Summary
As the value of clinical imaging is expanded through retrospective analyses, it is imperative that all efforts are made to optimize validity. Such considerations for retrospective designs should prioritize factors like naturalistic conditions for observations and measurement replicability, while avoiding sample biases and reliance on strict clinical timelines. Valid methodological approaches are immanent for successful translation from retrospective observational designs into prospective pragmatic research with actionable potential. In particular, thousands of studies have sought to associate clinical outcomes to measures of body composition across diverse patient groups. Post-hoc use of computed tomography (CT) to quantify adiposity and lean tissue characteristics has most frequently involved just a single slice at the level of the third lumbar vertebrae (L3). Abundant in statistical significance and inconsistencies alike, such methods have yet to be implemented or deemed valuable for making real-world clinical decisions. We present herein a concerning perspective, for both magnitude and prevalence, of a widely overlooked source of data variability for this methodology: the hinderance of pants and other tightly fit clothing.
Keywords: Retrospective analysis, clinical imaging, clothing, data quality, skeletal muscle, adiposity
Introduction
Retrospective analysis of computed tomography (CT) imaging has been widely utilized in clinical populations as the “gold standard” method for quantifying body composition and tissue volumes.1 Thousands of published studies across the last 30 years suggest a concerningly high heterogeneity for statistical associations involving skeletal muscle and adiposity across patient populations that represent all types of cancer, COPD, and recently COVID-19.2-8 Like most clinical datasets, the extensive presence of confounds, inconsistencies, and missing data tend to complicate post-hoc imaging analyses.9 In addition to obvious data artifact, ample threats to study validity can be well concealed by lengthy patient charts, co-occurring factors, and methodological limitations. In the absence of a highly controlled environment, we neglect to consider the multiplicity of factors that can influence naturally occurring data, and thus, real-world utility of findings.9,10 Most importantly, we often fail to rehumanize collections of datapoints to understand patterns, compound clinical effects, and limitations experienced by both the clinical team and patient that maximize the value of post-hoc conclusions.
Most studies utilize a single axial slice at the level of the third lumbar vertebrae (L3) for estimating body composition.11 Justification for use of the L3 slice specifically is rarely discussed, though it has been suggested to have the strongest correlation to full body composition.12,13 Notably, the L3 slice is minimally affected by anatomical variation that stems from differing position of the arms and legs, making it seemingly consistent across individuals. Further, this region is rarely affected by foreign structures (i.e. joint replacements, port-systems, implantable cardioverter defibrillators, etc.) aside from the occasional spinal implant.
Despite the L3 slice having the greatest anatomical consistency among populations, consistent views of this lumbar level require imaging of the abdominopelvic region or “full-body” scans, as chest or thorax windows alone often fail to caudally extend to L3. As such, many studies have attempted to utilize axial slices from the more commonly imaged thoracic and upper lumbar regions, 14 though these rarely demonstrate full-body associations superior to that of L3.13
Single slice surface area (SA) and attenuation (radiodensity) of skeletal muscle, intramuscular adipose tissue (IMAT), subcutaneous adipose tissue (SAT), and visceral adipose tissue (VAT) are the most widely quantified variables. These tissue characteristics are suggested to represent tissue quantity and quality, and have been associated with functional and clinical outcomes including survival, treatment toxicity, and cachexia.2-4,15-18 These variables are often acquired using semi-automated segmentation of an axial image based upon pre-defined radiodensity ranges, or via manual segmentation, potentially increasing inter-study variability.11 Single slice variables can be used on their own or in combination with other variables; for example, many established cut points for sarcopenia utilize skeletal muscle index (SMI), which is defined as single slice skeletal muscle SA normalized to body height.11 Skeletal muscle gauge (SMG) is also commonly used, defined as the product of SMI and mean attenuation of the skeletal muscle.19 Similarly, multiple equations have been developed for estimation of full-body fat mass (FM) and fat-free mass (FFM) using these variables.20 Characterization of the described tissues via CT has more frequently been used to stratify patient cohorts cross-sectionally, with fewer studies longitudinally tracking the same cohort of patients using these methods.
Commonly reported protocols for L3 slice segmentation involve a single axial DICOM image, retrospectively obtained, provided to the research team and further analyzed using software such as Slice-O-Matic, ImageJ, Osirix, or directly within PACS.11 Under this methodology, concern for how a single axial slice relates to the rest of the body is not considered. Can we afford to ignore the considerable displacement of tissues in this region frequently caused by clothing, namely pants?
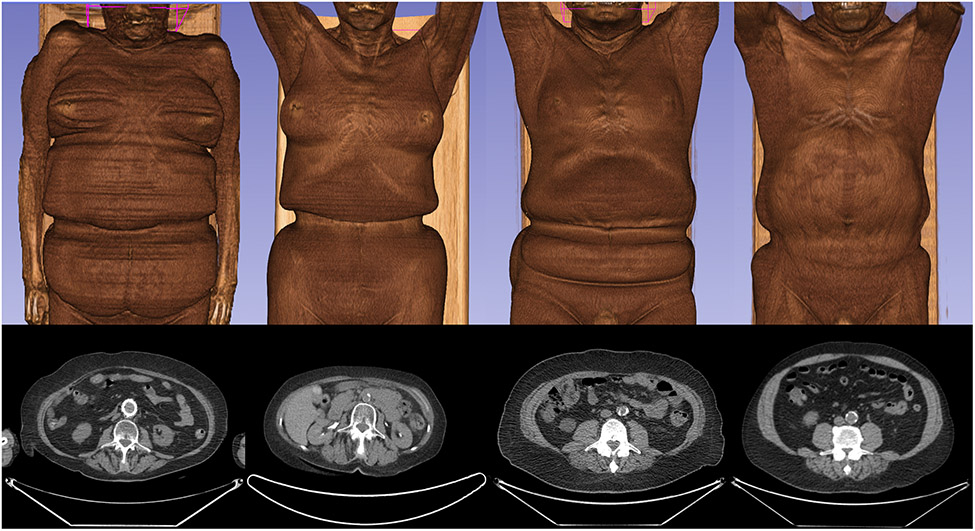
A critical issue when utilizing retrospective data for post-hoc analysis is consideration for the context in which the data were initially obtained.9 To clarify, virtually no CT images are clinically obtained for the primary purpose of assessing body composition or tissue volumes. In the context of standard of care procedures that involve CT imaging, patients may not be instructed to remove clothing if it does not hinder the objective of the test. In oncological PET/CT imaging, which is becoming increasingly common, clothing is unlikely to impede the ability to monitor for metastases, and thus, is often not removed. This seemingly insignificant influence of clothing has yet to be studied or even discussed as a limitation for this widely utilized research method. As demonstrated in Figure 1, two-dimensional perspectives from the L3 axial slice may not indicate any reason for concern; however, the depicted L3 axial slice exists within the affected region in each of these four patients.
Figure 1. 2D & 3D Representations of Patients Imaged with Clothing.
Four patients (two female patients on the left, two male patients on the right) depicted as 3D reconstructions and as 2D axial views from the L3 level using the 3D Slicer software.21
Prevalence of Clothing Interference
To assess the prevalence of clothing artifacts at the waist, including at the L3 level, all public-access full-body and skull-to-thigh PET/CT scans from The Cancer Imaging Archive (TCIA) were visually inspected.22 1026 scans were evaluated across 23 open-access TCIA datasets (466M/560F, age 62.0±11.7y, BMI 27.3±5.6 kg/m2); of the scans assessed, 96.2%, the presence of pants or undergarments was apparent in 96.2% of scans, though noticeable tissue displacement existed in 72.7%. Further, 60.8% of scans had an apparent presence of pants or undergarments specifically at the mid-L3 level, including 51.1% with noticeable tissue displacement. Breakdowns of clothing artifact prevalence by patient diagnosis and for each TCIA dataset are provided in Tables 1 and S1, respectively.
Table 1.
Open Access PET/CT Scans Reviewed from TCIA
| Diagnosis | # Scans Reviewed |
# Scans with Apparent Pants |
# Scans with Tissue Displacement from Pants |
Dataset Citations |
||
|---|---|---|---|---|---|---|
| Any | L3 | Any | L3 | |||
| Breast Cancer | 218 | 215 | 165 | 162 | 145 | 22-25 |
| Endocrine Cancers | 1 | 1 | 0 | 1 | 0 | 22,26 |
| Gastrointestinal Cancers | 2 | 2 | 1 | 2 | 1 | 22,27,28 |
| Gynecological Cancers | 79 | 74 | 63 | 67 | 56 | 22,29-32 |
| Lung Cancers | 663 | 633 | 366 | 461 | 295 | 22,33-42 |
| Non-cancer | 4 | 4 | 4 | 4 | 4 | 22,43 |
| Prostate Cancer | 26 | 25 | 7 | 22 | 6 | 22,44-46 |
| Skin & Soft Tissue Cancers | 24 | 24 | 12 | 18 | 11 | 22,47-50 |
| Urological Cancers | 9 | 9 | 6 | 9 | 6 | 22,51,52 |
| Total | 1026 | 987 (96.2%) | 624 (60.8%) | 746 (72.7%) | 524 (51.1%) | |
Full-body and skull-to-thigh PET/CT scans reviewed from datasets available from The Cancer Imaging Archive (TCIA). Of the reviewed scans, the number of scans classified as suspected to have clothing present (apparent pants) and the number of scans with pants causing noticeable displacement of tissues are displayed; each are quantified across any location, as well as specifically at the level of the third lumbar vertebrae (L3).
The sexually dimorphic influence of clothing should be particularly emphasized. Notable tissue displacement from pants or undergarments was more common in females (436 of 560 scans, 77.9%) than males (310 of 466 scans, 66.5%). This effect, however, is particularly evident at the L3 level, the region most widely utilized for body composition; at the L3 slice, 69.6% of females demonstrated tissue displacement, compared to only 28.8% of males. This sexually dimorphic effect is likely due to the tendency for women to wear their pants higher than men, as well as sex differences in the regional distribution of adipose tissue. When apparent in males examined herein, lower-body clothing appeared to begin around the L4/L5 vertebral level and extend into the sacrum, which was substantially lower than most females.
Additional relationships with clothing artifact also existed for age and BMI. Compared to scans unrestricted by lower-body clothing, scans with pant-related tissue displacement were from a significantly older cohort of patients (p<0.0001, 3.52y mean difference), including at the L3 level (p=0.001, 2.43y mean difference). Similarly, scans restricted by lower-body clothing also represented a patient cohort with significantly higher BMI (p<0.0001, 2.53 kg/m2 mean difference), including at the L3 level (p<0.0001, 2.26 kg/m2 mean difference). Increasing BMI tended to correspond with overall severity of clothing-associated tissue displacement for both males and females.
It should be noted that the L3 level is not the only area of concern for this type of interference. Though less common, use of the umbilicus and various thoracic vertebrae have also been explored as potential landmarks in this type of research.53-56 Herein, we identified 51.3% of scans to have notable tissue displacement at the level of the umbilicus (526 of 1026 scans), and thoracic restriction resulting from the bra was present in 28.0% of female scans (157 of 403 scans).
Magnitude of Clothing Interference
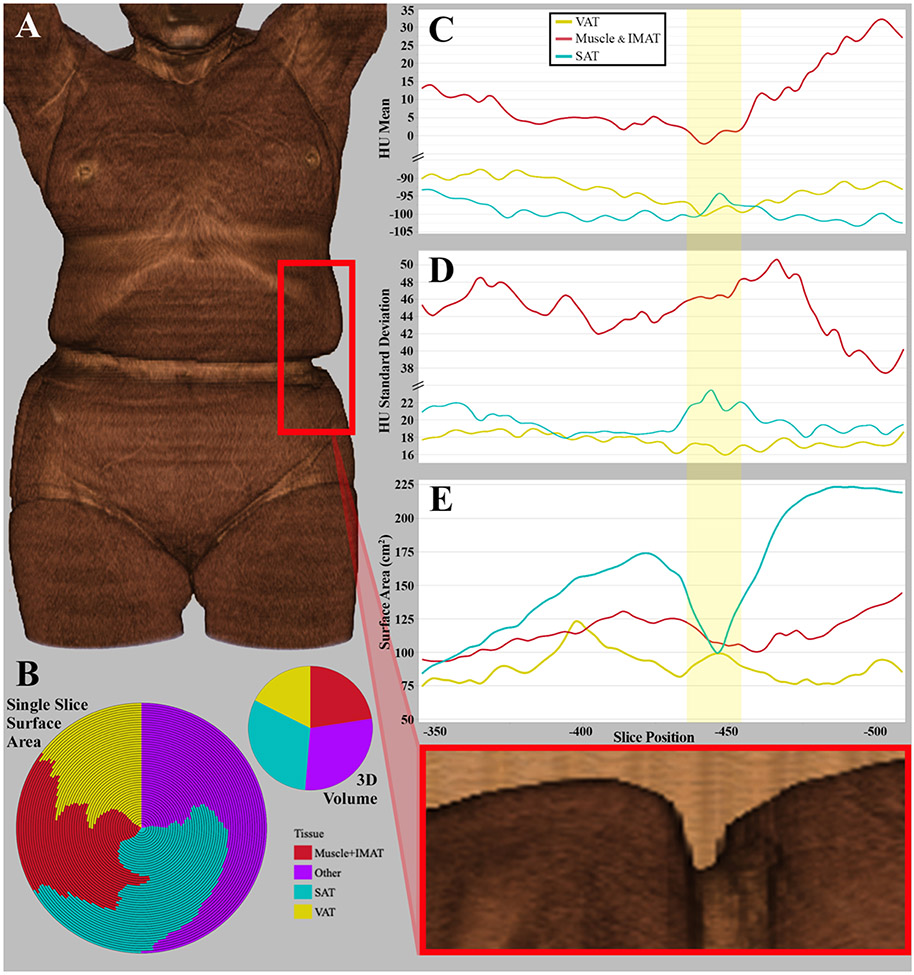
To further assess the impact that clothing restriction may have on measured variables, tissue volumes for a single patient were segmented across affected slices using Slice-O-Matic (version 6.6); following the Alberta protocol,57 skeletal muscle (-29 to 150 HU), IMAT (-190 to -30 HU), VAT (-150 to -50 HU), and SAT (-190 to -30 HU) were segmented across 51 consecutive axial slices, spanning a vertical distance of 163 mm. Skeletal muscle and IMAT were further combined to represent all tissue within the muscle fascia. Total surface area (SA), encompassing all tissues, was also examined for each slice.
Slice-by-slice comparisons are demonstrated in Figure 2, including representations of key tissue proportions (Figure 2B), attenuation mean and standard deviation (SD; Figure 2C-D), and SA (Figure 2E). Compared to tissue proportions across the 51-slice volume, single slice SA of each tissue was highly variable across near slices (Figure 2B). Relative tissue SA quantified from slices with the lowest and highest proportions have a range of 24.1% for SAT (16.6–40.7%), 11.5% for VAT (13.7-25.2%), and 10.5% for combined muscle and IMAT tissues (18.1-28.5%).
Figure 2A-E. Tissue Morphology by Influence of Pants.
A) 3D rendering of a female patient (78 years old, BMI of 23.9). B) Volumetric proportions of key tissues used in body composition evaluations, representing the high variability in relative tissue proportions from 51 consecutive axial slices (left) and the volumetric tissue proportion across all combined slices (right). C) Slice-by-slice representation of average attenuation (HU) by tissue. D) Slice-by-slice representation of attenuation standard deviation (HU) by tissue. E) Slice-by-slice representation of surface area (cm2) by tissue.
While not as drastic as shifts to SA, changes also appear to exist for averages and variability (SD) in tissue radiodensity. VAT and muscle tissues experience absolute minima for mean attenuation within the bodily region experiencing the greatest clothing constriction (Figure 2C). This region, highlighted in yellow in Figure 2C-E, also reflects the maximum SD of SAT radiodensity (Figure 2D). Average single slice SAT attenuation had a range of 10 HU (-103.3 to -93.3 HU), 12.8 HU for VAT (-100.4 to -87.6 HU), and varied by 34.2 HU within the muscle fascia (-2.1 to 32.1 HU).
The most compelling evidence surrounding clothing artifact comes from sizable changes to tissue SA (Figure 2E). Compared to the slice with the lowest SA, the largest slice was 49.4% greater in total 2D area (20 slices, 65.4 mm distance). This relative difference in SA had substantial variability for all key tissues; compared to the slice with the lowest SA, the slice with the largest SA was 165·6% greater in SAT SA (45 slices, 147.2 mm), 66.1% greater in VAT (16 slices, 52.4 mm), and 54.8% greater in muscle and IMAT tissues combined (49 slices, 160.23 mm). Considerable changes to tissue SA occurred in only a few slices; compared to the slice most affected by pant restriction, there was a 61.5% and 79.9% greater SAT SA on the axial image five slices above and below it, respectively (16.35 mm distance). Across the same distance, VAT SA reflected a decrease of up to 15.4%, whereas muscle increased by up to 16.8%.
Conclusion
Taken together, the high slice-to-slice intraindividual variability for these widely used variables presents a difficult challenge for post-hoc analyses. SMI cut points of 39 and 41 across the L3 to L5 region are commonly used to stratify females for sarcopenia;5,11,58 the patient case presented herein can be classified as above or below either of these thresholds based solely upon slice selection between L3 and L5. Relative changes alone that occur to tissues over the span of a few slices provide reason for concern, but these effects may be even further compounded among calculated metrics, such as SMG, that considers both skeletal muscle SA and attenuation from a given axial slice.
Not only can single slice selection influence stratification to patient cohorts in cross-sectional analyses but it could also affect the reliability of repeated measures analyses in tracking longitudinal changes. On top of any real change to tissues, differences in the location or magnitude of clothing interference between scans could artificially distort, inflate or mask any real change within tissues.
The high prevalence of tissue displacement by result of clothing, especially in females, has the potential to substantially effect clinical research findings, as well as their applications, when utilizing methods that involve only a single axial slice in the lumbar region. These data emphasize the importance of performing data quality assessments regarding real-world context and variability within clinical presentation of data. As software advancements continue, methodology to account for the potential influence of clothing interference should be inherent in all study designs; namely, the use of volumetric segmentation or standardized regions/volumes of interest (ROI/VOI) across patients should be prioritized.
Supplementary Material
Supplementary Table S1
Utilized Datasets from TCIA
Full-body and skull-to-thigh PET/CT scans reviewed from each dataset available from The Cancer Imaging Archive (TCIA). Of the reviewed scans, the number of scans classified as suspected to have clothing present (apparent pants) and the number of scans with pants causing noticeable displacement of tissues are provided; each are quantified across any location and at the level of the third lumbar vertebrae (L3). [CM, cutaneous melanoma; EC, endometrial carcinoma; HCC, hepatocellular carcinoma; NSCLC, non-small cell lung cancer; PDAC, pancreatic ductal adenocarcinoma; RCC, renal cell carcinoma; SCC, squamous cell carcinoma; TCIA, The Cancer Imaging Archive; UC, urothelial carcinoma]
References
- 1.Sheean P, Gonzalez MC, Prado CM, McKeever L, Hall AM, Braunschweig CA. American Society for Parenteral and Enteral Nutrition Clinical Guidelines: The Validity of Body Composition Assessment in Clinical Populations. J Parenter Enteral Nutr. 2020; 44: 12–43. doi: 10.1002/jpen.1669. [DOI] [PubMed] [Google Scholar]
- 2.Cheng E, Kirley J, Cespedes Feliciano EM, Caan BJ. Adiposity and cancer survival: a systematic review and meta-analysis. Cancer Causes Control. 2022; 33 (10): 1219–1246. doi: 10.1007/s10552-022-01613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsui R, Watanabe J, Banno M, Inaki N, Fukunaga T. Association of visceral adipose tissue with postoperative outcome in upper gastrointestinal cancer: A systematic review and meta-analysis. Am J Clin Nutr. 2022; 116 (6): 1540–1552. doi: 10.1093/ajcn/nqac273. [DOI] [PubMed] [Google Scholar]
- 4.Kazemi-Bajestani SM, Mazurak VC, Baracos V. Computed tomography-defined muscle and fat wasting are associated with cancer clinical outcomes. Semin Cell Dev Biol. 2016; 54: 2–10. doi: 10.1016/j.semcdb.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Hilmi M, Jouinot A, Burns R, et al. Body composition and sarcopenia: The next-generation of personalized oncology and pharmacology? Pharmacol Ther. 2019; 196: 135–159. doi: 10.1016/j.pharmthera.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson JM, Orsso CE, Nourouzpour S, et al. Computed tomography-based body composition measures in COPD and their association with clinical outcomes: A systematic review. Chron Respir Dis. 2022; 19: 14799731221133387. doi: 10.1177/14799731221133387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montes-Ibarra M, Orsso CE, Limon-Miro AT, et al. Prevalence and clinical implications of abnormal body composition phenotypes in patients with COVID-19: A systematic review. Am J Clin Nutr. 2023; 117 (6): 1288–1305. doi: 10.1016/j.ajcnut.2023.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedrikovetski S, Seow W, Kroon HM, Traeger L, Moore JW, Sammour T. Artificial intelligence for body composition and sarcopenia evaluation on computed tomography: A systematic review and meta-analysis. Eur J Radiol. 2022; 149: 110218. doi: 10.1016/j.ejrad.2022.110218. [DOI] [PubMed] [Google Scholar]
- 9.Huebschmann AG, Leavitt IM, Glasgow RE. Making Health Research Matter: A Call to Increase Attention to External Validity. Annu Rev Public Health. 2019; 40: 45–63. doi: 10.1146/annurev-publhealth-040218-043945. [DOI] [PubMed] [Google Scholar]
- 10.Subbiah V. The next generation of evidence-based medicine. Nat Med. 2023; 29 (1): 49–58. doi: 10.1038/s41591-022-02160-z. [DOI] [PubMed] [Google Scholar]
- 11.Amini B, Boyle SP, Boutin RD, Lenchik L. Approaches to Assessment of Muscle Mass and Myosteatosis on Computed Tomography: A Systematic Review. J Gerontol A Biol Sci Med Sci. 2019; 74 (10): 1671–1678. doi: 10.1093/gerona/glz034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mourtzakis M, Prado CM, Lieffers JR, Reiman T, McCargar LJ, Baracos VE. A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl Physiol Nutr Metab. 2008; 33: 997–1006. doi: 10.1139/H08-075. [DOI] [PubMed] [Google Scholar]
- 13.Hong JH, Hong H, Choi YR, et al. CT analysis of thoracolumbar body composition for estimating whole-body composition. Insights Imaging. 2023; 14 (1): 69. doi: 10.1186/s13244-023-01402-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz AA, Zhou L, Young TP, et al. Chest CT measures of muscle and adipose tissue in COPD: Gender-based differences in content and in relationships with blood biomarkers. Acad Radiol. 2014; 21 (10): 1255–1261. doi: 10.1016/j.acra.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cespedes Feliciano EM, Chen WY, Lee V, et al. Body Composition, Adherence to Anthracycline and Taxane-Based Chemotherapy, and Survival After Nonmetastatic Breast Cancer. JAMA Oncol. 2020; 6 (2): 264–270. doi: 10.1001/jamaoncol.2019.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huh J, Park B, Lee H, et al. Prognostic Value of Skeletal Muscle Depletion Measured on Computed Tomography for Overall Survival in Patients with Non-Metastatic Breast Cancer. J Breast Cancer. 2020; 23 (1): 80–92. doi: 10.4048/jbc.2020.23.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim EY, Lee HY, Kim YS, et al. Prognostic significance of cachexia score assessed by CT in male patients with small cell lung cancer. Eur J Cancer Care (Engl). 2018; 27 (1). doi: 10.1111/ecc.12695. [DOI] [PubMed] [Google Scholar]
- 18.Park MJ, Cho JM, Jeon KN, et al. Mass and fat infiltration of intercostal muscles measured by CT histogram analysis and their correlations with COPD severity. Acad Radiol. 2014; 21 (6): 711–717. doi: 10.1016/j.acra.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Marquardt JP, Roeland EJ, Van Seventer EE, et al. Percentile-based averaging and skeletal muscle gauge improve body composition analysis: Validation at multiple vertebral levels. J Cachexia Sarcopenia Muscle. 2022; 13 (1): 190–202. doi: 10.1002/jcsm.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacoste Jeanson A, Dupej J, Villa C, Bruzek J. Body composition estimation from selected slices: Equations computed from a new semi-automatic thresholding method developed on whole-body CT scans. PeerJ. 2017; 5: e3302. doi: 10.7717/peerj.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012; 30 (9): 1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J Digit Imaging. 2013; 26 (6): 1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinahan P, Muzi M, Bialecki B, Coombs L. Data from ACRIN-FLT-Breast, Version 2 [Dataset]. The Cancer Imaging Archive. 2017. doi: 10.7937/K9/TCIA.2017.ol20zmxg. [DOI] [Google Scholar]
- 24.Kostakoglu L, Duan F, Idowu MO, et al. A Phase II Study of 3'-Deoxy-3'-18F-Fluorothymidine PET in the Assessment of Early Response of Breast Cancer to Neoadjuvant Chemotherapy: Results from ACRIN 6688. J Nucl Med. 2015; 56 (11): 1681–1689. doi: 10.2967/jnumed.115.160663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloch BN, Jain A, Jaffe CC. BREAST-DIAGNOSIS [Dataset]. The Cancer Imaging Archive. 2015. doi: 10.7937/K9/TCIA.2015.SDNRQXXR. [DOI] [Google Scholar]
- 26.Kirk S, Lee Y, Roche C, Bonaccio E, Filippini J, Jarosz R. The Cancer Genome Atlas Thyroid Cancer Collection (TCGA-THCA), Version 3 [Dataset]. The Cancer Imaging Archive. 2016. doi: 10.7937/K9/TCIA.2016.9ZFRVF1B. [DOI] [Google Scholar]
- 27.Erickson BJ, Kirk S, Lee Y, et al. The Cancer Genome Atlas Liver Hepatocellular Carcinoma Collection (TCGA-LIHC), Version 5 [Dataset]. The Cancer Imaging Archive. 2016. doi: 10.7937/K9/TCIA.2016.IMMQW8UQ. [DOI] [Google Scholar]
- 28.National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Pancreatic Ductal Adenocarcinoma Collection (CPTAC-PDA), Version 13 [Dataset]. The Cancer Imaging Archive. 2018. doi: 10.7937/K9/TCIA.2018.SC20FO18. [DOI] [Google Scholar]
- 29.National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Uterine Corpus Endometrial Carcinoma Collection (CPTAC-UCEC), Version 10 [Dataset]. The Cancer Imaging Archive. 2019. doi: 10.7937/K9/TCIA.2018.3R3JUISW. [DOI] [Google Scholar]
- 30.Mayr N, Yuh WTC, Bowen S, et al. Cervical Cancer – Tumor Heterogeneity: Serial Functional and Molecular Imaging Across the Radiation Therapy Course in Advanced Cervical Cancer, Version 1 [Dataset]. The Cancer Imaging Archive. 2023. doi: 10.7937/ERZ5-QZ59. [DOI] [Google Scholar]
- 31.Bowen SR, Yuh WTC, Hippe DS, et al. Tumor radiomic heterogeneity: Multiparametric functional imaging to characterize variability and predict response following cervical cancer radiation therapy. J Magn Reson Imaging. 2018; 47 (5): 1388–1396. doi: 10.1002/jmri.25874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erickson BJ, Mutch D, Lippmann L, Jarosz R. The Cancer Genome Atlas Uterine Corpus Endometrial Carcinoma Collection (TCGA-UCEC), Version 4 [Dataset]. The Cancer Imaging Archive. 2016. doi: 10.7937/K9/TCIA.2016.GKJ0ZWAC. [DOI] [Google Scholar]
- 33.Kinahan P, Muzi M, Bialecki B, Herman B, Coombs L. Data from the ACRIN 6668 Trial NSCLC-FDG-PET, Version 2 [Dataset]. The Cancer Imaging Archive. 2019. doi: 10.7937/tcia.2019.30ilqfcl. [DOI] [Google Scholar]
- 34.Machtay M, Duan F, Siegel BA, et al. Prediction of survival by [18F]fluorodeoxyglucose positron emission tomography in patients with locally advanced non-small-cell lung cancer undergoing definitive chemoradiation therapy: results of the ACRIN 6668/RTOG 0235 trial. J Clin Oncol. 2013; 31 (30): 3823–3830. doi: 10.1200/JCO.2012.47.5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhavi P, Patel S, Tsao AS. Data from Anti-PD-1 Immunotherapy Lung [Dataset]. The Cancer Imaging Archive. 2019. doi: 10.7937/tcia.2019.zjjwb9ip. [DOI] [Google Scholar]
- 36.National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Lung Squamous Cell Carcinoma Collection (CPTAC-LSCC), Version 14 [Dataset]. The Cancer Imaging Archive. 2018. doi: 10.7937/K9/TCIA.2018.6EMUB5L2. [DOI] [Google Scholar]
- 37.National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Lung Adenocarcinoma Collection (CPTAC-LUAD), Version 12 [Dataset]. The Cancer Imaging Archive. 2018. doi: 10.7937/K9/TCIA.2018.PAT12TBS. [DOI] [Google Scholar]
- 38.Bakr S, Gevaert O, Echegaray S, et al. Data for NSCLC Radiogenomics, Version 4 [Dataset]. The Cancer Imaging Archive. 2017. doi: 10.7937/K9/TCIA.2017.7hs46erv. [DOI] [Google Scholar]
- 39.Bakr S, Gevaert O, Echegaray S, et al. A radiogenomic dataset of non-small cell lung cancer. Sci Data. 2018; 5. doi: 10.1038/sdata.2018.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology. 2012; 264 (2): 387–396. doi: 10.1148/radiol.12111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albertina B, Watson M, Holback C, et al. The Cancer Genome Atlas Lung Adenocarcinoma Collection (TCGA-LUAD), Version 4 [Dataset]. The Cancer Imaging Archive. 2016. doi: 10.7937/K9/TCIA.2016.JGNIHEP5. [DOI] [Google Scholar]
- 42.Kirk S, Lee Y, Kumar P, et al. The Cancer Genome Atlas Lung Squamous Cell Carcinoma Collection (TCGA-LUSC), Version 4 [Dataset]. The Cancer Imaging Archive. 2016. doi: 10.7937/K9/TCIA.2016.TYGKKFMQ. [DOI] [Google Scholar]
- 43.Saltz J, Saltz M, Prasanna P, et al. Stony Brook University COVID-19 Positive Cases [Dataset]. The Cancer Imaging Archive. 2021. doi: 10.7937/TCIA.BBAG-2923. [DOI] [Google Scholar]
- 44.Kurdziel KA, Shih JH, Apolo AB, et al. The kinetics and reproducibility of 18F-sodium fluoride for oncology using current PET camera technology. J Nucl Med. 2012; 53 (8): 1175–1184. doi: 10.2967/jnumed.111.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurdziel KA, Apolo AB, Lindenberg L, et al. Data From NaF PROSTATE [Dataset]. The Cancer Imaging Archive. 2015. doi: 10.7937/K9/TCIA.2015.ISOQTHKO. [DOI] [Google Scholar]
- 46.Zuley ML, Jarosz R, Drake BF, et al. The Cancer Genome Atlas Prostate Adenocarcinoma Collection (TCGA-PRAD), Version 4 [Dataset]. The Cancer Imaging Archive. 2016. doi: 10.7937/K9/TCIA.2016.YXOGLM4Y. [DOI] [Google Scholar]
- 47.National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Cutaneous Melanoma Collection (CPTAC-CM), Version 10 [Dataset]. The Cancer Imaging Archive. 2018. doi: 10.7937/K9/TCIA.2018.ODU24GZE. [DOI] [Google Scholar]
- 48.National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Sarcomas Collection (CPTAC-SAR), Version 10 [Dataset]. The Cancer Imaging Archive. 2019. doi: 10.7937/tcia.2019.9bt23r95. [DOI] [Google Scholar]
- 49.Vallieres M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015; 60 (14): 5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
- 50.Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities (Soft-tissue-Sarcoma) [Dataset]. The Cancer Imaging Archive. 2015. doi: 10.7937/K9/TCIA.2015.7GO2GSKS. [DOI] [PubMed] [Google Scholar]
- 51.Kirk S, Lee Y, Lucchesi FR, et al. The Cancer Genome Atlas Urothelial Bladder Carcinoma Collection (TCGA-BLCA), Version 8 [Dataset]. The Cancer Imaging Archive. 2016. doi: 10.7937/K9/TCIA.2016.8LNG8XDR. [DOI] [Google Scholar]
- 52.Linehan M, Gautam R, Kirk S, et al. The Cancer Genome Atlas Cervical Kidney Renal Papillary Cell Carcinoma Collection (TCGA-KIRP), Version 4 [Dataset]. The Cancer Imaging Archive. 2016. doi: 10.7937/K9/TCIA.2016.ACWOGBEF. [DOI] [Google Scholar]
- 53.Okamura A, Watanabe M, Yamashita K, et al. Implication of visceral obesity in patients with esophageal squamous cell carcinoma. Langenbecks Arch Surg. 2018; 403 (2): 245–253. doi: 10.1007/s00423-017-1643-0. [DOI] [PubMed] [Google Scholar]
- 54.Li XT, Tang L, Chen Y, Li YL, Zhang XP, Sun YS. Visceral and subcutaneous fat as new independent predictive factors of survival in locally advanced gastric carcinoma patients treated with neo-adjuvant chemotherapy. J Cancer Res Clin Oncol. 2015; 141 (7): 1237–1247. doi: 10.1007/s00432-014-1893-y. [DOI] [PubMed] [Google Scholar]
- 55.Iwase T, Sangai T, Nagashima T, et al. Impact of body fat distribution on neoadjuvant chemotherapy outcomes in advanced breast cancer patients. Cancer Med. 2016; 5 (1): 41–48. doi: 10.1002/cam4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vangelov B, Bauer J, Kotevski D, Smee RI. The use of alternate vertebral levels to L3 in computed tomography scans for skeletal muscle mass evaluation and sarcopenia assessment in patients with cancer: A systematic review. Br J Nutr. 2022; 127 (5): 722–735. doi: 10.1017/S0007114521001446. [DOI] [PubMed] [Google Scholar]
- 57.Steele S, Lin F, Le TL, et al. Segmentation and Linear Measurement for Body Composition Analysis using Slice-O-Matic and Horos. J Vis Exp. 2021; (169). doi: 10.3791/61674. [DOI] [PubMed] [Google Scholar]
- 58.Cimsit C, Kursun M, Demircioglu O, Dilber F, Demirtas CO, Ergenc I. Radiological Quantification of Sarcopenic Obesity and its Role in Chronic Liver Disease Severity. Acad Radiol. 2023; 30 Suppl 1: S124–S131. doi: 10.1016/j.acra.2023.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1
Utilized Datasets from TCIA
Full-body and skull-to-thigh PET/CT scans reviewed from each dataset available from The Cancer Imaging Archive (TCIA). Of the reviewed scans, the number of scans classified as suspected to have clothing present (apparent pants) and the number of scans with pants causing noticeable displacement of tissues are provided; each are quantified across any location and at the level of the third lumbar vertebrae (L3). [CM, cutaneous melanoma; EC, endometrial carcinoma; HCC, hepatocellular carcinoma; NSCLC, non-small cell lung cancer; PDAC, pancreatic ductal adenocarcinoma; RCC, renal cell carcinoma; SCC, squamous cell carcinoma; TCIA, The Cancer Imaging Archive; UC, urothelial carcinoma]