: 8 min of sonication.
| Gene ID and functional category | Gene log2FCa | Descriptionb | Sample |
|---|---|---|---|
| Transports | |||
| Phosphotransferase system (PTS) | |||
| LBCZ_RS12590 | 4.04 | fructose-specific PTS transporter subunit EIIC | T, ST |
| LBCZ_RS13975 | 3.18 | PTS mannitol transporter subunit IICBA | T, ST |
| LBCZ_RS00290 | 2.36 | glucose PTS transporter subunit IIA | T, ST |
| LBCZ_RS13605 | 2.14 | lactose-specific PTS transporter subunit EIIC | T, ST |
| LBCZ_RS10155 | 1.92 | beta-glucoside-specific PTS transporter subunit IIABC | T, ST |
| LBCZ_RS09730 | 1.69 | PTS transporter subunit IIBCA | T |
| LBCZ_RS09655 | 1.28 | PTS transporter subunit EIIC | T, ST |
| LBCZ_RS02285 | 1.27 | PTS cellobiose transporter subunit IIC | T, ST |
| LBCZ_RS13310 | 1.06 | mannose/fructose/sorbose PTS transporter subunit IIA | T |
| Major Facilitator Superfamily | |||
| LBCZ_RS09005 | −1.02 | MFS transporter (Major Facilitator Superfamily) | ST |
| LBCZ_RS10135 | −1.03 | DHA2 family efflux MFS transporter permease subunit | ST |
| Carbohydrate metabolism | |||
| Glycolysis | |||
| LBCZ_RS12595 (pfkB) | 3.75 | 1-phosphofructokinase | T, ST |
| LBCZ_RS02130 (ldh) | 1.42 | L-lactate dehydrogenase | T, ST |
| Pyruvate metabolism | |||
| LBCZ_RS06080 (pdhA) | 2.11 | pyruvate dehydrogenase (acetyl-transferring E1 component subunit alpha) | T, ST |
| LBCZ_RS10870 (ppdk) | 1.24 | pyruvate, phosphate dikinase | T, ST |
| LBCZ_RS09930 (spxb) | 1.03 | pyruvate oxidase | T, ST |
| Nucleotide metabolism | |||
| Purine metabolism | |||
| LBCZ_RS08450 (purE) | 2.72 | 5-carboxyaminoimidazole ribonucleotide mutase | T, ST |
| LBCZ_RS08445 (purK) | 2.38 | 5-carboxyaminoimidazole ribonucleotide synthase | T, ST |
| LBCZ_RS00770 (guaB) | 1.83 | IMP dehydrogenase | T, ST |
| LBCZ_RS08430 (purQ) | 1.78 | phosphoribosylformylglycinamidine synthase subunit PurQ | T, ST |
| Nucleotide metabolism | |||
| Purine metabolism | |||
| LBCZ_RS08415 (purM) | 1.37 | phosphoribosylformylglycinamidine cyclo-ligase | T |
| LBCZ_RS08425 (purL) | 1.27 | phosphoribosylformylglycinamidine synthase subunit PurL | T, ST |
| LBCZ_RS11860 | 1.25 | redoxin domain-containing protein | T, ST |
| LBCZ_RS08420 (purF) | 1.24 | amidophosphoribosyltransferase | T |
| LBCZ_RS08400 (purD) | 1.09 | phosphoribosylamine--glycine ligase | T, ST |
| LBCZ_RS08405 (purH) | 1.08 | bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase | T |
| LBCZ_RS10635 (nrdJ) | 1.06 | ribonucleoside-triphosphate reductase, adenosylcobalamin-dependent | T |
| LBCZ_RS08410 (ndk) | 1.04 | phosphoribosylglycinamide formyltransferase | T |
| Pyrimidine metabolism | |||
| LBCZ_RS06785 (pyrR) | −1.69 | bifunctional pyr operon transcriptional regulator/uracil phosphoribosyltransferase PyrR | T,ST |
| LBCZ_RS06755 (pyrD) | −2.97 | dihydroorotate dehydrogenase | T, ST |
| LBCZ_RS06780 (pyrP) | −3.00 | solute carrier family 23 protein | T, ST |
| LBCZ_RS06750 (pyrF) | −3.05 | orotidine-5′-phosphate decarboxylase | T, ST |
| LBCZ_RS06760 (carB) | −3.23 | carbamoyl-phosphate synthase large subunit | T, ST |
| LBCZ_RS06775 (pyrB) | −3.27 | aspartate carbamoyltransferase catalytic subunit | T, ST |
| LBCZ_RS06765 (carA) | −3.42 | carbamoyl phosphate synthase small subunit | T, ST |
| LBCZ_RS06770 (pyrC) | −3.66 | dihydroorotase | T, ST |
| Mobilome: prophages and transposons | |||
| LBCZ_RS04465 | −1.96 | phage holin | T, ST |
| LBCZ_RS01885 | −4.89 | phage portal protein | T, ST |
| LBCZ_RS01960 | −4.99 | phage holin | T, ST |
| LBCZ_RS03480 | −5.02 | phage portal protein | T, ST |
| LBCZ_RS03495 | −5.08 | head–tail connector protein | T, ST |
| LBCZ_RS01895 | −5.97 | phage major capsid protein | T, ST |
| LBCZ_RS03920 | −6.04 | phage tail tape measure protein | T, ST |
| LBCZ_RS03955 | −6.08 | phage holin | T, ST |
| LBCZ_RS01905 | −6.22 | phage head closure protein | T, ST |
| LBCZ_RS03490 | −6.25 | phage major capsid protein | T, ST |
| LBCZ_RS03875 | −6.33 | phage major capsid protein | T, ST |
| LBCZ_RS03500 | −6.44 | phage head closure protein | T, ST |
| LBCZ_RS03865 | −6.48 | phage portal protein | T, ST |
| LBCZ_RS01900 | −6.61 | head–tail connector protein | T, ST |
| LBCZ_RS03735 | −6.75 | phage/plasmid primase, P4 family | T, ST |
| LBCZ_RS03330 | −7.02 | phage antirepressor KilAC domain-containing protein | T, ST |
| Translation | |||
| LBCZ_RS11810 (rplV) | −1.02 | 50S ribosomal protein L22 | T |
| LBCZ_RS11720 (rspK) | −1.04 | 30S ribosomal protein S11 | T |
| LBCZ_RS11710 (rplQ) | −1.13 | 50S ribosomal protein L17 | T |
| LBCZ_RS11880 (rpsG) | −1.13 | 30S ribosomal protein S7 | T |
| LBCZ_RS10605 (rplK) | −1.16 | 50S ribosomal protein L11 | T |
| LBCZ_RS11815 (rpsS) | −1.19 | 30S ribosomal protein S19 | T |
| LBCZ_RS11775 (rplE) | −1.24 | 50S ribosomal protein L5 | T |
| LBCZ_RS11745 (rplO) | −1.27 | 50S ribosomal protein L15 | T |
| LBCZ_RS11725 (rpsM) | −1.29 | 30S ribosomal protein S13 | T |
| LBCZ_RS11785 (rplN) | −1.32 | 50S ribosomal protein L14 | T |
| LBCZ_RS11885 (rpsL) | −1.35 | 30S ribosomal protein S12 | T, ST |
| LBCZ_RS07590 (rpsP) | −1.43 | 30S ribosomal protein S16 | T, ST |
| LBCZ_RS11760 (rplR) | −1.48 | 50S ribosomal protein L18 | T |
| LBCZ_RS11810 (rplV) | −1.02 | 50S ribosomal protein L22 | T |
| LBCZ_RS11720 (rspK) | −1.04 | 30S ribosomal protein S11 | T |
| LBCZ_RS11710 (rplQ) | −1.13 | 50S ribosomal protein L17 | T |
| LBCZ_RS11880 (rpsG) | −1.13 | 30S ribosomal protein S7 | T |
| LBCZ_RS10605 (rplK) | −1.16 | 50S ribosomal protein L11 | T |
| LBCZ_RS11795 (rpmC) | −1.52 | 50S ribosomal protein L29 | T |
| LBCZ_RS11790 (rpsQ) | −1.53 | 30S ribosomal protein S17 | T |
| LBCZ_RS11800 (rplP) | −1.57 | 50S ribosomal protein L16 | T |
| LBCZ_RS11750 (rpmD) | −1.60 | 50S ribosomal protein L30 | T |
| LBCZ_RS15045 (rpmJ) | −1.61 | 50S ribosomal protein L36 | T |
| LBCZ_RS11770 (rpsH) | −1.73 | 30S ribosomal protein S8 | T |
| LBCZ_RS11765 (rplF) | −1.78 | 50S ribosomal protein L6 | T |
| LBCZ_RS11780 (rplX) | −1.79 | 50S ribosomal protein L24 | T |
| LBCZ_RS11755 (rpsE) | −1.81 | 30S ribosomal protein S5 | T |
Gene log2FC: fold change of T-C, ST-C > 1 and < -1, FDR < 0.05 means up- and down-regulation;
Function were assigned from the KEGG pathways.
3.4.1. Membrane transport
Genes involved in the synthesis of PTS are up-regulated upon sonication treatments, both in T and ST, with a fold change ranging from 1.06 to 4.04. According to KEGG database, these genes belong to the macro-category of environmental processing information and to the sub-category of membrane transport. Indeed, two genes encoding the Major Facilitator Superfamily (MFS) are down-regulated only after the more intense treatment (ST). These genes are categorized within the comprehensive Brite hierarchies, specifically within the protein families of signalling and cellular process, ultimately aligning with the group of transporters.
LAB show a metabolic perturbation as general stress-response mechanism under challenging environment [24]. The persistence of these microorganisms under environmental stress strictly depends on their ability to transport and metabolize carbon sources. Membrane transport is a vital cellular process by which molecules such as ions, nutrients, and signaling molecules cross biological membranes. This dynamic mechanism, facilitated by specialized proteins, ensures the controlled movement of substances in and out of cells, maintaining cellular homeostasis and supporting various physiological functions. Carbohydrates transport and utilization in LAB is mediated by numerous proteins. These proteins are members of the ATP-binding cassette (ABC) superfamily, the Phosphotransferase System (PTS), or the Major Facilitator Superfamily (MFS). The PTS is a complex and widespread bacterial transport involved in the coupled uptake and phosphorylation of sugars. PTS are constituted of two generic cytoplasmatic components, EI and HPr, which are common to all PTS of carbohydrates. Indeed, the EII subunit is an integral membrane protein, and it is substrate-specific. These translocator proteins are coded by a single gene (IIABC) or separated genes (IIA, IIB and IIC) [30]. PTS sugar transport is also a fast sugar uptake system. Under a stress condition, the synthesis of PTS transporters favor the survival of bacteria. The competitive advantage of these transporters has been suggested previously [31]. In addition, under stress environment bacteria accumulate compatible solute to enable cell growth and division. Therefore, we can hypothesize that the overexpression of genes encoding PTS is a survival strategy to ensure enough nutrients to maintain vital functions. In addition, some of the up-regulated PTS genes found in T and ST encode for sugar-specific transporters, like fructose, lactose, mannose, and sorbose. It has been underlined that the synthesis of PTS for the uptake of sugars different than glucose is inhibited in optimal environments [24]. Thus, in Lc. casei ATCC 393 the expression of PTS-genes is increased to cope with the ultrasound-induced stress.
The MFS is the largest superfamily of secondary transporters found in bacteria. MFS are employed by bacteria to transport a variety of molecules, carbohydrates, ions, lipids, amino acids, peptides and so on. Structurally, MFS are constituted of 12 transmembrane helices (TMs) organized in two folded domains, the N and C domains, each containing six consecutive TMs. The physiological function attributed to these transporters are related to the uptake of nutrients and to the efflux of toxic compounds. Therefore, MFS are vital in bacteria adaptation and survival under stress [32]. Our data show a specific stress response induced in Lc. casei ATCC 393 upon 8-min of sonication. In fact, the down-regulated genes encoding for the MFS are found only in ST. MFS are involved in the maintenance of intracellular pH by regulating the ions and protons transports. MFS also work as efflux pumps playing a key role in defence mechanisms towards environmental stress. Moreover, MFS also contribute to the transport of metabolic intermediates and by-products and, thus, to the cell metabolic flexibility. Our results are not in agreement with what previously reported by Kumar et al. [33]. Although upregulation of MFS is a common strategy used by bacteria to cope with stress and dynamic environments, we found a controversial result. Considering the role of MFS and the specific down-regulation of MFS-genes, we hypothesize that 8 min of sonication did not induce the formation of toxic compounds in Lc. casei ATCC 393.
3.4.2. Carbohydrate metabolism
We identified that some genes up-regulated in T and ST belonged to carbohydrate metabolism according to KEGG classification. Among the five genes identified in this category, two are directly involved in the glycolytic pathway (pfkB, ldh), while three are involved in pyruvate metabolism (spxB, ppdk, pdhA). DGE analysis revealed a 1.03–3.75 fold up-regulation compared to untreated cells.
Carbon from the external environment can enter several metabolic pathways that are interconnected in the complex networks of central carbon metabolism. Bacteria use central carbon metabolism to obtain the building blocks of cellular components and obtain the necessary amount of energy to sustain cellular processes [34]. Ultrasound has multiple sites of action on bacteria. In addition to the physical and chemical damages of the cell components, ultrasound also decrease the ATP level [35]. Thus, the cell needs enough energy to simultaneously repair the ultrasound-induced damage, maintain intracellular homeostasis and cellular metabolism [25]. Therefore, we speculate that ultrasound-treated probiotic could enhance the synthesis of enzymes involved in the carbohydrate metabolism to increase the level of ATP.
Deeper classification allowed us to further sub-group the carbohydrate metabolism-related DEGs in the glycolytic pathway or pyruvate metabolism. In our previous study also performed on Lc. casei ATCC 393 [36], we showed that 6 and 8 min of sonication induced a transient and a prolonged attenuation effect, respectively. The critical evaluation of the data collected in the previous and current studies allow us to have a deeper understanding of effects of ultrasound on this probiotic strain at metabolic level. The rate of acidification is directly related to the flux of sugars into the glycolytic pathway [37]. Although there is an increase in genes involved in PTS synthesis and pyruvate metabolism, there is also a slower production of lactic acid (acidification). Under environmental stress, among all the strategies used by LAB to survive, one overall response strategy is to select an alternative fate for pyruvate [24]. Therefore, our results suggest that cells increase the production of pyruvate upon ultrasound exposure and select an alternative fate for it rather than glycolysis.
3.4.3. Nucleotide metabolism
Genes involved in nucleotide metabolic pathways are also reported in Table 2. Genes involved in purine nucleotide de novo biosynthesis purRDHNMFQCKE and guaB were up-regulated with a fold change ranging from 1.04 to 2.72. We also found an over-expression of the genes nrdJ and ndK. The former is involved in purine nucleotide de novo biosynthesis and salvage. The latter, indeed, is involved in both pathways and in the alarmone guanosine tetraphosphate (p)ppGpp metabolism. In addition, one of the major groups of down-regulated DEGs was involved in de novo pyrimidine nucleotide metabolism. The fold change of pyrBCDEFPR and carAB genes was between −3.66 and −2.76, for both T and ST.
Nucleotide metabolism is intricately linked to the stress physiology of lactic acid bacteria (LAB). Nucleotides play a key role in bacteria physiology not only for their constitutive function as basic blocks of nucleic acids but also for their regulatory function of metabolic process and gene expression [38]. Interestingly, the gene expression profile across sonicated Lc. casei ATCC 393 differed between purine and pyrimidine biosynthesis. Similar results were reported by Raynaud et al. [39], in which authors studied the metabolic and transcriptomic adaptation of the strain L. lactis subsp. lactis biovar diacetylactis LD61 exposed to acid stress in skim milk. They found a similar pattern of regulation, and in fact, purine and pyrimidine pathways were overexpressed and repressed, respectively.
Purine nucleotides de novo biosynthesis occurs trough 10 step leading to the formation of the metabolic intermediate inosine monophosphate (IMP). The metabolite is further converted to adenosine monophosphate (AMP) or guanosine monophosphate (GMP). AMP and GMP can be further phosphorylated to produce adenosine triphosphate (ATP) and guanosine triphosphate (GTP). ATP and GTP are high-energy nucleotides that serve as primary carriers of chemical energy in cells. Under stress environment the cell increases the demand for supporting processes such as repair mechanisms, adaptation, defence responses, and basic cellular activities. The up-regulation of the purine nucleotide biosynthetic genes suggests the shift toward the synthesis of GTP and ATP and reduction of accumulation of IMP. ATP and GTP are also precursors of the ppGpp, that is a signalling molecule involved in the stringent response mechanism. Finally, in the salvage pathway external nucleotides are dephosphorylated and transported into the cell as nucleosides and finally phosphorylated by kinases [40]. The up-regulation of salvage pathway genes could be an additional stress-response of the probiotic to an increased purine nucleotide requirement. Therefore, we suppose that the increase in transcription of purine nucleotides and, thus, increase in the pool of purine nucleotides is one of the defense mechanisms activated in the probiotic Lc. casei ATCC 393 to cope with the several alterations induced by ultrasound.
Pyrimidine nucleotide is synthesized in LAB by the de novo biosynthesis of uridine monophosphate (UMP). In Lc. casei ATCC 393 sonicated, both T and ST, all the genes encoding for the catalytic enzymes of the pyrimidine biosynthesis are down-regulated.
The pyrimidine biosynthetic genes are grouped in a single operon on the chromosome of Lc. casei ATCC 393. The first gene on the operon is pyrR encoding a regulatory protein. Martinussen et al. [41] underlined the role of the pyrR gene in the expression of pyrimidine biosynthetic genes. The authors observed that a mutational inactivation of pyrR induced a 3- to 8-fold depression in all the genes that regulate the biosynthesis of pyrimidine, except pyrDa. Moreover, this mechanism is a conserved regulatory mechanism among LAB. Therefore, we hypothesize that a similar mutational event occurs in Lc. casei ATCC 393 upon ultrasound exposure induced the down-regulation of genes involved in pyrimidine metabolism.
3.4.4. Phage-related genes
Also genes derived from phage DNA were downregulated. This gene group is the second most enriched group among the downregulated DEGs, both in T and ST, with very high log2FC values. Apart from LCBZ_RS04465 (log2FC −1.96), the other 15 genes were downregulated with a fold change ranging from −4.89 to −7.02.
Many strains of LAB are lysogenic, meaning that they carry an integrated copy of one or more temperate phages (prophages) within their genome. The extra cytoplasmatic stress influence the lysogenic maintenance or the phage induction [24]. The phage induction leads to the excision of the prophage from the host chromosome, followed by replication of phage DNA and production of virions, ultimately resulting in lysis of the bacterial cell. Our results suggest that ultrasound stressor is not a major prophage activator. Moreover, the down-regulation of this gene cluster also suggest a tight control of the cell on the maintenance of the lysogenic status. A strictly control of phage induction was observed in L. lactis under various industrial environment [42].
3.4.5. Translation
DEGs related to both large and small ribosomal proteins synthesis were exclusively down-regulated in the 6 min treated cells. Specifically, we found the repression of genes rpsEGHKMQS involved in the biogenesis of the small ribosomal subunits, and of the genes rplEFKNOPQRVX and rpmCDJ involved in the biogenesis of the large ribosomal subunit. However, the genes rpsLP were also down-regulated in the 8 min treated sample. The fold change detected for these genes ranged from −1.81 to −1.02.
Functional classification of ultrasound affected genes revealed that ribosomal biogenesis plays a central role in Lc. casei ATCC 393 ultrasound stress response. Prokaryotic ribosomes consist of two subunits, the small subunit (30S) and the large subunit (50S), which together form the functional 70S ribosome. During the translation process, the messenger RNA (mRNA) is read by the ribosome, initiating the synthesis of proteins. The ultrasound stress induced by a 6 min exposure reduced the Lc. casei ATCC 393 capacity to synthesize new ribosome proteins. The decreased in ribosome biogenesis led to the reduction of the overall capacity for the translation process. It has been stressed by Njenga et al. [43] that bacteria under environmental stress redirect the cellular metabolism towards the synthesis of stress resistance factors while rapidly inhibiting the synthesis of ribosomal proteins. This mechanism is also explained by Zegarra et al. [44]. High abundance of ribosomes, high energy demanded for their biogenesis, and high cost of protein synthesis justify the decrease in the transcription of their related encoding genes.
The normal transcription of these gene after 8 min of sonication let us to hypothesize that a prolonged exposure to ultrasound induce the adaptation of the probiotic to the experimental condition. Moreover, we also suppose that the absence of variation in these gene in ST is justified by the more intense physical damages induced by ultrasound. Therefore, ST requires a normal synthesis of ribosomal proteins and, thus, a normal protein synthesis to counteract the ultrasound-induced damages. Besides, in Escherichia coli O157:H7 exposed to ultrasound was detected an over-expression of genes encoding for the ribosomal subunits to promptly synthesize stress response proteins [45].
3.4.6. Other DEGs cluster
Other minor gene clusters were also found as differentially expressed (Table 3). Such genes showed a minor involvement in terms of enrichment values and the genes count.
Table 3.
Other functional categories associated with Differentially Expressed Genes (DEG) in ultrasound-treated Lc. casei ATCC 393. T: 6 min of sonication; ST: 8 min of sonication.
| Gene ID and functional category | Gene log2FCa | Descriptionb | Sample |
|---|---|---|---|
| Genetic information processing | |||
| DNA repair | |||
| LBCZ_RS05805 (radC) | 1.90 | redox-sensing transcriptional repressor Rex | T, ST |
| LBCZ_RS08395 | 1.67 | CopY/TcrY family copper transport repressor | T |
| LBCZ_RS06270 (uvrC) | 1.18 | excinuclease ABC subunit UvrC | T, ST |
| LBCZ_RS10825 (radA) | 1.02 | DNA repair protein RadA | T |
| Replication and repair | |||
| LBCZ_RS09210 | −1.08 | IS30 family transposase | T |
| LBCZ_RS15870 | −1.19 | IS3 family transposase | T |
| LBCZ_RS12095 | −1.34 | IS30 family transposase | T, ST |
| LBCZ_RS09445 | −1.38 | IS30 family transposase | T, ST |
| LBCZ_RS14405 | −1.48 | IS30 family transposase | T, ST |
| LBCZ_RS12855 | −1.73 | IS30 family transposase | T |
| LBCZ_RS04675 | −4.02 | IS3 family transposase | T, ST |
| LBCZ_RS03855 | −4.46 | IS3 family transposase | T, ST |
| LBCZ_RS04440 | −4.91 | IS30 family transposase | T, ST |
| LBCZ_RS03890 | −6.08 | IS30 family transposase | T, ST |
| Cell wall biogenesis | |||
| LBCZ_RS00295 (murQ) | 3.41 | N-acetylmuramic acid 6-phosphate etherase | T, ST |
| LBCZ_RS09570 (wecB) | 1.40 | UDP-N-acetylglucosamine 2-epimerase(non-hydrolyzing) | ST |
| LBCZ_RS01180 (ngA) | 1.27 | N-acetylglucosamine-6-phosphate deacetylase | T |
| LBCZ_RS01965 | −5.40 | GH25 family lysozyme (bacteria cell wall degradation, hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues) | T, ST |
| LBCZ_RS03575 | −5.63 | T, ST | |
| LBCZ_RS03960 | −6.04 | T, ST | |
| Amino acid metabolism | |||
| LBCZ_RS08430 (purQ) | 1.78 | phosphoribosylformylglycinamidine synthase subunit PurQ | T, ST |
| LBCZ_RS10110 (tdcB) | 1.69 | bifunctional threonine ammonia-lyase/L-serine ammonia-lyase TdcB | T |
| LBCZ_RS06690 (hisB) | 1.37 | imidazoleglycerol-phosphate dehydratase HisB | T, ST |
| LBCZ_RS08420 (purF) | 1.24 | amidophosphoribosyltransferase | T |
| LBCZ_RS13225 (argH) | 1.13 | argininosuccinate lyase | T |
| LBCZ_RS10930 (proB) | 1.02 | glutamate 5-kinase | T |
| Environmental information processing | |||
| Signal transduction | |||
| LBCZ_RS08915 (citF) | −1.55 | citrate lyase subunit alpha | T |
| LBCZ_RS08930 (citC) | −1.57 | [citrate pro-3S-lyase] ligase | T |
| LBCZ_RS08910 (citX) | −1.64 | citrate lyase holo-[acyl-carrier protein] synthase | T |
| LBCZ_RS08925 (citD) | −1.70 | citrate lyase acyl carrier protein | T |
| Fatty acid biosynthesis | |||
| LBCZ_RS09775 (accA) | −1.24 | acetyl-CoA carboxylase biotin carboxyl carrier protein | T |
| LBCZ_RS09780 (fabF) | −1.45 | beta-ketoacyl-ACP synthase II | T |
| LBCZ_RS09765 (accC) | −1.67 | acetyl-CoA carboxylase biotin carboxylase subunit | T |
| LBCZ_RS09755 (accB) | −1.87 | carboxyltransferase subunit alpha | ST |
| Carbohydrates metabolism | |||
| LBCZ_RS09950 (sfsA) | −1.02 | DNA/RNA nuclease SfsA (sugar fermentation stimulation) | ST |
Gene log2FC: fold change of T-C, ST-C > 1 and < -1, FDR < 0.05 means up- and down-regulation;
Function were assigned from the KEGG pathways.
Genes encoding for genetic information processing were over expressed in sonicated samples. The gene uvrC and radA were up-regulated with a fold change of 1.18 and 1.02, respectively. The gene uvrC is a component of the nucleotide excision repair (NER) system used by bacterial cell to recognize and repair DNA damages while the gene radA encodes for the DNA repair protein (RadA) involved in the SOS-response. Interestingly, the gene uvrC was differentially expressed in both sonicated samples, while the radA was differentially expressed only in T. The over-expression of these genes suggest that ultrasound damages the DNA [23], [24]. It also seems that the exposure of Lc. casei ATCC 393 to ultrasound for 6 min activated a major transcriptional response that was not further observed after 8 min. Again, we suppose that an adaptation to the external stress was induced in the probiotic that lower the transcriptional response after 8 min of treatment.
Another cluster of DEGs is represented by genes encoding for IS3 and IS30 transposase families. Transposase catalyses the movement of transposons within the genome. The down-regulation of this cluster is a stress-response mechanism activated in the probiotic under ultrasound external stress. In fact, the repression of these genes and, thus, the reduced synthesis of transposase maintain the genomic integrity and stability [46]. Although transcriptomic analysis revealed the down-regulation of these genes both in T and ST, three genes (LBCZ_RS09210, LBCZ_RS1587, LBCZ_RS12855) were differentially expressed only in T. In addition, also some genes involved in the mechanisms of DNA integration and recombination were down-regulated upon ultrasound. These data highlight that ultrasound stress activate the transcriptional regulation of different genes involved in the preservation of DNA structure.
The transcription of genes involved in the cell wall biogenesis was also affected by ultrasound stress. This category is the only one in which we noted both up- and down-regulated DEGs. However, the up-regulation interested genes involved in the peptidoglycan turnover (murQ) and in the biosynthesis of main components of the cell wall (nagA, wecB). Indeed, the down-regulation interested genes encoding for a protein family that catalyse the degradation of the cell wall. Moreover, the fold change is of about 1 and −6 for the former and the latter cluster, respectively. Finally, murQ gene was differentially expressed by both sonicated samples, while nagA and wecB genes only by T and ST, respectively. Therefore, an obvious deduction is that murQ up-regulation is a general response to ultrasound stress, while the up-regulation of wecB and nagA depends on the time of the exposure. The down-regulated DEGs overlapped in the two sonicated samples. These variations in the gene expression may contribute to maintain the cell membrane function in Lc. casei ATCC 393 after ultrasound treatment. The cell wall and membrane play a crucial role in cell viability and function by performing various structural and functional tasks, including the regulation of stress resistance in LAB [25]. Thus, the transcriptional changes in these genes are supported by scientific evidence collected on general mechanisms of LAB stress response to different stressors.
An over-expression in genes involved in the amino acid biosynthesis and metabolism was also detected. DEGs (log2FC 1.02–1.78) in this category encode for enzymes involved in histidine metabolism (hisB), arginine biosynthesis (argH, purQ), arginine and proline metabolism (proB), alanine, aspartate and glutamate metabolism (purF, purQ), and valine, leucine and isoleucine metabolism (tdaB). Amino acid metabolism is involved in both vital processes and stress resistance mechanism in LAB [24]. Therefore, the probiotic may increase the expression of these genes to increase the availability of amino acids. Bacteria accumulate free amino acid to control the intracellular pH and maintain the cellular homeostasis [47] and to adapt to challenging environment [48], [49]. The role of amino acid metabolism in bacteria adaptation mechanism to challenging environment or treatment could justify the specific up-regulation of four of these genes only in the 6-min sonicated sample.
The lower intense treatment induced the transcriptional repression of the citCDFX genes categorized in the environmental information processing. These genes belong to the sub-category of signal transduction and are involved in the two-component regulatory mechanism [50]. We hypothesized that the down-regulation of the citCDFX genes is involved in a mechanism of homeostasis maintenance avoiding an overstimulation and ensuring an appropriate stress response to the external stimulus.
The probiotic lipid metabolism, specifically the fatty acid metabolism, was also affected by ultrasound treatment. The genes accBC and fabF were down-regulated in T, while the gene accB were down-regulated both in T and ST with a fold change ranging from −1.87 to −1.24. The genes accABC encode for the enzyme acetyl-CoA carboxylase and the gene fabF mediate the fatty acid elongation. The acetyl-CoA carboxylase is the key enzyme in the fatty acid biosynthesis. Bacteria exposed to external stress reprogram cellular processes in favor of the ones critical for the immediate survival. In addition, the accumulation of energy is preferred to its consumption in biosynthetic pathways [24], [25]. Therefore, the fatty acid biosynthesis seems to be not a key pathway for the survival of Lc. casei ATCC 393 during ultrasound exposure.
4. Conclusion
In this study we investigated the transcriptional response of Lc. casei ATCC 393 after two ultrasound treatments. RNA-Seq and DGE analysis revealed that the transcription of genes related to multiple important cellular processes was altered under ultrasound stress. The up-regulation of PTS-encoding genes favoured transmembrane transport and altered membrane permeability in ultrasound-treated samples. Considering also the up-regulation of genes related to carbohydrate metabolism, the capacity of the strain to accumulate ATP increased. ATP preservation was also the result of the over-expression of genes coding for the synthesis of its precursors, the purine nucleotides. In addition, the synthesis of ribosomal proteins was down-regulated, indicating that the biosynthetic capacity of some proteins decreased under ultrasound stress. Finally, the down-regulation of genes involved in the modification of DNA structure indicated that the transcriptional response of Lc. casei ATCC 393 preserved the integrity of DNA.
Considering that several pathways undergo transcriptional changes under ultrasound stress, we conclude that the attenuation of probiotic acidification capacity and the modification of its phenotype depend on the overall metabolic perturbation. However, the construction of Lc. casei ATCC 393 mutants could be a possible strategy to properly identify the key pathway or network of pathways involved in the probiotic attenuation, among those highlighted in the present study.
CRediT authorship contribution statement
Irene Giordano: Writing – original draft, Investigation, Formal analysis, Data curation. Edoardo Pasolli: Data curation. Gianluigi Mauriello: Writing – review & editing, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Project funded under the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.3 - Call for proposals No. 341 of 15 March 2022 of Italian Ministry of University and Research funded by the European Union – NextGenerationEU; Project code PE00000003, Concession Decree No. 1550 of 11 October 2022 adopted by the Italian Ministry of University and Research, CUP D93C22000890001, Project title “ON Foods - Research and innovation network on food and nutrition Sustainability, Safety and Security – Working ON Foods”.
References
- 1.Bibi A., Xiong Y., Rajoka M.S.R., Mehwish H.M., Radicetti E., Umair M., Shoukat M., Khan M.K.I., Aadil R.M. Recent advances in the production of exopolysaccharide (EPS) from lactobacillus spp. and its application in the food industry: A review. Sustainability. 2021;13(22):12429. doi: 10.3390/su132212429. [DOI] [Google Scholar]
- 2.Mayo B., Rodríguez J., Vázquez L., Flórez A.B. Microbial interactions within the cheese ecosystem and their application to improve quality and safety. Foods. 2021;10(3):602. doi: 10.3390/foods10030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma H., Ozogul F., Bartkiene E., Rocha J.M. Impact of lactic acid bacteria and their metabolites on the techno-functional properties and health benefits of fermented dairy products. In Crit. Rev. Food Sci. Nutr. 2023;63(21):4819–4841. doi: 10.1080/10408398.2021.2007844. [DOI] [PubMed] [Google Scholar]
- 4.LeBlanc J.G., Levit R., Savoy de Giori G., de Moreno de LeBlanc A. Application of vitamin-producing lactic acid bacteria to treat intestinal inflammatory diseases. Appl. Microbiol. Biotechnol. 2020;104(8):3331–3337. doi: 10.1007/s00253-020-10487-1. [DOI] [PubMed] [Google Scholar]
- 5.Abdul Hakim B.N., Xuan N.J., Oslan S.N.H. A Comprehensive Review of Bioactive Compounds from Lactic Acid Bacteria: Potential Functions as Functional Food in Dietetics and the Food Industry. Foods. 2023;12(15):2850. doi: 10.3390/foods12152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prete R., Alam M.K., Perpetuini G., Perla C., Pittia P., Corsetti A. Lactic acid bacteria exopolysaccharides producers: A sustainable tool for functional foods. Foods. 2021;10(7):1653. doi: 10.3390/foods10071653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gustaw K., Niedźwiedź I., Rachwał K., Polak-Berecka M. New insight into bacterial interaction with the matrix of plant-based fermented foods. Foods. 2021;10(7):1603. doi: 10.3390/foods10071603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fusco V., Fanelli F., Chieffi D. Authenticity of probiotic foods and dietary supplements: A pivotal issue to address. Crit. Rev. Food Sci. Nutr. 2022;62(25):6854–6871. doi: 10.1080/10408398.2021.1907300. [DOI] [PubMed] [Google Scholar]
- 9.Hadj Saadoun J., Bertani G., Levante A., Vezzosi F., Ricci A., Bernini V., Lazzi C. Fermentation of agri-food waste: A promising route for the production of aroma compounds. Foods. 2021;10(4):707. doi: 10.3390/foods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soto L.P., Sirini N.E., Frizzo L.S., Zbrun M.V., Zimmermann J.A., Ruiz M.J., Rosmini M.R., Sequeira G.J., Miotti C., Signorini M.L. Lactic acid bacteria viability in different refrigerated food matrices: a systematic review and Meta–analysis. Crit. Rev. Food Sci. Nutr. 2023;63(33):12178–12206. doi: 10.1080/10408398.2022.2099807. [DOI] [PubMed] [Google Scholar]
- 11.Wendel U. Assessing Viability and Stress Tolerance of Probiotics—A Review. Front. Microbiol. 2022;12 doi: 10.3389/fmicb.2021.818468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendonça A.A., de Pinto-Neto W., P., da Paixão, G. A., Santos, D. da S., De Morais, M. A., & De Souza, R. B. Journey of the Probiotic Bacteria: Survival of the Fittest. Microorganisms. 2023;11(1):95. doi: 10.3390/microorganisms11010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar M., Yadav A.K., Verma V., Singh B., Mal G., Nagpal R., Hemalatha R. Bioengineered probiotics as a new hope for health and diseases: an overview of potential and prospects. Future Microbiol. 2016;11(4):585–600. doi: 10.2217/fmb.16.4. [DOI] [PubMed] [Google Scholar]
- 14.Sharma A., Gupta G., Ahmad T., Kaur B., Hakeem K.R. Tailoring cellular metabolism in lactic acid bacteria through metabolic engineering. J. Microbiol. Methods. 2020;170 doi: 10.1016/j.mimet.2020.105862. [DOI] [PubMed] [Google Scholar]
- 15.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., Calder P.C., Sanders M.E. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 16.Ballini A., Charitos I.A., Cantore S., Topi S., Bottalico L., Santacroce L. About functional foods: The probiotics and prebiotics state of art. Antibiotics. 2023;12(4):635. doi: 10.3390/antibiotics12040635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Speranza B., Campaniello D., Petruzzi L., Altieri C., Sinigaglia M., Bevilacqua A., Corbo M.R. The inoculation of probiotics in vivo is a challenge: Strategies to improve their survival, to avoid unpleasant changes, or to enhance their performances in beverages. Beverages. 2020;6(2):1–18. doi: 10.3390/beverages6020020. [DOI] [Google Scholar]
- 18.B. Speranza, A. Bevilacqua, M.R. Corbo, M. Sinigaglia, Starter Cultures in Food Production, in: John Wiley & Sons, Ltd (Eds.), Starter cultures in food production, Hoboken, NJ, USA, 2017, pp. xiii + 421.
- 19.Nicolau-Lapeña I., Lafarga T., Viñas I., Abadias M., Bobo G., Aguiló-Aguayo I. Ultrasound Processing Alone or in Combination with Other Chemical or Physical Treatments as a Safety and Quality Preservation Strategy of Fresh and Processed Fruits and Vegetables: A Review. Food Bioprocess Tech. 2019;12(9):1452–1471. doi: 10.1007/s11947-019-02313-y. [DOI] [Google Scholar]
- 20.Téllez-Morales, J. A., Hernández-Santo, B., & Rodríguez-Miranda, J. Effect of ultrasound on the techno-functional properties of food components/ingredients: A review. Ultrason. Sonochem. 61 (2020) 104787. Elsevier B.V. https://doi.org/10.1016/j.ultsonch.2019.104787. [DOI] [PubMed]
- 21.Beitia E., Gkogka E., Chanos P., Hertel C., Heinz V., Valdramidis V., Aganovic K. Microbial decontamination assisted by ultrasound-based processing technologies in food and model systems: A review. Compr. Rev. Food Sci. Food Saf. 2023;22(4):2802–2849. doi: 10.1111/1541-4337.13163. [DOI] [PubMed] [Google Scholar]
- 22.Bermudez-Aguirre D., Niemira B.A. Pasteurization of Foods with Ultrasound: The Present and the Future. Appl. Sci. 2022;12(20):10416. doi: 10.3390/app122010416. [DOI] [Google Scholar]
- 23.Zupanc, M., Pandur, Ž., Stepišnik Perdih, T., Stopar, D., Petkovšek, M., & Dular, M. (2019). Effects of cavitation on different microorganisms: The current understanding of the mechanisms taking place behind the phenomenon. A review and proposals for further research. Ultrason. Sonochem. 57 (2019) 147–165. https://doi.org/10.1016/j.ultsonch.2019.05.009. [DOI] [PubMed]
- 24.Papadimitriou K., Alegría Á., Bron P.A., de Angelis M., Gobbetti M., Kleerebezem M., Lemos J.A., Linares D.M., Ross P., Stanton C., Turroni F., van Sinderen D., Varmanen P., Ventura M., Zúñiga M., Tsakalidou E., Kok J. Stress Physiology of Lactic Acid Bacteria. Microbiol. Mol. Biol. Rev. 2016;80(3):837–890. doi: 10.1128/mmbr.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H., He M., Wu C. Cross protection of lactic acid bacteria during environmental stresses: Stress responses and underlying mechanisms. LWT. 2021;144 doi: 10.1016/j.lwt.2021.111203. [DOI] [Google Scholar]
- 26.O’Donnell S.T., Ross R.P., Stanton C. The Progress of Multi-Omics Technologies: Determining Function in Lactic Acid Bacteria Using a Systems Level Approach. Front. Microbiol. 2020;10 doi: 10.3389/fmicb.2019.03084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Den Berge K., Hembach K.M., Soneson C., Tiberi S., Clement L., Love M.I., Patro R., Robinson M.D. Annual Review of Biomedical Data Science RNA Sequencing Data: Hitchhiker’s Guide to Expression Analysis INTRODUCTION: OVERVIEW OF THE RNA SEQUENCING ASSAY. Annu. Rev. Biomed. Data Sci. 2019;2:139–173. doi: 10.1146/annurev-biodatasci-072018. [DOI] [Google Scholar]
- 28.Huang Y.R., Li L., Wei X.M., Li H.Z., Zeng J.Y., Kuang R. An investigation of mechanisms for the enhanced coagulation removal of Microcystis aeruginosa by low-frequency ultrasound under different ultrasound energy densities. Ultrason. Sonochem. 2020;69 doi: 10.1016/j.ultsonch.2020.105278. [DOI] [PubMed] [Google Scholar]
- 29.Fenouil R., Pradel N., Belahbib H., Roumagnac M., Bartoli M., Ben Hania W., Denis Y., Garel M., Tamburini C., Ollivier B., Summers Z., Armougom F., Dolla A. Adaptation Strategies to High Hydrostatic Pressures in Pseudothermotoga species Revealed by Transcriptional Analyses. Microorganisms. 2023;11(3):773. doi: 10.3390/microorganisms11030773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monedero V., Mazé A., Boël G., Zúñiga M., Beaufils S., Hartke A., Deutscher J. The phosphotransferase system of Lactobacillus casei: Regulation of carbon metabolism and connection to cold shock response. J. Mol. Microbiol. Biotechnol. 2006;12(1–2):20–32. doi: 10.1159/000096456. [DOI] [PubMed] [Google Scholar]
- 31.Castro R., Neves A.R., Fonseca L.L., Pool W.A., Kok J., Kuipers O.P., Santos H. Characterization of the individual glucose uptake systems of Lactococcus lactis: Mannose-PTS, cellobiose-PTS and the novel GlcU permease. Mol. Microbiol. 2009;71(3):795–806. doi: 10.1111/j.1365-2958.2008.06564.x. [DOI] [PubMed] [Google Scholar]
- 32.Yan N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 2013;38(3):151–159. doi: 10.1016/j.tibs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S., Lekshmi M., Parvathi A., Ojha M., Wenzel N., Varela M.F. Functional and structural roles of the major facilitator superfamily bacterial multidrug efflux pumps. Microorganisms. 2020;8(2):266. doi: 10.3390/microorganisms8020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papagianni M. Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb. Cell Fact. 2012;11(50):1–13. doi: 10.1186/1475-2859-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., Ma L., Liao X., Liu D., Lu X., Chen S., Ye X., Ding T. Ultrasound-induced Escherichia coli O157:H7 cell death exhibits physical disruption and biochemical apoptosis. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giordano I., Mauriello G. Ultrasound Attenuation Improves Some Surface Properties of the Probiotic Strain Lacticaseibacillus casei ATCC 393. Microorganisms. 2023;11(1):142. doi: 10.3390/microorganisms11010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinussen J., Solem C., Holm A.K., Jensen P.R. Engineering strategies aimed at control of acidification rate of lactic acid bacteria. Curr. Opin. Biotechnol. 2013;24(2):124–129. doi: 10.1016/j.copbio.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Kilstrup M., Hammer K., Jensen P.R., Martinussen J. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol. Rev. 2005;29(3):555–590. doi: 10.1016/j.femsre.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Raynaud, S., Perrin, R., Cocaign-Bousquet, M., & Loubiere, P. Metabolic and transcriptomic adaptation of Lactococcus lactis subsp. lactis biovar diacetylactis in response to autoacidification and temperature downshift in skim milk. Appl. Environ. Microbiol, 71(12) (2005) 8016–8023. https://doi.org/10.1128/AEM.71.12.8016-8023.2005. [DOI] [PMC free article] [PubMed]
- 40.Torrents E. Ribonucleotide reductases: Essential enzymes for bacterial life. Front. Cell. Infect. Microbiol. 2014;4:52. doi: 10.3389/fcimb.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinussen J., Schallert J., Andersen B., Hammer K. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol. 2001;183(9):2785–2794. doi: 10.1128/JB.183.9.2785-2794.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho C.H., Stanton-Cook M., Beatson S.A., Bansal N., Turner M.S. Stability of active prophages in industrial Lactococcus lactis strains in the presence of heat, acid, osmotic, oxidative and antibiotic stressors. Int. J. Food Microbiol. 2016;220:26–32. doi: 10.1016/j.ijfoodmicro.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Njenga R., Boele J., Öztürk Y., Koch H.G. Coping with stress: How bacteria fine-tune protein synthesis and protein transport. J. Biol. Chem. 2023;299(9) doi: 10.1016/j.jbc.2023.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zegarra V., Bedrunka P., Bange G., Czech L. How to save a bacterial ribosome in times of stress. Semin. Cell Dev. Biol. 2023;136:3–12. doi: 10.1016/j.semcdb.2022.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Li J., Liu D., Ding T. Transcriptomic analysis reveal differential gene expressions of Escherichia coli O157:H7 under ultrasonic stress. Ultrason. Sonochem. 2021;71 doi: 10.1016/j.ultsonch.2020.105418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darmon E., Leach D.R.F. Bacterial Genome Instability. Microbiol. Mol. Biol. r. 2014;78(1):1–39. doi: 10.1128/mmbr.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaucher F., Bonnassie S., Rabah H., Marchand P., Blanc P., Jeantet R., Jan G. Review: Adaptation of beneficial propionibacteria, lactobacilli, and bifidobacteria improves tolerance toward technological and digestive stresses. Front. Microbiol. 2019;10:841. doi: 10.3389/fmicb.2019.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broadbent J.R., Larsen R.L., Deibel V., Steele J.L. Physiological and transcriptional response of Lactobacillus casei ATCC 334 to acid stress. J. Bacteriol. 2010;192(9):2445–2458. doi: 10.1128/JB.01618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan, N., Liu, L., Shin, H. dong, Chen, R. R., Zhang, J., Li, J., Du, G., Shi, Z., & Chen, J. Systems-level understanding of how Propionibacterium acidipropionici respond to propionic acid stress at the microenvironment levels: Mechanism and application. J. Biotech. 167(1) (2013) 56–63. https://doi.org/10.1016/j.jbiotec.2013.06.008. [DOI] [PubMed]
- 50.Monedero V., Revilla-Guarinos A., Zúñiga M. Physiological Role of Two-Component Signal Transduction Systems in Food-Associated Lactic Acid Bacteria. Adv. Appl. Microbiol. 2017;99:1–51. doi: 10.1016/bs.aambs.2016.12.002. [DOI] [PubMed] [Google Scholar]