Abstract
Per- and poly-fluoroalkyl substances (PFAS) are extensively used in commerce leading to their prevalence in the environment. Due to their chemical stability, PFAS are considered persistent and bioaccumulative; they are frequently detected in both the environment and humans. Because of this, PFAS as a class (composed of hundreds to thousands of chemicals) are contaminants of very high concern. Little information is available for the vast majority of PFAS, and regulatory agencies lack safety data to determine whether exposure limits or restrictions are needed. Cell-based assays are a pragmatic approach to inform decision-makers on potential health hazards; therefore, we hypothesized that a targeted battery of human in vitro assays can be used to determine whether there are structure-bioactivity relationships for PFAS, and to characterize potential risks by comparing bioactivity (points of departure) to exposure estimates. We tested 56 PFAS from 8 structure-based classes in concentration response (0.1–100 μM) using six human cell types selected from target organs with suggested adverse effects of PFAS – human induced pluripotent stem cell (iPSC)-derived hepatocytes, neurons, and cardiomyocytes, primary human hepatocytes, endothelial and HepG2 cells. While many compounds were without effect; certain PFAS demonstrated cell-specific activity highlighting the necessity of using a compendium of in vitro models to identify potential hazards. No class-specific groupings were evident except for some chain length- and structure-related trends. In addition, margins of exposure (MOE) were derived using empirical and predicted exposure data. Conservative MOE calculations showed that most tested PFAS had a MOE in the 1–100 range; ~20% of PFAS had MOE<1, providing tiered priorities for further studies. Overall, we show that a compendium of human cell-based models can be used to derive bioactivity estimates for a range of PFAS, enabling comparisons with human biomonitoring data. Furthermore, we emphasize that establishing structure-bioactivity relationships may be challenging for the tested PFAS.
Keywords: PFAS, in vitro, grouping, risk characterization, margin of exposure
1. Introduction
Per- and poly-fluoroalkyl substances (PFAS) are used ubiquitously in commerce because of their water and oil repellant, temperature resistant, and other desired properties. Their uses range from consumer products such as food packaging, cookware, and clothing to industrial applications in semi-conductor manufacturing and fire suppression (e.g., in aqueous film forming foams). As a result of their ubiquitous use and releases into the environment through landfills and industrial discharges, PFAS have been detected in almost all tested environmental and human samples (National Academies of Sciences Engineering and Medicine 2022). Exposures to some PFAS have been linked to adverse effects in humans (Carlson et al. 2022; Radke et al. 2022; Zahm et al. 2023); however, there are little to no data on the potential health hazards for the vast majority of these chemicals (U.S. Environmental Protection Agency 2021a, b). Thus, to better understand PFAS as a class, governmental agencies are refining the inventories, improving exposure assessment approaches, and identifying data gaps pertaining to the risk assessment needs (European Commission 2020; U.S. Environmental Protection Agency 2021a; U.S. National Science and Technology Council 2023).
Based on the environmental and human health hazard data for a limited number of PFAS, including perflurooctanoic acid (PFOA) and perfluorooctane sulfonic acid (PFOS), United States EPA and some state agencies have instituted health-based guidance values and restrictions (i.e., drinking water limits); further, efforts are underway to add a number of PFAS to the Superfund hazardous substances inventory (U.S. Environmental Protection Agency 2023). In the European Union (EU), acceptable food levels for four PFAS have been proposed in 2020 (Efsa Panel on Contaminants in the Food Chain et al. 2020). Recently, several EU member states submitted a proposal to implement a ban on the manufacturing and use of all PFAS as a class by 2030. The proposal is currently being evaluated by the European Chemicals Agency. (European Chemicals Agency 2023).
Despite the attention to PFAS by the general public and the regulatory agencies around the world, the vast majority of PFAS are very data-poor substances and active research efforts are underway to understand both exposure and the potential environmental and human health effects of a broader set of PFAS (Carlson et al. 2022; OECD 2022). The current approach to prioritize PFAS for additional toxicity testing largely relies on chemical structure-based groupings (Buck et al. 2011; Patlewicz et al. 2019; Patlewicz et al. 2022). In addition, grouping by mechanism of action, chain length, and other physicochemical descriptors have been proposed with the effort to prioritize PFAS for testing and the development of predictive in silico models (U.S. National Science and Technology Council 2023). International efforts are underway to compile information on these subclasses (i.e., chemical properties, bioaccumulation, and transformation patterns, uses, regulatory status, etc.), data that could be used, if groupings can be scientifically justified, for read-across to PFAS that lack data (OECD 2022).
There are thousands of PFAS in various chemical inventories and their hazard characterization by testing individual substances using traditional animal-based methods is not feasible (National Academies of Sciences Engineering and Medicine 2022; U.S. Environmental Protection Agency 2021a). Therefore, cell-based models are a sensible approach to evaluate a large number of PFAS in a time efficient manner in order to characterize their potential effects to human health and the environment (U.S. Environmental Protection Agency 2021a; U.S. National Science and Technology Council 2023). Indeed, several recent studies have applied a range of new approach methods (NAMs) to test a large (dozens to over a hundred) number of PFAS (Carstens et al. 2023; Houck et al. 2023; Houck et al. 2021; Reardon et al. 2021) using several human cell-based models. However, because many additional tissue/cell types have been shown (or suggested) to be potential targets for PFAS toxicity in humans (National Academies of Sciences Engineering and Medicine 2022), additional studies are needed.
One approach to broaden biological coverage is to use a large compendium of tests and cell-based models, such as those in ToxCast/Tox21 testing programs (Harrill et al. 2021; Richard et al. 2021). An alternative is to use a targeted battery of human cell-based assays that has been shown to be informative for deriving protective points of departure (PODs), determining structure-bioactivity relationships, and characterizing potential risks by comparing bioactivity to exposure estimates (Chen et al. 2020; Chen et al. 2021; Cordova et al. 2023; House et al. 2021). The cell types in the latter approach include some of the organs that have been shown to be affected by PFAS – the liver, cardiovascular system, and the central nervous system. Therefore, this study tested a hypothesis that a targeted battery of human cell-based assays can be used to determine whether there are structure-bioactivity relationships for PFAS, and to characterize potential risks by comparing bioactivity (point of departure) to exposure estimates. We used a diverse set of 56 PFAS from 8 structure-based classes and collected data in concentration response (0.1 to 100 μM) in 6 human cell types – human induced pluripotent stem cell (iPSC)-derived hepatocytes, neurons, cardiomyocytes, as well as primary human hepatocytes, endothelial and HepG2 cells.
2. Experimental Methods
2.1. Chemicals
Test PFAS (n = 56, Table 1) were obtained from a chemical repository at MRIGlobal (Kansas City, MO) that was assembled from the following vendors: Matrix Scientific (Elgin, SC), SynQuest Laboratories (Alachua, FL), Apollo Scientific (Bredbury, UK), TCI America (Portland, OR), Oakwood Products (Estill, SC), Sigma-Aldrich (St. Louis, MO), MuseChem (Fairfield, NJ), or BOC Sciences (Shirley, NY). Purity of each compound was determined by MRIGlobal and substances were shipped in 1 mL vials, with Teflon-lined screw-cap vials under Argon gas headspace. The chemicals were selected to represent a broad spectrum of PFAS subclasses and chain lengths (Table S1 and Figure S1). PFAS were assigned into subclasses using side-chain/head group descriptors for the classification as previously suggested (Buck et al. 2011; Buck et al. 2021). Chemicals were randomized with respect to the position on the test plates to avoid potential selection bias with respect to sub-class assignments. Upon receipt, chemical stocks were stored at −80○C until use. Tissue-culture grade dimethyl sulfoxide (DMSO, CAS# 67-68-5, sc-358801) was obtained from Santa Cruz Biotechnology (Dallas, TX). Assay-specific positive controls were obtained from various vendors (Table S2).
Table 1.
PFAS substances used in these studies. Additional chemicals details can be found in Table S1.
| PFAS Category | Chemical Name (Abbreviated Name) | CAS No. | Supplier | Lot No. | Purity*(%) |
|---|---|---|---|---|---|
| Alcohol | 1-Pentafluoroethylethanol (PFBOH) | 374-40-3 | Matrix Scientific | S20E | 99.0 |
| Alcohol | 3-(Perfluoro-2-butyl)propane-1,2-diol (PFHp2OH) | 125070-38-4 | SynQuest Laboratories | 00006922 | 99.7 |
| PFECA | Perfluoro-3,6-dioxaoctane-1,8-dioic acid (PFHx2Et2OA) | 55621-21-1 | SynQuest Laboratories | Q15B-10 | N/A |
| PFAN | Nonafluoropentanamide (PFNAM) | 13485-61-5 | SynQuest Laboratories | 00004660 | 100 |
| PFCA | 3-(Perfluoroisopropyl)-2-propenoic acid (PFIpOA) | 243139-64-2 | Apollo Scientific | AS483671 | 96.9 |
| PFAN | Octafluoroadipamide (PFO2AM) | 355-66-8 | SynQuest Laboratories | Q18A-83 | 98.1 |
| Alcohol | Dodecafluoroheptanol (7H 6:1 FTOH) | 335-99-9 | TCI America | WLPPG | 98.1 |
| PFCA | 3,3-Bis(trifluoromethyl)-2-propenoic acid (TFPrOA) | 1763-28-6 | SynQuest Laboratories | 00000111 | 97.1 |
| PFECA | Perfluoro(4-methoxybutanoic) acid (PFMBA) | 863090-89-5 | SynQuest Laboratories | Q10A-107 | 97.9 |
| PFSA | Perfluorobutanesulfonic acid (PFBS) | 375-73-5 | TCI America | B36UC | 95.8 |
| PFECA | Perfluoro-3,6-dioxaheptanoic acid (NFDHA) | 151772-58-6 | SynQuest Laboratories | Q162-70 | 97.4 |
| Alcohol | Heptafluorobutanol (HpFBOH) | 375-01-9 | TCI America | WN7ZI | 98.6 |
| PFSA | 4:2 Fluorotelomer sulfonic acid (4:2 FTS) | 757124-72-4 | SynQuest Laboratories | 237800 | 95.6 |
| Alcohol | Hexafluoroamylene glycol (CFHx2OH) | 376-90-9 | TCI America | M52SK | 100 |
| PFAN | Heptafluorobutyramide (PFHpAM) | 662-50-0 | SynQuest Laboratories | 00016753 | 100 |
| PFAN | Perfluoropentanamide (PFPAM) | 355-81-7 | Apollo Scientific | AS483010 | 99.4 |
| PFCA | 2,2,3,3,4,4-Hexafluorobutanoic acid (4H-PFBA) | 679-12-9 | SynQuest Laboratories | 00017311 | 97.5 |
| PFCA | Ammonium perfluorooctanoate (NH4PFOA) | 3825-26-1 | SynQuest Laboratories | 425300 | 97.3 |
| PFCA | Perfluorononanoic acid (PFNA) | 375-95-1 | Oakwood Products | G30K | 95.1 |
| Alcohol | 2-Aminohexafluoropropan-2-ol (AmFPrOH) | 31253-34-6 | SynQuest Laboratories | Q192-57 | 98.1 |
| n:2 FTOH | 4:2 Fluorotelomer alcohol (4:2 FTOH) | 2043-47-2 | SynQuest Laboratories | Q154-127 | 100 |
| PFECA | Perfluoro-3,6,9-trioxatridecanoic acid (PFPE-6) | 330562-41-9 | SynQuest Laboratories | 00001078 | 98.2 |
| Alcohol | 1H,1H,5H-Perfluoropentanol (PFPOH) | 355-80-6 | Sigma-Aldrich | MKCD4243 | 99.5 |
| n:2 FTOH | 8:2 Fluorotelomer alcohol (8:2 FTOH) | 678-39-7 | Sigma-Aldrich | MKBL4812V | 99.4 |
| PFCA | Perfluorohexanoic acid (PFHxA) | 307-24-4 | TCI America | TKU3I | 97.1 |
| n:2 FTOH | 6:2 Fluorotelomer alcohol (6:2 FTOH) | 647-42-7 | Sigma-Aldrich | MKCD3356 | 99.7 |
| PFCA | Perfluorobutanoic acid (PFBA) | 375-22-4 | Apollo Scientific | AS475966 | 97.4 (isomers) |
| PFCA | Perfluorooctanoic acid (PFOA) | 335-67-1 | TCI America | F7P2F | 99.7 |
| PFSA | Potassium perfluorobutanesulfonate (PFBS-K) | 29420-49-3 | Sigma-Aldrich | S87103 | 100 |
| PFCA | 2H,2H,3H,3H-Perfluorooctanoic acid (5:3 FTCA) | 914637-49-3 | SynQuest Laboratories | 00010397 | 95.4 |
| PFAN | Perfluorohexanesulfonamide (PFHxSA) | 41997-13-1 | SynQuest Laboratories | Q156-19 | 97.6 (sum of isomers) |
| n:2 FTSA | 6:2 Fluorotelomer sulfonic acid (6:2 FTS) | 27619-97-2 | SynQuest Laboratories | 512700 | 97.8 |
| n:2 FTSA | 8:2 Fluorotelomer sulfonic acid (8:2 FTS) | 39108-34-4 | SynQuest Laboratories | 523500 | 95.5 |
| PFCA | 9-Chloro-perfluorononanoic acid (Cl-PFNA) | 865-79-2 | SynQuest Laboratories | 00008888 | 97.8 |
| PFCA | Perfluoroundecanoic acid (PFUnDA) | 2058-94-8 | Oakwood Products | 002265L20H | 99.6 |
| PFECA | Perfluoro-4-isopropoxybutanoic acid (PFPE-1) | 801212-59-9 | SynQuest Laboratories | 279800 | 99.3 |
| PFSA | Perfluorooctanesulfonic acid (PFOS) | 1763-23-1 | SynQuest Laboratories | 375600 | 98.8 |
| PFSA | 2,2,2-Trifluoroethyl perfluorobutanesulfonate (ET-PFBS) | 79963-95-4 | SynQuest Laboratories | Q11A-55 | 98.7 |
| PFECA | Methyl perfluoro(3-(1-ethenyloxypropan-2-yloxy)propanoate) (MePF2ETOA) | 63863-43-4 | Apollo Scientific | AS437089 | 100 |
| Alcohol | 1-(Perfluorofluorooctyl)propane-2,3-diol (PFUd2OH) | 94159-84-9 | SynQuest Laboratories | 320700 | 99.9 |
| Other (FTCA) | 3:3 Fluorotelomer carboxylic acid (3:3 FTCA) | 356-02-5 | SynQuest Laboratories | Q18B-51 | 100 |
| PFCA | 3-Perfluoroheptylpropanoic acid (7:3 FTCA) | 812-70-4 | SynQuest Laboratories | 563200 | 96.0 |
| PFAN | N-Methyl-N-(2-hydroxyethyl)perfluorooctanesulfonamide (MeFOSE) | 24448-09-7 | MuseChem | M19J05027 | 94.6 |
| PFCA | Perfluoroheptanoic acid (PFHpA) | 375-85-9 | Apollo Scientific | AS480225 | 99.5 |
| PFCA | Perfluoropropanoic acid (PFPrA) | 422-64-0 | Apollo Scientific | AS468359 | 98.7 |
| PFCA | Perfluoropentanoic acid (PFPeA) | 2706-90-3 | SynQuest Laboratories | 00015707 | 99.7 |
| n:2 FTOH | 6:1 Fluorotelomer alcohol (6:1 FTOH) | 375-82-6 | SynQuest Laboratories | 00012609 | 100 |
| PFAN | Perfluorooctanamidine (PFOAMD) | 307-31-3 | BOC Sciences | BS19S10081 | 98.1 |
| PFECA | Perfluoro-3-methoxypropanoic acid (PFMPA) | 377-73-1 | Apollo Scientific | AS473550 | 99.3 |
| PFAN | Perfluorooctanamide (PFOAM) | 423-54-1 | SynQuest Laboratories | Q4A-24 | 99.2 |
| PFSA | Perfluorohexanesulfonic acid (PFHxS) | 355-46-4 | SynQuest Laboratories | 334800 | 95.3 |
| Other (PFPA) | ((Perfluorooctyl)ethyl)phosphonic acid (8:2 FTPA) | 80220-63-9 | SynQuest Laboratories | 00017193 | 97.3 |
| Other (non-PFAA perfluoroalkyls) | 3H,3H-Perfluoro-2,4-hexanedione (PFHx2ON) | 20825-07-4 | SynQuest Laboratories | 00017405 | 96.2 |
| PFCA | Octafluoroadipic acid (PFHx2OA) | 336-08-3 | TCI America | 5AGFD | 99.9 |
| PFCA | Perfluorodecanoic acid (PFDA) | 335-76-2 | Sigma-Aldrich | MKCF2877 | 96.4 |
| Alcohol | Fluorinated triethylene glycol monomethyl ether (C7F3ETOH) | 147492-57-7 | Apollo Scientific | AS463658 | 98.2 |
Purity as determined by MRIGlobal (Kansas City, MO).
2.2. Cells and Biological Reagents
The human cell types for in vitro experiments were selected to represent various organ/tissues that have been shown previously to demonstrate adverse effects of PFAS (Burnett et al. 2021; Liao et al. 2012; Marques et al. 2022; Ojo et al. 2020; Tukker et al. 2020). Selected cell types included: (i) iCell hepatocytes 2.0 (iCell Heps, Cat# C1023, FujiFilm Cellular Dynamics, Madison, WI), (ii) iCell GABAneurons (iCell Neurons, Cat# C1012, FujiFilm Cellular Dynamics), (iii) iCell cardiomyocytes (iCell Cardios, Cat# C1006, FujiFilm Cellular Dynamics), (iv) human umbilical vein endothelial cells (HUVECs, Cat# C2517A, Lonza, Walkersville, MD), (v) primary human hepatocytes (PHHs, Cat# HMCPIS; Gibco, Waltham, MA), and (vi) HepG2 cells (HepG2s, Cat# HB-8065; ATCC, Manassas, VA). Formulations for both plating and maintenance media were cell-specific and are described in detail in Supplemental Text. Briefly, cell-specific media and supplements for iCells were obtained from FujiFilm Cellular Dynamics as part of the cell-specific kits or purchased as indicated in the protocol. HUVECs were cultured in EGM-2 medium (Cat# CC-3156, Lonza) supplemented with the EGM™−2 BulletKits™ (Cat# CC-4176, Lonza) were from Lonza (Walkersville, MD). PHHs were cultured in William’s E Medium, no phenol red (Cat# A1217601, Gibco) and HepG2s were cultured in DMEM (Cat# 11965–092, Gibco). Additional reagents that were used for cell culture experiments included penicillin-streptomycin (Cat# 15140–122, Gibco), TrypLE Express Enzyme (1×)™ (Cat# 12605–010, Gibco), B-27™ medium supplement (Cat# 17504–044, Gibco), and gentamicin (Cat# 157060, Gibco). Fetal bovine serum (FBS, Cat# 97068–085) was obtained from VWR (Radnor, PA). Tissue culture grade 384-well plates (Cat# 3764) were obtained from Corning (Kennebunk, ME). Gelatin (Cat# G1890–500G), Trypan Blue 0.4% solution (Cat# T8154–100ML), and D-PBS (Cat# D8537) were obtained from Sigma-Aldrich (St. Louis, MO).
The justifications for the selected cell-specific phenotypes evaluated in this study have been previously detailed (Gerets et al. 2012; Grimm et al. 2015; Iwata et al. 2017; Kato et al. 2022; Sirenko et al. 2014). Details of cell type-specific phenotyping are included in Supplemental Text. Briefly, cells were cultured in 384-well plates and exposed to 56 PFAS in concentration response (0.1 to 100 μM); the chemicals were prepared originally in master plates in 100% cell-culture grade DMSO and diluted 100-fold to yield 2× working solutions in 1% DMSO. The final concentration in assay wells following addition of test chemicals was 0.1, 1, 10, and 100 μM in 0.5% DMSO (v/v). The amount of DMSO was consistent with previous studies that show no effects from DMSO across all cell type-specific phenotypes. Chemical exposure times were cell-specific and have been tested in previous studies (Chen et al. 2020). The 384-well plate design (Figure S2) included all 56 PFAS in concentration response ranging from 0.1 to 100 μM. The chemicals were assigned an ID and their location on the plate was randomized. Intra-plate replicate chemicals were included to evaluate reproducibility, as well as negative controls including media and vehicle-treated wells – DMSO (0.5%), and cell-type specific positive controls (see Supplemental Text and Table S2 for the list).
2.3. Cytotoxicity Assays
High-content live cell imaging was used to evaluate cytotoxicity in all cell types following the chemical exposures (specific for each in vitro model, see Supplemental Text for details) using an ImageXpress Micro Confocal High-Content Imaging System (Molecular Devices, San Jose, CA). The cells were stained with various fluorescent dyes as follows. Hoechst 33342 (Cat# H3570, Invitrogen, Carlsbad, CA) was used to stain nuclei, Calcein AM Green (Cat# C3100MP, Invitrogen) for cytoplasm. ATP production was evaluated in some cell types (iCell Neurons and HUVECs) using the CellTiter-Glo reagent (Cat# G9241, Promega, Madison, WI) and the FLIPR Tetra Imaging Device and Screenworks 4.0 software (Molecular Devices). The high-content imaging was done using the DAPI, FITC, and TRITC filters at 10× and 20× magnifications. The embedded MultiWavelength Cell Scoring software was used as the imaging processing software to quantify the images for the various cytotoxic phenotypes of interest. The ATP production data were extracted using the FLIPR Tetra Screenworks 4.0 software. All data were then used for concentration response modeling to derive PODs (see below).
2.4. Functional Phenotypic Assays
Cell type-specific functional phenotypes were evaluated in each model as described in Table 2 and Table S2. Specific details for each cell type are listed in Supplemental Text and have been reported previously. Mitochondrial integrity and intensity (MitoTracker Orange, Cat# M7510, Invitrogen), and lipid accumulation (HCS LipidTOX Deep Red, Cat# H34477, Invitrogen) for the hepatocyte models (iCell Heps, PHHs, and HepG2s) and neurite outgrowth phenotypes for the iCell Neurons were evaluated using high-content imaging. Angiogenesis phenotypes for the HUVECs were evaluated by seeding the cells in an extracellular gel matrix, allowing for 3D tubule formation, this was then followed by chemical exposures and high-content imaging. The EarlyTox Cardiotoxicity Kit was used to look at the Ca2+ flux of the iCell Cardios which was measured using high-throughput kinetic imaging on the FLIPR Tetra (Molecular Devices). The embedded MultiWavelength Cell Scoring, Neurite Outgrowth, Granularity and Angiogenesis Tube Formation Modules were used as the imaging processing software to quantify the images for the various phenotypes of interest.
Table 2.
Cell types and cell type-specific phenotypes evaluated in this study. See Supplemental Text for experimental details and culture conditions.
| Cell type | Exposure duration | Functional Phenotypes (and abbreviations) | Cytotoxic Phenotypes (and abbreviations) |
|---|---|---|---|
| iCell Neurons | 72 h | Total Outgrowth (TotOut) Mean Outgrowth (MeanOut) Total Processes (TotProc) Total Branches (TotBranch) Cells with Significant Growth (CWG) |
Cell Number (TotCell) Mitochondrial Integrity (MitoInteg) Cytoplasmic Integrity (CytoInteg) Total Cell Body Area (CBA) ATP Production (ATP) |
| iCell Cardios | 90 min | QT Prolongation (QT Prol) Positive Chronotrope (NegChron) Negative Chronotrope (PosChron) Asystole (Asys) |
Cell Number (TotCell) |
| HUVECs | 18 or 24h | Mean Tube Length (MTL) Total Tube Length (TTL) Total Tube Area (TTA) |
Cell Number (TotCell) Mitochondrial Integrity (MitoInteg) Mitochondrial Intensity (MitoIntens) Cytoplasmic Integrity (CytoInteg) Nuclei Mean Area (NMA) ATP Production (ATP) |
| iCell Heps | 48 h | Mitochondrial Integrity (MitoInteg) Mitochondrial Intensity (MitoIntens) Granularity (Gran) Total Granule Area (TGA) |
Cell Number (TotCell) Nuclei Intensity (NucInt) All Cell Mean Area (CMA) Cytoplasmic Integrity (CytoInteg) |
| PHHs | |||
| HepG2s |
2.5. Assay Quality Control Evaluation
To evaluate experimental reproducibility, each cell type was seeded in multiple 384-well plates and each chemical was tested on each plate. Chemicals were added to plates with cells using the automated liquid handling of the FLIPR Tetra (Molecular Devices). After the culture period and assays, the raw phenotypic data was extracted from the various software types, including ImageXpress application modules (MultiWavelength Cell Scoring, Neurite Outgrowth, Angiogenesis Tube Formation, and Granularity) on the MetaXpress, as well as Screenworks 4.0 software (Molecular Devices) using pre-determined algorithms in each software package. All data were normalized to plate-specific vehicle controls (0.5% DMSO-treated wells). The vehicle controls were screened for outliers, and values were removed if any were identified outside of the inter-quartile range of the controls. Supplementary quality control accounted for the coefficient of variability (CV, %) of the vehicle control samples; if the CV% was above 20%, manual screening was done to identify any anomalies. Additional quality control was assessed for each phenotype by evaluating any differences between the negative controls (vehicle-control vs. media), cell-specific positive controls and looking at both the intra-plate and inter-plate reproducibility. First, quality control data (positive and negative controls) were evaluated for reproducibility and expected effects; only when the quality criteria were met (see above), analyses of PFAS effects were conducted.
2.6. Concentration-Response Modeling
Normalized data from each phenotype was used as the input for the concentration-response modeling to derive a POD for each chemical, cell type and phenotype. The normalized data for each treatment were fitted to a curve with a nonlinear logistic function using whole-batch data processing and a standardized dose-response modeling protocol as previously detailed (Sirenko et al. 2017). Generally, the POD for each phenotype was defined as the effective concentration at which the fitted curve exceeds a 10% relative change from vehicle-treated controls. The iCell Cardios functional phenotypes used a 5% relative increase or decrease from controls as the POD for positive/negative chronotrope and QT prolongation, and a 95% relative decreasing change from controls as the POD for asystole based on the human relevance of these in vitro phenotypes as detailed elsewhere (Blanchette et al. 2020).
2.7. Data Integration Using Toxicological Priority Index (ToxPi) Approach
Toxicological Priority Index Graphical User Interface (ToxPi GUI, https://toxpi.org/, last accessed October 1, 2023) was used to integrate and visualize the phenotypic data (Marvel et al. 2018; Reif et al. 2013). The PODs from all 48 phenotypes from 6 in vitro models were used, and as instructed by the ToxPi protocol, the PODs were inversely scaled from 0 – 1, with 0 representing the highest POD values (no effect) and 1 representing the lowest POD (highest observed bioactivity). The scores were weighted proportionately based on the number of phenotypes for each cell type (i.e. iCell Neurons – 10 phenotypes, iCell Cardios – 5 phenotypes). These scores were then used to determine an overall ranking across all cell types and on a cell-to-cell basis to determine what cells were the most bioactive.
2.8. Margin of Exposure Estimates
The ratio of the observed in vitro effects to the estimated human exposure predicted estimates were evaluated to calculate the margin of exposure (MOE) to determine the overall risk of the tested PFAS. The MOE was calculated using the most sensitive chemical-specific POD across all phenotypes and comparing it to the highest exposure value. Exposure data was taken from the CompTox Dashboard (https://comptox.epa.gov/dashboard/, Version 2.2.1, last accessed on June 28, 2023) (Williams et al. 2017) using the predicted exposure levels or values detected in human biological matrices from published PFAS biomonitoring studies. Specifically, the exposure data were either (i) extracted from the EPA’s CompTox Dashboard predictions (based on the General-Exposure-Predictions-2022-05-16 dataset that was generated using the R package SEEM3) (Wambaugh et al. 2013), or (ii) obtained through a literature search of blood concentrations of the tested PFAS in humans (Table S3). For the latter, the full chemical name, CAS-RN, and abbreviated names (Table 1) were searched in Google (https://www.google.com/) and PubMed (https://pubmed.ncbi.nlm.nih.gov/). Resulting studies were evaluated for relevance (i.e., containing blood levels of PFAS of interest) to levels in human blood, serum, or plasma. Data from a total of 65 studies were deemed relevant and are listed in Table S3. From the available data across all studies, the 5th and 95th percentile values were calculated to generate a comparable range to that of the exposure predicted values (50th – 95th percentile) from the CompTox Dashboard. Then, if a chemical had multiple studies or both predicted and measured value, the highest exposure values were used as a conservative approach to calculate the MOE. Specifically, exposure estimates were first converted into human steady-state plasma concentrations (Css), assuming median toxicokinetic parameters using the function “calc_analytic_css” with the three-compartment steady state model in the httk R package (Pearce et al. 2017). If data was not available in httk, protein binding values were used as previously reported (Kreutz et al. 2023; Smeltz et al. 2023) under the assumptions of no hepatic clearance and renal clearance only via glomerular filtration. The MOE for each substance was calculated by dividing the POD from the most sensitive cell-specific phenotype by the most conservative (i.e., highest) Css estimate which is converted from the 95th percentile of human exposure using equation:
2.9. Comparisons to Other In Vitro Studies
As a benchmark comparison to the in vitro PFAS bioactivity data collected herein, the most-sensitive human, cell-based assay AC50 values were extracted from the Tox21 database, available through the CompTox Dashboard (last accessed on June 15, 2023 using version 2.2.1) (Williams et al. 2017). If the lowest reported AC50 value was from a cell-free assay and other values were available from human cell-based assays, then the lowest cell-based assay AC50 value was used. Also, in the case that the lowest POD was from an animal-based in vitro assay, then the lowest human cell-based assay was used.
In addition to comparing the PFAS to other human in vitro studies, we also made comparisons to in vitro data on a diverse set (n=42) of industrial chemicals (Chen et al. 2020). Of the 6 cell types tested, 4 were used in both studies (iCell Neurons, iCell Cardios, iCell Heps, and HUVECs) and the phenotypic data were used as the input for the comparison. We utilized ToxPi (as previously described) to integrate and visualize the data across both chemical datasets. ToxPi ranks were assigned to each of the chemicals used in this analysis.
2.10. Correlation Analyses and Cross-Validated Predictions for Bioactivity using Chemical Descriptors and Physicochemical Properties of PFAS
In addition to the hierarchal clustering based on subclass and subgroup and phenotype attributes, we conducted correlation analyses and cross-validation predictions. Our primary objective was to thoroughly evaluate the efficacy of incorporating a diverse array of chemical molecular descriptors as a predictive tool to discern bioactivity patterns. For these analyses, we used Saagar descriptors (Sedykh et al. 2021), a collection of diverse chemical sub-structures (i.e., local “motifs” that are searched and counted in each query molecule), and physicochemical properties predicted using the OPERA modeling suite (Mansouri et al. 2018). Saagar descriptors were derived from basic chemical functionalities, including metrics such as alkyl halogen counts, di-halogen atom pairs, etc. The overall hit-count matrix for Saagar (834) and physicochemical (9 OPERA, molecular weight and carbon chain-length) features for tested PFAS chemicals is provided in Table S4 and the bioactivity data matrix is provided in Table S5.
Saagar chemical descriptors and physicochemical properties were examined for associations with bioactivity phenotypes, using a combination of pairwise analyses and cross-validated predictions using an established penalized regression approach. As each set of descriptors is of independent interest, we performed analyses using the physicochemical descriptors, using the Saagar descriptors, and then using the combination of physicochemical and Saagar descriptors. Within each analysis set, p-value adjustment (Holm 1979) was performed for the corresponding number of tests. We considered an adjusted p<0.05 as statistically significant.
Among the original 48 bioactivity phenotypes, 7 did not vary (i.e., were not affected by PFAS) across the 56 PFAS and were removed. We then computed minimum PODs across the phenotypes (i) within phenotypes corresponding to each cell type, and (ii) overall (among all cell types), resulting in a final set of 48 bioactivity phenotypes for these analyses. Additional data reduction was performed for Saagar descriptors because the cross-validated prediction method requires at least two samples with a feature to differ from the remaining samples. After applying this filter, 123 Sagaar descriptors were retained for cross-validated analysis. We used this same set of descriptors for the pairwise association analysis, as descriptors failing the variation criterion have almost no power to detect associations.
For pairwise correlation analyses, we calculated Spearman rank correlations for each set of Saggar descriptors and physicochemical properties vs. the 48 bioactivity phenotypes, with two-sided p-values adjusted for the multiple testing (Benjamini and Hochberg 1995).
For cross-validated prediction analyses, we used an approach previously reported by (Chen et al., 2021) to determine if a diverse set of chemical features could be used predict bioactivity. The approach uses multivariate ridge regression-based prediction with a common penalty parameter across features to avoid overfitting, and n-fold cross-validation. As the procedure does not use nested tuning parameter selection, to guard against any subtle overfitting, 10,000 permutations of the procedure were performed for each set of descriptors, resulting in an empirical p-value for the correlation between the cross-validated predictions of each phenotype and the actual observed phenotype values. Multiple testing correction used Holm’s method (denoted padj) corrected for the 48 bioactivity phenotypes (Holm 1979).
3. Results
3.1. Phenotyping of the human cells treated with PFAS in dose-response.
Overall, this study used the PFAS bioactivity data to identify potential grouping strategies, and to understand the potential margins between exposure and effects to inform risk management. Specifically, we used a targeted battery of 6 human organotypic cell types to test 56 diverse PFAS from various subclasses (Figure 1). First, cell-specific phenotypes (Table 2) were collected to represent both functional cytotoxicity endpoints and concentration response modeling performed to calculate PODs. Second, the PODs were used to determine both overall and cell type-specific bioactivity of each PFAS, and to test if the bioactivity was structure-dependent. For the latter, we used unsupervised hierarchical clustering, as well as machine learning-based supervised analysis to predict bioactivity for each substance using chemical descriptors (Sedykh et al. 2021) and physicochemical properties (Mansouri et al. 2018). Finally, we calculated chemical-specific MOEs from in vitro PODs.
Figure 1. Overall study outline.
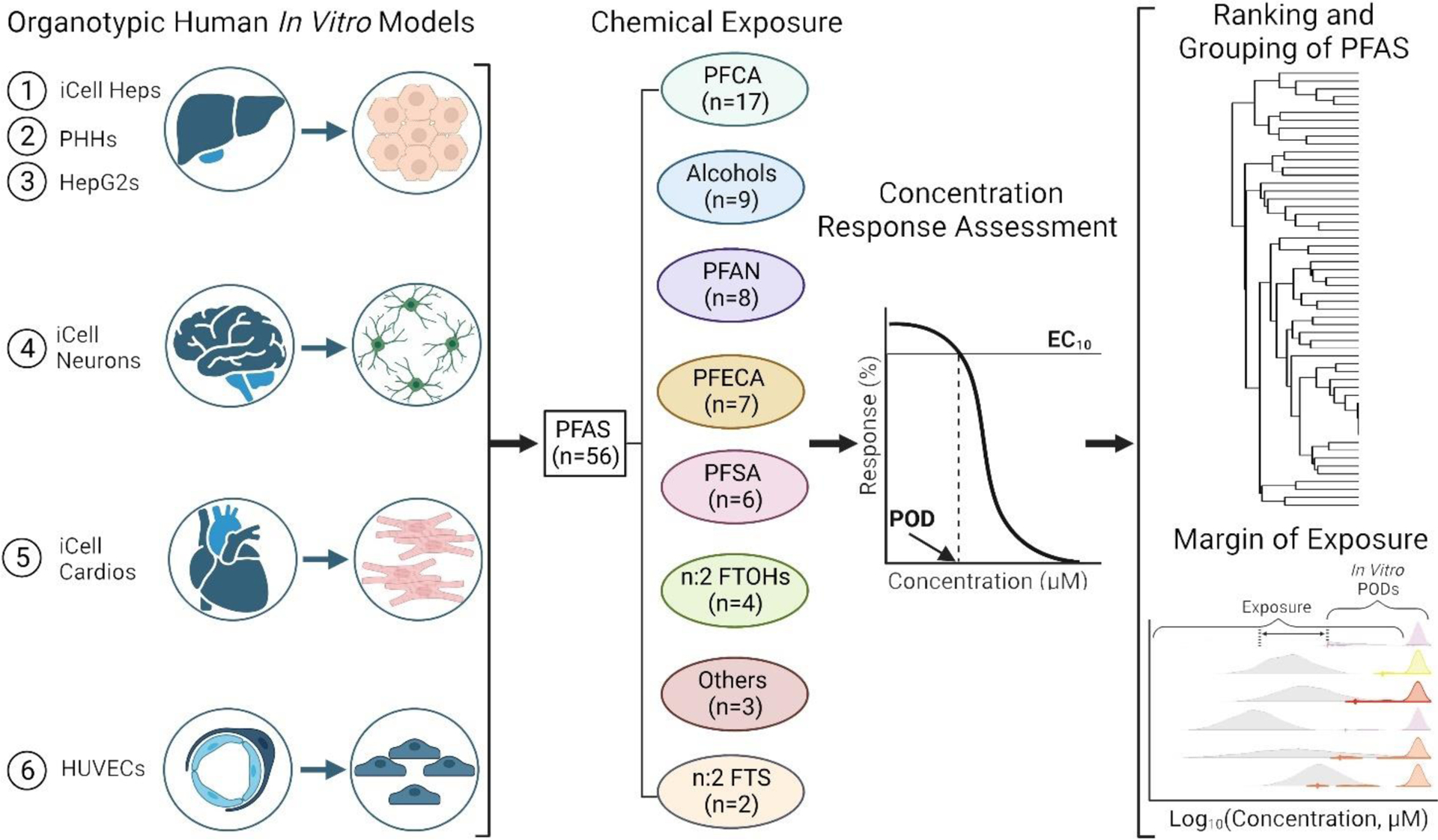
A total of 6 in vitro models were used as a representative battery to assess the effects of 56 PFAS. All cell types were exposed to PFAS, and cell-specific phenotypic endpoints were assessed and used for concentration-response modeling to derive PODs. The PODs were subsequently used to evaluate structure-bioactivity relationships and to determine chemical-specific MOE.
To ensure functionality and reproducibility, a rigorous quality control assessment was conducted prior to any concentration response modeling. Cell-specific positive control treatments were used for each cell type (see Supplemental Text), and compared to the negative controls, both vehicle (DMSO 0.5%) and media. As demonstrated in Figure 2, the vehicle control and media samples had no effect across all cells. Conversely, the cell-specific positive controls exhibited a concentration dependent effect on the respective model. All phenotypic quality control data can be found in Figures S3–S5 and Table S6.
Figure 2. Cell-specific reproducibility of the phenotypic assays in each tested cell type.
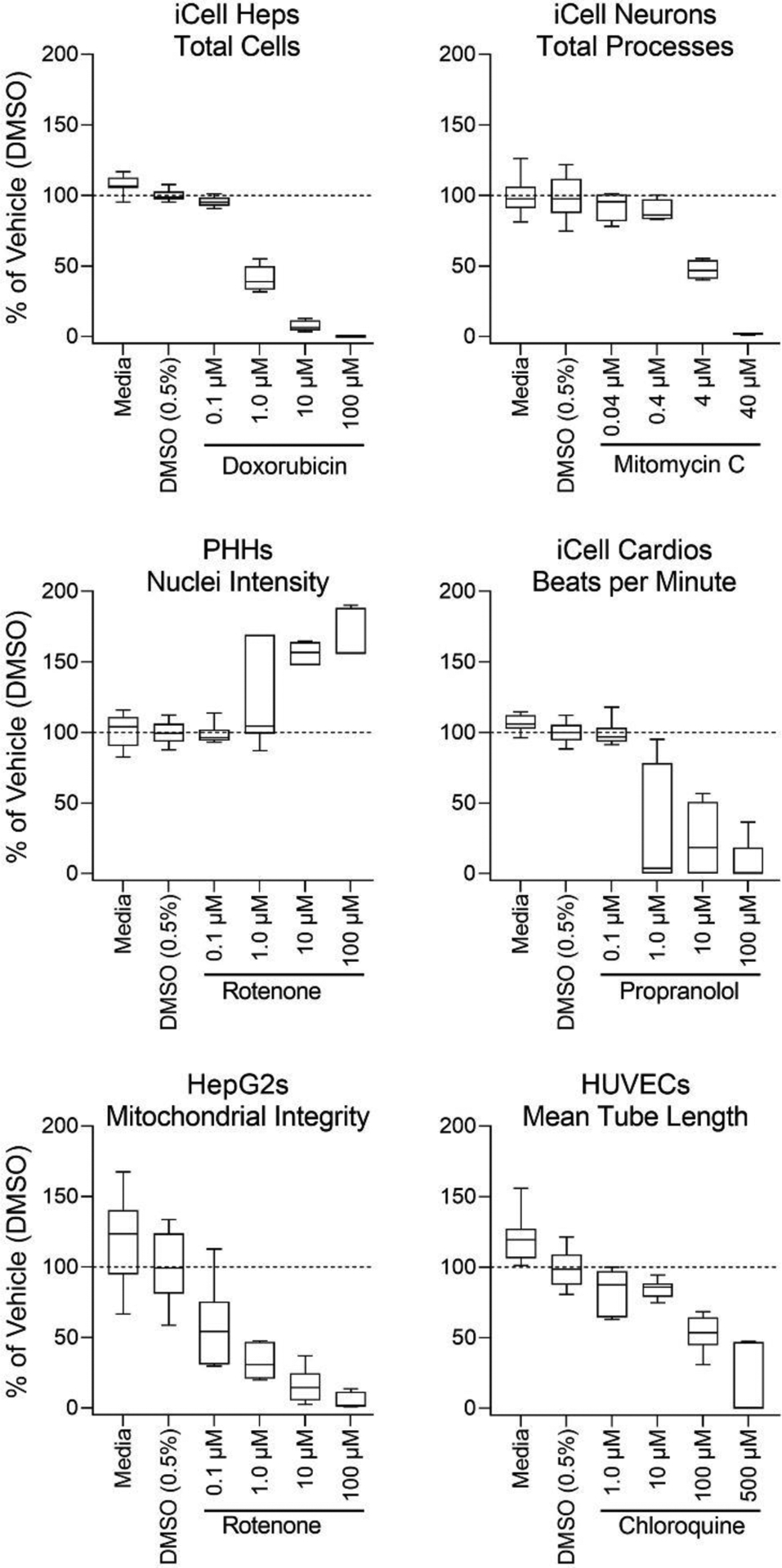
Quality control assessments for a representative phenotype from each in vitro model. Cell-specific positive controls were screened in concentration-response and compared to the negative controls; media and DMSO (0.5%). Cell type specific quality control distribution is represented by box-and-whisker plots with the interquartile range (boxes) and the 10th and 90th percentiles (whiskers). Dotted lines indicate the mean of DMSO controls.
Concentration response analyses were conducted across six cell types with a total of 48 phenotypes (Table 2). When no effect of a chemical was observed, the highest tested concentration (100 μM) was designated as the POD. Figure 3A shows the PODs for each tested PFAS grouped by structure-based class. Within each class a wide range of bioactivity was observed as demonstrated by the range of PODs. For example, within the class of perfluorocarboxylic acids (PFCA), some substances were bioactive (i.e., perfluoroundecanoic acid (PFUnDA)) and others had no effect in any of the tested cell types (i.e., perfluorobutanoic acid (PFBA)). The bioactivity also varied by cell type, as indicated in Figure 3B and Figure S6. To illustrate which cell types were contributing to the bioactivity in Figure 3A, the active phenotypes (i.e., PODs <100 μM) were assembled as a stacked bar by cell type. Of all possible phenotype/chemical combinations (n=2,688), only 230 (8.6%) were active.
Figure 3. Overall bioactivity across all in vitro models.
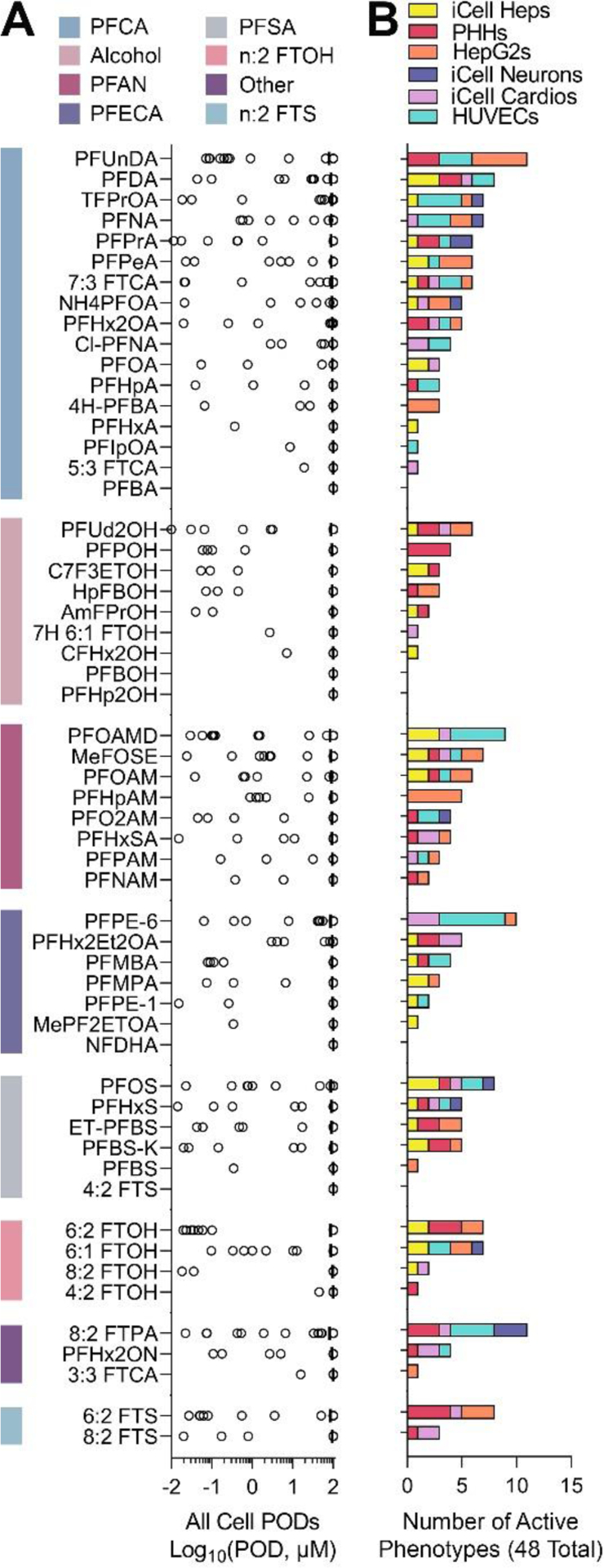
(A) Phenotypic PODs (n = 48) from all cell types are shown for all 56 PFAS tested (chemicals are arranged by sub-class (indicated by a color bar), and within each sub-class by decreasing median POD). (B) Stacked bar plots represent the number of cell-specific “active” (i.e. lower than the highest tested concentration) phenotypes for each PFAS. The colors indicate cell types (see legend).
Figure 4 shows what cell types were most sensitive to tested PFAS. Among 56 tested compounds, 51 were bioactive in at least one cell type (Figure 4A). PFAS were active most often in PHHs, iCell Heps, HepG2, HUVECs, and iCell Cardios (23 to 28 PFAS per cell type). In iCell Neurons, 9 PFAS exhibited bioactivity. Figure 4B shows that with respect to the potency in bioactivity (judged by the POD being less than the top concentration tested), the median for each PFAS was equal to the highest concentration tested; however, in all cell types there was a wide range of bioactivity across PFAS. Overall, iCell Heps were the most sensitive cell type as indicated by the interquartile range in bioactivity.
Figure 4. POD comparisons across different in vitro datasets.
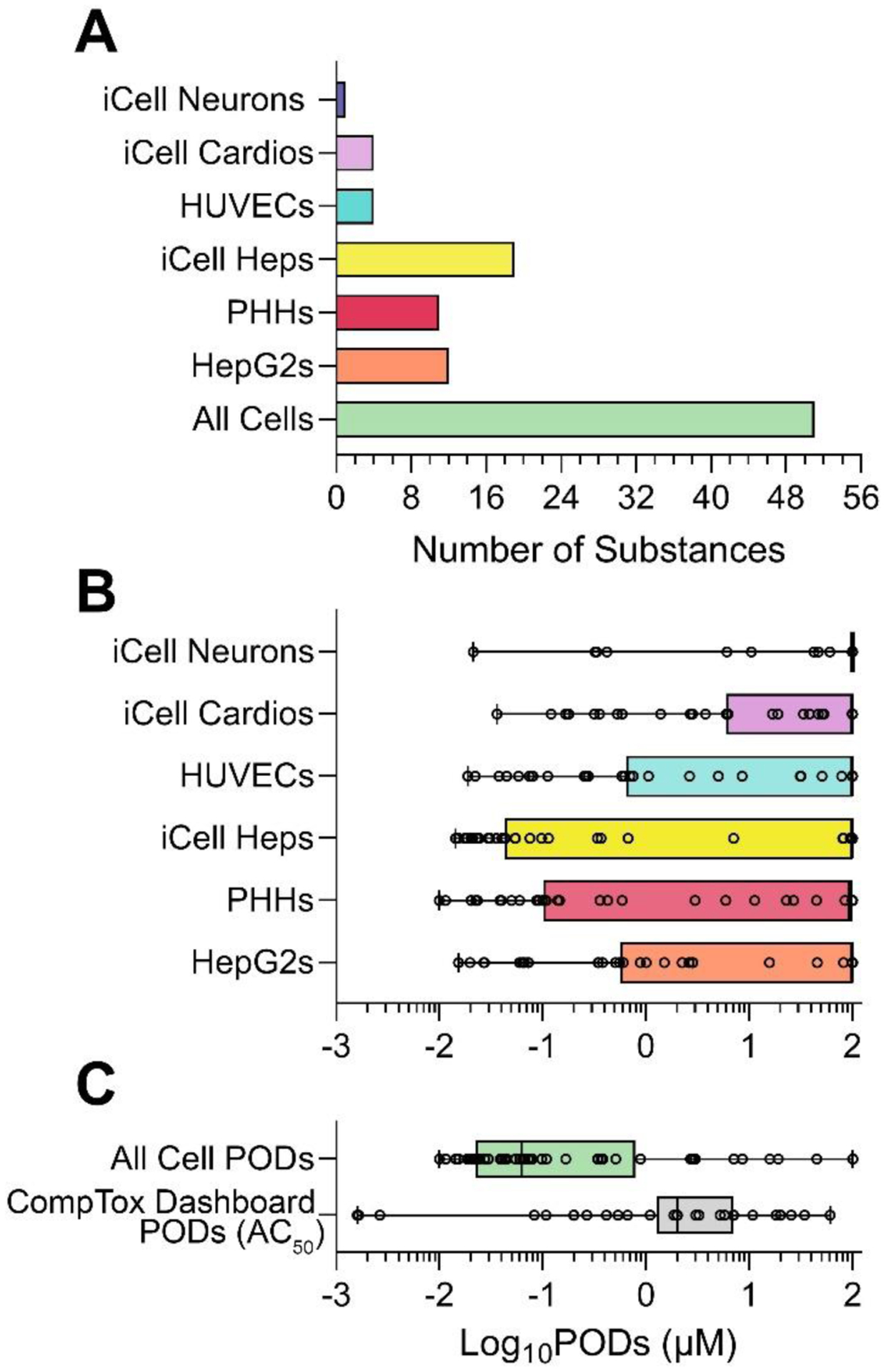
(A) Number of PFAS (out of 56 total) that were active for at least one of the phenotypes (in each cell type as indicated by colors). (B) Cell-type specific POD distributions for all 56 tested PFAS are shown as box and whisker plots (individual values are shown as dots, boxes are inter-quartile range and whiskers are min-max values). (C) Comparison of the lowest PODs from this study (in any cell type) for all 56 tested PFAS to the PODs (i.e. AC50) for the same PFAS from all human cell-based assays available from the CompTox Dashboard as of November 1, 2023.
3.2. Comparison of the PFAS effects in a targeted battery of 6 human organotypic cell types to those from the CompTox Dashboard.
We also examined how the data from this study compared to the bioactivity of the same PFAS in a larger in vitro test battery available on the CompTox Dashboard (https://comptox.epa.gov/dashboard/) (21 human cell types and 55 phenotypes). For this analysis we selected the lowest bioactive PODs for each PFAS across 6 cell types and 48 phenotypes in this study. For the EPA CompTox Dashboard data, we selected the lowest AC50 values for the same 56 PFAS from all available human cell-based assays. Figure 4C shows the box-and-whiskers plot of the PODs from this study and those from the dashboard. Overall, the targeted battery of 6 cell types yielded more protective (i.e., lower) PODs when comparing the mean values between datasets; however, this comparison should be interpreted with caution because the testing ranges and POD types were different among datasets.
To rank PFAS based on their overall bioactivity, we integrated the PODs across 6 cell types and all phenotypes using a ToxPi approach (Marvel et al. 2018; Reif et al. 2013). Figure 5 shows the overall ranking of PFAS using ToxPi where each sector in the pie chart represents a cell type and was weighted proportional to the number of phenotypes in that cell type (iCell Heps (n=8), PHHs (n=8), HepG2s (n=8), iCell Neurons (n=10), iCell Cardios (n=5), HUVECs (n=9)). The stacked bars show a cumulative ToxPi score for each PFAS and the contribution from each cell type. The chemicals were then ranked based on the ToxPi score; a wide range of cumulative bioactivity was observed. The most potent PFAS in this analysis had a score of less than 0.5 on a 0 to 1 scale, so it is noteworthy that no substance was consistently bioactive across all cell types, further indicating cell-specific effects of PFAS. The corresponding chemical subclass for each PFAS is shown by a colored square to the left of the substance’s abbreviation; it is evident that the ToxPi rankings did not show trends consistent with structure-based subclasses, as shown by the interspersed order of subclasses. The ToxPi scores and the contributions from each cell type are provided in Table S7.
Figure 5. ToxPi Ranking by individual cell type and overall score.
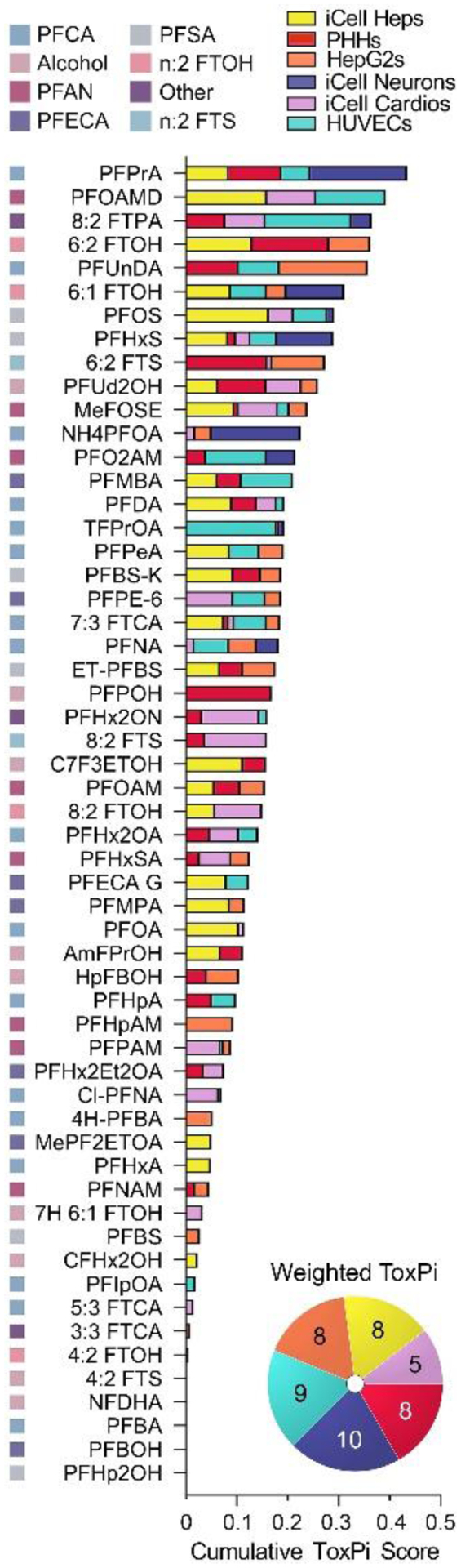
Chemical-specific ToxPi scores for all 56 tested PFAS (their chemical sub-class is indicated by a colored square (see legend) were arranged from the highest to lowest bioactivity. Stacked bars show the contribution of cell-specific (cell types are indicated by colors as indicated in the legend) weighted ToxPi scores. A pie chart in the bottom right indicates the number of phenotypes included in ToxPi calculation for each cell type (colors).
3.3. Comparison of the similarity in PFAS effects based on structural and physicochemical properties
A leading current approach to group PFAS is based on their structure and properties (Buck et al. 2011; OECD 2022). To further test if bioactivity was chemical subclass-specific, we used unsupervised clustering analysis. Figure 6 shows no clear groupings in unsupervised hierarchal clustering of both phenotypes and chemicals based on the PODs. This result may be driven by the fact that for most chemical/phenotype pairings, no effects were observed (see Figure 3). Some of the tested PFAS exhibited similar effects across different liver cell models (PHHs, iCell Heps and HepG2s) supporting previous observations that the liver is a target organ for PFAS (Fenton et al. 2021).
Figure 6. Hierarchal clustering bioactivity heatmap.
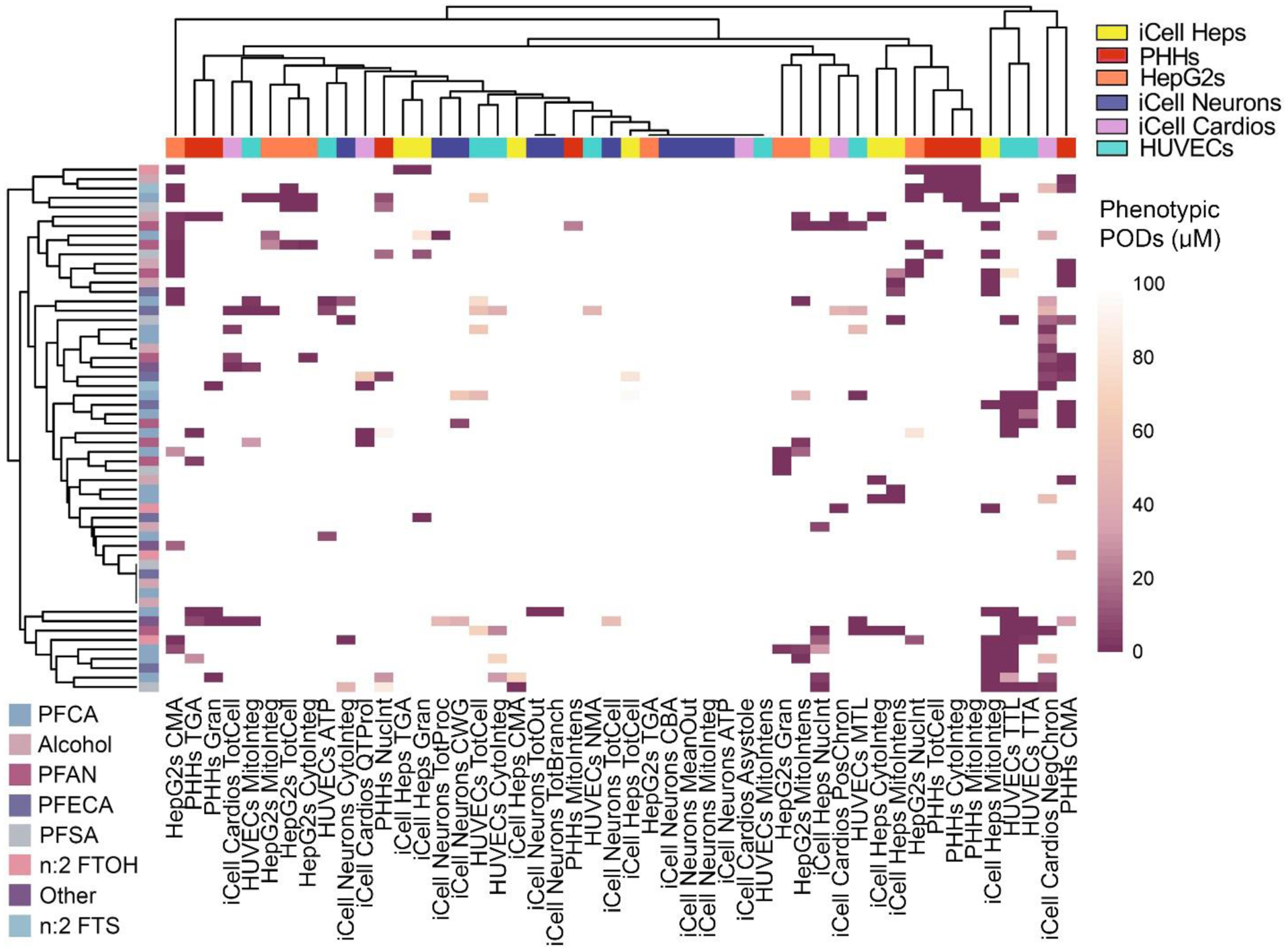
A heatmap shows the PODs (colored by a gradient as indicated in a color bar, with lighter colors representing higher PODs (lower bioactivity/less bioactive), and darker colors, representing lower (more bioactive) PODs). Both chemicals (rows, PFAS subclasses are indicated by colors as indicated in the legend) and phenotypes (columns, cell types are indicated by colors as indicated in the legend) were clustered (average linkage method).
In addition, because unsupervised clustering analysis with all bioactivity data was unsuccessful in demonstrating PFAS subclass groupings, we investigated whether PFAS structural descriptors or physicochemical properties are correlated with, or could be used to predict, bioactivity. First, we used physicochemical properties of PFAS calculated from the OPERA suite (Mansouri et al. 2018) because some previous studies showed that carbon chain length, molecular weight and other properties may be associated with trends in observed in vitro bioactivity (Amstutz et al. 2022; Feng et al. 2023; Goodrum et al. 2021; Houck et al. 2023; Houck et al. 2021; Zhang et al. 2014). Upon calculating all pair-wise correlations (Spearman rank) among all in vitro phenotypes and physicochemical properties and adjusting for multiple comparisons, we found that only the minimum PODs from all iCell Cardios-derived phenotypes were significantly negatively correlated (ρ = −0.54 to −0.57; padj<0.05) with molecular weight, carbon chain length and octanol-water partition coefficient, with the latter three being highly correlated with each other (ρ = 0.84 to 0.92). For pairwise correlations with Saagar descriptors, 11 phenotype-descriptor combinations were significant (Table 3 and Table S8). While some of the Saagar descriptors that were significantly negatively correlated (presence of the feature in a molecule indicated greater effect, i.e. lower POD) with the phenotypes represented chemical structural features, others code for the features indicative of the carbon chain length. Finally, using the combined physicochemical properties and Saagar descriptors, 13 phenotype-descriptor combinations were significant – the same 11 Saagar descriptors (Table 3 and Table S9), as well as molecular weight and carbon chain length. All details and statistical results of these analyses can be found in Tables S8–S13 and Figures S7–S9.
Table 3.
Chemical structure descriptors that were significantly correlated with bioactivity.
| Descriptora | Structureb | Descriptor Meaning | ρc | P adj d | Chemicals with Descriptor | Cell Type (Phenotype) |
|---|---|---|---|---|---|---|
| SGR10703 |
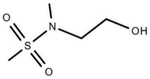
|
N-attached double bonded heteroatoms | −0.70 | 1.08E-05 | MeFOSE PFHxSA | PHHs (Mitochondrial Intensity) |
| SGR10587 | Sulfonamide | |||||
| SGR10099 | Heteroatom-nitrogen bond | |||||
| SGR10668 | Heteroatom-bonded methyl group | −0.70 | 1.08E-05 | MeFOSE MePF2EtOA | ||
| SGR10199 | Two oxygens, 5 bonds apart | −0.57 | 3.11E-02 | MeFOSE PFBOH MePF2EtOA | ||
| SGR10032 | Methyl group | −0.57 | 3.11E-02 | MeFOSE, PFPE-1, PFMBA | ||
| SGR10343 |
|
Any primary amine | −0.56 | 4.17E-02 | AmFPrOH, PFHxSA, PFOAMD | iCell Heps (Cytoplasmic Integrity) |
| SGR10704 |
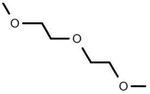
|
Polyethers | −0.69 | 2.84E-05 | PFPE-6, C7F3ETOH | HUVECs (All Nuclei Mean Area) |
| SGR10013 | Any carbon | −0.57 | 2.46E-02 | All tested PFAS (C # varies) | iCell Cardio (Min POD) | |
| SGR10029 | Any heteroatom | −0.57 | 2.79E-02 | All tested PFAS (heteroatom # varies) | ||
| SGR10308 | H-bond acceptors | −0.57 | 3.18E-02 | All tested PFAS (# of H-bond acceptors varies) |
Saagar descriptors can be found in Table S4.
Combined (top) or individual Saagar descriptor structures are shown, where applicable. Individual Saagar descriptors listed herein can be visualized using the hyperlinks in Table S8.
Spearman (ρ) correlation coefficients are shown.
Multiple comparison adjustment was done using Holm–Bonferroni method (see Methods section).
For the cross-validated prediction analyses, only the cardiomyocyte minimum POD could be significantly predicted from either the physicochemical properties (ρ = 0.57 between observed and predicted values, padj = 0.007), or the combination of Saagar descriptors and physicochemical properties (ρ = 0.51, padj = 0.049). For the Saagar descriptors alone, no phenotype associations were significant, which we attribute to the implicit need to correct for choice of tuning parameter, as well as the inclusion of numerous potentially uninformative descriptors in the prediction model.
3.4. Risk characterization of PFAS using data from a targeted battery of 6 human organotypic cell types
Because bioactivity was observed for several PFAS, we reasoned that the in vitro PODs could be used as a conservative quantitative estimate of hazard and thus, used to estimate risk. Therefore, to put the observed bioactivity data into the context of risk assessment, the PODs were compared to human exposure levels using predicted and measured PFAS blood levels to calculate the MOE. Figure 7 (left) shows (i) predicted (density plots) and (ii) reported (whiskers plot) blood levels with the highest (95th percentile) exposure indicated as a triangle, and (iii) the lowest POD (filled circles, colored by the cell type) for each of the tested PFAS. Except for several PFAS, exposure ranges extend several orders of magnitude for both predicted and reported measured data; the latter was available for only one half of tested PFAS. Unfortunately, some tested PFAS had neither exposure data type available, preventing risk characterization analysis. Next, the ratios between the most sensitive chemical-specific POD and the most conservative exposure value are plotted as the MOE. Using the most conservative assumptions, we found that 12 of 56 tested compounds had an MOE<1 indicating potential human health concern at current exposure levels, albeit not all of these have actual measured blood levels. The majority of tested PFAS had MOEs between 1 and 100; 16 of 56 had MOEs above 100. All values used to derive the MOE and the MOEs are reported in Table S14.
Figure 7. Risk characterization of PFAS.
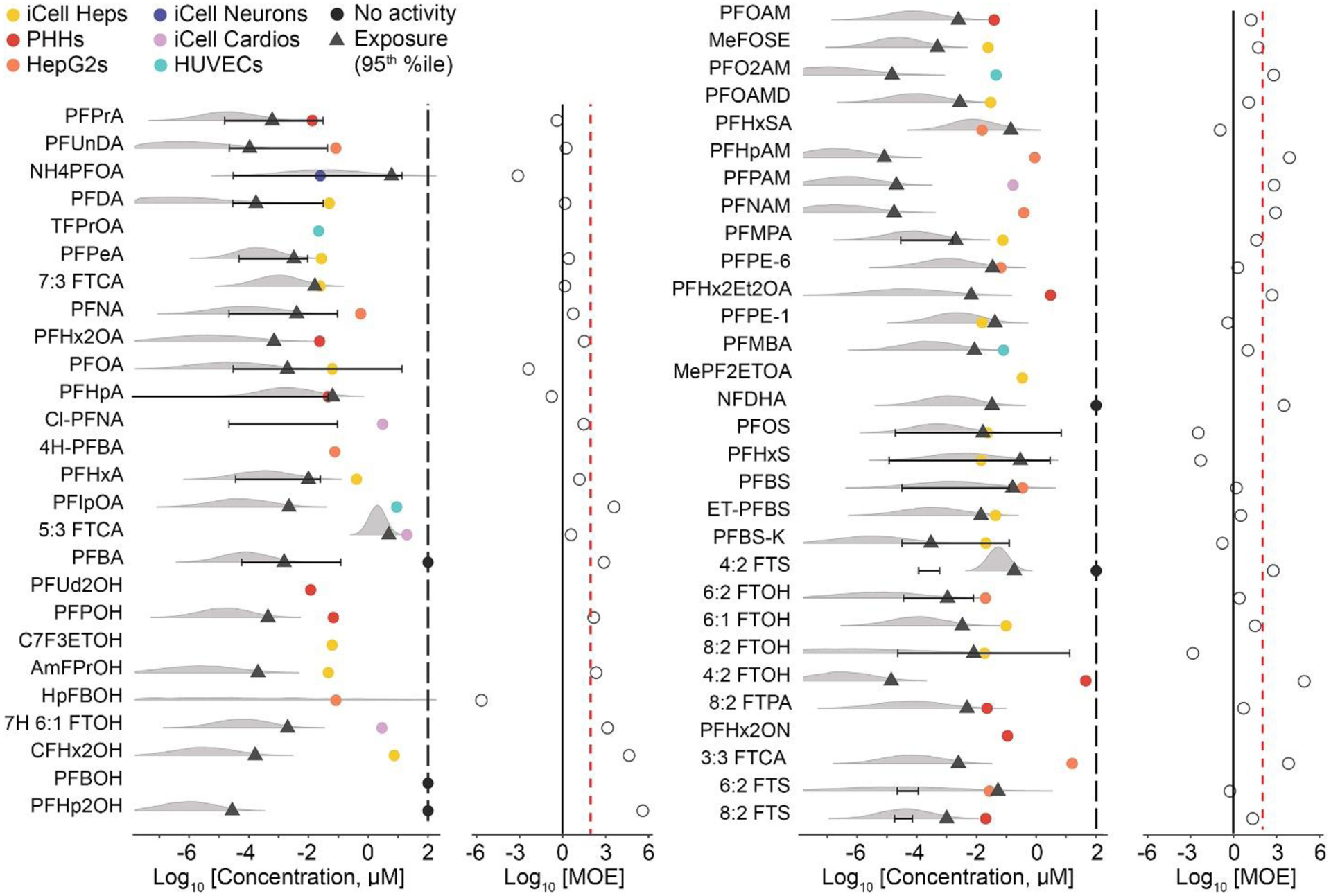
Margin of Exposure (MOE) for each PFAS was calculated by combining all phenotypic PODs and using the most sensitive POD (most bioactive POD) and dividing it by chemical-specific exposure data (using exposure predicted data or human blood levels). The grey density plots represent the uncertainty distribution for the modeled population median exposure reported on the CompTox Dashboard, with diamonds representing the predicted 95th upper percentile; black lines/error bars represent the 5th to 95th percentiles for measured blood levels in humans reported in the literature. Colored dots represent the cell type (see legend) from which the most sensitive phenotypic POD was derived, black dots denote chemicals that were without effect across all models, and vertical black lines indicate the median POD across all PODs. The ratio between exposure and bioactivity is demonstrated as the MOE (on a log10 scale, open circles), the vertical red dashed line represents a margin of 100 and the vertical solid line represents a margin of 1.
Finally, to place the PFAS bioactivity as reported in our study in context with in vitro effects of other industrial chemicals, we compared our results to those previously reported using a similar targeted battery (Chen et al. 2020). Specifically, the observed PFAS bioactivity was compared to in vitro data from the previous study of 42 chemicals (polycyclic aromatic hydrocarbons (PAHs), heavy metals, high production volume chemicals (HPV), pesticides, and phthalates) in 4 of the 6 cell types that overlapped with our study (Chen et al. 2020). Figure 8 illustrates these comparisons using both ToxPi and MOE approaches. An overall ToxPi integrating POD data for PFAS and the 42 industrial chemicals using matching cell types and phenotypes shows the cumulative rank and potency for both chemical sets combined (Figure 8A and Table S15). The top 9 chemicals with the highest ranking in this analysis were industrial chemicals (Table S15). The top-ranked PFAS in this comparison was perfluorooctanamidine (PFOAMD); which had the 2nd highest ranking in the analysis that included all 6 cell types (Figure 5). Furthermore, of the top 30 chemicals with the highest ToxPi scores, only 4 were PFAS, with each being from different subclasses. Overall, the vast majority of PFAS were in the bottom two thirds of all compounds indicating relatively low bioactivity in terms of in vitro (nominal) concentration as compared to other industrial chemicals. In addition to integrating the POD datasets using ToxPi, MOEs were derived when exposure data were available using similarly conservative approaches as detailed for Figure 7. Even though PFAS were relatively low-ranked for the bioactivity, a larger fraction of them had MOEs<1 (Figure 8B) indicating that their potential risk is driven by higher estimates of exposure and/or bioaccumulation, as compared to the industrial chemicals. The bioactivity (removing HepG2s and PHHs) and exposure data used to derive the MOE and the corresponding MOEs are provided in Table S16 and S17.
Figure 8. Potency comparison using ToxPi to rank PFAS with other industrial chemicals.
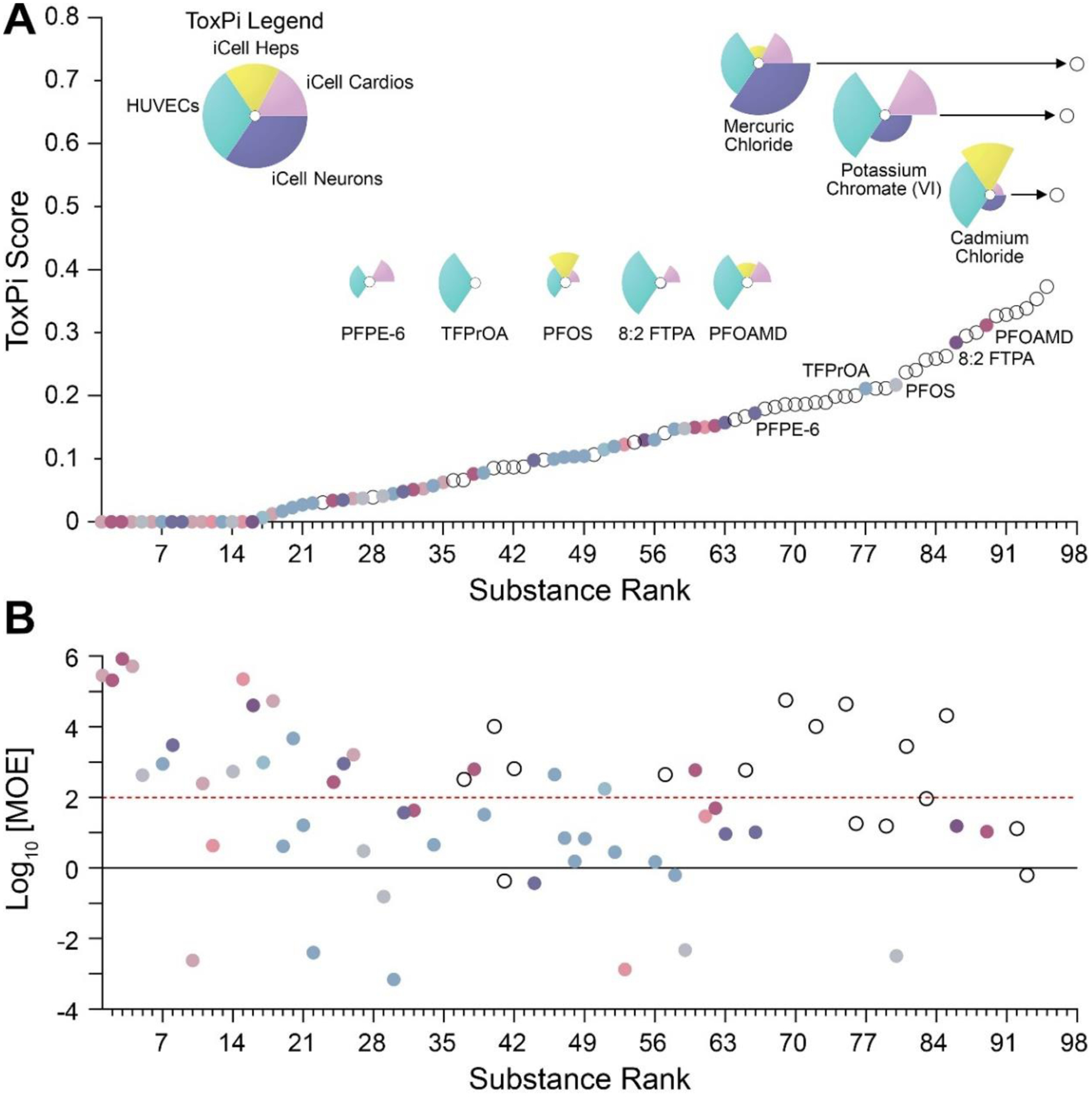
(A) In vitro bioactivity data from 4 overlapping/common cell types (see ToxPi legend in the top left) were used to compare bioactivity of PFAS (colored circles by sub-class) and 42 diverse environmental chemicals (open circles). An overall ToxPi score integrating both datasets was used as the means to compare bioactivity among different chemical classes. Representative ToxPi plots are shown for top-3 overall bioactive compounds and top-5 bioactive PFAS. (B) Margins of exposure (MOEs) for the chemicals in panel (A) were calculated using the lowest POD across all overlapping cell types. PFAS chemicals are colored, and other chemicals are shown as open circles. Horizontal lines (red dashed and black solid) represent MOEs of 100 and 1, respectively.
4. Discussion
We hypothesized that bioactivity measured in a targeted battery of human cell-based assays can be used not only to determine whether there are structure-bioactivity-based relationships for PFAS, but also to characterize and prioritize the potential risks of these chemicals by comparing the bioactivity to exposure estimates. We tested 56 PFAS from 8 structure-based subclasses in concentration-response across 6 human cell types. The bioactivity data was used to derive PODs which were subsequently applied to group and rank PFAS. Also, the in vitro data was compared to (i) in vitro data on PFAS from other studies, (ii) in vitro data on other chemicals, and (iii) predicted exposure data to derive chemical-specific MOEs. Overall, our study aimed to address several over-arching challenges in the safety evaluation of PFAS – we tested a compendium of cell types to determine potential effects across multiple tissues, evaluated similarity of the effects within and across chemical subclasses, determined whether effects can be correlated with structural features, and used in vitro PODs and exposure estimates to perform risk characterization.
Many in vitro studies with PFAS focused on a limited number of compounds and specific cell types; more recently, the potential adverse effects of PFAS have been tested using larger (dozens to hundreds) number of compounds but still focused on particular tissue or cell type (Carlson et al. 2022; Carstens et al. 2023; Houck et al. 2021; Patlewicz et al. 2019). While targeted assays are informative for identifying potential organ-specific and mechanistic events, many PFAS are known to operate by multiple mechanisms or their targets are yet to be identified (Goodrum et al. 2021). Therefore, to gain a more comprehensive understanding of the potential adverse effects across multiple tissues it is crucial to not only broaden the spectrum of tested PFAS but also to incorporate organotypic in vitro assays from different tissues. Our study showed that effects of PFAS are highly cell type-specific and future in vitro testing should include different cell-based models. It is challenging, however, to decide a priori what cell-based models should be used. One approach is to have as broad coverage as possible using the ToxCast approach (Richard et al. 2021; Richard et al. 2016). Another approach is to rely on smaller in vitro test batteries that use fewer cell types but still assess a wide range of relevant phenotypic endpoints and have comparable performance to that of ToxCast (Chen et al. 2020). Our study applied the latter approach and showed that many compounds had tissue-specific effects and thus future studies should include broad-coverage assays.
To enable studies of potential adverse effects across various tissues and organs, it is crucial to prioritize and group PFAS for future testing. Indeed, approaches for PFAS grouping have received heightened attention in the past decade and it is generally agreed that a better understanding of the relationships between physical and chemical properties and potential adverse effects is needed (ETH Zurich Safety and Environmental Technology Group 2017; Ritscher et al. 2018). For example, it was suggested that both elemental composition and molecular structure of PFAS can be used for grouping to inform chemical risk assessment (Anderson et al. 2022; Buck et al. 2011; Buck et al. 2021; Cousins et al. 2020). Additional suggested strategies include grouping by structural subclass, carbon chain length, and mode of action (Gaballah et al. 2020; Health Canada 2022; U.S. Environmental Protection Agency 2021a; U.S. National Science and Technology Council 2023). Overall, while a consensus is yet to emerge on what approach is most scientifically justifiable, there are many who strongly argue that all PFAS should not be grouped together as an entire chemical class (Anderson et al. 2022), even though such an approach is being advocated by a number of Member States in Europe (European Chemicals Agency 2023).
In this study, we tested 56 PFAS that were representative of a broad range of structural subclasses (Buck et al. 2011; Buck et al. 2021; OECD 2018). We used the chemical-specific PODs from various in vitro assays to identify structure-based similarities in effects. While we found that across the 6 cell types some PFAS had cell- and phenotype-specific effects, our data revealed no overall structure-related trends as demonstrated by hierarchal clustering. We also found that the majority chemical-assay test combinations (nearly two-thirds) were inactive at the concentrations tested herein (up to 100 μM). Additionally, no specific subclass exhibited consistently “high” or “low” bioactivity or ToxPi scores. We did find that some bioactivity was correlated with carbon chain length and molecular weight – the larger the number of carbons in a PFAS, the more likely it would have an effect in human iPSC-derived cardiomyocytes.
Other studies reported associations between structural features of PFAS and in vitro effects. In a study of 15 PFAS in HepG2s cells (Zhang et al. 2014) it was shown that the binding affinity of peroxisome proliferator-activated receptor gamma (PPARG) was also concordant with PFAS chain length. A study of 142 PFAS in HepG2 cells also reported that PPAR activation potency correlated with some physicochemical properties such as presence of the negatively charged structures and acidic hydrogen atoms, and that retinoid X receptor beta (RXRβ) activation potency correlated with molecular shape and presence of carboxylate groups (Houck et al. 2021). Shorter-chain PFAS (C4–C6) have lower binding affinity to various human nuclear receptors (i.e. PPAR-α), constitutive activated/androstane receptor, liver fatty acid binding protein, pregnane X receptor, and thyroid receptor) than the long-chain PFAS (>C6) (Goodrum et al. 2021). A study of 13 PFAS in HepG2s found that longer-chain compounds were more cytotoxic than the short-chain ones (Amstutz et al. 2022). Long-chain PFAS were also more cytotoxic and reduced the rate of maturation in a mouse oocyte in vitro model (Feng et al. 2023). In addition, a study of 147 PFAS identified a trend between molecular weight and bioactivity related to in vitro immunosuppression (Houck et al. 2023). A study of 160 PFAS reported that compounds with long carbon chains (C8 or greater), high carbon to fluorine ratio, or containing a carboxylic acid moiety were more likely to exhibit developmental neurotoxicity in vitro (Carstens et al. 2023).
While individual studies mentioned above reported associations, they were not entirely consistent in terms of which physicochemical properties of PFAS were informative, except for the carbon chain length that was identified by several reports. Our study found a similar significant trend for molecular weight and carbon chain length, including Saagar descriptors that indicate the number of carbons and heteroatoms (e.g., F) in each molecule. It is well established that individual PFAS have major differences in half-lives, persistence, as well as chemical properties that add complexity when attempting to group them using structure-based considerations (Fenton et al. 2021; Goodrum et al. 2021). Indeed, our attempt to use an unbiased approach to discover structure-bioactivity associations through a set of molecular substructures that have been used for read-across predictions (Sedykh et al. 2021) resulted in few significant associations. For example, presence of N-attached double bonded heteroatoms, sulfonamide moiety, heteroatom-nitrogen bond, heteroatom-bonded methyl group, two oxygens that were 5 bonds apart, a primary amine, or polyether moiety were significant indicators of higher effects (lower POD) for several liver and endothelial phenotypes. While the library of PFAS tested in this study may have been insufficient to discover associations that are generalizable to other PFAS, our results demonstrate that it may be challenging to establish a robust set of physicochemical and/or structural features of PFAS that would broadly predict their bioactivity across different cell types and molecular events. Even though larger datasets may enhance the strength of yet to be identified associations, confidence in the widespread applicability of the potential structure-bioactivity relations for grouping and read-across may still be uncertain due to potential for high residual variability.
Although our study did not find many structure-bioactivity relationships among tested PFAS, our data does show that in vitro bioactivity data can be used to rank and prioritize PFAS for additional studies. We found that most tested compounds had no effects across any of the tested cell types. We also found that in comparison to other chemicals, the majority of PFAS were low ranked in terms of their bioactivity. These findings indicate that testing all PFAS of concern may be needed to identify the few that may have potentially adverse effects. Such testing would likely need be done in a limited, but diverse set of cell-based models because the number of chemicals to be tested will be in the hundreds, if not thousands. A targeted battery of broad coverage assays in six human cell types that was used herein could be one approach, especially because it has been used in studies of other chemicals (Chen et al. 2020), mixtures (Hsieh et al. 2021), and complex substances (House et al. 2021). It is also similar to the focused approaches proposed by others (Carmichael et al. 2022; Daston et al. 2022).
In addition, the in vitro concentration-response data could be used to derive candidate PODs and perform risk characterization of PFAS. Indeed, the number of studies reporting levels of PFAS in human (i.e. blood) and environmental samples (i.e. water, soil) is rapidly increasing which creates a need to address both hazard and risk of those exposures (National Academies of Sciences Engineering and Medicine 2022). To address these data gaps, we used the bioactivity data collected herein together with available exposure data to calculate the MOEs and inform risk-based decision-making. We found that most tested PFAS for which exposure estimates were also available have MOEs between 1 and 100, with ~20% that exhibited bioactivity below the estimated or measured blood concentrations. While these findings represent the most conservative estimates of risk because we used most sensitive indicators of bioactivity, lowest PODs from nominal PFAS concentrations (i.e. assuming that all compounds were 100% free in both blood and in vitro), and high-end estimates of exposures, they could nonetheless be used to prioritize PFAS for further testing or for regulatory action. Also, despite our efforts to integrate exposure and in vitro bioactivity, only 20 of the 56 tested PFAS had reported blood levels in humans, which provide more reliable MOE estimates. For other chemicals we had to rely on predicted exposures to derive PFAS-specific MOEs. Notably, about two-thirds of cases where both blood levels and exposure predictions were available, the upper 95th percentile blood level was above the upper 95th percentile prediction, sometimes by a factor > 1000. Thus, although predicted exposure data is a useful tool to estimate MOEs, measured human blood levels for a wide variety of PFAS are imperative to understand the MOE and reduce uncertainty, which spans many orders of magnitude for exposure predictions. Recently published expanded systematic evidence map for hundreds of PFAS may be an additional source of human exposure data (Shirke et al. 2024).
Furthermore, in order to use in vitro data for risk characterization, specifically, to inform the risk assessment of PFAS, additional studies are needed to provide sufficient data to inform quantitative in vitro to in vivo extrapolations (QIVIVE) for these chemicals. The need for additional data arises due to the variability in their affinity to proteins and large differences in protein contents between human blood and bioassays, as highlighted by (Qin et al. 2023). Additionally, it is evident that in vivo clearance, and thus the relationship between external and internal dose, varies greatly across PFAS. Thus, there is not only the need for more exposure data (DeLuca et al. 2022), but also for better characterization of their protein binding (Kreutz et al. 2023; Smeltz et al. 2023) as well as more importantly their overall clearance.
Our study has several additional limitations that must be acknowledged. Although we used broad coverage assays by incorporating 6 cell types and various phenotypic endpoints, there were some biological pathways and target organs for PFAS that were not included, specifically, the immune system and kidneys. Recent epidemiological, animal, and in vitro studies have shown that exposure to certain PFAS can result in adverse effects on the immune system while other PFAS exhibited no immunotoxic effects (Corsini et al. 2011; Ehrlich et al. 2023; National Toxicology Program 2016; Steenland et al. 2010). Epidemiological studies have reported that exposure to PFAS [mainly PFOS, PFOA, and perfluorohexanesulfonic acid (PFHxS)] affects the immune system, specifically causing immunosuppression in children (von Holst et al. 2021). Potential for immunotoxicity of PFAS has also been reported with various in vitro models (Gimenez-Bastida et al. 2015; Houck et al. 2023; Zhou et al. 2017).
Finally, it is known that there are many PFAS in the environment, thus humans are exposed to many PFAS, likely at low doses, rather than a single chemical throughout day-to-day activities (Kumar et al. 2020). In addition, accounting for vulnerable populations and sensitive populations is often overlooked but is critical to ensure public health protection (Zeise et al. 2013). Testing environmentally-relevant mixtures, using concentrations that are relevant to human exposures, repeat doses to recapitulate chronic exposures, and addressing inter-individual variability are all scenarios that present in vitro study challenges. Recent studies of PFAS mixtures provide important initial clues that effects may be additive (Addicks et al. 2023; Maddalon et al. 2023); therefore, additional studies of PFAS effects on other cell types are needed. Finally, although the MOE was applied to put the testing concentrations into perspective with the exposure levels, the in vitro chemical exposures were delivered as a single-dose and tested chemical-by-chemical. Therefore, future studies can incorporate these challenges to better capture real-world exposure scenarios to PFAS.
In summary, our data demonstrates that structure-based groupings could not be supported with the in vitro bioactivity data obtained in this study. However, our study shows the feasibility and utility of an approach using bioactivity measured in a targeted compendium of human in vitro models to (i) rank and prioritize chemicals for additional testing, as well as (ii) compare or integrate with other environmental chemicals, and (iii) characterize potential risk by comparison with predicted and measured exposure data. Overall, this study demonstrates that a targeted battery of broad coverage assays in six human cell types is a sensible approach to inform risk-based decision making and identify potential target organs for PFAS.
Supplementary Material
Acknowledgements
We thank US EPA for providing the compounds tested in this study. We are grateful for Dr. Michael DeVito (US EPA) for useful discussions. This work was supported, in part, by grants from the National Institute of Environmental Health Sciences (P42 ES027704 and T32 ES026568) and US Environmental Protection Agency (STAR RD83580201 and RD84045001). This publication contents are solely the responsibility of the grantee and do not necessarily represent the official views of the funding agencies. Further, funding agencies do not endorse the purchase of any commercial products or services mentioned in the publication.
Abbreviations
- PFAS
Per- and polyfluoroalkyl substances
- MOE
Margin(s) of exposure
- PFOA
Perfluorooctanoic acid
- PFOS
Perfluorooctane sulfonic acid
- EU
European Union
- NAMs
New Approach Methodologies
- iPSC
induced pluripotent stem cells
- DMSO
Dimethyl sulfoxide
- iCell Heps
human iPSC-derived hepatocytes
- iCell Neurons
human iPSC-derived neurons
- iCell Cardios
human iPSC-derived cardiomyocytes
- HUVECs
Human umbilical vein endothelial cells
- PHHs
Primary human hepatocytes
- FBS
Fetal bovine serum
- CV
Coefficient of variability
- POD
Point(s) of departure
- ToxPi
Toxicological Priority Index
- ToxPi GUI
Toxicological Priority Index Graphical User Interface
- Css
Steady-state plasma concentration(s)
- AC50
Active concentration at 50% of maximal activity
- OPERA
Open (quantitative) structure-activity/property relationship app
- PFCA
Perfluorocarboxylic acids
- PFUnDA
Perfluoroundecanoic acid
- PFBA
Perfluorobutanoic acid
- PFOAMD
Perfluorooctanamidine
- PPARG
Peroxisome proliferator-activated receptor gamma
- RXRβ
Retinoid X receptor beta
- PPAR-α
Peroxisome proliferator-activated receptor alpha
- QIVIVE
Quantitative in vitro to in vivo extrapolations
- PFHxS
Perfluorohexanesulfonic acid
References
- Addicks GC, Rowan-Carroll A, Reardon AJF, Leingartner K, Williams A, Meier MJ, Moffat I, Carrier R, Lorusso L, Wetmore BA, Yauk CL and Atlas E (2023) Per- and polyfluoroalkyl substances (PFAS) in mixtures show additive effects on transcriptomic points of departure in human liver spheroids. Toxicol Sci 194, 38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amstutz VH, Cengo A, Gehres F, Sijm D and Vrolijk MF (2022) Investigating the cytotoxicity of per- and polyfluoroalkyl substances in HepG2 cells: A structure-activity relationship approach. Toxicology 480, 153312. [DOI] [PubMed] [Google Scholar]
- Anderson JK, Brecher RW, Cousins IT, DeWitt J, Fiedler H, Kannan K, Kirman CR, Lipscomb J, Priestly B, Schoeny R, Seed J, Verner M and Hays SM (2022) Grouping of PFAS for human health risk assessment: Findings from an independent panel of experts. Regul Toxicol Pharmacol 134, 105226. [DOI] [PubMed] [Google Scholar]
- Benjamini Y and Hochberg Y (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met 57, 289–300. [Google Scholar]
- Blanchette AD, Burnett SD, Grimm FA, Rusyn I and Chiu WA (2020) A Bayesian Method for Population-wide Cardiotoxicity Hazard and Risk Characterization Using an In Vitro Human Model. Toxicol Sci 178, 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck RC, Franklin J, Berger U, Conder JM, Cousins IT, de Voogt P, Jensen AA, Kannan K, Mabury SA and van Leeuwen SP (2011) Perfluoroalkyl and polyfluoroalkyl substances in the environment: terminology, classification, and origins. Integr Environ Assess Manag 7, 513–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck RC, Korzeniowski SH, Laganis E and Adamsky F (2021) Identification and classification of commercially relevant per- and poly-fluoroalkyl substances (PFAS). Integr Environ Assess Manag 17, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett SD, Blanchette AD, Chiu WA and Rusyn I (2021) Cardiotoxicity Hazard and Risk Characterization of ToxCast Chemicals Using Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes from Multiple Donors. Chem Res Toxicol 34, 2110–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LM, Angrish M, Shirke AV, Radke EG, Schulz B, Kraft A, Judson R, Patlewicz G, Blain R, Lin C, Vetter N, Lemeris C, Hartman P, Hubbard H, Arzuaga X, Davis A, Dishaw LV, Druwe IL, Hollinger H, Jones R, Kaiser JP, Lizarraga L, Noyes PD, Taylor M, Shapiro AJ, Williams AJ and Thayer KA (2022) Systematic Evidence Map for Over One Hundred and Fifty Per- and Polyfluoroalkyl Substances (PFAS). Environ Health Perspect 130, 56001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael PL, Baltazar MT, Cable S, Cochrane S, Dent M, Li H, Middleton A, Muller I, Reynolds G, Westmoreland C and White A (2022) Ready for regulatory use: NAMs and NGRA for chemical safety assurance. ALTEX 39, 359–366. [DOI] [PubMed] [Google Scholar]
- Carstens KE, Freudenrich T, Wallace K, Choo S, Carpenter A, Smeltz M, Clifton MS, Henderson WM, Richard AM, Patlewicz G, Wetmore BA, Paul Friedman K and Shafer T (2023) Evaluation of Per- and Polyfluoroalkyl Substances (PFAS) In Vitro Toxicity Testing for Developmental Neurotoxicity. Chem Res Toxicol 36, 402–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Liu Y, Wright FA, Chiu WA and Rusyn I (2020) Rapid hazard characterization of environmental chemicals using a compendium of human cell lines from different organs. ALTEX 37, 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Lloyd D, Zhou YH, Chiu WA, Wright FA and Rusyn I (2021) Risk Characterization of Environmental Samples Using In Vitro Bioactivity and Polycyclic Aromatic Hydrocarbon Concentrations Data. Toxicol Sci 179, 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova AC, Klaren WD, Ford LC, Grimm FA, Baker ES, Zhou YH, Wright FA and Rusyn I (2023) Integrative Chemical-Biological Grouping of Complex High Production Volume Substances from Lower Olefin Manufacturing Streams. Toxics 11, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini E, Avogadro A, Galbiati V, dell’Agli M, Marinovich M, Galli CL and Germolec DR (2011) In vitro evaluation of the immunotoxic potential of perfluorinated compounds (PFCs). Toxicol Appl Pharmacol 250, 108–116. [DOI] [PubMed] [Google Scholar]
- Cousins IT, DeWitt JC, Gluge J, Goldenman G, Herzke D, Lohmann R, Miller M, Ng CA, Scheringer M, Vierke L and Wang Z (2020) Strategies for grouping per- and polyfluoroalkyl substances (PFAS) to protect human and environmental health. Environ Sci Process Impacts 22, 1444–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daston GP, Mahony C, Thomas RS and Vinken M (2022) Assessing Safety Without Animal Testing: The Road Ahead. Toxicol Sci 187, 214–218. [DOI] [PubMed] [Google Scholar]
- DeLuca NM, Minucci JM, Mullikin A, Slover R and Cohen Hubal EA (2022) Human exposure pathways to poly- and perfluoroalkyl substances (PFAS) from indoor media: A systematic review. Environ Int 162, 107149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efsa Panel on Contaminants in the Food Chain, Schrenk D, Bignami M, Bodin L, Chipman JK, Del Mazo J, Grasl-Kraupp B, Hogstrand C, Hoogenboom LR, Leblanc JC, Nebbia CS, Nielsen E, Ntzani E, Petersen A, Sand S, Vleminckx C, Wallace H, Barregard L, Ceccatelli S, Cravedi JP, Halldorsson TI, Haug LS, Johansson N, Knutsen HK, Rose M, Roudot AC, Van Loveren H, Vollmer G, Mackay K, Riolo F and Schwerdtle T (2020) Risk to human health related to the presence of perfluoroalkyl substances in food. EFSA J 18, e06223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich V, Bil W, Vandebriel R, Granum B, Luijten M, Lindeman B, Grandjean P, Kaiser AM, Hauzenberger I, Hartmann C, Gundacker C and Uhl M (2023) Consideration of pathways for immunotoxicity of per- and polyfluoroalkyl substances (PFAS). Environ Health 22, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ETH Zurich Safety and Environmental Technology Group. (2017) International Workshop Supporting the Dialogue Between Science and Policy on PFASs. Zurich, Switzerland. [Google Scholar]
- European Chemicals Agency. (2023) Annex XV Restriction Report - Per- and polyfluoroalkyl substances (PFASs) European Chemicals Agency Helsinki, Finland. [Google Scholar]
- European Commission. (2020) Commission Staff Working Document: Poly- and perfluoroalkyl substances (PFAS). European Commission Brussels, Belgium. [Google Scholar]
- Feng J, Soto-Moreno EJ, Prakash A, Balboula AZ and Qiao H (2023) Adverse PFAS effects on mouse oocyte in vitro maturation are associated with carbon-chain length and inclusion of a sulfonate group. Cell Prolif 56, e13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton SE, Ducatman A, Boobis A, DeWitt JC, Lau C, Ng C, Smith JS and Roberts SM (2021) Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research. Environ Toxicol Chem 40, 606–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballah S, Swank A, Sobus JR, Howey XM, Schmid J, Catron T, McCord J, Hines E, Strynar M and Tal T (2020) Evaluation of Developmental Toxicity, Developmental Neurotoxicity, and Tissue Dose in Zebrafish Exposed to GenX and Other PFAS. Environ Health Perspect 128, 47005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerets HH, Tilmant K, Gerin B, Chanteux H, Depelchin BO, Dhalluin S and Atienzar FA (2012) Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol Toxicol 28, 69–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Bastida JA, Surma M and Zielinski H (2015) In vitro evaluation of the cytotoxicity and modulation of mechanisms associated with inflammation induced by perfluorooctanesulfonate and perfluorooctanoic acid in human colon myofibroblasts CCD-18Co. Toxicol In Vitro 29, 1683–1691. [DOI] [PubMed] [Google Scholar]
- Goodrum PE, Anderson JK, Luz AL and Ansell GK (2021) Application of a Framework for Grouping and Mixtures Toxicity Assessment of PFAS: A Closer Examination of Dose-Additivity Approaches. Toxicol Sci 179, 262–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm FA, Iwata Y, Sirenko O, Bittner M and Rusyn I (2015) High-Content Assay Multiplexing for Toxicity Screening in Induced Pluripotent Stem Cell-Derived Cardiomyocytes and Hepatocytes. Assay Drug Dev Technol 13, 529–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill JA, Everett LJ, Haggard DE, Sheffield T, Bundy JL, Willis CM, Thomas RS, Shah I and Judson RS (2021) High-Throughput Transcriptomics Platform for Screening Environmental Chemicals. Toxicol Sci 181, 68–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canada Health. (2022) Objective for Canadian Drinking Water Quality Per- and Polyfluoroalkyl Substances. In: W.a.A.Q. Bureau (Ed), Ottawa, ON [Google Scholar]
- Holm S (1979) A Simple Sequentially Rejective Multiple Test Procedure. Scandinavian Journal of Statistics 6, 65–70. [Google Scholar]
- Houck KA, Friedman KP, Feshuk M, Patlewicz G, Smeltz M, Clifton MS, Wetmore BA, Velichko S, Berenyi A and Berg EL (2023) Evaluation of 147 perfluoroalkyl substances for immunotoxic and other (patho)physiological activities through phenotypic screening of human primary cells. ALTEX 40, 248–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck KA, Patlewicz G, Richard AM, Williams AJ, Shobair MA, Smeltz M, Clifton MS, Wetmore B, Medvedev A and Makarov S (2021) Bioactivity profiling of per- and polyfluoroalkyl substances (PFAS) identifies potential toxicity pathways related to molecular structure. Toxicology 457, 152789. [DOI] [PubMed] [Google Scholar]
- House JS, Grimm FA, Klaren WD, Dalzell A, Kuchi S, Zhang SD, Lenz K, Boogaard PJ, Ketelslegers HB, Gant TW, Wright FA and Rusyn I (2021) Grouping of UVCB substances with new approach methodologies (NAMs) data. ALTEX 38, 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh NH, Chen Z, Rusyn I and Chiu WA (2021) Risk Characterization and Probabilistic Concentration-Response Modeling of Complex Environmental Mixtures Using New Approach Methodologies (NAMs) Data from Organotypic in Vitro Human Stem Cell Assays. Environ Health Perspect 129, 17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Klaren WD, Lebakken CS, Grimm FA and Rusyn I (2017) High-Content Assay Multiplexing for Vascular Toxicity Screening in Induced Pluripotent Stem Cell-Derived Endothelial Cells and Human Umbilical Vein Endothelial Cells. Assay Drug Dev Technol 15, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Lim AY, Sakolish C, Valdiviezo A, Moyer HL, Hewitt P, Bajaj P, Han G and Rusyn I (2022) Analysis of reproducibility and robustness of OrganoPlate(R) 2-lane 96, a liver microphysiological system for studies of pharmacokinetics and toxicological assessment of drugs. Toxicol In Vitro 85, 105464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutz A, Clifton MS, Henderson WM, Smeltz MG, Phillips M, Wambaugh JF and Wetmore BA (2023) Category-Based Toxicokinetic Evaluations of Data-Poor Per- and Polyfluoroalkyl Substances (PFAS) using Gas Chromatography Coupled with Mass Spectrometry. Toxics 11, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Boobis AR and Moretto A (2020) Test and Risk Assessment Strategies for combined exposure to multiple chemicals. Food Chem Toxicol 144, 111607. [DOI] [PubMed] [Google Scholar]
- Liao Y, Wang J, Huang QS, Fang C, Kiyama R, Shen H and Dong S (2012) Evaluation of cellular response to perfluorooctane sulfonate in human umbilical vein endothelial cells. Toxicol In Vitro 26, 421–428. [DOI] [PubMed] [Google Scholar]
- Maddalon A, Pierzchalski A, Kretschmer T, Bauer M, Zenclussen AC, Marinovich M, Corsini E and Herberth G (2023) Mixtures of per- and poly-fluoroalkyl substances (PFAS) reduce the in vitro activation of human T cells and basophils. Chemosphere 336, 139204. [DOI] [PubMed] [Google Scholar]
- Mansouri K, Grulke CM, Judson RS and Williams AJ (2018) OPERA models for predicting physicochemical properties and environmental fate endpoints. J Cheminform 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques E, Pfohl M, Wei W, Tarantola G, Ford L, Amaeze O, Alesio J, Ryu S, Jia X, Zhu H, Bothun GD and Slitt A (2022) Replacement per- and polyfluoroalkyl substances (PFAS) are potent modulators of lipogenic and drug metabolizing gene expression signatures in primary human hepatocytes. Toxicol Appl Pharmacol 442, 115991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvel SW, To K, Grimm FA, Wright FA, Rusyn I and Reif DM (2018) ToxPi Graphical User Interface 2.0: Dynamic exploration, visualization, and sharing of integrated data models. BMC Bioinf 19, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences Engineering and Medicine. (2022) Guidance on PFAS Exposure, Testing, and Clinical Follow-Up, The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- National Toxicology Program. (2016) NTP Monograph: Immunotoxicity Associated with Exposure to Perfluorooctanoic Acid or Perfluorooctane Sulfonate. U.S. Department of Health and Human Services, Research Triangle Park, NC. [Google Scholar]
- OECD. (2018) Toward a New Comprehensive Global Database of Per-and Polyfluoroalkyl Substances (PFASs): Summary Report on Updating the OECD 2007 List of Per and Polyfluoroalkyl Substances (PFASs). Environment Directorate, ORGANISATION FOR ECONOMIC COOPERATION AND DEVELOPMENT, Paris, France. [Google Scholar]
- OECD. (2022) Fact Cards of Major Groups of Per- and Polyfluoroalkyl Substances (PFAS). Environmental Directorate, ORGANISATION FOR ECONOMIC CO-OPERATION AND DEVELOPMENT, Paris, France. [Google Scholar]
- Ojo AF, Peng C and Ng JC (2020) Combined effects and toxicological interactions of perfluoroalkyl and polyfluoroalkyl substances mixtures in human liver cells (HepG2). Environ Pollut 263, 114182. [DOI] [PubMed] [Google Scholar]
- Patlewicz G, Richard AM, Williams AJ, Grulke CM, Sams R, Lambert J, Noyes PD, DeVito MJ, Hines RN, Strynar M, Guiseppi-Elie A and Thomas RS (2019) A Chemical Category-Based Prioritization Approach for Selecting 75 Per- and Polyfluoroalkyl Substances (PFAS) for Tiered Toxicity and Toxicokinetic Testing. Environ Health Perspect 127, 14501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patlewicz G, Richard AM, Williams AJ, Judson RS and Thomas RS (2022) Towards reproducible structure-based chemical categories for PFAS to inform and evaluate toxicity and toxicokinetic testing. Comput Toxicol 24, 100250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RG, Setzer RW, Strope CL, Wambaugh JF and Sipes NS (2017) httk: R Package for High-Throughput Toxicokinetics. J Stat Softw 79, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Henneberger L, Huchthausen J, Konig M and Escher BI (2023) Role of bioavailability and protein binding of four anionic perfluoroalkyl substances in cell-based bioassays for quantitative in vitro to in vivo extrapolations. Environ Int 173, 107857. [DOI] [PubMed] [Google Scholar]
- Radke EG, Wright JM, Christensen K, Lin CJ, Goldstone AE, Lemeris C and Thayer KA (2022) Epidemiology Evidence for Health Effects of 150 per- and Polyfluoroalkyl Substances: A Systematic Evidence Map. Environ Health Perspect 130, 96003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon AJF, Rowan-Carroll A, Ferguson SS, Leingartner K, Gagne R, Kuo B, Williams A, Lorusso L, Bourdon-Lacombe JA, Carrier R, Moffat I, Yauk CL and Atlas E (2021) Potency Ranking of Per- and Polyfluoroalkyl Substances Using High-Throughput Transcriptomic Analysis of Human Liver Spheroids. Toxicol Sci 184, 154–169. [DOI] [PubMed] [Google Scholar]
- Reif DM, Sypa M, Lock EF, Wright FA, Wilson A, Cathey T, Judson RR and Rusyn I (2013) ToxPi GUI: an interactive visualization tool for transparent integration of data from diverse sources of evidence. Bioinformatics 29, 402–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AM, Huang R, Waidyanatha S, Shinn P, Collins BJ, Thillainadarajah I, Grulke CM, Williams AJ, Lougee RR, Judson RS, Houck KA, Shobair M, Yang C, Rathman JF, Yasgar A, Fitzpatrick SC, Simeonov A, Thomas RS, Crofton KM, Paules RS, Bucher JR, Austin CP, Kavlock RJ and Tice RR (2021) The Tox21 10K Compound Library: Collaborative Chemistry Advancing Toxicology. Chem Res Toxicol 34, 189–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin MT, Wambaugh JF, Knudsen TB, Kancherla J, Mansouri K, Patlewicz G, Williams AJ, Little SB, Crofton KM and Thomas RS (2016) ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem Res Toxicol 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- Ritscher A, Wang Z, Scheringer M, Boucher JM, Ahrens L, Berger U, Bintein S, Bopp SK, Borg D, Buser AM, Cousins I, DeWitt J, Fletcher T, Green C, Herzke D, Higgins C, Huang J, Hung H, Knepper T, Lau CS, Leinala E, Lindstrom AB, Liu J, Miller M, Ohno K, Perkola N, Shi Y, Smastuen Haug L, Trier X, Valsecchi S, van der Jagt K and Vierke L (2018) Zurich Statement on Future Actions on Per- and Polyfluoroalkyl Substances (PFASs). Environ Health Perspect 126, 84502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedykh AY, Shah RR, Kleinstreuer NC, Auerbach SS and Gombar VK (2021) Saagar-A New, Extensible Set of Molecular Substructures for QSAR/QSPR and Read-Across Predictions. Chem Res Toxicol 34, 634–640. [DOI] [PubMed] [Google Scholar]
- Shirke AV, Radke EG, Lin C, Blain R, Vetter N, Lemeris C, Hartman P, Hubbard H, Angrish M, Arzuaga X, Congleton J, Davis A, Dishaw LV, Jones R, Judson R, Kaiser JP, Kraft A, Lizarraga L, Noyes PD, Patlewicz G, Taylor M, Williams AJ, Thayer KA and Carlson LM (2024) Expanded Systematic Evidence Map for Hundreds of Per- and Polyfluoroalkyl Substances (PFAS) and Comprehensive PFAS Human Health Dashboard. Environ Health Perspect 132, 26001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko O, Grimm FA, Ryan KR, Iwata Y, Chiu WA, Parham F, Wignall JA, Anson B, Cromwell EF, Behl M, Rusyn I and Tice RR (2017) In vitro cardiotoxicity assessment of environmental chemicals using an organotypic human induced pluripotent stem cell-derived model. Toxicol Appl Pharmacol 322, 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko O, Hesley J, Rusyn I and Cromwell EF (2014) High-content high-throughput assays for characterizing the viability and morphology of human iPSC-derived neuronal cultures. Assay Drug Dev Technol 12, 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeltz M, Wambaugh JF and Wetmore BA (2023) Plasma Protein Binding Evaluations of Per- and Polyfluoroalkyl Substances for Category-Based Toxicokinetic Assessment. Chem Res Toxicol 36, 870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenland K, Fletcher T and Savitz DA (2010) Epidemiologic evidence on the health effects of perfluorooctanoic acid (PFOA). Environ Health Perspect 118, 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukker AM, Bouwman LMS, van Kleef R, Hendriks HS, Legler J and Westerink RHS (2020) Perfluorooctane sulfonate (PFOS) and perfluorooctanoate (PFOA) acutely affect human alpha(1)beta(2)gamma(2L) GABA(A) receptor and spontaneous neuronal network function in vitro. Sci Rep 10, 5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency. (2021a) National PFAS Testing Strategy: Identification of Candidate Per- and Polyfluoroalkyl Substances (PFAS) for Testing. U.S. Environmental Protection Agency; Washington, DC. [Google Scholar]
- U.S. Environmental Protection Agency. (2021b) PFAS Strategic Roadmap: EPA’s Commitments to Action 2021–2024. U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- U.S. Environmental Protection Agency. (2023) PFAS National Primary Drinking Water Regulation Rulemaking. United States Environmental Protection Agency; Washington, DC. [Google Scholar]
- U.S. National Science and Technology Council. (2023) Per- and Polyfluoroalkyl Substances (PFAS) Report. Executive Office of the President of the United States, Washington, DC. [Google Scholar]
- von Holst H, Nayak P, Dembek Z, Buehler S, Echeverria D, Fallacara D and John L (2021) Perfluoroalkyl substances exposure and immunity, allergic response, infection, and asthma in children: review of epidemiologic studies. Heliyon 7, e08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambaugh JF, Setzer RW, Reif DM, Gangwal S, Mitchell-Blackwood J, Arnot JA, Joliet O, Frame A, Rabinowitz J, Knudsen TB, Judson RS, Egeghy P, Vallero D and Cohen Hubal EA (2013) High-throughput models for exposure-based chemical prioritization in the ExpoCast project. Environ Sci Technol 47, 8479–8488. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, Patlewicz G, Shah I, Wambaugh JF, Judson RS and Richard AM (2017) The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahm S, Bonde JP, Chiu WA, Hoppin J, Kanno J, Abdallah M, Blystone CR, Calkins MM, Dong GH, Dorman DC, Fry R, Guo H, Haug LS, Hofmann JN, Iwasaki M, Machala M, Mancini FR, Maria-Engler SS, Moller P, Ng JC, Pallardy M, Post GB, Salihovic S, Schlezinger J, Soshilov A, Steenland K, Steffensen IL, Tryndyak V, White A, Woskie S, Fletcher T, Ahmadi A, Ahmadi N, Benbrahim-Tallaa L, Bijoux W, Chittiboyina S, de Conti A, Facchin C, Madia F, Mattock H, Merdas M, Pasqual E, Suonio E, Viegas S, Zupunski L, Wedekind R and Schubauer-Berigan MK (2023) Carcinogenicity of perfluorooctanoic acid and perfluorooctanesulfonic acid. Lancet Oncol. [DOI] [PubMed] [Google Scholar]
- Zeise L, Bois FY, Chiu WA, Hattis D, Rusyn I and Guyton KZ (2013) Addressing human variability in next-generation human health risk assessments of environmental chemicals. Environ Health Perspect 121, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren XM, Wan B and Guo LH (2014) Structure-dependent binding and activation of perfluorinated compounds on human peroxisome proliferator-activated receptor gamma. Toxicol Appl Pharmacol 279, 275–283. [DOI] [PubMed] [Google Scholar]
- Zhou X, Dong T, Fan Z, Peng Y, Zhou R, Wang X, Song N, Han M, Fan B, Jia J and Liu S (2017) Perfluorodecanoic acid stimulates NLRP3 inflammasome assembly in gastric cells. Sci Rep 7, 45468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


