Abstract
Poliovirus infectious RNA can be synthesized in vitro using phage DNA-dependent RNA-polymerases. These synthetic transcripts contain several extra nucleotides at the 5′ end, which are deleted during replication to generate authentic viral genomes. We removed those 5′-end extra nucleotides utilizing a hammerhead ribozyme to produce transcripts with accurate 5′ ends. These transcripts replicate substantially more rapidly in cell culture, demonstrating no lag before replication; they also replicate more efficiently in Xenopus laevis oocytes and in in vitro translation-replication cell extracts. In both systems, an exact 5′ end is necessary for synthesis of positive-strand RNA but not negative-strand RNA.
The successful reconstruction of a positive-stranded RNA virus genome as an infectious cDNA clone allowed the use of reverse genetics to study the replication cycle of these viruses (29). The system was improved by the use of viral RNA synthesized in vitro by DNA-dependent RNA polymerases using biologically active cDNA clones as templates (1, 36). A number of positive- and negative-strand RNA viruses require exact termini for successful replication. Addition and deletion of sequences at the 5′ and 3′ ends often have deleterious effects on replication or are repaired at low frequency in vivo to generate authentic ends (reviewed in reference 9).
The effect of nonauthentic ends on in vitro transcripts during replication has been studied for several members of the family Picornaviridae. For example, the nonviral guanosine residues at the 5′ ends of poliovirus and mengovirus in vitro transcripts are removed during replication in tissue culture cells, resulting in authentic 5′ ends (12, 17, 36). In vitro-transcribed coxsackievirus B3 RNA missing the two 5′-terminal uridine residues regains these nucleotides during replication, and it has been proposed that the addition takes place during positive-strand RNA synthesis (21). These observations suggest that the well-conserved enterovirus 5′-UU terminus plays an essential role during genome replication.
Poliovirus, a member of the family Picornaviridae and genus Enterovirus, contains a single plus-strand RNA genome of approximately 7.5 kb. There are two noncoding regions, at the 5′ and 3′ ends, flanking an open reading frame of 6,528 nucleotides (16, 35). Poliovirus RNA is translated into a single polypeptide which is processed into its smaller, functional proteins mainly by the viral proteases 3Cpro and 3CDpro, with two additional cleavages by 2Apro (16, 19, 35).
Poliovirus RNA replication follows a strategy common to all positive-stranded RNA viruses: the viral genome is transcribed into a complementary RNA (negative strand), which in turn is used as a template to synthesize new strands of genomic RNA. The virus-encoded RNA-dependent RNA polymerase (3Dpol) catalyzes the synthesis of both strands. However, because 3Dpol is a primer-dependent enzyme, several other viral and cellular factors are likely to be involved in the initiation process of RNA synthesis. Genetic analysis has implicated most of the nonstructural viral proteins in RNA synthesis (7, 10, 15, 20, 22, 24). Uridylation of the viral polypeptide VPg has been postulated to be an essential step during initiation of poliovirus replication. This reaction can be mediated by 3Dpol in vitro in the presence of poly(A) (28). VPg-pUpU has been proposed to act as a primer for both negative- and positive-strand RNA synthesis and can be detected covalently attached to each strand in virally infected cells. In addition, initiation of positive-strand RNA synthesis requires a ribonucleoprotein complex that forms at the 5′ end of the positive strand, which contains the cellular factor poly(rC) binding protein and the precursor of the viral proteinase- and polymerase (3CD)-containing polypeptides (2, 3, 18). This complex participates in trans initiation of positive-strand synthesis (2). Many aspects of this mechanism have yet to be worked out in detail.
Although poliovirus in vitro transcripts containing extra nucleotides at the 5′ end are able to initiate the replication cycle in tissue culture cells, they do not replicate efficiently in alternative experimental systems. Positive-strand RNA synthesis was not detected in experiments using Xenopus laevis oocytes injected with poliovirus in vitro transcripts, though negative-strand RNA synthesis was demonstrated (12; J. Herold and R. Andino, unpublished observation). Similarly, scant (33, 34) or no detectable (5, 6) positive-strand RNA was synthesized in a cell-free translation-replication system.
We show here that the 5′ end of the input genome is crucial for efficient positive-strand RNA synthesis during poliovirus replication. Taking advantage of a cis-active ribozyme, we were able to synthesize viral RNAs with authentic 5′ ends in vitro. These transcripts replicated in tissue culture cells with improved kinetics, were capable of replicating in X. laevis oocytes, and replicated in a cell-free replication system comparably to virion RNA.
MATERIALS AND METHODS
Virus and cells.
HeLa S3 cells (ATCC CCL 2.2) were grown either (i) in tissue culture flasks in Dulbecco's modified Eagle medium–nutrient mixture F-12 (Ham) (1:1), supplemented with 2 mM l-glutamine, 100 U of penicillin and streptomycin per ml, and 10% newborn calf serum or (ii) in suspension in suspension minimal essential medium (Joklik modified) supplemented with 2 mM l-glutamine, 100 U of penicillin and streptomycin per ml, and 10% fetal calf serum. 293 cells (ATCC CRL 1573) were used for transfection experiments and cultured in minimal essential medium Eagles with Earle's balanced salt solution, supplemented with 2 mM l-glutamine, 100 U of penicillin and streptomycin per ml, and 10% newborn calf serum. Poliovirus type 1 (Mahoney) was grown in HeLa S3 cells.
Oligonucleotides.
The following oligonucleotides were used in this study: I, 5′-TGCAGCGCGCAGGCCTTAATACGACTCACTATAGGGTGTTTTAA-3′; II, 5′-ATCAGTTAAAACACCCTATAGTGAGTCGTATTAAGGCCTGCGCGCTGCA-3′; III, 5′-CTGATGAGGCCGAAAGGCCGAAAACCCGGTATCCCGGGTTCTTAAAACAGCTCTGGGGTTG-3′; IV, 5′-CCAGAGCT GTTTTAAGAACCCGGGATACCGGGTTTTCGGCCTTTCGGCCTC-3′; V, 5′-CTGATGAGGCCGAAAGGCCGAAAACCCGGTATCCCGGGTTGTTAAAACAGCTCTGGGGTTG-3′; VI, 5′-CCAGAGCTGTTTTAACAACCCGGGATACCGGGTTTTCGGCCTTTCGGCCTC-3′; VII, 5′-TACCCACCCCAGAGGCCCACGTGGCGCGCACGT-3′; VIII, 5′-ACGTGCGCGCCACGTGGGCCTCTGGGGTGGGTACAA-3′; and IX, 5′-GGCGTACAAGGGTACCGCAA-3′.
Plasmids.
Oligonucleotides I to VIII were 5′ phosphorylated with T4 polynucleotide kinase. Equimolar amounts of oligonucleotide pairs I/II, III/IV, V/VI, and VII/VIII were combined and annealed by heating to 94°C in 10 mM Tris-HCl (pH 7.5)–50 mM NaCl and slowly cooling to 20°C. Pairs I/II, III/IV, and VII/VIII (representing a T7 RNA polymerase promoter, a hammerhead ribozyme, the 43 5′-terminal nucleotides of the poliovirus type 1 [Mahoney] genome and restriction sites for cloning purposes) or I/II, V/VI, and VII/VIII (like the other pairs, but containing a mutated hammerhead ribozyme) were ligated and cloned into a pBluescript II vector, resulting in pBlue rib(+) and pBlue rib(−), respectively. After sequence analysis, the inserts were cloned in front of cDNA copies of either the Mahoney strain of poliovirus (pXpA) or a luciferase-expressing, poliovirus-derived replicon (pRLuc31 [2]), resulting in prib(+) or (−)XpA and prib(+) or (−)RLuc.
In vitro transcription.
Poliovirus-specific transcripts were obtained using a T7 Megascript transcription kit (Ambion, Austin, Tex.) after linearization of the plasmid DNA. Fifty microcuries of [α-33P]UTP (10 μCi/μl; Amersham Pharmacia Biotech, Piscataway, N.J.) were added to transcription reactions to obtain uniformly radiolabeled transcripts.
Primer extension.
Oligonucleotide IX (complementary to nucleotides 60 to 80 of the poliovirus genomic RNA) was 5′-end labeled with 33P using T4 polynucleotide kinase. Subsequently, 10 ng was used as a primer in a 20-μl reverse transcription reaction (10 mM dithiothreitol, 1 mM each deoxynucleoside triphosphate [dNTP], 50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, 200 U of Superscript II reverse transcriptase [Life Technologies]) with either 1 μg of poliovirus genomic RNA isolated from virions, rib(+)XpA-RNA, rib(−)XpA-RNA, or XpA-RNA as templates. After 30 min of incubation at 37°C, the reaction products were denatured and separated on denaturing 6% polyacrylamide gels and visualized by autoradiography.
RNA transfection.
293 cells were trypsinized, washed twice with phosphate-buffered saline, and adjusted to 4 × 106 cells/ml. Then 800-μl aliquots were electroporated in 0.4-ml cuvettes with 20 μg of RNA, using a Electro Cell Manipulator 600 (BTX Inc., San Diego, Calif.) with the following settings: 300 V, 1,000 μF, 24 Ω. Subsequently, 10 volumes of medium was added, and depending on the experiment, 2 × 105, 1 × 106, or 2 × 106 cells were plated per 10-cm2 dish and incubated at 37°C in a 5% CO2 incubator. Guanidinium hydrochloride (Sigma Chemical Co., St. Louis, Mo.) was added to the medium to a final concentration of 2 mM when indicated.
Luciferase expression.
Replicon-transfected cells (5 × 105) were scraped off, washed once with phosphate-buffered saline, and then lysed in 200 μl of cell culture lysis reagent (Promega, Madison, Wis.). Luciferase activity in 10 μl of lysate was determined in a luminometer.
Isolation and analysis of intracellular poly(A)+ RNA.
Poly(A)+ RNA was isolated from 106 RNA-transfected cells or 2 × 106 33P-labeled-RNA-transfected cells, using oligo(dT)25-Dynabeads (Dynal A.S., Oslo, Norway) (32). The RNAs were separated by electrophoresis in a 2.2 M formaldehyde–0.8% agarose gel. The gel was dried at 60°C under vacuum and hybridized with 33P-5′-end-labeled oligonucleotide IX as described elsewhere (25).
Replication in X. laevis oocytes.
RNA (1 μg/μl; 25 ng/oocyte), together with HeLa cell S10 cell lysate (25 nl/oocyte), was injected into X. laevis oocytes as described elsewhere (13). [α-33P]UTP (10 μCi/μl; 250 nCi/oocyte) was injected after 1 h. Total RNA was isolated from 3, 6, 9, and 12 h of incubation at 30°C in the presence of actinomycin D (50 μg/ml), analyzed after poly(A)+ selection in native Tris-borate-EDTA (TBE)-agarose gels, and detected by autoradiography after drying the gel at 60°C under vacuum.
Replication in cell extracts.
Preparation of HeLa cell S10 extracts and initiation factors has been described in detail (4). The ability of in vitro-transcribed RNA to replicate in vitro was tested by method III (4), with some minor modifications. Briefly, 1 μg of RNA was mixed with 25 μl of S10 extract, 10 μl of initiation factors, 5 μl of 10× NTP/energy mix (10 mM ATP, 2.5 mM GTP, 2.5 mM CTP, 600 mM potassium acetate, 300 mM creatine phosphate, 155 mM HEPES-KOH [pH 8.0], 4 mg of creatine kinase per ml [Boehringer Mannheim, Mannheim, Germany]), and 1 μl of 100 mM guanidinium hydrochloride in a total volume of 50 μl and then incubated at 30°C for 4 h. After centrifugation at 15,000 × g for 15 min, the supernatant was discarded and the preinitiation complexes were resuspended in 50 μl of labeling mix (40 μl of S10 extract, previously dialyzed for 3 h against 1,000 volumes of dialysis buffer [40 mM HEPES {pH 8.0}, 120 mM potassium acetate, 5.5 mM magnesium acetate, 10 mM potassium chloride, 6 mM dithiothreitol], 5 μl of [α-33P]UTP [10 μCi/μl] and 5 μl of 10× NTP/energy mix). Aliquots of 10 μl were removed after 15, 30, 60, and 90 min and mixed with 190 μl of TENSK buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 100 mM NaCl, 1% sodium dodecyl sulfate, 200 μg of proteinase K per ml). After a 20-h incubation at 30°C, proteins were extracted with 1 volume of phenol-chloroform (1/1, vol/vol), and the RNA was precipitated with ethanol. Subsequently, the radiolabeled RNAs were separated in native TBE-agarose gels and detected by autoradiography after drying the gel at 60°C under vacuum.
RESULTS
Construction of a cis-active hammerhead ribozyme.
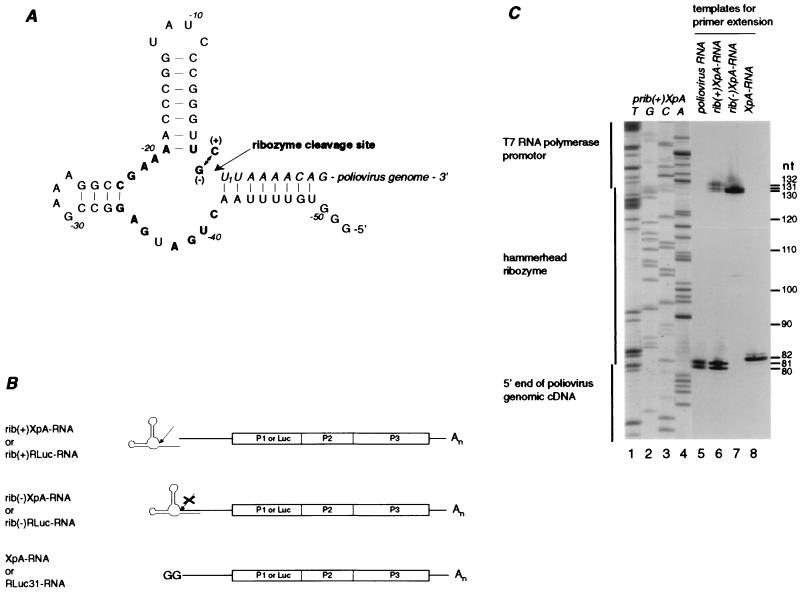
To examine whether poliovirus requires a precise 5′ end for efficient replication, a cDNA copy of a cis-active hammerhead ribozyme (8, 31) (Fig. 1A), was cloned between the T7 RNA polymerase promoter and the 5′-terminal sequences of a full-length poliovirus cDNA (pXpA) or a luciferase-expressing poliovirus replicon cDNA (pRLuc31). The resulting plasmids were named prib(+)XpA and prib(+)RLuc, respectively (Fig. 1B). As a control, we constructed two molecular clones, prib(−)XpA and prib(−)RLuc, that differ from prib(+)XpA and prib(+)RLuc only at an essential cytosine at position −1 of the hammerhead ribozyme, which was mutated to guanosine (Fig. 1A). This mutation yields an inactive form of the ribozyme.
FIG. 1.
A poliovirus-specific cis-acting hammerhead ribozyme. (A) Predicted secondary structure of the cis-active hammerhead ribozyme attached to the 5′ end of the poliovirus genome. Core residues of the ribozyme are shown in bold; poliovirus sequences are in italic. Numbering refers to the first poliovirus nucleotide U as 1. The active ribozyme contains a cytosine at position −1; the inactive form has a guanosine. (B) Schematic presentation of the in vitro transcripts used throughout this study. Either poliovirus type 1 (Mahoney) transcripts (XpA) or luciferase-expressing replicon RNA (RLuc) were used. Constructs containing an active ribozyme at the 5′ end are referred to as rib(+); those containing the inactive form are indicated as rib(−). The parental constructs contain two extra guanosine residues at the 5′ end upon transcription. (C) 5′-end analysis of in vitro transcripts by primer extension. Radiolabeled oligonucleotide IX was hybridized to virion RNA or in vitro-transcribed RNA and extended with reverse transcriptase. The reaction products were analyzed on a denaturing 6% polyacrylamide gel. Lanes 1 to 4, sequencing reaction using the plasmid prib(+)XpA as a template and oligonucleotide IX as sequencing primer; lanes 5 to 8, primer extension using virion RNA (lane 5), rib(+)XpA-RNA (lane 6), rib(−)XpA-RNA (lane 7), or XpA-RNA (lane 8) as the template.
In vitro transcripts were analyzed by primer extension to determine whether the ribozyme generated authentic poliovirus 5′ ends. Using poliovirus RNA as a template, two reaction products were detected (Fig. 1C, lane 5): one with the expected length of 80 nucleotides corresponding to the authentic 5′ end of the genomic RNA, and a second comprised of 81 nucleotides. As previously described, this second product acquired the additional nucleotide in a non-template-dependent fashion during reverse transcription (11). An identical double band was detected when rib(+)XpA-RNA was examined (Fig. 1C, lane 6), suggesting that a large fraction of rib(+)XpA-RNA is autocatalytically cleaved (compare 80/81-nucleotide reaction products with reaction products with a length of around 130 derived from the uncleaved RNA) to generate authentic 5′ ends. In contrast, when rib(−)XpA-RNA, carrying an inactive form of the ribozyme, was used as a template, the reaction product was comprised of approximately 130 nucleotides (Fig. 1C, lane 7). An 82-nucleotide product, corresponding to the first 80 nucleotides of the genomic RNA plus two additional 5′-end guanydyl residues required for efficient T7 RNA polymerase activity, was detected with XpA-RNA (Fig. 1C, lane 8).
Thus, in vitro-synthesized poliovirus transcripts containing an active hammerhead ribozyme at the 5′ end generated up to 80% of transcripts with authentic poliovirus 5′ ends.
In vitro transcripts with authentic 5′ ends replicate more efficiently after transfection in tissue culture cells.
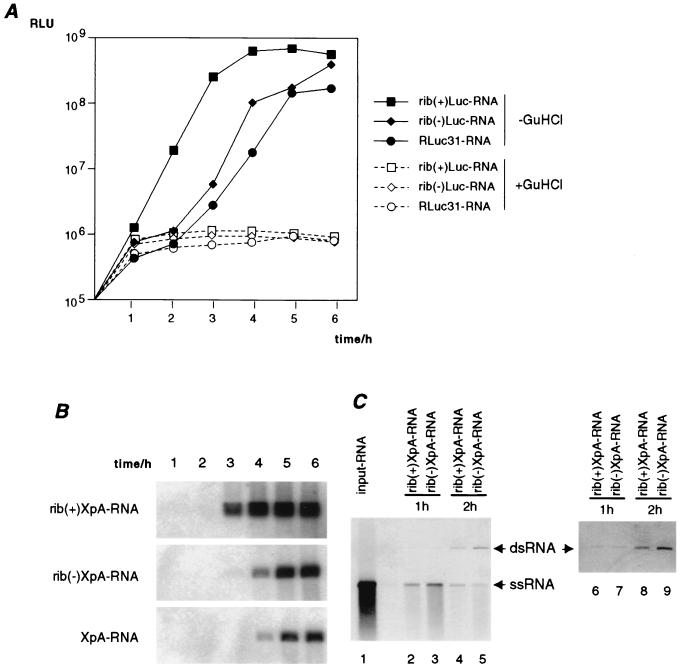
To examine the effect of the 5′ ends on replication, in vitro-transcribed rib(+)RLuc-RNA, rib(−)RLuc-RNA, or RLuc31-RNA was electroporated into 293 cells. Cells were harvested every hour during the first 6 h after transfection to determine luciferase activity, which can be taken as an indication of the replication efficiency of each construct. While the luciferase activity in cells transfected with rib(+)RLuc-RNA increased exponentially immediately, the luciferase activity in rib(−)RLuc-RNA- or RLuc31-RNA-transfected cells show a sigmoid curve with a delay of approximately 1.5 h (Fig. 2A). At 2 to 3 h posttransfection, the luciferase activity in rib(−)RLuc-RNA or RLuc31-RNA starts to increase exponentially until it reaches levels comparable to those for rib(+)RLuc-RNA-transfected cells at 6 h posttransfection (Fig. 2A). The initial luciferase activity detected during the first 2 h in cells transfected with rib(−)RLuc-RNA or RLuc31-RNA was derived from translation of the input RNA, because luciferase activity of cells incubated with 2 mM guanidinium hydrochloride, a potent inhibitor of poliovirus replication, is comparable to that of untreated cells during the first 2 h (Fig. 2A). Thus, the extra sequences present in rib(−)RLuc-RNA or RLuc31-RNA delay replication in tissue culture cells by 1 to 2 h.
FIG. 2.
Replication of in vitro transcripts in tissue culture cells. (A) Luciferase expression in replicon RNA-transfected 293 cells. rib(+)RLuc-RNA, rib(−)RLuc-RNA, or RLuc31-RNA was transfected into 293 cells, and the luciferase activity (relative light units [RLU]) corresponding to 2.5 × 104 cells was measured every hour for 6 h. The cells were kept either in the presence (open symbols) or absence (closed symbols) of 2 mM guanidinium hydrochloride (GuHCl). RNA transfections were repeated at least twice. Error bars have been omitted since the range of variation was smaller than the symbol size. (B) Accumulation of positive-strand RNA in transfected 293 cells. rib(+)XpA-RNA, rib(−)XpA-RNA, or XpA-RNA was transfected into 293 cells, and poly(A)+ RNA was isolated from 105 cells every hour for 6 h. The RNA was separated by electrophoresis in denaturing agarose gels. The gels were dried and hybridized with oligonucleotide IX. (C) Negative-strand RNA synthesis in radiolabeled RNA-transfected 293 cells. 33P-labeled (107 cpm/μg) rib(+)XpA-RNA (lanes 2 and 4) or rib(−)XpA-RNA (lanes 3 and 5) was transfected into 293 cells, and poly(A)+ RNA was isolated after 1 (lanes 2 and 3) and 2 (lanes 4 and 5) h. The RNA was analyzed by electrophoresis in native agarose gels and detected by autoradiography after exposure for 2 days (lanes 1 to 5) or 7 days (lanes 6 to 9). Lane 1, radiolabeled rib(+)XpA-RNA (104 cpm).
To confirm these results, we measured the amount of positive-strand RNA produced after transfection by Northern blotting. Newly synthesized positive-strand RNA was detected 3 h posttransfection in cells transfected with rib(+)XpA-RNA (Fig. 2B). In contrast, it took 4 to 5 h to accumulate similar amounts of transcript in cells transfected with rib(−)XpA-RNA or XpA-RNA (Fig. 2B). Primer extension and sequence analysis of RNA isolated at 6 h posttransfection demonstrated that the viral transcripts that accumulated in cells transfected with rib(−)XpA-RNA and XpA-RNA have authentic poliovirus 5′ ends (data not shown).
Next, we determined whether an authentic 5′ end is necessary for efficient initiation of negative-strand RNA synthesis in vivo. Uniformly 33P-labeled rib(+)XpA-RNA or rib(−)XpA-RNA was transfected into 293 cells, and poly(A)+ RNA was isolated at 1 and 2 h posttransfection. At 2 h, double-stranded RNA (dsRNA) molecules were readily detected in both rib(+)XpA-RNA- and rib(−)XpA-RNA-transfected cells (Fig. 2C, lanes 4 and 5). On a longer exposure, we also observed equal levels of dsRNAs for each construct at 1 h posttransfection (Fig. 2C, lanes 6 and 7). This dsRNA corresponds to the replicative-form (RF) RNA and is a direct measurement of negative-strand RNA synthesis (14). On the other hand, the single-stranded (ssRNA) detected corresponds to input RNA that either has not yet been used as a template or has been already displaced from the RF during subsequent positive-strand RNA synthesis. It is possible that the block of positive-strand RNA synthesis is responsible for the slightly higher amounts of RF accumulated 2 h after transfection of rib(−)XpA-RNA (Fig. 2C, lanes 4 and 5). This result shows that negative-strand RNA synthesis in vivo is not affected by the two extra nucleotides at the 5′ end of the positive strand, and that the defect in replication is specific for positive-strand RNA synthesis.
In vitro transcripts with authentic 5′ ends replicate in alternative experimental systems.
X. laevis oocytes, as well as a cell-free replication system, have been previously established as alternative experimental systems to study poliovirus translation and replication (13, 26). However, in vitro transcripts do not replicate with the same efficiency as virion RNA in these systems. In fact, in vitro transcripts do not replicate at all in X. laevis oocytes. In order to determine whether the relative replicative inefficiency of in vitro transcripts is due to the extra sequences at their 5′ ends, we tested X. laevis oocytes and translation-replication cell extracts with ribozyme-containing poliovirus transcripts.
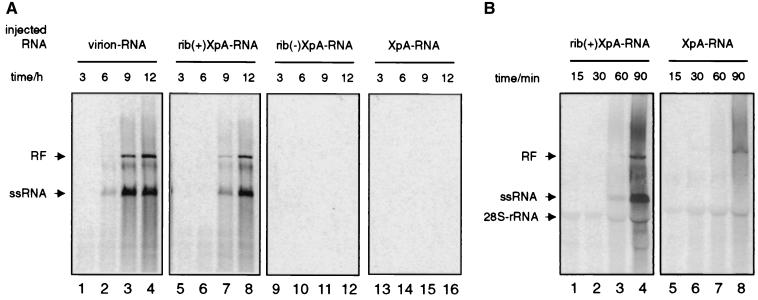
Virion-derived RNA, rib(+)XpA-RNA, rib(−)XpA-RNA, or XpA-RNA was injected into X. laevis oocytes together with HeLa S10 cell extract. [α-33P]UTP was coinjected after 1 h, and RNA was isolated after 3, 6, 9, and 12 h. Newly synthesized viral RNA can be detected as early as 6 h after injection of virion RNA (Fig. 3A, lanes 1 to 4). For rib(+)XpA-RNA, a delay of approximately 3 h was observed (Fig. 3A, lanes 5 to 8). Under these conditions, no products could be detected when rib(−)XpA-RNA or XpA-RNA was injected (Fig. 3A, lanes 9 to 16), indicating that the additional nucleotides at the 5′ end of the input RNA prevent successful amplification. It has been previously shown by microinjecting uniformly 32P-labeled XpA-RNA that the in vitro transcripts can be efficiently transcribed into negative-strand RNA in X. laevis oocytes (14). Here we were unable to detect any incorporation of [α-P33]UTP into RNA, possibly because the pool of endogenous ribonucleotides in these cells lowered the effective concentration of [α-33P]UTP.
FIG. 3.
RNA replication of in vitro transcripts in alternative replication systems. (A) RNA replication in X. laevis oocytes. Virion RNA (lanes 1 to 4), rib(+)XpA-RNA (lanes 5 to 8), rib(−)XpA-RNA (lanes 9 to 12), or XpA-RNA (lanes 13 to 16) was injected into X. laevis oocytes together with [α-33P]UTP, and total RNA was isolated at 3, 6, 9, and 12 h postinjection. After poly(A) selection, the RNA was analyzed on native agarose gels and detected by autoradiography. (B) RNA replication in translation-replication extracts. rib(+)XpA-RNA (lanes 1 to 4) and rib(−) XpA-RNA (lanes 5 to 8) were used to program a cell extract. After 4 h of incubation at 30°C in the presence of 2 mM guanidinium hydrochloride, preinitiation complexes were isolated by centrifugation at 15,000 × g. Preinitiation complexes were resuspended in labeling mix containing [α-33P]UTP, and total RNA was prepared at 15, 30, 45, and 60 min. The RNAs were separated on native agarose gels and detected by autoradiography.
Finally, we used a cell-free poliovirus replication system (4, 26) to study both positive- and negative-strand RNA synthesis. Extracts were programmed with rib(+)XpA-RNA or XpA-RNA; preinitiation complexes were obtained (see Materials and Methods), resuspended in labeling mix, and further incubated at 30°C. Newly synthesized ssRNA, as well as double-stranded RF RNA, is readily detectable when rib(+)XpA-RNA has been used to program the lysate (Fig. 3B, lanes 1 to 4). The ratio of positive-strand RNA to RF was calculated to be 20 to 1 and is indistinguishable from a reaction programmed with virion RNA (data not shown) and close to the ratio observed in poliovirus-infected cells (27). Interestingly, RF RNA but no ssRNA species were generated when XpA-RNA was used. These results are consistent with observations made by Barton et al. (5, 6) that show that negative-strand RNA synthesis in the cell-free replication system is not affected by additional sequences at the 5′ end of the positive-strand RNA. Also, the data presented in Fig. 3B are consistent with the results obtained in intact cells (Fig. 2C), and they establish that an authentic 5′ end is essential for the initiation of positive-strand RNA synthesis under the in vitro experimental conditions.
DISCUSSION
We designed a cis-active hammerhead ribozyme that was cloned in front of poliovirus-specific cDNAs. Transcripts derived from these plasmids show improved replication kinetics in tissue culture cells compared to transcripts that contained extra sequences at their 5′ ends, since the latter were delayed in the onset of positive-strand RNA synthesis. However, at later time points poliovirus RNA accumulated to similar levels irrespective of the nature of the initial 5′ end. This is in accordance with results obtained by Sarnow (30) and many others when measuring the titer produced after transfection of virion RNA or in vitro transcripts with two extra guanosines at their 5′ ends into tissue culture cells. An early report showed that increasing the length of the 5′-end extensions decrease the infectivity of in vitro-transcribed poliovirus RNA (36). We did not observe such a correlation with our constructs [compare the levels of replication of RLuc31/XpA-RNA and rib(−)RLuc/rib(−)XpA]. It therefore seems likely that the primary sequence of a long 5′-end extension determines the impact on RNA replication.
The luciferase data suggest a reinterpretation of previous studies of poliovirus replication using subgenomic replicons. With true 5′ ends, there is no initial translation phase of the input viral genome for 2 to 3 h before a switch to a replicative state as reported earlier (2). Replication begins early after infection detectable in about an hour. Thus, the biphasic expression of luciferase from replicon RNAs observed was not due to the necessity to accumulate viral proteins involved in replication, but rather reflects the need to remove extra nucleotides at the 5′ end. The use of cis-acting ribozymes to produce authentic 5′ ends might be also useful for generating infectious transcripts that initiate more faithfully replication of other, noncapped, positive-stranded RNA viruses infecting plants and animals.
As also shown by others, nonauthentic 5′ ends are repaired during poliovirus replication (17, 36), and several possible repair mechanisms could be envisioned: (i) the 5′ end of the input RNA has to be trimmed by a 5′-exonucleolytic activity to the correct length before initiation of negative-strand RNA synthesis can occur; (ii) the 3′ end of the newly synthesized negative-strand RNA is trimmed by a 3′-exonucleolytic activity, so that initiation of positive strand can occur at the correct initiation site; and (iii) priming of positive-strand RNA synthesis by VPg-pUpU can occur internally, at much lower efficiency, generating an authentic genomic 5′ end in newly synthesized positive strands. Experiments presented here show that negative-strand RNA synthesis is not affected by extra nucleotides at the 5′ end of the input RNA. Therefore, we consider the first of the proposed repair mechanisms to be unlikely. Since the length of the 5′ extension in the range investigated here (2 or 50 nucleotides) had no influence on the duration of the delay, we suggest that internal priming of initiation of positive-strand RNA synthesis by VPg-pUpU is the mechanism for generating perfect 5′ ends. However, further experiments are needed to ultimately define the step in the replication cycle where authentic 5′ ends are generated.
Based on our results, we postulate here a functional difference in the initiation process of positive- and negative-strand RNA synthesis during poliovirus replication. On the one hand, we have shown that efficient positive-strand RNA synthesis depends on a very precise 5′ end, suggesting that VPg-pUpU priming takes place at the extreme 3′ end of the template strand. On the other hand, the dependence on authentic 3′ ends of the positive-strand RNA is much less stringent, in vitro as well as in vivo (5, 30). Negative-strand RNA synthesis seems not to be initiated at the extreme 3′ end but internally, leaving a protruding poly(A) tail on the RF. Our conclusion is based on the fact that we were able to isolate RF RNA by poly(A)+ selection after a first round of negative-strand RNA synthesis in the absence of positive-strand RNA synthesis (Fig. 2C). It is in principle possible that a small subset of RF lacks a poly(A) tail and thus escapes detection. However, the levels of this hypothetical subset must be very low given that the ratio of RF to ssRNA found after poly(A)+ selection of RNA derived from X. laevis oocytes (Fig. 3A, lane 8) closely matches the ratio found in vivo (27) and in vitro (Fig. 3B, lane 4). This is in accordance with previous findings showing that RF RNA isolated at later time points in infection also contains protruding poly(A) tails (23). Although there is a clear difference in the requirements for the initiation of positive- and negative-strand RNA synthesis, the detailed mechanisms and factors involved remain to be elucidated.
While virion RNA has been successfully used to study translation and replication in X. laevis oocytes (13), in vitro transcripts failed to undergo a complete replication cycle since they were stalled at the level of the RF (14). To our knowledge, there have been only two reports describing the successful replication of poliovirus transcripts containing additional sequences at their 5′ ends in a cell-free translation-replication system (33, 34). In those experiments, the ratio of ssRNA to RF RNA synthesized in vitro was approximately 1 to 1, compared to 20 to 1 seen in virion RNA programmed extracts. These authors as well as others (5, 6) proposed that the extra nucleotides at the 5′ end might play a role in the reduced synthesis of positive-strand RNA in vitro. In the experiment described above, we did not detect any positive-strand RNA synthesis at all. We do not know why we failed to observe positive-strand RNA synthesis when transcripts with extra 5′ nucleotides were tested for replication in vitro, but it may have been due to slight differences in the cell extracts or the experimental procedure.
In conclusion, the use of a hammerhead ribozyme that produces in vitro transcripts with authentic 5′ ends which then replicate faithfully both in cells and in vitro, together with the possibility to study the fate of the input RNA, might turn out to be a powerful tool for further detailed investigations of the cis-acting sequences and trans-acting factors functional in the initiation of poliovirus RNA synthesis.
ACKNOWLEDGMENTS
We are grateful to Shane Crotty for useful comments on the manuscript.
This work was supported by funds provided by Public Health Service grant AI40085 to R.A. J.H. is supported by the Deutsche Akademie der Naturforscher Leopoldina, grant BMBF-LPD 9801-2.
REFERENCES
- 1.Ahlquist P, French R, Janda M, Loesch-Fries L S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc Natl Acad Sci USA. 1984;81:7066–7070. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 4.Barton D J, Black E P, Flanegan J B. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton D J, Morasco B J, Flanegan J B. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 1996;275:35–57. doi: 10.1016/s0076-6879(96)75005-x. [DOI] [PubMed] [Google Scholar]
- 6.Barton D J, Morasco B J, Flanegan J B. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J Virol. 1999;73:10104–10112. doi: 10.1128/jvi.73.12.10104-10112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein H D, Sarnow P, Baltimore D. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J Virol. 1986;60:1040–1049. doi: 10.1128/jvi.60.3.1040-1049.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birikh K R, Heaton P A, Eckstein F. The structure, function and application of the hammerhead ribozyme. Eur J Biochem. 1997;245:1–16. doi: 10.1111/j.1432-1033.1997.t01-3-00001.x. [DOI] [PubMed] [Google Scholar]
- 9.Boyer J C, Haenni A L. Infectious transcripts and cDNA clones of RNA viruses. Virology. 1994;198:415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- 10.Burns C C, Lawson M A, Semler B L, Ehrenfeld E. Effects of mutations in poliovirus 3Dpol on RNA polymerase activity and on polyprotein cleavage. J Virol. 1989;63:4866–4874. doi: 10.1128/jvi.63.11.4866-4874.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark J M. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duke G M, Palmenberg A C. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J Virol. 1989;63:1822–1826. doi: 10.1128/jvi.63.4.1822-1826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamarnik A V, Andino R. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 1996;15:5988–5998. [PMC free article] [PubMed] [Google Scholar]
- 14.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giachetti C, Semler B L. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J Virol. 1991;65:2647–2654. doi: 10.1128/jvi.65.5.2647-2654.1991. . (Errata, 65:3972 and 65:5653.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanecak R, Semler B L, Anderson C W, Wimmer E. Proteolytic processing of poliovirus polypeptides: antibodies to polypeptide P3-7c inhibit cleavage at glutamine-glycine pairs. Proc Natl Acad Sci USA. 1982;79:3973–3977. doi: 10.1073/pnas.79.13.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmon S A, Richards O C, Summers D F, Ehrenfeld E. The 5′-terminal nucleotides of hepatitis A virus RNA, but not poliovirus RNA, are required for infectivity. J Virol. 1991;65:2757–2760. doi: 10.1128/jvi.65.5.2757-2760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 19.Hellen C U, Krausslich H G, Wimmer E. Proteolytic processing of polyproteins in the replication of RNA viruses. Biochemistry. 1989;28:9881–9890. doi: 10.1021/bi00452a001. [DOI] [PubMed] [Google Scholar]
- 20.Kirkegaard K. Genetic analysis of picornaviruses. Curr Opin Genet Dev. 1992;2:64–70. doi: 10.1016/S0959-437X(05)80324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klump W M, Bergmann I, Muller B C, Ameis D, Kandolf R. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J Virol. 1990;64:1573–1583. doi: 10.1128/jvi.64.4.1573-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn R J, Tada H, Ypma-Wong M F, Semler B L, Wimmer E. Mutational analysis of the genome-linked protein VPg of poliovirus. J Virol. 1988;62:4207–4215. doi: 10.1128/jvi.62.11.4207-4215.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen G R, Dorner A J, Harris T J, Wimmer E. The structure of poliovirus replicative form. Nucleic Acids Res. 1980;8:1217–1229. doi: 10.1093/nar/8.6.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J P, Baltimore D. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J Virol. 1988;62:4016–4021. doi: 10.1128/jvi.62.11.4016-4021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinkoth J, Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984;138:267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- 26.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 27.Novak J E, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 29.Racaniello V R, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- 30.Sarnow P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott W G. Crystallographic analyses of chemically synthesized modified hammerhead RNA sequences as a general approach toward understanding ribozyme structure and function. Methods Mol Biol. 1997;74:387–391. doi: 10.1385/0-89603-389-9:387. [DOI] [PubMed] [Google Scholar]
- 32.Thiel V, Rashtchian A, Herold J, Schuster D M, Guan N, Siddell S G. Effective amplification of 20-kb DNA by reverse transcription PCR. Anal Biochem. 1997;252:62–70. doi: 10.1006/abio.1997.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Todd S, Towner J S, Semler B L. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology. 1997;229:90–97. doi: 10.1006/viro.1996.8416. [DOI] [PubMed] [Google Scholar]
- 34.Towner J S, Mazanet M M, Semler B L. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J Virol. 1998;72:7191–7200. doi: 10.1128/jvi.72.9.7191-7200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyoda H, Nicklin M J, Murray M G, Anderson C W, Dunn J J, Studier F W, Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45:761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 36.van der Werf S, Bradley J, Wimmer E, Studier F W, Dunn J J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]